Abstract
Biodegradation pathways of synthetic nitroaromatic compounds and anilines are well documented, but little is known about those of nitroanilines. We previously reported that the initial step in 5-nitroanthranilic acid (5NAA) degradation by Bradyrhizobium sp. strain JS329 is a hydrolytic deamination to form 5-nitrosalicylic acid (5NSA), followed by ring fission catalyzed by 5NSA dioxygenase. The mechanism of release of the nitro group was unknown. In this study, we subcloned, sequenced, and expressed the genes encoding 5NAA deaminase (5NAA aminohydrolase, NaaA), 5NSA dioxygenase (NaaB) and lactonase (NaaC), the key genes responsible for 5NAA degradation. Sequence analysis and enzyme characterization revealed that NaaA is a hydrolytic metalloenzyme with a narrow substrate range. The nitro group is spontaneously eliminated as nitrite concomitant with the formation of a lactone from the ring fission product of 5NSA dioxygenation. The elimination of the nitro group during lactone formation is a previously unreported mechanism for denitration of nitro aliphatic compounds.
INTRODUCTION
Nitro-substituted compounds are widely used as dyes, pesticides, synthetic intermediates, and explosives (29, 41). The research on biodegradation of nitro compounds has focused on synthetic chemicals, and a great deal is known about the biochemistry and molecular biology of the pathways. The degradation pathways for synthetic compounds appear to have evolved by recruitment and assembly of genes from other pathways in response to the recent introduction of such chemicals into the biosphere (7, 18, 45, 46). Over 200 natural nitro compounds are produced by a variety of microbes, plants, and animals (6, 20, 30), but little is known about their biodegradation mechanisms.
The presence of amino and nitro groups in aromatic compounds presents microorganisms with a more complex challenge than the presence of either group alone. We recently reported the biodegradation of the natural aminonitroaromatic compound 5-nitroanthranilic acid (5NAA) (32), produced by Streptomyces scabies, the causative agent of potato scab (22). In Bradyrhizobium sp. strain JS329, isolated from potato farm soil, the biodegradation of 5NAA was initiated by a deamination reaction catalyzed by a novel aminohydrolase (5NAA deaminase, NaaA) to produce 5-nitrosalicylic acid (5NSA), followed by an unusual ring fission dioxygenation prior to removal of the nitro group (32). The genes involved were localized on a 40-kb fosmid clone (32). The properties of 5NAA deaminase (NaaA) and the mechanism of removal of the nitro group were not established. The mechanisms of removal of the nitro groups from the ring fission products of picric acid and 2,6-dinitrotoluene (2,6-DNT) are long-standing mysteries (9, 15, 27). Therefore, the elucidation of the mechanism of nitro group removal from the ring cleavage product of 5NSA could serve as a precedent.
In this study, we characterized 5NAA deaminase and elucidated the mechanism of elimination of the nitro group from the ring fission product. Transition divalent metals are essential for the hydrolytic activity of 5NAA deaminase, and the nitro group is spontaneously removed as nitrite during the lactonization of the ring fission product. The enzymes and mechanisms involved in the biodegradation of 5NAA are unusual and distinct from those previously described for oxygenase-catalyzed removal of phenyl nitro and amino groups.
MATERIALS AND METHODS
Fosmid DNA sequencing and in silico analysis.
Fosmid pJS800 DNA (Table 1) was purified with a FosmidMAX DNA purification kit (Epicentre Biotechnologies, WI) and sent to the Emory GRA Genomic Center for 454 pyrosequencing. The reads were assembled with CLC Genomics Workbench. Open reading frames (ORFs) were identified by using GeneMark (3) (http://exon.biology.gatech.edu/) and then compared to the GenBank database by using BLASTP (1).
Table 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Description | Reference or source |
|---|---|---|
| Strains | ||
| Bradyrhizobium sp. strain JS329 | 5-Nitroanthranilic acid degrader | 32 |
| Escherichia coli EPI300 | Host strain for pJS800 and for Tn5 transposon mutants | Epicentre (Madison, WI) |
| E. coli Rosetta 2(DE3)pLysS | Cmr, host strain for pJS803, pJS804, pJS805, contains coding sequences for 7 rare codons | Novagen |
| Plasmids | ||
| pCC1FOS | Cmr, 8.1-kb fosmid vector for the construction of genomic library | Epicentre (Madison, WI) |
| pET21a | Ampr, 5,443-bp overexpression vector | Novagen |
| pJS800 | Cmr, 40-kb pCC1FOS containing naaA and naaB from JS329 (5NAA+ 5NSA+) | 32 |
| pJS803 | Ampr, 6,720-bp pET-21a containing the gene encoding 5NAA deaminase (NaaA) from Bradyrhizobium sp. JS329 | This study |
| pJS804 | Ampr, 6,066-bp pET-21a containing the gene encoding 5NSA dioxygenase (NaaB) from Bradyrhizobium sp. JS329 | This study |
| pJS805 | Ampr, 6,190-bp pET-21a containing the gene encoding lactonase (NaaC) from Bradyrhizobium sp. JS329 | This study |
| Primers | ||
| naaAFa | 5′-AAAGGATCCATGGCTGGAAGTAACGACGT-3′ | This study |
| naaARb | 5′-AAACTCGAGGGGCGTACGATTGCACAGAT-3′ | This study |
| naaBFa | 5′-AAAGGATCCATGAAATGGAGCAACAAAGA-3′ | This study |
| naaBRb | 5′-AAACTCGAGTTATTCGCCTTGCTTGAGAA-3′ | This study |
| naaCFa | 5′-AAAGGATCCATGGCAACTGAAACCATCGC-3′ | This study |
| naaCRb | 5′-AAACTCGAGTCACACCGTGCGCTTGC-3′ | This study |
| T7 | 5′-TAATACGACTCACTATAGGG-3′ | Genewiz (South Plainfield, NJ) |
| T7 term | 5′-GCTAGTTATTGCTCAGCGG-3′ | Genewiz (South Plainfield, NJ) |
Engineered BamHI recognition sites are underlined for forward primers.
