Abstract
Regulation of tau exon 10 splicing plays an important role in tauopathy. One of the cis elements regulating tau alternative splicing is a stem-loop structure at the 5′ splice site of tau exon 10. The RNA helicase(s) modulating this stem-loop structure was unknown. We searched for splicing regulators interacting with this stem-loop region using an RNA affinity pulldown-coupled mass spectrometry approach and identified DDX5/RNA helicase p68 as an activator of tau exon 10 splicing. The activity of p68 in stimulating tau exon 10 inclusion is dependent on RBM4, an intronic splicing activator. RNase H cleavage and U1 protection assays suggest that p68 promotes conformational change of the stem-loop structure, thereby increasing the access of U1snRNP to the 5′ splice site of tau exon 10. This study reports the first RNA helicase interacting with a stem-loop structure at the splice site and regulating alternative splicing in a helicase-dependent manner. Our work uncovers a previously unknown function of p68 in regulating tau exon 10 splicing. Furthermore, our experiments reveal functional interaction between two splicing activators for tau exon 10, p68 binding at the stem-loop region and RBM4 interacting with the intronic splicing enhancer region.
INTRODUCTION
Alternative splicing is one of most powerful posttranscriptional mechanisms for generating multiple gene products from individual gene loci, thereby contributing to genetic and proteomic diversity (7, 84, 86, 89). Numerous studies have provided evidence for cell-type-specific and developmentally regulated alternative splicing events (6, 87). Alternative splicing plays critical roles in the nervous system, where production of distinct splicing isoforms regulates multiple cellular processes, including cell fate determination, axon guidance, synaptogenesis, and neural transmission (reviewed in references 8, 30, 73, and 80). Defects or disruption in regulation of alternative splicing contributes to the pathogenesis of a large number of neurodegenerative disorders, such as amyotrophic lateral sclerosis (ALS), familial Alzheimer's disease (FAD), and frontotemporal dementia with Parkinsonism linked to chromosome 17 (FTDP-17, or frontotemporal lobar degeneration with tauopathy [FTLD-tau]) (reviewed in references 31, 40, 41, 42, 53, 71, and 96).
Tau, a microtubule-associated protein (MAP) enriched in axons, plays an important role in polymerization and stabilization of neuronal microtubules. Tau is critical for maintenance of neuronal cytoskeleton and axonal transport (reviewed in references 5 and 56). Human Tau protein is encoded by a single gene on chromosome 17q21 containing 16 exons with the formation of six splicing isoforms in the adult brain as a result of alternative inclusion of exons 2, 3, and 10 (3, 18, 51, 85, 106; reviewed in references 1 and 66). There are four microtubule-binding repeats in the human Tau protein, and they are encoded by exons 9, 10, 11, and 12, respectively (3, 51). tau exon 10 (E10), which encodes the second microtubule-binding domain, is alternatively spliced, resulting in Tau protein isoforms containing either three (E10−) or four (E10+) microtubule-binding domains referred to as three-repeat Tau (Tau3R) and four-repeat Tau (Tau4R), respectively (21, 42, 52). Tau4R and Tau3R show distinct activities in microtubule binding (43, 68, 69, 70, 100–103, 105). tau exon 10 alternative splicing undergoes developmental stage-specific regulation, with almost exclusive expression of Tau3R in the fetal brain and both isoforms being expressed equally (Tau4R/Tau3R ratio of 1:1) in the adult human brain (3, 4, 38, 60, 74, 88, 90).
FTLD-tau is a group of neurodegenerative diseases characterized by the presence of Tau-immunoreactive lesions in the frontotemporal regions and elsewhere, often with the formation of disease-specific tangles or other types of inclusion bodies containing hyperphosphorylated Tau protein (references 12 and 78 and references therein). FTLD-tau can be caused by either missense or splicing mutations in the tau gene (24, 49, 50, 54; reviewed in references 53 and 66). In some forms of familial FTLD-tau, splicing mutations disrupt tau exon 10 splicing regulation and alter the Tau4R/Tau3R ratio in the brain, leading to neurodegeneration (17, 19, 23, 24, 26, 43, 49, 55, 97, 110). Several studies have begun to reveal cis-acting regulatory elements and trans-acting factors that regulate tau exon 10 splicing (9, 26, 27, 28, 37, 45, 60, 61, 67, 106). One of the cis-acting elements is a stem-loop structure downstream of the 5′ splice site of exon 10 (45, 54, 60, 103, 104). Biochemical and structural studies suggest that this stem-loop structure may promote exon 10 exclusion by preventing U1snRNP binding to the 5′ splice site (60, 64, 103, 104). However, RNA helicases that interact with the stem structure and regulate tau exon 10 splicing have not previously been reported.
In this study, we have developed an RNA affinity pulldown-coupled mass spectrometry (MS) approach to identify regulatory factors interacting with the stem-loop structure at the 5′ splice site of tau exon 10. One of the proteins identified is DEAD/H box polypeptide 5 (DDX5), also known as RNA helicase p68. We show that p68 regulates tau exon 10 splicing by interacting with the stem-loop region, destabilizing the stem structure, and facilitating U1snRNP binding to this 5′ splice site. Our biochemical experiments demonstrate that p68 interacts with an intronic splicing activator, RNA binding motif protein 4 (RBM4), thereby stimulating tau exon 10 inclusion.
MATERIALS AND METHODS
RNA affinity pulldown-coupled mass spectrometry.
Biotinylated RNA oligonucleotides corresponding to the tau gene containing the last 24 nucleotides of exon 10 and the first 24 nucleotides of intron 10 were synthesized together with a control RNA with random sequence (Dharmacon). In RNA affinity pulldown experiments, 0.5 pmol of biotinylated RNA oligonucleotide was incubated in a 500-μl reaction mixture containing 100 microliters of HeLa cell nuclear extract under splicing conditions (20 mM HEPES, 72 mM KCl, 1.5 mM MgCl2, 1.6 mM magnesium acetate [MgOAc], 0.5 mM dithiothreitol [DTT], 4 mM glycerol, 1 mM ATP, 200 units of RNasin; Promega) at 30°C for 30 min. After incubation, 100 μl of preequilibrated magnetic streptavidin beads (Dynal Corporation) was added to the reaction mixture and incubated for 1 h at room temperature. After three washes with reaction buffer (20 mM HEPES, 72 mM KCl, 1.5 mM MgCl2, 1.6 mM MgOAc, 0.5 mM DTT, 4 mM glycerol, 1 mM ATP, 200 units of RNasin; Promega), the resin-bound proteins were eluted with 100 μl of 7 M urea and were analyzed by liquid chromatography-tandem mass spectrometry (LC- MS/MS).
Following elution, the samples were analyzed at the proteomics facility of the National Institute of Biological Sciences. After trypsin digestion, the peptide mixtures were separated by either a single C18 reversed-phase column or a two-dimensional high-performance liquid chromatography (2D-HPLC) setup (cation exchange column coupled to C18 reversed-phase column). The eluted peptides were ionized and directly sprayed into an LTQ tandem mass spectrometer (ThermoFisher Scientific). The MS data were processed on an in-house Sequest (29) cluster to generate a protein list for each sample. The protein lists were then compared to determine the proteins that specifically interact with the tau exon 10 stem-loop RNA transcript.
Plasmids, antibodies, and purified proteins.
The wild-type (wt) tau9-11 and tau10-11 minigene constructs or corresponding mutant minigene constructs containing a C-to-T single-nucleotide mutation in intron 10 at the +14 position, named the DDPAC mutation, were described previously (60). Mammalian expression plasmids for hemagglutinin (HA)-tagged wild-type or LGLD mutant p68 (76) and Flag-RBM4 and Flag-Raver (44, 76) were previously described. Bacterial expression constructs for His-tagged wild-type p68 and LGLD mutant p68 were previously described (52, 75, 107). Monoclonal anti-Flag antibody was purchased from Sigma. Monoclonal anti-HA antibody was purchased from Covance.
Wild-type or LGLD mutant 6×His-tagged p68 protein was purified as previously described using HisTrap and Akta purifier (GE) (107).
Cell culture, transfection, and alternative splicing assay.
HEK293 cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal calf serum and transfected as previously described with 1 to 3 μg of DNA containing the tau minigene and plasmids expressing different candidate splicing regulators (60, 72). Cells were harvested 48 h after transfection, and RNA was extracted using an RNAspin mini-RNA isolation kit (GE). tau exon 10 alternative splicing products or other transcripts were detected by reverse transcription-PCR (RT-PCR) following reverse transcription with Superscript II (Invitrogen), as previously described (62). In RNA interference (RNAi) experiments, 1 to 3 μg of either control or p68-specific interfering RNA (with the nucleotide sequence described in reference 47) together with 1 μg tau minigene was cotransfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) genes was detected as internal controls using corresponding specific primers. PCR products were separated by electrophoresis using 6% polyacrylamide gels. PCR bands were quantified using a BAS 5000 Phosphoimager (Fuji Films).
RNA-protein interactions.
