Abstract
Mass spectrometry analysis of Streptococcus pneumoniae bacteriophage Cp-1 identified a total of 12 proteins, and proteome-wide yeast two-hybrid screens revealed 17 binary interactions mainly among these structural proteins. On the basis of the resulting linkage map, we suggest an improved structural model of the Cp-1 virion.
TEXT
Few proteome-wide studies of bacteriophages are currently available, in contrast to their bacterial hosts or eukaryotic viruses. A comprehensive analysis of the coliphage T7 interactome was the only such study for many years (1). We recently demonstrated for the virulent Streptococcus pneumoniae phage Dp-1 (Siphoviridae family) that a combination of mass spectrometry (MS) analysis of mature bacteriophage particles coupled with proteome-wide yeast two-hybrid (Y2H) screens is a powerful strategy to obtain insights into the assembly and structure of phage virions (17). While MS analysis of phage virions is able to identify structural proteins, Y2H screening determines binary protein-protein interactions, including transient interactions during virion morphogenesis.
Here we investigated another virulent Streptococcus pneumoniae phage, namely, the Podoviridae phage Cp-1 by using both methods. Phage Cp-1 belongs to the proposed Nanovirinae subfamily of the Podoviridae family (8). Of note, phage Cp-1 was originally classified within the φ29-like genus of the Podoviridae family (13) due to sequence similarities. However, it was recently excluded because of a different genome anatomy and transcriptional organization (8, 11). Thus, phages Cp-1 and φ29 are now considered representatives of separate genera (that bear their names) within the Nanovirinae subfamily of the Podoviridae family. Pneumophage Cp-1 has a very short tail that it uses to infect unencapsulated S. pneumoniae strains. Its double-stranded DNA genome was previously sequenced (19,345 bp), annotated, and found to encode 28 putative proteins (11, 15, 16) (see Table S1 in the supplemental material).
Identification of structural proteins.
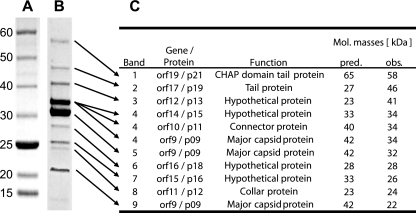
To identify the structural proteins of phage Cp-1, mature virions were first isolated by two rounds of ultracentrifugation on a CsCl gradient. The purified complete phage particles were sonicated, boiled in loading buffer, and separated on a 4 to 15% precast gradient SDS-polyacrylamide gel followed by Coomassie blue staining (Fig. 1). Nine protein bands were clearly visible on the SDS-polyacrylamide gel (in addition to two faint ones). When analyzed by liquid chromatography-tandem mass spectrometry (LC-MS-MS) at the proteomic platform of the Centre de Génomique du Québec, these nine protein bands were mapped to nine Cp-1 structural proteins, including one band (band 4) that contained a mix of three protein fragments (Fig. 1). The two main protein bands (bands 4 and 5) included p09 (major capsid protein), most likely processed in two different forms after endopeptidase cleavage by p14, the product of orf13, as previously described (12). The other seven bands contained p11 (connector protein), p12 (collar protein), p13, p15, p16, p18, p19 (tail protein), and p21 (tail protein). The remaining two faint bands contained fragments of p08, p09, p10, p12, and p14 (data not shown). The same phage preparation was also directly digested with trypsin and analyzed by mass spectrometry as described previously (17). A total of 12 phage proteins were detected from the complete CsCl-purified phage sample, including those previously identified by SDS-PAGE as well as p04, p08, and p20 (see Table S2 in the supplemental material). The terminal protein p04 is known to be covalently linked to the genome ends and plays a role in replication-dependent protein priming through loading the DNA polymerase onto the DNA molecule. This protein usually does not enter the SDS-polyacrylamide gel and was detected only with the purified phage sample (10). The putative scaffolding protein (p08) was highly prevalent and had good protein sequence coverage in the complete purified phage sample. The p08 protein was also identified on the SDS-polyacrylamide gel but only in small amounts and in a fragmented state (data not shown). In Bacillus subtilis phage φ29, the p08 homolog Gp7 was shown to be involved in prohead formation but absent from mature virions (2). It is likely that small amounts of the scaffolding protein remain associated with the mature virion, although the biological significance of this association remains unknown. The p20 protein (encoded by orf18) was detected as a set of unique peptides in the CsCl-purified phage sample, but it was not detected on the SDS-polyacrylamide gel (see Table S2 in the supplemental material). All identified structural proteins are encoded by genes in the gene cluster that has been previously suggested to encode structural proteins (orf8 to orf19) (11). Of note, only complete phage particles could be observed in the CsCl-purified phage sample when visualized by electron microscopy (data not shown).
Fig. 1.
