Abstract
Pseudomonas aeruginosa uses the quaternary amine choline as a carbon source, osmoprotectant, and macromolecular precursor. The importance of choline in P. aeruginosa physiology is highlighted by the presence of multiple known and putative choline transporters encoded within its genome. This report describes the relative roles of three choline transporters, the ABC transporter CbcXWV and two symporters, BetT1 and BetT3, in P. aeruginosa growth on choline under osmotic conditions that are physiologically relevant to eukaryotic hosts. The increased lag phases exhibited by the ΔbetT1 and ΔbetT1 ΔbetT3 mutants relative to the wild type upon transfer to medium with choline as a sole carbon source suggested roles for BetT1 and BetT3 in cells newly exposed to choline. BetT3 and CbcXWV, but not BetT1, were sufficient to support growth on choline. betT1 and betT3 expression was regulated by the repressor BetI and choline, whereas cbcXWV expression was induced by the activator GbdR and glycine betaine. The data support a model in which, upon transfer to a choline-based medium, the glycine betaine derived from choline taken up by BetT1 and BetT3 promotes subsequent GbdR-mediated cbcXWV induction. Furthermore, growth data indicated that the relative contributions of each transporter varied under different conditions, as BetT1 and CbcXWV were the primary choline transporters under hypo-osmolar conditions whereas BetT3 was the major choline transporter under hyperosmolar conditions. This work represents the first systematic approach to unravel the mechanisms of choline uptake in P. aeruginosa, which has the most complex bacterial choline uptake systems characterized to date.
INTRODUCTION
Pseudomonas aeruginosa utilizes the quaternary ammonium compound (QAC) choline in a variety of biological processes. Choline can serve as a sole carbon, nitrogen, and energy source (29). Once inside the cell, choline is catabolized through the action of BetA and BetB (4) to yield glycine betaine (GB) which, along with other choline catabolites, can function in the regulation of genes encoding virulence determinants, including the hemolytic phospholipase C (PlcH) and phosphorylcholine phosphatase (PchP) (40). Choline can be used as a potent osmoprotectant due to its conversion to the compatible solute GB (8, 10, 27). Choline also serves as a biosynthetic precursor for phosphatidylcholine (44), a membrane phospholipid often present in bacteria that form intimate associations with eukaryotic hosts (34). The choline derivative phosphorylcholine (ChoP) is an important outer surface component for virulence in several pathogens of the human respiratory tract (9, 36), and the decoration of surface-exposed Ef-Tu with ChoP in P. aeruginosa has been implicated in its interaction with host cells (3).
P. aeruginosa cannot synthesize choline de novo and thus must acquire it from its environment. Potential infection sites in the lungs, urinary tract, skin, and eyes contain choline, and choline can be derived from abundant host molecules, such as phosphatidylcholine, acetylcholine, and phosphorylcholine (21, 23, 26, 46, 47). Choline may be similarly plentiful in plants, some of which also serve as hosts for P. aeruginosa (24), and is known to be released from plant roots and from dead and decaying plant and animal tissues (2, 15).
Gram-negative bacteria mainly use two types of transporters for the uptake of choline and related quaternary amine compounds: the binding protein-dependent ATP-binding cassette (ABC) transporters and the betaine-choline-carnitine transporters (BCCTs). These are typically energized by ATP and proton/sodium-motive force, respectively (45, 48). Previously, we characterized the full set of choline transporters present in the phytopathogen Pseudomonas syringae. This set includes one BCCT family transporter, BetT, and two ABC transporters, OpuC and CbcXWV. OpuC and BetT function primarily in uptake for osmoprotection under hyperosmolar conditions, whereas CbcXWV functions mainly in uptake for catabolism under hypo-osmolar conditions. The core CbcWV transporter components interact not only with CbcX but also with two additional periplasmic solute binding proteins, BetX and CaiX, which are encoded elsewhere in the genome and bind GB and carnitine, respectively (7). Orthologs of the P. syringae BetT, CbcXWV, and OpuC transporters are highly conserved among fluorescent pseudomonads, including P. aeruginosa, and expression of the P. aeruginosa homologs of betT (betT3), cbcXWV, and opuCA complemented a choline transport-deficient P. syringae strain (6, 7).
Choline uptake in P. aeruginosa is more complex than in P. syringae due to the presence of not only OpuC (PA14_13580 to PA14_13600) and CbcVWX (PA14_71000 to PA14_71030), but also three BCCTs, designated BetT1, BetT2, and BetT3 (PA14_70980, PA14_12990, and PA14_69850, respectively). We previously showed that BetT2 and BetT3 transport GB and choline, respectively, and confer osmoprotection when expressed in P. syringae in a hyperosmolar environment (6). In this report, we examine the roles of P. aeruginosa choline transporters in choline-dependent growth and uptake at an osmolarity of 280 mOsm/kg/H2O, which is relevant to a mammalian host (13). Our results support a model of coordinated regulation of the expression and activity of CbcVWX, BetT1, and BetT3. Two transcriptional regulators, the activator GbdR (40) and the repressor BetI (17), contribute to the control of transporter expression. In addition, this is the first report characterizing BetT1 activity. We also demonstrate dramatic changes in the relative contributions of the transporters to choline uptake at distinct osmolarities. Together, these studies provide insight into how P. aeruginosa, a pathogen of multiple species, including animals and plants, acquires choline, particularly at an osmolarity likely relevant to many tissues in its eukaryotic hosts (13).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. The P. aeruginosa and Escherichia coli strains were maintained on LB medium at 37°C. A modified morpholinepropanesulfonic acid (MOPS) minimal medium (22) that contains 75 mM NaCl (referred to as MOPS-P) was used to achieve a physiologically relevant osmolarity, 280 mOsm/kg/H2O (13). -21C medium, with an osmolarity of 95 mOsm/kg/H2O (12), was used as the low-osmolarity medium, and MOPS minimal medium with 750 mM NaCl and an osmolarity of 1,505 mOsm/kg/H2O was used as a hyperosmolar medium (MOPS-H). Glucose, pyruvate, choline, or GB was added as the sole source of carbon at 20 mM. Pyruvate or glucose was used as a carbon source (10, 28) when preparing inocula for growth curve analyses in media with choline or GB. The osmolality of the medium was determined using an osmometer (Osmomette A; Precision Systems Inc., Sudbury, MA). When necessary, antibiotics were used at the following concentrations (in μg ml−1): gentamicin (75) and kanamycin (500) for P. aeruginosa and gentamicin (15) and carbenicillin (75) for E. coli. All liquid cultures were grown at 37°C with vigorous aeration.
Table 1.
