Abstract
ATP participates in many cellular metabolic processes as a major substrate to supply energy. Many systems for acidic resistance (AR) under extremely acidic conditions have been reported, but the role of ATP has not been examined. To clarify whether or not ATP is necessary for the AR in Escherichia coli, the AR of mutants deficient in genes for ATP biosynthesis was investigated in this study. The deletion of purA or purB, each of which encodes enzymes to produce AMP from inosinate (IMP), markedly decreased the AR. The content of ATP in these mutants decreased rapidly at pH 2.5 compared to that of the wild type. The AR was again decreased significantly by the mutation of adk, which encoded an enzyme to produce ADP from AMP. The DNA damage in the purA and purB mutants was higher than that in the wild type. These results demonstrated that metabolic processes that require ATP participate in survival under extremely acidic conditions, and that one such system is the ATP-dependent DNA repair system.
INTRODUCTION
Since the normal human stomach averages pH 2 for approximately 2 h after it becomes empty, to survive in the mammalian host both commensal and pathogenic enteric bacteria have resistance systems to protect themselves against acidic stress (5, 32, 36).
Four acidic resistance (AR) systems that are induced under different conditions have been proposed for Escherichia coli (5). Acidic resistance system 1 (AR1), which is induced in cells grown to stationary phase in a moderately acidic medium, requires the sigma factor RpoS (3, 27) and the cyclic AMP receptor protein CRP (2). The underlying mechanism remains unclear. The other three systems depend on the presence of specific amino acids. The second AR system (AR2) is a glutamate-dependent system that requires two glutamate decarboxylases (GadA and GadB) and a putative glutamate/γ-amino- butyric acid (GABA) antiporter, GadC (3, 8, 31). AR3 is an arginine-dependent system. It is induced by low pH under anaerobic conditions, and it requires arginine decarboxylase (AdiA) and arginine/agmatine antiporter (AdiC) (6, 10). AR4 is a lysine-dependent system that requires lysine decarboxylase (CadA) and a lysine/cadaverin antiporter (CadB) (22, 40).
In addition to these enzymes, multiple global regulators, such as H-NS, CysB, SspA, and HU (1, 7, 19, 34, 35), small RNAs (DsrA and GadY) (17, 25), topoisomerase I (37), and Asr (33) have been reported to have some roles in AR, either directly or indirectly. Furthermore, some small molecules, such as indole (9) and CO2 (38), induced AR. These reports have suggested that multiple metabolic processes besides amino acid decarboxylation are required for survival under acidic conditions.
The maintenance of energy is required for many metabolic processes, including the biosynthesis of cellular materials, the membrane transport of ions and organic compounds, DNA repair, cell division and cell motility, and the degradation of macromolecules. E. coli has two major energy sources, ATP and the proton-motive force. The latter is generated via the respiratory chain and is used mainly for ATP synthesis and various membrane transports. Since E. coli can survive at low pH without an oxygen supply, ATP may be more important for survival under extremely acidic conditions. We found in this study that the deletion of genes required for ATP biosynthesis decreased the ATP level and the AR of cells growing logarithmically. These results suggested that the ATP-dependent system plays an important role in the survival of E. coli under extremely acidic conditions in addition to amino acid decarboxylation. Our results also demonstrated that a DNA repair system was one such ATP-dependent system.
MATERIALS AND METHODS
Bacterial strains and culture media.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli was grown at 37°C in EG medium, i.e., minimal E medium (11) containing 0.4% glucose. The medium pH was adjusted by the addition of NaOH or HCl. LB medium also was used as a rich medium. Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 25 μg/ml.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| W3110 | λ− F− derived from E. coli K-12 | 11 |
| BW25113 | lacIqrrnBT14 ΔlacZWJ16hsdR514 | 4 |
| ΔaraBADAH33 ΔrhaBADLD78 | ||
| JW4135 | BW25113 purA::Kmr | Keio collectionb |
| MP402 | BW25113 purB::Kmr | 26 |
| JW2461 | BW25113 purC::Kmr | Keio collection |
| JW0511 | BW25113 purK::Kmr | Keio collection |
| JW2541 | BW25113 purL::Kmr | Keio collection |
| JW2491 | BW25113 guaA::Kmr | Keio collection |
| JW5401 | BW25113 guaB::Kmr | Keio collection |
| JW2651 | BW25113 nrdF::Kmr | Keio collection |
| JW4197 | BW25113 nrdD::Kmr | Keio collection |
| JW2788 | BW25113 recB::Kmr | Keio collection |
| TH1559 | K12 recBrecC sbcB | 23 |
| IH125 | W3110 adk(ts) | NIGb |
| SE1796 | W3110 purA::Kmr | This study, W3110 × P1(JW4135) |
| SE1888 | W3110 purB::Kmr | This study, W3110 × P1(MP402) |
| SE1797 | W3110 purC::Kmr | This study, W3110 × P1(JW2461) |
| SE1831 | W3110 purK::Kmr | This study, W3110 × P1(JW0511) |
| SE1832 | W3110 purL::Kmr | This study, W3110 × P1(JW2541) |
| SE1887 | W3110 guaA::Kmr | This study, W3110 × P1(JW2491) |
| SE1833 | W3110 guaB::Kmr | This study, W3110 × P1(JW5401) |
| SE1929 | W3110 recB::Kmr | This study, W3110 × P1(JW2788) |
| Plasmids | ||
| ppurA | pNTR-SD-purA | Mobile plasmid collectionb |
| ppurB | pNTR-SD-purB | Mobile plasmid collection |
Kmr, resistant to kanamycin.
