Abstract
Bacterial sporulation in Gram-positive bacteria results in small acid-soluble proteins called SASPs that bind to DNA and prevent the damaging effects of UV radiation. Orthologs of Bacillus subtilis genes encoding SASPs can be found in many sporulating and nonsporulating bacteria, but they are noticeably absent from spore-forming, Gram-negative Myxococcus xanthus. This is despite the fact that M. xanthus can form UV-resistant spores. Here we report evidence that M. xanthus produces its own unique group of low-molecular-weight, acid-soluble proteins that facilitate UV resistance in spores. These M. xanthus-specific SASPs vary depending upon whether spore formation is induced by starvation inside cell aggregations of fruiting bodies or is induced artificially by glycerol induction. Molecular predictions indicate that M. xanthus SASPs may have some association with the cell walls of M. xanthus spores, which may signify a different mechanism of UV protection than that seen in Gram-positive spores.
INTRODUCTION
When Myxococcus xanthus is starved for nutrients on solid media, hundreds of thousands of cells begin to aggregate and construct a macroscopic fruiting body. Inside this multicellular structure, individual rod-shaped cells begin to differentiate into spherical, dormant spores that are resistant to many types of environmental stress (43, 44). In addition to carbon starvation, it is possible to induce sporulation in liquid-grown M. xanthus cultures by adding glycerol (7). When added to log-phase cultures, glycerol induces individual rod-shaped vegetative cells to undergo rapid and synchronous conversion into spores. Glycerol-induced spores (glycerol spores) assume many of the morphological changes and stress-resistance properties associated with fruiting-body spores (44). Both spore types contain protein U (19), and both sporulation processes induce β-lactamase activity (31). Furthermore, mutation and gene expression studies have revealed a number of loci required for both glycerol-induced and starvation-induced sporulation (21, 29). However, multiple differences in the molecular compositions of the two spore types have been found. Glycerol-induced spores have thinner protective layers (15, 20) and lack the spore coat proteins S and C and intracellular protein W, which are seen in fruiting-body spores (16, 26, 32). While fruiting-body spores each contain two copies of their chromosomes, glycerol-induced spores have variable numbers of chromosome copies, which likely reflect the various replication states of vegetatively growing cells when glycerol induction was initiated (35, 47). Glycerol-induced spores have less intracellular trehalose but more ribosomes than their starvation-induced counterparts (25, 51).
In comparison to their vegetative counterparts, mature M. xanthus spores (called myxospores or microcysts) are substantially more resistant to environmental factors such as heat, desiccation, and UV light (44). M. xanthus spores are structurally complex, with several distinct compartments visible by transmission electron microscopy (TEM) (16, 46). The innermost compartment of a myxospore is the core, which is surrounded by inner and outer membranes, followed by an electron-dense cortex and an outer spore coat.
Studies of spore formation in Gram-positive bacteria can offer only limited clues about how sporulation occurs in Gram-negative bacteria. This is primarily due to two reasons. First, sporulation is an inherently different process between the two groups of bacteria, as a Gram-positive endospore forms inside the protective environment of the mother cells which shields the developing spore from osmotic pressure. Gram-negative spores, however, must maintain the integrity of their cell walls to counter osmotic pressure as they morph from vegetative cells to spherical spores in the absence of a protective mother cell. A second limitation to using Gram-positive sporulation characteristics to formulate hypotheses for Gram-negative sporulation is the paucity of homologs for sporulation genes in myxobacteria. Bacillusanthracis is a model organism extensively used for the study of spore formation. Expression studies suggest that over 500 proteins may be involved in endospore formation in B. anthracis (22), but scarcely any homologs for these proteins exist in M. xanthus. Although it is likely that M. xanthus also uses a large variety of proteins to construct a spore, only a few spore-specific proteins have been identified and shown to play roles in spore development within fruiting bodies (10, 14, 16, 26, 32). Furthermore, most of these M. xanthus sporulation proteins are not required for stress resistance of the spores. In recent studies, we identified the following four proteins that are important for M. xanthus sporulation and stress resistance: CbgA, MspA, MspB, and MspC (4, 46). Using proteome comparisons between vegetative cells and fruiting-body spores, our laboratory recently identified three myxospore proteins, named MspA, MspB, and MspC, that are important for stress resistance (4). Strains lacking mspA, mspB, or mspC formed starvation-induced spores that were more sensitive to heat and SDS detergent than wild-type (WT) spores. However, msp mutant fruiting-body spores showed no defects in resistance to UV light.
UV resistance in Gram-positive spores, such as those produced by Bacillus subtilis, has been studied extensively. B. subtilis spores are 5- to 50-fold more resistant to UV radiation than actively growing cells (41). Much of this enhanced UV resistance is attributed to the action of small acid-soluble proteins called SASPs (36). About a dozen different genes encoding SASPs (called ssp genes) have been discovered in B. subtilis. Orthologs of SASPs have been found in other spore-forming Gram-positive bacteria (3, 8, 23, 24, 33) as well as in non-spore-forming bacteria (48), but no SASP orthologs were found in the M. xanthus genome using pBLAST searches with E value cutoffs of <0.37. SASPs can comprise 5 to 20% of the total spore protein in B. subtilis and are capable of saturation binding of spore DNA (28). These proteins are devoid of secondary structures, which allows for solubility at low pH (11). However, SASPs become significantly α-helical upon binding to DNA (38, 45). In addition to SASPs undergoing conformational changes, SASP binding induces a conformational change in DNA from the B form to the A form (28, 39). Bound SASPs can protect spore DNA from a number of DNA-damaging agents/events such as DNase, H2O2, hydroxyl radicals, hydrolytic deamination, and restriction enzyme digestion (37, 40, 42). The UV photochemistry of B. subtilis spores changes with the presence of SASPs. With SASPs present, the primary photoproduct is the spore photoproduct (SP; thymidyl-thymidine adduct), while in the absence of SASPs, pyrimidine dimers are the primary photoproducts. SPs are much less lethal in Gram-positive spores than pyrimidine dimers, and these SPs are rapidly repaired during spore germination.