Engineered XhoI recognition sites are underlined for reverse primers.
Transposon mutagenesis and screening.
The fosmid clone pJS800 (32), containing genes whose products are able to release nitrite from both 5NAA and 5NSA, was subjected to Tn5 transposon mutagenesis with an Ez-Tn5 <KAN-2> insertion kit (Epicentre Biotechnologies, Madison, WI) according to the manufacturer's protocol. Colonies able to grow on LB agar supplemented with chloramphenicol (12.5 μg/ml) and kanamycin (50 μg/ml) were transferred to 96-well plates containing LB liquid medium supplemented with chloramphenicol (12.5 μg/ml), kanamycin (50 μg/ml), 5NAA (100 μM), and 1× CopyControl fosmid induction solution (Epicentre Biotechnologies, Madison, WI) at 30°C for 17 h. After growth, microplates were centrifuged and supernatants were removed. BLK liquid medium (5) (200 μl) supplemented with 5NSA (200 μM) and 1× CopyControl fosmid induction solution was added to each well, followed by incubation for 48 h at 30°C. Fosmid clones were screened colorimetrically for nitrite release from 5NSA. Mutants unable to release nitrite from 5NSA were sequenced with Ez-Tn5 <KAN-2>-specific outward-reading primers (Ez-Tn5 <KAN-2> FP-1 forward primer and Ez-Tn5 <KAN-2> RP-1 reverse primer; Epicentre Biotechnologies, Madison, WI) by Nevada Genomics Center (Reno, NV).
Cloning of 5NAA deaminase (NaaA), 5NSA dioxygenase (NaaB), and lactonase (NaaC).
naaA, naaB, or naaC was amplified by PCR using the appropriate primers (Table 1) (Integrated DNA Technologies, Coralville, IA) and GoTaq flexi DNA polymerase (Promega, Madison, WI) for naaA and naaB or DeepVent proofreading polymerase (New England BioLabs, Ipswich, MA) for naaC. The purified PCR products were ligated into BamHI and XhoI sites of the pET-21a vector (Invitrogen Corp., Carlsbad, CA). The resulting recombinant plasmids, pJS803, pJS804, and pJS805, were transformed into E. coli DH5α (New England BioLabs, Ipswich, MA) (Table 1) to maintain the plasmid or into E. coli Rosetta 2(DE3)pLysS competent cells (Novagen) (Table 1) for overexpression according to the manufacturers' protocols. The resulting inserts (Table 1) were sequenced with the primers T7 and T7 term by Genewiz, Inc. (New Jersey) to verify the absence of mutations.
Overexpression of naaA, naaB, and naaC.
Single colonies of E. coli Rosetta 2(pJS803), Rosetta 2(pJS804), or Rosetta 2(pJS805) were transferred into LB medium supplemented with ampicillin (100 μg/ml) and glucose (1%) and incubated for 17 h at 37°C with shaking. Cells were subcultured into fresh medium (1:20 [vol/vol]) and incubated until the optical density at 600 nm (OD600) was 0.5 to 0.8. Expression of the genes was induced by adding isopropyl β-d-1-thiogalactopyranoside (IPTG; 200 μg/ml) and incubating at 30°C with shaking for 2 to 3 h or by adding IPTG (100 μg/ml) and incubating at 24°C with shaking for 12 h for the subsequent purification of NaaA. The induced cells were harvested by centrifugation (10,000 × g) for 10 min at 4°C, washed with ice-cold potassium phosphate buffer (pH 7.0, 20 mM), and stored at −80°C until used.
Enzyme assays.
Cell pellets were suspended in ice-cold potassium phosphate buffer (20 mM, pH 7.0) and passed twice through a French pressure cell at 20,000 lb/in2. Cell debris was removed by centrifugation (20,000 × g, 4°C, 20 min). Enzyme assays were carried out at 30°C in potassium phosphate (pH 7.0, 20 mM) for crude extracts or 44°C in HEPES buffer (pH 7.4, 50 mM) for purified NaaA. The assay mixtures contained 0.01 to 1.5 mg of protein/ml and 5NAA (100 to 200 μM), 5NSA (400 to 500 μM), or lactones (less than 50 μM). Mn2+ (1 mM) was added for the enzyme assays with purified NaaA (200 μg/ml), but external cofactors were not required for other assays. After appropriate intervals, trifluoroacetic acid (TFA) was added (1:100 [vol/vol]) to stop the reactions. The acidified reaction mixtures were clarified by centrifugation before high-performance liquid chromatography (HPLC) analysis or colorimetric assays.
Purification of 5-nitroanthranilic acid aminohydrolase (NaaA).
Cells of E. coli Rosetta 2(pJS803) were resuspended in HEPES buffer (pH 7.5, 50 mM) containing imidazole (50 mM), and cell extracts were prepared as described above. The crude extract containing heterologously expressed NaaA was loaded onto a Ni2+ nitrilotriacetic acid (NTA) affinity column (HiTrap chelating HP; GE Healthcare) equilibrated with HEPES buffer (pH 7.5, 50 mM) containing imidazole (50 mM). After a washing with the same buffer, protein was eluted with HEPES buffer (pH 7.5, 50 mM) containing imidazole (200 mM).
Preparation of lactone [2-oxo-3-(5-oxofuran-2-ylidene)propanoic acid].