Obtained from the National BioResource Project (NIG, Japan) for E. coli.
Measurement of AR.
The AR of the logarithmic-phase cells was measured as previously described (1). After the cells had been precultured overnight in LB medium with antibiotics if necessary, the cells were diluted 1,000-fold with EG medium at pH 7.5 and cultured at 37°C until the optical density at 600 nm (OD600) reached 0.3 to 0.4. For the adaptation to acidic pH, cells collected by centrifugation at 5,000 × g for 5 min were suspended with a 2-fold volume of EG medium at pH 5.5 and then incubated for 4 h under anaerobic culture conditions. The adapted cells were washed with fresh EG medium at pH 5.5 and then suspended with a 40-fold volume of EG medium at pH 2.5. After incubation at 37°C for 1 to 2 h, the cells were diluted with phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, and 1.4 mM KH2PO4, pH 7.4) and spread on LB agar plates. Colonies appearing after overnight culture at 37°C were counted, and viability was expressed as the percentage of viable cell numbers out of the total cell number before the acidic challenge.
Measurement of the ATP content.
After E. coli cells had been cultured as indicated, the cells were chilled on ice and then centrifuged at 10,000 × g for 5 min at 4°C. The pellets were treated with the solution containing 20 mM glycine, 50 mM MgSO4, 4 mM EDTA, and 50% methanol at pH 7.4 for 30 min at 70°C (21) and then were centrifuged at 10,000 × g for 5 min. The ATP content of the supernatant was measured using a luminometer (Turner Designs, Inc.) as described previously (16). Luciferase and standard ATP were purchased from Sigma Chemical Co.
Genomic DNA damage test.
Genomic DNA damage was measured as described previously (12) with modifications. The genomic DNA was extracted using the standard method (30). One μg of the isolated chromosomal DNA was digested with Bal31 nuclease (0.2 U) at 30°C for 30 min. After the enzyme had been inactivated at 75°C for 10 min, the mixture was chilled on ice, and the resulting DNA fragments were analyzed using 0.8% (wt/vol) agarose gel electrophoresis and ethidium bromide staining.
Intracellular pH measurement.
Internal pH was determined by the distribution of salicylic acid between the outside and inside the cells as described previously (13). After the cells had been cultured in EG medium at pH 5.5 for 4 h, the cells collected by centrifugation at 10,000 × g for 5 min were suspended in EG medium at pH 5.5 or 2.5 at approximately 1 × 109 cells per ml, and [14C]salicylic acid (10 μM; 0.2 μCi/ml) was added as an indicator. After incubation at 37°C for the times indicated, 1 ml of the medium was centrifuged at 10,000 × g for 5 min through the oil mixture (laurylbromide-liquid paraffin). The radioactivity of the supernatant and the pellet were measured to obtain the indicator concentrations outside and inside the cells, respectively. The amount of protein in the pellet was measured, and the radioactivity of the pellet was divided by the water content of the pellet calculated from the protein content of the pellet. The intracellular pH (pHi) was calculated by the following equation: pHi = log{([A]in/[A]out)(10pKa +10pHout) − 10pKa}, where [A]in and [A]out are concentrations inside and outside the cells, respectively, and the pKa of salicylic acid used was 2.89.
Other methods.
Transduction with P1kc (18), transformation with CaCl2 (30), and plasmid isolation (30) were performed as described previously. Internal levels of K+ and Na+ were measured as described previously (24). Protein was measured as described previously (20), and bovine serum albumin was used as a standard.
RESULTS
The effect of the deletion of genes for purine nucleotide biosynthesis on AR and intracellular ATP level.
E. coli cells grown to stationary phase have been used mainly for measurements of AR, and such cells may be more resistant to various stresses (28). To minimize the responses to stresses other than acidic stress in the stationary phase, cells growing exponentially were used in this study.