Like Gram-positive spores, Gram-negative spores are also more resistant to UV radiation than their vegetative cell counterparts. However, the mechanism for this UV resistance involves proteins different than known SASPs of Gram-positive spores. Here we report the appearance of low-molecular-weight, acid-soluble proteins in the spores of M. xanthus that are associated with enhanced resistance to UV light. These proteins appear to differ depending upon whether M. xanthus spores were generated in a starvation-dependent process (fruiting bodies) or starvation-independent process (glycerol induction). Whereas these proteins do not seem as plentiful as SASPs in Gram-positive spores, these myxospore proteins do seem to share the same properties of intrinsic protein disorder and protection from lethal effects of UV radiation.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1. The wild-type strain DK1622 (17) was used as the parental strain from which mutants were generated. The sapA (mxan_7407) gene was inactivated in strain DK1622 by single-crossover recombination of a kanamycin-resistant plasmid containing an internal PCR-generated fragment of sapA, as previously described (2, 46). Briefly, PCR primers 5′-CAAACAACTGGAAGAACCTGG-3′ (forward primer) and 5′-GTTCACCTCCACGTCATCGC-3′ (reverse primer) were used to amplify a 317-bp internal region of the mxan_7407 gene that was cloned into plasmid pCR2.1-TOPO (Invitrogen) to create plasmid pJD0619. M. xanthus strain DK1622 was electroporated with purified pJD0619 before selecting for single-crossover-event cells on CTTYE agar (1% Casitone, 0.5% yeast extract, 10 mM Tris-HCl [pH 8.0], 1 mM KH2PO4, 8 mM MgSO4, 1.5% agar) with kanamycin (40 μg/ml). Insertion of pJD0619 into the sapA gene was confirmed by PCR and Southern blot analysis (data not shown), and the confirmed sapA mutant was named JD0619. Fruiting-body development occurred on TPM agar, as previously described (2, 4). Induction of spore formation using 0.5 M glycerol was performed as previously described (7).
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| DK1622 | Wild-type motility and development | 17 |
| AG681 | Plasmid insert carried by mspA; Kanr | 4 |
| AG710 | Plasmid insert carried by mspB; Kanr | 4 |
| AG810 | Plasmid insert carried by mspC; Kanr | 4 |
| JD0619 | pJD0619::sapA (plasmid insert carried by sapA) | This study |
| Plasmids | ||
| pCR2.1-TOPO | Kanr cloning vector | Invitrogen |
| pJD0619 | 317-bp fragment extending from bp 74 to 588 of the sapA gene | This study |
Preparation of protein lysates and one-dimensional SDS-PAGE analysis.
M. xanthus cells were grown in CTTYE to Klett readings of 100 before pelleting cells and lysing them by vortexing with 0.1-mm-diameter glass beads in dithiothreitol (DTT) lysis buffer, as previously described (4). Whole cells and large-cell debris was removed by centrifugation at 12,000 × g for 5 min at 4°C before saving clarified supernatants for precipitation. Trichloroacetic acid (TCA) precipitation of myxococcal proteins was prepared using a modification of the procedure originally described by Natarajan et al. (30). Briefly, TCA was added to clarified lysates to a final concentration of 2.5% (wt/vol) before incubation on ice for 45 min and then centrifugation for 20 min at 12,000 × g and 4°C. The insoluble pellet was considered the 2.5% TCA-insoluble fraction, and the supernatant was saved and considered the 2.5% TCA-soluble fraction. Further sequential precipitations of 5% and 25% TCA were performed. TCA-precipitated pellets were washed with 250 μl ice-cold acetone containing 5% HCl, aspirated, dried for 2 min in a 40°C heat block, suspended in 1× SDS sample buffer (125 mM Tris base, 20% glycerol, 2% SDS, 2% β-mercaptoethanol, 0.001% bromophenol blue), and underwent 12% SDS-polyacrylamide gel electrophoresis (PAGE). Resulting gels were stained with Coomassie brilliant blue, and protein bands were photographed and excised for identification.
Identification of separated proteins.
Coomassie blue-stained protein bands were excised from 12% polyacrylamide gels and destained for 2 h in a solution of 50% methanol plus 5% glacial acetic acid in distilled water. Gel bands were dehydrated with acetonitrile, followed by reduction and alkylation with 10 mM DTT plus 50 mM iodoacetamide in 100 mM NH4HCO3, dehydrated, rehydrated in 100 mM NH4HCO3, dehydrated again, and digested with trypsin (20 ng/μl) in ice-cold 50 mM NH4HCO3. Samples were incubated overnight at 37°C with 20 μl of 50 mM NH4HCO3. After this incubation, the solutions containing the digested peptides were desalted and concentrated using C18 ZipTip pipette tips (Millipore). Samples were analyzed by matrix-assisted laser desorption/ionization (MALDI) using the Voyager DE RP system (Applied Biosystems). To identify the proteins, the Mascot database (Matrix Science) was searched for monoisotopic peptide masses between 700 and 4,000 Da detected in the samples.
Electron microscopy.
Scanning electron microscopy (SEM) of fruiting bodies of wild-type M. xanthus and the mutant strain JD0619 lacking sapA was performed as previously described (4). Samples were analyzed by a Hitachi S570 SEM, with images being captured with the PCI quartz imaging program.
Stress resistance assays.
Vegetatively growing cells and fruiting-body and glycerol-induced spores were assayed for UV resistance at a wavelength of 254 nm and an intensity of 31 μW/cm2, as previously described (4). Light microscopy of methylene blue-stained cells was performed to ensure a lack of clumping to cells prior to UV exposure. After cells were subjected to increasing exposure to UV light, they were allowed to recover on CTTYE agar plates with or without kanamycin sulfate (40 μg/ml) before enumeration of survivors by counting the numbers of CFU (numbers of CFU/ml). Spores were also subjected to the stresses of sonication, including heat, SDS, and lysozyme, as previously described (4, 46).
