Abstract
Formate dehydrogenases (FDHs) are enzymes that catalyze the formate oxidation to carbon dioxide and that contain either Mo or W in a mononuclear form in the active site. In the present work, the influence of Mo and W salts on the production of FDH by Desulfovibrio alaskensis NCIMB 13491 was studied. Two different FDHs, one containing W (W-FDH) and a second incorporating either Mo or W (Mo/W-FDH), were purified. Both enzymes were isolated from cells grown in a medium supplemented with 1 μM molybdate, whereas only the W-FDH was purified from cells cultured in medium supplemented with 10 μM tungstate. We demonstrated that the genes encoding the Mo/W-FDH are strongly downregulated by W and slightly upregulated by Mo. Metal effects on the expression level of the genes encoding the W-FDH were less significant. Furthermore, the expression levels of the genes encoding proteins involved in molybdate and tungstate transport are downregulated under the experimental conditions evaluated in this work. The molecular and biochemical properties of these enzymes and the selective incorporation of either Mo or W are discussed.
INTRODUCTION
Sulfate-reducing bacteria (SRB) are anaerobic prokaryotes that have a major economic impact in biocorrosion processes of ferrous metals and water pollution (18, 21, 22). These organisms use electrons donors, such as hydrogen, organic acids, or alcohols, to reduce sulfate to sulfide (43).
Desulfovibrio species are sulfate-reducing bacteria in which the oxidation of formate to carbon dioxide is catalyzed by Mo- or W-containing formate dehydrogenases (FDHs). This reaction provides electrons that are transferred probably through the periplasmic cytochrome network to the cytoplasm to be used for sulfate reduction (19). The formate dehydrogenases identified in Desulfovibrio species belong to the dimethyl sulfoxide (DMSO) reductase family and have been isolated as heterodimeric (1, 6) or heterotrimeric (9, 41) proteins. The α subunit of these enzymes (∼90 kDa) contains the active site and one [4Fe-4S] cluster. The active site is constituted by a Mo or a W atom coordinated by two pyranopterin molecules, a selenocysteine (typical in this type of FDH), and a sulfur atom. The β subunit (∼29 kDa) contains three additional [4Fe-4S] clusters, which together with the heme groups harbored in the third subunit (∼16 kDa) of some FDHs form part of the enzyme electron transfer pathway (7, 20, 26, 37). FDHs have been isolated as Mo-containing proteins from Desulfovibrio desulfuricans ATCC 27774 (9, 14, 36) and Desulfovibrio vulgaris Hildenborough (12, 40, 41) and as a W-containing protein from Desulfovibrio gigas (1, 32, 33). In contrast, Desulfovibrio alaskensis NCIMB 13491 cells grown in medium C (31) produce an FDH which can incorporate either Mo or W (6). Analysis of the genome of D. alaskensis G20 (http://genome.jgi-psf.org), closely related to D. alaskensis NCIMB 13491 (8, 35), shows at least three periplasmic FDHs encoded in the chromosome of this organism, corresponding to the locus tags Dde_3512-Dde_3514, Dde_0716-Dde_0718, and Dde_0813-0814. Note that the D. alaskensis G20 genome corresponds to that deposited as D. desulfuricans G20. The reclassification of D. desulfuricans G20 as D. alaskensis G20 has been proposed but not yet officially adopted (29). Amino acid sequence alignment of the proteins encoded by these genes shows a high identity percentage (48 to 75%) with the homologous proteins isolated and characterized from other Desulfovibrio species. Likewise, D. alaskensis G20 FDHs appear as heterodimeric soluble enzymes composed of α and β subunits. Three major features are found in the amino acid sequence of the α subunit: a twin-arginine motif [RRXF(L/I)K] characteristic of Sec-independent export to periplasm (2, 38), a molybdopterin dinucleotide-binding region, and the motif CXXCXnCXmC, known to coordinate [4Fe-4S] clusters. The β subunit contains three additional [4Fe-4S] binding motifs, which is in agreement with the [4Fe-4S] centers observed in the crystallographic structure of the homologous D. gigas FDH (32, 33).
Although the synthesis of the Mo and W pyranopterin cofactors (MoCo and WCo, respectively) present in FDHs and other closely related enzymes has been extensively studied (3, 25, 39), the selective incorporation of either Mo or W in the enzyme cofactor is not understood yet. Several proteins are involved in the metal insertion step of this biosynthetic pathway, with MoeA being one of them (27, 28). The function proposed for the MoeA protein is to form a complex with the adenylylated metal-binding pterin (MPT), which is hydrolyzed in the presence of either molybdate or tungstate to incorporate the metal into the cofactor (23). The presence of two moeA genes, sharing approximately 40% sequence similarity in numerous bacterial genomes, gave rise to the hypothesis that these proteins would have Mo- or W-selective activities (3, 4). The analysis of the D. alaskensis G20 genome also reveals two moeA-like genes sharing 32.5% sequence identity (Dde_0230 and Dde_3228, respectively), suggesting that this hypothesis may also be proposed for this organism.
The metal (Mo or W) ion incorporated into the cofactor is transported into the cells as molybdate and tungstate ions by a protein system belonging to the ABC transporter family. This system is composed of a periplasmic component responsible for recognition and binding of molybdate (protein A), a transmembrane pore (protein B) which enables the entry of the substrate into the cell, and a cytoplasmic protein responsible for the energy of the transport (protein C) (17, 24, 39). D. alaskensis G20 contains a Mo transport system encoded by the mod gene cluster, which contains the Dde_3518, Dde_3519, and Dde_3520 genes (modA, modB, and modC, respectively). Similarly, the proteins putatively involved in tungstate transport are encoded by the tup genes, which include the Dde_0234, Dde_0233, and Dde_0232 genes (tupA, tupB, and tupC, respectively).
This work aims to understand the effects of Mo or W supplementation on the gene expression levels and activity profiles of the different FDHs in D. alaskensis NCIMB 13491. In addition, the effects of molybdate and tungstate on the expression levels of the genes coding for the proteins involved in Mo and W transport and their insertion in the cofactor were investigated. The results show that the presence of each metal in the culture medium has remarkable effects on both gene expression and biochemical properties of FDHs.
