Abstract
Erysipelothrix rhusiopathiae is a Gram-positive bacterium that represents a new class, Erysipelotrichia, in the phylum Firmicutes. The organism is a facultative intracellular pathogen that causes swine erysipelas, as well as a variety of diseases in many animals. Here, we report the first complete genome sequence analysis of a member of the class Erysipelotrichia. The E. rhusiopathiae genome (1,787,941 bp) is one of the smallest genomes in the phylum Firmicutes. Phylogenetic analyses based on the 16S rRNA gene and 31 universal protein families suggest that E. rhusiopathiae is phylogenetically close to Mollicutes, which comprises Mycoplasma species. Genome analyses show that the overall features of the E. rhusiopathiae genome are similar to those of other Gram-positive bacteria; it possesses a complete set of peptidoglycan biosynthesis genes, two-component regulatory systems, and various cell wall-associated virulence factors, including a capsule and adhesins. However, it lacks many orthologous genes for the biosynthesis of wall teichoic acids (WTA) and lipoteichoic acids (LTA) and the dltABCD operon, which is responsible for d-alanine incorporation into WTA and LTA, suggesting that the organism has an atypical cell wall. In addition, like Mollicutes, its genome shows a complete loss of fatty acid biosynthesis pathways and lacks the genes for the biosynthesis of many amino acids, cofactors, and vitamins, indicating reductive genome evolution. The genome encodes nine antioxidant factors and nine phospholipases, which facilitate intracellular survival in phagocytes. Thus, the E. rhusiopathiae genome represents evolutionary traits of both Firmicutes and Mollicutes and provides new insights into its evolutionary adaptations for intracellular survival.
INTRODUCTION
The phylum Firmicutes consists of Gram-positive bacteria with low genomic G+C contents. It includes many important pathogens, including Bacillus, Staphylococcus, and Streptococcus species, and bacteria beneficial to humans, including Lactobacillus and Lactococcus species. Firmicutes was previously described as consisting of three classes, Bacilli, Clostridia, and Mollicutes (14). However, Mollicutes, which are represented by the genus Mycoplasma, have recently been removed from the phylum Firmicutes and placed in a newly created phylum, Tenericutes, because these species lack rigid cell walls and there is no alternative marker except 16S rRNA sequences to support retention of Mollicutes in Firmicutes (23). On the other hand, the family Erysipelotrichaceae, which consists of Gram-positive walled bacteria that were previously classified as Mollicutes, was retained in Firmicutes as a member of a newly generated class, Erysipelotrichia, which comprises a single order and a single family: the order Erysipelotrichales and the family Erysipelotrichaceae. The family Erysipelotrichaceae is composed of eight genera: Erysipelothrix, Allobaculum (a newly described genus), Bulleidia, Catenibacterium, Coprobacillus, Holdemania, Solobacterium, and Turicibacter. The last six genera were transferred from other families to Erysipelotrichaceae (23).
Erysipelothrix rhusiopathiae, a Gram-positive, non-spore-forming, rod-shaped bacterium, is the standard species of the genus Erysipelothrix and comprises the genus, along with Erysipelothrix tonsillarum and Erysipelothrix inopinata (53). E. rhusiopathiae is ubiquitous in nature and has been isolated from many species of wild and domestic mammals, birds, reptiles, amphibians, and fish (58). The organism can grow either aerobically or anaerobically. As has been observed for Mesoplasma species in Mollicutes (41), it does not grow at all or grows very slowly if the medium is not supplemented with 5 to 10% serum or 0.1% Tween 80 (polyoxyethylene sorbitan), even in a nutrient-rich medium, such as brain heart infusion (BHI) medium (11).
E. rhusiopathiae is generally regarded as an opportunistic animal pathogen that causes a variety of diseases in many species of birds and mammals, including humans. It is best known as a facultative intracellular pathogen that causes swine erysipelas, which may occur as acute septicemia or chronic endocarditis and polyarthritis (58). The organism has a capsule (52). Interestingly, it can survive inside polymorphonuclear leukocytes and macrophages if it is phagocytosed (51). The primary survival strategy in phagocytes appears to be escape from reactive oxidative metabolites generated by phagocytic cells; however, the precise mechanisms for intracellular survival of the organism are unknown (45, 51).
Here, we report the results of genome sequencing of E. rhusiopathiae strain Fujisawa, the first complete genome sequence of a bacterium belonging to the class Erysipelotrichia. Fujisawa is a highly virulent strain originally isolated from a diseased pig and has been extensively used in studies of E. rhusiopathiae pathogenesis (32, 47, 49–52, 55). This genome analysis provides new insights into how the organism has evolved and adapted as an intracellular pathogen.
MATERIALS AND METHODS
DNA sequencing and annotation, and data analyses.
The E. rhusiopathiae strain Fujisawa was grown at 37°C in BHI (Becton, Dickinson and Company, Baltimore, MD) supplemented with 0.1% Tween 80 (pH 8.0), and the genomic DNA was prepared as described previously (50). The genome sequence was determined at Dragon Genomics Center Co. Ltd. (Mie, Japan) by a combination of three sequence technologies: the Genome Sequencer 20 (GS20) (Roche), SOLiD (sequencing by oligonucleotide ligation and detection) (Applied Biosystems), and Genome Analyzer II (GAII) (Illumina) systems. The genomic DNA was first sequenced with GS20 to generate a draft sequence. Then, a total of 140 million SOLiD reads (a 25-bp sequence for each read) produced from the 1.5-kb and 5.5-kb paired-end libraries were mapped to the GS20 sequence to correct sequence errors. At this stage, the depth of coverage from high-quality reads was approximately 23.5-fold. The sequences obtained by GS20 and SOLiD were further compared with data obtained by GAII, and unmatched sequences and gap sequences in the contigs were corrected or closed by PCR amplification followed by Sanger sequencing.
Finally, three gaps, all of which were derived from rRNA (rrn) operons, remained to be closed. Because the organization of these rrn operons was too complex to be closed by a simple procedure, we constructed a fosmid library. Using the fosmid clones containing each rrn locus, we determined the copy number of rrn operons and the gene composition (16S-23S-5S rRNA) of each locus by Southern hybridization analysis, PCR, and Sanger sequencing with primer walking. This series of analyses revealed that one locus contains a single rrn operon (16S-23S-5S rRNA) and that the other two loci contain two and four operons, respectively, in a tandem organization. Therefore, we were unable to determine the complete nucleotide sequences of these six rrn operons. However, by careful inspection of the Sanger sequencing data from these two rrn loci, we found no peak patterns suggestive of the presence of sequence heterogeneity between copies. Therefore, we conclude that the rrn sequences in each of the two loci are identical to those of the single rrn operon.
Potential protein-coding sequences (CDSs) of greater than 150 bp were identified using the gene prediction software MGA (MetaGeneAnnotator) (36). Short CDSs of less than 150 bp in intergenic regions were identified by the IMCGE (in silico Molecular Cloning Genomics Edition) software (37) with a BLASTP search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) as a guide. tRNA and rRNA genes were identified by tRNAscan-SE 1.23 (22) and RNAmmer 1.2 (20), respectively. Functional annotation of each CDS was made according to the results from the BLASTP search against the NCBI RefSeq database (release 34) (40). For the metabolic pathway analysis, the KEGG (Kyoto Encyclopedia of Genes and Genomes) database (18; http://www.genome.jp/kegg/) was used.
Phylogenetic analysis.
A phylogenetic tree based on 16S rRNA gene sequences was constructed as described previously (56). A genomic phylogenetic tree was constructed from alignments of the concatenated protein sequences of 31 universal genes, which are all involved in translation and have been shown to be conserved in all of the analyzed bacterial species (6). Briefly, multiple-sequence alignments of each COG (cluster of orthologous groups) were created using MUSCLE (8). The sequence alignments were concatenated and poorly aligned, and divergent regions were removed using Gblocks (4). Based on the resulting final sequence alignments, a phylogenic tree was constructed by the maximum-likelihood method using the MEGA 5 program (http://www.megasoftware.net/).
Genome comparison.
Genome sequences of other Erysipelotrichia strains, including E. rhusiopathiae strain ATCC 19414, all of which are incomplete or unfinished, were obtained from the National Center for Biotechnology Information (NCBI database) (http://www.ncbi.nlm.nih.gov/genomes/MICROBES/microbial_taxtree.html) in February 2011. E. rhusiopathiae strain-specific CDSs were identified by bidirectional best-hit analysis between the genomes of strains Fujisawa and ATCC 19414 with a threshold of >90% amino acid identity and >60% aligned length coverage of a query sequence. Conservation of the CDSs of Fujisawa in other Erysipelotrichia strains (Solobacterium moorei, Coprobacillus sp., Holdemania filiformis, Catenibacterium mitsuokai, Bulleidia extructa, Erysipelotrichaceae bacterium, Clostridium ramosum, Clostridium spiroforme, Eubacterium biforme, and Eubacterium dolichum) for which draft genome sequences were available was examined by BLASTP analysis with an E value threshold of 10−10.