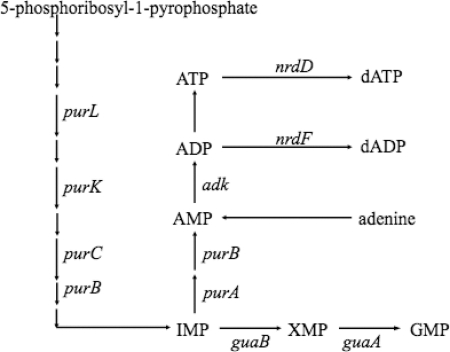
ATP was produced from inorganic phosphate and ADP that was synthesized through metabolic pathways as described in Fig. 1. We first examined the effect of the deletion of genes for AMP synthesis from inosinate (IMP), adenylosuccinate synthetase (purA), or adenylosuccinate lyase (purB) on AR. The purA and purB mutants were unable to grow without the addition of adenine or adenosine in EG medium at pH 7.5 and 5.5. To decrease the ATP level during the acidic challenge at pH 2.5, we designed the following experimental protocol. These mutants first were cultured in EG medium at pH 7.5 containing 0.1 mM adenine until the OD600 reached 0.3. After the cells had been washed with EG medium at pH 5.5, the cells were suspended in the same medium at pH 5.5 without the addition of adenine. The resulting cells grew in EG medium at pH 5.5 for at least 6 h at a lower rate than the wild type. We therefore adapted the cells at pH 5.5 for 4 h in the absence of adenine, and the survival at pH 2.5 was measured. In addition to the purA and purB mutants, the survival of the mutants deficient in genes described in Fig. 1 was examined.
Fig. 1.
Metabolic pathway for purine nucleotide synthesis.
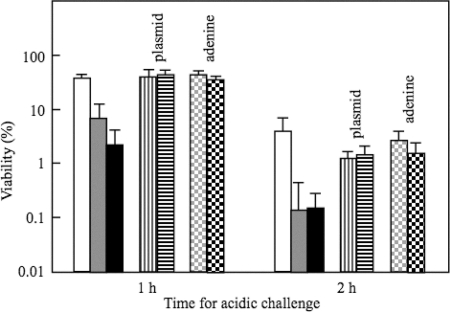
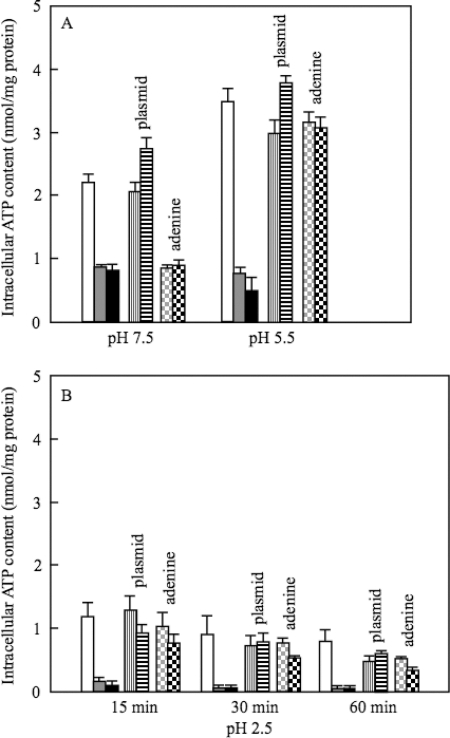
The survival rates after 1 h of challenge at pH 2.5 were decreased 5-fold and 15-fold in purA and purB mutants, respectively (Fig. 2). For the wild-type strain W3110, the internal level of ATP was increased during the adaptation at pH 5.5 and decreased after the cells had been transferred to medium (pH 2.5) (Fig. 3). The ATP content in the purA or purB mutant was lower at pH 5.5 and decreased more rapidly at pH 2.5 compared with those of W3110 (Fig. 3). The plasmids having purA or purB complemented the decrease in the AR (Fig. 2) and the ATP content of the deficient mutants (Fig. 3). When 0.1 mM adenine was added to the culture medium at pH 5.5 for the adaptation, the AR and ATP contents of the purA and purB mutants were recovered (Fig. 2 and 3).
Fig. 2.
Survival of purA and purB mutants at pH 2.5. After the cells had been grown in EG medium at pH 7.5 until the OD600 reached 0.3, the cells were harvested and suspended in EG medium at pH 5.5. After the cells had been grown for 4 h at pH 5.5 under anaerobic conditions, the cells were challenged at pH 2.5 for 1 and 2 h under anaerobic conditions, and the viable cells were counted. Adenine (0.1 mM) was added to EG medium at pH 7.5 for the growth of purA and purB mutants. Adenine also was added at pH 5.5 but not at pH 2.5 for bars labeled adenine. Isopropyl-β-d-thiogalactopyranoside (0.25 mM) was added when the cells containing plasmids were cultured. Symbols: open bars, W3110; gray bars, W3110 purA; black bars, W3110 purB; vertical striped bars, W3110 purA containing ppurA; horizontal striped bars, W3110 purB containing ppurB; gray dotted bars, W3110 purA with the addition of adenine; black dotted bars, W3110 purB with the addition of adenine. The average values and standard deviations obtained from three experiments using separate cultures are represented.
