Abstract
During spore formation, Bacillus subtilis divides asymmetrically, resulting in two cells with different fates. Immediately after division, the transcription factor σF becomes active in the smaller prespore, followed by activation of σE in the larger mother cell. We recently showed that a delay in σE activation resulted in the novel phenotype of two spores (twins) forming within the same mother cell. Mother cells bearing twins are substantially longer than mother cells with single spores. Here we explore the regulation of the growth and DNA replication of the mother cell. We find that length correlates with chromosome number in the mother cell. We show that replication and growth could occur after asymmetric division in mother cells with no active σE. In contrast, when σE was active, replication and growth ceased. In growing mother cells, with no active σE, Spo0A-directed transcription levels remained low. In the presence of active σE, Spo0A-directed gene expression was enhanced in the mother cells. Artificial Spo0A activation blocked mother cell growth in the absence of σE. Spo0A activation blocked growth even in the absence of SirA, the Spo0A-directed inhibitor of the initiation of replication. Together, the results indicate that the burst of Spo0A-directed expression along with the activation of σE provides mechanisms to block the DNA replication and growth of the mother cell.
INTRODUCTION
The formation of spores by Bacillus subtilis is a primitive system of cell differentiation that is triggered by nutrient depletion (37). An early stage in the process is an asymmetrically located division, which divides the bacterium into two unequal cells: the larger mother cell and the smaller prespore (also called the forespore). When first formed, the prespore contains only the origin-proximal 30% of a chromosome; the remainder of the chromosome is then translocated into the prespore, leaving another copy of the chromosome in the mother cell (55). The prespore becomes engulfed by the mother cell and develops into a mature spore; the mother cell is necessary for spore formation but ultimately lyses. Ordinarily, chromosome replication is completed before asymmetric division, and there is little or no growth of the mother cell (3, 10, 22, 51). However, we have found that in some genetic backgrounds, DNA replication may continue, and the mother cell may continue to grow. These events occur when prespore-to-mother cell signaling is delayed, conditions that can lead to the formation of two spores (twins), rather than one, within one mother cell (13). Here we explore the controls of DNA replication and growth in the mother cell.
A fundamental feature of spore formation is the existence of distinct programs of gene expression in the two cell types that depend on different RNA polymerase sigma factors. The first sigma factor to become active, σF, does so in the prespore immediately after septum formation (21, 24). The σF regulon includes about 50 genes (47, 53). Expression of one of these genes, spoIIR, is critical to the activation of σE in the mother cell (28, 31). The SpoIIR protein is thought to act across the division septum via SpoIIGA to cause cleavage of the pro-sequence from pro-σE and hence to activate σE; activation of σE occurs exclusively in the mother cell (21, 27, 28, 31). Inactivation of the structural genes for σF, σE, or SpoIIR results in an “abortively disporic” phenotype in which septa are formed at both poles of the cell, but development proceeds no further; in this circumstance, each prespore receives a chromosome, while the mother cell becomes anucleate (14, 37, 44). The second asymmetric division is largely accounted for by the finding that expression of particular σE-directed genes is required to prevent the formation of the second septum and to allow normal spore development (1, 12, 39). Delaying and attenuating spoIIR expression can also lead to many organisms displaying this phenotype, although a few sporulate successfully (29, 57). Delaying spoIIR expression with no attenuation can result in some organisms displaying an additional phenotype: twin spore formation (13). During twin spore formation, there were extra rounds of chromosome replication, with the result that both developing prespores had a chromosome, and the mother cell also had one or more chromosomes. In studies by time lapse microscopy, twin-spore-forming organisms were observed to be generated via a process termed “sporulation escape” (13). Escape occurred after an asymmetrically located sporulation septum had formed and σF become active in the prespore. It involved DNA replication and growth in the mother cell. In these “escaping” mother cells, σE was not active; the mother cells could proceed to symmetric division and further growth or to asymmetric division, activation of σE, and the formation of twin spores (13).
During vegetative growth, B. subtilis divides at midcell with remarkable accuracy (32). Chromosome replication is intimately tied to the vegetative cell division cycle (52). The formation of spores by B. subtilis involves a modified cell division (25). However, this division is distinct from vegetative division in several substantive ways, indicating sporulation-specific controls: it is triggered by starvation; the division occurs near the cell pole (37); the structure of the septum is very different (24); chromosome partitioning lags dramatically behind septation (55, 56); chromosome replication does not continue after the division (3, 51); the division occurs under conditions of nutrient limitation, and there is little or no subsequent cell growth (3). As mentioned above, we have found that growth and replication can continue in some genetic backgrounds. Here we show that growth of the mother cell, the larger cell formed after the asymmetric division, is blocked by σE activation in the mother cell. It is also blocked by enhanced activity of Spo0A, the master transcription factor for the entry into spore formation (4, 19); this effect is distinct from the Spo0A-directed block of DNA replication that is mediated by SirA (40, 51).
MATERIALS AND METHODS
Media.
B. subtilis was grown in modified Schaeffer's sporulation medium (MSSM) or on Schaeffer's sporulation agar as described previously (7, 38). When required, the medium was supplemented with chloramphenicol at 5 μg/ml, erythromycin at 1.5 μg/ml, neomycin at 3.5 μg/ml, spectinomycin at 100 μg/ml, or tetracycline at 10 μg/ml. Escherichia coli was grown on Luria-Bertani lysogeny broth agar, containing 100 μg ampicillin/ml when required.
Strains.
The B. subtilis strains used in this study are listed in Table 1. B. subtilis 168 strain BR151 (trpC2 metB10 lys-3) was the parental strain; all the strains listed had its auxotrophic mutations.
Table 1.
B. subtilis strains used
| Straina | Relevant genotypeb | Source or reference |
|---|---|---|
| SL10257 | thrC::PspoIIQ-gfp | Lab stock |
| SL12420 | PspoIIG-gfp@PspoIIG | Lab stock |
| SL12475 | thrC::PspoIID-gfp | Lab stock |
| SL14118 | spoIIR::spc thrC::PspoIIQ-gfp | This study |
| SL14123 | spoIIRdelay | 13 |
| SL14130 | spoIIRdelayspo0J-gfp@spo0J | This study |
| SL14240 | spoIIR::spc spo0J-gfp@spo0J | This study |
| SL14519 | spoIIRdelayspo0J (359°)::pAT15 (lacO cassette cat) thrC::Ppen-gfpmut2′–′lacIΔ11 mls) | This study |
| SL14826 | spoIIRdelayspo0J-gfp@spo0J thrC::PspoIID-gfp | This study |
| SL14922 | spoIIRdelay PspoIIG-gfp@PspoIIG | This study |
| SL15001 | thrC::PspoIID-gfp spoIIP::tet | This study |
| SL15002 | thrC::PspoIID-gfp sirA::tet | This study |
| SL15261 | spoIIGB::erm spo0J-gfp@spo0J | This study |
| SL15268 | spoIIR::spc PspoIIG-gfp@PspoIIG | This study |
| SL15277 | PspoIIG-gfp@PspoIIGsirA::tet | This study |
| SL15322 | thrC::PspoIID-gfp sirA::tet spoIIP::kan | This study |
| SL15325 | spoIIR::spc PspoIIG-gfp@PspoIIGyabA::cat | This study |
| SL15326 | PspoIIG-gfp@PspoIIGyabA::cat | This study |
| SL15337 | thrC::PspoIID-gfp spoIIP::tet yabA::cat | This study |
| SL15347 | thrC::Psda-gfp | This study |
| SL15373 | PspoIIG-gfp@PspoIIGthrC::Pspac-kinA | This study |
| SL15376 | spoIIR::spc PspoIIG-gfp@PspoIIGthrC::Pspac-kinA | This study |
| SL15439 | thrC::PspoIID-gfp yabA::cat sirA::tet | This study |
| SL15449 | spoIIRdelaythrC::Psda-gfp | This study |
All strains are in the genetic background of B. subtilis 168 strain BR151 (trpC2 lys-3 metB10). They have all its auxotrophic markers.