Nucleotide sequence accession number.
The genome sequence reported in this paper has been deposited in the DDBJ/GenBank/EMBL databases (accession no. AP012027).
RESULTS AND DISCUSSION
General genomic features.
The general features of the genome of E. rhusiopathiae strain Fujisawa are summarized in Table 1. The origin of replication (nucleotide position 1) was assigned to a region showing a clear G+C skew transition and containing the dnaA gene accompanied by several DnaA boxes. The genome contains 1,704 CDSs. Of these, biological functions were assigned to 1,332 CDSs, 327 CDSs showed sequence similarities to proteins of unknown function, and the remaining 45 had no significant database match. The genome contains only seven recognizable pseudogenes (four caused by frameshift and three caused by point mutations).
Table 1.
General features of the genome of E. rhusiopathiae strain Fujisawa
| Feature | Value |
|---|---|
| Genome size (bp) | 1,787,941 |
| Overall G+C content (%) | 36.6 |
| G+C content of CDS (%) | 36.7 |
| No. of CDSs | 1704 |
| No. of pseudogenes | 7 |
| No. of rRNA operons (16S-23S-5S) | 7 |
| No. of tRNA genes | 55 |
| No. of prophage | 1 |
| No. of plasmids | 0 |
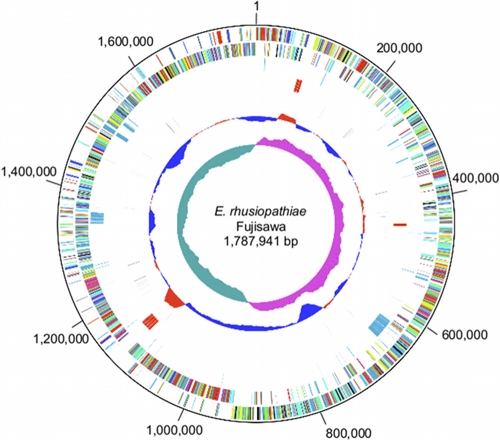
Like many other members of Firmicutes with lower genome G+C contents, the majority (75.6%) of genes are located on the leading strand. The genome contains seven rrn operons with the typical order of 16S, 23S, and 5S rRNA genes and 55 tRNA genes, including cognates for all amino acids (Fig. 1). The seven rrn operons are located at three loci and show a unique genetic organization: in the two loci (nucleotide positions 78525 to 89100 and 1117642 to 1139038), two and four operons, respectively, which probably have identical sequences (see Materials and Methods), are located in tandem.
Fig. 1.
Circular representation of the genome of E. rhusiopathiae strain Fujisawa. Beginning with the outer region, the circle shows (i) nucleotide positions in base pairs, (ii and iii) predicted CDSs transcribed on the forward (clockwise) (ii) and reverse (counterclockwise) (iii) DNA strands, (iv) positions of Fujisawa strain-specific genes not present in the genome of strain ATCC 19414, (v) rRNA operon(s), (vi) tRNA genes, (vii) percent G+C content (red and blue, respectively, represent regions with higher and lower G+C contents compared to the average value for the entire genome), and (viii) the G+C skew curve. The color of each CDS was assigned according to the COG functional grouping (http://www.ncbi.nlm.nih.gov/COG/).
We identified a 36.5-kb prophage (named PP_Erh_Fujisawa) including 49 genes (ERH_0581 through ERH_0629) but no CRISPR (clustered regularly interspaced short palindromic repeats). We found 22 intact insertion sequence (IS) elements and two truncated IS elements, none of which were inserted into CDSs of known function. These IS elements were classified into six types (named ISErh1 to ISErh6); all types were newly identified, with ISErh2 being the predominant type (eight intact copies) (Table 2). Most of the IS elements belong to the IS30 and IS3 families, but one type (ISErh3) is unclassified. Notably, the two IS30 family members, ISErh2 and ISErh4, are associated with atypically long (23- to 41-bp) direct repeats (DRs). Within the IS30 family, such long DRs have so far been detected only in IS1630 (19 to 26 bp) of Mycoplasma fermentans (3) and ISMbov6 (22 to 36 bp) of Mycoplasma bovis (24). The predicted transposases of both ISErh2 and ISErh4 show the highest homology (32.4% and 32.0% amino acid sequence identity, respectively) to that of IS1630 of M. fermentans.
Table 2.
Insertion sequences identified in E. rhusiopathiae strain Fujisawa
| Name | No. | IS family | No. of CDSs | Size (bp) | Length of DR (bp) |
|---|---|---|---|---|---|
| ISErh1 | ERH_IS01 | 3 | 2 | 1,280 | 3 |
| ISErh1 | ERH_IS02 | 3 | 2 | 1,280 | 3 |
| ISErh1 | ERH_IS06 | 3 | 2 | 1,280 | 3 |
| ISErh1 | ERH_IS14 | 3 | 2 | 1,280 | 4 |
| ISErh1 | ERH_IS22 | 3 | 2 | 1,280 | 3 |
| ISErh2 | ERH_IS03 | 30 | 1 | 1,031 | −a |
| ISErh2 | ERH_IS08 | 30 | 1 | 1,031 | 27 |
| ISErh2 | ERH_IS09 | 30 | 1 | 1,031 | 29 |
| ISErh2 | ERH_IS15 | 30 | 1 | 1,031 | 32 |
| ISErh2 | ERH_IS17 | 30 | 1 | 1,031 | 23 |
| ISErh2 | ERH_IS18 | 30 | 1 | 1,031 | 24 |
| ISErh2 | ERH_IS23 | 30 | 1 | 1,031 | 30 |
| ISErh2 | ERH_IS24 | 30 | 1 | 1,031 | 31 |
| ISErh3 | ERH_IS04 | ?b | 3 | 1,351 | 6 |
| ISErh3 | ERH_IS07c | ? | 3 | 1,176 | 6 |
| ISErh4 | ERH_IS05 | 30 | 1 | 1,032 | 30 |
| ISErh4 | ERH_IS10 | 30 | 1 | 1,032 | 26 |
| ISErh4 | ERH_IS11 | 30 | 1 | 1,032 | 37 |
| ISErh4 | ERH_IS16 | 30 | 1 | 1,032 | 26 |
| ISErh4 | ERH_IS19 | 30 | 1 | 1,032 | 41 |
| ISErh5 | ERH_IS12 | 3 | 2 | 1,247 | − |
| ISErh5 | ERH_IS13 | 3 | 2 | 1,247 | − |
| ISErh5 | ERH_IS20c | 3 | 2 | 1,206 | − |
| ISErh6 | ERH_IS21 | 3 | 2 | 1,240 | − |
−, not identified.
?, unclassified; both copies appear to contain internal deletions.
Truncated IS.
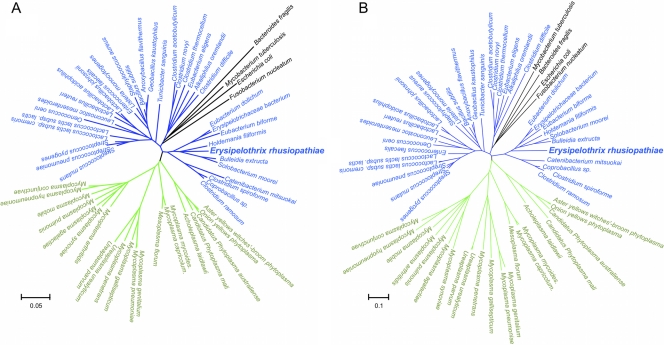
Phylogenetic position of E. rhusiopathiae.
We constructed a genomic phylogenetic tree using the 31 universal genes (6) that are conserved among all analyzed bacterial species and compared it with a tree that was constructed using 16S rRNA sequences. In this analysis, we included all Erysipelotrichia strains, draft sequences of which were available, to better understand the phylogenetic position, not only of E. rhusiopathiae, but also of the class Erysipelotrichia. In both trees (Fig. 2A and B), E. rhusiopathiae and other Erysipelotrichia strains, with the exception of Turicibacter sanguinis, clustered together, forming a cluster distinct from other Firmicutes species and placed at the position closest to Mollicutes. These results indicate a very close phylogenetic relationship between Erysipelotrichia, including E. rhusiopathiae, and Mollicutes. Furthermore, our results may raise the possibility that Erysipelotrichia should be separated from Firmicutes and classified as a distinct phylum, as Mollicutes were moved to a separate phylum, Tenericutes (23). In contrast, T. sanguinis was placed in both trees at a very distant position from other Erysipelotrichia strains and clustered together with other Firmicutes species, suggesting that the species should be separated from Erysipelotrichia.
Fig. 2.
Phylogenetic position of E. rhusiopathiae. Phylogenetic trees based on 16S rRNA gene sequences (A) and on concatenated protein sequence alignments derived from 31 universal protein families (B) are shown. Species are colored according to the current taxonomy: the phylum Firmicutes, blue; Mollicutes, green; others, black. E. rhusiopathiae is indicated by boldface. The scale bar represents the expected number of changes per sequence position. A bootstrap test with 1,000 replicates was used to estimate the confidence of the branching patterns of the trees. Branches corresponding to partitions reproduced in less than 50% of bootstrap replicates are collapsed.