Fig. 3.
ATP content of purA and purB mutants at pH 5.5 and 2.5. The same strains as those used in Fig. 2 were cultured as described in the legend of Fig. 2. The cells were harvested at the indicated times at pH 2.5. The ATP contents were measured as described in Materials and Methods. Symbols are the same as those for Fig. 2. The average values and standard deviations obtained from three experiments using separate cultures are represented.
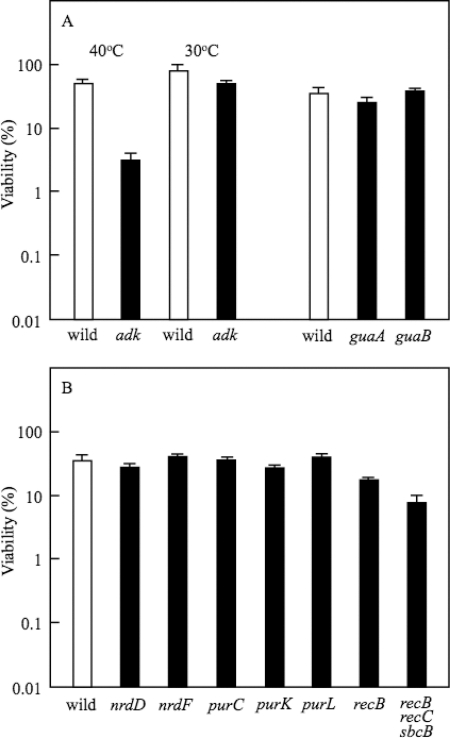
For further confirmation of the suggestion that ATP biosynthesis is required for AR in E. coli, the effect of the deletion of adenylate kinase, encoded by adk, which synthesizes ADP from AMP, was investigated. The ADP biosynthesis is indispensable for growth, and adenylate kinase is the sole enzyme to produce ADP. We therefore used a temperature-sensitive mutant. W3110 adk(ts) was grown in EG medium at pH 7.5 and 30°C and then adapted in EG medium at pH 5.5 and 40°C to inactivate adenylate kinase. The acidic challenge at pH 2.5 was carried out at 37°C. After the acidic challenge, the viable cell count was carried out at 30°C. A control experiment was carried out at 30°C for all steps. The AR was markedly decreased by the inactivation of adenylate kinase (Fig. 4 A). The ATP level of the mutant adapted at 40°C was 0.275 ± 0.010 nmol/mg protein after the 1-h challenge at pH 2.5, while the level for the mutant adapted at 30°C was 0.670 ± 0.030 nmol/mg protein after the same challenge. Since E. coli has multiple enzymes to produce ATP from ADP and ATP synthesis is essential for growth, the effect of the deletion of these genes could not be examined.
Fig. 4.
Survival of various mutants at pH 2.5. W3110, W3110 adk(ts), W3110 guaA, W3110 guaB, BW25113 nrdD, BW25113 nrdF, W3110 purC, W3110 purK, W3110 purL, W3110 recB, and TH1559 (recB, recC, and sbcB) were used. After the cells had been grown in EG medium at pH 7.5 until the OD600 reached 0.3, the cells were harvested and suspended in EG medium at pH 5.5. After the cells had been grown for 4 h at pH 5.5, the cells were challenged at pH 2.5 for 1 h and the viable cells were counted. Adenine (0.1 mM) was added only to medium of pH 7.5 for the growth of the pur mutants. For the bars labeled 40°C, the cells were cultured at 30°C in medium at pH 7.5, adapted at 40°C in medium at pH 5.5, and challenged at 37°C in medium at pH 2.5. After the acidic challenge at pH 2.5, the cells were cultured on LB agar plates at 30°C. For the bars labeled 30°C, the cells were cultured at 30°C for all steps. The average values and standard deviations obtained from three experiments using separate cultures are represented.
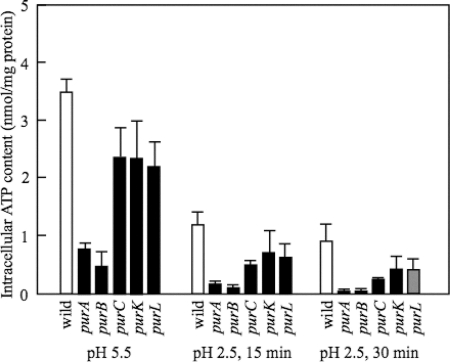
In contrast to genes for AMP biosynthesis, IMP dehydrogenase (guaB) and GMP synthetase (guaA), enzymes for GMP synthesis from IMP had no significant role in AR (Fig. 4A). Furthermore, the survival of the mutants deficient in ribonucleoside-triphosphate reductase (nrdD) and ribonucleoside-diphosphate reductase (nrdF) was similar to that of the wild type (Fig. 4B). The deletion of genes for IMP biosynthesis, such as purL, purC, and purK, did not cause a significant decrease in AR (Fig. 4B). The ATP level of these mutants was higher than that of the purA or purB mutant but lower than that of the wild type (Fig. 5). These results suggested that the ATP level is important for survival at pH 2.5.