Induced cells of E. coli Rosetta 2(pJS804) were washed with phosphate buffer, suspended in the same phosphate buffer supplemented with 5NSA (500 μM), and incubated with shaking at 30°C until production of lactone (see compounds I and II in Fig. 3 and 4) was complete. Cells were removed by centrifugation (20,000 × g, 4°C, 10 min), and the supernatant was stored at −80°C until used in enzyme assays. Approximately 85% of the lactone remained after 15 days of storage at −80°C.
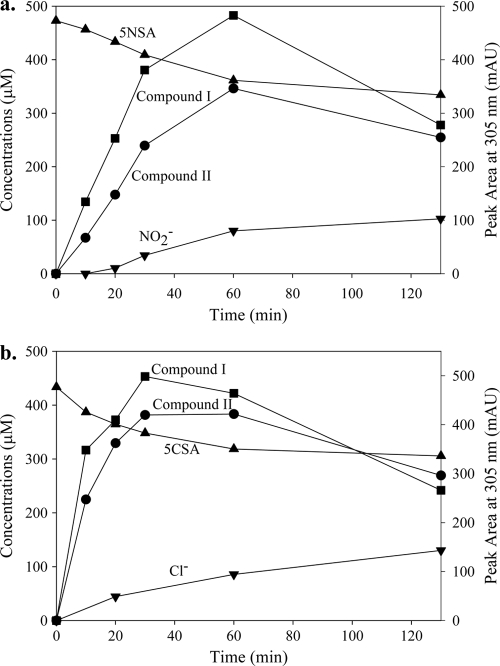
Fig. 3.
Biotransformation of 5NSA (a) and 5CSA (b) by cell extracts from E. coli Rosetta 2(pJS804). Reaction mixtures contained 1.3 mg/ml of protein. The concentrations (left) correspond to 5NSA or 5CSA and NO2− (a) or Cl− (b), while the peak area at 305 nm (right) corresponds to compounds I and II. mAU, milli-absorbance unit.
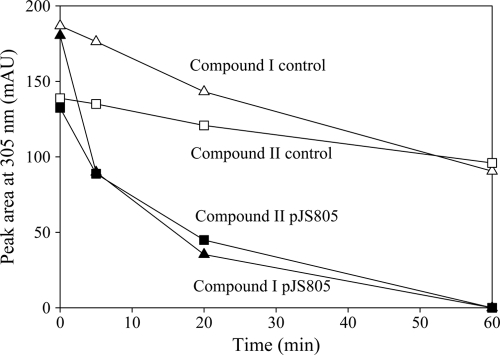
Fig. 4.
Biotransformation of lactones by cell extracts from E. coli Rosetta 2(pJS805). Reaction mixtures contained 0.015 mg of protein/ml. ▪, compound II; ▴, compound I; □, compound II for the abiotic control; ▵, compound I for the abiotic control.
Transformation of 5NSA by intact cells.
E. coli EPI300(pJS800) was grown in LB medium supplemented with chloramphenicol (12.5 μg/ml), 1× CopyControl fosmid autoinduction solution (Epicentre Biotechnologies, Madison, WI), and 5NAA (100 μM) at 30°C with shaking for 17 h. Induced cells were harvested by centrifugation (10,000 × g, 10 min, 4°C), washed twice with ice-cold potassium phosphate buffer (pH 7.0, 20 mM), and suspended in the same phosphate buffer. 5NSA was added to initiate the reaction, and the mixture was incubated at 30°C with shaking. Samples were taken at appropriate intervals, mixed with TFA (1:100 [vol/vol]) to stop the reactions, and clarified by centrifugation at 16,100 × g for 3 min prior to HPLC analysis.
Analytical methods.
5-Chlorosalicylic acid (5CSA) and lactone [cis and trans isomers of 2-oxo-3-(5-oxofuran-2-ylidene)propanoic acid] were analyzed by HPLC using the method described previously for 5NAA and 5NSA (32). 5CSA and the lactones were monitored at 305 nm (retention times [RT] were 3.5 min for lactone 1, 4.6 min for lactone 2, and 8.4 min for 5CSA). Fumarate was analyzed by HPLC with an ion exclusion column (ICSep ICE-ION-310 FAST Transgenomic, 6.5 mm by 150 mm) with an isocratic mobile phase of 1.74 mM H2SO4 at a flow rate of 0.3 ml/min over a period of 15 min and detected at 210 nm (RT, 12.1 min).
Ammonia (31) and nitrite (40) were measured as reported previously. Chloride was analyzed according to a colorimetric method (2). Protein concentration was determined with a Pierce bicinchoninic acid (BCA) protein assay kit (Rockford, IL).
Chemicals.
5-Chlorosalicylic acid (5CSA) was from TCI America (Portland, OR). Fumaric acid was from Chem Service (West Chester, PA). 5-Nitroanthranilic acid (5NAA) was from Sigma-Aldrich (Milwaukee, WI). 5-Nitrosalicylic acid (5NSA) was from Eastman Kodak (Rochester, NY).
Nucleotide sequence accession number.
The nucleotide sequence of the DNA fragment containing the complete operon involved in the biodegradation of 5NAA was deposited in GenBank under the accession number GU188569.
RESULTS
In silico analysis of the genes involved in 5NAA degradation by Bradyrhizobium sp. strain JS329.