MATERIALS AND METHODS
Bacteria, culture media, and growth conditions.
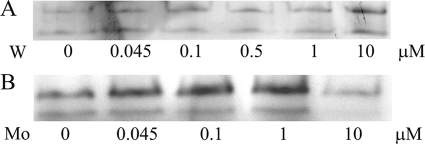
D. alaskensis NCIMB 13491 cells were used in all the experiments performed in this work. Since the genome of this subspecies is not reported, the genomic analysis was performed on the basis of the genome of the closely related D. alaskensis G20 strain (http://genome.jgi-psf.org). All experiments were performed in duplicate by using independent batches of media under the same conditions. The FDH activity gels showed in Fig. 1 correspond to one of the replicates.
Fig. 1.
Influence of W and Mo on the FDH activity of soluble extract of D. alaskensis NCIMB 13491. The soluble extracts were obtained from cells grown until the end of the exponential phase (OD600 of 0.5 to 0.6) in modified medium C supplemented with different concentrations of Na2WO4·2H2O (A) or Na2MoO4·2H2O (B). Metal concentrations indicated in each lane correspond to the metal added to the medium. Note that this is not the final concentration of W, as the medium contains 0.27 ± 0.12 μM W without metal supplementation. The total amounts of proteins loaded in each lane under tungstate and molybdate supplementation conditions were 10 μg and 45 μg, respectively.
D. alaskensis NCIMB 13491 was grown at 37°C under anaerobic conditions in a modified version of medium C from Postgate (31). The medium was composed of 3.6 mM KH2PO4, 18.7 mM NH4Cl, 31.7 mM Na2SO4, 0.24 mM MgSO4·7H2O, 0.27 mM CaCl2·2H2O, 0.1% (wt/vol) yeast extract, 14.4 μM FeSO4·7 H2O, 1.02 mM sodium citrate dihydrate, 0.43 mM NaCl, and 25.5 mM lactate and 4.5 mM formate as energy and carbon sources. The pH was adjusted to 7.5 with NaOH. This medium, which was shown to be appropriate for FDH production, is here named modified medium C.
The effect of Mo and W supplementation on the FDH production was evaluated by adding to the modified medium C 45 nM, 100 nM, 500 nM, 1 μM, and 10 μM either Na2MoO4·2H2O or Na2WO4·2H2O.
Soluble extract preparation and activity tests.
D. alaskensis NCIMB 13491 cells in each condition described above were collected at the end of the exponential growth phase (optical density at 600 nm [OD600] of 0.5 to 0.6) by centrifugation (7,000 × g for 15 min at 4°C), and the pellet was resuspended in 10 mM Tris-HCl buffer (pH 7.6) to a cell density of 0.1 g cells (wet weight) per ml. To prepare soluble extracts, cell suspension was subjected to four freeze-thaw cycles and centrifuged at 7,000 × g for 40 min. Supernatant was subsequently used for activity measurements. Total protein concentration for each extract was determined by using the bicinchoninic acid (BCA) kit from Sigma-Aldrich. In-gel formate dehydrogenase activity was detected by running equal amounts of total proteins from each soluble extract on 7.5% native polyacrylamide gels under native conditions (10 μg and 45 μg of total proteins were loaded for the W and Mo supplementation conditions, respectively). This normalization procedure was validated by SDS-PAGE. After electrophoresis, the activity assay was performed at 37°C under anaerobic conditions by bubbling argon in all solutions. Gels were placed in rubber-stoppered tubes containing 10 mM HCOONa, 130 mM β-mercaptoethanol, and 60 mM Tris-HCl buffer (pH 8.0) and incubated for 10 min. Then, methyl viologen was added (7.5 mM final concentration) to develop blue activity bands. Finally, to fix the activity bands, 2,3,5-triphenyltetrazolium chloride (Sigma-Aldrich) was added to a final concentration of 7.5 mM, and the gels were incubated for an additional 15 min.
Steady-state activity assays were performed under anaerobic conditions at 37°C in a final volume of 1.1 ml by monitoring spectrophotometrically the reduction of benzyl viologen at 555 nm as previously described (36). Each assay contained 300 nM Mo/W-FDH or 10 nM W-FDH (see Results). In order to compare the values obtained for the W-FDH, the specific activity of the D. gigas W-FDH was determined using the activity test described in reference 36.
Enzyme purification.
D. alaskensis NCIMB 13491 was anaerobically cultured in modified medium C containing either 1 μM Na2MoO4·2H2O or 10 μM Na2WO4·2H2O. Cells were collected by centrifugation at the end of the exponential growth phase (OD600 of 0.5 to 0.6). Soluble extracts were obtained from cells resuspended in 10 mM Tris-HCl buffer (pH 7.6) and disrupted with a French press at 9,000 lb/in2. Broken cells were centrifuged at 10,000 × g for 45 min at 4°C, and the supernatants were further centrifuged at 180,000 × g for 60 min at 4°C.
All purification steps were performed at 4°C and pH 8.0. Soluble fractions were dialyzed overnight against 5 mM Tris-HCl and loaded onto a DE-52 cellulose resin equilibrated with 5 mM Tris-HCl. Elution was performed with a linear gradient (5 mM to 500 mM Tris-HCl) in 5 column volumes. Soluble extracts from cells cultured under either Mo or W excess conditions showed FDH activity in fractions eluted at 350 mM Tris-HCl, but only the condition corresponding to Mo supplementation showed additional enzyme activity in fractions eluted at 200 mM Tris-HCl. The protein pools containing FDH were independently concentrated and loaded onto a Source 15Q column equilibrated with 10 mM Tris-HCl buffer. Elution was performed using a linear gradient to 50 mM Tris-HCl/500 mM NaCl in 10 column volumes. Fractions containing FDH were pooled, dialyzed, concentrated, and loaded into a Resource Q column (6 ml; GE Healthcare) equilibrated with 50 mM potassium phosphate buffer (KPB). Elution was performed using a linear gradient to 50 mM KPB-500 mM NaCl in 25 column volumes. FDH-active samples were pooled, concentrated, and loaded onto a Superdex 200 column equilibrated with 50 mM KPB-150 mM NaCl. After this purification step, the enzymes that eluted at 350 mM Tris-HCl from the first DE-52 column were considered pure as judged by SDS-PAGE and UV-visible spectroscopy.