Different RNA probes were prepared and radiolabeled by in vitro transcription in the presence of [α-32P]UTP (3,000 Ci/mmol; MP Biomedical) using T7 RNA polymerase from corresponding linearized DNA templates as described previously (60, 61). Wild-type or DDPAC mutant (containing a single-nucleotide C-to-T change at position +14 of intron 10) tau RNA oligonucleotides containing the last 24 residues of exon 10 and the first 24 residues of intron 10 of the human tau gene were end labeled using polynucleotide kinase (Roche) in the presence of [γ-32P]ATP (7,000 Ci/mmol; MP Biomedical). For UV cross-linking, ∼20 fmol of RNA probe was incubated at 30°C with 100 μl of cell lysates in a 150-μl reaction mixture under in vitro splicing conditions (60, 61). The samples were irradiated on ice under a 254-nm UV lamp for 10 min (120,000 microjoules/cm2) using a Stratalinker 2000 UV cross-linker (Stratagene). The probes were digested in equal volumes of RNase A (5 mg/ml; Sigma) at 30°C for 20 min, except for experiments in which the end-labeled probes were used. For immunoprecipitation, following the RNase treatment, samples were incubated with monoclonal M2 anti-Flag antibody (Sigma) at 4°C for 2 h. Protein A/G agarose beads were then added with further incubation and gentle mixing. Following washing, the immunoprecipitated RNA-bound proteins were eluted and resolved on SDS-PAGE followed by autoradiography.
Oligonucleotide-directed RNase H cleavage assay.
[α-32P]UTP-labeled wild-type and DDPAC tau pre-mRNA transcripts containing exon 10-intron 10 sequence were used in this assay, as illustrated in Fig. 5 and 6. The RNA probes were incubated at 37°C for 60 min in standard splicing buffer in the presence of 0.5 U of RNase H (Roche) in 25-μl reaction mixtures with 50 fmol of an oligonucleotide (5′-GAAGGTACTCACACTGCC-3′) complementary to the 5′ splice site of exon 10 together with the purified recombinant wild-type or LGLD mutant p68 protein as described earlier (60). Cleaved RNA products were separated on 6% polyacrylamide denaturing gels containing 8 M urea followed by autoradiography and quantification using a PhosphorImager as described previously (60). The efficiency of RNase H cleavage was calculated by dividing the band intensities of the cleaved fragments by the band intensities of the total RNA fragments in each lane.
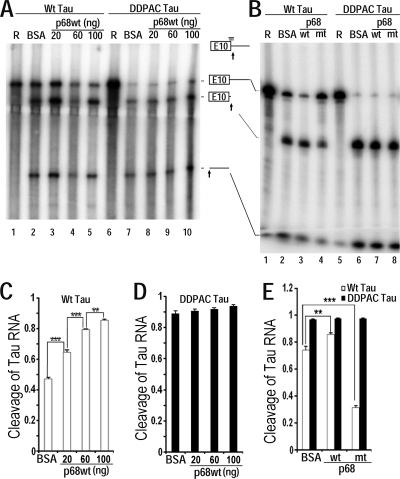
Fig. 5.
Oligonucleotide-directed RNase H cleavage assay shows that p68 changes the conformation of the stem-loop region at the 5′ splice site of tau exon 10. (A and B) Radiolabeled wild-type tau pre-mRNA transcript containing exon 10 and intron 10 (A, lanes 1 to 5; B, lanes 1 to 4) or the DDPAC mutant transcript (A, lanes 6 to 10; B, lanes 5 to 8) was incubated with RNase H and the oligonucleotide complementary to the 5′ splice site at 37°C for 30 min under splicing conditions in the presence of purified p68 protein: wild-type p68 (A, lanes 3 to 5 and 8 to 10; B, lanes 3 and 7) or LGLD mutant p68 (B, lanes 4 and 8) or BSA control (A, lanes 2 and 7; B, lanes 2 and 6). After incubation, the RNA was isolated, separated by denaturing gel electrophoresis, and detected by autoradiography. Panel A lanes 1 and 6 and panel B lanes 1 and 5 contain corresponding input RNA transcripts used. The tau RNA transcript and its RNase H cleavage products are illustrated in the diagram between panels A and B, with the arrows indicating the cleavage sites and the horizontal bar above the exon 10-intron 10 junction representing the DNA oligomer which formed base pairing (vertical bars) with the tau RNA sequence. (C to E) Quantification of data from lanes 2 to 5 in panel A (C), lanes 7 to 10 in panel A (D), and the corresponding lanes in panel B (E) of cleavage of tau RNA as measured using a PhosphorImager. Data are presented as the ratios of total cleavage products to the corresponding total transcripts in each lane ± standard error (***, P < 0.0001; **, P < 0.001).
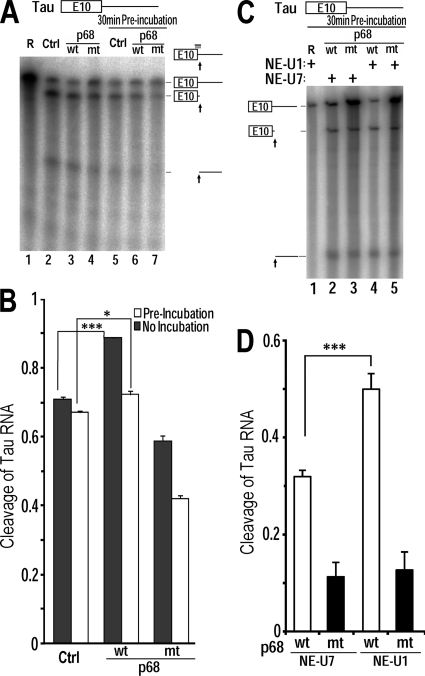
Fig. 6.
RNA helicase p68 promotes U1snRNP binding to the 5′ splice site of tau exon 10. (A) Radiolabeled wild-type tau pre-mRNA transcript was incubated at 30°C in HeLaNE in the presence of purified wild-type p68 (wt, lanes 3 and 6) or LGLD mutant p68 (mt, lanes 4 and 7) or the BSA control (Ctrl, lanes 2 and 5) for either 0 min (lanes 2 to 4) or 30 min (lanes 5 to 7). Following incubation, RNase H (0.5 U) and the DNA oligomer complementary to the 5′ splice site of tau exon 10 were added and the reaction mixture was incubated at 37°C for 30 min. The RNA cleavage products were analyzed by denaturing gel electrophoresis. (B) Quantification of the cleavage of the tau RNA transcript was measured as the ratio of total amounts of cleavage products to the corresponding total transcripts in each lane ± standard error (***, P < 0.0001; *, P < 0.005). (C) tau RNA transcripts were incubated in the presence of purified p68 protein, wt p68 (lanes 2 and 4) or LGLD mutant p68 (lanes 3 and 5), at 30°C for 30 min with either U1snRNP-depleted HeLaNE (NE-U1; lanes 4 and 5) or U7 snRNP-depleted HeLaNE (NE-U7; lanes 2 and 3). Following incubation, the oligonucleotide complementary to the 5′ splice site of tau exon 10 was added along with RNase H, and the incubation was continued for another 30 min at 37°C. The RNA cleavage products were then analyzed by denaturing gel electrophoresis. (D) Quantification of the cleavage of tau RNA transcript was measured as the ratio of total cleavage products to the corresponding total transcript in each lane ± standard error (***, P < 0.0001).
U1snRNP depletion and U1snRNP protection assay.
U1snRNP was depleted using oligonucleotide-targeted RNase H cleavage as described in our previous study (60). Briefly, HeLa cell nuclear extract was incubated at 30°C for 1 h in the presence of RNase H and the oligonucleotide 5′-CCAGGTAAGTAT-3′ complementary to the 5′ end of U1snRNA (reference 60 and references within). As a control, U7-depleted nuclear extract was obtained by incubation with RNase H and a U7snRNA-specific oligonucleotide. The U1snRNP protection assay was carried out under splicing reaction conditions with 50,000 cpm of 32P-labeled tau pre-mRNA transcripts with nontreated, U1-depleted, or U7-depleted nuclear extracts at 30°C for 0 or 20 min. RNase H (0.4 U) and 20 pmol (molar excess) of the oligonucleotide complementary to the 5′ splice site of exon 10 (5′-GAAGGTACTCACACTGCC-3′) were then added to 25-μl splicing reaction mixtures (60), and incubation was continued for 30 min at 37°C to completely cleave the pre-mRNA transcripts that were not protected. Each reaction mixture was then separated on 6% polyacrylamide denaturing gels containing 8 M urea followed by autoradiography and quantification using a PhosphorImager as described previously (60). The efficiency of RNase H cleavage was calculated by dividing the band intensities of the cleaved fragments by the total band intensities of the total RNA fragments in each lane.
Immunoprecipitation and Western blot analysis.
HEK293 cells transfected with plasmids expressing HA-tagged wild-type p68 or LGLD mutant p68 or Flag-tagged RBM4 or Raver were lysed with a lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 1 mM DTT, 1× protease inhibitor mixture [Roche Molecular Biochemicals]) as described previously (65). Lysates were immunoprecipitated with specific antibodies and protein A/G agarose beads for 2 h at 4°C. Fifty percent of the immunoprecipitated proteins and 20% of the cell lysate used for the pulldown were separated using SDS-PAGE and detected by Western blotting using corresponding specific antibodies with an enhanced chemiluminescence kit (GE Healthcare).
RESULTS
DDX5/RNA helicase p68 interacts with the tau exon 10 stem-loop structure.