Strains and plasmids
| Strain or plasmid | Description | Strain no. | Source or reference |
|---|---|---|---|
| P. aeruginosa PA14 strains | |||
| WT | P. aeruginosa PA14 wild type | DH122 | 25 |
| ΔcbcV | In-frame deletion mutant of cbcV | DH1578 | This study |
| ΔT1 (ΔbetT1) | In-frame deletion mutant of betT1 | DH1579 | This study |
| ΔT3 (ΔbetT3) | In-frame deletion mutant of betT3 | DH1580 | This study |
| ΔcbcVΔT1 | In-frame deletions of cbcV and betT1 | DH1584 | This study |
| ΔcbcVΔT3 | In-frame deletions of cbcV and betT3 | DH1581 | This study |
| ΔT1ΔT3 | In-frame deletions of betT1 and betT3 | DH1583 | This study |
| T1::pMQ | betT1::pMQ89 disruption mutant; Gmr | DH1593 | This study |
| ΔT3T1::pMQ | In-frame deletion mutant of betT3, betT1::pMQ89 disruption mutant; Gmr | DH1597 | This study |
| ΔcbcVΔT3T1::pMQ | ΔVΔT3 with a betT1::pMQ89 disruption mutation; Gmr | DH1596 | This study |
| ΔgbdR | In-frame deletion of gbdR | DH466 | 42 |
| betI::TnM | betTI::TnM; Gmr (mutant ID no. 42999) | DH1575 | 19 |
| Plasmids | |||
| pUCP22 | High-copy-number Pseudomonas-stabilized vector; Gmr | 32 | |
| pEX18Gm | Integrating vector in P. aeruginosa; Gmr | 31 | |
| pMQ89 | Suicide vector in P. aeruginosa; Gmr | 33 | |
| pME6041 | Broad-host-range vector; Kmr | 7 | |
| pcbcXWV | pME6041 with cbcXWV from P. aeruginosa PAO1; Kmr | 7 | |
| pbetT3 | pME6041 with betT3 from P. aeruginosa PAO1; Kmr | 6 | |
| pbetT1 | pUCP22 with PA14 betT1 ORF as defined by start site in original PAO1/PA14 annotations; Gmr | This study | |
| pbetT1N | pUCP22 with PA14 betT1 ORF as defined by alternative upstream start site; Gmr | This study |
Generation of mutant strains and complementation constructs.
In-frame deletion constructs for cbcV, betT1, and betT3 were generated using splice overlap extension (SOE) PCR to amplify and splice the 1-kb regions immediately upstream and downstream of each locus. The primers (listed in Table S1 of the supplemental material) were designed to delete the regions between the first and last few amino acids of each open reading frame (ORF). The amplified fragment for each deletion construct was cloned into the pEX18-Gm vector, and the deletion mutations in P. aeruginosa were obtained by recombination, as described previously (31). Briefly, the pEX18-Gm constructs were transformed into E. coli strain S17/λpir and then conjugated into the recipient P. aeruginosa strain. Single-crossover (SCO) derivatives were selected by growth on gentamicin and were then plated on LB medium containing 5% sucrose with no NaCl to obtain double recombinants. The various in-frame deletion mutants were identified by PCR. SCO mutants of betT1 were created by using plasmids based on pMQ89 as described previously (33). The primers (listed in Table S1 of the supplemental material) were designed to amplify a 500-bp region within betT1, and the amplicon was cloned into pMQ89. Plasmids were propagated in E. coli S17/λpir. The pMQ89 derivatives were conjugated into P. aeruginosa, and SCO mutants were selected by growth on gentamicin. All of the mutants generated in this study grew like the parental strain in medium containing pyruvate as the sole carbon source (see Fig. S3A in the supplemental material). Complementing plasmids for betT1 were created using the pUCP22 vector (43). To create pBetT1, the original predicted ORF of betT1 was amplified using primers betT1Fw (5′-GTGTTCTTCGGCTCGACCGC-3′) and betT1Rev (5′-TCAGGTCTGGTGCTGGGTTGG-3′). To create pBetT1N, which had an alternative start site 54 bp upstream of the annotated start site, the betT1 ORF was amplified using primers betT1AltFw (5′-ATGAGTACTGCTTCGCCAATA-3′) and betT1Rev (5′-TCAGGTCTGGTGCTGGGTTGG-3′). The amplicons were cloned into PCR2.1 (Invitrogen), excised from PCR2.1 by XbaI and HindIII, and ligated into pUCP22.
Growth curve analysis.
Strains were grown overnight in MOPS-P medium with either 20 mM glucose or 20 mM pyruvate as the sole carbon source. Stationary-phase cultures were centrifuged, and the cell pellets were washed twice with MOPS buffer and resuspended in an equal volume of the same buffer. The assay media were inoculated at an initial optical density at 600 nm (OD600) of 0.015 and grown at 37°C with vigorous shaking. Aliquots were taken at the indicated time points, and the optical densities (OD600) were measured using a spectrophotometer (Spectramax M2; Molecular Devices, Sunnyvale, CA).
Transport assays.
[methyl-14C]choline (specific activity of 55 mCi/mmol) was obtained from American Radiolabeled Chemicals, Inc. (St. Louis, MO). For the E. coli MKH13 derivatives, choline uptake was measured in M63 medium (22) as described previously (5, 6); for P. aeruginosa PA14 derivatives, choline uptake was similarly evaluated following the transfer of cells from overnight cultures grown in MOPS-P medium with 20 mM glucose into MOPS-P medium with or without 20 mM choline as inducer and grown at 37°C for 6 h. The cells were harvested and the pellets were washed three times in an iso-osmolar buffer, M9 medium (22) without glucose, before resuspension in MOPS-P medium to an OD600 of 1.0. A 5-μl aliquot of [14C]choline (final concentration, 1 mM) was added to 0.5 ml of cells, followed by shaking for 5 min, and uptake was terminated by centrifugation at 13,000 × g. Pellets were washed once with MOPS-P buffer and resuspended in 2 ml of ScintiVerse BD (Fisher Scientific, Fair Lawn, NJ). The amount of radiolabel in the cells was determined using a liquid scintillation counter (Tri-Carb liquid scintillation analyzer, model 2100TR; Packard Instrument Co., Meriden, CT), as previously described (5), and expressed as the initial uptake rate (in nmol choline/min/mg of protein).
RNA isolation and quantitative real-time PCR.
Cells were grown as indicated, and RNA was isolated using an RNeasy kit (Qiagen). RNA was treated with 2 μl of RQ1 DNase (Promega) for 1 h at 37°C, followed by phenol-cholorform extraction. The resulting RNA was subjected to PCR to verify the absence of contaminating DNA and quantified using a Nanodrop spectrophotometer. cDNA was synthesized using SuperScript III (Invitrogen), 750 ng of starting RNA, and a 5′-NSNSNSNSNS-3′ primer instead of random hexamers. The regimen for cDNA synthesis was 25°C for 5 min, 52°C for 60 min, and 70°C for 15 min. The primers used are listed in Table S1 of the supplemental material. The PCR regimen was 94°C for 5 min and 40 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s. Quantitative real-time reverse transcription-PCR (qRT-PCR) was conducted with Power SYBR green and AmpliTaq Gold DNA polymerase according to the manufacturer's instructions (Applied Biosystems). The resulting threshold cycle (CT) values were calculated by using the ABI Prism 7700 sequence detection system and analyzed using the relative standard curve method (for separate tubes) as described in ABI user bulletin no. 2. Briefly, amplification profiles of each of the primer pairs were generated in duplicate with a 10-fold dilution series of pooled stock cDNA. A standard curve was generated for each primer pair by regression analysis as a basis for further quantification of target transcripts in each sample. The rpoD transcript served as a stable housekeeping control transcript for all the experiments (30). The induction of mRNA was determined from the threshold values, which were normalized for rpoD expression. A dissociation curve step was included at the end of each PCR to verify that only a single product was amplified.