@ indicates that the fusion has been introduced by single-crossover (Campbell-like) recombination.
Strain SL14123 contained spoIIR::spc and ppsB::PspoIIQ-spoIIR (13), which was introduced by double crossover at ppsB using linearized pVK317 as the donor; this combination of spoIIR manipulations is referred to below as spoIIRdelay. SL14519 was derived from SL14123 using DNA from DCL693. Strain DCL693 was kindly provided by Alan Grossman (Massachusetts Institute of Technology). It contains thrC::Ppen-gfpmut2′-′lacIΔ11 (gene product referred to as GFP-LacI) mls; it also has pAT15, harboring tandem repeats of lacO, integrated by single crossover at the origin-proximal spo0J locus (48, 54). Strain SL14130 was derived from SL14123 using DNA from strain MMB354 (kindly provided by Alan Grossman); MMB354 contains the translational fusion spo0J-gfp integrated by single crossover at spo0J. SL12475 was transformed with DNA from LR69 to yield SL15002; strain LR69 contains a sirA-null mutation and was kindly provided by Richard Losick (Harvard University). Strain SL12420 was transformed with DNA from AI109 to yield SL15326; strain AI109 contains a yabA-null mutation and was kindly provided by Alan Grossman. To construct Psda-gfp, a 500-bp DNA region that contains the promoter of sda (49) was amplified by PCR from BR151 chromosomal DNA and was cloned upstream of gfp into pVK370, which was kindly provided by Vasant Chary. The construct was introduced by double crossover into the thrC locus of BR151 to yield SL15347. In order to construct Pspac-kinA, a 1.8-kb DNA region that contains the kinA open reading frame (ORF), including an enhanced ribosome binding site (RBS) (9), was amplified by PCR from BR151 chromosomal DNA and was cloned downstream of the Pspac promoter into pVK11 (kindly provided by Vasant Chary). The construct was introduced by double crossover into the thrC locus of SL12420 to yield SL15373. The PspoIIG-gfp, PspoIID-gfp, spoIIR::spc, spoIIGB::erm, and spoIIP::tet constructs were kindly provided by Vasant Chary. Plasmids were constructed in E. coli DH5α, and their structures were confirmed by restriction enzyme digestions and also by PCR.
Fluorescence microscopy.
The conditions used for the growth and imaging of the samples were essentially those described previously (3, 7). Cell lengths were measured using the ImageJ program, version 1.36b (http://rsb.info.nih.gov/ij/).
Time lapse studies.
Exponentially growing bacteria were inoculated onto a thin semisolid matrix of heated MSSM supplemented with 1.5% low-melting-point agarose attached to a microscope slide. These slides were prepared as described previously (50). For membrane staining, FM4-64 (Invitrogen) was added at a concentration of 0.4 μg/ml to the agarose pad before the addition of bacteria. For strains carrying a Pspac-kinA construct, the inducer isopropyl-β-d-thiogalactopyranoside (IPTG) was added at a 1 mM concentration to the agarose pad before the addition of bacteria. Microscope slides were monitored with a TCS SP5 confocal microscope system (Leica) contained within a temperature-controlled environmental chamber set at 30°C. Fluorescent images were recorded at specified intervals. To prevent phototoxicity, the excitation light for the green fluorescent protein (GFP) and FM4-64 signals was limited to 10%. During time lapse studies, GFP levels were quantitated with the Leica TCS SP5 confocal system software.
Assessment of the stages of engulfment.
The names of morphological intermediates in the process of engulfment are essentially those suggested by Illing and Errington (26) and used in reference 3. The stages were visualized using the membrane stain FM4-64 (45). The spoIIRdelay strains showed different developmental fates within the same population. Specific types were assigned to classes as follows: stage 0 organisms to the aseptate class, stage IIi and IIii organisms to the monoseptate class, stage III and III+ organisms to the monoengulfment class, stage IIi/IIi and IIii/IIii organisms to the biseptate class, and stage IIiii/IIiii, III/III, and III+/III+ organisms to the biengulfment class.
Other methods.
The methods used for the transformation of B. subtilis and for sporulation by exhaustion in MSSM, as well as other methods, were essentially those described previously (6). The initiation of spore formation in MSSM is taken to occur at the end of exponential growth.
RESULTS
Mother cells of twin-forming organisms have increased length.
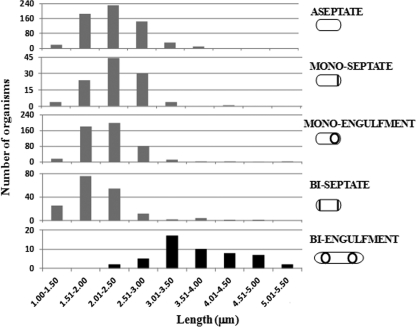
Twin spore formation was achieved by delaying the activation of σE. For this purpose, spoIIR was relocated to a terminus-proximal position and was expressed from the strong σF-directed spoIIQ promoter, a construct referred to below as spoIIRdelay (13). Under liquid sporulation conditions, spoIIRdelay strains contained a mixture of monosporic (normal spore-forming), abortively disporic (not proceeding to spore formation), and twin-forming organisms, as well as nonsporulating bacteria. We measured the lengths of the mother cells of organisms present at various stages of sporulation in samples taken at different times from batch cultures of a spoIIRdelay strain (SL14123) (Fig. 1; the different cell types are represented schematically). Twin-forming organisms were significantly longer than monosporic or abortively disporic cells within the same population. The mother cells of twin-forming organisms (the “biengulfment” class) had lengths between 3 and 5 μm. In contrast, most mother cells of monoseptate and monoengulfment organisms had lengths between 1.5 and 3 μm. Mother cells of biseptate organisms were also predominantly in the 1.5- to 3-μm range; the biseptate organisms could be either abortively disporic or intermediates in twin formation. There was also a small subpopulation (3 to 5%) of monosporic and biseptate organisms whose mother cells had lengths comparable to those of twin-forming organisms (>3 μm). For the parental spoIIR+ strain (SL10257), most (311/326 [95%]) monoseptate and monoengulfment organisms observed had mother cells with lengths between 1.5 and 3 μm, similar to those for the spoIIRdelay strain; for a spoIIR-null strain (SL14118), almost all (148/150) monoseptate or biseptate organisms had mother cells <3 μm long. No biengulfment organisms were observed for either the parental or the spoIIR-null strain, consistent with the requirement of the spoIIRdelay manipulation for twin formation. We conclude that twin formation involves elongated mother cells.
