Abstract
Calmodulin is a universal calcium-sensing protein that has pleiotropic effects. Here we show that calmodulin inhibits a new SCF (Skp1–Cullin–F-box) E3 ligase component, FBXL2. During Pseudomonas aeruginosa infection, SCF (FBXL2) targets the key enzyme, CCTα, for its monoubiquitination and degradation, thereby reducing synthesis of the indispensable membrane and surfactant component, phosphatidylcholine. P. aeruginosa triggers calcium influx and calcium-dependent activation of FBXL2 within the Golgi complex, where it engages CCTα. FBXL2 through its C terminus binds to the CCTα IQ motif. FBXL2 knockdown increases CCTα levels and phospholipid synthesis. The molecular interaction of FBXL2 with CCTα is opposed by calmodulin, which traffics to the Golgi complex, binds FBXL2 (residues 80 to 90) via its C terminus, and vies with the ligase for occupancy within the IQ motif. These observations were recapitulated in murine models of P. aeruginosa-induced surfactant deficiency, where calmodulin gene transfer reduced FBXL2 actions by stabilizing CCTα and lessening the severity of inflammatory lung injury. The results provide a unique model of calcium-regulated intermolecular competition between an E3 ligase subunit and an antagonist that is critically relevant to pneumonia and lipid homeostasis.
INTRODUCTION
Protein ubiquitination is a key posttranslational modification that regulates diverse physiologic processes (28). The conjugation of ubiquitin to a target protein is orchestrated by a series of enzymatic reactions involving an E1 ubiquitin-activating enzyme, ubiquitin transfer from an E1-activating enzyme to an E2-conjugating enzyme, and last, generation of an isopeptide bond between the substrate's ε-amino lysine and the carboxy terminus of ubiquitin, which is catalyzed by an E3 ubiquitin ligase (29). The behavior of ubiquitinated proteins within cells depends, in part, upon whether proteins are ligated with monomeric, multimeric, or polymeric ubiquitin. For example, conjugation of one (monoubiquitination) or multiple (multiubiquitination) ubiquitin molecules to lysines within target proteins serves as an important endocytic signal for internalization and targeting of various proteins within the lysosomal/endocytic pathway (7). Polyubiquitinated (with at least four ubiquitin molecules) proteins most commonly are targeted for degradation within the 26S proteasome.
The biologic role of the majority of ubiquitin E3 ligases still remains enigmatic. Of the SCF (Skp1–Cullin 1–F-box) E3 ligase family, for example, only ∼6 from over 60 family members have been well studied. The SCF complex contains a multisubunit catalytic core consisting of Skp1, Cullin 1, and the E2-conjugating (Ubc) enzyme (14). A variable and integral adaptor subunit, termed the F-box protein, confers specificity for interaction to a variety of substrates through specific domain interactions (28). F-box proteins have two domains: an NH2-terminal F-box motif, which binds Skp1, and a carboxyl-terminal leucine-rich repeat (LRR) or WD motif, which recognizes substrates (2). SCF ligases regulate DNA repair, cell cycle progression, cell growth, and survival (28). There exists a limited understanding of potential SCF subunit competitive antagonists.
Calmodulin (CaM; 16.7 kDa) is a highly conserved calcium-sensing protein that antagonizes some proteinases and modulates stability of regulatory proteins (1, 16, 25). CaM binds its targets in a calcium-bound (holoCaM) or calcium-free (apopCaM) form, and thus its interactions with partners may be Ca2+ dependent or Ca2+ independent (25). Many CaM binding proteins harbor recognition motifs characterized by a basic amphipathic helix, moderate to high helical hydrophobic moment, and a net positive charge (25). Other motifs described include an IQ motif (I/LQXXXRGXXXR) and 1-8-14 and 1-5-10 CaM binding motifs (25). We have shown that CaM stabilizes a critical target, CTP:phosphocholine cytidylyltransferase (CCTα), by binding within a canonical IQ motif (3). CCTα is an indispensable regulatory enzyme needed for synthesis of phosphatidylcholine (PtdCho), which is utilized for formation of animal membranes, and of pulmonary surfactant (3). CCTα contains four functional regions within its primary structure, including nuclear localization, catalytic, membrane binding, and phosphorylation domains. We previously showed that CCTα levels in cells are critically dependent upon its rate of monoubiquitination, a sorting signal that targets the enzyme for its rapid degradation within the lysosome (4). The identity of the ubiquitin E3 ligase that catalyzes CCTα monoubiquitination is unknown.