Reductive genome evolution and metabolic capabilities.
Genome reduction has also taken place in many bacteria living in nutrient-rich environments, including lactic acid bacteria (25) and the members of Mollicutes (28). Among the sequenced members of Firmicutes, including lactic acid bacteria, the genome of E. rhusiopathiae is one of the smallest, indicating that reductive genome evolution occurred in the organism (Table 3).
Table 3.
Numbers of orthologous genes related to DNA repaira
| Species (strain) | Genome size (Mb) | GC content (%) | Phylum or class | No. |
||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | adaA | alkA | alkB | dinP | exoA (xth) | fpg | ligA | mfd | mpg | mutH | mutL | mutS | mutT | mutY | nfo | nth | ogt | pcrA | phr | polA | priA | radA | radC | recA | recD | recF | recG | recJ | recN | recO | recQ | recR | recU | rexA | rexB | ruvA | ruvB | ruvC | tag1 | umuC | ung | uvrA | uvrB | uvrC | ||||
| Alkaliphilus oremlandii (OhILAs) | 3.12 | 36.3 | Firmicutes | 43 | 1 | 2 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 2 | 1 | 1 | 1 | 1 | 2 | 2 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 2 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| Clostridium acetobutylicum (ATCC 824) | 3.94 | 30.9 | Firmicutes | 40 | 0 | 1 | 0 | 1 | 1 | 0 | 2 | 1 | 1 | 0 | 1 | 2 | 0 | 1 | 1 | 1 | 1 | 2 | 0 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 0 | 2 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| Clostridium difficile (630) | 4.29 | 29.1 | Firmicutes | 38 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 2 | 0 | 0 | 1 | 1 | 2 | 2 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 2 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 |
| Clostridium novyi (NT) | 2.55 | 28.9 | Firmicutes | 35 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 2 | 1 | 0 | 1 | 1 | 0 | 2 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 0 | 2 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| Clostridium thermocellum (ATCC 27405) | 3.84 | 39.0 | Firmicutes | 36 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 1 | 1 | 2 | 0 | 1 | 1 | 1 | 2 | 1 | 2 | 1 | 1 | 2 | 1 | 1 | 0 | 1 | 0 | 2 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 |
| Eubacterium eligens (ATCC 27750) | 2.14 | 37.7 | Firmicutes | 39 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 2 | 1 | 0 | 1 | 1 | 0 | 3 | 0 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 |
| Streptococcus mutans (UA159) | 2.03 | 36.8 | Firmicutes | 37 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 2 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
| Streptococcus pneumoniae (TIGR4) | 2.16 | 39.7 | Firmicutes | 33 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
| Streptococcus pyogenes (SF370) | 1.85 | 38.5 | Firmicutes | 36 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 2 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
| Anoxybacillus flavithermus (WK1) | 2.85 | 41.8 | Firmicutes | 38 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 2 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| Bacillus subtilis (168) | 4.22 | 43.5 | Firmicutes | 49 | 1 | 2 | 0 | 2 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 2 | 1 | 1 | 1 | 1 | 2 | 2 | 0 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 0 | 2 | 1 | 1 | 1 | 1 |
| Geobacillus kaustophilus (HTA426) | 3.54 | 52.1 | Firmicutes | 38 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| Listeria monocytogenes (EGD-e) | 2.94 | 38.0 | Firmicutes | 49 | 2 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 2 | 1 | 1 | 1 | 1 | 3 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 1 | 1 | 2 | 1 | 1 | 2 |
| Staphylococcus aureus (N315) | 2.81 | 32.8 | Firmicutes | 41 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 2 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| Enterococcus faecalis (V583) | 3.36 | 37.5 | Firmicutes | 47 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 2 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 | 0 | 1 | 5 | 1 | 1 | 1 | 1 |
| Lactobacillus acidophilus (NCFM) | 1.99 | 34.7 | Firmicutes | 37 | 0 | 0 | 0 | 1 | 2 | 1 | 1 | 1 | 1 | 0 | 1 | 2 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
| Lactobacillus brevis (ATCC 367) | 2.29 | 46.2 | Firmicutes | 41 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
| Lactobacillus casei (ATCC 334) | 2.90 | 46.6 | Firmicutes | 45 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 2 | 1 | 1 | 1 | 1 | 1 |
| Lactobacillus delbrueckii subsp. bulgaricus (ATCC 11842) | 1.86 | 49.7 | Firmicutes | 35 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 2 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 |
| Lactobacillus fermentum (IFO 3956) | 2.10 | 51.5 | Firmicutes | 36 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 2 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| Lactobacillus gasseri (ATCC 33323) | 1.89 | 35.3 | Firmicutes | 39 | 0 | 0 | 0 | 1 | 2 | 1 | 1 | 1 | 1 | 0 | 1 | 2 | 0 | 0 | 0 | 1 | 2 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| Lactobacillus helveticus (DPC 4571) | 2.08 | 37.1 | Firmicutes | 36 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 2 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
| Lactobacillus johnsonii (NCC 533) | 1.99 | 34.6 | Firmicutes | 39 | 0 | 0 | 0 | 1 | 2 | 1 | 1 | 1 | 1 | 0 | 1 | 2 | 1 | 0 | 0 | 1 | 2 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
| Lactobacillus plantarum (WCFS1) | 3.35 | 44.5 | Firmicutes | 42 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| Lactobacillus reuteri (JCM 1112) | 2.04 | 38.9 | Firmicutes | 38 | 0 | 0 | 0 | 0 | 2 | 1 | 1 | 1 | 1 | 0 | 1 | 2 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| Lactobacillus rhamnosus (GG) | 3.01 | 46.7 | Firmicutes | 45 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 2 | 1 | 1 | 1 | 1 | 1 |
| Lactobacillus sakei (23K) | 1.88 | 41.3 | Firmicutes | 40 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 2 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| Lactobacillus salivarius (UCC118) | 1.83 | 32.9 | Firmicutes | 35 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
| Leuconostoc mesenteroides (ATCC 8293) | 2.04 | 37.7 | Firmicutes | 40 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 2 | 2 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 1 | 2 | 1 | 1 | 1 | 1 |
| Lactococcus lactis subsp. cremoris (MG1363) | 2.53 | 35.8 | Firmicutes | 40 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 2 | 4 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| Lactococcus lactis subsp. lactis (IL1403) | 2.37 | 35.3 | Firmicutes | 41 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 2 | 3 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| Oenococcus oeni (PSU-1) | 1.78 | 37.9 | Firmicutes | 33 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 3 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| Erysipelothrix rhusiopathiae (Fujisawa) | 1.79 | 36.6 | Firmicutes | 34 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Acholeplasma laidlawii (PG-8A) | 1.50 | 31.9 | Mollicutes | 33 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 2 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 2 | 1 | 1 | 1 | 1 |
| “Candidatus Phytoplasma mali” (AT) | 0.60 | 21.4 | Mollicutes | 16 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| “Candidatus Phytoplasma australiense” | 0.88 | 27.4 | Mollicutes | 11 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| Aster yellows witches' broom phytoplasma (AYWB) | 0.71 | 26.9 | Mollicutes | 11 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| Onion yellows phytoplasma (OY-M) | 0.85 | 27.8 | Mollicutes | 11 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| Mesoplasma florum (L1; ATCC 33453) | 0.79 | 27.0 | Mollicutes | 17 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
| Mycoplasma agalactiae (PG2) | 0.88 | 29.7 | Mollicutes | 16 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 2 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| Mycoplasma arthritidis (158L3-1) | 0.82 | 30.7 | Mollicutes | 16 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| Mycoplasma conjunctivae (HRC/581T) | 0.85 | 28.5 | Mollicutes | 16 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| Mycoplasma gallisepticum (R) | 1.00 | 31.5 | Mollicutes | 15 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| Mycoplasma genitalium (G37) | 0.58 | 31.7 | Mollicutes | 13 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| Mycoplasma hyopneumoniae (232) | 0.89 | 28.6 | Mollicutes | 15 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| Mycoplasma mobile (163K) | 0.78 | 25.0 | Mollicutes | 15 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| Mycoplasma capricolum (ATCC 27343) | 1.01 | 23.8 | Mollicutes | 15 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| Mycoplasma mycoides (PG1) | 1.21 | 24.0 | Mollicutes | 16 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| Mycoplasma penetrans (HF-2) | 1.36 | 25.7 | Mollicutes | 17 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
| Mycoplasma pneumoniae (M129) | 0.82 | 40.0 | Mollicutes | 14 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| Mycoplasma pulmonis (UAB CTIP) | 0.96 | 26.6 | Mollicutes | 18 | 0 | 0 | 0 | 1 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| Mycoplasma synoviae (53) | 0.80 | 28.5 | Mollicutes | 15 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| Ureaplasma parvum (ATCC 700970) | 0.75 | 25.5 | Mollicutes | 15 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| Ureaplasma urealyticum (ATCC 33699) | 0.87 | 25.8 | Mollicutes | 14 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| Esherichia coli (K-12 MG1655) | 4.64 | 50.8 | Proteobacteria | 44 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Fusobacterium nucleatum (ATCC 25586) | 2.17 | 27.2 | Fusobacteria | 30 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 |
Orthologous genes were identified using the MBGD (Microbial Genome Database for Comparative Analysis) (http://mbgd.genome.ad.jp/).