The highest purity of the FDH samples eluted at 200 mM Tris-HCl from the first DE-52 column (see above) was achieved by a fifth purification step that included a MonoQ column (1 ml; GE Healthcare) equilibrated with 50 mM KPB and eluted to 50 mM KPB-300 mM NaCl in 25 column volumes.
Protein and metal quantification.
Protein quantification was performed using the BCA kit from Sigma-Aldrich with bovine serum albumin as the standard. Metal contents in both samples and media were determined through inductively coupled plasma atomic emission spectroscopy (ICP-AES) by using a Jobin-Yvon (Ultima) instrument. A tungsten standard solution from CRM (aqueous calibration solution) was used to quantify this metal, and multielement standard solution (Reagecon 23 ICP) was used for molybdenum. The concentration range of both standard solutions was 0.5 to 15.6 μM.
Molecular mass and cofactor content determination.
Molecular masses of the pure proteins were determined under nondenaturing conditions by gel filtration chromatography on a Superdex 200 column (GE Healthcare) connected to a high-performance liquid chromatography (HPLC) device (AKTA Basic; GE Healthcare). The elution buffer used was 50 mM KPB-150 mM NaCl (pH 7.0), and the column was calibrated by using proteins from 13 kDa to 669 kDa, all from GE Healthcare. Subunit composition and purity of the samples were evaluated by SDS-PAGE on 12.5% polyacrylamide gels. Molecular-mass standards (Fermentas) were used for calibration, and gels were stained with Coomassie brilliant blue G-250 (Merck).
The cofactor content was determined as previously published (13, 15). Briefly, FDH samples in sulfuric acid (3% [vol/vol]) were heated at 95°C for 10 min and centrifuged for 5 min at 10,000 × g. A volume of 100 μl was loaded in a reverse-phase column (Merck LiChrospher 100, 250 mm by 4 mm [inside diameter], RP-18e; 4 μm) connected to an HPLC device (AKTA Basic; GE Healthcare). Ammonium acetate (50 mM, pH 6.8) was used as eluent at a flow rate of 1 ml/min, and nucleotides were detected at 254 nm. Quantitative determinations were performed by loading fresh solutions of known mononucleotide amounts (Sigma-Aldrich) previously submitted to identical acid/heating treatment.
Determination of the N-terminal sequence of FDHs from D. alaskensis NCIMB 13491.
FDH samples (600 pmol) were loaded on a 12.5% SDS-polyacrylamide gel. After staining, gel pieces corresponding to each subunit were cut off and incubated in 500 μl of Millipore water at 4°C for 36 h. Gel was then removed, and the solution was concentrated to approximately 50 μl in a SpeedVac centrifuge (UniEquip, Martinsried, Germany; model UNIVAPO 100H) equipped with a refrigerated aspirator vacuum pump, model Unijet II. Subunits were sequenced by Edman degradation reaction on a protein sequencer (Applied Biosystems; model 491). Sequences were compared to the deduced amino acid sequences from the D. alaskensis G20 genome (http://genome.jgi-psf.org).
qRT-PCR.
D. alaskensis NCIMB 13491 cells were cultured at 37°C under anaerobic conditions in 100 ml of modified medium C as described above without metal addition (control) and supplemented with either 1 μM Na2MoO4·2H2O or 10 μM Na2WO4·2H2O. Once the OD600 reached 0.5 to 0.6, cells were chilled on ice for 10 min, harvested by centrifugation at 8,000 × g for 20 min at 4°C, and washed once with 1 ml of 10 mM Tris-HCl (pH 8.0). The pellets were subsequently used for RNA extraction and quantitative real-time PCR (qRT-PCR) experiments.
For total RNA isolation, cell pellets were resuspended in 200 μl of 10 mM Tris-HCl and 1 mM EDTA (pH 8.0) RNase-free buffer. Total RNA was isolated by using the High Pure RNA isolation kit (Roche Diagnostics) according to the manufacturer's instructions with an extra DNase I digestion step in order to eliminate contaminating DNA. RNA quality was checked by electrophoresis on agarose gels, and the absence of DNA contamination was confirmed by PCR. RNA was quantified spectrophotometry at 260 nm (NanoDrop 1000 ThermoScience). For cDNA synthesis, 10 μg of total RNA and 3 μg of random primers (Invitrogen) were mixed, heated to 70°C for 3 min, and placed on ice. The cDNA synthesis mix (50 mM Tris-HCl [pH 8.3], 40 mM KCl, 6 mM MgCl2, 10 mM dithiothreitol [DTT], and 0.3 mM deoxynucleoside triphosphate [dNTPs]) was then added. The reaction mix (30 μl) was incubated for 5 min at 25°C, and 300 units of Superscript II reverse transcriptase (Invitrogen) was then added. The reaction mix was incubated for 5 min at 25°C, for 1 h at 42°C, and finally for 15 min at 70°C for heat inactivation. The reaction mixture volume was then adjusted to 100 μl with UltraPure water. cDNAs were prepared from three independent cultures and were further used for qRT-PCR.
Quantification of cDNAs was carried out with the LightCycler system using the LightCycler FastStart DNA Masterplus SYBR green I kit (Roche), according to the manufacturer's instructions. Primers were designed on the basis of the D. alaskensis G20 genome (http://genome.jgi-psf.org) to specifically amplify a portion of each targeted gene (Table 1). The LightCycler was programmed for an initial step at 95°C for 480 s, followed by 45 thermal cycles at 95°C for 12 s, 59°C for 6 s, and 72°C for 20 s. Specificities of accumulated products were verified by using the melting-curve analysis. The relative expression software tool (REST) was used to calculate the relative expression of each gene in each condition (30), using the 16S RNA gene as the reference for normalization. Quantification was performed in triplicate on each cDNA preparation.
Table 1.