Fig. 1.
Mother cells of twin-forming organisms are longer. Strain SL14123 (spoIIRdelay) was induced to form spores in MSSM. Lengths are shown for the different classes of organisms present in sporulating cultures. Data for each class are combined from samples taken 5, 6, and 7 h after the end of exponential growth. The lengths of whole cells are given for aseptate organisms, and those of mother cells are given for the indicated classes of sporulating organisms (monoseptate, monoengulfment, biseptate, and biengulfment). Membranes were visualized by staining with FM4-64.
There is a correlation between the length of the mother cell and the number of chromosome origins present in it.
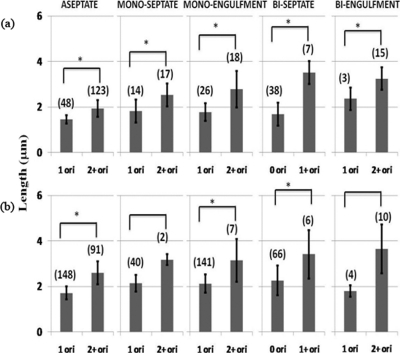
Usually, during the sporulation of B. subtilis, chromosome replication is completed by the time of the asymmetric division, and there are just two chromosomes: one in the mother cell and the other destined for the prespore but with the origin-distal 70% still in the mother cell (3, 51, 55). An increase in chromosome number is a predisposing factor for twin formation (13). Given that there is a correlation between cell length and chromosome replication during vegetative growth (22, 30, 46), we investigated the question of whether the enhanced length of mother cells of twin-forming strains is associated with an increase in the chromosome content. To estimate the chromosome number, we used fluorescent tags for the chromosome origin region. We measured the lengths of mother cells present at various stages of sporulation of spoIIRdelay strains containing Spo0J-GFP-tagged (SL14130) or GFP-LacI-tagged (SL14519) chromosomal origins (Fig. 2). These tags give a reasonable estimate of the chromosome number provided replication is complete, as is normally the case after the formation of the sporulation septum (3, 51); however, in the case of twin formers, they could provide an overestimate of completed chromosomes if replication is still continuing.
Fig. 2.
Correlation between length and the number of chromosome origins present in the mother cell. spoIIRdelay strains with Spo0J-GFP-tagged chromosomal origins (SL14130) (a) or with GFP-LacI-tagged chromosomal origins (SL14519) (b) were analyzed. Cultures were induced to form spores in MSSM. Samples were analyzed 4 h after the end of exponential growth. Membranes were visualized by staining with FM4-64. The figure shows the number of origin foci present in the entire cell for aseptate organisms and in the mother cell for sporulating organisms; prespores always had a single GFP focus. The lengths of whole organisms are given for aseptate organisms, and the lengths of mother cells are given for sporulating cells. Bars represent average lengths, and standard deviations are shown. The number of organisms measured for each group is given in parentheses. Asterisks indicate significant differences (P, <0.05 by the t test) in length between origin classes.
Aseptate (stage 0) organisms usually showed either one or two GFP foci; most of the bacteria containing two Spo0J-GFP foci were longer than those that contained one focus (1.9 ± 0.29 μm and 1.46 ± 0.18 μm, respectively; P, <0.05 by the t test) (strain SL14130) (Fig. 2a). A similar distinction was also apparent for the LacI-GFP fusion (strain SL14519) (Fig. 2b).
For organisms that had reached stage II of sporulation (septation), we compared the mother cell length to the number of origin foci in that compartment (in all organisms examined, the prespores contained a single GFP focus). A trend of increased length with an increased number of foci is apparent for all morphological classes (Fig. 2). For all cell types except biseptate cells, mother cells had predominantly either one or two foci; occasional mother cells had more than two foci, and these data were pooled with the data for two foci. The increase in length between cells with one focus and those with more than one focus was significant for all morphological classes of strain SL14130 (Fig. 2a). For biseptate organisms, mother cells often had no focus, indicating no DNA, which is typical of abortively disporic organisms; the difference in length between those with and those without a focus was significant (Fig. 2). The same general trend was suggested for strain SL14519, although the distinctions were less clear-cut (Fig. 2b). Collectively, the results point to a correlation between length and the number of origins contained in the mother cell.
Replication and growth of the mother cell could occur in the absence of active σE, but not when σE became active.
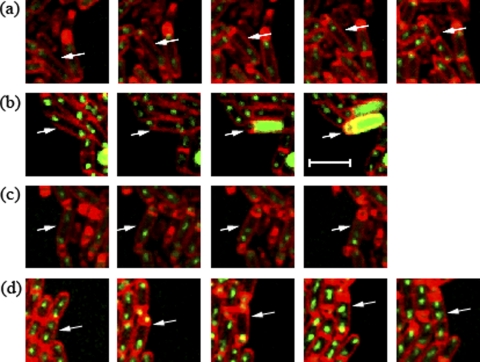
The results discussed above suggested that the spoIIRdelay manipulation could result in growth of the mother cell. We hypothesized that this mother cell growth is permitted because of the absence of σE activity as a consequence of the delay in spoIIR expression. We performed time lapse experiments in order to monitor the growth of individual mother cells. The spoIIRdelay strain used (SL14826) contained a σE-directed GFP reporter as well as Spo0J-GFP-tagged origins; if σE became active, its GFP signal was readily detected but swamped the signal from Spo0J-GFP. Under the time lapse conditions, by monitoring of bacteria in agarose pads, three classes of organisms that had undergone asymmetric division were observed: (i) monosporic organisms, which formed spores normally, with σE active in the mother cell and little or no growth of the mother cell; (ii) abortively disporic organisms, where the mother cell lacked a Spo0J-GFP focus (and, by inference, lacked a chromosome) and did not grow; and (iii) organisms that followed a process of “sporulation escape” described previously (13) (Table 2). During this escape process, replication and growth of the mother cell continued after the asymmetric division; subsequently, a septum was often formed in the middle of the mother cell, separating the organism into two daughter cells, which resumed vegetative growth. No σE activity was detected in these growing mother cells (Fig. 3a, arrow). In some cases, not studied here, growing mother cells divided asymmetrically and went on to form twin spores (13).
Table 2.
Impaired activation of σE permits mother cell growtha
| Strain | Relevant genotypeb | Frequency (%) of the following developmental fate after asymmetric divisionc: |
Total organisms scored | ||
|---|---|---|---|---|---|
| Monospore (no MC growth) | Abortively disporic (no MC growth) | Escape (MC growth) | |||
| SL14826 | spoIIRdelay | 64 | 13 | 22 | 90 |
| SL14130 | spoIIRdelay | 60 | 13 | 27 | 126 |
| SL14315 | spo+ | 100 | 0 | 0 | 64 |
| SL14240 | spoIIR | 0 | 37 | 63 | 103 |
| SL15261 | spoIIGB | 0 | 34 | 66 | 88 |
Bacteria were grown on top of a thin layer of agarose in such a way that single cells grew into sporulating microcolonies, which were monitored by time lapse microscopy (50). Fluorescence images were acquired at periodic intervals for FM4-64 (membrane staining) and GFP (origin location and/or σE activity [see the text]). Cell fates were recorded for all organisms that underwent asymmetric division.