The loss of DNA repair systems, a genomic feature that is often observed in bacteria showing genome reduction (30, 31), was evaluated and compared with DNA repair system loss in other bacteria in Firmicutes and Mollicutes. The E. rhusiopathiae genome contains 34 genes related to DNA repair functions (Table 3), which is a greater number of DNA repair-related genes than is present in Mollicutes, which varies from 11 to 33, and is similar to the number of genes in other Firmicutes with reduced genome sizes (Table 3), although these numbers do not always correlate with genome size. It may be noteworthy that, among the DNA repair-related genes that are highly conserved in Firmicutes but are absent in Mollicutes, four (radA, recD, rexA, and rexB homologs) are missing from E. rhusiopathiae.
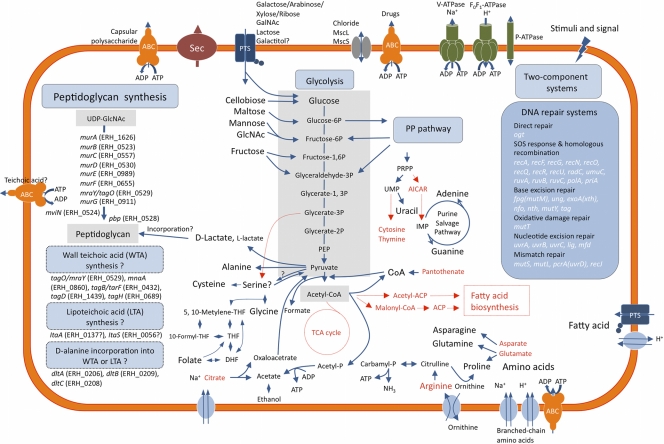
E. rhusiopathiae encodes a full set of enzymes for the glycolysis and pentose phosphate pathways; some enzymes (glucokinase, 6-phosphofructokinase, and phosphoglycerate mutase) are duplicated. However, as expected, the organism lacks numerous genes for many other metabolic pathways (Fig. 3 and Table 4). E. rhusiopathiae lacks all of the genes for the tricarboxylic acid cycle, with the exception of fumarate hydratase (Table 4). Importantly, the organism also lacks all of the genes for the biosynthesis of unsaturated and saturated fatty acids. This genomic feature—the complete lack of genes for fatty acid biosynthesis—is observed in the genomes of all Mollicutes species (with the exception of Acholeplasma laidlawii), but not in other bacteria in the phylum Firmicutes, although several gene losses in fatty acid biosynthetic pathways have been observed in some lactobacilli (25).
Fig. 3.
Overview of the basic metabolic pathways of E. rhusiopathiae. Pathways or steps for which no enzymes were identified are indicated in red, as are the compounds for which de novo synthetic pathways were not identified. The question marks indicate that particular uncertainties exist. ABC, ATP-binding cassette superfamily; Sec, secretion pathway; PTS, phosphotransferase system; GalNAc, N-acetylgalactosamine; GlcNAc, N-acetylglucosamine; MscL, large conductance mechanosensitive ion channel family; MscS, small conductance mechanosensitive ion channel family; PP pathway, pentose phosphate pathway; PRPP, phosphoribosyl-pyrophosphate; AICAR, 5′-phosphoribosyl-4-carboxamide-5-aminoimidazole; PEP, phosphoenolpyruvate; ACP, acyl carrier protein; TCA, tricarboxylic acid; THF, tetrahydrofolate; DHF, dihydrofolate.
Table 4.
Enzymes in the biosynthetic pathways of E. rhusiopathiae
| Category or compound (gene) | Locus tag(s) |
|---|---|
| Energy metabolism | |
| Glycolysis | |
| Glucokinase (glk) | ERH_0239, ERH_1009 |
| Glucose-6-phosphate isomerase (pgi) | ERH_0099 |
| 6-Phosphofructokinase (pfkA) | ERH_1011, ERH_1051 |
| Fructose-bisphosphate aldolase (fba) | ERH_1632 |
| Triose-phosphate isomerase (tpiA) | ERH_1335 |
| Glyceraldehyde 3-phosphate dehydrogenase (gapA) | ERH_1534 |
| Phosphoglycerate kinase (pgk) | ERH_1336 |
| Phosphoglycerate mutase (gpmA) | ERH_0241, ERH_0435, ERH_0457, ERH_1685 |
| Enolase (eno) | ERH_1334 |
| Pyruvate kinase (pyk) | ERH_1010 |
| Pentose phosphate pathway | |
| Glucose-6-phosphate 1-dehydrogenase (zwf) | ERH_0455 |
| 6-Phosphogluconolactonase (pgl) | ERH_0398 |
| 6-Phosphogluconate dehydrogenase (gnd) | ERH_0456 |
| Transketolase (tkt) | ERH_1578 |
| Ribose 5-phosphate isomerase (rpiB) | ERH_1609 |
| Ribulose-phosphate 3-epimerase (rpe) | ERH_1027 |
| TCA cycle | |
| Fumarate hydratase (fumC) | ERH_0730 |
| Amino acid biosynthesis | |
| Proline | |
| Glutamate 5-kinase (proB) | ERH_0054 |
| Glutamate-5-semialdehyde dehydrogenase (proA) | ERH_0055 |
| Pyrroline-5-carboxylate reductase (proC) | ERH_0057 |
| Glutamine | |
| Glutamine synthetase (glnA) | ERH_0836 |
| Alanine | |
| Alanine dehydrogenase (ald) | ERH_1073 |
| Asparagine | |
| Aspartate-ammonia ligase (asnA) | ERH_1353 |
| Cysteine | |
| Serine O-acetyltransferase (cysE) | ERH_0421 |
| Cysteine synthase A (cysK) | ERH_0469 |
| Serine | |
| l-Serine dehydratase (sdaA) | ERH_1322 |
| l-Serine dehydratase (sdaB) | ERH_1323 |
| Glycine | |
| Glycine hydroxymethyltransferase (glyA) | ERH_1608 |
| Arginine (incomplete pathway) | |
| Arginine deiminase (arcA) | ERH_0795 |
| Carbamate kinase (arcC) | ERH_0797 |
| Ornithine carbamoyltransferase (argF) | ERH_0796 |
| Cofactors, vitamins, prosthetic groups and carriers | |
| Pantothenate (incomplete pathway) | |
| 2-Dehydropantoate 2-reductase (panE/apbA) | ERH_1317 |
| CoA | |
| Type III pantothenate kinase (coaX) | ERH_0124, ERH_0188 |
| Phosphopantothenate-cysteine ligase (coaB) | ERH_0126 |
| Phosphopantothenoylcysteine decarboxylase (coaC) | ERH_0125 |
| Pantetheine-phosphate adenylyltransferase (coaD) | ERH_1118 |
| Dephospho-CoA kinase (coaE) | ERH_1014 |
| Folate (incomplete pathway) | |
| Folylpolyglutamate synthase (folC) | ERH_1520 |
| Dihydrofolate reductase (folA) | ERH_0263, ERH_0996 |
| Thiamine (incomplete pathway) | |
| Cysteine desulfurase | ERH_0498, ERH_0508 |
| Nucleoside-triphosphatase | ERH_0545 |
| Thiamine biosynthesis protein (thiI) | ERH_0509 |
It appears that the mechanism by which Tween 80 enhances the growth of E. rhusiopathiae does not involve oleic acid, which is the major ingredient of Tween 80, because we were unable to demonstrate growth enhancement of the organism after adding various quantities of oleic acid. Its growth was completely inhibited by concentrations of oleic acid greater than 0.001% (data not shown). This finding suggests that Tween 80 may merely aid in membrane transport or another nutrient utilization process.
E. rhusiopathiae also lacks many genes for amino acid biosynthesis; the organism can synthesize only seven amino acids (alanine, asparagine, glutamine, serine, cysteine, glycine, and proline) through de novo pathways or using intermediate molecules as derivatives. Moreover, all of the genes for the biosynthesis of biotin, riboflavin, ubiquinone, and menaquinone and some of the genes involved in the biosynthesis of pantothenate, thiamine, and folate are missing from the E. rhusiopathiae genome (Table 4).
In agreement with its poor biosynthetic capacities, E. rhusiopathiae devotes as much as 13.7% (minimally) of its genes to transport functions (Table 5), a level similar to that observed for lactic acid bacteria (13 to 18%) (21). In particular, E. rhusiopathiae contains a remarkably high percentage of primary active transporters (68.4%). This property is also shared with Mycoplasma species (39).
Table 5.