RESULTS
P. aeruginosa CbcXWV is essential for growth on GB but not choline at a physiologically relevant osmolarity.
To examine the roles of the P. aeruginosa transporters at an osmolarity that is likely encountered when living in association with mammalian hosts, we amended MOPS medium with NaCl to an osmolarity comparable to that of blood and tissue fluids (280 mOsm/kg/H2O), forming MOPS-P (13). In MOPS-P, the PA14 ΔcbcV mutant, which lacks the ATPase required for a functional CbcXWV transporter, did not grow on GB (Fig. 1 A), consistent with our previous observation that the core transporter CbcWV, in association with the periplasmic substrate binding proteins CbcX and BetX, functions as the major GB transporter for catabolism-associated uptake (7). In contrast, the ΔcbcV mutant grew on choline, where it exhibited a longer lag phase (9 h) relative to the wild type (WT; 4.5 h) (Fig. 1A). The growth defects of the ΔcbcV strain could be rescued by genetic complementation with cbcXWV expressed in trans (see Fig. S1A in the supplemental material). Following initiation of exponential growth, the doubling time of ΔcbcV was slightly longer than that of the wild type (3 h versus 2.4 h), and growth assays using ΔcbcV cells from cultures grown on choline verified that the delayed growth was not due to suppressor mutations (data not shown). These data indicate that one or more transporters in addition to the CbcXWV transporter contribute to growth on choline at a physiologically relevant osmolarity.
Fig. 1.
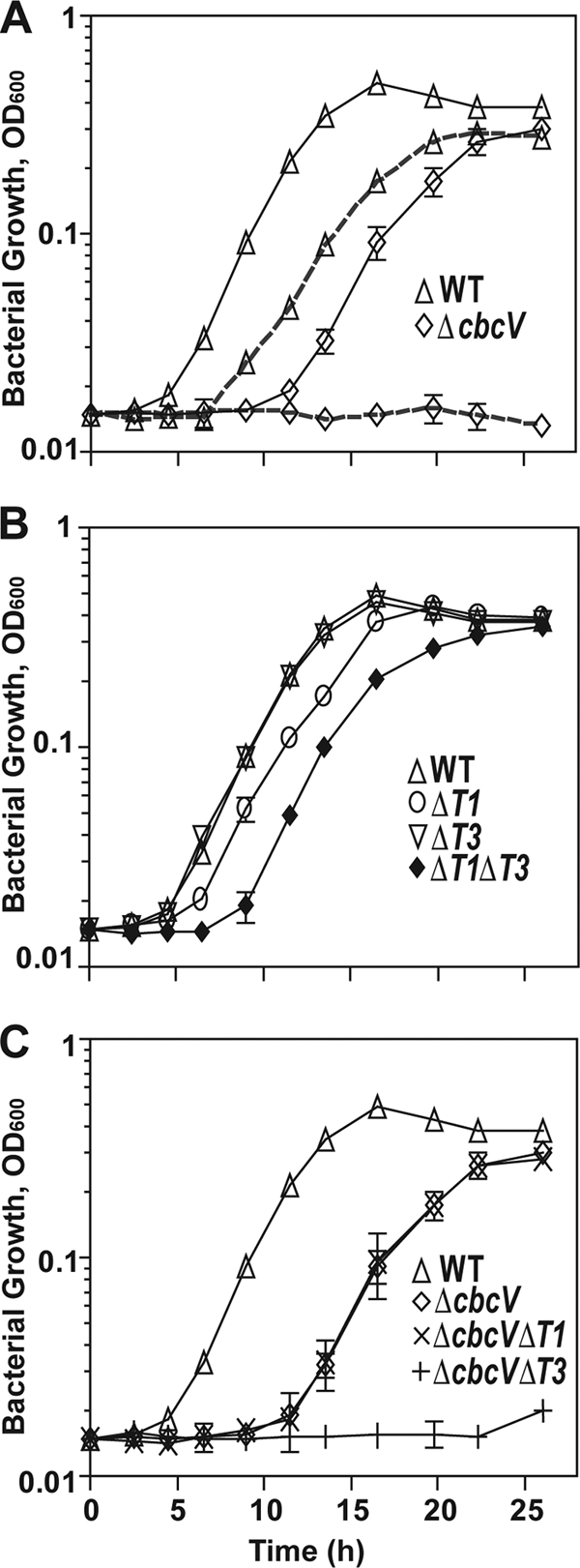
Growth curve analysis of PA14 and various transporter mutants on choline and glycine betaine. (A) Comparison of WT and ΔcbcV grown in MOPS-P medium amended with 20 mM either choline (solid lines) or glycine betaine (dashed lines). (B) Comparison of ΔbetT1, ΔbetT3, and ΔbetT1 ΔbetT3 grown in MOPS-P medium amended with 20 mM choline. (C) Comparison of ΔcbcV, ΔcbcV ΔbetT1, and ΔcbcV ΔbetT3 transporter mutants grown in MOPS-P medium amended with 20 mM choline. Values represent the mean optical densities at 600 nm from three independent replicates, and error bars depict the standard deviations. In some cases, the error bars are obscured by the symbol.
Despite the fact that choline is converted to GB as it is catabolized for growth (38, 41), wild-type cells exhibited a shorter lag phase when grown on choline than on GB as the sole source of carbon (4 h compared to 6.5 h) (Fig. 1A). However, after adaptation to the new carbon source, the growth rates were similar on both compounds. These results suggest that the initial transport rate for choline is faster than for GB.
BetT1 and BetT3 contribute to choline acquisition for catabolism at a physiologically relevant osmolarity.
To determine the full complement of choline transporters in P. aeruginosa that function under physiological conditions, we first analyzed the expression levels of known and putative QAC transporters in MOPS-P with glucose alone or glucose with either choline or GB as the growth substrate (Fig. 2). The cbcV transcript levels were ∼15- and 4-fold higher in medium with choline and GB, respectively, compared to the glucose-only control cultures (Fig. 2). The betT1 and betT3 transcripts were induced more than 20-fold in medium with choline compared to medium with glucose alone or glucose and GB (Fig. 2). Neither choline nor GB affected levels of betT2 and opuCA transcripts in MOPS-P medium (Fig. 2).
Fig. 2.
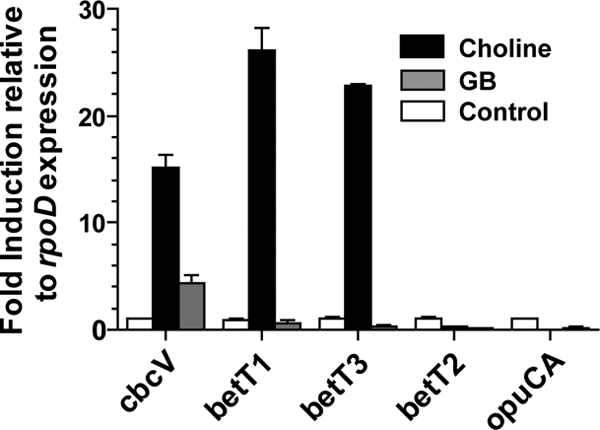
qRT-PCR analysis of expression of QAC transporters in the presence of choline and GB. The strains were grown in MOPS-P medium with 20 mM glucose and 10 mM choline or GB for 5.5 h. Medium without any quaternary ammonium compounds served as the control condition. The qRT-PCR data represent the means obtained from biological duplicates. Expression levels are shown relative to levels of the housekeeping gene rpoD. All ratios were normalized to expression levels in medium without QAC (set to 1). Error bars are the standard deviations.