Previous studies indicate that splicing mutations in the human tau gene that are predicted to destabilize the exon 10 stem-loop structure increase tau exon 10 inclusion, suggesting that this stem-loop region plays an important role in regulating tau exon 10 splicing (54, 60, 103, 104). This model also suggests that proteins influencing the stability of this stem-loop structure may regulate tau exon 10 splicing. To search for splicing regulators that interact with the tau exon 10 stem-loop structure, we established an RNA affinity pulldown-coupled mass spectrometry approach. As illustrated in Fig. 1A, a biotinylated RNA oligomer corresponding to the tau exon 10 stem-loop region was incubated under splicing conditions with HeLa nuclear extract (HeLaNE). RNA-protein complexes were isolated by affinity pulldown using streptavidin beads. Protein components of the RNA-protein complex were identified using liquid chromatography-coupled tandem mass spectrometry (LC-MS/MS). A random RNA oligonucleotide was used as an internal control to exclude proteins that nonspecifically bind RNA. Table 1 profiles the proteins identified by this RNA affinity pulldown approach. Candidate proteins of interest included alternative splicing regulators such as SAP62, TIA-1, Fox2, p68, and RBM25. We were particularly interested in p68, as it had been reported to be an alternative splicing regulator that functioned by influencing RNA secondary structures such as that in H-ras (13, 16).
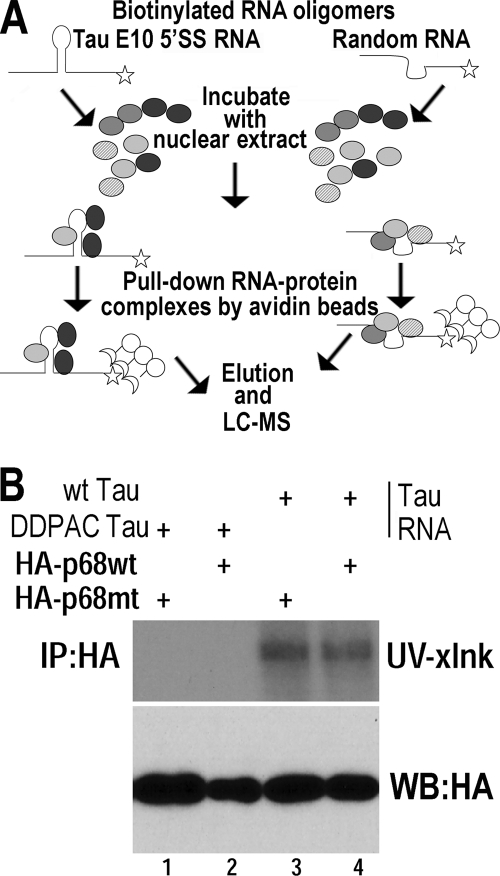
Fig. 1.
RNA helicase p68 interacts with the tau pre-mRNA at the 5′ splice site stem-loop region downstream of exon 10. (A) A diagram illustrating the RNA affinity pulldown approach. (B) UV cross-linking experiments were performed using cell lysates prepared from HEK293 cells expressing HA-tagged wild-type p68 (lanes 2 and 4) or LGLD mutant p68 (lanes 1 and 3). Wild-type (wt) and DDPAC tau RNA oligomers (lanes 3 and 4 and lanes 1 and 2, respectively) were radiolabeled and used in the UV cross-linking assay. Following UV cross-linking and RNase treatment, the reaction products were analyzed by SDS-PAGE and autoradiography.
Table 1.
Candidate proteins affinity purified with the tau exon 10 stem-loop structure-containing RNA
| Protein description and/or name | GI no. | Mass (Da) | x-corra | Sequence coverage (%) |
|---|---|---|---|---|
| DDX5/p68 | GI:4758138 | 69,148 | 3.71 | 5 |
| prp40 | GI:7106840 | 105,270 | 3.65 | 5 |
| hnRNPC | GI:13937888 | 35,598 | 3.35 | 18 |
| TIA | GI:6094480 | 42,460 | 3.21 | 8.5 |
| TIA-1-related protein, TIAR | GI:4507499 | 41,250 | 2.27 | 6.5 |
| RBM9/Fox2 | GI:6572238 | 41,360 | 3.68 | 6.5 |
| hnRNP M | GI:33874022 | 44,443 | 2.44 | 4 |
| SF3A, subunit 1 | GI:5032087 | 87,230 | 2.11 | 4 |
| SF3B subunit 1/SAP155 | GI:6912654 | 145,724 | 3.42 | 4 |
| SAP62 | GI:21361376 | 51,040 | 2.55 | 4 |
| U1-70K | GI:29568103 | 48,070 | 2.84 | 8 |
| RBM25 | GI:37545959 | 84,560 | 2.43 | 6 |
x-corr, cross correlation score.
To confirm that p68 interacted with tau pre-mRNA in the stem-loop region downstream of exon 10, UV cross-linking-coupled immunoprecipitation experiments were carried out. The wild-type (wt) tau stem-loop-containing RNA oligomer used in the RNA affinity pulldown experiment or a mutant tau stem-loop oligomer containing a C-to-T mutation at the intron 10 +14 position, named the DDPAC mutation (17, 54), was end labeled using [γ-32P]ATP and incubated with protein lysates prepared from HEK293 cells transfected with plasmids expressing either wild-type p68 or LGLD mutant p68. This p68 mutant lacks ATPase/helicase activity and contains an arginine (R)-to-leucine (L) change in the consensus sequence motif (V), RGLD, thus being named the LGLD mutant (75). As shown in Fig. 1B, both wild-type and mutant HA-p68 interacted with the wt tau stem-loop-containing RNA (lanes 3 and 4). However, no interaction was detected between the DDPAC mutant tau RNA and either wild-type or mutant p68 protein (Fig. 1B, lanes 1 and 2). These results suggest that p68 indeed interacts with wt tau RNA in the stem-loop region but not with the DDPAC mutant tau RNA containing a single-nucleotide change in the stem region that destabilizes the stem structure.
RNA helicase p68 stimulates tau exon 10 inclusion.
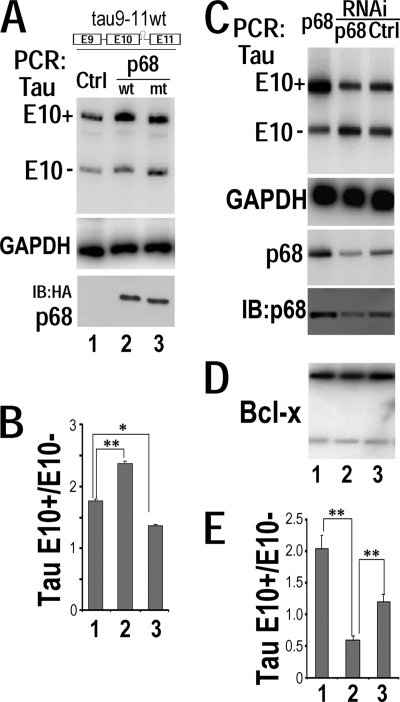
To examine the functional significance of the interaction of p68 with tau pre-mRNA at the exon 10 stem-loop region, we tested the effect of p68 on tau exon 10 splicing. HEK293 cells were cotransfected using a wild-type tau minigene containing exons 9, 10, and 11 (tau9-11wt [60]) together with either the vector control or wild-type or LGLD mutant p68 expression plasmid (Fig. 2A and B). The tau exon 10 alternative splicing products were detected using RT-PCR with specific primers as described earlier (60). Overexpression of wild-type p68 increased tau exon 10 inclusion compared to that in the vector control (Fig. 2A and B, lanes 1 and 2). Interestingly, overexpression of LGLD mutant p68 suppressed tau exon 10 inclusion, leading to increased formation of the isoform lacking exon 10 (E10−) (Fig. 2A and B, compare lanes 1 and 3), indicating that the RNA helicase activity is involved in the p68 activity in regulating tau exon 10 splicing. Next, we performed RNA interference (RNAi)-mediated knockdown of p68 to test the effect of the endogenously expressed p68 on tau exon 10 splicing. HEK293 cells were cotransfected using the tau9-11wt minigene together with p68-specific interfering RNA or control interfering RNA or wild-type p68 expression plasmid (Fig. 2C). Consistent with the previous studies in which p68 knockdown led to increased cell death (59, 96), it was difficult to drastically downregulate p68 expression without causing significant cytotoxicity. Under our conditions, without eliciting significant cell death, we were able to achieve approximately a 50% decrease in p68 transcript levels, as detected by RT-PCR with p68-specific primers and Western blotting (Fig. 2C, compare lanes 2 and 3). With RNAi-mediated reduction in p68 expression, the tau exon 10-containing splicing product (E10+) was significantly decreased with a concomitant increase in the tau E10− isoform (Fig. 2C and E, compare lanes 2 with lanes 1 and 3). On the other hand, alternative splicing of Bcl-x remained unaffected (Fig. 2D), demonstrating the specificity of p68 in regulating tau exon 10 alternative splicing.
Fig. 2.
RNA helicase p68 regulates tau exon 10 inclusion. (A and B) The corresponding plasmids expressing either the vector control (lanes 1), wild-type p68 (lanes 2), or LGLD mutant p68 (lanes 3) were cotransfected with the wild-type tau9-11 minigene (tau9-11wt) into HEK293 cells. (A) tau exon 10 (E10) alternative splicing was detected using RT-PCR, with GAPDH transcripts in the corresponding groups showing comparable amounts of RNA used in each reaction mixture. The levels of wt and LGLD mutant p68 protein in the corresponding reaction mixtures, as detected using Western blotting with anti-HA antibody, are shown. (B) Quantification of tau exon 10 splicing with data derived from six independent experiments. The graph shows the average ratios of tau E10+ to E10− transcripts ± standard errors (**, P < 0.001; *, P < 0.005). (C to E) The tau9-11wt minigene was cotransfected into HEK293 cells together either with the construct expressing wild-type p68 (lanes 1) or with interfering RNA specific for human p68 (lanes 2) or control interfering RNA (lanes 3). Alternative splicing products were detected by RT-PCR using corresponding specific primers. (C) tau exon 10 alternative splicing isoforms were detected by RT-PCR, with GAPDH transcript as the internal control. Levels of p68 RNA were detected by RT-PCR and immunoblotting. (D) Alternative splicing of the endogenous Bcl-x was not affected by p68 levels, as detected by RT-PCR using Bcl-x-specific primers. (E) Quantification of tau exon 10 splicing isoforms as shown in panel A. The graph shows the average ratios of E10+ to E10− transcripts ± standard errors, with data from six independent experiments (**, P < 0.001).