Transporter proteins of E. rhusiopathiae
| Family (% of proteins) | No. of transport systems (no. of genes involved) | Locus tag(s) |
|---|---|---|
| Channels (1.3) | ||
| Chloride channel | 1 (1) | ERH_1566 |
| Large conductance mechanosensitive ion channel family | 1 (1) | ERH_0678 |
| Small conductance mechanosensitive ion channel family | 1 (1) | ERH_0358 |
| Secondary transporters (13.7) | ||
| Major facilitator superfamily | 5 (5) | ERH_0034, ERH_0137, ERH_0232, ERH_0502, ERH_0783 |
| Amino acid transporter family | 5 (5) | ERH_0305, ERH_0316, ERH_0766, ERH_0842, ERH_1618 |
| Arginine/ornithine antiporter | 2 (2) | ERH_0798, ERH_1416 |
| Ethanolamine transporter family | 1 (1) | ERH_0866 |
| Cation diffusion facilitator family | 1 (1) | ERH_0032 |
| Resistance-nodulation-cell division superfamily | 1 (1) | ERH_0839 |
| Small multidrug resistance family | 3 (3) | ERH_0155, ERH_1251, ERH_1256 |
| Dicarboxylate/amino acid: cation (Na+ or H+) symporter family | 1 (1) | ERH_0776 |
| Citrate: cation symporter family | 1 (1) | ERH_0754 |
| K+ transporter (Trk) family | 1 (2) | [ERH_0373, ERH_0374] |
| Nucleobase: cation symporter family | 3 (3) | ERH_0078, ERH_0275, ERH_1333 |
| Chromate ion transporter family | 2 (4) | [ERH_0158, ERH_0159],[ERH_1595, ERH_1596] |
| Phosphate: Na+ symporter family | 2 (3) | ERH_0216, [ERH_1127, ERH_1128] |
| Primary active transporters (68.4) | ||
| ATP-binding cassette superfamily | ||
| Carbohydrate uptake transporter | 7 (14) | ERH_0199, [ERH_0226, ERH_0227, ERH_0228], [ERH_0413, ERH_0414], ERH_0417, [ERH_1083, ERH_1084, ERH_1085], [ERH_1244, ERH_1245, ERH_1246], ERH_1523 |
| Amino acid uptake transporter | 4 (11) | [ERH_0895, ERH_0896], [ERH_1350, ERH_1351, ERH_1352], [ERH_1517. ERH_1518, ERH_1519], [ERH_1666, ERH_1667, ERH_1668] |
| Ion uptake transporter | 7 (16) | [ERH_0024, ERH_0025], [ERH_0044, ERH_0045, ERH_0046, ERH_0047], [ERH_0202, ERH_0203], [ERH_0945, ERH_0946], [ERH_0977, ERH_0978, ERH_0979, ERH_0980], ERH_1017, ERH_1356 |
| Polyamine uptake transporter | 2 (8) | [ERH_0876, ERH_0877, ERH_0878, ERH_0879], [ERH_1495, ERH_1496, ERH_1497, ERH_1498] |
| Peptide/nickel uptake transporter | 1 (5) | [ERH_0442, ERH_0443, ERH_0444, ERH_0445, ERH_0446] |
| Cobalt/nickel uptake transporter | 1 (3) | [ERH_1134, ERH_1135, ERH_1136] |
| Thiamine uptake transporter | 1 (1) | ERH_1061 |
| Ion chelate uptake transporter | 2 (9) | [ERH_0496, ERH_0497, ERH_0498, ERH_0499, ERH_0500], [ERH_1367, ERH_1368, ERH_1369, ERH_1370] |
| Lipoprotein uptake transporter | 1 (1) | ERH_0873 |
| Quaternary amine uptake transporter | 1 (4) | [ERH_1627, ERH_1628, ERH_1629, ERH_1630] |
| Nitrate/sulfonate/bicarbonate uptake transporter | 1 (3) | [ERH_0294, ERH_0295, ERH_0296] |
| Drug exporter | 18 (27) | [ERH_0095, ERH_0096], [ERH_0113, ERH_0114], [ERH_0322, ERH_0323], [ERH_0426, ERH_0427], [ERH_0449, ERH_0450], [ERH_0689, ERH_0690], ERH_0710, ERH_0714, [ERH_0792, ERH_0793], ERH_0939, ERH_1112, ERH_1372, [ERH_1376, ERH_1377], ERH_1381, ERH_1387, [ERH_1411, ERH_1412], ERH_1508, ERH_1683 |
| Peptide exporter | 14 (19) | ERH_0026, [ERH_0465, ERH_0466], ERH_0704, ERH_0733, ERH_0874, ERH_0986, ERH_1182, ERH_1184, [ERH_1186, ERH_1187], ERH_1263, [ERH_1360, ERH_1361], ERH_1383, [ERH_1457, ERH_1458], [ERH_1491, ERH_1492] |
| F-type and V-type ATPase superfamily | 2 (16) | [ERH_0365, ERH_0366, ERH_0367, ERH_0368, ERH_0369, ERH_0370, ERH_0371, ERH_0372], [ERH_1042, ERH_1043, ERH_1044, ERH_1045, ERH_1046, ERH_1047, ERH_1048, ERH_1049] |
| P-type ATPase superfamily | 7 (7) | ERH_0074, ERH_1075, ERH_1200, ERH_1283, ERH_1307, ERH_1338, ERH_1401 |
| Uncharacterized ABC-type transporters | 8 (16) | [ERH_0409, ERH_0410, ERH_0411, ERH_0412], ERH_0819, ERH_0846, [ERH_1002, ERH_1003, ERH_1004], ERH_1052, ERH_1125, ERH_1186, [ERH_1477, ERH_1478, ERH_1479, ERH_1480] |
| Group translocations (16.2) | ||
| Phosphotransferase system (PTS) | 23 (38) | ERH_0005, ERH_0020, [ERH_0133, ERH_0134, ERH_0135], ERH_0219, ERH_0222, [ERH_0255, ERH_0256], [ERH_0282, ERH_0283, ERH_0284], ERH_0286, [ERH_0348, ERH_0349, ERH_0350, ERH_0351], ERH_0680, [ERH_0722, ERH_0723, ERH_0724], ERH_0814, ERH_0849, ERH_0851, ERH_1041, ERH_1120, ERH_1122, ERH_1145, ERH_1208, [ERH_1219, ERH_1220, ERH_1221], ERH_1327, [ERH_1393, ERH_1394, ERH_1395, ERH_1396], ERH_1399 |
| Unclassified (0.4) | ||
| Metal ion transporter (MIT) family | 1 (1) | ERH_1190 |
| Total | 132 (234) |
Virulence-associated genes. (i) Two-component signal transduction systems.
Most bacteria employ two-component signal transduction systems to regulate the expression of many genes in response to various changes in environmental conditions (54). In the E. rhusiopathiae genome, we identified a total of 15 genes encoding response regulators, 14 of which were adjacent to genes encoding cognate histidine kinases (Table 6). The numbers of two-component signal transduction systems in the genomes of Streptococcus pyogenes (12), Streptococcus pneumoniae (57), Listeria monocytogenes (15), Enterococcus faecalis (16), and Bacillus subtilis (10) are 13, 14, 16, 17, and 35, respectively. Compared to these bacteria, lactic acid bacteria contain fewer two-component systems (between five and nine) (9), and Mycoplasma species lack this system entirely, with the exception of Mycoplasma penetrans (42). A loss of regulatory systems is also an evolutionary pattern observed in many bacteria with reduced genome sizes (31). Considering the presence of many two-component systems in the E. rhusiopathiae genome compared to lactic acid bacteria and the important roles of these systems in stress responses, such as oxidative or acid stress responses (9), the two-component systems of E. rhusiopathiae may be very closely associated with its virulence.
Table 6.
Two-component systems in E. rhusiopathiae
| Kinase | Response regulator | Organizationa | Predicted function | Reference |
|---|---|---|---|---|
| ERH_0119 | ERH_0118 | RH | Similar to secretion stress response | 17 |
| ERH_0212 | ERH_0211 | RH | Unknown | |
| ERH_0230 | ERH_0231 | HR | Unknown | |
| ERH_0270 | ERH_0269 | RH | Unknown | |
| ERH_0310 | ERH_0309 | RH | Unknown | |
| ERH_0313 | ERH_0312 | RH | Unknown | |
| ERH_0805 | ERH_0806 | HR | Similar to AgrA family | 35 |
| ERH_0872 | ERH_0871 | RH | Unknown | |
| ERH_0920 | ERH_0921 | RH | Unknown | |
| ERH_0981 | ERH_0982 | RH | Similar to phosphatase assimilation | 26, 27 |
| ERH_1188 | ERH_1189 | RH | Unknown | |
| ERH_1193 | ERH_1194 | RH | Similar to LytTR family | 35 |
| ERH_1319 | Orphanb | |||
| ERH_1430 | ERH_1429 | RH | Unknown | |
| ERH_1493 | ERH_1494 | RH | Unknown |
Organization of each kinase-regulator pair on the E. rhusiopathiae chromosome (RH, 5′ response regulator-3′ histidine kinase; HR, 5′ histidine kinase-3′ response regulator).
