Abstract
The Y-family polymerases help cells tolerate DNA damage by performing translesion synthesis, yet they also can be highly error prone. One distinctive feature of the DinB class of Y-family polymerases is that they make single-base deletion errors at high frequencies in repetitive sequences, especially those that contain two or more identical pyrimidines with a 5′ flanking guanosine. Intriguingly, different deletion mechanisms have been proposed, even for two archaeal DinB polymerases that share 54% sequence identity and originate from two strains of Sulfolobus. To reconcile these apparent differences, we have characterized Dpo4 from Sulfolobus solfataricus using the same biochemical and crystallographic approaches that we have used previously to characterize Dbh from Sulfolobus acidocaldarius. In contrast to previous suggestions that Dpo4 uses a deoxynucleoside triphosphate (dNTP)-stabilized misalignment mechanism when creating single-base deletions, we find that Dpo4 predominantly uses a template slippage deletion mechanism when replicating repetitive DNA sequences, as was previously shown for Dbh. Dpo4 stabilizes the skipped template base in an extrahelical conformation between the polymerase and the little-finger domains of the enzyme. This contrasts with Dbh, in which the extrahelical base is stabilized against the surface of the little-finger domain alone. Thus, despite sharing a common deletion mechanism, these closely related polymerases use different contacts with the substrate to accomplish the same result.
INTRODUCTION
The Y-family of DNA polymerases was defined in 2001 (21), following the discovery that the gene responsible for the variant form of xeroderma pigmentosum encodes polymerase (Pol) eta, the seventh human polymerase identified (19, 30). The major function of these polymerases is in the replication of damaged DNA by the process known as translesion synthesis (TLS). The TLS polymerases are thought to rescue stalled replicative polymerases, synthesizing short stretches of DNA before falling off and allowing replicative polymerases to continue. Y-family enzymes cluster into six groups. Of these, the DinB polymerases comprise the only group that is found in eukaryotes, bacteria, and the archaea.
Like most other types of polymerases, the Y-family DNA polymerases have a structural architecture that consists of finger, palm, and thumb domains arranged to resemble a right hand. The topology of the palm domain, which contains the essential catalytic residues, places these enzymes into the “classical” polymerase superfamily (34). The relatively small sizes of the fingers and thumb domains, however, result in a spacious active site, which accounts for the high tolerance that the Y-family polymerases have for replicating damaged DNA bases and contributes to their low fidelity of DNA replication. All Y-family DNA polymerases possess a unique little-finger or polymerase-associated domain (LF/PAD) that is tethered to the polymerase thumb domain through a flexible polypeptide linker. The LF/PAD enhances DNA association by interacting with the upstream DNA duplex.
Although they provide cells with the ability to tolerate DNA damage, the TLS polymerases are more error-prone than replicative polymerases (13) and can substantially increase DNA mutation rates in cells. DinB polymerases make single-base deletions at high rates, especially in sequences where two or more identical pyrimidines are located with a guanosine base to the 5′ side, i.e., in a 5′-GPyPy-3′ sequence context (12, 23). The requirement for the guanosine in this “deletion hot spot” sequence correlates with deoxycytidine (dC) being incorporated more efficiently than the other bases, which is likely to be related to the specificity of the DinB polymerases for replicating damaged guanosine bases (11, 24, 33).
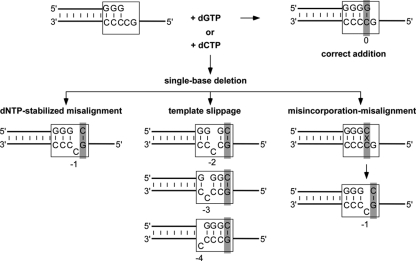
The following three different mechanisms (Fig. 1) have been proposed to explain how DNA deletions arise: template slippage, deoxynucleoside triphosphate (dNTP)-stabilized misalignment, and misinsertion-misalignment. Template slippage was first proposed to account for the observation that most deletions occur in repetitive sequences (26). The other two mechanisms, dNTP-stabilized misalignment (4, 7, 14) and misinsertion-misalignment (3, 15), were originally proposed to account for deletions in nonrepetitive sequences. Template slippage is unique in requiring isomerization of correctly paired primer and template bases prior to nucleotide addition. In dNTP-stabilized misalignment, only the single-stranded template DNA isomerizes, while in misincorporation-misalignment, isomerization of a base mispair occurs after nucleotide addition.
Fig. 1.
Single-base deletion mechanisms. The DinB polymerases create single-base deletion mutations at high rates when replicating “deletion hot spots,” repetitive sequences such as the one shown (boxed) that contain, in the template strand, two or more pyrimidines with a guanosine on the 5′ side. Incorporation of dCTP (instead of the next correct nucleotide, dGTP) is the first step in creating a single-base deletion that can occur via one of the following three different mechanisms: dNTP-stabilized misalignment, template slippage, or misincorporation-misalignment. The base positions indicated are numbered relative to the nucleotide located in the nascent base pair binding pocket, defined as position 0 (shaded in gray). See the text for additional information.