RNA helicase p68 interacts with RBM4, a tau exon 10 splicing activator.
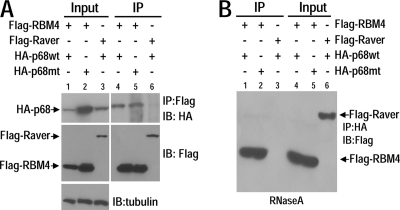
Using an expression cloning screening approach, we had previously identified RNA binding motif protein 4 (RBM4) as an intronic splicing activator of tau exon 10 splicing (65). RBM4 increases tau exon 10 inclusion by binding to an intronic splicing enhancer located 100 nucleotides downstream of the 5′ splice site of exon 10 (65); however, the mechanism of RBM4-mediated exon 10 splicing stimulation was unclear. Because p68 had a similar effect on tau exon 10 splicing, we investigated if RBM4 interacted with p68. We performed coimmunoprecipitation experiments using HEK293 cells cotransfected with epitope-tagged p68 and RBM4-HA-tagged p68 and Flag-tagged RBM4 (Fig. 3). RBM4 was coimmunoprecipitated with either wt or LGLD mutant p68 (Fig. 3A, lanes 4 and 5). This interaction with p68 was specific to RBM4, because another RNA binding motif-containing protein, Raver, was not coimmunoprecipitated with p68 (Fig. 3A, lane 6). To test if the interaction of RBM4 with p68 was dependent on RNA, we carried out coimmunoprecipitation experiments in the presence of RNase A. The specific association of both wt and LGLD mutant p68 with RBM4, but not with Raver, was detected in coimmunoprecipitation following treatment with RNase A, demonstrating that p68 specifically interacted with RBM4 in an RNA-independent manner (Fig. 3B).
Fig. 3.
RNA helicase p68 interacts with RBM4. An anti-Flag antibody (A) or anti-HA antibody (B) was used to immunoprecipitate (IP) protein complexes from HEK293 cell lysates coexpressing either Flag-RBM4 and wild-type HA-p68 (HA-p68wt) or Flag-RBM4 and LGLD mutant HA-p68 (HA-p68mt) or Flag-Raver and wild-type HA-p68, as indicated above each lane of the corresponding panels. The cell lysates used in panel B were treated with RNase A prior to immunoprecipitation. The expression of p68 or Raver proteins was detected by immunoblotting (IB) using anti-HA or anti-Flag antibodies, respectively.
RNA helicase p68-mediated stimulation of tau exon 10 inclusion is dependent on RBM4.
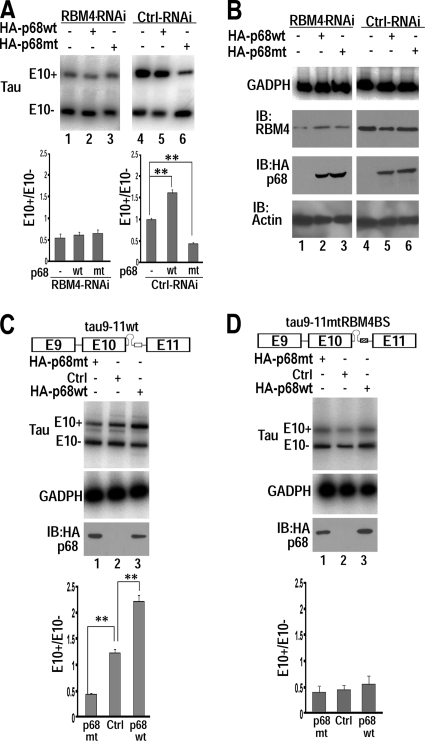
To examine the functional significance of the interaction between p68 and RBM4, we tested the effects of RBM4 on p68-mediated tau exon 10 regulation by knocking down RBM4 (Fig. 4). In cells treated with the control interfering RNA, overexpression of wild-type p68 stimulated, whereas the LGLD mutant p68 lacking ATPase/helicase activity inhibited, tau exon 10 inclusion (Fig. 4A and B, compare lanes 5 and 6 with lanes 4), similar to our previous observation (Fig. 2). When RBM4 was downregulated by RBM4-specific RNAi, effects of either wt or LGLD mutant p68 on tau exon 10 splicing were no longer detectable (Fig. 4A, compare lanes 2 and 3 with lane 1). These data indicate that the activity of p68 in regulating tau exon 10 splicing is RBM4 dependent.
Fig. 4.
RNA helicase p68-mediated tau exon 10 splicing stimulation is dependent on RBM4 and on interaction of RBM4 with tau pre-mRNA. (A and B) RBM4-specific interfering RNA (lanes 1 to 3) or control interfering RNA (lanes 4 to 6) was cotransfected into HEK293 cells with the tau9-11wt minigene and the vector control (lanes 1 and 4) or HA-tagged wt p68 (lanes 2 and 5) or LGLD mutant p68 (lanes 3 and 6). tau exon 10 alternative splicing was detected by RT-PCR, with quantification of data from six independent experiments. The graphs show the average ratios of tau E10+ to E10− transcripts ± standard errors (**, P < 0.001). Shown in panel B are the levels of internal control GAPDH transcript detected by RT-PCR or levels of RBM4 or wt or mutant p68 or actin proteins as detected by immunoblotting (IB) using anti-RBM4, anti-HA, or antiactin antibodies, corresponding to reactions in panel A, respectively. (C and D) The expression plasmid for wt p68 or LGLD mutant p68 or vector control (Ctrl) was cotransfected into HEK293 cells with either the wild-type tau9-11 minigene (tau9-11wt) (C) or the mutant tau minigene (tau9-11mtRBM4BS) in which the RBM4 binding site was mutated (D). tau exon 10 alternative splicing was assayed using RT-PCR, with GAPDH transcript as a loading control. Levels of wt or LGLD mutant p68 were detected by immunoblotting (IB) using anti-HA antibody. The graphs show the average ratios of tau E10+ to E10− transcripts ± standard errors, with data from 6 independent experiments (**, P < 0.001).
We further examined whether this activity of p68 was dependent on the interaction of RBM4 with the tau pre-mRNA. We made use of a mutant tau pre-mRNA minigene in which the RBM4 binding site was mutated, tau9-11mtRBM4BS (65). As shown in Fig. 4C and D, both wt p68-mediated stimulation of tau exon 10 inclusion and LGLD mutant p68-mediated suppression of tau exon 10 splicing were abolished when the interaction of RBM4 with tau pre-mRNA was disrupted by its binding site mutation (Fig. 4C and D, compare lanes 1 to 3 with lanes 4 to 6). These results indicate that p68-mediated exon 10 splicing activation is dependent on its interaction with RBM4 and on the ability of RBM4 to interact with the tau pre-mRNA.
RNA helicase p68 promotes conformational changes in tau exon 10 stem-loop structure.
To further investigate the mechanism underlying p68-mediated tau exon 10 splicing activation, we examined the effect of p68 on the tau exon 10 stem-loop structure. We had developed an oligonucleotide-directed RNase H cleavage assay in which a DNA oligomer complementary to the 5′ splice site downstream of tau exon 10 was used to examine conformational changes in this stem-loop region (60). We performed the oligonucleotide-directed RNase H cleavage assay using either wt tau pre-mRNA or DDPAC mutant tau transcript containing the single-nucleotide change that reduced the base pair interaction in the stem region. The RNase H cleavage assay was performed using either wt or DDPAC mutant tau transcripts in the presence of control bovine serum albumin (BSA) or purified p68 protein (Fig. 5). Incubation of wt p68 protein with wt tau RNA transcript led to a concentration-dependent increase in the cleavage of the wt tau RNA transcript (Fig. 5A, lanes 3 to 5 and lane 1; Fig. 5B, compare lanes 2 and 3), whereas LGLD mutant p68 decreased the cleavage of the wt tau RNA transcript (Fig. 5B, compare lanes 2 and 4). On the other hand, neither wt p68 nor LGLD mutant p68 changed the cleavage of DDPAC mutant tau transcript by RNase H (Fig. 5A, lanes 6 to 10; Fig. 5B, lanes 6 to 8). These results suggest that interaction of p68 with the tau stem structure may lead to a more open conformation in this region, allowing enhanced access of the DNA oligomer and increased RNase H cleavage. Once the stem structure is destabilized by the DDPAC mutation, the effect of p68 is no longer detectable. Also, the p68-mediated conformational change is dependent on the RNA helicase activity, because the helicase-dead LGLD mutant p68 decreased RNase H cleavage of the wt tau RNA.
RNA helicase p68 increases U1snRNP binding to the 5′ splice site of tau exon 10.