To characterize the contribution of the BetT1 and BetT3 transporters to growth, we tested the growth kinetics of ΔbetT1 and ΔbetT3 strains in MOPS-P medium with choline. ΔbetT1 cultures showed a longer lag than the wild type (Fig. 1B) but achieved a similar growth rate and final yield (Fig. 1B). The same phenotype was observed in a betT1 insertional mutant, betT1::pMQ89 (see Fig. S2 in the supplemental material), suggesting that this reduced growth on choline was not a result of an unintended deletion of promoter elements of the divergently transcribed betIBA locus, which encodes the genes involved in choline catabolism. These results suggest that BetT1 is involved in choline uptake in PA14 under physiologically relevant conditions. However, repeated attempts to complement the P. aeruginosa betT1 growth phenotype with a plasmid expressing the structural betT1 gene from PAO1 as identified in the original annotation (18) were unsuccessful. Furthermore, P. syringae GB4, a strain deficient in choline uptake, did not result in any choline uptake activity under low-salt (C. Chen and G. A. Beattie, unpublished data) or high-salt conditions (6). This apparent contradiction prompted us to evaluate whether the published ORF for betT1 was annotated correctly. Sequence analysis showed that the N-terminal amino acid sequence of PA5375 (BetT1) lacked 18 amino acids that were present in closely related homologs (accession numbers YP_002875176, NP_795000, and YP_001666502); moreover, a potential alternative start site was found 54 bp upstream of the annotated start site. To evaluate if these additional 18 amino acids are required for an active form of BetT1, we cloned both ORFs and expressed them in E. coli MKH13 (14), which is deficient in choline uptake. A clone expressing betT1 from the alternative start site (pbetT1N) was able take up [14C]choline in M63 medium with an osmolarity comparable to MOPS-P (5.0 ± 1.4 nmol/mg of protein in 10 min), whereas a clone expressing betT1 (pbetT1) was not (0.02 ± 0.01 nmol/mg of protein in 10 min).
When BetT1 (from pbetT1N) was expressed in MKH13, activity was detected even in a minimal medium lacking sodium amendment (data not shown), consistent with proton rather than sodium symport activity, as observed for other BCCT choline transporters (48). Furthermore, BetT1 uptake activity decreased in experiments in which the osmolarity of the assay buffer was increased by increments of 0.2 M NaCl (data not shown). No increase in uptake rate was observed as the choline availability increased from 10 μM to 1 mM (data not shown), suggesting that BetT1 is a low-capacity choline transporter, unlike the exceptionally high capacity of P. syringae BetT (6). BetT1 also appears to be a high-affinity transporter, based on its ability to take up a significant amount of choline (5 nmol/mg of protein in 10 min) when MKH13 cells expressing betT1 are provided with only 10 μM [13C]choline; this is in comparison to our previous finding of low-affinity transport by the P. syringae BetT transporter (6).
Unlike the delayed growth observed for ΔbetT1 cultures, a ΔbetT3 strain was not altered in growth compared to the wild type (Fig. 1B). To further analyze the relative roles of CbcXVW, BetT1, and BetT3 in choline transport, we made a series of double and triple mutants (listed in Table 1) and assessed their growth in medium with choline. The ΔbetT1 ΔbetT3 mutant showed a longer lag and grew at a slightly lower rate than the WT and single ΔbetT1 and ΔbetT3 mutants (Fig. 1B). Genetic complementation with betT1 (pbetT1N) (see Fig. S1B in the supplemental material) or betT3 (see Fig. S1C) expressed in trans restored the phenotype of the ΔbetT1 ΔbetT3 mutant to that of the relevant single mutants. The longer lag phase of the double mutant indicated that both BetT1 and BetT3 contribute to the early uptake of choline, i.e., to its uptake during the lag phase following a shift to choline as a carbon source, since the ΔbetT1 ΔbetT3 mutant exhibited a more profound defect in initiation of growth on choline than either single mutant.
To better assess the activities of BetT1 and BetT3 during growth on choline, the betT1 and betT3 genes were also deleted in a ΔcbcV background. The ΔcbcV ΔbetT1 double mutant exhibited a longer lag and slower growth rate (doubling time of 3 h), similar to that of the ΔcbcV mutant (Fig. 1C). However, the ΔcbcV ΔbetT3 mutant failed to grow with choline as the sole source of carbon even by 22 h (Fig. 1C), as did a triple mutant lacking all three transporters (see Fig. S2 in the supplemental material). All of the single and double mutants grew like the wild type in minimal medium with pyruvate, indicating that the defects are specific to choline uptake (see Fig. S3A in the supplemental material). These data suggest that the growth on choline observed in the ΔcbcV mutant background depends on BetT3 and not BetT1. Collectively, these studies show that CbcXWV, BetT1, and BetT3 can all contribute to choline uptake for catabolism under physiologically relevant osmolarities, but they also show that BetT1 is distinguished by its inability to support growth when CbcXWV and BetT3 are both absent (Fig. 1C), possibly due to a low transport capacity, its undetectable contribution to growth when CbcXWV and BetT3 are both present (Fig. 1B), and its ability to partially compensate for the loss of BetT3 when CbcXWV is present.
Both BetT1 and BetT3 contribute to early uptake of choline.
The lengthened lag phases of the ΔbetT1 and ΔbetT1 ΔbetT3 mutants following transfer to choline (Fig. 1B) suggest that their activities contribute to the acquisition of choline during this period. To evaluate this hypothesis, we measured the uptake rates of [14C]choline, with and without 6 h of preinduction with choline, of the various mutants in MOPS-P. In uninduced cells provided with a high concentration of choline (1 mM), choline uptake was significantly reduced in mutants lacking a functional betT3 gene (Fig. 3). Furthermore, the lack of a difference between the double (ΔbetT1 ΔbetT3) or triple (ΔcbcV ΔbetT3 betT1::pMQ89) mutants and the ΔbetT3 single mutant suggests that BetT3 was the major contributor to choline transport in uninduced cells under these conditions (Fig. 3). In cells incubated in medium with choline for 6 h, which would support induction of betT1, betT3, and cbcXWV transcription (Fig. 2), but prior to rapid growth on choline in wild-type cultures (Fig. 1), the wild type showed a 4-fold increase in choline transport activity (Fig. 3). Choline transport was decreased by 52% and 80% in the ΔbetT1 and ΔbetT3 mutants, respectively, and by 90% in the ΔbetT1 ΔbetT3 mutant and triple mutant (Fig. 3). The ΔcbcV mutant did not show a decrease in initial uptake rate of choline relative to the wild type under either condition (Fig. 3). These data show that basal choline uptake and early uptake after exposure to choline was primarily due to the activities of BetT1 and BetT3. We predict that the activities of BetT1 and BetT3 contribute to a subsequent increase in CbcXWV activity, either through increased cbcXWV transcription or CbcXWV activity, and this hypothesis is tested in the experiments described below.
Fig. 3.