Identification of the virion proteins of Cp-1 by SDS-PAGE and mass spectrometry. (A) Size marker (5- to 250-kDa prestained protein ladder; NEB). The numbers to the left of the gels indicate molecular masses in kilodaltons. (B) Migration of the structural Cp-1 proteins on a 4 to 15% precast gradient SDS-polyacrylamide gel. The arrows indicate the bands extracted for LC-MS-MS analysis. (C) Identification and characterization of the structural Cp-1 proteins detected by mass spectrometry. The predicted (pred.) and observed (obs.) molecular (Mol.) masses of the proteins are indicated.
Proteome-wide identification of binary protein interactions.
We cloned all 28 protein-coding Cp-1 open reading frames (ORFs) into the entry vectors pDONR207 and pDONR221 using the Gateway system (Invitrogen). The ORFs were then transferred into the expression vectors pDEST32/pDEST22 (Invitrogen) and pGBKT7g/pGADT7g (Clontech) (20). PCRs, plasmid selection, propagation, verification, and yeast transfection were done as described previously (17). Y2H screens were performed by screening individual bait strains (pDEST32 or pGBKT7g) against a Cp-1 prey matrix (ORFs in pDEST22 and pGADT7g, respectively) and then retested individually as reported previously (3, 4, 14, 17).
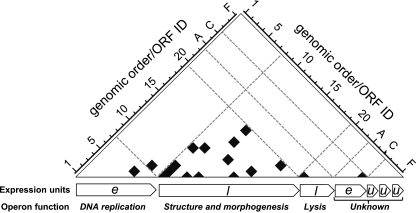
In total, we detected 17 binary interactions among 15 Cp-1 proteins (p05, p06, p08, p09, p11, p12, p13, p14, p15, p16, p18, p19, p21, p23, and p26). Thus, at least 53.6% of the 28 Cp-1 proteins are involved in mediating protein-protein interactions (PPIs). All interactions detected are shown in Fig. 2 and 3 and listed in Table S3 in the supplemental material. Cp-1 shows a highly modular pattern on the protein interaction level reflecting its genome structure (Fig. 2). Fourteen PPIs (out of 17) were detected exclusively between late structural proteins. This is in stark contrast to pneumococcal phage Dp-1 (Siphoviridae) which shows much more cross talk between proteins of different functional modules (17). Only two interactions were found between early gene products, namely, the DNA polymerase (p05), which interacts with the hypothetical protein p06, while hypothetical protein p26 appears to form a homomer. The only interaction detected between early and late gene products was identified between p06 (unknown function) and the possible scaffolding protein p08. The interactions between the gene products encoded by the morphogenesis gene cluster suggest that most Cp-1 protein interactions are involved in virion assembly or structure.
Fig. 2.
The genomic organization of protein interactions reveals a modular organization of the Cp-1 interactome. The gene order within the Cp-1 genome is plotted against the protein-protein interactions (PPIs). Detected PPIs are indicated by black boxes, and operon borders are indicated by dashed lines. The longest transcriptional units that have been experimentally validated were considered and are indicated below the matrix (11). Expression timing of the transcripts is indicated as follows: e, early; l, late; u, unknown. The generalized function of the operons is given. orfA to -F are localized on the reverse genome strand. For clarification of the matrix, all ORFs have been illustrated in a sequential order. Because of the differences between the previous published genome annotation (11) and the current GenBank annotation, the orf and protein nomenclature may differ in certain cases (e.g., orf14 corresponds to the p15 gene product; see NCBI reference sequence file NC_001825.1). A list of ORF and protein identifications (IDs) is provided in Table S1 in the supplemental material.
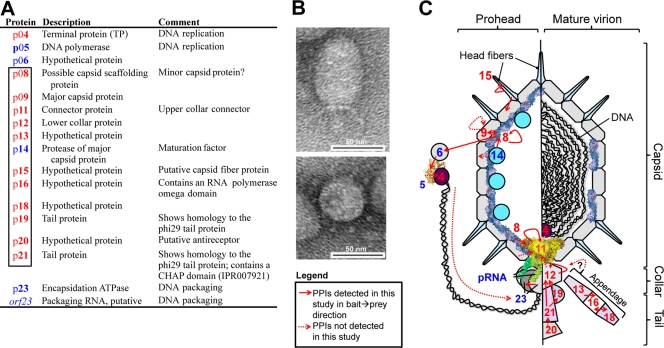
Fig. 3.