Primers used for the qRT-PCR experiments
| Primer | Sequencea | Target gene/locus tag |
|---|---|---|
| Dde0717left | 5′GATGCACAAGGAGCAGTTCA | W-fdhA/Dde_0717 |
| Dde0717right | 5′CGTCTTGATGTCGGAAGGAT | |
| Dde3513left | 5′TTCACCGTTCAGGGAAAAAC | Mo/W-fdhA/Dde_3513 |
| Dde3513right | 5′TGTCGGCCTTGCGTATATT | |
| Dde0813left | 5′TGTACTGCGCTTCGTACACC | fdh3A/Dde_0813 |
| Dde0813right | 5′GTACGTCGCCTGTCCATTTT | |
| Dde3518right | 5′CAGTTTGCCTGTGGAGGAAT | modA/Dde_3518 |
| Dde3518left | 5′ATGCTGTCACGTCTGTACGC | |
| Dde0234right | 5′GTCGCAGTTTTCACCCATTT | tupA/Dde_0234 |
| Dde0234left | 5′AGCACCGGTTCTTATGATGG | |
| Dde3519right | 5′ATCAGCCGTTCGTTGAAAAC | modB/Dde_3519 |
| Dde3519left | 5′TAGCAGACCCGCTGTACCTT | |
| Dde0233right | 5′GCAGGGCATAGACGATAAGC | tupB/Dde_0233 |
| Dde0233left | 5′CGGTAACCACCATGTCTGTG | |
| Dde0230right | 5′ATAACTTCGTCTCCGGTGGA | moeA1/Dde_0230 |
| Dde0230left | 5′CCGTTGAAATACGCAAATCC | |
| Dde3328right | 5′AAATGTGGGGGAGGAAAAAC | moeA2/Dde_3228 |
| Dde3228left | 5′CAAGCGCAATGTCTACCTGA | |
| Dde2231left | 5′AGTCCGAAGGTGAAATGCTG | rpoA/Dde2231 |
| Dde2231right | 5′CCTCATTTTGCTGCTTCCTC | |
| Dde16Sleft | 5′CGTGGGTAGCAAACAGGATT | rRNA 16S gene |
| Dde16Sright | 5′CCGGATGTCAAGCCTAGGTA |
Sequences were based on the closely related D. alaskensis G20 genome (http://genome.jgi-psf.org).
RESULTS
Influence of Mo and W ions on FDH activity.
The influence of Mo and W ions on FDH activity was evaluated by adding different concentrations of Na2MoO4·2H2O or Na2WO4·2H2O (from 45 nM to 10 μM) to the modified medium C. It is important to note that this medium without Mo/W addition contains 0.27 ± 0.12 μM W, probably included in the yeast extract, and no detectable Mo (ICP-AES detection limit, 10 nM). The gel activity pattern of W-supplemented cell cultures showed two FDH activity bands of similar intensity (Fig. 1A). In contrast, cells cultured in modified medium C supplemented with Mo showed an upper activity band more intense than the lower band (Fig. 1B). The maximal FDH activity was detected in the medium supplemented with 10 μM Na2WO4·2H2O, whereas the medium supplemented with Mo yielded the maximal activity using Na2MoO4·2H2O concentrations in the range of 0.045 to 1 μM (Fig. 1). The media containing the highest concentrations of metal (Mo and W) were chosen for the studies described below. These conditions guarantee the probable synthesis of MoCo over WCo (or vice versa) and the excess of Mo over W in the case of the molybdenum-supplemented medium (note that the modified medium C contains W on the order of 200 nM).
Protein purification, identification, and biochemical characterization.
The differences found in the FDH expression patterns suggested the production of more than one enzyme (Fig. 1). In order to identify and characterize the different FDHs produced by D. alaskensis NCIMB 13491, the enzymes were isolated from cells cultured in modified medium C supplemented with either 1 μM molybdate or 10 μM tungstate. As already stated in Materials and Methods, FDH activity was found in fractions eluted at 350 mM Tris-HCl from the first DE-52 cellulose chromatography column under Mo or W supplementation conditions. Nevertheless, when the culture medium was supplemented with Mo, an additional fraction that eluted at 200 mM Tris-HCl from the first chromatographic column also exhibited FDH activity.
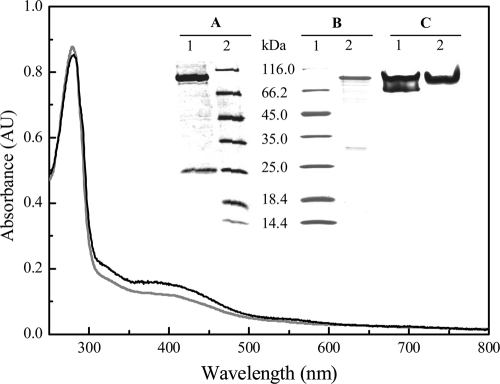
From cells cultured in the presence of W, a sample containing pure FDH was isolated as judged by SDS-PAGE (Fig. 2, inset, lane 1 in gel A) and the A400/A280 ratio of the UV-visible spectrum (Fig. 2, black line). The total yield from this purification process was 5 mg of pure protein per 300 liters of cell culture. This enzyme contains 1.1 ± 0.1 W atoms per heterodimer (no Mo was detected) and has a specific activity of 94 ± 6 U/mg. The as-purified protein (W-FDH) has a molecular mass of approximately 290 kDa as determined by gel filtration chromatography. This result, together with the determination of the molecular mass of the subunits, suggests that this protein is a dimer of a heterodimer composed of two subunits of approximately 109 kDa (α subunit) and 27 kDa (β subunit) (Table 2 and Fig. 2, inset, lane 1 in gel A).
Fig. 2.
UV-visible spectrum of W-FDH (black line) and Mo/W-FDH (gray line) isolated from D. alaskensis NCIMB 13491 cells. Inset: (A) SDS-PAGE of W-FDH (lane 1; 2 μg) and molecular weight markers (lane 2); (B) SDS-PAGE of molecular weight markers (line 1) and Mo/W-FDH (lane 2; 1 μg); (C) gel activity of the as-purified W-FDH (lane 1; 2 μg) and Mo/W-FDH (lane 2; 9 μg).
Table 2.
