Abstract
Listeria monocytogenes is a Gram-positive facultative intracellular bacterium that causes life-threatening diseases in humans. It grows and survives in environments of low oxygen tension and under conditions of strict anaerobiosis. Oxygen-limiting conditions may be an important factor in determining its pathogenicity. L. monocytogenes serovar 1/2a strain EGD-e has been employed intensively to elucidate the mechanisms of intracellular multiplication and virulence. Listeria possesses genes encoding class I aerobic and class III anaerobic ribonucleotide reductases (RNRs). The class III RNR consists of a catalytic subunit NrdD and an activase NrdG. Surprisingly, L. monocytogenes EGD-e, but not other L. monocytogenes strains or other listerial species, is unable to grow under strict anaerobic conditions. Inspection of listerial NrdD amino acid sequences revealed a six-amino acid deletion in the C-terminal portion of the EGD-e protein, next to the essential glycyl radical domain. Nevertheless, L. monocytogenes EGD-e can grow under microaerophilic conditions due to the recruitment of residual class Ia RNR activity. A three-dimensional (3D) model based on the structure of bacteriophage T4 NrdD identified the location of the deletion, which appears in a highly conserved part of the NrdD RNR structure, in the α/β barrel domain near the glycyl radical domain. The deleted KITPFE region is essential either for interactions with the NrdG activase or, indirectly, for the stability of the glycyl radical loop. Given that L. monocytogenes EGD-e lacks a functional anaerobic RNR, the present findings are relevant to the interpretation of studies of pathogenesis with this strain specifically, in particular under conditions of low oxygen tension.
INTRODUCTION
Listeria monocytogenes is a Gram-positive, facultative intracellular bacterium that can live as a saprophyte, primarily in decaying vegetation in soil and as a pathogen in the tissues of mammals and birds, in which it can cause life-threatening disease (16, 29, 60). L. monocytogenes infects a variety of phagocytic and nonphagocytic mammalian cells. Following internalization, the bacteria escapes from the vacuole/phagosome by membrane lysis into the cytosol, where it proliferates. Actin polymerization propels the bacterium to the host cell membrane, where it spreads to neighboring cells (10). The genes essential for these processes have been investigated thoroughly (60). L. monocytogenes has the capacity to cross three tight barriers, the intestinal, blood-brain, and fetoplacental barriers. These features are considered to be central to the pathophysiology of listeriosis.
During the last few years considerable information has been accumulated about L. monocytogenes metabolism and its mechanism of pathogenesis. However, relatively few studies have been performed to characterize growth, physiology, and virulence under defined conditions of low oxygen tension or under anaerobic conditions (reviewed in reference 36). In the anaerobic environment of the gastrointestinal tract, L. monocytogenes induces expression of numerous genes, among which are those responsible for production of vitamin B12 and ethanolamine utilization, suggesting that ethanolamine can, when oxygen is limited, serve as a carbon and nitrogen source for intracellular growth (6, 29). Low oxygen tension also facilitates increased expression and secretion of factors that promote in vitro adhesion to intestinal epithelial cells (8). Prior adaptation to low oxygen tension significantly enhances the in vivo infective potential of the L. monocytogenes clinical serovar 4b isolate Scott A (5). Moreover, an anaerobically grown strain, L. monocytogenes strain F4244 (wild type [WT], serovar 4b) exhibited greater translocation to the liver and spleen relative to aerobically grown organisms (8). These and other observations, for example the recent study of Marteyn et al. (38) of the modulation of Shigella virulence in response to available oxygen in vivo which explores the influence of virulence determinants in response to localized microenvironments in the host, in particular to various oxygen concentrations in the gastrointestinal tract, emphasize the importance of oxygen deprivation on the growth, survival, and pathogenesis of L. monocytogenes. In this report we focus on the nature of the listerial ribonucleotide reductase (RNR) system necessary for anaerobic growth. RNRs are essential enzymes that provide the sole de novo pathway for synthesis of the deoxyribonucleotides (deoxynucleoside triphosphates [dNTPs]) required for DNA synthesis (28). RNRs catalyze the controlled reduction of all four ribonucleotides (nucleoside triphosphates [NTPs]) to maintain a balanced pool of dNTPs during the cell cycle. Three major classes of RNRs have been characterized (41). Class I RNRs are oxygen-dependent enzymes that are divided into two main subclasses, Ia and Ib (28, 41). Class Ia RNRs are found in eukaryotes, in a wide range of bacteria, and in some Archaea; they are encoded by operons containing nrdA and nrdB genes that specify the NrdA subunit containing the catalytic and allosteric regulatory sites and the NrdB radical-generating subunit, respectively. Class Ib RNRs are encoded by operons consisting of nrdE and nrdF genes that specify the corresponding subunits NrdE and NrdF, respectively. Both class I RNRs require oxygen for the generation of the essential diferric tyrosyl radical cofactor, which initiates radical propagation to the active-site cysteine of NrdA or NrdE. In Escherichia coli and many enterobacteria, the class Ib RNR operon contains two additional genes: nrdH, coding for NrdH, an ∼9-kDa thio-disulfide redox protein that functions as a specific electron donor (55), and nrdI, coding for an ∼15-kDa flavodoxin protein NrdI that functions as an electron donor in the maintenance of the NrdF diferric-tyrosyl radical (13, 47). The NrdAB and NrdEF RNRs have limited sequence identity but share many catalytic properties (28). With few exceptions, all eukaryotes possess just the class Ia RNR. Class II RNRs are oxygen-independent enzymes encoded by the nrdJ gene and use coenzyme B12 (adenosylcobalamin) to generate a transient thiyl radical. The cofactor fulfills the function of the radical-generating subunit in class I enzymes. Class III RNRs are oxygen-sensitive enzymes encoded by nrdD, which often occurs in an operon containing nrdG, determining a specific iron-sulfur containing activase NrdG. The activase mediates cleavage of S-adenosylmethionine to generate a stable oxygen-sensitive glycyl radical close to the NrdD active site (28, 35, 41). In E. coli the glycyl radical is located on residue G681 of the 712-amino acid polypeptide (18, 56). The importance of the anaerobic RNR system for bacterial pathogenesis has already been suggested for Staphylococcus aureus by Masalha et al. (39), and more recently the Streptococcus sanguinis anaerobic RNR was suggested to be a virulence determinant in infective endocarditis (43) and in Proteus mirabilis to contribute to urinary tract infection and colonization of the bladder (7).