Because the stem-loop structure overlaps with the site for U1snRNP binding to the tau exon 10 5′ splice site, U1snRNP access to this 5′ splice site may be affected by the stem-loop structure. Consistent with this model, FTLD-tau mutations that destabilize this stem structure increase exon 10 inclusion (17, 53, 54, 60). Our results show that overexpression of p68 increased tau exon 10 splicing (Fig. 2) and that p68 affected the conformation of the stem-loop structure at the 5′ splice site of tau exon 10 (Fig. 5); hence, we investigated whether p68 influenced U1snRNP interaction with this 5′ splice site. We had previously established a U1 protection assay to measure U1snRNP binding to tau pre-mRNA at this 5′ splice site (60). In this assay, U1snRNP binding at the 5′ splice site of tau exon 10 was assessed as a measure of the U1snRNP-mediated protection of the tau RNA from RNase H cleavage following incubation of the tau RNA transcripts with HeLa nuclear extract (HeLaNE) either containing or lacking U1snRNP under splicing conditions. The radiolabeled wt tau RNA transcript was first preincubated with HeLaNE under splicing conditions for 0 or 30 min to recruit U1snRNP to the tau 5′ splice site (Fig. 6). The DNA oligomer complementary to the 5′ splice site was then added together with RNase H. The RNase H-cleaved and -protected RNA fragments were resolved by denaturing polyacrylamide gel electrophoresis and quantified with a PhosphorImager. As shown in Fig. 6A and B, without preincubation with HeLaNE (lanes 2 to 4), wt p68 protein enhanced RNase H cleavage (compare lane 3 with lane 2). This enhancement was significantly reduced upon preincubation of the tau RNA transcript with HeLaNE, suggesting that HeLaNE contained a factor(s) that protected this 5′ splice site from the binding of the DNA oligomer and RNase H cleavage. Consistent with the finding that LGLD mutant p68 interacted with wt tau RNA (Fig. 1B), this helicase-dead mutant appeared to stabilize the stem structure, making the tau RNA less accessible to RNase H cleavage, and the preincubation with HeLaNE further decreased tau RNA cleavage (Fig. 6A, lanes 4 and 7). We next tested the role of U1snRNP in protecting this 5′ splice site in tau pre-mRNA using the HeLaNE in which U1snRNP was depleted by RNase H with an oligonucleotide specific against U1snRNA, NE-U1 (60). Such a U1-depleted HeLaNE was splicing incompetent as confirmed using an in vitro splicing assay (60). As a control, HeLaNE depleted of U7snRNA was used. HeLaNE depleted in either U1snRNA or U7snRNA was used in the U1 protection assay together with the wt tau RNA transcript. Depletion of U1snRNA in the nuclear extract (NE-U1) led to significantly increased RNase H cleavage of the wt tau RNA compared with that in the reaction using the control U7-depleted nuclear extract (NE-U7) in the presence of wild-type p68 under the same preincubation conditions (Fig. 6C, compare lane 4 with lane 2; Fig. 6D). Interestingly, the LGLD mutant p68 seemed to interact with and protect the wt tau RNA from RNase H cleavage, and this protection was not affected by the depletion of U1snRNP from nuclear extract (Fig. 6C, compare lanes 3 and 5 with lanes 2 and 4; Fig. 6D). These results support our finding that p68 interacts with and destabilizes the stem structure, thereby enhancing U1snRNP recognition of the 5′ splice site downstream of tau exon 10.
DISCUSSION
Multiple lines of evidence suggest that disruption of tau exon 10 splicing regulation plays an important role in the pathogenesis of tauopathy. More than 30 mutations have been identified among FTDP-17 patients, many of which affect tau exon 10 splicing (26, 45, 54, 60, 61, 91, 92, 97; reviewed in references 2, 53, and 66). In addition, a number of studies have reported an altered Tau4R-to-Tau3R ratio or aberrant tau exon 10 splicing in a range of sporadic tauopathies, including progressive supranuclear palsy (PSP), corticobasal degeneration (CBD), multiple system tauopathy with dementia (MSTD), and argyrophilic grain disease (AGD), and even in some Alzheimer's patients, among others (20, 39, 101; reviewed in references 2, 12, 36, and 78 and references therein). It is interesting that a significant fraction of these patients show increased levels of Tau4R without detectable mutations in the tau gene. These studies underscore the significance of tau exon 10 splicing regulation in the maintenance and function of the nervous system and also suggest that factors other than tau mutation may play a role in tauopathy, including tau alternative splicing regulators.
Molecular studies from several groups, including ours, have begun to reveal cis elements and trans-acting factors regulating tau exon 10 alternative splicing. One of the cis elements involved is the stem-loop structure at the tau exon 10-intron 10 junction, which blocks the maximal access of U1snRNP to the 5′ splice site (45, 54, 60, 103, 104). Mutations that destabilize the base pairing of the stem structure lead to increased exon 10 inclusion (54, 60, 103, 104, 109). These observations suggest that modulation of the conformation or stability of this stem-loop structure may serve as a critical mechanism for tau exon 10 splicing regulation. Prior to this study, no protein factors had been identified to destabilize this stem structure and no RNA helicase had been found to modulate the function of this stem-loop region in tau splicing. RNA secondary structures have been implicated in regulation of alternative splicing in a number of genes (reviewed in references 10, 47, and 83). Understanding how RNA stem-loop structures are regulated by RNA helicases may have general implications for alternative splicing regulation.
In this study, we have developed an RNA affinity pulldown-coupled mass spectrometry approach to search for potential trans-acting factors that interact with the tau exon 10 stem-loop structure. This approach can be used for studying other important cellular processes involving regulatory RNA elements. With this approach, we have identified candidate trans-acting factors (Table 1), including p68, that interact with the tau pre-mRNA stem-loop region. ATP-dependent DEAD/H box-containing RNA helicase p68 is involved in a wide range of cellular processes from transcriptional to posttranscriptional gene regulation: transcription activation, pre-mRNA splicing, and rRNA, microRNA (miRNA), and ribosome biogenesis. Some of the p68-interacting proteins have been reported, including transcription factors (AF1, MyoD, and p53) and splicing regulatory proteins such as hnRNP A1, hnRNP H, and FUS-TLS, among others (11, 13, 14, 16, 22, 32, 33, 34, 35, 46, 57, 58, 75, 77, 93–95, 108; for a recent review, see reference 59).
Previous analyses of spliceosomal complexes have revealed p68 as a component of mammalian splicing machinery (15, 48, 63, 79). Detection of cross-linking of p68 to the U1snRNA-5′ splice site duplex led to the proposal of p68 as an essential pre-mRNA splicing factor involved in unwinding this RNA duplex (77). Our RNase H cleavage experiments using purified p68 protein show that p68 alone is sufficient for interacting with the wt tau pre-mRNA and also suggest that p68 may be recruited to the 5′ splice site prior to U1snRNP recruitment to change the secondary structure of tau pre-mRNA, thus enhancing U1snRNP binding to the 5′ splice site (Fig. 7). Our studies on the role of p68 in promoting tau exon 10 inclusion reveal that its ATPase/RNA helicase activity is essential for its splicing activity.
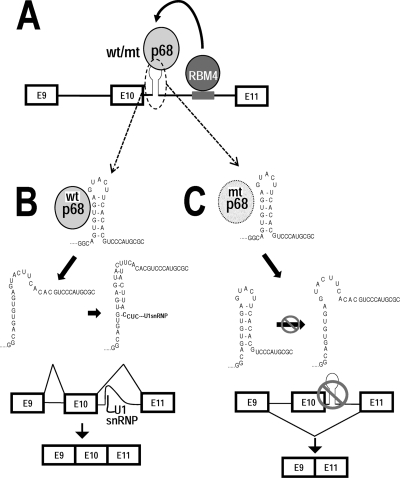
Fig. 7.
A model for RNA helicase p68 in regulation of tau exon 10 splicing. (A) A diagram illustrating the model for p68 in regulating tau exon 10 splicing. RNA helicase p68 binds to a stem-loop structure at the exon 10-intron 10 junction to tau pre-mRNA and interacts via protein-protein interaction with RBM4, which binds to an intronic splicing enhancer to promote tau exon 10 splicing. (B) The tau exon 10 stem-loop exists in a dynamic equilibrium. When the stem-loop is in a closed conformation, it sequesters the 5′ splice site and prevents the access of U1snRNP to the 5′ splice site, leading to exon 10 exclusion (reference 60 and this study). When the stem-loop is in an open conformation, U1snRNP interacts with the 5′ splice site and recruits the splicing machinery to utilize this splice site, thereby increasing tau exon 10 inclusion. Both wild-type and LGLD mutant p68 can bind to the stem-loop structure in the wild-type tau pre-mRNA. However, only the helicase-active wild-type p68 can open the stem structure, facilitating U1snRNP binding and promoting exon 10 splicing. The helicase-dead LGLD mutant p68 binds to and stabilizes the stem structure, reducing U1snRNP access to the 5′ splice site and thus suppressing exon 10 inclusion.
It has been suggested that p68 is responsible for ATP-dependent unwinding of the U1snRNA-5′ splice site duplex (75, 77). However, it was not clear whether p68 affects pre-mRNA splicing of all genes or certain specific targets. We have tested a number of alternative splicing substrates and found that not all splicing events are affected by p68, including Bcl-x (Fig. 2D) and others. This suggests that p68 may affect only selected alternative splicing events. Biochemical studies on CD44 and H-ras splicing show that p68 suppresses inclusion of alternative exons (13, 16, 46). In the case of CD44 splicing, the role of p68 in suppressing CD44 variable exon inclusion is suggested to be cotranscriptional, wherein p68 enhances transcription of androgen-responsive genes by binding to the androgen receptor response element (ARE) in the promoter regions of these genes. It was proposed that p68 might affect splicing factor recruitment to RNA, thereby influencing exon splicing (16). In the H-ras gene, inclusion or skipping of its alternative intron D exon (IDX) leads to formation of p19 or p21 isoforms, and p68 suppresses IDX inclusion by interacting with an intronic element that may contain an extended stem-loop structure (13, 46). In this case, p68 blocks the binding of hnRNP H to the intronic region in ras pre-mRNA and acts as a splicing suppressor (13).