Predictions of protein intrinsic disorder and phosphorylation.
Protein disorder for spore proteins was predicted by the algorithms PONDR VSL2 (34) and PONDR VL-XT (13). TMpred (transmembrane prediction) is an algorithm used to predict membrane-spanning regions (12), and PSIPRED (27) is an algorithm used to predict protein structure; both were applied to the myxospore acid-soluble proteins indentified here.
RESULTS
Acid precipitation identifies fruiting-body spore-specific proteins.
To investigate whether or not M. xanthus expressed acid-soluble proteins, concentrations of trichloroacetic acid (TCA) precipitations were performed on cell lysates from both vegetatively growing cells and from 5-day-old fruiting-body spores. SDS-PAGE analysis showed that all proteins in the vegetative cell lysates precipitated with 2.5% TCA (Fig. 1, lane 2), with no visible acid-soluble proteins present (Fig. 1, lanes 3 and 4). However, two low-molecular-weight proteins in fruiting-body spore lysates were soluble in 2.5% and 5% TCA (Fig. 1, lanes 7 and 8, respectively). The larger band in lanes 7 and 8 (Fig. 1, arrow) was named “small, acid-soluble protein A” (SapA) and was excised, digested with trypsin, and identified by peptide mass fingerprinting. The gene encoding this protein is mxan_7407 (NCBI reference sequence YP_635513), which is described in the recently published M. xanthus genome as encoding a hypothetical protein (9). The sapA gene product contains 207 amino acids, with a calculated molecular mass of 21.6 kDa and a pI value of 7.56. The only protein that shares any homology with SapA is another myxobacterial hypothetical protein in Stigmatella aurantiaca, predicted to have 184 amino acids sharing 39% identity with SapA from M. xanthus. The lower-molecular-mass SapB band shown in Fig. 1 (arrowhead) was identified as the gene product of mxan_3885, which is also listed in the M. xanthus annotated genome as a hypothetical protein.
Fig. 1.
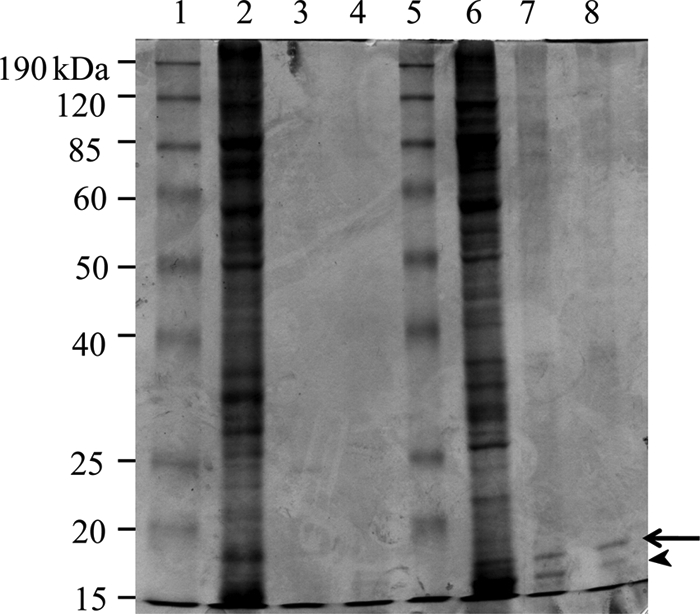
SDS-PAGE (12%) analysis of acid-soluble proteins from wild-type M. xanthus strain DK1622. Vegetative, liquid-grown cells (lanes 2 to 4) and 5-day-old fruiting-body spores (lanes 6 to 8) were washed in Tris-buffered saline, lysed by sonication, and subjected to precipitation by increasing concentrations of trichloracetic acid (TCA). Pelleted proteins were resuspended in running buffer and subjected to SDS-PAGE separation before Commassie blue staining. Protein pellets from 2.5% TCA (lanes 2 and 6), 5% TCA (lanes 3 and 7), and 25% TCA (lanes 4 and 8) are shown. Molecular mass standards are shown in lanes 1 and 5.
Fruiting body myxospores lacking SapA are more sensitive to UV radiation than WT spores.
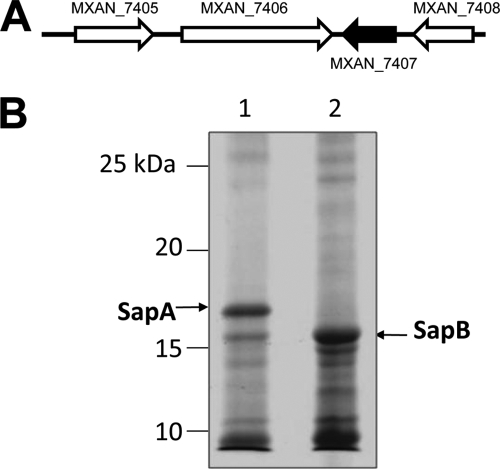
The sapA gene (mxan_7407) appears to be the last gene in an operon (Fig. 2 A). This gene was inactivated by homologous recombination with a plasmid carrying an internal PCR product of mxan_7407, which results in loss of SapA from the TCA-soluble fraction of the fruiting-body spore lysates (Fig. 2B, compare lane 2 to lane 1). Compared to the wild-type strain, the sapA mutant strain JD0619 did not show any alteration in growth rate in liquid CTTYE media (data not shown). However, JD0619 did show moderate alteration in fruiting-body formation on TPM starvation agar (Fig. 3). Compared to wild-type DK1622, fruiting bodies for JD0619 were smaller in size and appeared to have coccoidal spores less densely packed together (compare Fig. 3A to C with Fig. 3F to H). Transmission electron microscopy showed no obvious differences between the mutant and wild-type strains for spore ultrastructure (Fig. 3E and J). Because SapA is similar in both size and acid solubility to B. subtilis SASPs known to protect spores from UV damage, we compared the UV resistance levels of fruiting-body spores from wild-type and sapA mutant strains. While vegetatively growing cells of the two strains were equally sensitive to UV light, the number of surviving sapA mutant spores was nearly 50-fold less than that of WT spores (Fig. 4). This indicates that SapA plays an important role in UV resistance in M. xanthus fruiting-body spores. To determine if SapA enhanced resistance against other forms of stress, fruiting-body spores of DK1622 and JD0619 were further compared for survival. Whereas both spore types showed equal resistance levels to heat (55°C) and sonication (setting of 4.5 with a model 100 Sonic Dismembrator [Fisher]), there was a 3-fold decrease in the survival rate of JD0619 spores with regard to SDS resistance and a 2-fold decrease in lysozyme resistance (data not shown). This suggests that fruiting-body spores of the sapA mutant may have slightly altered surfaces to cause increased sensitivity to this detergent and enzyme.