All strains contained the Spo0J-GFP fusion for the visualization of chromosome origins. Strain SL14826 contained thrC::PspoIID-gfp for the visualization of σE activity.
MC, mother cell.
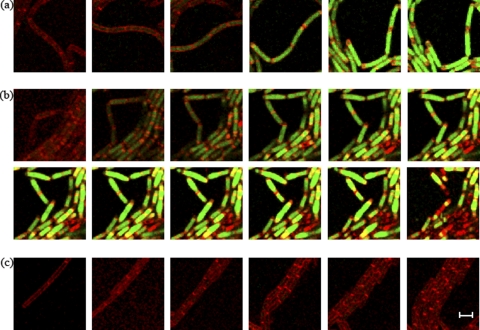
Fig. 3.
Replication and growth of the mother cell could commence in the absence of active σE. Arrows identify organisms showing the fates discussed in the text. (a and b) Time lapse images of a spoIIRdelay strain (SL14826) with Spo0J-GFP-tagged chromosomal origins and a PspoIID-gfp fusion for the visualization of σE activity. (a) σE is not active in the mother cell. (b) σE is active in the mother cell. (c) Images of a spoIIRdelay strain with Spo0J-GFP-tagged chromosomal origins (SL14130). The initiation of engulfment indicates that σE is active in the mother cell. (d) Images of a spoIIR::spc strain with Spo0J-GFP-tagged chromosomal origins (SL14240). Inactivation of spoIIR results in no σE activity in the mother cell. Membranes were visualized by staining with FM4-64. Images were taken every 30 min. Bar, 3 μm.
Because of the overinitiating effect of the Spo0J-GFP construct (13, 35), initiation of replication (inferred from the formation of bilobed Spo0J-GFP foci) occurred in many organisms soon after they had formed an asymmetric septum. As mentioned in the preceding paragraph, mother cells that did not have active σE grew and continued chromosome replication, as inferred from the separation of the bilobed origin foci. However, in mother cells that did activate σE, growth ceased (Fig. 3b, arrow). The σE-directed GFP signal swamped the Spo0J-GFP signal so that it was not possible to tell if the bilobed origin foci became separated, which would indicate continued replication. In order to monitor the behavior of the origins in mother cells with active σE, we also studied an isogenic strain (SL14130) that lacked the σE-directed GFP reporter, and we inferred σE activation by the initiation of engulfment, which requires σE activity (1). The proportions of the three classes in strain SL14130 were similar to those in SL14826 (Table 2). In strain SL14130, bilobed origin foci in the mother cells, though mobile, stayed close to each other when engulfment proceeded and did not become resolved into two well-separated foci. This behavior suggests that replication had started and then stalled; the mother cells did not grow (Fig. 3c, arrow). The results suggest that continued replication and growth of the mother cell ceased when σE became active in that compartment. Consistent with this suggestion, introduction of Spo0J-GFP into the parental spoIIR+ strain did not result in growing mother cells after asymmetric division, because σE was active in all sporulating bacteria (as assayed by the initiation of engulfment that occurred soon after septation) (strain SL14315) (Table 2). All mother cells of SL14315 contained either one focus or two closely attached origin foci when newly formed. As engulfment proceeded, there was no further initiation of DNA replication, and bilobed origin foci remained closely associated, suggesting that replication was blocked.
We also tested the behavior of Spo0J-GFP-tagged origins in a spoIIR-null strain in which σE is not active and a second polar septum is commonly formed. Time lapse studies with this strain revealed two fates: first, formation of abortively disporic organisms that showed an origin focus in each of the two prespores and no focus in the mother cell; second, mother cells that had two or more foci after the formation of a single septum (strain SL14240) (Table 2). The latter mother cells grew (Fig. 3d, arrow); often, a central division septum was eventually formed in the mother cell. A mutant in which the structural gene for σE, spoIIGB, was inactivated (strain SL15261) showed the same two phenotypes as the spoIIR mutant (Table 2). Collectively, these results suggest that mother cells containing a chromosome could grow after septation provided that σE was not active.
Role of spoIIP in controlling the growth of the mother cell.
Growing mother cells were observed only when σE activity was impaired. Plausibly, the product of a σE-directed gene inhibited the growth of the mother cell. A likely candidate was the spoIIP locus. A key role for spoIIP in growth control was suggested by a study of the commitment of organisms to continue forming spores despite the restoration of nutrients. In particular, mother cells of organisms at stage II of sporulation were able to resume growth when nutrients were added to them if spoIIP was inactivated, but not if spoIIP was functional (11). In order to investigate a potential role of spoIIP in the growth of the mother cell, studied here, we monitored a spoIIP-null strain containing a σE-directed GFP reporter (SL15001). With this strain, we observed no growing mother cells (0/320); in all organisms observed to divide asymmetrically, σE became active soon after septation (320/320), and sporulation was blocked at that stage. Similar results were obtained in overreplicating sirA spoIIP or yabA spoIIP double mutants containing a σE-directed GFP reporter (strains SL15322 and SL15337, for which 400/400 and 91/91 organisms, respectively, displayed σE activity and no growth). Thus, mother cell growth was blocked when σE was activated after septation, independently of SpoIIP. This result distinguishes the regulation of mother cell growth, described here, from the regulation of commitment (11).
Growing mother cells display low levels of Spo0A activity.
The results described above indicated an important role for σE in inhibiting replication and mother cell growth. It is also possible that another transcriptional regulator is involved; a likely candidate is Spo0A, the master regulator for entrance to spore formation. In addition to its role at the start of spore formation, active Spo0A continues to function in the mother cell at later stages of sporulation, when it accumulates to high levels (18, 19). Active Spo0A directs the expression of the gene encoding SirA, an inhibitor of the initiation of DNA replication (40, 51). Spo0A is also thought to suppress the expression of genes for known cell wall hydrolases and cell division proteins (2, 17). Thus, it is plausible that the high activity of Spo0A in the mother cell (19) leads to inhibition of DNA replication and suppression of growth.