Orphan, a response regulator gene that is not directly associated with a histidine kinase gene.
(ii) Cell wall synthesis.
E. rhusiopathiae possesses a full set of the peptidoglycan (PG) biosynthesis genes, which are relatively dispersed throughout the genome (Fig. 3). However, it is not clear whether all the genes involved in the complete biosynthetic pathways of wall teichoic acids (WTA) and lipoteichoic acids (LTA) are present. Furthermore, although the dlt operon encoding the enzymes that catalyze the incorporation of d-alanine residues into WTA or LTA is always composed of dltABCD genes in Gram-positive bacteria (34), the gene order is not conserved, and the dltD gene is missing in E. rhusiopathiae (Fig. 3), suggesting that the organism contains atypical WTA and/or LTA. The dltABCD genes have been detected in all Firmicutes bacteria with small genomes sequenced so far (19, 34), with the exception of Oenococcus oeni, which is a lactic acid bacterium with the smallest genome of all Firmicutes (Table 3). In the future, it will be worthwhile to intensively analyze the structures and chemical compositions of WTA and LTA of E. rhusiopathiae.
(iii) Capsular polysaccharide synthesis.
Immunological and microscopic approaches were used to demonstrate that E. rhusiopathiae produces a capsule that plays an important role in virulence (49, 51, 52); however, its chemical and biological properties are uncharacterized. We previously showed that in the representative acapsular mutant 33H6, which was generated by transposon mutagenesis with Tn916 (52), the transposon was inserted into a gene (now corresponding to ERH_0855) (48). The E. rhusiopathiae genome analysis revealed that this gene is located in a cluster of genes encoding seven proteins (ERH_0855 to ERH_0861) that appear to be involved in capsular polysaccharide biosynthesis (Table 7). Reverse transcription-PCR analysis of the intergenic regions of the seven genes indicates that these genes are transcribed as a polycistronic mRNA forming an operon (data not shown).
Table 7.
BLAST results for E. rhusiopathiae capsular polysaccharide biosynthetic homologs
| Locus tag | Size (aa)a | GenBank accession no. | Gene product | Bacterial species | Hit length/length of best-hit homolog (% aa sequence identity) |
|---|---|---|---|---|---|
| ERH_0855 | 442 | EEG30921 | Sugar transferase | Clostridium methylpentosum | 182/469 (38) |
| ERH_0856 | 590 | ADE82409 | O-antigen polymerase | Prevotella ruminicola | 90/344 (26) |
| ERH_0857 | 401 | EFI08982 | Glycosyltransferase | Bacteroides sp. 3119 | 174/407 (42) |
| ERH_0858 | 337 | EEI59831 | UDP-glucose 4-epimerase (CapD) | Enterococcus faecium | 238/335 (71) |
| ERH_0859 | 369 | EEV61046 | NAD-dependent epimerase/dehydratase | Enterococcus faecium | 224/370 (60) |
| ERH_0860 | 374 | EFF20431 | UDP-N-acetylglucosamine 2-epimerase | Enterococcus faecium | 299/374 (79) |
| ERH_0861 | 390 | EEK97598 | Glycosyltransferase | Bacillus cereus | 172/360 (47) |
aa, amino acids.
(iv) Protein secretion systems and surface-associated or extracellular enzymes/proteins.
Gram-positive bacteria produce a variety of extracellular or cell surface-associated proteins, many of which play important roles in virulence (13). In Gram-positive bacteria, these proteins could be translocated across the membrane via three pathways: the Sec (general secretory)-dependent pathway, the signal recognition particle (SRP)-dependent pathway, and the twin-arginine translocation (Tat) pathway (38).
E. rhusiopathiae encodes homologs for signal peptidase I, signal peptidase II (lipoprotein signal peptidase), SRP, and a full set of Sec proteins. In contrast, as seen in other members of Firmicutes with small genomes (Streptococcus species and lactic acid bacteria) and in Mollicutes (7), E. rhusiopathiae does not contain the Tat secretion system or any recognizable Tat substrates.
During the secretion process in Gram-positive bacteria, many secreted proteins are covalently attached to peptidoglycan through their carboxyl termini by transpeptidases, called sortases, to be exposed as surface proteins (33). E. rhusiopathiae contains a single sortase (ERH_0013) and 21 potential sortase substrates that contain a sortase recognition sequence (LPXTG) followed by a membrane-spanning hydrophobic domain and a positively charged tail (Table 8). They include various hydrolyzing enzymes, such as proteases and peptidases, and proteins that can potentially mediate host-bacterium interactions. Three of these proteins possess KXW repeat modules, as well, which are conserved in several adhesins of Gram-positive bacteria belonging to Firmicutes and have been shown to play important roles in biofilm formation (50).
Table 8.
Possible virulence factors of E. rhusiopathiae
| Locus tag | Gene | Predicted function and/or description | Motif/modulesa |
|---|---|---|---|
| Surface proteins | |||
| ERH_0075 | Collagen-binding protein | LPXTG | |
| ERH_0094 | spaA.1 | Unknown, protective antigen | GW |
| ERH_0150 | hylA | Hyaluronidase | LPXTG |
| ERH_0161 | Peptidase M14 | LPXTG | |
| ERH_0201 | Pectin lyase fold-containing protein | LPXTG | |
| ERH_0221 | Glycoside hydrolase, family 16 | LPXTG | |
| ERH_0260 | Proteinase | LPXTG | |
| ERH_0278 | Unknown | LPXTG | |
| ERH_0299 | nanH.1 | Neuraminidase | LPXTG |
| ERH_0407 | cbpA | Unknown, choline-binding protein | GW |
| ERH_0561 | Glycosyl hydrolase, family 85 | LPXTG | |
| ERH_0668 | rspA | Biofilm formation, protective antigen | LPXTG/KXW |
| ERH_0669 | rspB | Biofilm formation | LPXTG/KXW |
| ERH_0728 | Internalin-like | LPXTG | |
| ERH_0765 | hylB | Hyaluronidase | LPXTG |
| ERH_0768 | cbpB | Unknown, choline-binding protein | GW |
| ERH_0777 | Dipeptidase | LPXTG | |
| ERH_1139 | ushA | 5′-Nucleotidase | LPXTG |
| ERH_1210 | hylC | Hyaluronidase | LPXTG |
| ERH_1258 | Unknown | LPXTG | |
| ERH_1436 | Collagen-binding protein | LPXTG | |
| ERH_1454 | Unknown | LPXTG | |
| ERH_1472 | Internalin-like | LPXTG | |
| ERH_1687 | rspC | Biofilm formation | LPXTG/KXW |
| Antioxidant proteins | |||
| ERH_0162 | tpx | Thiol peroxidase | |
| ERH_0175 | ahpC | Alkyl-hydroperoxide reductase | |
| ERH_0356 | nrdH | Glutaredoxin | |
| ERH_0375 | trxA.1 | Thioredoxin | |
| ERH_1065 | sodA | Superoxide dismutase | |
| ERH_1311 | trxB.1 | Thioredoxin-disulfide reductase | |
| ERH_1345 | ahpD | Alkylhydroperoxide reductase | |
| ERH_1500 | trxA.2 | Thioredoxin | |
| ERH_1541 | trxB.2 | Thioredoxin-disulfide reductase | |
| Phospholipase | |||
| ERH_0072 | Patatin-like phospholipase | ||
| ERH_0083 | Phospholipase/Carboxylesterase family | ||
| ERH_0148 | pldB | Lysophospholipase | |
| ERH_0333 | cls | Cardiolipin synthetase | |
| ERH_0334 | Patatin-like phospholipase | ||
| ERH_0347 | Phospholipase/carboxylesterase | ||
| ERH_0388 | Phospholipase D | ||
| ERH_1214 | Lysophospholipase | ||
| ERH_1433 | Lysophospholipase | ||
| Hemolysins | |||
| ERH_0467 | Hemolysin-related protein | ||
| ERH_0649 | Hemolysin III | ||
| Other extracellular proteins/enzymes | |||
| ERH_0761 | nanH.2 | Neuraminidase | |
| ERH_1034 | Fibronectin-binding protein | ||
| ERH_1356 | Adhesin | ||
| ERH_1467 | Biofilm formation |
Of particular interest are three hyaluronidases (ERH_0150, ERH_0765, and ERH_1210) and one neuraminidase (ERH_0299) that contain LPXTG motifs. Cell surface associations of hyaluronidases and neuraminidase via the LPXTG motif have been shown only in S. pneumoniae (13). It is most likely that these four enzymes of E. rhusiopathiae are also surface associated. Notably, the organism encodes an additional neuraminidase (ERH_0761) that lacks the LPXTG motif and thus is probably extracellularly secreted.