Fig. 2.
Inactivation of sapA in M. xanthus. (A) Orientation of the sapA gene (mxan_7407) (dark arrow) in the M. xanthus chromosome relative to that of the surrounding genes (image from NCBI; http://www.ncbi.nlm.nih.gov). (B) Commassie blue-stained 12% SDS-PAGE gel showing 2.5% TCA-soluble proteins from 5-day-old fruiting bodies on TPM starvation agar. Lane 1, wild-type strain DK1622; lane 2, mutant strain JD0619. The positions of the SapA and SapB proteins are indicated by arrows.
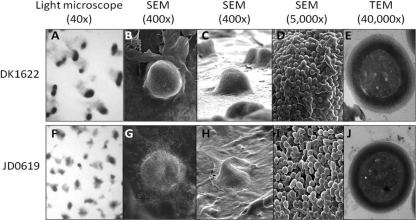
Fig. 3.
Comparison of fruiting bodies and spores between the WT strain (DK1622) and the sapA mutant strain (JD0619). Light microscopy, scanning electron microscopy (SEM), and transmission electron microscopy (TEM) images are shown.
Fig. 4.
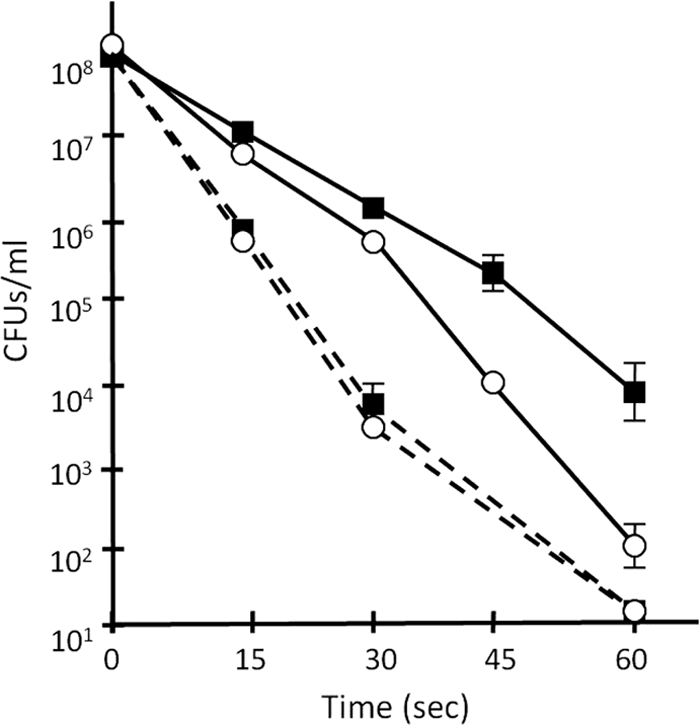
Cells lacking SapA are less resistant to UV exposure. Vegetatively growing cells (dashed lines) and fruiting-body spores (solid lines) were prepared from WT strain DK1622 (solid squares) and the sapA mutant strain (open circles) and subjected to UV irradiation before viable survivors on CTTYE agar plates were recovered. Error bars represent standard deviations.
Acid-soluble proteins of glycerol-induced spores are different from the acid-soluble proteins of fruiting-body spores.