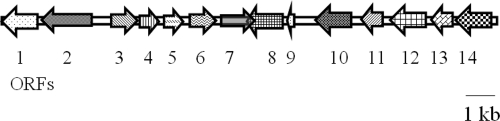
The genes involved in the conversion of 5NAA to small organic acids are located on a 40-kb fosmid clone (pJS800) (Table 1) from Bradyrhizobium sp. strain JS329 (32). To identify the genes involved and their arrangement, pJS800 was sequenced. The assembled sequence comprised three gene clusters, one of which (Fig. 1) contained naaA and naaB, identified previously as encoding 5NAA aminohydrolase (NaaA) and 5-nitrosalicylate dioxygenase (NaaB) (32). Based on BLASTP analysis, the deduced amino acid sequence of ORF2, encoding NaaA, is distantly related to the amino acid sequences of M20 peptidases (32) and contains metal-binding residues. ORF3, encoding NaaB, is distantly related to genes encoding salicylate dioxygenase, gentisate dioxygenase, and 1-hydroxy-2-naphthoic acid dioxygenase (32). It contains conserved histidine residues that are essential for binding the ferrous ion in the catalytic site (13, 32, 47). The deduced amino acid sequence of ORF5 contains a conserved domain in common with the amino acid sequences of dienelactone hydrolase and related enzymes (Clusters of Orthologous Groups of proteins identifier COG0412) (25), but the overall sequence has less than 33% identity to that of the nearest relative (Fig. 1). The deduced amino acid sequence of ORF6 has 68% amino acid identity to fumarylacetoacetate (FAA) hydrolase from Paracoccus denitrificans PD1222, based on BLASTX results. The enzyme contains conserved domains similar to those of the FAA hydrolase (pfam01557 [NCBI database]) and isomerase (TIGR02303 and TIGR02305 [NCBI database]), involved in keto-enol isomerization of 2-oxohept-3-ene-1,7-dioate (25). The findings are consistent with fumarate and a lactone as intermediates in the degradation pathway of 5NAA. The product of ORF8 is closely related to succinate dehydrogenase/fumarate reductase from Beggiatoa sp. PS (77% amino acid identity), which indicates that it could be responsible for the metabolism of fumarate. The other ORFs seem not to be involved in the biodegradation of 5NAA.
Fig. 1.
Map of the gene cluster containing the genes involved in the degradation of 5NAA by Bradyrhizobium sp. strain JS329. The amino acid identity to the best match from BLASTP search and the corresponding NCBI accession number are shown in parentheses as follows: 1, 4-hydroxybenzoate transporter (46%, ZP_00946270.1); 2, 5NAA deaminase (100%, GU188569.1); 3, 5NSA dioxygenase (100%, GU188569.1); 4, Rieske 2Fe-2S family protein (35%, ZP_01125630.1); 5, carboxymethylenebutenolidase (33%, NP_926347.1); 6, fumarylacetoacetate (FAA) hydrolase (NaaD) (68%, YP_913985.1); 7, unknown function; 8, succinate dehydrogenase/fumarate reductase (77%, ZP_01998497.1); 9, short-chain dehydrogenase/reductase (72%, YP_002496152.1); 10, 2-oxo-hept-3-ene-1,7-dioate hydratase (55%, ZP_05966920.1); 11, translation initiation inhibitor (47%, ZP_00946594.1); 12, 3-hydroxyanthranilate 3,4-dioxygenase (49%, YP_509389.1); 13, unknown function; and 14, ferredoxin reductase (40%, ZP_01224350.1).
Properties of NaaA.
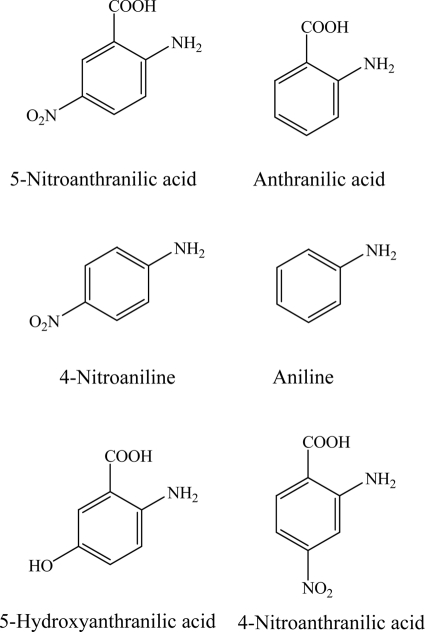
5NAA deaminase (NaaA) is responsible for an unusual hydrolytic deamination of 5NAA to produce 5NSA in Bradyrhizobium sp. JS329 (32). To test the substrate specificity and provide insight about why aminohydrolase and not dioxygenase enzymes catalyze the initial attack, crude extracts of E. coli EPI300(pJS800) were used to determine the activity toward several structural analogs of 5NAA (Fig. 2). No activity was detected with anthranilic acid, 4-nitroaniline, aniline, 5-hydroxyanthranilic acid, and 4-nitroanthranilic acid (Fig. 2), which indicates that both nitro and carboxyl groups in the meta position on the aromatic ring are essential for 5NAA deamination. The optimum pH and temperature (in phosphate buffer) were 7.2 and 42 to 45°C (testing range, 15 to 55°C). The addition of the metal ion chelators EDTA (1 mM), 2,2′-dipyridyl (1 mM), and o-phenanthroline (1 mM) inhibited the activity of NaaA by 99%, 70%, and 98%. The activity of NaaA was restored by the addition of divalent metal Co2+ (100%), Mn2+ (85%), Zn2+ (81%), Fe2+ (67%), or Ni2+ (39%) to a final concentration of 667 μM.
Fig. 2.
5NAA and its analogs.
With purified NaaA, Mn2+ (1 mM) stimulated enzyme activity more than the other divalent metal ions (data not shown), which suggested that Mn2+ plays a physiological role. NaaA contains the sequence HEAWH (amino acid residues 108 to 112), which is similar to the zinc binding motif HEXXH (24). A previous report indicated that an Mn2+-dependent peptidase had a tightly bound Zn2+ and a loosely bound Mn2+ (4), so it remains to be seen which metals play physiological roles with NaaA. The metal requirements and metal-binding residue(s) will be established rigorously when the structure of NaaA is determined. Ultrafiltration of the overexpressed NaaA did not inhibit the enzyme activity, consistent with our earlier observations with crude extracts from Bradyrhizobium sp. JS329 (32), which indicated that the divalent metal was bound to the enzyme.