Fig. 5.
ATP content of various mutants at pH 5.5 and 2.5. W3110, W3110 purA, W3110 purB, W3110 purL, W3110 purC, and W3110 purK were used. The cells were cultured as described in the legend to Fig. 4. They were harvested at the indicated times at pH 2.5. The ATP contents were measured as described in Materials and Methods. The average values and standard deviations obtained from three experiments using separate cultures are represented.
DNA damage in the purA, purB, and recB mutants at pH 2.5.
The data described above indicated that metabolic processes consuming ATP are required for survival at pH 2.5. Which metabolic process is required? The DNA repair system is a candidate system, because cells would not grow if DNA damage corrupts the integrity and accessibility of essential information in the genome. A variety of repair strategies has evolved to avoid the loss of DNA information, and the DNA repair systems need ATP as a substrate to supply energy (39, 41, 42). Rapid DNA damage was observed previously at acidic pH in mutants deficient in genes for DNA repair systems with a concomitant decrease in the AR (12), suggesting that the DNA repair processes are active for survival under acidic conditions. We found that the mutant deficient in recB, whose function requires ATP, decreased the AR of W3110 (Fig. 4B). Furthermore, TH1559, having multiple mutations (recB, recC, and sbcB), had a low survival rate at pH 2.5 (Fig. 4B). These data indicated that the DNA repair system contributes to the survival of E. coli cells in acidic conditions. Since RecB and RecC require ATP for their function, it can be assumed that the low ATP level attenuates the repair activity.
To clarify whether or not a low ATP level in the purA or purB mutant affects the DNA repair systems, we investigated the DNA damage using Bal31 as described previously (12). Bal31 cleaves DNA at nicks, gaps, single-stranded regions, or other lesions of duplex DNA, and hence DNA is fragmented by this enzyme if these DNA damages are not repaired. No significant DNA fragmentation was detected with Bal31 in the purA, purB, and recB mutants as well as W3110 at pH 5.5. The DNA damage was significantly increased in the purA and purB mutants compared to that of the parental strain at pH 2.5, and the increase was similar to that observed in the recB mutant. These results indicated that the maintenance of the ATP level was essential for protection against the DNA damage caused under acidic stress.
Since pHi regulation was proposed to be important for survival under acidic conditions, we measured the pHi of the purA and purB mutants showing low survival at acidic pH. pHi was not affected significantly by the deletion of purA (Table 2). pHi was decreased by the deletion of purB, but the decrease was less than 0.3 pH units (Table 2). The low ATP levels in these mutants may still be high enough for pHi regulation under acidic conditions. Alternatively, ATP may not be essential for pHi regulation under such conditions.
Table 2.
Cytoplasmic pH (pHi)a
| Strain | pHi |
|||
|---|---|---|---|---|
| pHo 5.5 at 15 min | pHo 2.5 at: |
|||
| 15 min | 30 min | 60 min | ||
| W3110 | 7.14 ± 0.11 | 4.06 ± 0.02 | 3.99 ± 0.02 | 3.76 ± 0.06 |
| W3110 purA | 7.02 ± 0.01 | 4.10 ± 0.25 | 3.93 ± 0.16 | 3.72 ± 0.02 |
| W3110 purB | 7.14 ± 0.16 | 3.88 ± 0.08 | 3.73 ± 0.13 | 3.60 ± 0.09 |
After the cells had been grown in EG medium at pH 7.5 until the OD600 reached 0.3, the cells were suspended with a 2-fold volume of EG medium at pH 5.5 and then cultured for 4 h. The cells were incubated with the indicator in EG medium at pH 5.5 or 2.5 for the times indicated, and then pHi was measured as described in Materials and Methods. pHo is the pH value of the medium.
Glutamate, arginine, and lysine enhanced the ATP concentration and AR.
We next examined whether or not ATP was required for amino acid-dependent AR (AR2, AR3, and AR4). When the cells were adapted at pH 5.5 and challenged at pH 2.5 in the presence of 0.5 mM glutamate, arginine, or lysine, AR was increased as reported previously (5), and the ATP level was higher than that of the cells adapted in the absence of these amino acids. The deletion of purA or purB decreased the AR in the presence of 0.5 mM glutamate, arginine, or lysine. It is now generally accepted that AR2 to AR4 systems enhance survival under acidic conditions via the maintenance of pHi homeostasis by amino acid decarboxylation. In addition, our results suggested that the increase in the ATP level is another function of AR2 to AR4 systems.