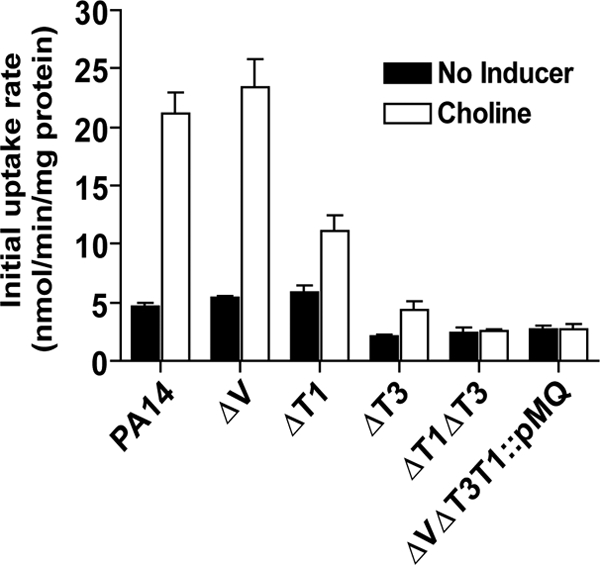
BetT1 and BetT3, but not CbcXWV, are involved in early choline uptake. Uptake of 1 mM [14C]choline by P. aeruginosa PA14 and various mutants is shown. Cells were grown in MOPS-P, incubated for 6 h in the presence or absence of 20 mM choline as an inducer, washed three times in an isomolar medium (M9 without glucose), and evaluated for uptake in MOPS-P containing 20 mM glucose in a 5-min assay. Black bars represent uninduced cultures, and white bars represent cultures preinduced with choline. Values are mean initial uptake rates ± standard errors of the means (n = 3 or 6).
cbcXWV transcription is controlled by GbdR in response to GB.
To evaluate the effects of BetT1 and BetT3 activity on cbcXWV expression, we investigated cbcXWV regulation. The cbcXWV genes are near the gene encoding the transcriptional regulator GbdR (see Fig. S4A in the supplemental material). We have previously demonstrated that GbdR, an AraC family transcription factor, is required for GB-induced expression of the phospholipase C gene plcH, the choline phosphatase gene pchP, and the GB catabolic genes through direct DNA binding to a GbdR box (40, 42). Analysis of the promoter region of cbcXWV revealed the presence of a GbdR recognition sequence (40) upstream of the cbcXWV genes (see Fig. S4A). To determine if GbdR regulated cbcXWV induction, the transcript levels of cbcV were analyzed in the wild-type and ΔgbdR mutant backgrounds after growth in medium with glucose alone or glucose and choline. Glucose was added to the medium to promote similar growth by the mutant strains and the wild type; glucose is known not to repress glycine betaine use via catabolite repression (10). cbcV expression in the wild type was induced over 15-fold by choline and was over 15-fold higher than in the ΔgbdR mutant (Fig. 4). These data suggest that the cbcXWV operon is part of the GbdR regulon. In contrast, transcript levels of betT1 and betT3 were largely unaffected when gbdR was inactivated; that is, although they were induced 26- and 22-fold by choline, respectively, they were only 2-fold higher in the wild type than the ΔgbdR mutant (Fig. 4). These results suggest that GbdR regulates the expression of cbcXWV but not betT1 and betT3.
Fig. 4.
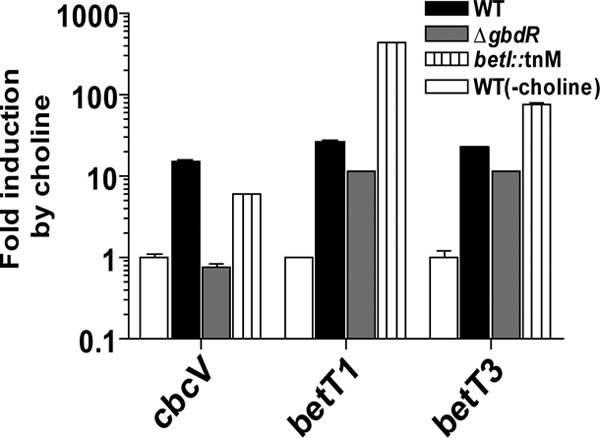
Regulation of expression of P. aeruginosa choline transporter genes. qRT-PCR analysis of expression of cbcV, betT1, and betT3 in WT, ΔgbdR, and betI::TnM in MOPS-P medium with 20 mM glucose and 10 mM choline after 5.5 h of growth. Medium without any quaternary ammonium compounds served as the control condition. The expression levels were normalized to rpoD transcript levels and are presented as the fold induction compared to levels in the absence of choline. Values are the means of biological duplicates ± the standard deviations.
Expression of betT1 and betT3 is derepressed by BetI in response to choline.
Similar to the genomic context of its homologs in E. coli and Sinorhizobium meliloti, betT1 is transcribed divergently from the betIBA genes (see Fig. S4B); in E. coli, BetI encodes a transcriptional repressor of both betT and betIBA (4, 16). The analysis of the P. aeruginosa betT1 promoter performed by Velasco-Garcia et al. (39) revealed a possible BetI binding site at bp −187 (see Fig. S4B). Analysis of the promoter region of betT3 also revealed the presence of a BetI binding site at bp −79 (see Fig. S4B). To determine if the BetI repressor regulates betT1 and betT3 expression, the transcript levels of betT1 and betT3 were analyzed in the betI::TnM mutant background in a medium with choline. betT1 and betT3 mRNA levels were more than 20- and 5-fold higher, respectively, in the betI mutant than in wild-type cells (Fig. 4). The expression of cbcV was only slightly reduced in a betI mutant (<2-fold) (Fig. 4). These data suggest that betT1 and betT3 expression levels are repressed by BetI and that choline promotes derepression.
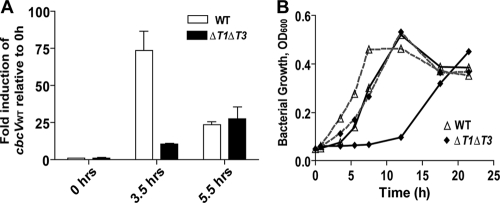
To determine if uptake of choline by BetT1 and BetT3 and the subsequent conversion of choline to GB enhanced the rate of cbcXWV induction following exposure to choline, we analyzed the transcript levels of the cbcV gene in wild-type and ΔbetT1 ΔbetT3 cells upon transfer from medium with pyruvate as the sole source of carbon to a medium with choline as the growth substrate. At 3.5 h after transfer, the wild-type and ΔbetT1 ΔbetT3 mutant cultures were at the same density and were just exiting the lag phase (Fig. 5 B). At this time, as shown in Fig. 5A, the transcript levels of cbcV had increased 75-fold in the wild type but only 15-fold in the ΔbetT1 ΔbetT3 mutant, indicating that BetT1 and BetT3 enhanced cbcXWV induction by choline. The relative levels of cbcV expression decreased to levels comparable to those in the ΔbetT1 ΔbetT3 mutant at 5.5 h after transfer into the medium with choline (Fig. 5A), perhaps due to the consumption of inducing compounds, such as GB and dimethylglycine, during catabolism (42).
Fig. 5.
Contributions of BetT1 and BetT3 to cbcV expression in PA14. (A) qRT-PCR analysis of expression of cbcV in the wild type and ΔbetT1 ΔbetT3. The strains were grown in MOPS-P medium amended with 20 mM choline, and RNA was extracted from aliquots removed at the indicated time points. Expression levels were normalized to rpoD transcript levels and are presented as the fold induction compared to levels in the WT at 0 h (uninduced). Values are the mean fold induction of biological duplicates ± the standard deviation. (B) Comparison of PA14 and ΔbetT1 ΔbetT3 grown in MOPS-P medium amended with 20 mM choline. The cells were pregrown in medium with pyruvate (solid lines) or glycine betaine (dashed lines). Values are mean optical densities at 600 nm (n = 3).
To evaluate if BetT1 and BetT3 contributed to the induction of the cbcXWV genes, we determined if pregrowth on GB, the inducing ligand of GbdR, would be sufficient to eliminate the differences in lag phase between the wild type and the ΔbetT1 ΔbetT3 mutant. As shown in Fig. 5B, the delay in growth of ΔbetT1 ΔbetT3 was abolished when the cells were preinduced with GB. Furthermore, after 3.5 h with GB rather than choline as a preinducer, cbcV transcript levels were similar in the wild type and the ΔbetT1 ΔbetT3 mutant, with the mutant having cbcV transcript levels that were 82 ± 1.8% (mean ± standard deviation) of those in the wild type in GB but only ∼5% of those in the wild type in choline (Fig. 5A).