Biochemical properties of FDHs isolated from D. alaskensis NCIMB 13491
| FDH | Metal content |
Mol mass (as isolated, kDa) | Mol mass of subunits (kDa)b |
Specific activity (U/mg) | ||
|---|---|---|---|---|---|---|
| Mo/heterodimer | W/heterodimer | α | β | |||
| W-FDH | NDa | 1.1 ± 0.1 | 290 | 109 | 27 | 94 ± 6 |
| Mo/W-FDH | 0.35 ± 0.02 | 0.53 ± 0.03 | 146 | 109 | 28 | 4.65 ± 0.07 |
ND, not detected.
Molecular mass calculated from the amino acid sequence analysis.
The N-terminal sequence of α and β subunits showed that W-FDH is encoded by the Dde_0716-0717 and Dde_0718 genes (here called W-fdhA and W-fdhB, respectively). It is important to note that the gene encoding the α subunit contains a UGA codon that is specifically decoded by the selenocysteine insertion machinery. This codon was annotated as a stop codon in the D. alaskensis G20 genome, which explains the splitting of the W-fdhA gene into two genes (Dde_0716 and Dde_0717). The gel activity pattern of the pure W-FDH showed two bands (Fig. 2, inset, lane 1 in gel C). To determine whether this activity pattern was an artifact or not, the bands were analyzed by SDS-PAGE, and the N-terminal sequences of the isolated α and β subunits of both bands were determined. The results showed that the α and β subunits of both bands are coded by the W-fdhA and W-fdhB genes, respectively, indicating that the gel activity profile is an artifact, likely due to the electrophoresis conditions. Moreover, the analysis of this protein by exclusion chromatography showed only one peak in the chromatogram (data not shown), which agrees with the presence of only the W-FDH. This is not the only case where two activity bands are observed for an isolated protein. For instance, Syntrophobacter fumaroxidans produces two W-containing FDHs, and one of them shows the same behavior on native PAGE (10).
The D. alaskensis NCIMB 13491 W-FDH was also isolated from the cells grown in medium supplemented with Mo.
As mentioned above, cells cultured in the presence of Mo yielded an extra FDH activity fraction that eluted at lower ionic strength (200 mM Tris HCl) from the first chromatographic column. The purification protocol described in Materials and Methods allowed the isolation of 5.5 mg of FDH from a 300-liter cell culture. This enzyme showed a specific activity of 4.65 ± 0.07 U/mg and was isolated as a heterodimer of approximately 146 kDa, constituted by an α and a β subunit (Fig. 2, inset, lane 2 in gel B). Activity gels of this enzyme showed a unique band (Fig. 2, inset, lane 2 in gel C), and metal content analysis detected 0.35 ± 0.02 Mo atoms and 0.53 ± 0.03 W atoms per heterodimer. This FDH is here named Mo/W-FDH. The amino acid sequence of the α and β subunits matched with the translated amino acid sequence of the Dde_3512-3513 and Dde_3514 genes (here called Mo/W-fdhA and Mo/W-fdhB, respectively). As for the W-FDH, the Mo/W-FDH contains a selenocysteine, explaining the genome annotation (Dde_3512 and Dde_3513) for the α subunit. The molecular masses of the α and β subunits calculated from the translated amino acid sequences are 109 and 28 kDa, respectively (Table 2). The biochemical properties of this enzyme indicated that it corresponds to the FDH isolated by Brondino et al. (6).
Both Mo/W-FDH and W-FDH proteins contain the guanidine form of the pyranopterin cofactor and exhibit UV-visible spectra with a shoulder at 320 nm and an absorbance band at 400 nm typical of proteins containing [4Fe-4S] metal clusters (Fig. 2). The isoelectric points calculated from the translated amino acid sequence were 6.92 and 7.84 for the W-FDH and the Mo/W-FDH, respectively. The differences between the pI values can explain the complete separation of both proteins by the anionic exchange chromatography column used as the first purification step. The biochemical properties of both FDHs are summarized in Table 2.
The third soluble FDH encoded by the Dde_0812-Dde_0813 gene (here called fdh3) was not detected under the growth conditions tested in this study.
Gene expression analysis.
The production of the enzymes was also evaluated at the transcriptional level by qRT-PCR. The expression levels of the genes Mo/W-fdhA, W-fdhA, and fdh3A were measured in D. alaskensis NCIMB 13491 grown in modified medium C without metal supplementation (control) and in the presence of either 1 μM Mo or 10 μM W.
When cells were grown in modified medium C without metal supplementation, transcripts corresponding to both W-fdhA and Mo/W-fdhA genes were detected. However, the amount of Mo/W-fdhA transcripts was about 6-fold lower than that of W-fdhA (data not shown). These results are in line with the W detected in the modified medium C.
In contrast, cells cultured in modified medium C supplemented with Mo showed a slight upregulation of the Mo/W-fdhA gene (Table 3), which is in agreement with the detection of a Mo/W-FDH different from the W-FDH isolated from cells cultured under W supplementation conditions. Under these Mo excess conditions, expression of the W-fdhA gene was downregulated about 4-fold (Table 3). On the other hand, addition of tungstate to the culture medium induced a strong downregulation of the Mo/W-fdhA gene (about 59 times) and had no significant effect on the W-fdhA gene (Table 3). No significant amounts of transcripts were detected for the fdh3A gene under the growth conditions tested, which suggests that this gene is either not expressed at all or expressed at a very low level. As a control, we followed the expression rate of the rpoA gene that was expected to be not affected by the presence of metal. No significant variation of the rpoA expression rate was observed whatever the metal supplementation (Table 3), reinforcing the relevance of the fdhs gene expression changes.
Table 3.
Relative gene expression as determined by qRT-PCRa
| Gene | Log2 ratio (±SD) |
|
|---|---|---|
| Mo supplementation | W supplementation | |
| Mo/W-fdhA (Dde_3513) | 1.70 (±0.23) | −5.90 (±0.20) |
| W-fdhA (Dde_0717 | −2.10 (±0.09) | −0.48 (±0.54) |
| modB (Dde_3519) | −2.49 (±0.30) | −11.11 (±0.56) |
| tupB (Dde_0233) | −1.24 (±0.63) | −7.65 (±0.36) |
| moeA1 (Dde_0230) | −0.67 (±0.08) | −3.23 (±0.59) |
| moeA2 (Dde_3228) | −0.40 (±0.59) | −0.46 (±0.60) |
| rpoA (Dde_2231) | +0.10 (±0.13) | −0.26 (±0.11) |
The abundance of transcripts was determined from cells cultured until the end of the exponential phase (OD600 of 0.5 to 0.6) in modified medium C supplemented with either 1 μM Mo or 10 μM W and in modified medium C without metal addition. Expression changes are given as log2 ratios (metal-supplemented modified medium C/modified medium C).