DinB polymerases have been suggested to use all three of these different mechanisms, despite displaying similar deletion specificities. Dbh has been shown to use a template slippage mechanism (6, 31), while Dpo4 has been proposed to use dNTP-stabilized misalignment (9, 12, 16). Earlier experiments indicated that DinB from Escherichia coli used a dNTP-stabilized misalignment mechanism (28), although more recent work shows that it uses a template slippage mechanism (10). Human polymerase kappa can use a misinsertion-misalignment mechanism in nonrepetitive sequences, while template slippage has been inferred as the mechanism used in repetitive sequences (20, 32).
To begin to understand the basis for the DinB polymerases using such a variety of different mechanisms, despite being homologous enzymes, we have characterized the behavior of Dpo4 on the repetitive sequence 5′-GCCCC-3′. This sequence is a deletion hot spot for Dbh, where one of the cytosines is skipped up to half the time this sequence is copied in vitro by Dbh (23). We have extensively characterized the structure and activity of Dbh as it copies this sequence (31). Using single-nucleotide incorporation assays and X-ray crystallography, we find that Dpo4, like Dbh, uses a template slippage mechanism when making a single-base deletion on this repetitive sequence and stabilizes the skipped template base in an extrahelical conformation.
MATERIALS AND METHODS
Protein expression and purification.
The gene encoding Dpo4 was amplified by PCR from Sulfolobus solfataricus P2 genomic DNA (ATCC 35092), simultaneously adding a C-terminal His6 tag, and then cloned into the expression vector pKKT7. Previous work has shown that this tag does not significantly alter the polymerization activity of Dpo4 (9), nor did it change its crystallization properties. The constructed plasmid pKKT7-Dpo4-CHis was then transformed into E. coli Rosetta(DE3)pLysS cells (Novagen). The transformed cells were grown in autoinduction medium (27) at 37°C, with shaking at 200 rpm, until the optical density at 600 nm (OD600) reached 1.0. The temperature was then reduced to 20°C for overnight expression. Cell pellets were harvested by centrifugation and resuspended in buffer NA (20 mM HEPES at pH 7.5, 100 mM NaCl, and 50 mM imidazole), lysed by sonication, and then heated at 75°C for 20 min. Subsequent steps in the purification were performed at 4°C.
The lysate was centrifuged for 1 h at 20,000 × g, and the supernatant was loaded onto a HiTrap chelating HP column (2 × 5 ml; GE Healthcare). Dpo4 was eluted from the column using a linear gradient of 0 to 100% buffer NB (buffer NA with 1 M imidazole). Fractions that contained Dpo4 were pooled and diluted with 5 volumes of buffer SA (20 mM HEPES at pH 7.5, 50 mM NaCl, 0.5 mM EDTA, and 1 mM dithiothreitol [DTT]) and then loaded onto a cation exchange HiTrap SP HP column (5 ml; GE Healthcare). Dpo4 was eluted using a linear gradient of 0 to 100% buffer SB (buffer SA with 1 M NaCl).
Fractions containing Dpo4 were pooled and concentrated to 30 mg/ml using a 10-kDa Amicon Ultra-15 centrifugal filter (Millipore), dialyzed into storage buffer (20 mM HEPES at pH 7.5, 100 mM NaCl, 0.5 mM EDTA, and 1 mM DTT), and kept at 4°C. Concentrations were determined by UV absorbance at 280 nm using a calculated extinction coefficient of 22,350 M−1 cm−1.
Polymerase assays.
Primer was synthesized with a 5′- 6-carboxyfluorescein (FAM) label and was annealed to the template in a solution containing 10 mM HEPES (pH 7.5) and 50 mM NaCl by heating it for 2 min at 95°C, incubating it for 5 min at 55°C, and then slowly cooling it to 25°C. Polymerase reaction mixtures contained 40 nM annealed primer-template DNA, 4 μM Dpo4, 20 mM HEPES (pH 7.0), 65 mM NaCl, 5 mM MgCl2, 1 mM DTT, and 1 mM dCTP or dGTP. Reaction mixtures were incubated at room temperature, and 10-μl aliquots were quenched after 1, 2, 4, 8, 12, or 20 min by mixing them with an equal volume of 80% formamide containing 100 mM EDTA, with bromophenol blue and xylene cyanol dyes. Samples were incubated at 95°C for 5 min just prior to electrophoresis on a 17.5% polyacrylamide (19:1 dilution)–7.5 M urea–1× Tris-borate-EDTA (TBE) gel that was preheated and run at 50°C. The amounts of FAM fluorescence in the unextended and extended primer bands were quantitated using a Typhoon 9400 scanner and ImageQuant software (GE Healthcare).