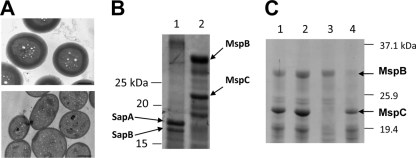
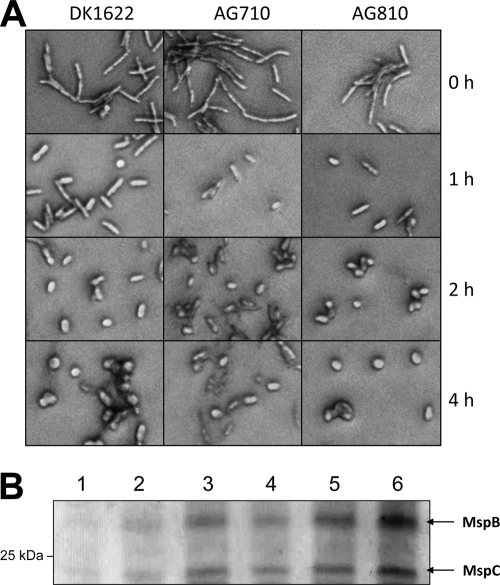
Just as starvation-induced fruiting-body spores were examined for acid-soluble proteins, we also examined glycerol-induced spore lysates. Figure 5 A shows ultrastructural comparisons between fruiting-body spores (top) and glycerol-induced spores (bottom) of a wild-type strain. Both spore types are similar in size and shape, with a noticeable difference being that glycerol-induced spores have a reduced electron-dense outer spore coat. Although SapA and SapB were present in acid-soluble fractions from fruiting-body spores (Fig. 5B, lane 1), both proteins are absent from acid-soluble fractions of glycerol-induced spores (Fig. 5B, lane 2). Two different and higher-molecular-weight, acid-soluble proteins were visible in the glycerol spore fraction, and they were identified by peptide mass fingerprinting as MspB and MspC (Fig. 5B, lane 2). These two proteins were recently discovered in our laboratory and characterized for their roles in spore stress resistance (4). To verify that these two acid-soluble proteins in glycerol-induced spores were MspB and MspC, mutant strains AG710 (mspB mutant) and AG810 (mspC mutant) were induced by 0.5 M glycerol for spore formation, and cell lysates were subjected to TCA precipitations (Fig. 5C). Glycerol spores of AG810 lack MspC in their acid-soluble protein fractions, and spores of AG710 lack MspB in their acid-soluble fractions (Fig. 5C, lanes 3 and 4, respectively). Because MspA was discovered with MspB and MspC under the same sporulation conditions (4), we examined acid-soluble proteins of the mspA mutant strain AG681 but found no difference in protein patterns from those of wild-type cells (Fig. 5C, compare lanes 1 and 2). The absence of mspB or mspC did not affect the rate of glycerol-induced formation of coccid-shaped spores over a 4-h period, as determined by light microscopy (Fig. 6 A). In wild-type DK1622 cells, glycerol induction of MspB and MspC expression is seen early, and these proteins accumulate with prolonged cell exposure to glycerol (Fig. 6B). Therefore, while the MspB and MspC proteins are part of glycerol spores, they are not required for the morphogenesis of glycerol-induced spores.
Fig. 5.
Glycerol-induced spores contain different acid-soluble proteins than spores from fruiting bodies. (A) TEM comparisons between fruiting-body spores (top) and glycerol-induced spores (bottom). Bars = 0.5 μm. (B) Coomassie blue staining and 12% SDS-PAGE showing 2.5% TCA-soluble proteins from 5-day-old fruiting-body spores (lane 1) and glycerol-induced spores (lane 2) of DK1622 cell lysates. (C) SDS-PAGE (12%) showing 2.5% TCA-soluble proteins from glycerol-induced spores of WT cells (strain DK1622; lane 1), the mspA mutant (strain AG681; lane 2), the mspC mutant (strain AG810, lane 3), and the mspB mutant (strain AG710; lane 4). Arrows indicate protein species identified by MALDI-TOF MS.
Fig. 6.
Glycerol-induced expression of MspB (mxan_2432) and MspC (mxan_6969) correlates with glycerol-induced spore formation. (A) Methylene blue-stained cells were photographed at ×1,000 using a phase-contrast microscope. Cells were exposed to 0.5 M glycerol for increasing lengths of time. (B) SDS-PAGE (12%) separation and Coomassie blue staining of 2.5% TCA-soluble proteins from DK1622 cells exposed to 0.5 M glycerol for 0 h (lane 1), 0.5 h (lane 2), 1 h (lane 3), 2 h (lane 4), 3 h (lane 5), and 4 h (lane 6). The positions of proteins MspB and MspC are indicated by arrows.
Glycerol-induced myxospores lacking MspB or MspC are more sensitive to UV radiation.
Previously we reported that fruiting-body myxospores lacking MspB or MspC are not altered in UV sensitivity (4). This is not the case, however, with glycerol-induced mutant spores. While vegetatively growing mutant strains show no difference in UV sensitivity from wild-type cells (Fig. 7, dashed lines), the absence of MspB or MspC from glycerol spores results in an almost 100-fold increase in UV sensitivity compared to that of corresponding wild-type spores (Fig. 7, solid lines).
Fig. 7.
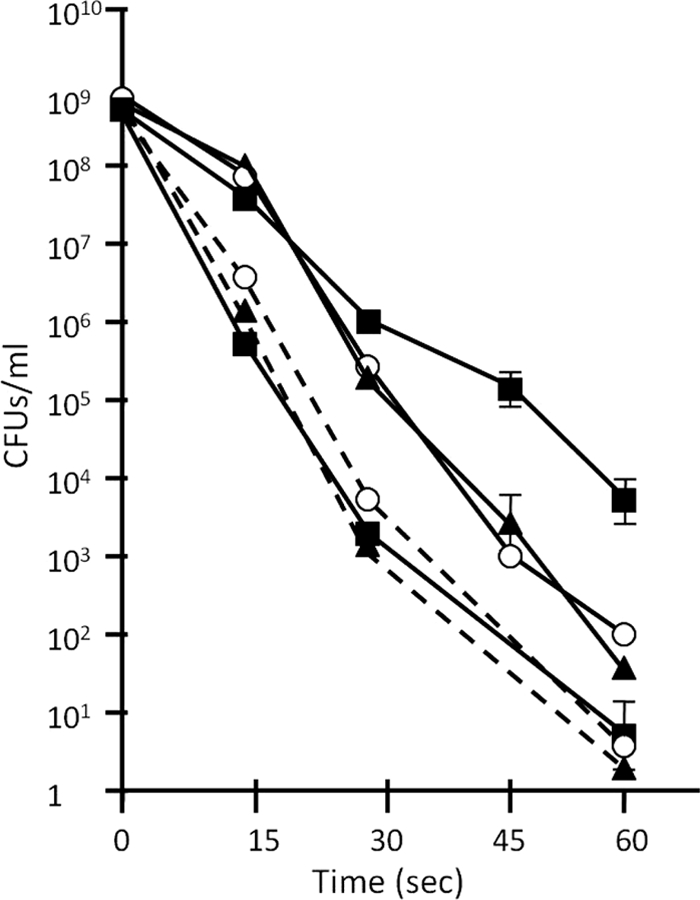
Glycerol-induced mutant spores are more sensitive to UV radiation than glycerol-induced WT spores. The survival rates of vegetative cells (dashed lines) and glycerol-induced spores (solid lines) of DK1622 (squares), AG710 (open circle), and AG810 (triangles) after increasing lengths of UV exposure were compared. Error bars represent standard deviations.