L. monocytogenes has not been well characterized in terms of its anaerobic growth. We supposed that, like most facultative aerobes, it contains a class III RNR (18). Bioinformatic analysis of L. monocytogenes and several other listerial species confirmed that all contain an open reading frame (ORF) predicted to encode the NrdD catalytic subunit of class III RNRs. We have characterized the L. monocytogenes anaerobic RNR system in the serovar 1/2a EGD-e strain, which is a commonly used model laboratory reference organism and which is intensively studied to elucidate mechanisms of intracellular multiplication and pathogenicity (19, 51). The L. monocytogenes EGD-e genome has been fully sequenced (21). Recently, global expression profiling of L. monocytogenes EGD-e has led to important new insights into the regulatory processes that operate in response to different environmental conditions, including gastrointestinal infection, transition from saprophytic growth to pathogenic infection in animals, adaptation from the extracellular to the intracellular environment, and response to stress agents (9, 59). Here we show that L. monocytogenes EGD-e, but not other L. monocytogenes strains or listerial species, contains a nonfunctional class III RNR due to a deletion in NrdD near the glycyl radical domain. The EGD-e strain cannot grow under strict anaerobic conditions; nevertheless, EGD-e can grow under conditions of low oxygen tension by recruitment of the class Ia RNR aerobic system.
MATERIALS AND METHODS
Bacterial strains, plasmids, culture conditions, and chemicals.
L. monocytogenes serovar 1/2a EGD-e is the sequenced strain (21), donated to us by T. Chakraborty (University of Giessen, Germany). It has been provided to T.C. by H. Hahn (Berlin). In the Berlin laboratory it was routinely propagated through mice (30). Other listerial species and strains used in this study are shown in Table 1. All strains were routinely cultured at 37°C in brain heart infusion (BHI) medium (Becton, Dickinson and Co.). E. coli strain XL1-Blue was used for plasmid construction, and E. coli strain BL21(λDE3) for protein overexpression. E. coli strains were cultured in Luria-Bertani (LB; Difco) broth and at 25°C or 37°C for protein overexpression. E. coli K-12 MG1655 and its isogenic menA mutant strain were grown in minimal medium on plates for evaluating anaerobic growth conditions in jars. pET14b and pET28a(+) expression vectors (Table 1) were used for protein overexpression. pKSV7 vector (54) was used for mutant construction, pGEM-T Easy vector (Promega) was used for cloning PCR products. Chemicals were purchased from Sigma Chemical Co. (St. Louis, MO) unless otherwise noted.
Table 1.
Bacterial strains, vectors, and plasmids
| Strain, vector, or plasmid construct | Relevant characteristic(s)a | Source and/or reference |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqlacZΔM15 Tn10(Tcr)] | Lab stock |
| BL21(λDF3) | F−ompT hsdSB(rB− mB−) gal dcm (λDE3) | Lab stock |
| MG1655 | F− λ−ilvG rfb-50 rph-1; referred to as the wild type | Lab stock |
| MG1655 menA | A null menA mutant; deficient for menaquinone (vitamin K2) biosynthesis; this mutation renders a strain unable to grow anaerobically | A gift of E. Ron (57) |
| Listeria spp. | ||
| L. monocytogenes EGD-e or ATCC BAA-679 | Serovar 1/2a, a wild-type strain; nrdD gene contains a 18-bp deletion in frame | Lab stock (21) |
| L. monocytogenes F2365 | Serovar 4b, a wild-type strain, contains an intact nrdD gene | Lab stock (40) |
| L. monocytogenes EGD-e ΔnrdDG | EGD-e containing deletion of 3′ end of nrdD and 5′ end of nrdG | This study |
| L. monocytogenes EGD-e nrdD+KITPFE | EGD-e containing a “corrected” nrdD gene | This study |
| L. monocytogenes F2365 nrdDΔKITPFE | F2365 containing an EGD-e-like 18-bp deletion in frame within nrdD gene | This study |
| L. grayi CLIP 12515 | Serovar 7, a wild-type strain | Würzburg Biozentrum collection |
| L. innocua CLIP 11262 | Serovar 6a, a wild-type strain | Würzburg Biozentrum collection |
| L. ivanovii PAM55 | Serovar 5, a wild-type strain | Würzburg Biozentrum collection |
| L. seeligeri SLCC 3954 | Serovar 1/2b, a wild-type strain | Würzburg Biozentrum collection |
| L. welshimeri SLCC 5334 | Serovar 6b, a wild-type strain | Würzburg Biozentrum collection |
| Vectors and constructs | ||
| pGEM-T Easy | Cloning PCR products, Apr | Promega |
| pET14b | Protein overexpression (N-terminal 6His tag), Apr | Novagen |
| pET28a(+) | Protein overexpression (C-terminal 6His tag), Kmr | Novagen |
| pKSV7 | Temp-sensitive oriCts-based vector for mutagenesis in Listeria; Cmr | Lab stock (54) |
| pKSV7::ΔnrdDG | pKSV7 containing an EGD-e nrdDG locus with a large internal deletion in frame; Cmr | This study |
| pKSV7::nrdD+KITPFE | pKSV7 containing an EGD-e modified nrdD (an insertion of 18 nucleotides encoding KITPFE) | This study |
| pKSV7::nrdDΔKITPFE | pKSV7 containing an-F2365 modified nrdD (an in-frame deletion of 18 nucleotides encoding KITPFE) | This study |
Abbreviations: Apr, Cmr, Kmr, and Tcr, resistance to ampicillin, chloramphenicol, kanamycin, and tetracycline, respectively.
Preparation of L. monocytogenes competent cells.
Overnight cultures of L. monocytogenes strains grown in BHI medium were diluted to a 0.1 absorbance at 600 nm in the same medium supplemented with 0.1 to 0.2% of glycine and incubated at 37°C, 250 rpm, to an absorbance of 0.5 at 600 nm. Penicillin G was added to a final concentration of 5 μg/ml, and the culture incubated to an absorbance of 0.7 at 600 nm. Cells were harvested by centrifugation at 4,000 × g for 10 min at 4°C, washed twice with SMHEM 3.5 buffer (925 mM sucrose, 3.5 mM MgCl2, 7 mM HEPES [pH 7.2]), and resuspended in 2.5 ml of the same buffer for aliquoting (100 μl/microtube). Bacteria were stored at −70°C.
Construction of mutants.