Modulating osmolarity alters the relative roles of the transporters for uptake of choline for catabolism.
Previously, P. aeruginosa has been shown to utilize choline as a sole carbon source under conditions of hypo- and hyperosmolarity (20). To gain better insight into the relative role of the transporters under the various conditions that P. aeruginosa might encounter, we examined growth of the various mutants on choline in hyper- and hypo-osmolar media. Growth assays showed that BetT3 plays a major role in growth on choline in hyperosmolar medium (MOPS-H) based on the finding that the wild type and ΔbetT1 and ΔcbcV mutants all had similar growth kinetics, a 9-h lag phase, and a doubling time of 3.4 h, whereas the ΔbetT3 and ΔbetT3 ΔcbcV mutants were severely compromised in growth (Fig. 6 A). In MOPS-H, choline is also used by P. aeruginosa as an osmoprotectant (see Fig. S3B in the supplemental material). The wild type and all of the mutants, including a triple mutant that lacks betT1, betT3, and cbcV, were able to use choline as an osmoprotectant in this medium (see Fig. S3B), demonstrating that other transporters contribute to P. aeruginosa osmoprotection.
Fig. 6.
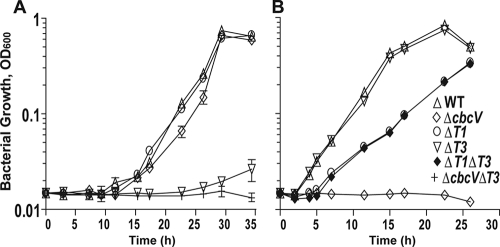
Growth curves of PA14 and various mutants on choline-containing media with high and low osmolarities. (A) Strains grown in hyperosmolar MOPS-H medium with 20 mM choline. (B) Strains grown in hypo-osmolar -21C medium with 20 mM choline. Values represent the mean optical densities at 600 nm from two independent replicates, and error bars, where visible, depict the standard deviations.
In hypo-osmolar medium with choline, the ΔcbcV mutant failed to grow and the ΔbetT1 mutant had a longer lag phase (5.4 h) and a slower growth rate (5.4 h) (Fig. 6B). In contrast, the ΔbetT3 mutant grew like the wild type, having a lag phase of 2 h and a doubling rate of 2.6 h (Fig. 6B), while the ΔbetT1 ΔbetT3 strain resembled ΔbetT1 in its growth kinetics (Fig. 6B). Based on these results, choline uptake under hypo-osmolar conditions is due to the activity of CbcXWV and BetT1.
DISCUSSION
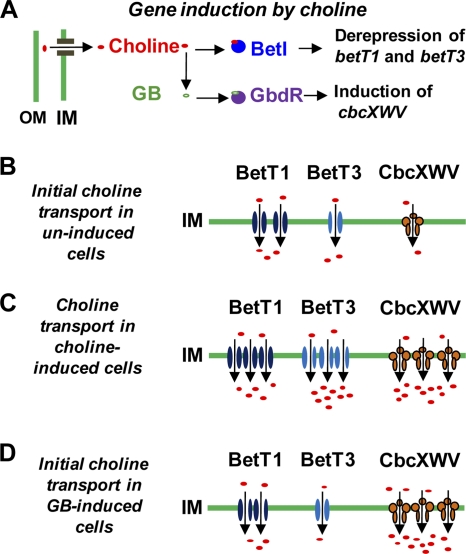
Our results demonstrate distinct roles for three P. aeruginosa choline transporters in supporting cellular growth on choline. These transporters differ in their contribution to growth not only at distinct osmolarities but also at different times following exposure to choline. In particular, immediately following the transfer of cells to conditions requiring choline catabolism at an osmolarity relevant to mammalian host tissues, the BetT1 and BetT3 transporters had the primary role in adaptation during the lag phase. Following adaptation, however, the CbcXWV transporter became increasingly important in its contribution to growth, while BetT1 and BetT3 diverged in their relative activities. Our results are consistent with the following model for the regulation and roles of these transporters in choline uptake for catabolism at a physiologically relevant osmolarity (Fig. 7). In cells that have not been induced by choline, the basal activities of BetT1 and BetT3 are higher than that of CbcXWV; thus, following exposure to choline, these transporters are primarily responsible for the initial choline uptake. The transported choline increases the expression of the betT1 and betT3 genes, at least in part via derepression of the betT1 and betT3 genes by BetI. The transported choline is converted to GB, which is an activating ligand for the transcriptional factor GbdR (40) to increase expression of cbcXWV. Lastly, as CbcXWV-mediated uptake increases, the relative contributions of BetT1 and BetT3 to uptake decrease, with the contribution of BetT1 decreasing to undetectable levels.
Fig. 7.
Model of the regulation and roles of the P. aeruginosa choline transporters in an environment with an osmolarity that is physiologically relevant to a eukaryotic host. (A) Choline promotes expression of betT1, betT3, and cbcXVW. Increased expression in response to choline and GB is dependent on the regulators BetI and GbdR, respectively, and choline is converted into GB by BetAB (1). (B) Following cellular exposure to an abundance of choline (e.g., >1 mM), the initial transport depends primarily on BetT1 and BetT3, with BetT1 initially contributing more than BetT3, possibly because of higher BetT1 protein levels in uninduced cells. (C) Adaption to the presence of abundant choline involves increasing expression of betT1 and betT3, as shown in panel A, with increasing choline uptake promoting the induction of cbcXWV. The contribution of CbcXWV to the total choline uptake increases as its protein levels increase, while the contribution of BetT3 relative to BetT1 increases due to the greater transport capacity of BetT3 (Fig. 3). Growth data indicated that either BetT3 or CbcXWV is sufficient for robust growth with choline as the sole carbon source. (D) Exposure of cells to abundant GB prior to choline greatly increases the contribution of CbcXWV to choline transport due to cbcXWV induction, and this circumvents the major roles for BetT1 and BetT3 in transport immediately following exposure to choline. OM, outer membrane; IM, inner membrane.
Several lines of evidence support a role for BetT1 and BetT3 in the initial induction of cbcXWV expression. First, the induction of cbcV in cells following transfer to a choline-based medium was delayed in the ΔbetT1 ΔbetT3 mutant relative to the wild type (Fig. 5A), consistent with a need for BetT1 and BetT3 during the initial adaption to this medium. Second, the lengthened lag period of the ΔbetT1 ΔbetT3 cells following transfer to a choline-based medium was significantly shortened by prior exposure to GB (Fig. 5B), consistent with early cbcXWV expression compensating for a BetT1/BetT3 deficiency. Third, measurements of the initial uptake rate of [14C]choline showed that CbcXWV exhibited little uptake activity in the absence of choline induction relative to the activity in its presence (Fig. 3).