DISCUSSION
Many systems are proposed to be involved in the AR of both logarithmic- and stationary-phase cells (5). The well-studied systems are amino acid-dependent systems. It has been reported that the amino acid-dependent systems enhance survival under acidic conditions via the consumption of cytoplasmic protons by amino acid decarboxylation (29). Why is such pHi regulation required for the survival of nongrowing cells under acidic stress? Some metabolic processes may work under such nongrowing conditions. In addition to amino acid-dependent induction, many studies have demonstrated that various genes participate in AR (5). Carbon dioxide, a substrate for nucleic acid and amino acid biosynthesis, induces AR (38), leading us to assume that nucleotide biosynthesis induces AR, but there has been no report to show the role of nucleotides in AR. We therefore examined the participation of purine nucleotide biosynthesis in survival under acidic conditions in the present study.
The present study with cells growing exponentially revealed that the survival of the cells under acidic stress required ATP in both the presence and absence of amino acids such as glutamate, arginine, and lysine. The ATP level increased during the adaptation of E. coli cells at pH 5.5 and decreased during acidic challenge at pH 2.5 (Fig. 3). The ATP level was low at pH 5.5 and ATP was lost rapidly at pH 2.5 in the purA and purB mutants, and these mutants showed a low survival rate at pH 2.5. The defect of the adenylate kinase activity decreased the ATP level and the AR. In contrast, the deletion of genes for IMP biosynthesis did not decrease the AR. The ATP levels of these mutants were higher than those of the purA and purB mutants but lower than that of the wild type. Furthermore, the requirement of genes for GMP or purine deoxyribonucleotide biosynthesis was suggested to be less significant for AR. These data implied that the ATP level is more important for survival under acidic conditions than the levels of other purine nucleotides and deoxynucleotides.
All pur mutants tested were unable to grow without the addition of adenine at pH 7.5, hence 0.1 mM adenine was added to EG medium at pH 7.5. The experimental conditions for the acidic adaptation and challenge were the same in all pur mutants, but the ATP level of the purA and purB mutants was lower than the level of the purC, purK, and purL mutants (Fig. 5). It may be possible that the level of IMP produced from adenine at pH 7.5 is enough for the maintenance of the ATP level required for survival at pH 2.5 in the purC, purK, and purL mutants. The other possibility is that E. coli has alternative enzymes or pathways functioning at acidic pH instead of PurC, PurK, and PurL. The functions of more than 2,000 genes in E. coli still are unknown.
It remains unclear why the ATP level of cells grown at pH 5.5 is higher than that at pH 7.5. The greater pH gradient at pH 5.5 might account for the high level of ATP. However, the membrane potential that drives ATP synthesis, together with the pH gradient, was low or inside positive at low pH (29). An alternative explanation is that many metabolic processes consuming ATP, such as the biosynthesis of macromolecules, decline at low pH because of the decrease in enzyme activities. In fact, the growth rate was low at pH 5.5.
ATP is a substrate to supply energy for various metabolic processes. Which ATP-dependent metabolic process supports the survival at pH 2.5? Jeong et al. (12) showed that mutants deficient in the genes required for DNA repair had low survival rates at low pH and that mutation caused more DNA damage. We found in the present study that the single deletions of recA, recD, sbcB, urvA, and urvB had no significant effect on the AR (data not shown), but the AR of the recB mutant was lower than that of the parent strain. Furthermore, multiple mutations (recB, recC, and sbcB) brought about low survival at pH 2.5 (Fig. 4). These data confirmed the suggestion that the DNA repair system is indispensable for survival in acidic conditions. The DNA damage analysis showed that the deletion of purA or purB caused more DNA damage in acidic conditions, suggesting that ATP keeps the DNA repair systems active.
E. coli has many other ATP-requiring systems, such as ion transport systems and macromolecule biosynthesis. We found that the intracellular levels of Na+ and K+ decreased at acidic pH, but the levels of these ions in the purA and purB mutants were almost the same as those of their parent strain (data not shown), suggesting that the low ATP level shown in the mutants is sufficient for the maintenance of the cytoplasmic levels of these cations in acidic conditions. The biosynthesis of macromolecules may not occur under nongrowing conditions at pH 2.5.
It was reported that the pHi of the wild type was 3.6 to 3.7 in medium of pH 2.3 to 2.4 without the addition of amino acid (29). In contrast, the pHi was 3.7 to 4.0 under our experimental conditions (Table 2). In the previous study, the cells grown to stationary phase were transferred to pH 2.5 medium and the pHi was measured. We adapted the cells in the logarithmic phase at pH 5.5, and the pHi was measured after the cells had been transferred to pH 2.5 medium. Therefore, cells growing logarithmically at pH 5.5 may have an increased ability to regulate pHi in the absence of amino acids.