The temperature-sensitive pKSV7 shuttle vector (54) was used for creating mutations in L. monocytogenes strains EGD-e and F2365. Briefly, the upstream and downstream flanking regions of the gene to be mutated were PCR amplified from genomic DNA, purified by gel electrophoresis, and cloned together into pKSV7. Vectors used in the constructions are listed in Table 1; see also Table S4 in the supplemental material for the primers used. The DNA fragments of 1 to 1.2 kb size were designed such that the 3′ end in the upstream flanking region is able to anneal to the 5′ end of the downstream flanking region. The DNA fragments contained suitable restriction endonuclease sites for cloning. The two DNA fragments were mixed and used as the template for a second PCR to create a larger fragment containing the expected mutation. PCR products were gel purified, digested with the appropriate restriction enzymes, ligated into pGEM-T Easy, and electroporated into E. coli XL1-Blue cells as previously described (48). Transformants were isolated on LB plates supplemented with 100 μg/ml ampicillin, 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and 80 μg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactoside). Positive transformants were detected using blue-white screening and colony PCR. DNA inserts were sequenced to verify their integrity. pGEM constructs were digested with the appropriate restriction enzymes, and L. monocytogenes DNA inserts purified and ligated into the digested pKSV7 vector. The resulting construct was electroporated into E. coli XL1-Blue and grown overnight at 30°C on LB plates supplemented with 100 μg/ml of ampicillin. Positive transformants were detected using colony PCR, and constructs were isolated. Constructs were introduced into L. monocytogenes competent cells by electroporation (2 to 5 μg plasmid DNA, 200 Ω, 2.5 μF, 2.5 Kv, using a Bio-Rad Gene Pulser II and 0.1-cm cuvettes). Transformants were selected for growth on BHI plates containing chloramphenicol (10 μg/ml) at 30°C for 2 days. The following procedure was used to create point, deletion, and substitution mutations in L. monocytogenes strains EGD-e and F2365. Mutant DNA fragments were introduced into the L monocytogenes chromosome by homologous recombination. Transformants were cultured on BHI medium supplemented with chloramphenicol at 30°C for 2 to 3 days, diluted, spread on plates of the same medium, and incubated for 2 to 3 days at 42°C, the nonpermissive temperature for pKSV7 replication. Several transformants were isolated on BHI plates containing chloramphenicol. Integration was confirmed in one transformant by PCR using primers flanking the pKSV7 multiple cloning site and primers flanking the insert. To select for segregation of the wild-type allele, integrants were cultured in liquid BHI medium without antibiotics for 2 to 3 days at 30°C, spread on BHI plates without antibiotics, and grown at 37°C overnight. Colonies were picked and plated on BHI plates with/without chloramphenicol (10 μg/ml), grown at 37°C overnight, and screened for loss of the antibiotic marker. Chloramphenicol-sensitive clones were analyzed by PCR and DNA sequencing to confirm the presence of the mutation and loss of the pKSV7 vector.
Anaerobic growth.
Anaerobic growth of bacteria on plates was performed using anaerobic jars containing dispensable anaerobic sachets and an anaerobic indicator (Oxoid, Hampshire, United Kingdom) as previously described (39). Two sachets were used per jar. Bacteria were serially diluted, plated on BHI plates with or without hydroxyurea (HU), and incubated for 3 days at 37°C. E. coli MG1655 and menA (57) strains were used to verify anaerobiosis. Fifty microliters of a 10−4 dilution of an overnight culture grown on minimal medium supplemented with 1 mM MgSO4 (Merck), 0.2% Casamino Acids (Sigma,), 0.5 mM tryptophan, 0.6% glycerol, 40 mM trimethylamine N-oxide dihydrate, and 5 μg/ml thiamine were incubated on plates in an anaerobic jar. Under the standard anaerobic conditions, E. coli MG1655 grows well on this medium, while the menA mutant is unable to grow. Anaerobic growth of L. monocytogenes in liquid media was performed in Wheaton serum bottles (100-ml capacity) containing 60 ml of BHI medium or in the case of prfA induction (62) LB supplemented with 50 mM MOPS (morpholinepropanesulfonic acid), 25 mM glucose-1-phosphate, and 0.2% activated charcoal, supplemented with 5.7 mM cysteine (Sigma) to scavenge traces of oxygen and 0.0001% of resazurin as a redox indicator (24, 39). The bottle contents were boiled for 5 min to remove dissolved oxygen in the medium, immediately purged with nitrogen for 5 min (0.75 atm), and capped prior to being autoclaved. Overnight aerobic and anaerobic cultures were subcultured (0.5 ml) in anaerobic bottles with or without 50 mM HU.
Bioinformatics.
Primary DNA sequence analyses and ORF searches were performed with the National Center for Biotechnology Information server ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) and the Clone Manager 7 program (Scientific & Educational Software, Durham, NC). The ATP-cone module of lmo2155 (NrdA) was identified using several domain motifs, including InterPro IPR005144 (http://www.ebi.ac.uk/interpro/IEntry?ac=IPR005144), PROSITE PS51161 (http://www.expasy.org/cgi-bin/nicedoc.pl?PS51161), and Pfam PF03477 (http://pfam.sanger.ac.uk/family?acc=PF03477). Deduced amino acid sequences of L. monocytogenes EGD-e RNR proteins were used as BLAST (1) queries to mine public databases, including those at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) and the Broad Institute (http://www.broadinstitute.org/). A putative L. monocytogenes NrdI (lmo2153) protein was identified using the Simple Modular Architecture Tool (SMART) (32) (http://smart.embl-heidelberg.de/), the Pfam protein family database (17) (http://pfam.sanger.ac.uk), the integrated resource of Protein Domains (InterPro) (26) (http://www.ebi.ac.uk/interpro/), the database of protein families and domains PROSITE (52) (http://www.expasy.ch/prosite/), and the SUPERFAMILY database of structural and functional annotation for all proteins and genomes (22) (http://supfam.cs.bris.ac.uk/SUPERFAMILY/). The parameters for molecular mass, theoretical pI, and amino acid composition were computed using the ProtParam Tool on the ExPASy server (http://www.expasy.org/tools/protparam.html). Pairwise and multiple sequence alignments were performed with the ClustalW program (25) using the EBI ClustalW2 server (http://www.ebi.ac.uk/Tools/clustalw2/index.html). Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 5 (58) (http://www.megasoftware.net/).
Modeling.
Three-dimensional homology models for NrdD from L. monocytogenes strains 1/2a EDG-e and HCC23 were generated using the Modeler program (15) through the ModWeb server (https://modbase.compbio.ucsf.edu/scgi/modweb.cgi). The best template identified by the server was the crystal structure of NrdD from bacteriophage T4 (33), Protein Data Bank [PDB] ID 1H7B. Residues 126 to 685 of the listerial proteins could be modeled based on residues 26 to 585 of T4 NrdD. The sequence identity was 50%, and the model was of good quality as judged by relevant criteria. When the structures were aligned using Pymol (http://www.pymol.org), the root mean square deviation in C-alpha positions between the EGD-e model and the template was 0.16 Å for 425 positions, while it was 0.14 Å for 423 positions in the HC233 model.
RESULTS AND DISCUSSION
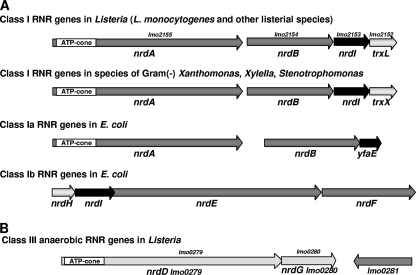
Listeria contains genes encoding class I and class III RNRs.