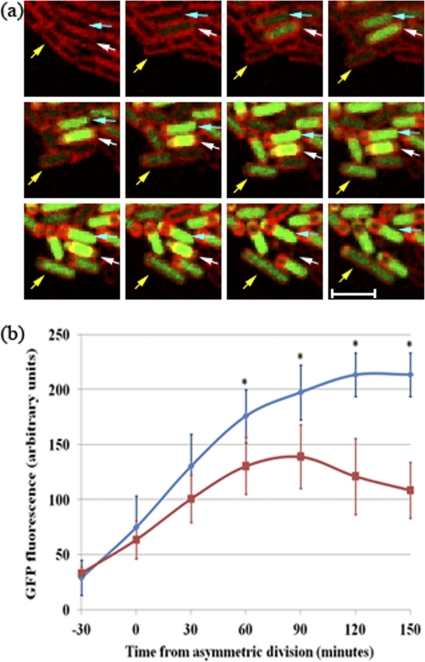
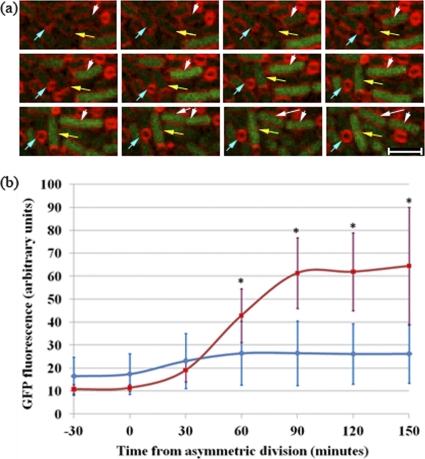
To test for a role of Spo0A in mother cell growth, we used time lapse microscopy to examine the levels of Spo0A-directed transcription in mother cells of organisms displaying the different phenotypes observed in spoIIRdelay strains. For this purpose, we used a Spo0A-directed GFP reporter (PspoIIG-gfp; strain SL14922). We again observed three fates after asymmetric division (Fig. 4a, arrows): monospore formers (blue arrows), abortively disporic organisms (white arrows), and growing mother cells (yellow arrows) (Table 3). All organisms showed some Spo0A-directed GFP expression before the formation of the asymmetric septum. Soon after septation, GFP expression was enhanced in the mother cells of monospore formers and became stronger as engulfment proceeded (Fig. 4b). This burst of Spo0A-directed GFP expression in mother cells of monosporic organisms has been reported previously (19). In growing mother cells, Spo0A-directed GFP expression also increased after septation, but not to the extent observed in mother cells of monospore-forming organisms. Moreover, after this early increase, GFP levels decreased as the mother cell grew (Fig. 4b). Growing mother cells that displayed this reduced Spo0A activity often divided, and the resulting daughter cells resumed vegetative growth. Similar results were obtained using another Spo0A-directed GFP reporter, PspoIIE-gfp (data not shown). Mother cells of abortively disporic organisms become anucleate, making interpretation of transcription data difficult; they were not included in the analysis.
Fig. 4.
Spo0A activity is impaired in growing mother cells. (a) Time lapse images of a spoIIRdelay strain (SL14922) with PspoIIG-gfp for the visualization of Spo0A-directed gene expression. The panel shows examples of a monosporic organism (blue arrows), an abortively disporic organism that eventually lyses (white arrows), and an organism with a growing mother cell that gives rise to two daughter cells that continue to grow vegetatively (yellow arrows). Membranes were visualized by staining with FM4-64. Images were taken every 30 min. Bar, 3 μm. (b) Quantitation of GFP levels present in the mother cell compartments of organisms with different phenotypes. The time lapse experiment illustrated in panel a was analyzed. GFP levels were analyzed for 25 monosporic organisms (blue) and 5 organisms whose mother cells grew after asymmetric division (red); for the latter group, growth became apparent about 90 min after septation. Engulfment was completed 60 to 90 min after the asymmetric division of monosporic organisms. The average fluorescence values at each time are shown with standard deviations. Asterisks indicate significant differences (P, <0.05 by the t test) between the two classes.
Table 3.
Cell fates and Spo0A activity in mother cellsa
| Strain | Relevant genotypeb | Frequency (%) of the following developmental fate after asymmetric divisionc: |
Total organisms scored | ||
|---|---|---|---|---|---|
| Monospore (no MC growth) | Abortively disporic (no MC growth) | Escape (MC growth) | |||
| SL14922 | spoIIRdelay | 74 | 21 | 5 | 273 |
| SL15268 | spoIIR | 0 | 84 | 16 | 104 |
| SL12420 | spo+ | 100 | 0 | 0 | 269 |
| SL15325 | spoIIR yabA | 0 | 25 | 75 | 81d |
| SL15326 | spo+ yabA | 100 | 0 | 0 | 152 |
Bacteria were grown on top of a thin layer of agarose in such a way that single cells grew into sporulating microcolonies, which were monitored by time lapse microscopy (50). Fluorescence images were acquired at periodic intervals for FM4-64 (membrane staining) and GFP (Spo0A activity [see the text]). Cell fates were recorded for all organisms that underwent asymmetric division.
All strains contained PspoIIG-gfp@spoIIG for the visualization of Spo0A activity. Reduced activity was displayed in growing mother cells of strains SL14922, SL15268, and SL15325.
MC, mother cell.
Twenty other bacteria displayed strong Spo0A activity but did not undergo sporulation division and eventually lysed; they were not considered further.
Growing mother cells exhibiting reduced levels of Spo0A-directed GFP were also observed for the spoIIR-null background (strain SL15268) (Table 3), along with abortively disporic organisms that displayed enhanced GFP levels. We observed no examples of growing mother cells in the parental spoIIR+ strain (strain SL12420) (Table 3). In that strain, after septum formation, Spo0A-directed GFP expression increased rapidly in the mother cells of all organisms that formed spores. Similar results were obtained using a second Spo0A-directed reporter (PspoIIE-gfp) for both the parental and the spoIIR-null background (data not shown).
High frequencies of growing mother cells with impaired Spo0A activity were observed in strains with mutations causing overinitiation of DNA replication.
The results described above suggested that when growth of the mother cell occurred, Spo0A-directed gene transcription remained at a low level in that compartment. They also suggested that mother cell growth was associated with DNA replication and that introduction of a mutation causing overinitiation of replication led to increased frequencies of growing mother cells in both spoIIRdelay and spoIIR-null strains. We wanted to test whether such enhanced replication affected Spo0A activity in growing mother cells by introducing a yabA-null mutation into a spoIIR-null mutant. The yabA locus encodes an inhibitor of DNA replication, and its deletion induces overinitiation of replication during vegetative growth (23, 34). The strain showed an enhanced frequency of growing mother cells after asymmetric division (strain SL15325, compared to the parental spoIIR mutant, SL15268 [Table 3]). Further, growth of the mother cells was accompanied by decreased Spo0A-directed GFP expression (data not shown). In contrast, growing mother cells were not observed in an isogenic yabA spoIIR+ strain (strain SL15326) (Table 3); with this strain, only monosporic organisms were observed, and GFP expression was enhanced in the mother cell soon after septation. Rare mother cells of the yabA spoIIR+ strains had increased length but were derived from long vegetative bacteria, and the mother cells did not grow; they also displayed enhanced Spo0A activity (data not shown).
A link between overreplication in these growing mother cells and the impaired Spo0A activity that they display could be the sporulation inhibitor protein Sda (5). Transcription of sda is induced prior to the initiation of DNA replication, and it results in reduced Spo0A-P levels because of direct inhibition of the kinase KinA by Sda (42). Therefore, we tested to see if sda transcription was increased in growing mother cells, using a Psda-gfp transcriptional fusion in a spoIIRdelay strain. We again observed three fates after asymmetric division: monospore-forming organisms (78/126), abortively disporic organisms (30/126), and organisms with growing mother cells (18/126). For this strain, growing mother cells showed a significant increase in GFP expression over nongrowing mother cells of monosporic organisms (SL15449) (Fig. 5a and b), although there was considerable variability between organisms; such variability in sda expression in stationary-phase organisms has been described previously (49). Growing mother cells that displayed increased sda expression were also observed for a spoIIR-null mutant, along with abortively disporic organisms showing lower sda expression (data not shown). No growing mother cells were observed for the isogenic spoIIR+ strain; only monosporic organisms were seen, and those also displayed heterogeneity in Psda-gfp expression, similar to that of the spoIIRdelay strain (data not shown).