SDS-PAGE of the purified NaaA revealed a single band with a molecular mass of 45 kDa (see Fig. S1 in the supplemental material), consistent with the in silico prediction (46.3 kDa). The reaction kinetics of pure NaaA were determined by linear regression on the Lineweaver-Burk transformation; the Km value for 5NAA was 335.1 μM, and the Vmax value was 181.5 nmol 5NSA/min/mg protein.
Mutagenesis of pJS800.
Nitrite was released during the oxidation of 5NSA by E. coli EPI300(pJS800) and when Bradyrhizobium sp. JS329 grew on 5NAA or 5NSA (32). To identify the gene involved in removal of the nitro group as nitrite, fosmid pJS800 was subjected to Tn5 transposon mutagenesis. Mutants that lost the ability to release nitrite during the transformation of 5NSA were sequenced. Tn5 transposons were inserted only into naaB, which is responsible for the ring cleavage of 5NSA (32). The results suggested that the release of nitrite takes place during the ring fission reaction or spontaneously after ring fission.
Transformation of 5NSA by enzymes in cell extracts.
In Bradyrhizobium sp. JS329, 5NSA is oxidized by 5-nitrosalicylate dioxygenase (NaaB), encoded by ORF3 (Fig. 1), whose activity was characterized previously (32). NaaB did not oxidize 4-chloro-/nitro-catechol. 5-Nitrosalicylate dioxygenase is distantly related to gentisate dioxygenase from Oligotropha carboxidovorans OM5 (32). In Pseudaminobacter salicylatoxidans, salicylate dioxygenase catalyzes the conversion of 5-chlorosalicylate to a ring fission product that undergoes spontaneous lactonization that is accompanied by the elimination of Cl− (13). We tested whether NaaB catalyzed similar reactions with 5-nitro- and 5-chlorosalicylate. Transformation of 5NSA or 5CSA in cell extracts from E. coli Rosetta 2(pJS804) was accompanied by the accumulation of nitrite or chloride and two unknown products (Fig. 3) with similar UV spectra (Amax ≈ 300 nm) (see Fig. S2 in the supplemental material).
Formation of maleylpyruvate by cell extracts from E. coli Rosetta 2(pJS805).
The spectra and behavior of the two unknown compounds formed from 5NSA by JS804 were identical to those of the lactone previously reported to be produced from 5-chlorosalicylate by salicylate dioxygenase (13). The products were not further transformed by enzymes in cell extracts of the clone and slowly decomposed.
pJS805 was constructed to express ORF5 (naaC), the putative lactone hydrolase. When extracts of E. coli Rosettaz(pJS805) cells were incubated with the unknown compounds produced from 5NSA or 5CSA, both compounds disappeared rapidly (Fig. 4), accompanied by an increase of absorbance at 332 nm (see Fig. S3 in the supplemental material), which is characteristic of the formation of maleylpyruvate (Amax = 330 to 334 nm) (10, 47). The results are consistent with a dioxygenase-catalyzed ring opening followed by spontaneous lactonization and nitrite elimination and enzyme-catalyzed hydrolysis of the lactone to produce maleylpyruvate (13).
Subsequent steps in the pathway.
Maleylpyruvate is the key intermediate in the biodegradation of 5-substituted salicylates, including 5-hydroxysalicylate (gentisate) (47) and 5-halosalicylates (13). The conversion of maleylpyruvate to central intermediates is well established for a variety of bacteria (47) and involves either (i) isomerization to fumarylpyruvate followed by hydrolysis to fumarate and pyruvate or (ii) direct hydrolysis to maleate and pyruvate (16). To determine which reaction is involved in the lower pathway of 5NSA degradation, E. coli EPI300(pJS800) was incubated with 5NSA. Transient accumulation of fumarate during the transformation (see Fig. S4 in the supplemental material) and the presence of the gene encoding the fumarylacetoacetate hydrolase ortholog (ORF6) (Fig. 1) suggest that maleylpyruvate is transformed in Bradyrhizobium sp. JS329 via fumarylpyruvate. Maleylpyruvate isomerase is a key enzyme involved in the metabolism of gentisate. Bradyrhizobium sp. JS329 grows on gentisate (data not shown), and the gentisate dioxygenase is induced during the growth of JS329 on 5NAA (32), so the maleylpyruvate isomerase must be present in JS329. The gene was not experimentally identified on the fosmid, but ORF6 is a likely candidate. The orthologs of the genes encoding maleylpyruvate isomerase are present in the sequenced genomes of different Bradyrhizobium strains (NCBI accession number YP_001242339.1, NP_766749.1, or YP_001202853.1). The potential for enzymes of the alternative pathway to be encoded elsewhere in the genome was not investigated.
DISCUSSION
The biodegradation mechanism of 5NAA is a departure from the typical dioxygenase-catalyzed removal of amino (44), nitro (29), and carboxyl (11) substituents from the benzene ring. Amino functional groups on aliphatic and heterocyclic compounds can be removed by hydrolases (23, 37); however, to our knowledge, 5NAA deaminase is the first hydrolase that can catalyze the hydrolytic removal of an amino group attached to the benzene ring. It seems likely that the -NO2 and -COOH groups synergistically polarize 5NAA due to the electronic effect, which makes the amino group of 5NAA equivalent to an “imino group” and, thus, facilitates the hydrolysis. The effects might similarly inhibit dioxygenase attack.