E. rhusiopathiae possesses another type of surface protein with glycine-tryptophan (GW) dipeptide repeat modules, which attach surface proteins to the cell wall through noncovalent interactions (59). In addition to the major surface protective antigen SpaA.1 protein (ERH_0094), E. rhusiopathiae strain Fujisawa contains two surface proteins (ERH_0407 and ERH_0768) with the GW dipeptide repeat modules. Thus, like many other Gram-positive pathogens (13), E. rhusiopathiae elaborates a number of cell surface components and extracellular products to interact with its target cells and initiate infection.
(v) Putative virulence factors required for intracellular survival.
E. rhusiopathiae is vulnerable to oxidative stress, and the escape from reactive oxygen species (ROS) is a major strategy for the intracellular survival of the organism (51). Genome analysis revealed that E. rhusiopathiae possesses nine CDSs encoding enzymes that potentially confer ROS resistance. They include a superoxide dismutase, two thioredoxins, two thioredoxin reductases, a thiol peroxidase, and a glutaredoxin (Table 8). Although E. rhusiopathiae lacks catalase, the organism may have evolved these redundant defense mechanisms against oxidative stress within host cells. Furthermore, the two alkyl-hydroperoxide reductases may be important in protecting the organism against damage caused by nitric oxide (5).
In addition to these anti-ROS proteins, E. rhusiopathiae contains additional enzymes that potentially help the organism survive within phagocytic cells (Table 8). Phospholipases are considered virulence factors in many intracellular pathogens (43). The E. rhusiopathiae genome contains at least nine CDSs encoding proteins with sequence homology to phospholipase family proteins, including phospholipase D and patatin-like phospholipases. Patatin, a storage protein found in potatoes, also has a phospholipase activity (29). E. rhusiopathiae possesses an extremely high number of phospholipolytic enzymes compared to other intracellular bacteria (1). The abundance of these enzymes of E. rhusiopathiae may be a reflection of the lack of fatty acid biosynthesis pathways. Efficient acquisition of fatty acids from the host membrane may be achieved by their combined actions. These phospholipases may allow the organism to escape from phagosomes into the cytoplasm by disrupting the phagosomal membrane (43). In fact, electron microscopic observation revealed that the organism multiplies predominantly within the cytoplasm of macrophages at the inoculation sites of mice (46), suggesting that the cytoplasm serves as a privileged in vivo niche for E. rhusiopathiae, which allows the organism to circumvent host immune responses. Antioxidant enzymes and phospholipases could also modulate immunological cell-signaling pathways (2, 44). Thus, the organism may regulate host cell functions by these enzymes for successful intracellular survival.
Genome comparison.
By bidirectional best-hit analysis with the draft sequence of E. rhusiopathiae strain ATCC 19414, we found that 1,594 (93.5%) CDSs of Fujisawa, including the genes for capsular polysaccharide biosynthesis (ERH_0855 to ERH_0861), are conserved in ATCC 19414. The orthologs exhibited a high level of sequence similarity (99.3% amino acid sequence identity on average). The result of dot plot analysis also revealed a high level of genomic synteny of the two genomes (see Fig. S1 in the supplemental material). These data indicate that the genomic backbone is highly conserved between the two strains.
In ATCC 19414, 71 out of the 1,645 CDSs were strain specific (see Table S1 in the supplemental material). In Fujisawa, 110 CDSs were strain specific (see Table S2 in the supplemental material). These Fujisawa-specific CDSs include the 46 genes on PP_Erh_Fujisawa (ERH_0581 through ERH_0626) and 23 genes that were also clustered in the genome (ERH_1273 through ERH_1295) (Fig. 1; see Table S2 in the supplemental material). Because this gene cluster contains genes for an integrase and a replication protein, as well as several IS elements, it is most likely that it represents an integrative element, although direct-repeat sequences were not found at the boundaries. This integrative element, designated IE_Erh_Fujisawa, includes the genes for a restriction-modification system and a heavy-metal transport system. Moreover, Fujisawa appears to have a strain-specific pathway for polysaccharide biosynthesis (ERH_1439 through ERH_1444). The G+C contents of most of the strain-specific CDSs in the genomes of Fujisawa and ATCC 19414 differ significantly from those of other parts of the genome (see Tables S1 and S2 in the supplemental material), suggesting that they have been acquired by each of the strains through horizontal gene transfer.
We further analyzed conservation of the Fujisawa CDSs in other Erysipelotrichia strains. We excluded the T. sanguinis strain from this analysis because it is very likely that the species is not a true member of Erysipelotrichia, as mentioned above in the discussion of the phylogenetic position of E. rhusiopathiae. This analysis revealed that between 52 and 61% of the CDSs of Fujisawa are conserved in each of these strains (see Table S3 in the supplemental material) and that a total of 625 CDSs (37%) are shared by all Erysipelotrichia strains examined. Although this analysis is not complete because the genome sequence information on all these Erysipelotrichia strains consists of draft sequences with various levels of sequence quality, these 625 CDSs can be regarded as roughly representing the core gene set of the bacterial group. It might be worth mentioning that it appears that these Erysipelotrichia strains also lack the dltABCD operon.
Conclusions.
This study describes the reductive genome revolution of E. rhusiopathiae and its unique strategy for intracellular parasitism. The phylogenetic study using the genome sequence information revealed that Erysipelotrichia strains, including E. rhusiopathiae, form a cluster distinct from other Firmicutes and are phylogenetically closest to Mollicutes. The genomic features of E. rhusiopathiae also represent evolutionary traits of both Firmicutes and Mollicutes. Like Mollicutes, during genome reduction, E. rhusiopathiae lost the genes necessary for fatty acid biosynthesis pathways and evolved redundant phospholipolytic enzymes, which may be utilized for fatty acid acquisition in vivo. Furthermore, the organism possesses many antioxidant factors, suggesting that E. rhusiopathiae specifically adapted to the intracellular environments of phagocytic cells and showing that its evolutionary adaptation is unique in the phylum Firmicutes. Comparative analysis with a draft genome sequence of another E. rhusiopathiae strain (ATCC 19414) revealed that these genomic features are highly conserved in the strain.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Vincent A. Fischetti for critically reading the manuscript and to Ken Kurokawa for helpful suggestions on the phylogenetic data analysis.
This work was supported by a Research and Development Projects for Application in Promoting New Policy of Agriculture Forestry and Fisheries grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (to Y.S.).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org.
Published ahead of print on 8 April 2011.
REFERENCES
- 1. Banerji S., Aurass P., Flieger A. 2008. The manifold phospholipases A of Legionella pneumophila—identification, export, regulation, and their link to bacterial virulence. Int. J. Med. Microbiol. 298:169–181 [DOI] [PubMed] [Google Scholar]
- 2. Bogdan C., Rollinghoff M., Diefenbach A. 2000. Reactive oxygen and reactive nitrogen intermediates in innate and specific immunity. Curr. Opin. Immunol. 12:64–76 [DOI] [PubMed] [Google Scholar]
- 3. Calcutt M. J., Lavrrar J. L., Wise K. S. 1999. IS1630 of Mycoplasma fermentans, a novel IS30-type insertion element that targets and duplicates inverted repeats of variable length and sequence during insertion. J. Bacteriol. 181:7597–7607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540–552 [DOI] [PubMed] [Google Scholar]
- 5. Chen L., Xie Q. W., Nathan C. 1998. Alkyl hydroperoxide reductase subunit C (AhpC) protects bacterial and human cells against reactive nitrogen intermediates. Mol. Cell 1:795–805 [DOI] [PubMed] [Google Scholar]
- 6. Ciccarelli F. D., et al. 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 311:1283–1287 [DOI] [PubMed] [Google Scholar]
- 7. Dilks K., Rose R. W., Hartmann E., Pohlschroder M. 2003. Prokaryotic utilization of the twin-arginine translocation pathway: a genomic survey. J. Bacteriol. 185:1478–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Edgar R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. El-Sharoud W. M. 2005. Two-component signal transduction systems as key players in stress responses of lactic acid bacteria. Sci. Prog. 88:203–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fabret C., Feher V. A., Hoch J. A. 1999. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J. Bacteriol. 181:1975–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Feist H., Flossmann K. D., Erler W. 1976. Investigations on the nutritional requirements of erysipelas bacteria. Arch. Exp. Veterinarmed. 30:49–57 [PubMed] [Google Scholar]