Overexpression of wild-type p68 increased tau exon 10 inclusion, whereas LGLD mutant p68 lacking ATPase/helicase activity decreased exon 10 splicing (Fig. 2A), suggesting that the ATPase/helicase activity of p68 is required for its function in regulating tau exon 10 splicing and that the mutant p68 may bind to and stabilize the stem structure, making the 5′ splice site less accessible to splicing machinery. Such ATPase/helicase-dependent regulatory activity of p68 in modulating alternative splicing has not been reported for other previously studied splicing target genes of p68.
Our results show, for the first time, that p68 promotes the recruitment of U1snRNP by binding to a stem-loop structure at the 5′ splice site and activates tau exon 10 inclusion in an ATPase/helicase-dependent manner (Fig. 7). Therefore, the role of p68 in tau exon 10 splicing regulation and the underlying mechanism reported in this study are different from that of p68 in regulating alternative splicing of either H-ras or CD44 genes.
Many tau splicing mutations associated with FTDP-tau, such as DDPAC, increase exon 10 splicing by destabilizing the stem-loop structure (60, 103, 104). Consistent with this, lengthening the stem region or binding of antibiotics that increase the stability of the stem structure decreases tau exon 10 inclusion (25, 64, 98, 109). Using RNase H cleavage and U1 protection assays, we probed potential conformational changes in the stem-loop region (Fig. 5 and 6). Our results suggest that the wild-type p68 may destabilize the stem structure, releasing the 5′ splice site and making it more accessible for U1snRNP interaction, thereby increasing exon 10 inclusion. In contrast, the ATPase/helicase-dead LGLD mutant p68 decreased tau RNA cleavage, suggesting stabilization of the stem-loop structure (Fig. 5). These results are consistent with the splicing changes observed in transfected cells. On the other hand, DDPAC mutant tau RNA did not show any changes in RNase H cleavage or U1snRNP protection, consistent with the UV cross-linking data showing that DDPAC mutant tau RNA did not interact with either wild-type or mutant p68. This further supports the idea that the interaction with tau pre-mRNA and the splicing activation activity of p68 are dependent on the presence of a more stable stem structure at the exon 10-intron 10 junction of tau pre-mRNA. However, these results do not preclude other potential p68-mediated effects such as remodeling of RNA-protein and protein-protein interaction at the 5′ splice site of tau exon 10 as possible mechanisms of action for p68.
RNA binding protein RBM4 is a multifunction protein containing an RNA recognition motif (82). RBM4 plays a role in regulating alternative splicing of alpha-tropomyosin, survival of motor neuron gene 2 (SMN2), SRp20 reporter genes, and tau exon 10 (65, 76, 81). An expression cloning screening of an adult human brain cDNA library together with biochemical studies identified RBM4 as a regulator of tau exon 10 splicing (65). RBM4 promotes tau exon 10 splicing by interacting with an intronic splicing enhancer element 100 nucleotides downstream of exon 10 (65). In the current study, we demonstrate that RBM4 interacts with RNA helicase p68 in an RNA-independent manner (Fig. 3). However, the splicing activation function of p68 is dependent on RBM4 binding to the intronic element downstream of the 5′ splice site (Fig. 4). These results suggest that RBM4 might function to recruit p68 to a nearby splice site, thus explaining the necessity for RNA interaction. Alternatively, recruiting p68 to RBM4-containing protein complexes might allow p68 to function by remodeling protein-protein interactions as reported earlier for H-ras and CD44 splicing, the RNAi pathway, and cellular differentiation (13, 16, 22, 46, 99, 104).
Studies of RBM4 activity in regulating pre-mRNA splicing of other substrates such as alpha-tropomyosin suggested possible recruitment of U1snRNP (76). Our studies suggest that RBM4-mediated exon splicing enhancement may be mediated by its interaction with p68. Our data demonstrate functional interaction between two splicing activators of tau exon 10, p68 binding to the stem-loop region at the 5′ splice site and RBM4 interacting with the intronic splicing enhancer in tau exon 10 splicing. This also suggests a potential selective mechanism by which RNA helicases such as p68 might influence only specific alternative splicing events. However, the details of the interaction and mechanism require further study. Taken together, our work uncovers a previously unknown mechanism in which trans-acting regulators p68 and RBM4 cooperate to promote inclusion of tau exon 10.
ACKNOWLEDGMENTS
We thank members of the Wu laboratory for helpful discussions and Eileen Bigio for critical reading and suggestions.
This work was supported by the National Institutes of Health (R01 AG17518 to J.Y.W.).
Footnotes
Published ahead of print on 22 February 2011.
REFERENCES
- 1. Andreadis A. 2005. Tau gene alternative splicing: expression patterns, regulation and modulation of function in normal brain and neurodegenerative diseases. Biochim. Biophys. Acta 1739:91–103 [DOI] [PubMed] [Google Scholar]
- 2. Andreadis A. 2006. Misregulation of tau alternative splicing in neurodegeneration and dementia. Prog. Mol. Subcell. Biol. 44:89–107 [DOI] [PubMed] [Google Scholar]
- 3. Andreadis A., Brown W. M., Kosik K. S. 1992. Structure and novel exons of the human tau gene. Biochemistry 31:10626–10633 [DOI] [PubMed] [Google Scholar]
- 4. Aniello F., Couchie D., Bridoux A. M., Gripois D., Nunez J. 1991. Splicing of juvenile and adult tau mRNA variants is regulated by thyroid hormone. Proc. Natl. Acad. Sci. U. S. A. 88:4035–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Avila J., Lim F., Moreno F., Belmonte C., Cuello A. C. 2002. Tau function and dysfunction in neurons: its role in neurodegenerative disorders. Mol. Neurobiol. 25:213–231 [DOI] [PubMed] [Google Scholar]
- 6. Black D. L. 2000. Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell 103:367–370 [DOI] [PubMed] [Google Scholar]
- 7. Black D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72:291–336 [DOI] [PubMed] [Google Scholar]
- 8. Black D. L., Grabowski P. J. 2003. Alternative pre-mRNA splicing and neuronal function. Prog. Mol. Subcell. Biol. 31:187–216 [DOI] [PubMed] [Google Scholar]
- 9. Broderick J., Wang J., Andreadis A. 2004. Heterogeneous nuclear ribonucleoprotein E2 binds to tau exon 10 and moderately activates its splicing. Gene 331:107–114 [DOI] [PubMed] [Google Scholar]
- 10. Buratti E., Baralle F. E. 2004. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol. Cell. Biol. 24:10505–10514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Buszczak M., Spradling A. C. 2006. The drosophila P68 RNA helicase regulates transcriptional deactivation by promoting RNA release from chromatin. Genes Dev. 20:977–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cairns N. J., et al. 2007. Neuropathologic diagnostic and nosologic criteria for frontotemporal lobar degeneration: consensus of the Consortium for Frontotemporal Lobar Degeneration. Acta Neuropathol. 114:5–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Camats M., Guil S., Kokolo M., Bach-Elias M. 2008. P68 RNA helicase (DDX5) alters activity of cis- and trans-acting factors of the alternative splicing of H-Ras. PLoS One 3:e2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Caretti G., Lei E. P., Sartorelli V. 2007. The DEAD-box p68/p72 proteins and the noncoding RNA steroid receptor activator SRA: eclectic regulators of disparate biological functions. Cell Cycle 6:1172–1176 [DOI] [PubMed] [Google Scholar]
- 15. Chen Y. I., et al. 2007. Proteomic analysis of in vivo-assembled pre-mRNA splicing complexes expands the catalog of participating factors. Nucleic Acids Res. 35:3928–3944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Clark E. L., et al. 2008. The RNA helicase p68 is a novel androgen receptor coactivator involved in splicing and is overexpressed in prostate cancer. Cancer Res. 68:7938–7946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Clark L. N., et al. 1998. Pathogenic implications of mutations in the tau gene in pallido-ponto-nigral degeneration and related neurodegenerative disorders linked to chromosome 17. Proc. Natl. Acad. Sci. U. S. A. 95:13103–13107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Collet J., et al. 1997. Developmentally regulated alternative splicing of mRNAs encoding N-terminal tau variants in the rat hippocampus: structural and functional implications. Eur. J. Neurosci. 9:2723–2733 [DOI] [PubMed] [Google Scholar]
- 19. Connell J. W., et al. 2001. Effects of FTDP-17 mutations on the in vitro phosphorylation of tau by glycogen synthase kinase 3beta identified by mass spectrometry demonstrate certain mutations exert long-range conformational changes. FEBS Lett. 493:40–44 [DOI] [PubMed] [Google Scholar]
- 20. Conrad C., et al. 2007. Single molecule profiling of tau gene expression in Alzheimer's disease. J. Neurochem. 103:1228–1236 [DOI] [PubMed] [Google Scholar]