Crystallization and structure determination.
Primer and template DNA oligonucleotides (Table 1) were annealed in a solution containing 10 mM Tris (pH 7.5) and 50 mM NaCl by being heated for 2 min at 95°C, incubated for 5 min at 55°C, and then slow cooled to 25°C. The Dpo4 T-3 complex was prepared at room temperature by combining 150 μM Dpo4, 180 μM DNA, and 1 mM dCTP in 20 mM HEPES (pH 7.0), 5 mM Ca(OAc)2, 85 mM NaCl, 0.25 mM EDTA, and 1 mM DTT (final concentrations). The Dpo4 TT-4 complex was prepared in similar way, but the concentrations of Dpo4 and DNA were increased to 200 μM and 240 μM, respectively. Crystals were grown at room temperature by hanging-drop vapor diffusion after mixing 2 μl of the complex with 2 μl of a well solution containing 18% polyethylene glycol 3350 (PEG 3350), 100 mM HEPES (pH 6.6), 100 mM Ca(OAc)2, and 2.5% glycerol for the T-3 complex or 12% PEG 3350, 100 mM MES (morpholinoethanesulfonic acid)-Tris (pH 6.0), 100 mM Ca(OAc)2, and 2.5% glycerol for the TT-4 complex. The crystals were stabilized and cryoprotected by the addition of a solution containing 20% PEG 3350, 100 mM Ca(OAc)2, 25% glycerol, 1 mM dCTP, and either 100 mM HEPES (pH 7.5) for the T-3 complex or 100 mM MES-Tris (pH 5.3) for the TT-4 complex. The crystals were then flash cooled in liquid nitrogen.
Table 1.
Oligonucleotides
| Primer or template | Sequencea |
|---|---|
| T0 primer | 5′-(FAM)-AGGCACTGATC GGG-3′ |
| T0 template | 3′-CCGTGACTAG CCCCG CATT-5′ |
| T-1 primer | 5′-(FAM)-AGGCACTGATC GGG-3′ |
| T-1 template | 3′-CCGTGACTAG CCCTG CATT-5′ |
| T-2 primer | 5′-(FAM)-AGGCACTGATC GG G-3′ |
| T-2 template | 3′-CCGTGACTAG CCTCG CATT-5′ |
| T-3 primer | 5′-(FAM)-AGGCACTGATC G GG-3′ |
| T-3 template | 3′-CCGTGACTAG CTCCG CATT-5′ |
| T-4 primer | 5′-(FAM)-AGGCACTGATC GGG-3′ |
| T-4 template | 3′-CCGTGACTAG TCCCG CATT-5′ |
| X-T-3 primer | 5′-GGCACTGATC G GG-3′ |
| X-T-3 template | 3′-CCGTGACTAG CTCCG CATT-5′ |
| X-TT-4 primer | 5′-GGCACTGATC AGG-3′ |
| X-TT-4 template | 3′-CCGTGACTAG TTCCG CATT-5′ |
All oligonucleotides were purchased from Integrated DNA Technologies. FAM, 6-carboxyfluorescein. Nucleotide differences from the sequence of T0 are shown in boldface.
X-ray diffraction data were collected at Brookhaven National Laboratory (BNL), National Synchrotron Light Source (NSLS) beamline X29, and were processed and scaled using HKL2000 (22). The structures were solved by molecular replacement with the program Phaser (18), using the protein structure from a standard Dpo4 ternary complex (Protein Data Bank [PDB] accession no. 1JX4) (29), and were refined using Refmac (5) and PHENIX (1), alternating with cycles of manual rebuilding using Coot (8). Some figures were made using PyMOL (see Fig. 3 and 4) (25).
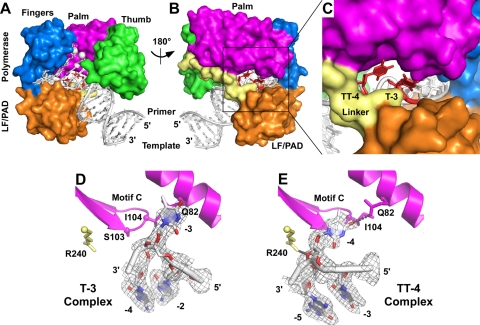
Fig. 3.