A virion model of Cp-1. (A) The 12 Cp-1 gene products that were identified as structural proteins by LC-MS-MS are shown in red, while the proteins in blue undergo binary protein interactions with structural proteins. Gene products shown in blue were not identified by MS. The rectangle in the first column highlights gene products encoded by the structural operon. Protein annotations were adopted from Martin et al. (10–12). (B) Electron micrograph of a mature Cp-1 virion particle. A lateral view (top image) and top down view (bottom image) are shown. (C) A schematic model of the Cp-1 virion. Proteins are indicated by symbols or by the three-dimensional (3D) structure of φ29 homologs. PDB entries are given in Table S1 in the supplemental material. The numbers are the gene product number. The left section indicates interactions that presumably occur during prohead formation; the right section illustrates additional interactions that may play a role in the mature virion collar and tail. PPIs that were not detected in the Y2H screens but that are expected to take place in vivo are indicated by dashed arrows. pRNA, packaging RNA.
Cp-1 virion model.
Using our MS and Y2H results as well as some similarities to other phages such as Bacillus φ29 (11, 18, 19, 21, 22), we propose a Cp-1 virion model (Fig. 3). Protein p14 encodes the major capsid protein-specific protease (12) and was not identified as a structural component in this study, indicating that this protein plays a role exclusively during the maturation process. While we could not detect an interaction between p14 and the major capsid protein (p09), the predicted scaffolding protein p08 interacted with both proteins, suggesting that the propeptide of the major capsid protein might be cleaved by the p08-p14 protein complex.
The scaffolding protein p08 also interacted with the connector p11, suggesting that the connector might be anchored within the capsid via p08 during prohead formation, similar to phage φ29 (7, 9). One further PPI of p08 was found with the hypothetical protein p06 that also interacts with the DNA polymerase p05. Hence, p06 could play a role in DNA replication or packaging, again similar to phage φ29. Here, the terminal protein-DNA complex is directed to the packaging motor presumably via an interaction with the packaging ATPase Gp16 bound to the prohead, playing a role in initiation of DNA packaging (5, 6).
Another interesting interaction was detected between the hypothetical protein p15 and the major capsid protein of Cp-1. p15 was identified in this study as a structural virion component, and its central 61 amino acids exhibit low sequence similarity to the phage φ29 capsid fiber protein Gp8.5. The specific interaction of p15 with the major capsid protein suggests that this protein may be the Cp-1 fiber-like protein located on the capsid (Fig. 3). Since p15 also interacts with itself, we predict the capsid fiber to be organized as homomeric units.
The tail and collar of Cp-1 are probably composed of at least five structural proteins (p11, p12, p19, p20, and p21) which show multiple interactions (Fig. 3; see Table S3 in the supplemental material). Interacting proteins p19 and p21 are homologous to the N and C termini of the φ29 tail protein (Gp9), respectively (11). Both of them also interact with p12 (collar protein). The position of the p20 protein is likely to be similar to its φ29 homolog Gp13, which has been shown to be in the distal part of the tail knob (22). Cp-1 p20 protein contains the enzymatic domains for the lysis of the peptidoglycan (lysozyme and metallopeptidase) and thus might be a tail-associated cell wall-degrading enzyme, although no interaction was detected under the conditions used here. While it has been previously suggested that p20 is involved in host recognition (11), we could not detect any similarity to other receptor-binding proteins by using PBlast, PsiBlast, FASTA, or HHPred alignment. While p20 has weak similarity to CHAP domains, the p21 tail protein clearly contains a CHAP domain between residues 69 and 131. CHAP domain proteins are named after their cysteine histidine-dependent amidohydrolase-peptidase activity involved in bacterial cell wall processing (for a recent review on S. aureus phage CHAP proteins, see reference 23). It is thus tempting to infer that p20 and p21 are involved in the processing of the host cell wall.
The exact positions and functions of the hypothetical proteins p13, p16, and p18 in the virion remain unclear, although we could confirm all of them as structural proteins. These three proteins also exhibit PPIs to each other, but a direct link to other virion components was not found (Fig. 3C). We speculate that these three proteins may be involved in the collar appendage of Cp-1, but this hypothesis requires further support.
Conclusions.
Although a few proteins (endolysin, protease) of phage Cp-1 have been extensively studied, its biology is hardly understood. By combining two proteomic approaches, we have identified the structural proteins and binary protein interactions among Cp-1 proteins. We certainly could not detect all protein-protein interactions, as any particular Y2H system detects only a fraction of all interactions (4). Nonetheless, the results of this study provide the first insights into the composition and organization of the Cp-1 virion and a basis for the detailed functional analysis of Cp-1 proteins as well as the assembly process of the virion and possibly other related phages.
Supplementary Material
Acknowledgments
We are grateful to Denise Tremblay for providing the electron micrograph.
This project was supported by the Landesstiftung Baden-Württemberg of Germany (P.U.), the Karlsruhe House of Young Scientists (R.H.), and the J. Craig Venter Institute (R.H. and P.U.), the Canadian Institutes of Health Research (Team Grant - Emerging: Novel Alternatives to Antibiotics) (S.M.), and European Union grant HEALTH-F3-2009-223101 (P.U.).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 22 April 2011.