No significant decrease in pHi was observed in the purA and purB mutants (Table 2). The mechanism for the maintenance of the pH gradient remains unclear. It has been clarified that the pH gradient is generated by the F-type H+ ATPase in enterococci (14). The same mechanism might work in E. coli, as proposed by Richard and Foster (29). However, the ATP hydrolysis activity of the H+ ATPase was negligible at pH of less than 5, and the Km for ATP was 0.6 mM in E. coli (15). ATP at 0.6 mM corresponds to approximately 1.8 nmol ATP per mg protein. Therefore, the ATPase may have difficulty extruding protons at acidic pH. It is possible that E. coli has an unidentified system for pH homeostasis. In any case, the pHi regulation is essential for survival under acidic stress. In addition, our present results suggested that the ATP-dependent repair system has an essential role in the AR.
ACKNOWLEDGMENTS
We thank T. Horie for the use of the illuminator, I. Yamato for the gift of TH1559, and George D. Markham for the gift of MP402.
Footnotes
Published ahead of print on 8 April 2011.
REFERENCES
- 1. Bi H., Sun L., Fukamachi T., Saito H., Kobayashi H. 2009. HU participates in expression of a specific set of genes required for growth and survival at acidic pH in Escherichia coli. Curr. Microbiol. 58:443–448 [DOI] [PubMed] [Google Scholar]
- 2. Castanie-Cornet M. P., Foster J. W. 2001. Escherichia coli acid resistance: cAMP receptor protein and a 20 bp cis-acting sequence control pH and stationary phase expression of the gadA and gadBC glutamate decarboxylase genes. Microbiology 147:709–715 [DOI] [PubMed] [Google Scholar]
- 3. Castanie-Cornet M. P., Penfound T. A., Smith D., Elliott J. F., Foster J. W. 1999. Control of acid resistance in Escherichia coli. J. Bacteriol. 181:3525–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Foster J. W. 2004. Escherichia coli acid resistance: tales of an amateur acidophile. Nat. Rev. Microbiol. 2:898–907 [DOI] [PubMed] [Google Scholar]
- 6. Gong S., Richard H., Foster J. W. 2003. YjdE (AdiC) is the arginine:agmatine antiporter essential for arginine-dependent acid resistance in Escherichia coli. J. Bacteriol. 185:4402–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hansen A. M., et al. 2005. SspA is required for acid resistance in stationary phase by downregulation of H-NS in Escherichia coli. Mol. Microbiol. 56:719–734 [DOI] [PubMed] [Google Scholar]
- 8. Hersh B. M., Farooq F. T., Barstad D. N., Blankenhorn D. L., Slonczewski J. L. 1996. A glutamate-dependent acid resistance gene in Escherichia coli. J. Bacteriol. 178:3978–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hirakawa H., Hayashi-Nishino M., Yamaguchi A., Nishino K. 2010. Indole enhances acid resistance in Escherichia coli. Microb. Pathog. 49:90–94 [DOI] [PubMed] [Google Scholar]
- 10. Iyer R., Williams C., Miller C. 2003. Arginine-agmatine antiporter in extreme acid resistance in Escherichia coli. J. Bacteriol. 185:6556–6561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jensen K. F. 1993. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J. Bacteriol. 175:3401–3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jeong K. C., Hung K. F., Baumler D. J., Byrd J. J., Kaspar C. W. 2008. Acid stress damage of DNA is prevented by Dps binding in Escherichia coli O157:H7. BMC Microbiol. 8:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kashket E. R. 1985. The proton motive force in bacteria: a critical assessment of methods. Annu. Rev. Microbiol. 39:219–242 [DOI] [PubMed] [Google Scholar]
- 14. Kobayashi H. 1985. A proton-translocating ATPase regulates pH of the bacterial cytoplasm. J. Biol. Chem. 260:72–76 [PubMed] [Google Scholar]
- 15. Kobayashi H., Anraku Y. 1972. Membrane-bound adenosine triphosphatase of Escherichia coli. I. Partial purification and properties. J. Biochem. 71:387–399 [PubMed] [Google Scholar]
- 16. Lasko D. R., Wang D. I. 1993. In situ fermentation monitoring with recombinant firefly luciferase. Biotechnol. Bioeng. 42:30–36 [DOI] [PubMed] [Google Scholar]
- 17. Lease R. A., Smith D., McDonough K., Belfort M. 2004. The small noncoding DsrA RNA is an acid resistance regulator in Escherichia coli. J. Bacteriol. 186:6179–6185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lennox E. S. 1955. Transduction of linked genetic characters of the host by bacteriophage P1. Viology 1:190–206 [DOI] [PubMed] [Google Scholar]
- 19. Lochowska A., et al. 2004. Identification of activating region (AR) of Escherichia coli LysR-type transcription factor CysB and CysB contact site on RNA polymerase alpha subunit at the cysP promoter. Mol. Microbiol. 53:791–806 [DOI] [PubMed] [Google Scholar]