Predictions of acid-soluble protein structures indicate that they have disordered regions and are membrane associated.
Computer programs have been used to predict the level of secondary structure disorder within proteins. Disorder means an absence of predicted secondary structures like α-helices and β-sheets. PONDR (Predictor of Naturally Disordered Regions) is a neural network predictor that was trained with over 1,000 proteins with known intrinsic disorder that has been experimentally confirmed by X-ray crystallography and nuclear magnetic resonance (NMR) analyses (5, 6). The predictor looks at sliding windows of 9 to 21 amino acids, factoring in amino acid composition, hydrophobicity, charge, and other sequence attributes. A PONDR score of 0.0 is an ideal prediction of order, while a score of 1.0 is an ideal prediction of disorder. Any score greater than 0.5 is considered to predict disorder. PONDR analysis predicts regions of order containing α-helices and β strands and large contiguous regions of predicted disorder (see Fig. S1, S2, S3, and S4 in the supplemental material). SapA (see Fig. S2), MspB (see Fig. S3), and MspC (see Fig. S4) each contain potential transmembrane regions, as predicted by TMpred. Recently, it has been shown that sites of protein phosphorylation reside in disordered regions, and this suggests that intrinsic disorder surrounding phosphorylation sites may be a prerequisite for phosphorylation (18, 34). Depp (Disorder enhanced phosphorylation predictor) analysis uses a neural network predictor trained with over 1,500 known (i.e., experimentally confirmed) phosphorylation sites and takes advantage of the observation that residues adjacent to protein phosphorylation sites have sequence attributes similar to those of residues of intrinsic disorder regions. Depp analysis reveals that MspB has 20 Ser and 14 Thr residues in predicted disordered regions (see Fig. S3 in the supplemental material) and that MspC has 5 Ser and 15 Thr residues in disordered regions (see Fig. S4). SapA has five Ser residues in predicted disordered regions (see Fig. S2).
DISCUSSION
Sporulation appears to be such an important phenomenon in the bacterial domain that groups as divergent as Gram-positive and Gram-negative bacteria perform it, albeit through two very different processes. Despite differences in the mechanisms, however, both groups produce spores with UV resistance properties that are appreciably greater than those for vegetatively growing cells. Found in soils around the globe, myxobacteria clearly live in environments exposed to UV radiation. Examination of UV resistance in vegetative M. xanthus cells reveals the possibility that the yellow pigment of vegetative M. xanthus colonies provides some degree of UV protection (1). M. xanthus spores are about 80-fold more resistant to UV light than their vegetative counterparts (44); however, the molecular basis for this enhanced UV resistance in M. xanthus spores is unknown. Whereas an extensive series of studies have examined UV resistance in Gram-positive spores, there are no recognizable homologs for α/β-type SASPs in M. xanthus (no M. xanthus proteins with BLAST E values of <0.38 when the genome was searched with SASP sequences). This is despite the observation that myxospores clearly have enhanced UV resistance compared to that of their vegetative cell counterparts. Evidence is shown here that proteins exist in M. xanthus spores that are absent from vegetative cells and that these proteins are linked to sensitivity to UV radiation. Comparisons of mutant and wild-type strains indicate that SapA contributes to UV resistance in fruiting-body spores (Fig. 4), while MspB and MspC facilitate UV resistance in glycerol-induced spores (Fig. 7). These findings raise a series of questions concerning the function of these proteins that potentially play vital roles in myxobacteria. If SapA, MspB, and MspC are found to be associated with the protective layers of myxospores and not with the spore cytoplasm, then they may signify that a completely different mechanism of UV protection exists in Gram-negative spores than that seen in Gram-positive spores. Unlike the SASPs involved in UV protection in B. subtilis, acid-soluble proteins from myxospores appear to be membrane associated based upon hydropathy analysis (see Fig. S2, S3, and S4 in the supplemental material). However, any such membrane associations for SapA, MspB, and MspC will have to be empirically determined.
MspB and MspC were first identified by proteomic analysis of fruiting-body spores (4), so it was unexpected that they would be present in the TCA-soluble fraction of glycerol spores but not of fruiting-body spores (Fig. 5B). There are at least three possible hypotheses for the absence of MspB and MspC from the acid-soluble fraction of fruiting-body spores. First, it is possible that the relative levels of mspB and mspC expression are much greater in glycerol spores than in fruiting-body spores. Recently, it was shown through microarray analysis that mspC expression increases more than 200-fold during glycerol-induced sporulation (29). Second, it is possible that in fruiting-body spores, MspB and MspC are complexed with proteins or cell structures that precipitate with TCA, while this is not the case in glycerol spores. Lastly, it is possible that MspB and MspC can exist in two different conformational states, with a structured conformation (i.e., precipitated by TCA) in fruiting-body spores and an intrinsically disordered conformation (i.e., soluble in TCA) in glycerol spores. The second and third hypotheses are not incompatible, since disordered proteins will often acquire tertiary structures upon binding with other molecules. SapA and SapB proteins may be missing from the acid-soluble fraction of glycerol spores because they are not expressed in these spores or because they exist in ordered conformations in glycerol spores that result in their precipitation by TCA.