- 21. Crowther T., Goedert M., Wischik C. M. 1989. The repeat region of microtubule-associated protein tau forms part of the core of the paired helical filament of Alzheimer's disease. Ann. Med. 21:127–132 [DOI] [PubMed] [Google Scholar]
- 22. Davis B. N., Hilyard A. C., Lagna G., Hata A. 2008. SMAD proteins control DROSHA-mediated microRNA maturation. Nature 454:56–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dawson H. N., Cantillana V., Chen L., Vitek M. P. 2007. The tau N279K exon 10 splicing mutation recapitulates frontotemporal dementia and parkinsonism linked to chromosome 17 tauopathy in a mouse model. J. Neurosci. 27:9155–9168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. DeTure M., et al. 2000. Missense tau mutations identified in FTDP-17 have a small effect on tau-microtubule interactions. Brain Res. 853:5–14 [DOI] [PubMed] [Google Scholar]
- 25. Donahue C. P., Muratore C., Wu J. Y., Kosik K. S., Wolfe M. S. 2006. Stabilization of the tau exon 10 stem loop alters pre-mRNA splicing. J. Biol. Chem. 281:23302–23306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. D'Souza I., et al. 1999. Missense and silent tau gene mutations cause frontotemporal dementia with parkinsonism-chromosome 17 type, by affecting multiple alternative RNA splicing regulatory elements. Proc. Natl. Acad. Sci. U. S. A. 96:5598–5603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. D'Souza I., Schellenberg G. D. 2000. Determinants of 4-repeat tau expression. Coordination between enhancing and inhibitory splicing sequences for exon 10 inclusion. J. Biol. Chem. 275:17700–17709 [DOI] [PubMed] [Google Scholar]
- 28. D'Souza I., Schellenberg G. D. 2006. Arginine/serine-rich protein interaction domain-dependent modulation of a tau exon 10 splicing enhancer: altered interactions and mechanisms for functionally antagonistic FTDP-17 mutations Delta280K and N279K. J. Biol. Chem. 281:2460–2469 [DOI] [PubMed] [Google Scholar]
- 29. Eng. J. K., McCormack A. L., Yates J. R., III 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976–989 [DOI] [PubMed] [Google Scholar]
- 30. Fairbrother W. G., Lipscombe D. 2008. Repressing the neuron within. Bioessays 30:1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Forman M. S., Trojanowski J. Q., Lee V. M. 2004. Neurodegenerative diseases: a decade of discoveries paves the way for therapeutic breakthroughs. Nat. Med. 10:1055–1063 [DOI] [PubMed] [Google Scholar]
- 32. Fukuda T., et al. 2007. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat. Cell Biol. 9:604–611 [DOI] [PubMed] [Google Scholar]
- 33. Fuller-Pace F. V. 2006. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 34:4206–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fuller-Pace F. V., Jacobs A. M., Nicol S. M. 2007. Modulation of transcriptional activity of the DEAD-box family of RNA helicases, p68 (Ddx5) and DP103 (Ddx20), by SUMO modification. Biochem. Soc. Trans. 35:1427–1429 [DOI] [PubMed] [Google Scholar]
- 35. Fuller-Pace F. V., Ali S. 2008. The DEAD box RNA helicases p68 (Ddx5) and p72 (Ddx17): novel transcriptional co-regulators. Biochem. Soc. Trans. 36:609–612 [DOI] [PubMed] [Google Scholar]
- 36. Gallo J. M., Noble W., Martin T. R. 2007. RNA and protein-dependent mechanisms in tauopathies: consequences for therapeutic strategies. Cell. Mol. Life Sci. 64:1701–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gao L., Wang J., Wang Y., Andreadis A. 2007. SR protein 9G8 modulates splicing of tau exon 10 via its proximal downstream intron, a clustering region for frontotemporal dementia mutations. Mol. Cell. Neurosci. 34:48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gao Q. S., et al. 2000. Complex regulation of tau exon 10, whose missplicing causes frontotemporal dementia. J. Neurochem. 74:490–500 [DOI] [PubMed] [Google Scholar]
- 39. Glatz D. C., et al. 2006. The alternative splicing of tau exon 10 and its regulatory proteins CLK2 and TRA2-BETA1 changes in sporadic Alzheimer's disease. J. Neurochem. 96:635–644 [DOI] [PubMed] [Google Scholar]
- 40. Goedert M. 2005. Tau gene mutations and their effects. Mov. Disord. 20(Suppl. 12):S45–S52 [DOI] [PubMed] [Google Scholar]
- 41. Goedert M., Spillantini M. G. 2001. Tau gene mutations and neurodegeneration. Biochem. Soc. Symp. 2001(67):59–71 [DOI] [PubMed] [Google Scholar]
- 42. Goedert M., Spillantini M. G., Jakes R., Rutherford D., Crowther R. A. 1989. Multiple isoforms of human microtubule-associated protein tau: sequences and localization in neurofibrillary tangles of Alzheimer's disease. Neuron 3:519–526 [DOI] [PubMed] [Google Scholar]
- 43. Goode B. L., Chau M., Denis P. E., Feinstein S. C. 2000. Structural and functional differences between 3-repeat and 4-repeat tau isoforms. Implications for normal tau function and the onset of neurodegenerative disease. J. Biol. Chem. 275:38182–38189 [DOI] [PubMed] [Google Scholar]
- 44. Gromak N., et al. 2003. The PTB interacting protein raver1 regulates alpha-tropomyosin alternative splicing. EMBO J. 22:6356–6364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Grover A., et al. 1999. 5′ splice site mutations in tau associated with the inherited dementia FTDP-17 affect a stem-loop structure that regulates alternative splicing of exon 10. J. Biol. Chem. 274:15134–15143 [DOI] [PubMed] [Google Scholar]
- 46. Guil S., et al. 2003. Roles of hnRNP A1, SR proteins, and p68 helicase in c-H-ras alternative splicing regulation. Mol. Cell. Biol. 23:2927–2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hallegger M., Llorian M., Smith C. W. 2010. Alternative splicing: global insights. FEBS J. 277:856–866 [DOI] [PubMed] [Google Scholar]
- 48. Hartmuth K., et al. 2002. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl. Acad. Sci. U. S. A. 99:16719–16724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hasegawa M., Smith M. J., Goedert M. 1998. Tau proteins with FTDP-17 mutations have a reduced ability to promote microtubule assembly. FEBS Lett. 437:207–210 [DOI] [PubMed] [Google Scholar]
- 50. Hasegawa M., Smith M. J., Iijima M., Tabira T., Goedert M. 1999. FTDP-17 mutations N279K and S305N in tau produce increased splicing of exon 10. FEBS Lett. 443:93–96 [DOI] [PubMed] [Google Scholar]
- 51. Himmler A. 1989. Structure of the bovine tau gene: alternatively spliced transcripts generate a protein family. Mol. Cell. Biol. 9:1389–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Huang Y., Liu Z. R. 2002. The ATPase, RNA unwinding, and RNA binding activities of recombinant p68 RNA helicase. J. Biol. Chem. 277:12810–12815 [DOI] [PubMed] [Google Scholar]
- 53. Hutton M. 2001. Missense and splice site mutations in tau associated with FTDP-17: multiple pathogenic mechanisms. Neurology 56:S21–S25 [DOI] [PubMed] [Google Scholar]
- 54. Hutton M., et al. 1998. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 393:702–705 [DOI] [PubMed] [Google Scholar]
- 55. Iijima-Ando K., Iijima K. 2010. Transgenic Drosophila models of Alzheimer's disease and tauopathies. Brain Struct. Funct. 214:245–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Iqbal K., Liu F., Gong C. X., Alonso Adel C., Grundke-Iqbal I. 2009. Mechanisms of tau-induced neurodegeneration. Acta Neuropathol. 118:53–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jacobs A. M., et al. 2007. SUMO modification of DEAD box protein p68 modulates its transcriptional activity and promotes its interaction with HDAC1. Oncogene 26:5866–5876 [DOI] [PubMed] [Google Scholar]
- 58. Jalal C., Uhlmann-Schiffler H., Stahl H. 2007. Redundant role of DEAD box proteins p68 (Ddx5) and p72/p82 (Ddx17) in ribosome biogenesis and cell proliferation. Nucleic Acids Res. 35:3590–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Janknecht R. 2010. Multi-talented DEAD-box proteins and potential tumor promoters: p68 RNA helicase (DDX5) and its paralog, p72 RNA helicase (DDX17). Am. J. Transl. Res. 2:223–234 [PMC free article] [PubMed] [Google Scholar]
- 60. Jiang Z., Cote J., Kwon J. M., Goate A. M., Wu J. Y. 2000. Aberrant splicing of tau pre-mRNA caused by intronic mutations associated with the inherited dementia frontotemporal dementia with parkinsonism linked to chromosome 17. Mol. Cell. Biol. 20:4036–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jiang Z., et al. 2003. Mutations in tau gene exon 10 associated with FTDP-17 alter the activity of an exonic splicing enhancer to interact with Tra2 beta. J. Biol. Chem. 278:18997–19007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jiang Z. H., Zhang W. J., Rao Y., Wu J. Y. 1998. Regulation of Ich-1 pre-mRNA alternative splicing and apoptosis by mammalian splicing factors. Proc. Natl. Acad. Sci. U. S. A. 95:9155–9160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jurica M. S., Licklider L. J., Gygi S. R., Grigorieff N., Moore M. J. 2002. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 8:426–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kalbfuss B., Mabon S. A., Misteli T. 2001. Correction of alternative splicing of tau in frontotemporal dementia and parkinsonism linked to chromosome 17. J. Biol. Chem. 276:42986–42993 [DOI] [PubMed] [Google Scholar]