A search of the complete L. monocytogenes serovar 1/2a strain EGD-e genome (accession number NC_003210) (21) for genes encoding putative RNR proteins was carried out by the use of standard BLAST procedures (1) via the NCBI server (http://www.ncbi.nlm.nih.gov/BLAST). Two gene clusters that are annotated as containing ORFs predicted to belong to class Ia and class III of the RNR family of proteins were found (Fig. 1). One gene cluster consisting of lmo2155 and lmo2154 is predicted to encode the class Ia RNR NrdA- and NrdB-like proteins (Fig. 1A). The listerial class I RNR large subunits contain an ATP-cone domain in their N-terminal regions similar to that present in the NrdA subunit of class Ia RNRs of enterobacteria and shown to regulate overall enzyme activity by interaction with ATP/dATP (2). Immediately downstream of lmo2154 are found two genes, lmo2153 and lmo2152, which are predicted to encode putative flavodoxin (NrdI-like) and thioredoxin (TrxL)-like proteins, respectively, suggesting that all four genes are likely to form an operon. Reverse transcriptase (RT) PCR analysis of the nrdABI-trxL gene cluster revealed that nrdA, nrdB, nrdI-like, and trxL genes are cotranscribed and form an operon (see Fig. S1 in the supplemental material). The lmo2151 gene immediately downstream of trxL is predicted to encode a MazG-like protein (63) which might be cotranscribed with nrdABI-trxL as well (see Fig. S1). The deduced amino acid sequence of Lmo2153 resembles that of the E. coli flavodoxin NrdI, which is involved in the maintenance of the diferric-tyrosyl radical cofactor (4, 12, 13). Thioredoxin-like Lmo2152 was denoted as TrxL to distinguish it from the TrxA ortholog encoded by lmo1233. Class I RNR enzyme activity was characterized employing NrdA, NrdB, NrdI, and TrxL Ni affinity purified His6-tagged recombinant proteins (see Fig. S2A to F in the supplemental material). RNR activity was assayed by monitoring the conversion of [3H]CDP to [3H]dCDP (14). The results establish that the L. monocytogenes aerobic RNR conforms to the typical pattern observed for the class Ia RNR subdivision (28). TrxL, the thioredoxin-like protein encoded by the class Ia RNR operon, can serve as an electron donor to the NrdAB reaction. In the presence of 1 to 2 mM dithiothreitol (DTT), TrxL significantly stimulated RNR activity (see Fig. S3 in the supplemental material), suggesting that in vivo it can function with thioredoxin reductase and NADPH as an electron donor for the aerobic class I RNR, in a manner similar to that of E. coli and S. aureus NrdH (39, 45).
Fig. 1.
Organization of the class I and III RNR genes of L. monocytogenes. (A) Organization of the L. monocytogenes EGD-e class I nrdABI-trxL RNR genes and comparison with some other bacterial class Ia and Ib RNR gene organizations. (B) Organization of the L. monocytogenes EGD-e class III nrdDG RNR genes.
A similar gene organization is found in the genomes of other L. monocytogenes strains and Listeria species, including nonpathogenic L. innocua (21) and L. welshimeri (23). Systematic analysis of other bacterial genomes revealed that several species of the family Xanthomonadaceae (gammaproteobacteria), plant-pathogenic Xanthomonas and Xylella (44, 53), and Stenotrophomonas maltophilia, an opportunistic human pathogen causing nosocomial infections (50), contain a class I RNR gene cluster similar to that in Listeria species (Fig. 1A). Phylogenetic analysis demonstrates that bacteria possessing the nrdABI-trxL operon comprise a well-defined clade that is well separated from other class Ia and Ib RNRs (see Fig. S4 in the supplemental material). Besides an unusual nrdABI-trxL locus, genomes of all listerial species contain a second gene cluster that encodes the “classic” anaerobic class III RNR proteins NrdD (reductase) and NrdG (activase) (Fig. 1B).
The L. monocytogenes EGD-e class III RNR system does not support anaerobic growth due to a deletion in NrdD.
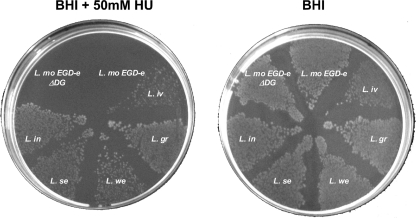
L. monocytogenes is a facultative anaerobe able to grow in aerobic and anaerobic conditions. We therefore expected that L. monocytogenes EGD-e would grow under anaerobic conditions and that growth would be unaffected by hydroxyurea (HU), which is a specific inhibitor of the class I RNR scavenging the free oxygen-dependent tyrosyl radical (46). Figure 2 shows that L. monocytogenes EGD-e (WT) does indeed grow on BHI in solid medium under anaerobic conditions. An E. coli menA mutant strain that is unable to grow anaerobically served as a control for anaerobiosis (57). To verify that growth is dependent on a functional class III RNR, we created an in-frame deletion in the nrdDG genes (Material and Methods) and were surprised to find that the mutant strain grew almost as well as the WT (Fig. 2). We therefore presumed that growth under anaerobic conditions was due to the class Ia RNR. To test this hypothesis, we supplemented BHI plates with 50 mM HU and observed that growth of EGD-e and the ΔnrdD mutant was totally abolished, indicating that under the anaerobic conditions employed EGD-e growth was due to the activity of the class Ia RNR. In contrast, L. monocytogenes strain F2365 (serovar 4b) is able to grow under the same anaerobic conditions in the presence of HU (not shown). Moreover, the listerial species L. ivanovii, L. welshimeri, L. innocua, L. seeligeri, and L. grayi are all also able to grow anaerobically in the presence of HU (Fig, 2). These observations imply that the L. monocytogenes EGD-e class III RNR is nonfunctional.
Fig. 2.
Anaerobic growth of Listeria spp. in solid medium. Listeria species were grown on BHI plates with and without 50 mM HU. Strains tested were L. monocytogenes EGD-e WT (L. mo EGD-e), an isogenic mutant possessing a deletion in nrdDG (L. mo EGD-e ΔDG), L. innocua (L. in), L. seeligeri (L. se), L. welshimeri (L. we), L. grayi (L. gr), and L. ivanovii (L. iv). L. monocytogenes EGD-e WT and the nrdDG mutant strain failed to grow under anaerobic conditions in the presence of HU. Anaerobic conditions were verified using E. coli MG1655 (WT) and its menA mutant strain grown on the minimal M9 medium; in anaerobic conditions the menA mutant is unable to grow.