Crystal structures of Dpo4 with extrahelical template bases. Superimposed structures of the Dpo4 T-3 and TT-4 ternary complexes are shown as a front overview (A), back overview (B), and view of the gap between the polymerase domain and the LF/PAD (C). The structures of the individual bulged template bases are shown for the T-3 complex (D) and the TT-4 complex (E), together with surrounding protein structures that are mentioned in the text. The gray mesh shows 2Fo-Fc electron density contoured at 1.0 sigma. Dpo4 protein domains are colored as follows: palm (magenta), thumb (green), fingers (blue), LF/PAD (orange), linker (pale yellow). (A to C) The extrahelical nucleotides are colored red, with the remaining DNA shown in white. (A) The white spheres are the two calcium ions bound at the polymerase active site. (D and E) The small spheres mark the positions of the C-alpha atoms of the amino acids shown in stick representation, and the DNA is colored by atom type, as follows: oxygen (red), nitrogen (blue), carbon and phosphate (white). Gray dotted lines indicate hydrogen bonds.
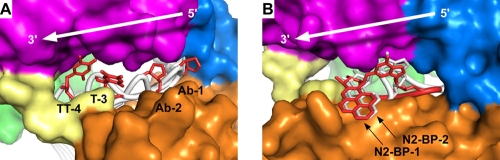
Fig. 4.
Comparison of extrahelical nucleotides displaced on the minor groove side of duplex DNA when bound to Dpo4. Superposition of Dpo4 crystal structures containing bulged abasic sites (17) and undamaged thymines (this work) (A) and N2-BP-dG-damaged DNA (2) (B). Structures are numbered according to their position along the DNA on the 3′ side of the templating base. Protein coloring is the same as described in the legend to Fig. 3. (A) Bulged nucleotides are colored red, and the surrounding DNA is white. (B) DNA from the N2-BP-1 structure is colored red, and DNA from the N2-BP-2 structure is white. White arrows indicate the direction that DNA translocates along the protein as Dpo4 extends DNA synthesis. The PDB accession numbers for the structures shown are as follows: Ab-1, 1S0N; Ab-2, 1S0O; N2-BP-1, 2IA6; and N2-BP-2, 2IBK.
Protein structure accession numbers.
The coordinates and structure factors for both complexes have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ, under accession no. 3QZ7 (T-3 complex) and 3QZ8 (TT-4 complex).
RESULTS
Dpo4 uses a template slippage deletion mechanism.
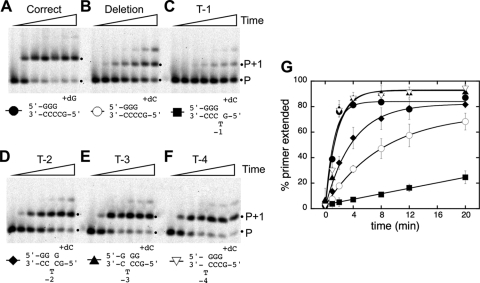
We first measured the rates of single-nucleotide incorporation by Dpo4 on a primer-template DNA containing the repetitive template sequence 5′-GCCCC-3′ (primer-template T0) (Table 1). On this DNA, incorporation of deoxyguanosine (dG) yields a correct extension product, while incorporation of dC initiates a single-base deletion. Under the single-turnover conditions used, dGTP was added at an observed rate of 0.81 min−1 (Fig. 2 A and G), and dCTP was added at a rate of 0.12 min−1 (Fig. 2B and G). Thus, Dpo4 can initiate a single-base deletion in this sequence context at a rate approximately 6-fold lower than that of correct extension.
Fig. 2.
Incorporation of single nucleotides by Dpo4 on primer-template DNAs containing single-base deletion hot spot sequences. The extent of nucleotide incorporation over time was measured for substrates containing the deletion hot spot sequence shown in Fig. 1 and variants in which each of the four consecutive cytosines was replaced by thymine. (A) T0 plus dGTP, for correct extension (black circles); (B) T0 plus dCTP, which initiates a single-base deletion (open circles); (C) T-1 (black squares); (D) T-2 (black diamonds); (E) T-3 (black triangles); (F) T-4 (open inverted triangle), all plus dCTP. (C to F) Substrates contain bulged bases at the −1, −2, −3, and −4 positions, respectively. Dpo4 (4 μM) and primer-template DNA (40 nM) were premixed to allow maximal complex formation, and then the reactions were initiated with the addition of 1 mM indicated nucleotide. Time points (left to right in each panel) were taken after 0, 1, 2, 4, 8, 12, and 20 min. For simplicity, only the hot spot region of the primer-template DNA for each gel is shown; full sequences are listed in Table 1. (G) Graph showing the percentage of primer extension as a function of time. Symbols correspond to those described for panels A to F. Error bars represent the standard deviations from three independent reactions. Black dots mark the migration positions of unextended (P) and extended (P+1) primers when the reaction products were separated on denaturing polyacrylamide gels. The second minor product band could result from the slow addition of a second nucleotide by one of several mechanisms, as follows: mispairing (A to F), dNTP-stabilized misalignment (see template sequence in Table 1) (A), or primer-template slippage (forming a C-C or C-T mispair instead of having a bulged template base) followed by correct extension (B to F).