Traditionally, studies of intracellular signaling cascades in bacteria have focused upon the reversible phosphorylations of histidine and aspartate residues in two-component signaling systems. However, there is a growing appreciation of the prevalence of eukaryotic-like serine/threonine kinases in bacteria (49, 50). The M. xanthus genome has revealed upwards of 99 potential protein serine/threonine kinases (PSTKs), which exceeds the number of PSTKs seen in other bacteria (for example, B. subtilis has only 1 PSTK). The complexity of PSTKs in M. xanthus is likely required for the intricate signaling cascades needed for cell movement, development, sporulation, and germination. Although the MspB and MspC proteins lack any homology to eukaryotic PSTKs, these two proteins may be involved in phosphorylation because their numerous Ser and Thr residues reside in predicted disordered regions. However, the roles of these residues in protein function await analysis by site-directed mutagenesis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Christine Davitt for assistance with TEM analysis and Gerhard Munske for assistance with MALDI-TOF MS (time of flight mass spectrometry) identification of proteins. We are especially appreciative for helpful suggestions from Anthony Garza and David Dutton.
This research was supported by internal funds from the University of Minnesota Duluth.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 22 April 2011.
REFERENCES
- 1. Burchard R. P., Burchard A. C., Parish J. H. 1977. Pigmentation phenotype instability in Myxococcus xanthus. Can. J. Microbiol. 23:1657–1662 [DOI] [PubMed] [Google Scholar]
- 2. Caberoy N. B., Welch R. D., Jakobsen J. S., Slater S. C., Garza A. G. 2003. Global mutational analysis of NtrC-like activators in Myxococcus xanthus: identifying activator mutants defective for motility and fruiting body development. J. Bacteriol. 185:6083–6094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cabrera-Martinez R. M., Mason J. M., Setlow B., Waites W. M., Setlow P. 1989. Purification and amino acid sequence of two small, acid-soluble proteins from Clostridium bifermentans spores. FEMS Microbiol. Lett. 52:139–143 [DOI] [PubMed] [Google Scholar]
- 4. Dahl J. L., et al. 2007. Identification of major sporulation proteins of Myxococcus xanthus using a proteomic approach. J. Bacteriol. 189:3187–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dunker A. K., et al. 2008. The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genomics 9(Suppl. 2):S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dunker A. K., et al. 2001. Intrinsically disordered protein. J. Mol. Graph. Model. 19:26–59 [DOI] [PubMed] [Google Scholar]
- 7. Dworkin M., Gibson S. M. 1964. A system for studying microbial morphogenesis: rapid formation of microcyst in Myxococcus xanthus. Science 146:243–244 [DOI] [PubMed] [Google Scholar]
- 8. Fliss E. R., Setlow P. 1985. Genes for Bacillus megaterium small, acid-soluble proteins: nucleotide sequence of two genes and their expression during sporulation. Gene 35:151–157 [DOI] [PubMed] [Google Scholar]
- 9. Goldman B. S., et al. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. U. S. A. 103:15200–15205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gollop R., Inouye M., Inouye S. 1991. Protein U, a late-developmental spore coat protein of Myxococcus xanthus, is a secretory protein. J. Bacteriol. 173:3597–3600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hayes C. S., Alarcon-Hernandex E., Setlow P. 2001. N-terminal amino acid residues mediate protein-protein interactions between DNA-bound alpha/beta-type small, acid-soluble spore proteins from Bacillus species. J. Biol. Chem. 276:2267–2275 [DOI] [PubMed] [Google Scholar]
- 12. Hofmann K., Stoffel W. 1993. TMbase—a database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 374:166 [Google Scholar]
- 13. Iakoucheva L. M., Brown C. J., Lawson J. D., Obradovic Z., Dunker A. K. 2002. Intrinsic disorder in cell-signaling and cancer-associated proteins. J. Mol. Biol. 323:573–584 [DOI] [PubMed] [Google Scholar]
- 14. Inouye S., Franceschini T., Inouye M. 1983. Structural similarities between the development-specific protein S and a Gram-negative bacterium, Myxococcus xanthus, and calmodulin. Proc. Natl. Acad. Sci. U. S. A. 80:6829–6833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Inouye M., Inouye S., Zusman D. R. 1979. Gene expression during development of Myxococcus xanthus: pattern of protein synthesis. Dev. Biol. 68:579–591 [DOI] [PubMed] [Google Scholar]
- 16. Inouye M., Inouye S., Zusman D. R. 1979. Biosynthesis and self-assembly of protein S, a development specific protein of Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 76:209–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kaiser D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 76:5952–5956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim J. H., Lee J., Oh B., Kimm K., Koh I. 2004. Prediction of phosphorylaton sites using SVMs. Bioinformatics 20:3179–3184 [DOI] [PubMed] [Google Scholar]
- 19. Komano T., Inouye S., Inouye M. 1980. Patterns of proteins production in Myxoccus xanthus during spore formation induced by glycerol, dimethyl sulfoxide, and phenethyl alcohol. J. Bacteriol. 144:1076–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kottel R. H., Bacon K., Clutter D., White D. 1975. Coats from Myxococcus xanthus: characterization and synthesis during myxospore differentiation. J. Bacteriol. 124:550–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Licking E., Gorski L., Kaiser D. 2000. A common step for changing cell shape in fruiting body and starvation-independent sporulation of Myxococcus xanthus. J. Bacteriol. 182:3553–3558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu H., et al. 2004. Formation and composition of the Bacillus anthracis endospore. J. Bacteriol. 186:164–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Loshon C. A., Genest P. C., Setlow B., Setlow P. 1999. Formaldehyde kills spores of Bacillus subtilis by DNA damage and small, acid-soluble spore proteins of the alpha/beta-type protect spores against this damage. J. Appl. Microbiol. 87:8–14 [DOI] [PubMed] [Google Scholar]