Although NaaA shows no significant identity to any biochemically characterized enzymes, the properties of 5NAA deaminase resemble those of the M20 family of peptidases (14, 35, 38) or acetylornithine deacetylase (17) in the requirement for transition metal ions, conserved metal-binding residues, inhibition by EDTA and 1,10-phenanthroline, and activity at relatively high temperature. By analogy to the well-characterized dipeptidase (19) and acetylornithine deacetylase (39) mechanisms, our hypothesis for the mechanism of 5NAA deamination is that the divalent metal ion serves to stabilize the conformation of the enzyme and activate water for the hydrolytic reaction. Determination of the structure and mechanism of 5NAA deaminase is currently in progress.
The biodegradation of 5NSA is one of several examples where a nitrophenol serves as the ring fission substrate without prior removal of the nitro group (9, 27, 32). Determination of the downstream pathway not only reveals the complete degradation pathway of the natural nitroaniline (5NAA) but also sheds light on the potential mechanism involved in the degradation of other important synthetic nitro compounds, such as picric acid and 2,6-DNT. The fact that Tn5 insertion into naaB disrupted nitrite release indicates that the ring fission dioxygenase is involved in the denitration but does not establish the mechanism. The mechanism of salicylate dioxygenase-catalyzed elimination of Cl− from 5-chlorosalicylate is well established, however (13). Salicylate 1,2-dioxygenase from Pseudaminobacter salicylatoxidans catalyzes the oxidative ring cleavage of 5-halosalicylates and produces the corresponding lactone, with the simultaneous release of hydrogen halides (HX), followed by abiotic hydrolysis of the lactone to form maleylpyruvate (12, 13).
Although salicylate dioxygenase did not attack 5NSA (13), the 5NSA dioxygenase clearly catalyzes an analogous reaction with both 5NSA and 5-chlorosalicylate. The products gave identical UV spectra (see Fig. S2 in the supplemental material) to each other and to the lactone produced from 5-halosalicylate by salicylate dioxygenase (13). When the lactone carries an alkene side chain, it can exist as cis and trans isomers with similar spectra but different retention times in HPLC (36). In this work, the interconversion of the isomers might have been enhanced by the acid in the HPLC solvent. No attempt was made to assign configurations to the isomers.
Therefore, according to our results and by analogy to those of previous studies, the initial ring fission product of 5NSA dioxygenation (not detected by HPLC) was spontaneously and rapidly cyclized to form lactones, accompanied by the release of NO2− (Fig. 5). A substantial amount of previous work (20) suggests that the nitro group is removed from nitro compounds as nitrite by monooxygenases (21, 29), dioxygenases (29, 42), hydrolases (33), or reductases that attack the aromatic ring (9, 29, 34). To our knowledge, this is the first report that the nitro group can be spontaneously released as nitrite during the formation of lactones from ring fission products.
Fig. 5.
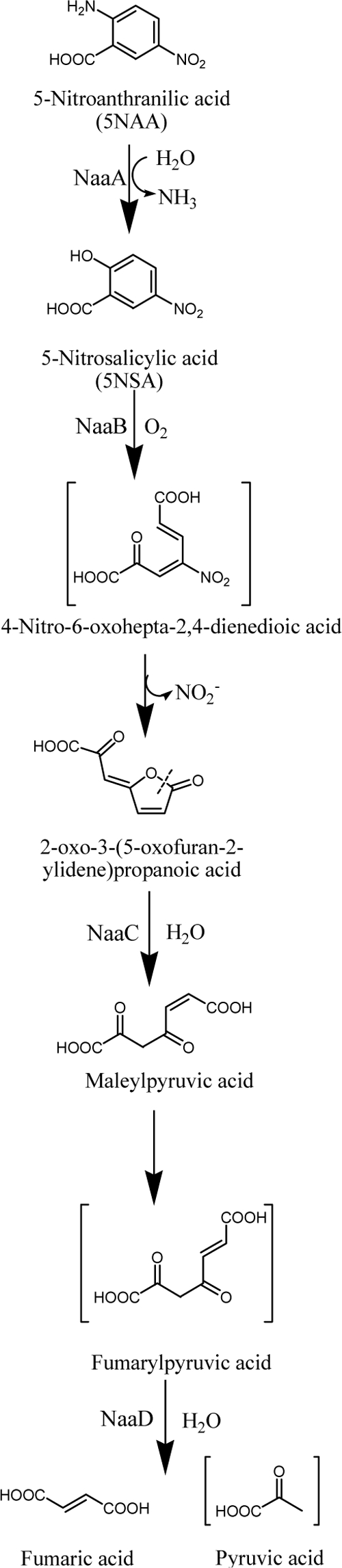
Proposed degradation pathway for 5NAA in Bradyrhizobium sp. strain JS329.
It now seems that salicylate (13, 32) and a wide variety of 5-substituted salicylates (8, 13, 43), including 5-amino-, 5-halo-, 5-methyl-, 5-nitro-, and 5-hydroxysalicylate can be transformed by enzymes related to salicylate/gentisate 1,2-dioxygenases to open the aromatic rings. When the 5 position is replaced with a halogen or nitro group, the spontaneous lactone formation is accompanied by the elimination of HX or HNO2. A recently described exception involves the biodegradation of 5-chlorosalicylate initiated by salicylate 1-hydroxylase-catalyzed oxidation to produce 5-chlorocatechol (26). Based on sequence analysis, biochemistry studies, and analogy with the mechanism of 5-chlorosalicylate degradation by salicylate dioxygenase (13), we propose a biodegradation pathway for 5NAA in Bradyrhizobium sp. JS329 (Fig. 5). The initial reaction is a hydrolytic removal of the amino group to form 5NSA, which is then oxidized by 5NSA dioxygenase to open the aromatic ring. The resulting ring fission product undergoes spontaneous lactonization accompanied by the removal of the nitro group as nitrite. The lactone ring opening is catalyzed by lactone hydrolase to form maleylpyruvate, which is in turn hydrolyzed to produce intermediates of central metabolism.