- 20. Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265–275 [PubMed] [Google Scholar]
- 21. Maharjan R. P., Ferenci T. 2003. Global metabolite analysis: the influence of extraction methodology on metabolome profiles of Escherichia coli. Anal. Biochem. 313:145–154 [DOI] [PubMed] [Google Scholar]
- 22. Meng S. Y., Bennett G. N. 1992. Nucleotide sequence of the Escherichia coli cad operon: a system for neutralization of low extracellular pH. J. Bacteriol. 174:2659–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nakamura H., Yamato I., Anraku Y., Lemieux L., Gennis R. B. 1990. Expression of cyoA and cyoB demonstrates that the CO-binding heme component of the Escherichia coli cytochrome o complex is in subunit I. J. Biol. Chem. 265:11193–11197 [PubMed] [Google Scholar]
- 24. Ohyama T., Igarashi K., Kobayashi H. 1994. Physiological role of the chaA gene in sodium and calcium circulations at a high pH in Escherichia coli. J. Bacteriol. 176:4311–4315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Opdyke J. A., Kang J. G., Storz G. 2004. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 186:6698–6705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pimkin M., Pimkina J., Markham G. D. 2009. A regulatory role of the Bateman domain of IMP dehydrogenase in adenylate nucleotide biosynthesis. J. Biol. Chem. 284:7960–7969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Price S. B., et al. 2000. Role of rpoS in acid resistance and fecal shedding of Escherichia coli O157:H7. Appl. Environ. Microbiol. 66:632–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ramírez Santos J., Contreras F. G., Gómez E. M. C. 2005. Stationary phase in Escherichia coli. Rev. Latinoam. Microbiol. 47:92–101 [PubMed] [Google Scholar]
- 29. Richard H., Foster J. W. 2004. Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J. Bacteriol. 186:6032–6041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 31. Sayed A. K., Foster J. W. 2009. A 750 bp sensory integration region directs global control of the Escherichia coli GadE acid resistance regulator. Mol. Microbiol. 71:1435–1450 [DOI] [PubMed] [Google Scholar]
- 32. Seputiene V., Daugelavicius A., Suziedelis K., Suziedeliene E. 2006. Acid response of exponentially growing Escherichia coli K-12. Microbiol. Res. 161:65–74 [DOI] [PubMed] [Google Scholar]
- 33. Seputiene V., et al. 2003. Molecular characterization of the acid-inducible asr gene of Escherichia coli and its role in acid stress response. J. Bacteriol. 185:2475–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shi X., Bennett G. N. 1994. Effects of rpoA and cysB mutations on acid induction of biodegradative arginine decarboxylase in Escherichia coli. J. Bacteriol. 176:7017–7023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shi X., Bennett G. N. 1994. Plasmids bearing hfq and the hns-like gene stpA complement hns mutants in modulating arginine decarboxylase gene expression in Escherichia coli. J. Bacteriol. 176:6769–6775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Smith J. L. 2003. The role of gastric acid in preventing foodborne disease and how bacteria overcome acid conditions. J. Food Prot. 66:1292–1303 [DOI] [PubMed] [Google Scholar]
- 37. Stewart N., et al. 2005. Loss of topoisomerase I function affects the RpoS-dependent and GAD systems of acid resistance in Escherichia coli. Microbiology 151:2783–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sun L., Fukamachi T., Saito H., Kobayashi H. 2005. Carbon dioxide increases acid resistance in Escherichia coli. Lett. Appl. Microbiol. 40:397–400 [DOI] [PubMed] [Google Scholar]
- 39. Tubbs J. L., et al. 2009. Flipping of alkylated DNA damage bridges base and nucleotide excision repair. Nature 459:808–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vazquez-Juarez R. C., et al. 2008. CadA negatively regulates Escherichia coli O157:H7 adherence and intestinal colonization. Infect. Immun. 76:5072–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wagner K., Moolenaar G. F., Goosen N. 2010. Role of the two ATPase domains of Escherichia coli UvrA in binding non-bulky DNA lesions and interaction with UvrB. DNA Repair (Amsterdam) 9:1176–1186 [DOI] [PubMed] [Google Scholar]
- 42. Wu C. G., Bradford C., Lohman T. M. 2010. Escherichia coli RecBC helicase has two translocase activities controlled by a single ATPase motor. Nat. Struct. Mol. Biol. 10:1210–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]