We then used variations of this sequence, in which each of the four cytosines was individually changed to thymine, to shed light on the deletion mechanism used by Dpo4. Because repetitive sequences can adopt multiple conformations (Fig. 1), these individual base substitutions (Table 1) were designed to favor the single, lower-energy conformation that has an unpaired T rather than the other conformations that would contain a G-T mispair (31). On the T-1 substrate, dCTP incorporation by Dpo4 was very slow (∼0.01 min−1) (Fig. 2C and G), with just 20% of the primer extended after 20 min. The nucleotide incorporation rate increased as the T was moved further away from the primer-template junction, with an intermediate rate of incorporation on the T-2 substrate (0.26 min−1) (Fig. 2D and G) and rates comparable to correct extension on the T-3 and T-4 substrates (0.58 min−1 and 0.62 min−1, respectively) (Fig. 2E to G).
The relative nucleotide incorporation rates on these substrates (T-4 ∼ T-3 > T-2 ≫ T-1) are what would be expected if Dpo4 uses a template slippage deletion mechanism, in which the skipped template base is two or more bases away from the nascent base pair. For either of the other deletion mechanisms, nucleotide incorporation on the T-1 substrate would be expected to be the fastest, since the thymine substitution at position −1 could adopt either the unpaired or mispaired conformation that these mechanisms use. Instead, the nucleotide incorporation rate on the T-1 substrate (∼0.01 min−1) is at least 10-fold lower than that required to account for the rate of dC addition that was observed on the T0 substrate (0.12 min−1), ruling out both the dNTP-stabilized misalignment and misinsertion-misalignment mechanisms as the primary deletion mechanism. To the extent that a second deletion mechanism is used by Dpo4, it is most likely that a dNTP-stabilized misalignment mechanism is the minor alternative, since nucleotide misincorporation rates in a nonrepetitive sequence were lower when the next templating base was not complementary to the incoming nucleotide (i.e., when neither template slippage nor dNTP misalignment were allowed) (9).
Dpo4 prefers to bind to DNA with an extrahelical base at the −4 position but can also accommodate a bulged base at the −3 position.
We have determined two crystal structures, T-3 and TT-4, of Dpo4 in ternary complexes with primer-template DNA and incoming nucleotides that show how extrahelical template bases are bound by the polymerase at the −3 and −4 positions (Fig. 3). In the T-3 complex, the DNA substrate (X-T-3) (Table 1) contains a single T substitution in the −3 position, while in the TT-4 complex, the DNA substrate (X-TT-4) (Table 1) contains T substitutions at both the −3 and −4 positions, with a single A in the primer strand that could potentially pair with either of the T's. Both structures contain dCTP as the incoming nucleotide and have been refined to high resolution limits of 2.0 Å (Table 2). The enzymes in the two complexes superimpose with a root mean square deviation (RMSD) of 0.21 Å (over 340 C-alpha atoms).
Table 2.
Data collection and refinement statistics
| Statisitics | Value(s)a |
|
|---|---|---|
| Dpo4 T-3 (PDB accession no. 3QZ7) | Dpo4 TT-4 (PDB accession no. 3QZ8) | |
| Data collection | ||
| Space group | P21212 | P21212 |
| No. of complexes in asymmetric unit | 1 | 1 |
| Unit cell dimensions (a, b, c [Å]) | 98.71, 102.46, 52.74 | 98.91, 102.69, 52.62 |
| Beamline/wavelength (Å) | X29A/1.0809 | X29A/1.0809 |
| Resolution range (Å) | 30–2.0 (2.03–2.00) | 25–2.0 (2.03–2.00) |
| No. of measured/unique reflections | 202,817/33,358 | 235,391/35,093 |
| Avg redundancy | 6.1 (1.9) | 6.7 (2.6) |
| Completeness (%) | 90.0 (31.9) | 94.7 (58.2) |
| Rmerge (%)b | 5.0 (34.6) | 7.0 (46.9) |
| I/σ(Ι) | 32.6 (1.6) | 26.0 (1.3) |
| Refinement | ||
| Resolution range (Å) | 25–2.0 | 25–2.0 |
| Rwork (%)c | 22.7 | 20.1 |
| Rfree (%)d | 26.7 | 24.2 |
| No. of nonhydrogen atoms | 3,421 | 3,636 |
| No. of protein atoms/ DNAs/water molecules/Ca2+ ions | 2,743/622/53/3 | 2,824/660/148/4 |
| Avg B-factors (Å2) | ||
| Protein/DNA/water/Ca2+ | 60.0/60.8/55.1/68.6 | 42.4/44.5/45.2/51.1 |
| RMSD | ||
| Bond length (Å)/bond angle (°) | 0.012/1.59 | 0.011/1.52 |
| Ramachandran plot (%) | ||
| Preferred regions/ allowed regions/ outliers | 97.1/2.6/0.3 | 97.1/2.9/0.0 |
Values for the outermost resolution shells are given in parentheses.