The expression levels of genes involved in molybdate transport (modA and modB) and tungstate (tupA and tupB) were also evaluated. When cells were cultured in modified medium C without metal supplementation, the expression levels of modA and modB genes were similar. In contrast, the expression levels of tupA and tupB genes were strongly different, with tupA being about 230-fold more expressed than tupB (data not shown). These data suggest that modA and modB genes belong to the same transcriptional unit (modABC), as commonly predicted for these gene clusters. In contrast, the different expression level of tupA and tupB genes indicated that they do not belong to the same transcriptional unit, as also predicted by in silico analysis. In light of these results, we decided to use modB and tupB as markers for the expression level of the molybdate and tungstate transport systems, respectively. Comparison of modB and tupB expression in cells cultured in modified medium C without metal supplementation revealed that the modB gene was about 22-fold less expressed than the tupB gene (not shown). The addition of molybdate to the culture medium induced a significant downregulation of the modB gene (6 times), while tupB gene expression was only very slightly affected. In contrast, a strong repression of both genes was observed upon tungstate addition, with modB being the most repressed gene (Table 3).
The effects of both ions on the expression levels of genes coding for putative MoeA proteins (moeA1/Dde_0230 and moeA2/Dde_3228 [Fig. 3 C and E, respectively]), which are proposed to be involved in the incorporation of the metal into the FDH cofactor, were also tested. The results showed that moeA1 was significantly downregulated in the presence of tungstate (about 9 times), whereas the expression level of the moeA2 was not significantly affected upon Mo or W supplementation (Table 3).
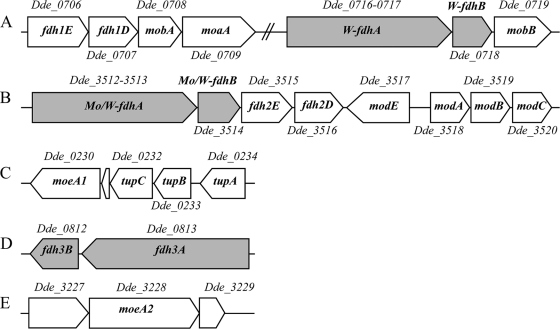
Fig. 3.
Gene organization of FDHs in D. alaskensis G20, including the elements situated in the surrounding area of the fdh genes that are putatively involved in molybdenum transport, biosynthesis, and incorporation of the metal cofactor. (A) fdh1E and fdh1D genes encode proteins likely involved in FDH formation, the mobA gene codes a protein that catalyzes the conversion of MPT to molybdopterin guanine dinucleotide, the moaA gene codes a protein that catalyzes the conversion of GTP into cyclic pyranopterin monophosphate (precursor Z), and the W-fdhA and W-fdhB genes encode the α and β subunits of the W-FDH, respectively. The Dde_0719 gene encodes a protein homologous to MobB which binds GTP and has weak GTPase activity (11). (B) The Mo/W-fdhA and Mo/W-fdhB genes encode the α and β subunits of the Mo/W-FDH. The modABC genes encode the molybdate uptake transport system. modE encodes a protein that regulates the modABC operon expression (16, 17, 34). (C) The Dde_0230 gene encodes a protein homologous to MoeA, which is involved in Mo/W insertion into the MPT. Dde_0232-Dde_0234 genes encode an ABC-type tungstate transport system (TupA, TupB, and TupC), similar to the modABC gene cluster. (D) fdh3A and fdh3B genes encode a third putative periplasmic FDH. (E) The Dde_3228 gene encodes a protein homologous to MoeA. The Dde_3227 and Dde_3229 genes encode a DNA-binding protein and a phospholipid-binding protein, respectively.
DISCUSSION
Molybdenum and tungsten occurrence in the active site of FDHs is a very interesting issue in this family of enzymes. Despite the fact that these metal atoms share similar chemical properties, it is still unknown why highly homologous enzymes that catalyze the same reaction and that are isolated from closely related organisms incorporate either Mo or W in the active site (1, 5, 9, 36, 42).
Analysis of the genome of D. alaskensis G20 revealed that this organism encodes at least three FDHs, all of them composed of one α and one β subunit (Fig. 3). In this work, we showed that D. alaskensis NCIMB 13491 grown in modified medium C supplemented with W produces one of these FDHs, the W-FDH encoded by the W-fdh genes. Cells cultured in Mo-supplemented medium produce, in addition to the W-FDH, the Mo/W-FDH encoded by the Mo/W-fdh genes. Remarkably, Mo/W-FDH from D. alaskensis NCIMB 13491 was the first FDH example able to incorporate either Mo or W (6).
The slight upregulation of the Mo/W-fdh genes on Mo supplementation (Table 3) suggests that the Mo/W-FDH enzyme, despite W incorporation, should preferentially work with Mo. This hypothesis is supported by the strong downregulation of the Mo/W-fdh genes in cells grown under W supplementation conditions (Table 3). In addition, the specific activity of the Mo/W-FDH (4.65 ± 0.07 U/mg) is at least one order of magnitude smaller than those determined for Mo-FDHs from other Desulfovibrio species (9, 36, 41), which indicates a low efficiency for catalysis.
In contrast to the Mo/W-FDH, the enzyme encoded by the W-fdh genes incorporates exclusively W. This is supported by the fact that only tungsten was detected in the as-purified enzyme independently of the metal added to the culture medium and also by the downregulation of the W-fdh genes observed in the presence of an excess of Mo (Table 3). The specific activity determined for the W-FDH (94 ± 6 U/mg) is comparable with that determined for the homologous D. gigas W-FDH (77 U/mg).