Fig. 5.
sda expression is increased in growing mother cells. (a). Time lapse images of a spoIIRdelay strain (SL15449) containing a Psda-gfp fusion for the visualization of sda transcription. Blue arrows indicate an example of a monosporic organism. Yellow and white arrows indicate two examples of organisms with mother cells growing after septation. In both examples, the prespore eventually detached from the growing mother cell, as observed previously (13). Membranes were visualized by staining with FM4-64. Images were taken every 30 min. Bar, 3 μm. (b) Quantitation of GFP levels present in the mother cells of different phenotypes of SL15449. The experiment illustrated in panel a was analyzed. GFP levels were analyzed for 26 monosporic organisms (blue) and 6 organisms with their mother cells growing after asymmetric division (red). Engulfment was completed 60 to 90 min after the asymmetric division in monosporic organisms. Mother cell growth was first observed about 90 min after septation for escaping mother cells. Asterisks indicate significant increases (P, <0.05 by the t test) in sda transcription in escaping mother cells.
High levels of Spo0A activity block mother cell growth.
The data presented above indicated that growing mother cells displayed reduced Spo0A activity. We next wanted to test if artificial activation of Spo0A would suppress the growth of mother cells. We induced high levels of active Spo0A during exponential growth by expressing kinA (which encodes a phosphorelay kinase) from an IPTG-inducible promoter (20). Exponentially growing bacteria of the parental or spoIIR background (strains SL15373 and SL15376, respectively) with an inducible Pspac-kinA fusion and a Spo0A-directed GFP reporter were added to an agarose pad containing the inducer; their behavior was monitored by time lapse microscopy. Initially, chains of growing organisms were observed for both strains. GFP expression increased gradually, and the bacteria then divided asymmetrically. This division was followed by engulfment and spore formation for the parental strain and by the formation of a second polar septum for the spoIIR mutant. No growth of any mother cell of either strain was observed (0/210 and 0/246 for SL15373 and SL15376, respectively; examples are shown in Fig. 6a and b); eventually, the parental strain sporulated efficiently, whereas the spoIIR mutant lysed. In the absence of the inducer, no asymmetric divisions were observed in the time frame of the experiments for either strain, and organisms continued to grow vegetatively, dividing symmetrically (the spoIIR mutant is shown in Fig. 6c). Thus, under these conditions, Spo0A activation caused asymmetric division and blocked the growth of the mother cell.
Fig. 6.
Induction of high levels of active Spo0A blocks growth. A Pspac-kinA fusion construct was used to induce a high level of Spo0A activity during exponential growth; strains contained a PspoIIG-gfp fusion construct for the visualization of Spo0A-directed transcription. Bacteria of the parental and spoIIR backgrounds in early-exponential growth were placed on agarose pads containing the inducer IPTG (1 mM). After 2 h, their behavior was monitored by time lapse microscopy. (a) Images of the parental strain (SL15373). GFP levels increased gradually, and monosporic organisms were observed at high frequencies. In all the monosporic organisms tested, there was no growth of the mother cell. (b) Images of a spoIIR-null strain (SL15376). After induction, GFP levels increased gradually, and asymmetric septation was observed; this was followed by the formation of a second polar septation. In all these biseptate organisms tested, there was no growth of the mother cell. (c) Images of the spoIIR-null strain (SL15376), which continues to grow vegetatively in the absence of the inducer. Membranes were visualized by staining with FM4-64. Images were taken every 30 min. Bar, 3 μm.
The effect of Spo0A on growth could be a secondary consequence of its effect on DNA replication, via sirA. Spo0A activates sirA transcription; the SirA protein inhibits DNA replication during entry into sporulation by targeting the initiating factor DnaA (17, 40, 51). To test for an effect of sirA on mother cell growth, we observed by time lapse microscopy sirA-null strains containing a σE- or Spo0A-directed GFP reporter. There was no mother cell growth in any sporulating organism of either strain (strains SL15002 and SL15277; 0/200 and 0/110 mother cells studied, respectively). Rare long mother cells were observed in the sirA populations. However, as with the yabA strain discussed earlier, these organisms were derived from bacteria that were already long before the asymmetric sporulation division; once formed, the mother cells did not grow, and where tested, they displayed typical enhanced σE- and Spo0A-directed GFP expression. A yabA sirA double mutant, strain SL15439 (which presumably displays a high degree of DNA overreplication), showed similar behavior: no mother cell was observed to grow (0/74). Thus, inhibition of mother cell growth resulting from increased Spo0A activity is not simply an indirect consequence of the block in DNA replication caused by Spo0A-mediated sirA expression.
DISCUSSION
In this study, we investigated the regulation of the growth and DNA replication of the mother cell during spore formation. We observed that in spoIIR mutants, some mother cells continued to grow after asymmetric division. The growing mother cells did not have active σE; they underwent extra rounds of chromosome replication; and they displayed reduced Spo0A activity. Further, the low Spo0A activity was critical to continued growth and replication. The block of growth caused by Spo0A is distinct from its SirA-mediated block of the initiation of DNA replication. In contrast to the mother cells, the prespores did not grow under the conditions employed, except as part of normal spore development, and they displayed normal σF activity; they are not considered further. We discuss first the role of σE and then that of Spo0A.
Growth of mother cells has been studied previously in the context of “commitment”: after spore formation has initiated, have organisms become committed to forming spores when they are transferred to a nutrient-rich medium, or do the mother cells (and prespores) resume growth? Several lines of evidence have suggested that commitment occurs soon after asymmetric sporulation division (15, 16) and depends on activation of the early cell-specific sigma factors σF and σE (36). In particular, σF-directed genes control the commitment of the prespore, whereas the commitment of the mother cell depends on the σE-directed transcription of spoIIP (11). There are clear morphological similarities between mother cells growing because of loss of commitment and those that we observed in spoIIR-impaired (and hence σE-impaired) mutants. However, we did not observe any mother cell growth in spoIIP spoIIR+ populations. This result distinguishes the regulation of mother cell growth under the nutrient-depleted sporulation conditions studied here from the regulation of commitment in nutrient-rich medium. However, in both situations, activation of σE is critical to “lock in” the mother cell to the nongrowing, spore-forming pathway. The σE-directed signal to block mother cell growth overrode the effects of DNA overreplication, which would otherwise favor continued growth.
Our results show that enhanced Spo0A activity also blocked the growth of the mother cell, separately from any σE-directed growth control. Spo0A is the master regulator for the entry to spore formation, and its activity is needed for the formation of both σF and σE (4). It has a second role in the newly formed mother cell, where its activity increases substantially and is critical to successful spore formation (19). This increase probably occurs after σE becomes active (8, 13) but does not depend on σE activity (discussed below). In agreement with a role for Spo0A in preventing growth, its activity was lower in growing mother cells than in nongrowing mother cells.
We found that growth of the mother cell was consistently associated with DNA replication in the mother cell. First, there was a correlation between the number of chromosome origins and the length of the mother cell in sporulating organisms in batch cultures. Second, origin regions were observed to duplicate and to separate in time lapse studies of growing mother cells. Third, the frequency of growing mother cells was increased by deletion of yabA, which encodes an inhibitor of the initiation of replication. Continued DNA replication is detrimental to spore formation (49), and in time lapse studies, the mother cells often “escaped” back to vegetative growth, although some escaping mother cells stopped replicating and went on to form twin spores (13).