Rmerge = ΣhklΣj|Ij,hkl − [&lang]IhklÅ|/ΣhklΣjIj,hkl, where Ij,hkl is the integrated intensity of a given reflection.
Rwork = Σhkl|Fobserved − Fcalculated|/ΣhklFobserved.
Rfree was calculated using 5% of the data randomly omitted from the refinement.
In the T-3 complex, the T at position −3 is in an extrahelical conformation, bulged out on the minor groove side of the duplex, between the N-terminal polymerase domain and the C-terminal LF/PAD. The base itself packs against Gln82 of the polymerase palm domain, near the base of the fingers (Fig. 3D).
In the TT-4 complex, the T at position −4 is in an extrahelical conformation, also bulged out on the minor groove side of the duplex, between the N-terminal polymerase domain and the C-terminal LF/PAD (Fig. 3E). In this case, the bulged base packs against Ser103 and Ile104, the two residues immediately preceding the catalytic aspartate (Asp105) in the conserved “motif C” beta turn in the palm, and O4 of the bulged T hydrogen bonds with Gln82.
In both structures, the phosphate on the 3′ side of the bulged base protrudes into the minor groove of the DNA duplex. In the T-3 complex, the extrahelical phosphate is not stabilized by direct protein contacts, but in the TT-4 complex, the phosphate has electrostatic interactions with Arg240 (Fig. 3E). Contacts to the other phosphates in DNA are virtually identical to those formed by Dpo4 with standard primer-template duplexes lacking bulged bases (16, 29).
The TT-4 complex suggests that the extrahelical base is more stable at position −4 than at position −3, since there is no evidence in the electron density maps of an alternate conformation in which the −3 position is extrahelical, even though the A in the primer strand could potentially pair with the T at either position −3 or position −4. The additional stability of the position −4 bulge could arise from the contacts with Gln82 and/or Arg240. Any differences in stability, however, do not alter the rate of nucleotide incorporation under single-turnover conditions where the primer-template DNA is incubated with saturating amounts of Dpo4 prior to nucleotide addition (Fig. 2G).
DISCUSSION
Conserved template slippage mechanism arising from nonconserved polymerase-DNA contacts.
We conclude from the data presented here that Dpo4 uses a template slippage mechanism when making single-base deletion mutations in repetitive DNA sequences. Previous work on Dbh (6, 31) and E. coli DinB (10) indicates that they also use a template slippage mechanism. The archaeal and bacterial DinB polymerases, therefore, share a conserved mechanism when copying the kinds of repetitive sequences in which deletions most frequently occur.
Remarkably, however, the template slippage mechanism does not arise from conserved contacts between the polymerase and the skipped template base in these two archaeal enzymes. The polymerases contact the skipped template bases using completely different regions of the protein: Dpo4 sandwiches the extrahelical template bases (at positions −3 and −4) between the polymerase domain and the LF/PAD, while Dbh stabilizes these extrahelical bases on the surface of the LF/PAD (31).
Dpo4 and Dbh bind the DNA in such different ways because their LF/PADs are in very different positions relative to their polymerase domains. In Dpo4, the LF/PAD is in contact with the fingers of the polymerase. In Dbh, the LF/PAD is rotated approximately 50° away from the polymerase domain, resulting in a much wider gap between the two domains (31). Even though the residues of Dpo4 that directly contact the bulged base (Gln82 and Arg240) are conserved in Dbh (as Glu82 and Lys241), the Dbh residues are located approximately 10 Å away from the DNA, and so they cannot make the same contacts to the substrate.
Even though the archaeal polymerases do not use the same protein-DNA contacts to stabilize a skipped template base, other DinB orthologs may do so. For example, biochemical characterization of E. coli DinB suggests that this polymerase, like Dpo4, prefers when the extrahelical template base is at position −4, suggesting that DinB may stabilize the extrahelical base between the polymerase domain and the LF/PAD, as does Dpo4 (9a).
Structural implications for mutagenesis and translesion synthesis.
The T-3 and TT-4 complexes described here, together with structures of Dpo4 with N2-benzo[a]pyrene-dG (N2-BP-dG) adducts (2) and abasic sites (17), show how both damaged and undamaged bases interact with Dpo4 on the minor groove side of the duplex after having passed through the polymerase active site (Fig. 4). In the four structures in which the extrahelical nucleotide is either an abasic site (Ab-1 and Ab-2) or an undamaged pyrimidine (T-3 and TT-4), the ribose group adopts the same conformation, with the phosphate on the 3′ side displaced into the minor groove (Fig. 4A). This arrangement suggests that the extrahelical nucleotide could translocate from position −1 to position −4 without either the DNA or the protein having to undergo any significant conformational changes.