The Mo and W transport systems in D. alaskensis G20 are encoded by the mod and tup genes (Fig. 3B and C, respectively). Analysis of the gene expression levels showed that both mod and tup genes are downregulated in the presence of either molybdate or tungstate in the culture medium. However, tup genes appeared to be more expressed than mod genes in modified medium C without metal addition (data not shown). This differential gene expression is more important when the culture medium is supplemented with W. These data are in agreement with the biochemical results, which showed that W-FDH was produced in all cases analyzed in this paper. The downregulation of mod and tup genes on metal supplementation is also in line with the regulation of the modABC operon reported for Escherichia coli (17). In this organism, the expression of such operon is repressed under micromolar concentrations of molybate and induced only under conditions of molybdate starvation.
Formate dehydrogenases from D. alaskensis NCIMB 13491 constitute an interesting system to study the genes involved in the biosynthesis and incorporation of the Mo and W cofactors in enzymes. In the present work, it was demonstrated that the production of FDH isoforms is influenced by the Mo or W concentration in the culture medium, but the analysis of the gene expression levels of moeA1 and moeA2 did not give clear clues on the selectivity of MoeA proteins for a given metal ion. This differential regulation of genes encoding respiration processes and genes involved in Mo/W transport and cofactor biosynthesis is in line with data reported for E. coli (34). Additional work, including gene knockout experiments, is necessary to obtain deeper insight into the processes regulating the Mo/W cofactor insertion in enzymes.
ACKNOWLEDGMENTS
This work was supported by Fundação para a Ciência e Tecnologia (PDCT/QUI/57701/2004). C.S.M. thanks FCT for grant SFRH/BD/32478/2006.
We thank Marta R. Santos (Chemistry Department, FCT, UNL) for the N-terminal sequencing of FDH subunits, Carla Rodrigues (Chemistry Department, FCT, UNL) for inductively coupled plasma (ICP) analysis, and Yann Denis (IMM Transcriptomic Facilities) and Marie-Claire Durand (IMM-CNRS) for helpful discussion and technical support of the qRT-PCR experiments, respectively.
C.D.B. is a member of the CONICET (Argentina).
Footnotes
Published ahead of print on 8 April 2011.
REFERENCES
- 1. Almendra M. J., et al. 1999. Purification and characterization of a tungsten-containing formate dehydrogenase from Desulfovibrio gigas. Biochemistry 38:16366–16372 [DOI] [PubMed] [Google Scholar]
- 2. Berks B. C., et al. 2000. A novel protein transport system involved in the biogenesis of bacterial electron transfer chains. Biochim. Biophys. Acta 1459:325–330 [DOI] [PubMed] [Google Scholar]
- 3. Bevers L. E., Hagedoorn P. L., Hagen W. R. 2009. The bioinorganic chemistry of tungsten. Coord. Chem. Rev. 253:269–290 [Google Scholar]
- 4. Bevers L. E., et al. 2008. Function of MoaB proteins in the biosynthesis of the molybdenum and tungsten cofactors. Biochemistry 47:949–956 [DOI] [PubMed] [Google Scholar]
- 5. Boyington J. C., Gladyshev V. N., Khangulov S. V., Stadtman T. C., Sun P. D. 1997. Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster. Science 275:1305–1308 [DOI] [PubMed] [Google Scholar]
- 6. Brondino C. D., et al. 2004. Incorporation of either molybdenum or tungsten into formate dehydrogenase from Desulfovibrio alaskensis NCIMB 13491; EPR assignment of the proximal iron-sulfur cluster to the pterin cofactor in formate dehydrogenases from sulfate-reducing bacteria. J. Biol. Inorg. Chem. 9:145–151 [DOI] [PubMed] [Google Scholar]
- 7. Brondino C. D., Rivas M. G., Romão M. J., Moura J. J. G., Moura I. 2006. Structural and electron paramagnetic resonance (EPR) studies of mononuclear molybdenum enzymes from sulfate-reducing bacteria. Acc. Chem. Res. 39:788–796 [DOI] [PubMed] [Google Scholar]
- 8. Chhabra S. R., et al. 2006. Global analysis of heat shock response in Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 188:1817–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Costa C., Teixeira M., LeGall J., Moura J. J. G., Moura I. 1997. Formate dehydrogenase from Desulfovibrio desulfuricans ATCC 27774: isolation and spectroscopic characterization of the active sites (heme, iron-sulfur centers and molybdenum). J. Biol. Inorg. Chem. 2:198–208 [Google Scholar]
- 10. de Bok F. A. M., et al. 2003. Two W-containing formate dehydrogenases (CO2-reductases) involved in syntrophic propionate oxidation by Syntrophobacter fumaroxidans. Eur. J. Biochem. 270:2476–2485 [DOI] [PubMed] [Google Scholar]
- 11. Eaves D. J., Palmer T., Boxer D. H. 1997. The product of the molybdenum cofactor gene mobB of Escherichia coli is a GTP-binding protein. Eur. J. Biochem. 246:690–697 [DOI] [PubMed] [Google Scholar]
- 12. ElAntak L., Dolla A., Durand M. C., Bianco P., Guerlesquin F. 2005. Role of the tetrahemic subunit in Desulfovibrio vulgaris Hildenborough formate dehydrogenase. Biochemistry 44:14828–14834 [DOI] [PubMed] [Google Scholar]
- 13. Frunzke K., Heiss B., Meyer O., Zumft W. G. 1993. Molybdopterin guanine dinucleotide is the organic moiety of the molybdenum cofactor in respiratory nitrate reductase from Pseudomonas stutzeri. FEMS Microbiol. Lett. 113:241–246 [Google Scholar]
- 14. George G. N., Costa C., Moura J. J. G., Moura I. 1999. Observation of ligand-based redox chemistry at the active site of a molybdenum enzyme. J. Am. Chem. Soc. 121:2625–2626 [Google Scholar]