We found that BetT1 and BetT3, which share 53% identity in amino acid sequence, exhibit distinct characteristics. For example, at a physiologically relevant osmolarity, BetT3 was sufficient for growth on choline in a ΔcbcV mutant background, whereas BetT1 was not (Fig. 1C). Transport assays also suggested greater uptake activity for BetT3 than BetT1 under these conditions (Fig. 3). Despite the greater contribution of BetT3 to growth, loss of BetT1, but not BetT3, resulted in a detectable lengthening of the lag phase following transfer to a choline-based medium. This may reflect a more rapid increase in betT1 expression in the presence of choline, although this has not yet been tested. BetT1 and BetT3 also differ in their response to external osmolarity, with BetT1 as well as CbcXWV serving as the major transporters contributing to growth on choline in a hypo-osmolar environment (Fig. 6B) and BetT3 as the dominant transporter during growth on choline in a hyperosmolar environment (Fig. 6A). Previously, we showed that choline uptake by P. syringae BetT, which is 53% and 79% identical to BetT1 and BetT3, respectively, increases with increasing osmolarity and that both osmoregulation and activity in high-salt environments depend on the presence of a C-terminal tail (6). Domain analysis of E. coli BetT also demonstrated the importance of the C-terminal tail for the osmoregulation of transport activity (37). The fact that BetT3 is salt activated (5) but BetT1 is not (data not shown) may be due to the fact that BetT1 lacks the cytoplasmic C-terminal tail, which follows the 12th transmembrane helix in P. syringae BetT and P. aeruginosa BetT3. The absence of the osmoregulatory tail may make P. aeruginosa BetT1 into a transporter that no longer requires salt for activation but with lower activity than BetT3. Our finding that ΔcbcV ΔbetT1 still grows with choline as the sole source of carbon in a medium at a physiologically relevant osmolarity but ΔcbcV ΔbetT3 cannot grow (Fig. 1C) is consistent with BetT1 having a lower transport activity.
Luchessi et al. (20) demonstrated that P. aeruginosa can grow on choline in a high-salt medium, and here we identified BetT3 as the key transporter involved in growth on choline under hyperosmolar conditions (Fig. 6A). It is not yet clear why other transporters involved in osmoprotection, such as the OpuC transporter (5), or catabolism, such as CbcXWV, do not support growth at high osmolarity (Fig. 6A). The OpuC transporter has been shown to transport choline for osmoprotection in P. syringae (5). Although the ΔcbcV ΔbetT3 betT1::pMQ89 triple mutant is defective in using choline as a carbon source (see Fig. S2 in the supplemental material), it is only slightly impaired in its ability to use choline as an osmoprotectant (see Fig. S3B in the supplemental material), suggesting a role for the P. aeruginosa OpuC homolog (PA3888 to -3891) in osmoprotection. Based on the low-capacity transport activity demonstrated by the P. syringae OpuC transporter (5), we propose that OpuC-mediated transport is not sufficient to support growth based on choline catabolism. In contrast, the CbcXWV transporter likely does not support growth at high osmolarity because of its inhibitory effects on CbcXWV activity, such as on the choline binding to the periplasmic binding protein or protein-protein interactions, or on the expression of cbcXWV. A previous study demonstrated that hyperosmolarity inhibits the activity of various ABC transporters (11).
Previously, we described the role of the GbdR transcription factor and its ligand GB in the transcriptional regulation of phospholipase C (plcH), phosphorylcholine phosphatase (pchP), and multiple genes involved in GB catabolism (40, 42). Here, we showed that cbcXWV is part of the GbdR regulon and that under conditions predicted to be relevant to host tissues, cbcXWV expression required BetT1 and BetT3 uptake to provide the GB coinducer for GbdR-mediated regulation. Thus, we predict that the uptake of choline by BetT1 and BetT3 is important in vivo. A recent microarray study by Son et al. investigating expression profiles of P. aeruginosa genes in sputum from cystic fibrosis patients showed high expression levels of transporter genes betT1 and betT3 genes along with the cbcXWV operon. In fact, the study found these choline transporter genes to be among the most highly expressed genes in the cystic fibrosis patient sputum, along with genes encoding lipases, amino acid and fatty acid degradation enzymes, and the betIBA operon, which is involved in conversion of choline to GB (35). This supports the hypothesis that choline is actively acquired during lung infections and that these transporters play an important role in colonization of the lungs. A complete understanding of choline uptake and the utilization of mutants unable to acquire choline in pathogenesis models will allow us to determine the importance of this abundant resource for P. aeruginosa virulence.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant P20-RR018787 from the IDeA Program of the National Center for Research Resources (to D.A.H.), by the Cystic Fibrosis Foundation Research Development Program (STANTO07R0), and by National Science Foundation grant MCB-0920156 (to G.A.B.).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 8 April 2011.
REFERENCES
- 1. Andresen P. A., Kaasen I., Styrvold O. B., Boulnois G., Strom A. R. 1988. Molecular cloning, physical mapping and expression of the bet genes governing the osmoregulatory choline-glycine betaine pathway of Escherichia coli. J. Gen. Microbiol. 134:1737–1746 [DOI] [PubMed] [Google Scholar]
- 2. Anthoni U., Christophersen C., Hougaard L., Nielsen P. H. 1991. Quaternary ammonium compounds in the biosphere: an example of a versatile adaptive strategy. Comp. Biochem. Physiol B: Comp. Biochem. 99:1–18 [Google Scholar]
- 3. Barbier M., et al. 2008. Novel phosphorylcholine-containing protein of Pseudomonas aeruginosa chronic infection isolates interacts with airway epithelial cells. J. Infect. Dis. 197:465–473 [DOI] [PubMed] [Google Scholar]
- 4. Boyd L. A., et al. 1991. Characterization of an Escherichia coli gene encoding betaine aldehyde dehydrogenase (BADH): structural similarity to mammalian ALDHs and a plant BADH. Gene 103:45–52 [DOI] [PubMed] [Google Scholar]
- 5. Chen C., Beattie G. A. 2007. Characterization of the osmoprotectant transporter OpuC from Pseudomonas syringae and demonstration that cystathionine-β-synthase domains are required for its osmoregulatory function. J. Bacteriol. 189:6901–6912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen C., Beattie G. A. 2008. Pseudomonas syringae BetT is a low-affinity choline transporter that is responsible for superior osmoprotection by choline over glycine betaine. J. Bacteriol. 190:2717–2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen C., Malek A. A., Wargo M. J., Hogan D. A., Beattie G. A. 2010. The ATP-binding cassette transporter Cbc (choline/betaine/carnitine) recruits multiple substrate-binding proteins with strong specificity for distinct quaternary ammonium compounds. Mol. Microbiol. 75:29–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cosquer A., et al. 1999. Nanomolar levels of dimethylsulfoniopropionate, dimethylsulfonioacetate, and glycine betaine are sufficient to confer osmoprotection to Escherichia coli. Appl. Environ. Microbiol. 65:3304–3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cundell D. R., Gerard N. P., Gerard C., Idanpaan-Heikkila I., Tuomanen E. I. 1995. Streptococcus pneumoniae anchor to activated human cells by the receptor for platelet-activating factor. Nature 377:435–438 [DOI] [PubMed] [Google Scholar]