- 24. Magill N. G., Loshon C. A., Setlow P. 1990. Small, acid-soluble, spore proteins and their genes from two species of Sporosarcina. FEMS Micobiol. Lett. 60:293–297 [DOI] [PubMed] [Google Scholar]
- 25. McBride M. J., Zusman D. R. 1989. Trehalose accumulation in vegetative cells and spores of Myxococcus xanthus. J. Bacteriol. 171:6383–6386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McCleary W. R., Esmon B., Zusman D. R. 1991. Myxococcus xanthus protein C is a major spore surface protein. J. Bacteriol. 173:2141–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McGuffin L. J., Bryson K., Jones D. T. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404–405 [DOI] [PubMed] [Google Scholar]
- 28. Mohr S. C., Sokolov N. V., He C. M., Setlow P. 1991. Binding of small acid-soluble spore proteins from Bacillus subtilis changes the confirmation of DNA from B to A. Proc. Natl. Acad. Sci. U. S. A. 88:77–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Müller F.-D., Treuner-Lange A., Heider J., Huntley S. M., Higgs P. I. 2010. Global transcriptome analysis of sporulation in Myxococcus xanthus reveals a locus necessary for cell differentiation. BMC Genomics 11:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Natarajan S., Xu C., Caperna T. J., Garrett W. M. 2005. Comparison of protein solubilization methods suitable for proteomic analysis of soybean seed proteins. Anal. Biochem. 342:214–220 [DOI] [PubMed] [Google Scholar]
- 31. O'Connor K., Zusman D. R. 1997. Starvation-independent sporulation in Myxococcus xanthus involves the pathway for β-lactamase induction and provides a mechanism for competitive cell survival. Mol. Microbiol. 24:839–850 [DOI] [PubMed] [Google Scholar]
- 32. Otani M., et al. 1998. Protein W, a spore-specific protein in Myxococcus xanthus, formation of a large electron-dense particle in a spore. Mol. Microbiol. 30:57–66 [DOI] [PubMed] [Google Scholar]
- 33. Quirk P. G. 1993. A gene encoding a small, acid-soluble spore protein from alkaliphilic Bacillus firmus OF4. Gene 125:81–83 [DOI] [PubMed] [Google Scholar]
- 34. Romero P., Obradovic Z., Dunker A. K. 2004. Natively disordered proteins: functions and predictions. Appl. Bioinformatics 3:105–113 [DOI] [PubMed] [Google Scholar]
- 35. Rosario C. J., Singer M. 2007. The Myxococcus xanthus developmental program can be delayed by inhibition of DNA replication. J. Bacteriol. 189:8793–8800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Setlow B., Atluri S., Kitchel R., Koziol-Dube K., Setlow P. 2006. Role of dipicolinic acid in resistance and stability of spores of Bacillus subtilis with or without DNA-protective alpha/beta-type small, acid-soluble proteins. J. Bacteriol. 188:3740–3747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Setlow B., Setlow P. 1993. Binding of small, acid-soluble spore proteins to DNA plays a significant role in the resistance of Bacillus subtilis spores to hydrogen peroxide. Appl. Environ. Microbiol. 59:3418–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Setlow B., Setlow P. 1995. Binding of DNA protects alpha/beta-type, small, acid-soluble spore proteins of Bacillus and Clostridium species against digestion by their specific protease as well as by other proteases. J. Bacteriol. 177:4149–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Setlow B., Sun D., Setlow P. 1992. Interaction between DNA and alpha/beta-type small, acid-soluble spore proteins: a new class of DNA-binding protein. J. Bacteriol. 174:2312–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Setlow P. 1992. DNA in dormant spores of Bacillus species is in an A-like conformation. Mol. Microbiol. 6:563–567 [DOI] [PubMed] [Google Scholar]
- 41. Setlow P. 2001. Resistance of spores of Bacillus species to ultraviolet light. Environ. Mol. Mutagen. 38:97–104 [DOI] [PubMed] [Google Scholar]
- 42. Sohail A., Hayes C. S., Divvela P., Setlow P., Bhagwat A. S. 2002. Protection of DNA by alpha/beta-type small, acid-soluble proteins from Bacillus subtilis spores against cytosine deamination. Biochemistry 41:11325–11330 [DOI] [PubMed] [Google Scholar]
- 43. Sudo S., Dworkin M. 1973. Comparative biology of prokaryotic resting cells. Adv. Microb. Physiol. 9:153–224 [DOI] [PubMed] [Google Scholar]
- 44. Sudo S. Z., Dworkin M. 1969. Resistance of vegetative cells and microcysts of Myxococcus xanthus. J. Bacteriol. 98:883–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sussman M. D., Setlow P. 1991. Cloning, nucleotide sequence, and regulation of the Bacillus subtilis gpr gene, which codes for the protease that initiates degradation of small, acid-soluble proteins during spore germination. J. Bacteriol. 173:291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tengra F. K., Dahl J. L., Dutton D., Coyne L., Garza A. G. 2006. Identification and characterization of a cortex biosynthesis gene involved in Myxococcus xanthus spore stress resistance. J. Bateriol. 188:8299–8302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tzeng L., Singer M. 2005. DNA replication during sporulation in Myxococcus xanthus fruiting bodies. Proc. Natl. Acad. Sci. U. S. A. 102:14428–14433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vocero-Villeta A. M., Schilling D. M., Fliss E. R. 1991. Nonsporulating bacterial species contain DNA sequences homologous to the Bacillus spore-specific C-proteins gene. Genomics 9:290–297 [DOI] [PubMed] [Google Scholar]
- 49. Wehenkel A., et al. 2008. Mycobacterial Ser/Thr protein kinases and phosphatases: physiological roles and therapeutic potential. Biochim. Biophys. Acta 1784:193–202 [DOI] [PubMed] [Google Scholar]
- 50. Zhang C. C., Jang J., Sakr S., Wang L. 2005. Proteins phosphorylation on Ser, Thr, and Tyr residues in cyanobacteria. J. Mol. Microbiol. Biotechnol. 9:154–166 [DOI] [PubMed] [Google Scholar]
- 51. Zusman D. 1980. Genetic approaches to the study of development in the Myxobacteria. In Leighton T., Parish H. (ed.), Molecular genetics of development, p. 41–78 Academic Press, New York, NY [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.