Comparison of the predicted L. monocytogenes EGD-e NrdD amino acid sequence with those of some 25 L. monocytogenes strains, including F2365 (see Table S2 in the supplemental material), revealed that EGD-e NrdD alone contains a deletion of six amino acids—KITPFE—in the C-terminal part of the polypeptide (Fig. 3). Furthermore, all other listerial species, including L. grayi, L. innocua, L. ivanovii, L. seeligeri, and L. welshimeri, contain ORFs coding for an intact NrdD protein. Based on the PROSITE profile of the glycine radical domain GLY_RADICAL_2 (PS51149) and on the reports of Luttringer et al. (37) on the Zn center in the E. coli anaerobic ribonucleotide reductase, the position of the deletion lies close to the glycyl radical-containing domain (see below), indicating that it is likely to be critical for the formation of the stable radical necessary for enzyme activity.
Fig. 3.
Multiple sequence alignment of NrdD proteins. (Top) The ATP-cone and glycyl radical-containing domains of L. monocytogenes EGD-e NrdD, which are according to PF03477 and PF01128, respectively. Amino acid numbering corresponds to that for L. monocytogenes EGD-e NrdD (accession number NP_463810). The six-amino acid deletion in EGD-e NrdD occurs between residues 588 and 589. Two pairs of zinc-binding cysteines are located between residues 642 and 681. (Bottom) Partial amino acid sequence of an alignment of NrdD proteins from Listeria and selected bacteria. The position in Listeria species of the KITPFE region that is deleted in EGD-e is marked in light gray. The start region of the glycyl radical domains in bacteriophage T4 and E. coli NrdD, according to Luttringer et al. (37), is shown in dark gray; that deduced for the L. monocytogenes NrdD is shown as well. The glycyl radical domain in L. monocytogenes starts at position R587. The KITPFE amino acid deletion is located at the beginning of the domain. Asterisks, colons, and dots indicate identical, highly similar, and weakly similar residues, respectively.
To test experimentally the hypothesis that the KITPFE deletion in L. monocytogenes EGD-e is responsible for creating a nonfunctional class III RNR, we introduced an 18-nucleotide DNA fragment into the EGD-e nrdD gene to generate the precise coding sequence present in L. monocytogenes F2365 and in other listerial nrdD genes. In a reciprocal experiment, we deleted the corresponding 18-nucleotide DNA fragment coding for KITPFE in strain F2365. The constructions were performed using a temperature-sensitive replication vector, pKSV7, as described in Materials and Methods. The pairs of isogenic strains EGD-e nrdDΔKITPFE (WT) and EGD-e nrdD+KITPFE and strains F2365 nrdD+KITPFE (WT) and F2365 nrdDΔKITPFE were tested for growth in BHI medium under aerobic and anaerobic conditions.
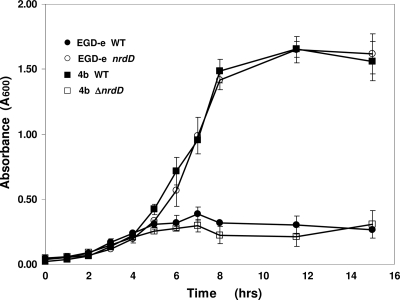
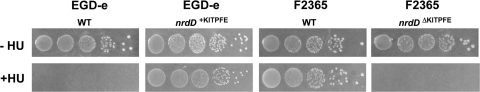
In BHI liquid medium there was no discernible difference in the growth of the 4 strains under aerobic conditions as judged by the rates and extents of growth (data not shown). Figure 4 shows growth in liquid BHI medium under anaerobic conditions. F2365 grew well and reached an absorbance (A600) of 1.6 after 8 h, whereas EGD-e grew poorly and reached an A600 of 0.3. The two complemented strains behaved in the opposite way; EGD-e nrdD+KITPFE grew equally well as F2365, while F2364 nrdDΔKITPFE grew poorly and reached an A600 of 0.3. The four strains were also tested for growth on BHI plates in anaerobic jars with and without HU (Fig. 5). In the absence of HU all four strains grew equally well on the plates (Fig. 5, top). When the plates were supplemented with 50 mM HU, both strains carrying the KITPFE deletion, EGD-e nrdDΔKITPFE (WT) and F2365 nrdDΔKITPFE, failed to grow, whereas both strains possessing the KITPFE sequence, EGD-e nrdD+KITPFE and F2365 nrdD+KITPFE, grew equally well and to the same extent as they did in the absence of HU (Fig. 5, bottom). These observations establish that the L. monocytogenes nrdD gene is essential for anaerobic growth. They recall similar studies with E. coli, Lactococcus lactis, and S. aureus in which the growth deficiency of an nrdD mutant was apparent only when strict anaerobic growth conditions were employed (20, 27, 39).
Fig. 4.
Anaerobic growth of L. monocytogenes EGD-e WT, EGD-e nrdD+KITPFE, F2365 WT, and F2365 nrdDΔKITPFE in liquid medium. Aerobic and anaerobic overnight cultures of all four strains were used to inoculate Wheaton serum anaerobic bottles containing BHI, resazurin, and cysteine and the liquid cultures grown at 37°C (see Materials and Methods). EGD-e WT and F2365 nrdDΔKITPFE possess a mutant NrdD protein; EGD-e nrdD+KITPFE and F2365 nrdDΔKITPFE contain a functional NrdD protein (38). The data shown are representative of two experiments; error bars show the average variation.
Fig. 5.
Anaerobic growth of L. monocytogenes EGD-e WT, EGD-e nrdD+KITPFE, F2365 WT, and F2365 nrdDΔKITPFE on solid BHI medium in the presence or absence of HU. Exponentially growing cultures were aliquoted on BHI plates with or without 50 mM HU. Plates were incubated for 48 h at 37°C in an anaerobic jar.
Our original observation of the growth of L. monocytogenes EGD-e (WT) on BHI plates in an anaerobic jar is most likely due to the residual activity of the class I RNR under the microaerophilic conditions existing rather than strict anaerobiosis. Thus, E. coli, L. lactis, and S. aureus nrdD mutants were found to grow at very low oxygen concentrations but failed to grow when strict anaerobic conditions were defined (20, 27, 39). Possibly, expression of the class I RNR genes may be upregulated at a low oxygen concentration. In Pseudomonas aeruginosa the class Ia RNR activity is significantly increased under microaerophilic conditions (61).
The L. monocytogenes NrdD deletion is located next to the glycyl radical domain.