Spore formation is linked to DNA replication via Spo0A in at least two ways, through SirA and through Sda. Spo0A activates sirA transcription, and SirA blocks further initiation of DNA replication by binding to DnaA (41, 51). Inactivation of sirA causes overinitiation of replication (40, 51). However, it did not relieve the Spo0A-dependent blockage of mother cell growth. Thus, although SirA has a clear role in Spo0A-directed control of DNA replication (40, 51), that role is not sufficient to explain the Spo0A-directed control of growth. The relationship of Sda to Spo0A and DNA replication is more complex (5). Sda inhibits KinA and so blocks activation of Spo0A by the phosphorelay. The Sda protein is unstable, so the effects of its induction are short-lived (43). It is thought that the rise in the DnaA-ATP concentration prior to the initiation of DNA replication induces sda transcription, and transcription stops once replication has initiated, concomitant with the loss of DnaA-ATP (49, 51); in time lapse studies, loss of Spo0A repressor activity followed bursts of sda expression (49). These results led us to suggest the following sequence in growing mother cells: events leading to the initiation of DNA replication result in increased Sda expression; this increased Sda expression could then account for the observed decrease in Spo0A activity; reduced Spo0A activity could, in turn, allow further DNA replication and growth to occur. Our results from the monitoring of sda transcription are consistent with such a scheme, in that sda transcription appeared to increase before Spo0A activity decreased in growing mother cells. However, there was considerable scatter in the data for sda (as noted previously in attempts to monitor sda expression in stationary-phase bacteria [47]), so that the evidence of a role for Sda in controlling mother cell growth is at best suggestive. Spo0A has also been shown to repress the transcription of several genes that play roles in DNA replication, such as dnaA, dnaG, dnaN, and holB (33). However, some of the repression effects were small and were not detected under conditions of growth in minimal medium (33). Moreover, induction of Spo0A activity in a sirA mutant did not block the initiation of DNA replication (40, 51). Consequently, the Spo0A-regulated SirA and Sda proteins are thought to be the major factors linking DNA replication and spore formation.
In the scheme outlined above, initiation of DNA replication is the first step that can lead to mother cell growth. Usually, organisms enter sporulation having only two chromosomes, and DNA replication is terminated before sporulation division (3, 51). However, as a consequence of noise in the system, a few sporulating organisms contain more than two chromosomes, and their DNA is replicating (13). This minor replicating population is increased when mutations that induce overreplication (such as yabA, sirA, or spo0J-gfp mutations) are introduced. In agreement with this explanation, we observed a low frequency of overreplicating and growing mother cells in spoIIR and spoIIGB strains; the frequency was increased when overreplication mutations were introduced into those backgrounds. However, growing mother cells were not observed when σE was active. We favor the possibility that σE-mediated repression of growth is exercised by ensuring that Spo0A activity remains high in the mother cell. Consistent with this possibility, the initial increase in Spo0A activity in the mother cell occurs independently of σE activation, but any subsequent decrease in Spo0A activity is seen only in the absence of σE activity. The questions of how σE ensures continued high Spo0A activity and how Spo0A blocks the growth of the mother cell remain unanswered.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant GM43577 from the National Institutes of Health.
We thank Vasant Chary and Monica Busuioc for many helpful discussions.
Footnotes
Published ahead of print on 8 April 2011.
REFERENCES
- 1. Abanes-De Mello A., Sun Y. L., Aung S., Pogliano K. 2002. A cytoskeleton-like role for the bacterial cell wall during engulfment of the Bacillus subtilis forespore. Genes Dev. 16:3253–3264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ben-Yehuda S., Losick R. 2002. Asymmetric cell division in Bacillus subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell 109:257–266 [DOI] [PubMed] [Google Scholar]
- 3. Bogush M., Xenopoulos P., Piggot P. J. 2007. Separation of chromosome termini during sporulation of Bacillus subtilis depends on SpoIIIE. J. Bacteriol. 189:3564–3572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burbulys D., Trach K. A., Hoch J. A. 1991. Initiation of sporulation in Bacillus subtilis is controlled by a multicomponent phosphorelay. Cell 64:545–552 [DOI] [PubMed] [Google Scholar]
- 5. Burkholder W. F., Kurtser I., Grossman A. D. 2001. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell 104:269–279 [DOI] [PubMed] [Google Scholar]
- 6. Chary V. K., Piggot P. J. 2003. Postdivisional synthesis of the Sporosarcina ureae DNA translocase SpoIIIE either in the mother cell or in the prespore enables Bacillus subtilis to translocate DNA from the mother cell to the prespore. J. Bacteriol. 185:879–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chary V. K., Xenopoulos P., Piggot P. J. 2006. Blocking chromosome translocation during sporulation of Bacillus subtilis can result in prespore-specific activation of σG that is independent of σE and of engulfment. J. Bacteriol. 188:7267–7273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chary V. K., Xenopoulos P., Eldar A., Piggot P. J. 2010. Loss of compartmentalization of σE activity need not prevent formation of spores by Bacillus subtilis. J. Bacteriol. 192:5616–5624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de Jong I. G., Veening J. W., Kuipers O. P. 2010. Heterochronic phosphorelay gene expression as a source of heterogeneity in Bacillus subtilis spore formation. J. Bacteriol. 192:2053–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dunn G., Jeffs P., Mann N. H., Torgersen D. M., Young M. 1978. The relationship between DNA replication and the induction of sporulation in Bacillus subtilis. J. Gen. Microbiol. 108:189–195 [Google Scholar]
- 11. Dworkin J., Losick R. 2005. Developmental commitment in a bacterium. Cell 121:401–409 [DOI] [PubMed] [Google Scholar]
- 12. Eichenberger P., Fawcett P., Losick R. 2001. A three-protein inhibitor of polar septation during sporulation in Bacillus subtilis. Mol. Microbiol. 42:1147–1162 [DOI] [PubMed] [Google Scholar]
- 13. Eldar A., et al. 2009. Partial penetrance facilitates developmental evolution in bacteria. Nature 460:510–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Errington J. 1993. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol. Rev. 57:1–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Freese E. B., Cooney P., Freese E. 1975. Conditions controlling commitment of differentiation in Bacillus megaterium. Proc. Natl. Acad. Sci. U. S. A. 72:4037–4041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fréhel C., Ryter A. 1969. Réversibilité de la sporulation chez Bacillus subtilis. Ann. Inst. Pasteur (Paris) 117:297–311 [PubMed] [Google Scholar]
- 17. Fujita M., Gonzalez-Pastor J. E., Losick R. 2005. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J. Bacteriol. 187:1357–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fujita M., Losick R. 2002. An investigation into the compartmentalization of the sporulation transcription factor σE in Bacillus subtilis. Mol. Microbiol. 43:27–38 [DOI] [PubMed] [Google Scholar]