- 12. Ferretti J. J., et al. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. U. S. A. 98:4658–4663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fischetti V. A. 2006. Surface proteins on gram-positive bacteria, p. 12–25 In Fischetti V. A., Novick R. P., Ferretti J. J., Portnoy D. A., Rood J. I. (ed.), Gram-positive pathogens, 2nd ed. ASM Press, Washington, DC [Google Scholar]
- 14. Garrity G. M., et al. 6 March 2007, posting date. Part 8, The Bacteria: phylum Firmicutes: class Mollicutes, p. 317–332 In Garrity G. M. (ed.), The taxonomic outline of Bacteria and Archaea, release 7.7. http://www.taxonomicoutline.org/ [Google Scholar]
- 15. Glaser P., et al. 2001. Comparative genomics of Listeria species. Science 294:849–852 [DOI] [PubMed] [Google Scholar]
- 16. Hancock L., Perego M. 2002. Two-component signal transduction in Enterococcus faecalis. J. Bacteriol. 184:5819–5825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hyyryläinen H. L., et al. 2001. A novel two-component regulatory system in Bacillus subtilis for the survival of severe secretion stress. Mol. Microbiol. 41:1159–1172 [DOI] [PubMed] [Google Scholar]
- 18. Kanehisa M. 1997. A database for post-genome analysis. Trends Genet. 13:375–376 [DOI] [PubMed] [Google Scholar]
- 19. Kovács M., et al. 2006. A functional dlt operon, encoding proteins required for incorporation of d-alanine in teichoic acids in gram-positive bacteria, confers resistance to cationic antimicrobial peptides in Streptococcus pneumoniae. J. Bacteriol. 188:5797–5805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lagesen K., et al. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35:3100–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lorca G. L., et al. 2007. Transport capabilities of eleven gram-positive bacteria: comparative genomic analyses. Biochim. Biophys. Acta 1768:1342–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lowe T. M., Eddy S. R. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ludwig W., Schleifer K. H., Whitman W. B. 2009. Revised road map to the phylum Firmicutes, p. 1–13 In Vos P. D., Garrity G. M., Jones D., Krieg N. R., Ludwig W. (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 3 Springer-Verlag, New York, NY [Google Scholar]
- 24. Lysnyansky I., et al. 2009. Molecular characterization of newly identified IS3, IS4 and IS30 insertion sequence-like elements in Mycoplasma bovis and their possible roles in genome plasticity. FEMS Microbiol. Lett. 294:172–182 [DOI] [PubMed] [Google Scholar]
- 25. Makarova K., et al. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. U. S. A. 103:15611–15616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Makino K., Shinagawa H., Amemura M., Nakata A. 1986. Nucleotide sequence of the phoB gene, the positive regulatory gene for the phosphate regulon of Escherichia coli K-12. J. Mol. Biol. 190:37–44 [DOI] [PubMed] [Google Scholar]
- 27. Makino K., Shinagawa H., Amemura M., Nakata A. 1986. Nucleotide sequence of the phoR gene, a regulatory gene for the phosphate regulon of Escherichia coli. J. Mol. Biol. 192:549–556 [DOI] [PubMed] [Google Scholar]
- 28. Merhej V., Royer-Carenzi M., Pontarotti P., Raoult D. 2009. Massive comparative genomic analysis reveals convergent evolution of specialized bacteria. Biol. Direct. 4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mignery G. A., Pikaard C. S., Park W. D. 1988. Molecular characterization of the patatin multigene family of potato. Gene 62:27–44 [DOI] [PubMed] [Google Scholar]
- 30. Moran N. A. 2002. Microbial minimalism: genome reduction in bacterial pathogens. Cell 108:583–586 [DOI] [PubMed] [Google Scholar]
- 31. Moran N. A., Wernegreen J. J. 2000. Lifestyle evolution in symbiotic bacteria: insights from genomics. Trends Ecol. Evol. 15:321–326 [DOI] [PubMed] [Google Scholar]
- 32. Nakato H., Shinomiya K., Mikawa H. 1986. Effect of C3 depletion on the genesis of thrombocytopenia induced in rats by Erysipelothrix rhusiopathiae. Scand. J. Haematol. 37:18–24 [DOI] [PubMed] [Google Scholar]
- 33. Navarre W. W., Schneewind O. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Neuhaus F. C., Baddiley J. 2003. A continuum of anionic charge: structures and functions of D-alanyl-teichoic acids in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:686–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nikolskaya A. N., Galperin M. Y. 2002. A novel type of conserved DNA-binding domain in the transcriptional regulators of the AlgR/AgrA/LytR family. Nucleic Acids Res. 30:2453–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Noguchi H., Taniguchi T., Itoh T. 2008. MetaGeneAnnotator: detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes. DNA Res. 15:387–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ohyama A., et al. 2006. Bioinformatics tool for genomic era; a step towards the in silico experiments—focused on molecular cloning. J. Comp. Aid. Chem. 7:102–115 [Google Scholar]
- 38. Pallen M. J., Chaudhuri R. R., Henderson I. R. 2003. Genomic analysis of secretion systems. Curr. Opin. Microbiol. 6:519–527 [DOI] [PubMed] [Google Scholar]
- 39. Paulsen I. T., Nguyen L., Sliwinski M. K., Rabus R., Saier M. H., Jr 2000. Microbial genome analyses: comparative transport capabilities in eighteen prokaryotes. J. Mol. Biol. 301:75–100 [DOI] [PubMed] [Google Scholar]
- 40. Pruitt K. D., Tatusova T., Maglott D. R. 2007. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35:D61–D65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rose D. L., Tully J. G., Bove J. M., Whitcomb R. F. 1993. A test for measuring growth responses of mollicutes to serum and polyoxyethylene sorbitan. Int. J. Syst. Bacteriol. 43:527–532 [DOI] [PubMed] [Google Scholar]
- 42. Sasaki Y., et al. 2002. The complete genomic sequence of Mycoplasma penetrans, an intracellular bacterial pathogen in humans. Nucleic Acids Res. 30:5293–5300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schmiel D. H., Miller V. L. 1999. Bacterial phospholipases and pathogenesis. Microbes Infect. 1:1103–1112 [DOI] [PubMed] [Google Scholar]
- 44. Schwarzer N., et al. 1998. Two distinct phospholipases C of Listeria monocytogenes induce ceramide generation, nuclear factor-kappa B activation, and E-selectin expression in human endothelial cells. J. Immunol. 161:3010–3018 [PubMed] [Google Scholar]
- 45. Shimoji Y. 2000. Pathogenicity of Erysipelothrix rhusiopathiae: virulence factors and protective immunity. Microbes Infect. 2:965–972 [DOI] [PubMed] [Google Scholar]
- 46. Shimoji Y. 2004. Erysipelothrix rhusiopathiae, p. 111–116 In Gyles G. L., Prescott J. F., Songer J. G., Thoen C. O. (ed.), Pathogenesis of bacterial infections in animals, 3rd ed. Blackwell Publishing, Ames, IA [Google Scholar]
- 47. Shimoji Y., Asato H., Sekizaki T., Mori Y., Yokomizo Y. 2002. Hyaluronidase is not essential for the lethality of Erysipelothrix rhusiopathiae infection in mice. J. Vet. Med. Sci. 64:173–176 [DOI] [PubMed] [Google Scholar]
- 48. Shimoji Y., Mori Y., Hyakutake K., Sekizaki T., Yokomizo Y. 1998. Use of an enrichment broth cultivation-PCR combination assay for rapid diagnosis of swine erysipelas. J. Clin. Microbiol. 36:86–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shimoji Y., Mori Y., Sekizaki T., Shibahara T., Yokomizo Y. 1998. Construction and vaccine potential of acapsular mutants of Erysipelothrix rhusiopathiae: use of excision of Tn916 to inactivate a target gene. Infect. Immun. 66:3250–3254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shimoji Y., et al. 2003. Adhesive surface proteins of Erysipelothrix rhusiopathiae bind to polystyrene, fibronectin, and type I and IV collagens. J. Bacteriol. 185:2739–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shimoji Y., Yokomizo Y., Mori Y. 1996. Intracellular survival and replication of Erysipelothrix rhusiopathiae within murine macrophages: failure of induction of the oxidative burst of macrophages. Infect. Immun. 64:1789–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shimoji Y., Yokomizo Y., Sekizaki T., Mori Y., Kubo M. 1994. Presence of a capsule in Erysipelothrix rhusiopathiae and its relationship to virulence for mice. Infect. Immun. 62:2806–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stackebrandt E. 2009. Genus I. Erysipelothrix, p. 1299–1306 In Vos P. D., Garrity G. M., Jones D., Krieg N. R., Ludwig W. (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 3 Springer-Verlag, New York, NY [Google Scholar]
- 54. Stock J. B., Ninfa A. J., Stock A. M. 1989. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Rev. 53:450–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Takahashi T., Hirayama N., Sawada T., Tamura Y., Muramatsu M. 1987. Correlation between adherence of Erysipelothrix rhusiopathiae strains of serovar 1a to tissue culture cells originated from porcine kidney and their pathogenicity in mice and swine. Vet. Microbiol. 13:57–64 [DOI] [PubMed] [Google Scholar]
- 56. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 57. Throup J. P., et al. 2000. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol. Microbiol. 35:566–576 [DOI] [PubMed] [Google Scholar]
- 58. Wood R. L. 1999. Erysipelas, p. 419–430 In Straw B. E., D'Allaire S., Mengelng W. L., Taylor D. J. (ed.), Diseases of swine, 8th ed. Iowa State University Press, Ames, IA [Google Scholar]
- 59. Yother J., White J. M. 1994. Novel surface attachment mechanism of the Streptococcus pneumoniae protein PspA. J. Bacteriol. 176:2976–2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.