- 15. Gremer L., Meyer O. 1996. Characterization of xanthine dehydrogenase from the anaerobic bacterium Veillonella atypica and identification of a molybdopterin-cytosine-dinucleotide-containing molybdenum cofactor. Eur. J. Biochem. 238:862–866 [DOI] [PubMed] [Google Scholar]
- 16. Grunden A. M., Ray R. M., Rosentel J. K., Healy F. G., Shanmugam K. T. 1996. Repression of the Escherichia coli modABCD (molybdate transport) operon by ModE. J. Bacteriol. 178:735–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grunden A. M., Shanmugam K. T. 1997. Molybdate transport and regulation in bacteria. Arch. Microbiol. 168:345–354 [DOI] [PubMed] [Google Scholar]
- 18. Hamilton W. A. 1998. Bioenergetics of sulphate-reducing bacteria in relation to their environmental impact. Biodegradation 9:201–212 [DOI] [PubMed] [Google Scholar]
- 19. Heidelberg J., et al. 2004. The genome sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Nat. Biotechnol. 22:554–559 [DOI] [PubMed] [Google Scholar]
- 20. Hille R. 2002. Molybdenum and tungsten in biology. Trends Biochem. Sci. 27:360–367 [DOI] [PubMed] [Google Scholar]
- 21. Lee A., Newman D. 2003. Microbial iron respiration: impacts on corrosion processes. Appl. Microbiol. Biotechnol. 62:134–139 [DOI] [PubMed] [Google Scholar]
- 22. Lee W., Lewandowski Z., Nielsen P. H., Hamilton W. A. 1995. Role of sulfate-reducing bacteria in corrosion of mild-steel—a review. Biofouling 8:165–194 [Google Scholar]
- 23. Llamas A., Otte T., Multhaup G., Mendel R. R., Schwarz G. 2006. The mechanism of nucleotide-assisted molybdenum insertion into molybdopterin—a novel route toward metal cofactor assembly. J. Biol. Chem. 281:18343–18350 [DOI] [PubMed] [Google Scholar]
- 24. Maupin-Furlow J. A., et al. 1995. Genetic analysis of the modABCD (molybdate transport) operon of Escherichia coli. J. Bacteriol. 177:4851–4856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mendel R. R. 2007. Biology of the molybdenum cofactor. J. Exp. Bot. 58:2289–2296 [DOI] [PubMed] [Google Scholar]
- 26. Moura J. J. G., Brondino C. D., Trincao J., Romao M. J. 2004. Mo and W bis-MGD enzymes: nitrate reductases and formate dehydrogenases. J. Biol. Inorg. Chem. 9:791–799 [DOI] [PubMed] [Google Scholar]
- 27. Nichols J. D., Rajagopalan K. V. 2002. Escherichia coli MoeA and MogA. J. Biol. Chem. 277:24995–25000 [DOI] [PubMed] [Google Scholar]
- 28. Nichols J. D., Rajagopalan K. V. 2005. In vitro molybdenum ligation to molybdopterin using purified components. J. Biol. Chem. 280:7817–7822 [DOI] [PubMed] [Google Scholar]
- 29. Pereira I. A. C., Haveman S. A., Voordouw G. 2007. Biochemical, genetic and genomic characterization of anaerobic electron transport pathways in sulphate-reducing Delta proteobacteria, p. 215–240In Barton L. L., Hamilton W. A. (ed.), Sulphate-reducing bacteria: environment and engineered systems, 1st ed. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 30. Pfaffl M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Postgate J. 1984. The sulphate-reducing bacteria, 2nd ed., p. 24–40 Cambridge University Press, London, United Kingdom [Google Scholar]
- 32. Raaijmakers H., et al. 2002. Gene sequence and the 1.8 angstrom crystal structure of the tungsten-containing formate dehydrogenase from Desulfolvibrio gigas. Structure 10:1261–1272 [DOI] [PubMed] [Google Scholar]
- 33. Raaijmakers H., et al. 2001. Tungsten-containing formate dehydrogenase from Desulfovibrio gigas: metal identification and preliminary structural data by multi-wavelength crystallography. J. Biol. Inorg. Chem. 6:398–404 [DOI] [PubMed] [Google Scholar]
- 34. Rech. S., Deppenmeier U., Gunsalus R. P. 1995. Regulation of the molybdate transport operon, modABCD, of Escherichia coli in response to molybdate availability. J. Bacteriol. 177:1023–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rivas M. G., et al. 2009. Molybdenum induces the expression of a protein containing a new heterometallic Mo-Fe cluster in Desulfovibrio alaskensis. Biochemistry 48:873–882 [DOI] [PubMed] [Google Scholar]
- 36. Rivas M. G., Gonzalez P. J., Brondino C. D., Moura J. J. G., Moura I. 2007. EPR characterization of the molybdenum(V) forms of formate dehydrogenase from Desulfovibrio desulfuricans ATCC 27774 upon formate reduction. J. Inorg. Biochem. 101:1617–1622 [DOI] [PubMed] [Google Scholar]
- 37. Romão M. J. 2009. Molybdenum and tungsten enzymes: a crystallographic and mechanistic overview. Dalton Trans. 2009:4053–4068 [DOI] [PubMed] [Google Scholar]
- 38. Sargent F., et al. 1998. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 17:3640–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schwarz G., Mendel R. R., Ribbe M. W. 2009. Molybdenum cofactors, enzymes and pathways. Nature 460:839–847 [DOI] [PubMed] [Google Scholar]
- 40. Sebban C., Blanchard L., Bruschi M., Guerlesquin F. 1995. Purification and characterization of the formate dehydrogenase from Desulfovibrio vulgaris Hildenborough. FEMS Microbiol. Lett. 133:143–149 [DOI] [PubMed] [Google Scholar]
- 41. Sebban-Kreuzer C., Dolla A., Guerlesquin F. 1998. The formate dehydrogenase-cytochrome c(553) complex from Desulfovibrio vulgaris Hildenborough. Eur. J. Biochem. 253:645–652 [DOI] [PubMed] [Google Scholar]
- 42. Sevcenco A.-M., et al. 2010. Molybdenum incorporation in tungsten aldehyde oxidoreductase enzymes from Pyrococcus furiosus. J. Bacteriol. 192:4143–4152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Thauer R., Stackebranndt E., Hamilton W. 2007. Energy metabolism and phylogenetic diversity of sulphate-reducing bacteria, p. 533.In Barton L., Hamilton W. (ed.), Sulphate-reducing bacteria. Environmental and engineered systems. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]