- 10. Diab F., et al. 2006. Succinate-mediated catabolite repression control on the production of glycine betaine catabolic enzymes in Pseudomonas aeruginosa PAO1 under low and elevated salinities. Microbiology 152:1395–1406 [DOI] [PubMed] [Google Scholar]
- 11. Fox M. A., White J. P., Hosie A. H., Lodwig E. M., Poole P. S. 2006. Osmotic upshift transiently inhibits uptake via ABC transporters in gram-negative bacteria. J. Bacteriol. 188:5304–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Halverson L. J., Firestone M. K. 2000. Differential effects of permeating and nonpermeating solutes on the fatty acid composition of Pseudomonas putida. Appl. Environ. Microbiol. 66:2414–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hendry E. B. 1962. The osmotic pressure and chemical composition of human body fluids. Clin. Chem. 8:246–265 [PubMed] [Google Scholar]
- 14. Kappes R. M., Kempf B., Bremer E. 1996. Three transport systems for the osmoprotectant glycine betaine operate in Bacillus subtilis: characterization of OpuD. J. Bacteriol. 178:5071–5079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kortstee G. J. 1970. The aerobic decomposition of choline by microorganisms. I. The ability of aerobic organisms, particularly coryneform bacteria, to utilize choline as the sole carbon and nitrogen source. Arch. Mikrobiol. 71:235–244 [PubMed] [Google Scholar]
- 16. Lamark T., et al. 1991. DNA sequence and analysis of the bet genes encoding the osmoregulatory choline-glycine betaine pathway of Escherichia coli. Mol. Microbiol. 5:1049–1064 [DOI] [PubMed] [Google Scholar]
- 17. Lamark T., Rokenes T. P., McDougall J., Strom A. R. 1996. The complex bet promoters of Escherichia coli: regulation by oxygen (ArcA), choline (BetI), and osmotic stress. J. Bacteriol. 178:1655–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lewenza S., Gardy J. L., Brinkman F. S., Hancock R. E. 2005. Genome-wide identification of Pseudomonas aeruginosa exported proteins using a consensus computational strategy combined with a laboratory-based PhoA fusion screen. Genome Res. 15:321–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liberati N. T., et al. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. U. S. A. 103:2833–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lucchesi G. I., Pallotti C., Lisa A. T., Domenech C. E. 1998. Constitutive choline transport in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 162:123–126 [DOI] [PubMed] [Google Scholar]
- 21. Magoon M. W., et al. 1983. Subfractionation of lung surfactant. Implications for metabolism and surface activity. Biochim. Biophys. Acta 750:18–31 [DOI] [PubMed] [Google Scholar]
- 22. Neidhardt F. C., Bloch P. L., Smith D. F. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pesin S. R., Candia O. A. 1982. Acetylcholine concentration and its role in ionic transport by the corneal epithelium. Invest. Ophthalmol. Vis. Sci. 22:651–659 [PubMed] [Google Scholar]
- 24. Plotnikova J. M., Rahme L. G., Ausubel F. M. 2000. Pathogenesis of the human opportunistic pathogen Pseudomonas aeruginosa PA14 in Arabidopsis. Plant Physiol. 124:1766–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rahme L. G., et al. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899–1902 [DOI] [PubMed] [Google Scholar]
- 26. Rennick B. R. 1981. Renal tubule transport of organic cations. Am. J. Physiol. 240:F83–F89 [DOI] [PubMed] [Google Scholar]
- 27. Sage A. E., Vasil A. I., Vasil M. L. 1997. Molecular characterization of mutants affected in the osmoprotectant-dependent induction of phospholipase C in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 23:43–56 [DOI] [PubMed] [Google Scholar]
- 28. Sage A. E., Vasil M. L. 1997. Osmoprotectant-dependent expression of plcH, encoding the hemolytic phospholipase C, is subject to novel catabolite repression control in Pseudomonas aeruginosa PAO1. J. Bacteriol. 179:4874–4881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Salvano M. A., Lisa T. A., Domenech C. E. 1989. Choline transport in Pseudomonas aeruginosa. Mol. Cell. Biochem. 85:81–89 [DOI] [PubMed] [Google Scholar]
- 30. Savli H., et al. 2003. Expression stability of six housekeeping genes: a proposal for resistance gene quantification studies of Pseudomonas aeruginosa by real-time quantitative RT-PCR. J. Med. Microbiol. 52:403–408 [DOI] [PubMed] [Google Scholar]
- 31. Schweizer H. D. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. Biotechniques 15:831–834 [PubMed] [Google Scholar]
- 32. Schweizer H. P. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97:109–121 [DOI] [PubMed] [Google Scholar]
- 33. Shanks R. M., Caiazza N. C., Hinsa S. M., Toutain C. M., O'Toole G. A. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl. Environ. Microbiol. 72:5027–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sohlenkamp C., Lopez-Lara I. M., Geiger O. 2003. Biosynthesis of phosphatidylcholine in bacteria. Prog. Lipid Res. 42:115–162 [DOI] [PubMed] [Google Scholar]
- 35. Son M. S., Matthews W. J., Jr., Kang Y., Nguyen D. T., Hoang T. T. 2007. In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect. Immun. 75:5313–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Swords W. E., et al. 2000. Non-typeable Haemophilus influenzae adhere to and invade human bronchial epithelial cells via an interaction of lipooligosaccharide with the PAF receptor. Mol. Microbiol. 37:13–27 [DOI] [PubMed] [Google Scholar]
- 37. Tondervik A., Strom A. R. 2007. Membrane topology and mutational analysis of the osmotically activated BetT choline transporter of Escherichia coli. Microbiology 153:803–813 [DOI] [PubMed] [Google Scholar]
- 38. Velasco-Garcia R., Gonzalez-Segura L., Munoz-Clares R. A. 2000. Steady-state kinetic mechanism of the NADP+ and NAD+ dependent reactions catalysed by betaine aldehyde dehydrogenase from Pseudomonas aeruginosa. Biochem. J. 352:675–683 [PMC free article] [PubMed] [Google Scholar]
- 39. Velasco-Garcia R., et al. 2006. Betaine aldehyde dehydrogenase from Pseudomonas aeruginosa: cloning, over-expression in Escherichia coli, and regulation by choline and salt. Arch. Microbiol. 185:14–22 [DOI] [PubMed] [Google Scholar]
- 40. Wargo M. J., Ho T. C., Gross M. J., Whittaker L. A., Hogan D. A. 2009. GbdR regulates Pseudomonas aeruginosa plcH and pchP transcription in response to choline catabolites. Infect. Immun. 77:1103–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wargo M. J., Hogan D. A. 2009. Identification of genes required for Pseudomonas aeruginosa carnitine catabolism. Microbiology 155:2411–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wargo M. J., Szwergold B. S., Hogan D. A. 2008. Identification of two gene clusters and a transcriptional regulator required for Pseudomonas aeruginosa glycine betaine catabolism. J. Bacteriol. 190:2690–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. West S. E., Schweizer H. P., Dall C., Sample A. K., Runyen-Janecky L. J. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81–86 [DOI] [PubMed] [Google Scholar]
- 44. Wilderman P. J., Vasil A. I., Martin W. E., Murphy R. C., Vasil M. L. 2002. Pseudomonas aeruginosa synthesizes phosphatidylcholine by use of the phosphatidylcholine synthase pathway. J. Bacteriol. 184:4792–4799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wood J. M. 2007. Bacterial osmosensing transporters. Methods Enzymol. 428:77–107 [DOI] [PubMed] [Google Scholar]
- 46. Wright J. R., Clements J. A. 1987. Metabolism and turnover of lung surfactant. Am. Rev. Respir. Dis. 136:426–444 [DOI] [PubMed] [Google Scholar]
- 47. Wright J. R., Hawgood S. 1989. Pulmonary surfactant metabolism. Clin. Chest Med. 10:83–93 [PubMed] [Google Scholar]
- 48. Ziegler C., Bremer E., Krämer R. 2010. The BCCT family of carriers: from physiology to crystal structure. Mol. Microbiol. 78:13–34 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.