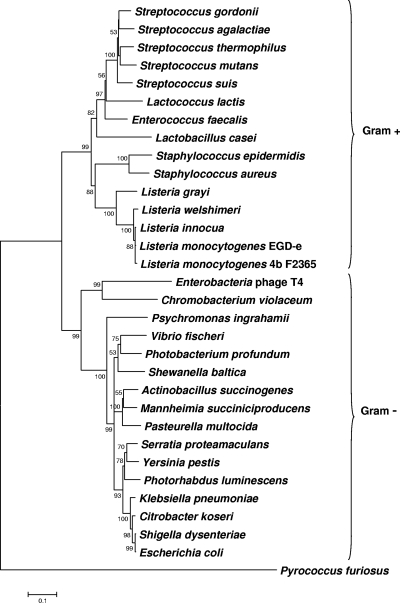
The L. monocytogenes EGD-e NrdD protein contains 710 amino acids. It shares 53% and 63% sequence identity with the E. coli and L. lactis orthologs, respectively, and about 50% identity with the bacteriophage T4 NrdD. Phylogeny of the L. monocytogenes NrdD protein with other bacterial class III RNR NrdD proteins shows that it clusters together with NrdDs of Gram-positive bacteria and is well separated from NrdDs of Gram-negative bacteria (Fig. 6). Figure 3, top, shows the EGD-e NrdD (Lmo0279) domain structure according to the Pfam database (2, 17). The N-terminal portion of the protein, comprising amino acids 9 to 96, exhibits high sequence identity (E value of 2.4e−22) with the ATP-cone regulatory domain found in the NrdA subunit of class Ia RNR proteins (2). The NrdD C-terminal portion contains the glycyl radical domain (55) consisting of residues 637 to 686. The glycyl radical is located on G680, corresponding to G681 in E. coli NrdD and G580 of bacteriophage T4 NrdD. Four conserved cysteine residues in a CXXC(X)14CXXC motif (34, 37) in the glycyl radical domain upstream of the glycyl radical site, involved in Zn binding, are conserved in the listerial NrdD protein.
Fig. 6.
Molecular phylogeny of bacterial NrdD proteins. The neighbor-joining (NJ) method was applied to estimate relationships among aligned sequences by using the MEGA 4 program. Pyrococcus furiosus was used as the outgroup in the tree. Bootstrap analysis using NJ was conducted with 500 replicates, and values above 50% are given at nodes. Bacterial names and accession numbers of NrdD proteins are shown in Table S5 in the supplemental material.
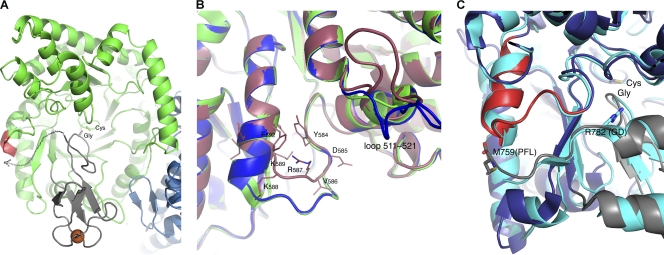
We have attempted to locate the KITPFE deletion site by employing a three-dimensional (3D) homology model of the EGD-e NrdD protein based on the structure established for the bacteriophage T4 NrdD (18) (Fig. 7), the only existing 3D structure for an anaerobic class III RNR protein. Surprisingly, as shown in Fig. 7A, it turns out that the deleted sequence is not a part of the C-terminal glycyl radical domain—either the metal-binding part or the loop containing the glycyl radical—as predicted by the protein domain databases. However, it is found nearby at the beginning of the second-last helix in the (α/β)10 barrel domain characteristic of the RNR family, a very highly conserved part of the structure. The deletion occurs between the two K residues in the sequence FHYDVRKKIDF. This deletion is likely to shorten significantly the loop between the end of this helix and the preceding β-strand. These are important interaction areas for the extreme C terminus of both pyruvate formate lyase and glycerol dehydratase (GD) (49), and we suppose that a similar interaction may occur in NrdD, although we have been unable to model it until now, as the last 20 residues of T4 NrdD are disordered in all the crystal structures (33). This might be because the C terminus is ordered only when in complex with the NrdG activase, or at some other specific point in the activation cycle.
Fig. 7.
Three-dimensional homology modeling of L. monocytogenes NrdD. (A) Mapping of the six-amino acid deletion onto the structure of NrdD from bacteriophage T4. This NrdD is shown in green, with the C-terminal domain in a darker shade. The second NrdD monomer of the dimeric enzyme is shown in blue. The deletion sequence is marked in red. The Gly radical site (G580, mutated to Ala in this structure) and the radical cysteine can also be seen, in stick representation. A plausible position for the C-terminal residue is sketched. (B) Comparison of the crystal structure of T4 NrdD (green) with the homology models for L. monocytogenes NrdD from strains HCC23 (blue) and EGD-e (dark pink). Side chain packing for the shortened loop in the EGD-e model, as suggested by Modeler, is shown. However, this is only one of several plausible packings that can be generated. The nearby loop t511-521, which is 10 residues longer in L. monocytogenes than in T4, is labeled. (C) Importance of interactions of the extreme C terminus of glycyl radical proteins with the deletion region. The structure of pyruvate formate lyase is shown in dark blue, with its C terminus in gray. The structure of glycerol dehydratase is shown in light blue. The loop and helix homologous to the deletion region in NrdD are highlighted in red. The extreme C-terminal amino acid of PFL (M759) and a residue important for conformational changes of the C terminus in activation of GD (R782) are shown as sticks and labeled.
In the EGD-e NrdD model, the shortening of the loop and the helix (the latter by about one turn) due to the deletion is evident (Fig. 7B). In contrast, when the NrdD protein of L. monocytogenes strain HCC23 possessing a complete NrdD (accession number YP_002351305) was modeled, the structure was essentially identical to that of bacteriophage T4, apart from a few variations in side-chain conformations (not shown). The deletion is one of only a few deviations between the two structures. There is also a 10-residue insertion relative to the T4 template structure in the nearby loop (residues 511 to 521 in Listeria) preceding the long helix associated with strand H of the α/β barrel, which could also be involved in interactions with the C terminus (Fig. 7B). However, since the insertion is common to both Listeria strains modeled, it cannot be responsible for differences in activity between them.
The homology model suggests that the remaining residues in EGD-e NrdD repack into a different compact conformation in that area that shifts the loop significantly. Pro486, which defines the N terminus of the α-helix, is one of the residues deleted in EGD-e NrdD. If we assume that the C terminus of NrdD interacts with the N terminus of this helix, then the presence of a secondary-structure-breaking proline there is very important, which explains its extreme conservation in all bacterial NrdD sequences. Our current hypothesis is that the KITPFE region is either essential for interactions with the NrdG activase or, indirectly, for the stability of the glycyl radical loop. Figure 7C shows the C terminus of the visible part of NrdD, which points in the general direction of this helix. There are 15 more residues in the sequence than we can see in the structure, so it is possible that interactions are made with the C terminus under certain conditions, perhaps when the activase is bound. Indeed, in pyruvate formate lyase (3, 31), the carboxylate group of the extreme C terminus interacts with the end of the equivalent helix (Fig. 7C). Significantly, a very similar interaction is found in GD (42). Moreover, a minimal point mutation (R782K) in the same three-dimensional region of glycerol dehydratase results in a tight, unproductive complex with the GD activase protein, critically reducing enzyme activity and implying that conformational changes in this area are very important for activation (42).
Concluding remarks.