The biodegradation pathways of synthetic compounds seem to have evolved from those of natural analogs (7, 18, 45, 46). Only a few biodegradation pathways have been established for natural nitro compounds (20), so our current understanding stops far short of enabling predictions about the evolution of specific pathways. Recent work, including this study, suggests that detailed characterization of the metabolic pathways and enzymes can uncover new metabolic diversity (32), reveal the genes that have enabled the rapid and recent evolution of pathways for the degradation of synthetic nitro compounds, and provide insight about the most fruitful area of inquiry to attack the problem of the many unknown ORFs (32) and incorrectly annotated genes (28).
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Sena Yoo for technical assistance, Derek Boyd, Andreas Stolz, and Raquel L. Lieberman for helpful discussion, and Shirley Nishino for reviewing the manuscript.
This work was supported by the Defense Threat Reduction Agency and the U.S. Army Research Office, grant W911NF-07-1-0077.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 15 April 2011.
REFERENCES
- 1. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bergmann J. G., Sanik J. 1957. Determination of trace amounts of chlorine in naphtha. Anal. Chem. 29:241–243 [Google Scholar]
- 3. Besemer J., Borodovsky M. 2005. GeneMark: Web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 33:W451–W454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Broder D. H., Miller C. G. 2003. DapE can function as an aspartyl peptidase in the presence of Mn2+. J. Bacteriol. 185:4748–4754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bruhn C., Lenke H., Knackmuss H. J. 1987. Nitrosubstituted aromatic compounds as nitrogen source for bacteria. Appl. Environ. Microbiol. 53:208–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buckingham J. 1992. Dictionary of natural products. Chapman and Hall, London, United Kingdom [Google Scholar]
- 7. Cases I., de Lorenzo V. 2001. The black cat/white cat principle of signal integration in bacterial promoters. EMBO J. 20:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Crawford R. L., Olson P. E., Frick T. D. 1979. Catabolism of 5-chlorosalicylate by a Bacillus isolated from the Mississippi River. Appl. Environ. Microbiol. 38:379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ebert S., Rieger P. G., Knackmuss H. J. 1999. Function of coenzyme F420 in aerobic catabolism of 2,4,6-trinitrophenol and 2,4-dinitrophenol by Nocardioides simplex FJ2-1A. J. Bacteriol. 181:2669–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fuenmayor S. L., Wild M., Boyes A. L., Williams P. A. 1998. A gene cluster encoding steps in conversion of naphthalene to gentisate in Pseudomonas sp. strain U2. J. Bacteriol. 180:2522–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Haddad S., Eby D. M., Neidle E. L. 2001. Cloning and expression of the benzoate dioxygenase genes from Rhodococcus sp. strain 19070. Appl. Environ. Microbiol. 67:2507–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hintner J. P., et al. 2001. Direct ring fission of salicylate by a salicylate 1,2-dioxygenase activity from Pseudaminobacter salicylatoxidans. J. Bacteriol. 183:6936–6942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hintner J. P., Reemtsma T., Stolz A. 2004. Biochemical and molecular characterization of a ring fission dioxygenase with the ability to oxidize (substituted) salicylate(s) from Pseudaminobacter salicylatoxidans. J. Biol. Chem. 279:37250–37260 [DOI] [PubMed] [Google Scholar]
- 14. Hirose J., et al. 2001. Characterization of the metal-substituted dipeptidyl peptidase III (rat liver). Biochemistry 40:11860–11865 [DOI] [PubMed] [Google Scholar]
- 15. Hofmann K. W., Knackmuss H. J., Heiss G. 2004. Nitrite elimination and hydrolytic ring cleavage in 2,4,6-trinitrophenol (picric acid) degradation. Appl. Environ. Microbiol. 70:2854–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hopper D. J., Chapman P. J., Dagley S. 1968. Enzymatic formation of D-malate. Biochem. J. 110:798–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Javid-Majd F., Blanchard J. S. 2000. Mechanistic analysis of the argE-encoded N-acetylornithine deacetylase. Biochemistry 39:1285–1293 [DOI] [PubMed] [Google Scholar]
- 18. Johnson G. R., Spain J. C. 2003. Evolution of catabolic pathways for synthetic compounds: bacterial pathways for degradation of 2,4-dinitrotoluene and nitrobenzene. Appl. Microbiol. Biotechnol. 62:110–123 [DOI] [PubMed] [Google Scholar]
- 19. Jozic D., et al. 2002. Crystal structure of the dinuclear zinc aminopeptidase PepV from Lactobacillus delbrueckii unravels its preference for dipeptides. Structure 10:1097–1106 [DOI] [PubMed] [Google Scholar]
- 20. Ju K. S., Parales R. E. 2010. Nitroaromatic compounds, from synthesis to biodegradation. Microbiol. Mol. Biol. Rev. 74:250–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kadiyala V., Spain J. C. 1998. A two-component monooxygenase catalyzes both the hydroxylation of p-nitrophenol and the oxidative release of nitrite from 4-nitrocatechol in Bacillus sphaericus JS905. Appl. Environ. Microbiol. 64:2479–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. King R. R., Lawrence C. H., Calhoun L. A. 1998. Unusual production of 5-nitroanthranilic acid by Streptomyces scabies. Phytochemistry 49:1265–1267 [Google Scholar]