- 19. Fujita M., Losick R. 2003. The master regulator for entry into sporulation in Bacillus subtilis becomes a cell-specific transcription factor after asymmetric division. Genes Dev. 17:1166–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fujita M., Losick R. 2005. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 19:2236–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harry E. J., Pogliano K., Losick R. 1995. Use of immunofluorescence to visualize cell-specific gene expression during sporulation in Bacillus subtilis. J. Bacteriol. 177:3386–3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hauser P. M., Errington J. 1995. Characterization of cell cycle events during the onset of sporulation in Bacillus subtilis. J. Bacteriol. 177:3923–3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hayashi M., Ogura Y., Harry E. J., Ogasawara N., Moriya S. 2005. Bacillus subtilis YabA is involved in determining the timing and synchrony of replication initiation. FEMS Microbiol. Lett. 247:73–79 [DOI] [PubMed] [Google Scholar]
- 24. Hilbert D. W., Piggot P. J. 2004. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol. Mol. Biol. Rev. 68:234–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hitchins A. D., Slepecky R. A. 1969. Bacterial sporulation as a modified procaryotic cell division. Nature 223:804–807 [DOI] [PubMed] [Google Scholar]
- 26. Illing N., Errington J. 1991. Genetic regulation of morphogenesis in Bacillus subtilis: roles of σE and σF in prespore engulfment. J. Bacteriol. 173:3159–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ju J., Luo T., Haldenwang W. G. 1997. Bacillus subtilis Pro-σE fusion protein localizes to the forespore septum and fails to be processed when synthesized in the forespore. J. Bacteriol. 179:4888–4893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Karow M. L., Glaser P., Piggot P. J. 1995. Identification of a gene, spoIIR, that links the activation of σE to the transcriptional activity of σF during sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 92:2012–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Khvorova A., Chary V. K., Hilbert D. W., Piggot P. J. 2000. The chromosomal location of the Bacillus subtilis sporulation gene spoIIR is important for its function. J. Bacteriol. 182:4425–4429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee P. S., Lin D. C., Moriya S., Grossman A. D. 2003. Effects of the chromosome partitioning protein Spo0J (ParB) on oriC positioning and replication initiation in Bacillus subtilis. J. Bacteriol. 185:1326–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Londoño-Vallejo J. A., Stragier P. 1995. Cell-cell signaling pathway activating a developmental transcription factor in Bacillus subtilis. Genes Dev. 9:503–508 [DOI] [PubMed] [Google Scholar]
- 32. Migocki M. D., Freeman M. K., Wake R. G., Harry E. J. 2002. The Min system is not required for precise placement of the midcell Z ring in Bacillus subtilis. EMBO Rep. 3:1163–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Molle V., et al. 2003. The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 50:1683–1701 [DOI] [PubMed] [Google Scholar]
- 34. Noirot-Gros M. F., et al. 2006. Functional dissection of YabA, a negative regulator of DNA replication initiation in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 103:2368–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ogura Y., Ogasawara N., Harry E. J., Moriya S. 2003. Increasing the ratio of Soj to Spo0J promotes replication initiation in Bacillus subtilis. J. Bacteriol. 185:6316–6324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Parker G. F., Daniel R. A., Errington J. 1996. Timing and genetic regulation of commitment to sporulation in Bacillus subtilis. Microbiology 142:3445–3452 [DOI] [PubMed] [Google Scholar]
- 37. Piggot P. J., Coote J. G. 1976. Genetic aspects of bacterial endospore formation. Bacteriol. Rev. 40:908–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Piggot P. J., Curtis C. A. 1987. Analysis of the regulation of gene expression during Bacillus subtilis sporulation by manipulation of the copy number of spo-lacZ fusions. J. Bacteriol. 169:1260–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pogliano J., et al. 1999. A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol. Microbiol. 31:1149–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rahn-Lee L., Gorbatyuk B., Skovgaard O., Losick R. 2009. The conserved sporulation protein YneE inhibits DNA replication in Bacillus subtilis. J. Bacteriol. 191:3736–3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rahn-Lee L., Merrikh H., Grossman A. D., Losick R. 2011. The sporulation protein SirA inhibits the binding of DnaA to the origin of replication by contacting a patch of clustered amino acids. J. Bacteriol. 193:1302–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rowland S. L., et al. 2004. Structure and mechanism of action of Sda, an inhibitor of the histidine kinases that regulate initiation of sporulation in Bacillus subtilis. Mol. Cell 13:689–701 [DOI] [PubMed] [Google Scholar]
- 43. Ruvolo M. V., Mach K. E., Burkholder W. F. 2006. Proteolysis of the replication checkpoint protein Sda is necessary for the efficient initiation of sporulation after transient replication stress in Bacillus subtilis. Mol. Microbiol. 60:1490–1508 [DOI] [PubMed] [Google Scholar]
- 44. Setlow B., et al. 1991. Condensation of the forespore nucleoid early in sporulation of Bacillus species. J. Bacteriol. 173:6270–6278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sharp M. D., Pogliano K. 1999. An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc. Natl. Acad. Sci. U. S. A. 96:14553–14558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sharpe M. E., Hauser P. M., Sharpe R. G., Errington J. 1998. Bacillus subtilis cell cycle as studied by fluorescence microscopy: constancy of cell length at initiation of DNA replication and evidence for active nucleoid partitioning. J. Bacteriol. 180:547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Steil L., Serrano M., Henriques A. O., Völker U. 2005. Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis. Microbiology 151:399–420 [DOI] [PubMed] [Google Scholar]
- 48. Teleman A. A., Graumann P. L., Lin D. C., Grossman A. D., Losick R. 1998. Chromosome arrangement within a bacterium. Curr. Biol. 8:1102–1109 [DOI] [PubMed] [Google Scholar]
- 49. Veening J. W., Murray H., Errington J. 2009. A mechanism for cell cycle regulation of sporulation initiation in Bacillus subtilis. Genes Dev. 23:1959–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Veening J. W., et al. 2008. Bet-hedging and epigenetic inheritance in bacterial cell development. Proc. Natl. Acad. Sci. U. S. A. 105:4393–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wagner J. K., Marquis K. A., Rudner D. Z. 2009. SirA enforces diploidy by inhibiting the replication initiator DnaA during spore formation in Bacillus subtilis. Mol. Microbiol. 73:963–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang J. D., Levin P. A. 2009. Metabolism, cell growth and the bacterial cell cycle. Nat. Rev. Microbiol. 7:822–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang S. T., et al. 2006. The forespore line of gene expression in Bacillus subtilis. J. Mol. Biol. 358:16–37 [DOI] [PubMed] [Google Scholar]
- 54. Webb C. D., et al. 1997. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of Bacillus subtilis. Cell 88:667–674 [DOI] [PubMed] [Google Scholar]
- 55. Wu L. J., Errington J. 1994. Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science 264:572–575 [DOI] [PubMed] [Google Scholar]
- 56. Wu L. J., Errington J. 1998. Use of asymmetric cell division and spoIIIE mutants to probe chromosome orientation and organization in Bacillus subtilis. Mol. Microbiol. 27:777–786 [DOI] [PubMed] [Google Scholar]
- 57. Zupancic M. L., Tran H., Hofmeister A. E. 2001. Chromosomal organization governs the timing of cell type-specific gene expression required for spore formation in Bacillus subtilis. Mol. Microbiol. 39:1471–1481 [DOI] [PubMed] [Google Scholar]