In order to establish infection, L. monocytogenes, in common with all food-borne pathogens, must be capable of sensing and responding to the hostile environment of the human gastrointestinal tract which may include nutrient deprivation, acidity, iron limitation, bile, and oxygen limitation. It has been reported that L. monocytogenes can grow as a biofilm which contains oxygen gradients and anaerobic pockets (11). Here we show that L. monocytogenes serovar 1/2a EGD-e, but not other L. monocytogenes strains or listerial species, contains a nonfunctional anaerobic class III RNR system due to a deletion in the C-terminal portion of the protein, in the essential glycyl radical domain. The EGD-e strain has been employed extensively as a model laboratory listerial pathogen to elucidate the mechanisms of intracellular multiplication and virulence. Because L. monocytogenes EGD-e lacks a functional anaerobic RNR, it is relevant to ask whether the findings presented here affect interpretation of studies of pathogenesis, particularly those with conditions of low oxygen tension. Plausibly, EGD-e can continue to serve as a suitable reference strain as long as anaerobic/microaerophilic conditions are not used, which is the case in many in vitro or cell culture experiments. However, the EGD-e strain would not be suitable for studies such as gastrointestinal infections or environmental conditions of very low oxygen tension. Animal model studies are under way to clarify whether the anaerobic RNR system is a virulence determinant.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the German Federal Ministry for Education and Research in the framework of the ERA- NET PathoGenoMics, project SPATELIS (grant no. BMBF/PTJ 0313939), and partially by the EU- funded Network of Excellence EuroPathoGenomics (NoE EPG). D.T.L. was supported by a grant (2006-4387) from the Swedish Research Council (VR).
We thank Michaela Yanku and Batia Gorovitz-Harris for technical assistance.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 8 April 2011.
REFERENCES
- 1. Altschul S. F., Lipman D. J. 1990. Protein database searches for multiple alignments. Proc. Natl. Acad. Sci. U. S. A. 87:5509–5513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aravind L., Wolf Y. I., Koonin E. V. 2000. The ATP-cone: an evolutionarily mobile, ATP-binding regulatory domain. J. Mol. Microbiol. Biotechnol. 2:191–194 [PubMed] [Google Scholar]
- 3. Becker A., Kabsch W. 2002. X-ray structure of pyruvate formate-lyase in complex with pyruvate and CoA. How the enzyme uses the Cys-418 thiyl radical for pyruvate cleavage. J. Biol. Chem. 277:40036–40042 [DOI] [PubMed] [Google Scholar]
- 4. Boal A. K., Cotruvo J. A., Jr, Stubbe J., Rosenzweig A. C. 2010. Structural basis for activation of class Ib ribonucleotide reductase. Science 329:1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bo Andersen J., Roldgaard B. B., Christensen B. B., Licht T. R. 2007. Oxygen restriction increases the infective potential of Listeria monocytogenes in vitro in Caco-2 cells and in vivo in guinea pigs. BMC Microbiol. 7:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buchrieser C., Rusniok C., Kunst F., Cossart P., Glaser P. 2003. Comparison of the genome sequences of Listeria monocytogenes and Listeria innocua: clues for evolution and pathogenicity. FEMS Immunol. Med. Microbiol. 35:207–213 [DOI] [PubMed] [Google Scholar]
- 7. Burall L. S., et al. 2004. Proteus mirabilis genes that contribute to pathogenesis of urinary tract infection: identification of 25 signature-tagged mutants attenuated at least 100-fold. Infect. Immun. 72:2922–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burkholder K. M., et al. 2009. Expression of LAP, a SecA2-dependent secretory protein, is induced under anaerobic environment. Microbes Infect. 11:859–867 [DOI] [PubMed] [Google Scholar]
- 9. Camejo A., et al. 2009. In vivo transcriptional profiling of Listeria monocytogenes and mutagenesis identify new virulence factors involved in infection. PLoS Pathog. 5:e1000449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cossart P., Sansonetti P. J. 2004. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science 304:242–248 [DOI] [PubMed] [Google Scholar]
- 11. Costerton J. W., Lewandowski Z., Caldwell D. E., Korber D. R., Lappin-Scott H. M. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711–745 [DOI] [PubMed] [Google Scholar]
- 12. Cotruvo J. A., Jr., Stubbe J. 2010. An active dimanganese(III)-tyrosyl radical cofactor in Escherichia coli class Ib ribonucleotide reductase. Biochemistry 49:1297–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cotruvo J. A., Jr., Stubbe J. 2008. NrdI, a flavodoxin involved in maintenance of the diferric-tyrosyl radical cofactor in Escherichia coli class Ib ribonucleotide reductase. Proc. Natl. Acad. Sci. U. S. A. 105:14383–14388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Engstrom Y., Eriksson S., Thelander L., Akerman M. 1979. Ribonucleotide reductase from calf thymus. Purification and properties. Biochemistry 18:2941–2948 [DOI] [PubMed] [Google Scholar]
- 15. Eswar N., et al. 2006. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics Chapter 5:Unit 5.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Farber J. M., Peterkin P. I. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Finn R. D., et al. 2008. The Pfam protein families database. Nucleic Acids Res. 36:D281–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fontecave M., Mulliez E., Logan D. T. 2002. Deoxyribonucleotide synthesis in anaerobic microorganisms: the class III ribonucleotide reductase. Prog. Nucleic Acid Res. Mol. Biol. 72:95–127 [DOI] [PubMed] [Google Scholar]
- 19. Freitag N. E., Port G. C., Miner M. D. 2009. Listeria monocytogenes—from saprophyte to intracellular pathogen. Nat. Rev. Microbiol. 7:623–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Garriga X., et al. 1996. nrdD and nrdG genes are essential for strict anaerobic growth of Escherichia coli. Biochem. Biophys. Res. Commun. 229:189–192 [DOI] [PubMed] [Google Scholar]
- 21. Glaser P., et al. 2001. Comparative genomics of Listeria species. Science 294:849–852 [DOI] [PubMed] [Google Scholar]
- 22. Gough J., Chothia C. 2002. SUPERFAMILY: HMMs representing all proteins of known structure. SCOP sequence searches, alignments and genome assignments. Nucleic Acids Res. 30:268–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hain T., Steinweg C., Chakraborty T. 2006. Comparative and functional genomics of Listeria spp. J. Biotechnol. 126:37–51 [DOI] [PubMed] [Google Scholar]
- 24. Härtig E., Hartmann A., Schatzle M., Albertini A. M., Jahn D. 2006. The Bacillus subtilis nrdEF genes, encoding a class Ib ribonucleotide reductase, are essential for aerobic and anaerobic growth. Appl. Environ. Microbiol. 72:5260–5265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Higgins D. G., Thompson J. D., Gibson T. J. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266:383–402 [DOI] [PubMed] [Google Scholar]