- 23. Klein M., Kaltwasser H., Jahns T. 2002. Isolation of a novel, phosphate-activated glutaminase from Bacillus pasteurii. FEMS Microbiol. Lett. 206:63–67 [DOI] [PubMed] [Google Scholar]
- 24. Lipscomb W. N., Strater N. 1996. Recent advances in zinc enzymology. Chem. Rev. 96:2375–2434 [DOI] [PubMed] [Google Scholar]
- 25. Marchler-Bauer A., et al. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37:D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nikodem P., Hecht V., Schlomann M., Pieper D. H. 2003. New bacterial pathway for 4- and 5-chlorosalicylate degradation via 4-chlorocatechol and maleylacetate in Pseudomonas sp. strain MT1. J. Bacteriol. 185:6790–6800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nishino S. F., Paoli G. C., Spain J. C. 2000. Aerobic degradation of dinitrotoluenes and pathway for bacterial degradation of 2,6-dinitrotoluene. Appl. Environ. Microbiol. 66:2139–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nishino S. F., Shin K. A., Payne R. B., Spain J. C. 2010. Growth of bacteria on 3-nitropropionic acid as a sole source of carbon, nitrogen, and energy. Appl. Environ. Microbiol. 76:3590–3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nishino S. F., Spain J. C., He Z. 2000. Strategies for aerobic degradation of nitroaromatic compounds by bacteria: process discovery to field application. In Spain J. C., Hughes J. B., Knackmuss H.-J. (ed.), Biodegradation of nitroaromatic compounds and explosives. Lewis Publishers, Boca Raton, FL [Google Scholar]
- 30. Parry R., Nishino S., Spain J. 2011. Naturally-occurring nitro compounds. Nat. Prod. Rep. 28:152–167 [DOI] [PubMed] [Google Scholar]
- 31. Parsons T. R., Maita Y., Lalli C. M. 1984. A manual of chemical and biological methods for seawater analysis. Pergamon Press, Oxford, United Kingdom [Google Scholar]
- 32. Qu Y., Spain J. C. 2010. Biodegradation of 5-nitroanthranilic acid by Bradyrhizobium sp. strain JS329. Appl. Environ. Microbiol. 76:1417–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Qu Y., Spain J. C. 2011. Catabolic pathway for 2-nitroimidazole involves a novel nitrohydrolase that also confers drug resistance. Environ. Microbiol. 13:1010–1017 [DOI] [PubMed] [Google Scholar]
- 34. Ramos J. L., Gonzalez-Perez M. M., Caballero A., van Dillewijn P. 2005. Bioremediation of polynitrated aromatic compounds: plants and microbes put up a fight. Curr. Opin. Biotechnol. 16:275–281 [DOI] [PubMed] [Google Scholar]
- 35. Rudolf E., Girardet J. M., Bautz A. M., Dournon C. 1997. Purification and partial characterization of peptidase-1, a sex-linked enzyme in Pleurodeles waltl (urodele amphibian). Biochem. Cell Biol. 75:803–806 [PubMed] [Google Scholar]
- 36. Schmidt E., Knackmuss H. J. 1980. Chemical structure and biodegradability of halogenated aromatic compounds. Conversion of chlorinated muconic acids into maleoylacetic acid. Biochem. J. 192:339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Seffernick J. L., de Souza M. L., Sadowsky M. J., Wackett L. P. 2001. Melamine deaminase and atrazine chlorohydrolase: 98 percent identical but functionally different. J. Bacteriol. 183:2405–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Seo J. M., Ji G. E., Cho S. H., Park M. S., Lee H. J. 2007. Characterization of a Bifidobacterium longum BORI dipeptidase belonging to the U34 family. Appl. Environ. Microbiol. 73:5598–5606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shi D., Yu X., Roth L., Tuchman M., Allewell N. M. 2007. Structure of a novel N-acetyl-l-citrulline deacetylase from Xanthomonas campestris. Biophys. Chem. 126:86–93 [DOI] [PubMed] [Google Scholar]
- 40. Smibert R. M., Krieg N. R. 1994. Phenotypic characterization, p. 607–654 In Gerhardt P., Murray R. G. E., Wood W. A., Krieg N. R. (ed.), Methods for general and molecular bacteriology. ASM Press, Washington, DC [Google Scholar]
- 41. Spain J. C. 1995. Biodegradation of nitroaromatic compounds. Annu. Rev. Microbiol. 49:523–555 [DOI] [PubMed] [Google Scholar]
- 42. Spanggord R. J., Spain J. C., Nishino S. F., Mortelmans K. E. 1991. Biodegradation of 2,4-dinitrotoluene by a Pseudomonas sp. Appl. Environ. Microbiol. 57:3200–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stolz A., Nortemann B., Knackmuss H. J. 1992. Bacterial metabolism of 5-aminosalicylic acid. Initial ring cleavage. Biochem. J. 282(Pt. 3):675–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Urata M., et al. 2004. Genes involved in aniline degradation by Delftia acidovorans strain 7N and its distribution in the natural environment. Biosci. Biotechnol. Biochem. 68:2457–2465 [DOI] [PubMed] [Google Scholar]
- 45. van der Meer J. R. 2006. Evolution of catabolic pathways in Pseudomonas through gene transfer. In Levesque R. C., Ramos J.-L. (ed.), Pseudomonas: molecular biology of emerging issues, vol. 4. Springer, New York, NY [Google Scholar]
- 46. Wackett L. P. 2009. Questioning our perceptions about evolution of biodegradative enzymes. Curr. Opin. Microbiol. 12:244–251 [DOI] [PubMed] [Google Scholar]
- 47. Zhou N. Y., Fuenmayor S. L., Williams P. A. 2001. nag genes of Ralstonia (formerly Pseudomonas) sp. strain U2 encoding enzymes for gentisate catabolism. J. Bacteriol. 183:700–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.