Translocation may not be as straightforward for bulkier minor groove adducts. In the two structures of extrahelical N2-BP-dG adducts (N2-BP-1 and N2-BP-2) (Fig. 4B), the lesion maintains essentially the same contacts with the protein, despite being located either one (N2-BP-1) or two (N2-BP-2) bases away from the template base that occupies the nascent base pair binding pocket. In the N2-BP-1 structure, the damaged base is swung out of the DNA duplex, but because the adjacent bases are not stacked together (i.e., there is a gap in the duplex where the −1 base would normally be located), the backbone phosphates are only minimally displaced from their positions in a standard DNA duplex. In contrast, the DNA backbone is strongly distorted in the N2-BP-2 structure, with the 5′ phosphate of the extrahelical damaged base displaced into the minor groove of the duplex, so that the adjacent template can stack together in the DNA duplex. These structures suggest that difficulty in DNA translocation may increase the likelihood of a deletion occurring during TLS.
To avoid futile cycles of TLS and proofreading, a Y-family polymerase must extend DNA synthesis far enough past a DNA lesion so that the replicative polymerase can productively continue DNA synthesis rather than using its exonuclease activity to remove the nucleotides that were just added during TLS. For Dpo4, the critical stage of continuing DNA synthesis may be when the damaged DNA reaches position −5. At this position, the linker connecting the polymerase domain and the LF/PAD closes off the groove between the two domains that provides the space for minor groove adducts to transition stepwise from the −1 to −4 positions.
The ultimate outcome of TLS is influenced by the entire series of specific protein-DNA interactions that occur as a polymerase first encounters a lesion and as it continues DNA synthesis. As the wide range of Y-family polymerase structures show, variations in protein conformation and sequence impact TLS in ways that are not always straightforward to predict.
ACKNOWLEDGMENTS
We thank M. Jackson for constructing the clone of Dpo4, J. Foti and G. Walker for communication of data prior to publication, and the staff at BNL NSLS beamline X29 for assistance during data collection. We acknowledge the use of the Wadsworth Center's X-ray Crystallography Core.
This work was supported by NIH grant R01-GM080573 to J.D.P. Use of the BNL NSLS was supported by the U.S. Department of Energy.
Footnotes
Published ahead of print on 18 March 2011.
REFERENCES
- 1. Adams P. D., et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bauer J., et al. 2007. A structural gap in Dpo4 supports mutagenic bypass of a major benzo[a]pyrene dG adduct in DNA through template misalignment. Proc. Natl. Acad. Sci. U. S. A. 104:14905–14910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bebenek K., Kunkel T. A. 1990. Frameshift errors initiated by nucleotide misincorporation. Proc. Natl. Acad. Sci. U. S. A. 87:4946–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bloom L. B., et al. 1997. Fidelity of Escherichia coli DNA polymerase III holoenzyme. The effects of beta, gamma complex processivity proteins and epsilon proofreading exonuclease on nucleotide misincorporation efficiencies. J. Biol. Chem. 272:27919–27930 [DOI] [PubMed] [Google Scholar]
- 5. Collaborative Computational Project, Number 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 [DOI] [PubMed] [Google Scholar]
- 6. DeLucia A. M., Grindley N. D., Joyce C. M. 2007. Conformational changes during normal and error-prone incorporation of nucleotides by a Y-family DNA polymerase detected by 2-aminopurine fluorescence. Biochemistry 46:10790–10803 [DOI] [PubMed] [Google Scholar]
- 7. Efrati E., Tocco G., Eritja R., Wilson S. H., Goodman M. F. 1997. Abasic translesion synthesis by DNA polymerase beta violates the “A-rule”. Novel types of nucleotide incorporation by human DNA polymerase beta at an abasic lesion in different sequence contexts. J. Biol. Chem. 272:2559–2569 [DOI] [PubMed] [Google Scholar]
- 8. Emsley P., Lohkamp B., Scott W. G., Cowtan K. 2010. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66:486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fiala K. A., Suo Z. 2004. Pre-steady-state kinetic studies of the fidelity of Sulfolobus solfataricus P2 DNA polymerase IV. Biochemistry 43:2106–2115 [DOI] [PubMed] [Google Scholar]