REFERENCES
- 1. Bartel P. L., Roecklein J. A., SenGupta D., Fields S. 1996. A protein linkage map of Escherichia coli bacteriophage T7. Nat. Genet. 12:72–77 [DOI] [PubMed] [Google Scholar]
- 2. Bjornsti M. A., Reilly B. E., Anderson D. L. 1983. Morphogenesis of bacteriophage φ29 of Bacillus subtilis: oriented and quantized in vitro packaging of DNA protein gp3. J. Virol. 45:383–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cagney G., Uetz P., Fields S. 2000. High-throughput screening for protein-protein interactions using two-hybrid assay. Methods Enzymol. 328:3–14 [DOI] [PubMed] [Google Scholar]
- 4. Chen Y. C., Rajagopala S. V., Stellberger T., Uetz P. 2010. Exhaustive benchmarking of the yeast two-hybrid system. Nature Methods 7:667–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Grimes S., Anderson D. 1997. The bacteriophage phi29 packaging proteins supercoil the DNA ends. J. Mol. Biol. 266:901–914 [DOI] [PubMed] [Google Scholar]
- 6. Guo P., Peterson C., Anderson D. 1987. Initiation events in in-vitro packaging of bacteriophage phi 29 DNA-gp3. J. Mol. Biol. 197:219–228 [DOI] [PubMed] [Google Scholar]
- 7. Guo P. X., et al. 1991. Regulation of the phage phi 29 prohead shape and size by the portal vertex. Virology 183:366–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lavigne R., Seto D., Mahadevan P., Ackermann H. W., Kropinski A. M. 2008. Unifying classical and molecular taxonomic classification: analysis of the Podoviridae using BLASTP-based tools. Res. Microbiol. 159:406–414 [DOI] [PubMed] [Google Scholar]
- 9. Lee C. S., Guo P. 1995. Sequential interactions of structural proteins in phage φ29 procapsid assembly. J. Virol. 69:5024–5032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Martin A. C., Blanco L., Garcia P., Salas M., Mendez J. 1996. In vitro protein-primed initiation of pneumococcal phage Cp-1 DNA replication occurs at the third 3′ nucleotide of the linear template: a stepwise sliding-back mechanism. J. Mol. Biol. 260:369–377 [DOI] [PubMed] [Google Scholar]
- 11. Martin A. C., Lopez R., Garcia P. 1996. Analysis of the complete nucleotide sequence and functional organization of the genome of Streptococcus pneumoniae bacteriophage Cp-1. J. Virol. 70:3678–3687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Martin A. C., Lopez R., Garcia P. 1998. Pneumococcal bacteriophage Cp-1 encodes its own protease essential for phage maturation. J. Virol. 72:3491–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pecenkova T., Paces V. 1999. Molecular phylogeny of phi29-like phages and their evolutionary relatedness to other protein-primed replicating phages and other phages hosted by gram-positive bacteria. J. Mol. Evol. 48:197–208 [DOI] [PubMed] [Google Scholar]
- 14. Rajagopala S. V., Hughes K. T., Uetz P. 2009. Benchmarking yeast two-hybrid systems using the interactions of bacterial motility proteins. Proteomics 9:5296–5302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ronda C., Garcia J. L., Lopez R. 1989. Infection of Streptococcus oralis NCTC 11427 by pneumococcal phages. FEMS Microbiol. Lett. 53:187–192 [DOI] [PubMed] [Google Scholar]
- 16. Ronda C., Lopez R., Garcia E. 1981. Isolation and characterization of a new bacteriophage, Cp-1, infecting Streptococcus pneumoniae. J. Virol. 40:551–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sabri M., et al. 2011. Genome annotation and intraviral interactome of the Streptococcus pneumoniae virulent phage Dp-1. J. Bacteriol. 193:551–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Simpson A. A., et al. 2000. Structure of the bacteriophage phi29 DNA packaging motor. Nature 408:745–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tao Y., et al. 1998. Assembly of a tailed bacterial virus and its genome release studied in three dimensions. Cell 95:431–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Uetz P., et al. 2006. Herpesviral protein networks and their interaction with the human proteome. Science 311:239–242 [DOI] [PubMed] [Google Scholar]
- 21. Xiang Y., et al. 2006. Structural changes of bacteriophage phi29 upon DNA packaging and release. EMBO J. 25:5229–5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xiang Y., et al. 2008. Crystal and cryoEM structural studies of a cell wall degrading enzyme in the bacteriophage phi29 tail. Proc. Natl. Acad. Sci. U. S. A. 105:9552–9557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zou Y., Hou C. 2010. Systematic analysis of an amidase domain CHAP in 12 Staphylococcus aureus genomes and 44 staphylococcal phage genomes. Comput. Biol. Chem. 34:251–257 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.