- 26. Hunter S., et al. 2009. InterPro: the integrative protein signature database. Nucleic Acids Res. 37:D211–D215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jordan A., et al. 1996. The ribonucleotide reductase system of Lactococcus lactis. Characterization of an NrdEF enzyme and a new electron transport protein. J. Biol. Chem. 271:8779–8785 [DOI] [PubMed] [Google Scholar]
- 28. Jordan A., Reichard P. 1998. Ribonucleotide reductases. Annu. Rev. Biochem. 67:71–98 [DOI] [PubMed] [Google Scholar]
- 29. Joseph B., et al. 2006. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J. Bacteriol. 188:556–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kaufmann S. H., Simon M. M., Hahn H. 1979. Specific Lyt 123 cells are involved in protection against Listeria monocytogenes and in delayed-type hypersensitivity to listerial antigens. J. Exp. Med. 150:1033–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lehtiö L., Leppanen V. M., Kozarich J. W., Goldman A. 2002. Structure of Escherichia coli pyruvate formate-lyase with pyruvate. Acta Crystallogr. D Biol. Crystallogr. 58:2209–2212 [DOI] [PubMed] [Google Scholar]
- 32. Letunic I., et al. 2004. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 32:D142–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Logan D. T., Andersson J., Sjoberg B. M., Nordlund P. 1999. A glycyl radical site in the crystal structure of a class III ribonucleotide reductase. Science 283:1499–1504 [DOI] [PubMed] [Google Scholar]
- 34. Logan D. T., et al. 2003. A metal-binding site in the catalytic subunit of anaerobic ribonucleotide reductase. Proc. Natl. Acad. Sci. U. S. A. 100:3826–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lundin D., Torrents E., Poole A. M., Sjoberg B. M. 2009. RNRdb, a curated database of the universal enzyme family ribonucleotide reductase, reveals a high level of misannotation in sequences deposited to Genbank. BMC Genomics 10:589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lungu B., Ricke S. C., Johnson M. G. 2009. Growth, survival, proliferation and pathogenesis of Listeria monocytogenes under low oxygen or anaerobic conditions: a review. Anaerobe 15:7–17 [DOI] [PubMed] [Google Scholar]
- 37. Luttringer F., Mulliez E., Dublet B., Lemaire D., Fontecave M. 2009. The Zn center of the anaerobic ribonucleotide reductase from E. coli.. J. Biol. Inorg. Chem. 14:923–933 [DOI] [PubMed] [Google Scholar]
- 38. Marteyn B., et al. 2010. Modulation of Shigella virulence in response to available oxygen in vivo. Nature 465:355–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Masalha M., Borovok I., Schreiber R., Aharonowitz Y., Cohen G. 2001. Analysis of transcription of the Staphylococcus aureus aerobic class Ib and anaerobic class III ribonucleotide reductase genes in response to oxygen. J. Bacteriol. 183:7260–7272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nelson K. E., et al. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nordlund P., Reichard P. 2006. Ribonucleotide reductases. Annu. Rev. Biochem. 75:681–706 [DOI] [PubMed] [Google Scholar]
- 42. O'Brien J. R., et al. 2004. Insight into the mechanism of the B12-independent glycerol dehydratase from Clostridium butyricum: preliminary biochemical and structural characterization. Biochemistry 43:4635–4645 [DOI] [PubMed] [Google Scholar]
- 43. Paik S., et al. 2005. Identification of virulence determinants for endocarditis in Streptococcus sanguinis by signature-tagged mutagenesis. Infect. Immun. 73:6064–6074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Qian W., et al. 2005. Comparative and functional genomic analyses of the pathogenicity of phytopathogen Xanthomonas campestris pv. campestris. Genome Res. 15:757–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rabinovitch I., et al. 2010. Staphylococcus aureus NrdH redoxin is a reductant of the class Ib ribonucleotide reductase. J. Bacteriol. 192:4963–4972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Reichard P., Ehrenberg A. 1983. Ribonucleotide reductase—a radical enzyme. Science 221:514–519 [DOI] [PubMed] [Google Scholar]
- 47. Roca I., Ballana E., Panosa A., Torrents E., Gibert I. 2008. Fumarate and nitrate reduction (FNR) dependent activation of the Escherichia coli anaerobic ribonucleotide reductase nrdDG promoter. Int. Microbiol. 11:49–56 [PubMed] [Google Scholar]
- 48. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 49. Selmer T., Pierik A. J., Heider J. 2005. New glycyl radical enzymes catalysing key metabolic steps in anaerobic bacteria. Biol. Chem. 386:981–988 [DOI] [PubMed] [Google Scholar]
- 50. Senol E. 2004. Stenotrophomonas maltophilia: the significance and role as a nosocomial pathogen. J. Hosp. Infect. 57:1–7 [DOI] [PubMed] [Google Scholar]
- 51. Seveau S., Pizarro-Cerda J., Cossart P. 2007. Molecular mechanisms exploited by Listeria monocytogenes during host cell invasion. Microbes Infect. 9:1167–1175 [DOI] [PubMed] [Google Scholar]
- 52. Sigrist C. J., et al. 2010. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 38:D161–D166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Simpson A. J., et al. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. The Xylella fastidiosa Consortium of the Organization for Nucleotide Sequencing and Analysis. Nature 406:151–159 [DOI] [PubMed] [Google Scholar]
- 54. Smith K., Youngman P. 1992. Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie 74:705–711 [DOI] [PubMed] [Google Scholar]
- 55. Sun X., et al. 1993. A possible glycine radical in anaerobic ribonucleotide reductase from Escherichia coli: nucleotide sequence of the cloned nrdD gene. Proc. Natl. Acad. Sci. U. S. A. 90:577–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sun X., et al. 1996. The free radical of the anaerobic ribonucleotide reductase from Escherichia coli is at glycine 681. J. Biol. Chem. 271:6827–6831 [PubMed] [Google Scholar]
- 57. Suvarna K., Stevenson D., Meganathan R., Hudspeth M. E. 1998. Menaquinone (vitamin K2) biosynthesis: localization and characterization of the menA gene from Escherichia coli. J. Bacteriol. 180:2782–2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 59. Toledo-Arana A., et al. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950–956 [DOI] [PubMed] [Google Scholar]
- 60. Vázquez-Boland J. A., et al. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wu M., Guina T., Brittnacher M., Nguyen H., Eng. J., Miller S. I. 2005. The Pseudomonas aeruginosa proteome during anaerobic growth. J. Bacteriol. 187:8185–8190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zemansky J., et al. 2009. Development of a mariner-based transposon and identification of Listeria monocytogenes determinants, including the peptidyl-prolyl isomerase PrsA2, that contribute to its hemolytic phenotype. J. Bacteriol. 191:3950–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang J., Inouye M. 2002. MazG, a nucleoside triphosphate pyrophosphohydrolase, interacts with Era, an essential GTPase in Escherichia coli. J. Bacteriol. 184:5323–5329 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.