- 65. Kar A., Havlioglu N., Tarn W. Y., Wu J. Y. 2006. RBM4 interacts with an intronic element and stimulates tau exon 10 inclusion. J. Biol. Chem. 281:24479–24488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kar A., Kuo D., He R., Zhou J., Wu J. Y. 2005. Tau alternative splicing and frontotemporal dementia. Alzheimer Dis. Assoc. Disord. 19(Suppl. 1):S29–S36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kondo S., et al. 2004. Tra2 beta, SF2/ASF and SRp30c modulate the function of an exonic splicing enhancer in exon 10 of tau pre-mRNA. Genes Cells 9:121–130 [DOI] [PubMed] [Google Scholar]
- 68. Konzack S., Thies E., Marx A., Mandelkow E. M., Mandelkow E. 2007. Swimming against the tide: mobility of the microtubule-associated protein tau in neurons. J. Neurosci. 27:9916–9927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. LeBoeuf A. C., et al. 2008. FTDP-17 mutations in Tau alter the regulation of microtubule dynamics: an “alternative core” model for normal and pathological Tau action. J. Biol. Chem. 283:36406–36415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lee G., Neve R. L., Kosik K. S. 1989. The microtubule binding domain of tau protein. Neuron 2:1615–1624 [DOI] [PubMed] [Google Scholar]
- 71. Lee V. M., Giasson B. I., Trojanowski J. Q. 2004. More than just two peas in a pod: common amyloidogenic properties of tau and alpha-synuclein in neurodegenerative diseases. Trends Neurosci. 27:129–134 [DOI] [PubMed] [Google Scholar]
- 72. Li H.-S., et al. 1999. Vertebrate Slit, a secreted ligand for the transmembrane protein Roundabout, is a repellent for olfactory bulb axons. Cell 96:807–818 [DOI] [PubMed] [Google Scholar]
- 73. Li Q., Lee J. A., Black D. L. 2007. Neuronal regulation of alternative pre-mRNA splicing. Nat. Rev. Neurosci. 8:819–831 [DOI] [PubMed] [Google Scholar]
- 74. Lichtenberg-Kraag B., Mandelkow E. M. 1990. Isoforms of tau protein from mammalian brain and avian erythrocytes: structure, self-assembly, and elasticity. J. Struct. Biol. 105:46–53 [DOI] [PubMed] [Google Scholar]
- 75. Lin C., Yang L., Yang J. J., Huang Y., Liu Z. R. 2005. ATPase/helicase activities of p68 RNA helicase are required for pre-mRNA splicing but not for assembly of the spliceosome. Mol. Cell. Biol. 25:7484–7493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lin J. C., Tarn W. Y. 2005. Exon selection in alpha-tropomyosin mRNA is regulated by the antagonistic action of RBM4 and PTB. Mol. Cell. Biol. 25:10111–10121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Liu Z. R. 2002. p68 RNA helicase is an essential human splicing factor that acts at the U1 snRNA-5′ splice site duplex. Mol. Cell. Biol. 22:5443–5450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mackenzie I. R., et al. 2010. Nomenclature and nosology for neuropathologic subtypes of frontotemporal lobar degeneration: an update. Acta Neuropathol. 119:1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Makarov E. M., et al. 2002. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298:2205–2208 [DOI] [PubMed] [Google Scholar]
- 80. Makrides V., et al. 2003. Microtubule-dependent oligomerization of tau. Implications for physiological tau function and tauopathies. J. Biol. Chem. 278:33298–33304 [DOI] [PubMed] [Google Scholar]
- 81. Markus M. A., et al. 2006. WT1 interacts with the splicing protein RBM4 and regulates its ability to modulate alternative splicing in vivo. Exp. Cell Res. 312:3379–3388 [DOI] [PubMed] [Google Scholar]
- 82. Markus M. A., Morris B. J. 2009. RBM4: a multifunctional RNA-binding protein. Int. J. Biochem. Cell Biol. 41:740–743 [DOI] [PubMed] [Google Scholar]
- 83. Martinez-Contreras R., et al. 2007. HnRNP proteins and splicing control. Adv. Exp. Med. Biol. 623:123–147 [DOI] [PubMed] [Google Scholar]
- 84. Matlin A. J., Clark F., Smith C. W. 2005. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 6:386–398 [DOI] [PubMed] [Google Scholar]
- 85. Montejo de Garcini E., et al. 1992. Differentiation of neuroblastoma cells correlates with an altered splicing pattern of tau RNA. FEBS Lett. 299:10–14 [DOI] [PubMed] [Google Scholar]
- 86. Moore M. J., Silver P. A. 2008. Global analysis of mRNA splicing. RNA 14:197–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Moroy T., Heyd F. 2007. The impact of alternative splicing in vivo: mouse models show the way. RNA 13:1155–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Neve R. L., Harris P., Kosik K. S., Kurnit D. M., Donlon T. A. 1986. Identification of cDNA clones for the human microtubule-associated protein tau and chromosomal localization of the genes for tau and microtubule-associated protein 2. Brain Res. 387:271–280 [DOI] [PubMed] [Google Scholar]
- 89. Nilsen T. W., Graveley B. R. 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463:457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Oblinger M. M., Argasinski A., Wong J., Kosik K. S. 1991. Tau gene expression in rat sensory neurons during development and regeneration. J. Neurosci. 11:2453–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Pickering-Brown S., et al. 2000. Pick's disease is associated with mutations in the tau gene. Ann. Neurol. 48:859–867 [PubMed] [Google Scholar]
- 92. Pickering-Brown S. M., et al. 2002. Inherited frontotemporal dementia in nine British families associated with intronic mutations in the tau gene. Brain 125:732–751 [DOI] [PubMed] [Google Scholar]
- 93. Salzman D. W., Shubert-Coleman J., Furneaux H. 2007. P68 RNA helicase unwinds the human let-7 microRNA precursor duplex and is required for let-7-directed silencing of gene expression. J. Biol. Chem. 282:32773–32779 [DOI] [PubMed] [Google Scholar]
- 94. Sambasivan R., et al. 2009. The small chromatin-binding protein p8 coordinates the association of anti-proliferative and pro-myogenic proteins at the myogenin promoter. J. Cell Sci. 122:3481–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Shin S., Rossow K. L., Grande J. P., Janknecht R. 2007. Involvement of RNA helicases p68 and p72 in colon cancer. Cancer Res. 67:7572–7578 [DOI] [PubMed] [Google Scholar]
- 96. Skovronsky D. M., Lee V. M., Trojanowski J. Q. 2006. Neurodegenerative diseases: new concepts of pathogenesis and their therapeutic implications. Annu. Rev. Pathol. 1:151–170 [DOI] [PubMed] [Google Scholar]
- 97. Spillantini M. G., et al. 1998. Mutation in the tau gene in familial multiple system tauopathy with presenile dementia. Proc. Natl. Acad. Sci. U. S. A. 95:7737–7741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Stoilov P., Lin C. H., Damoiseaux R., Nikolic J., Black D. L. 2008. A high-throughput screening strategy identifies cardiotonic steroids as alternative splicing modulators. Proc. Natl. Acad. Sci. U. S. A. 105:11218–11223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Suzuki H. I., et al. 2009. Modulation of microRNA processing by p53. Nature 460:529–533 [DOI] [PubMed] [Google Scholar]
- 100. Tomoo K., et al. 2005. Possible role of each repeat structure of the microtubule-binding domain of the tau protein in in vitro aggregation. J. Biochem. 138:413–423 [DOI] [PubMed] [Google Scholar]
- 101. Umeda Y., et al. 2004. Alterations in human tau transcripts correlate with those of neurofilament in sporadic tauopathies. Neurosci. Lett. 359:151–154 [DOI] [PubMed] [Google Scholar]
- 102. Utton M. A., et al. 2002. The slow axonal transport of the microtubule-associated protein tau and the transport rates of different isoforms and mutants in cultured neurons. J. Neurosci. 22:6394–6400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Varani L., et al. 1999. Structure of tau exon 10 splicing regulatory element RNA and destabilization by mutations of frontotemporal dementia and parkinsonism linked to chromosome 17. Proc. Natl. Acad. Sci. U. S. A. 96:8229–8234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Varani L., Spillantini M. G., Goedert M., Varani G. 2000. Structural basis for recognition of the RNA major groove in the tau exon 10 splicing regulatory element by aminoglycoside antibiotics. Nucleic Acids Res. 28:710–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wei M. L., Andreadis A. 1998. Splicing of a regulated exon reveals additional complexity in the axonal microtubule-associated protein tau. J. Neurochem. 70:1346–1356 [DOI] [PubMed] [Google Scholar]
- 106. Wu J. Y., Kar A., Kuo D., Yu B., Havlioglu N. 2006. SRp54 (SFRS11), a regulator for tau exon 10 alternative splicing identified by an expression cloning strategy. Mol. Cell. Biol. 26:6739–6747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Yang L., Liu Z. R. 2004. Bacterially expressed recombinant p68 RNA helicase is phosphorylated on serine, threonine, and tyrosine residues. Protein Expr. Purif. 35:327–333 [DOI] [PubMed] [Google Scholar]
- 108. Yang L., Lin C., Liu Z. R. 2006. P68 RNA helicase mediates PDGF-induced epithelial mesenchymal transition by displacing Axin from beta-catenin. Cell 127:139–155 [DOI] [PubMed] [Google Scholar]
- 109. Zheng S., Chen Y., Donahue C. P., Wolfe M. S., Varani G. 2009. Structural basis for stabilization of the tau pre-mRNA splicing regulatory element by novantrone (mitoxantrone). Chem. Biol. 16:557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Zilka N., Korenova M., Novak M. 2009. Misfolded tau protein and disease modifying pathways in transgenic rodent models of human tauopathies. Acta Neuropathol. 118:71–86 [DOI] [PubMed] [Google Scholar]