- 9a. Foti J. J., Walker G. C. 2011. Efficient extension of slipped DNA intermediates by DinB is required to escape primer template realignment by DnaQ. J. Bacteriol. 193:2637–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Foti J. J., Delucia A. M., Joyce C. M., Walker G. C. 2010. UmuD(2) inhibits a non-covalent step during DinB-mediated template slippage on homopolymeric nucleotide runs. J. Biol. Chem. 285:23086–23095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jarosz D. F., Godoy V. G., Delaney J. C., Essigmann J. M., Walker G. C. 2006. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature 439:225–228 [DOI] [PubMed] [Google Scholar]
- 12. Kokoska R. J., Bebenek K., Boudsocq F., Woodgate R., Kunkel T. A. 2002. Low fidelity DNA synthesis by a Y family DNA polymerase due to misalignment in the active site. J. Biol. Chem. 277:19633–19638 [DOI] [PubMed] [Google Scholar]
- 13. Kunkel T. A. 2009. Evolving views of DNA replication (in)fidelity. Cold Spring Harb. Symp. Quant. Biol. 74:91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kunkel T. A. 1986. Frameshift mutagenesis by eucaryotic DNA polymerases in vitro. J. Biol. Chem. 261:13581–13587 [PubMed] [Google Scholar]
- 15. Kunkel T. A., Soni A. 1988. Mutagenesis by transient misalignment. J. Biol. Chem. 263:14784–14789 [PubMed] [Google Scholar]
- 16. Ling H., Boudsocq F., Woodgate R., Yang W. 2001. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell 107:91–102 [DOI] [PubMed] [Google Scholar]
- 17. Ling H., Boudsocq F., Woodgate R., Yang W. 2004. Snapshots of replication through an abasic lesion; structural basis for base substitutions and frameshifts. Mol. Cell 13:751–762 [DOI] [PubMed] [Google Scholar]
- 18. McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., Read R. J. 2005. Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 61:458–464 [DOI] [PubMed] [Google Scholar]
- 19. McDonald J. P., et al. 1999. Novel human and mouse homologs of Saccharomyces cerevisiae DNA polymerase eta. Genomics 60:20–30 [DOI] [PubMed] [Google Scholar]
- 20. Ohashi E., et al. 2000. Fidelity and processivity of DNA synthesis by DNA polymerase kappa, the product of the human DINB1 gene. J. Biol. Chem. 275:39678–39684 [DOI] [PubMed] [Google Scholar]
- 21. Ohmori H., et al. 2001. The Y-family of DNA polymerases. Mol. Cell 8:7–8 [DOI] [PubMed] [Google Scholar]
- 22. Otwinowski Z., Minor W. 1997. Processing of X-ray diffration data collected in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 23. Potapova O., Grindley N. D., Joyce C. M. 2002. The mutational specificity of the Dbh lesion bypass polymerase and its implications. J. Biol. Chem. 277:28157–28166 [DOI] [PubMed] [Google Scholar]
- 24. Rechkoblit O., et al. 2006. Stepwise translocation of Dpo4 polymerase during error-free bypass of an oxoG lesion. PLoS Biol. 4:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schrödinger LLC. 2010. The PyMOL molecular graphics system, version 1.3r1. Schrödinger, LLC, Portland, OR: http://www.pymol.org/pymol [Google Scholar]
- 26. Streisinger G., et al. 1966. Frameshift mutations and the genetic code. Cold Spring Harb. Symp. Quant. Biol. 31:77–84 [DOI] [PubMed] [Google Scholar]
- 27. Studier F. W. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41:207–234 [DOI] [PubMed] [Google Scholar]
- 28. Tippin B., Kobayashi S., Bertram J. G., Goodman M. F. 2004. To slip or skip, visualizing frameshift mutation dynamics for error-prone DNA polymerases. J. Biol. Chem. 279:45360–45368 [DOI] [PubMed] [Google Scholar]
- 29. Vaisman A., Ling H., Woodgate R., Yang W. 2005. Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis. EMBO J. 24:2957–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Washington M. T., Johnson R. E., Prakash S., Prakash L. 1999. Fidelity and processivity of Saccharomyces cerevisiae DNA polymerase eta. J. Biol. Chem. 274:36835–36838 [DOI] [PubMed] [Google Scholar]
- 31. Wilson R. C., Pata J. D. 2008. Structural insights into the generation of single-base deletions by the Y family DNA polymerase dbh. Mol. Cell 29:767–779 [DOI] [PubMed] [Google Scholar]
- 32. Wolfle W. T., Washington M. T., Prakash L., Prakash S. 2003. Human DNA polymerase kappa uses template-primer misalignment as a novel means for extending mispaired termini and for generating single-base deletions. Genes Dev. 17:2191–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zang H., et al. 2006. Efficient and high fidelity incorporation of dCTP opposite 7,8-dihydro-8-oxodeoxyguanosine by Sulfolobus solfataricus DNA polymerase Dpo4. J. Biol. Chem. 281:2358–2372 [DOI] [PubMed] [Google Scholar]
- 34. Zhou B. L., Pata J. D., Steitz T. A. 2001. Crystal structure of a DinB lesion bypass DNA polymerase catalytic fragment reveals a classic polymerase catalytic domain. Mol. Cell 8:427–437 [DOI] [PubMed] [Google Scholar]