Here we show that CCTα is monoubiquitinated in a Ca2+-dependent manner by the SCF-E3 ligase complex involving the orphan F-box protein FBXL2 and that this process is antagonized by CaM. Following its initial description (14), FBXL2 was shown to interact with hepatitis C virus (HCV) nonstructural protein 5A (NS5A), and this association was required for HCV RNA replication (33). NS5A is the only known target of FBXL2. However, the authentication of FBXL2 as a ubiquitin E3 ligase component and its molecular behavior as it is linked to regulation of fundamental cellular processes have not been investigated. Our results unveil an exquisite model of molecular interplay where CaM, via specific molecular sequence signatures, engages FBXL2 in response to Ca2+ signals, thereby attenuating its E3 ligase activity. The ability of CaM to oppose SCF (FBXL2)-mediated CCTα degradation was observed in a murine model of pneumonia, underscoring its pivotal role as a molecular self-defense mechanism for preserving membrane and surfactant phospholipid homeostasis during proinflammatory stress.
MATERIALS AND METHODS
Materials.
The sources of murine lung epithelial (MLE) cells, CCTα, CaM, Erk, 14-3-3, and glutathione S-transferase (GST) antibodies were described previously (3). Ubc3, Ubc5, ubiquitin, A23187, purified bovine CaM, and the calpain activity assay kit were purchased from Calbiochem (La Jolla, CA). Ubiquitin E1 activation enzyme and ubiquitin aldehyde were purchased from Boston Biochem (Boston, MA). Ubiquitin, Cullin 1, Skp1, and Rbx1 antibodies were purchased from Cell Signaling (Danvers, MA). The FBLX2 and cytokine antibodies and scrambled RNA and small interfering RNA (siRNA) to FBXL2 were from Santa Cruz Biotechnology (Santa Cruz, CA). The RCFP polyclonal antibody pAmCyan1-C1 and pZsYellow-C1 vector were purchased from Clontech (Mountain View, CA). The Ca2+ Green staining kits, cameleon baculovirus expression system, mouse monoclonal V5 antibody, rabbit monoclonal anti-Golgi 97 antibody, endoplasmic reticulum (ER) tracker dye, the To-Pro-3 nuclear staining kit, the pcDNA3.1D cloning kit, Escherichia coli One Shot competent cells, the pENTR directional TOPO cloning kits, and the Gateway mammalian expression system were purchased from Invitrogen (Carlsbad, CA). BD Talon purification and buffer kits were purchased from BD Biosciences (San Jose, CA). The F-box protein cDNAs were purchased from OpenBiosystems (Huntsville, AL). The mammalian two-hybrid systems were purchased from Stratagene (La Jolla, CA) and Clontech (Mountain View, CA). The gel extraction kit and QIAprep spin miniprep kits were from Qiagen (Valencia, CA). FuGene6 transfection reagent was purchased from Roche Diagnostics (Indianapolis, IN). Nucleofector transfection kits were from Amaxa (Gaithersburg, MD). Immobilized protein A/G beads were from Pierce (Rockford, IL). All DNA sequencing was performed by the University of Iowa DNA Core Facility.
Cell culture.
MLE cells were cultured in Dulbecco's modified Eagle medium–F-12 (Gibco) supplemented with 2 or 10% fetal bovine serum (DMEM-2 or -10). In some studies, cells were serum starved (DMEM–F-12) and infected with Pseudomonas aeruginosa PA103 at a multiplicity of infection (MOI) of 10 for 1 h or treated with A23187 at 10 nM for 4 h. In other experiments, cells were incubated with 20 mM NH4Cl, 1:1,000 leupeptin, or a 1:1,000 dilution of lactacystin for 24 h. Cell lysates were prepared by brief sonication in 150 mM NaCl, 50 mM Tris, 1.0 mM EDTA, 2 mM dithiothreitol (DTT), 0.025% sodium azide, and 1 mM phenylmethylsulfonyl fluoride (buffer A) at 4°C.
Expression of recombinant proteins and RNA inhibition (RNAi).
Cellular expression of plasmids was facilitated using the Amaxa nucleofector system, with transfection efficiencies of >90% (3). MLE cells (4 × 106) were plated in 100-mm dishes for 24 h, infected with an adenovirus-CaM (Ad-CaM) vector or an empty vector (Ad-Con) at an MOI of 40 for 12 h, followed by transfection with FBXL2 plasmid. For cameleon expression, 5 × 104 cells were plated in 96-well plates. In baculovirus studies, 2 × 105 cells were plated in 35-mm glass-bottom dishes for 24 h and then infected with baculovirus-cameleon following the manufacturer's instructions. Recombinant CCTα was expressed and purified as described previously (26). For siRNA studies, 1 × 106 cells were transfected using nucleofection with 0.2 nmol of scrambled RNA or FBXL2 siRNA and harvested after an additional 48 h.
Bacterial culture.
Pseudomonas aeruginosa PA103 and PA103 mutants were kindly provided by Tim Yahr (University of Iowa, Iowa City, IA). Inocula were freshly prepared prior to experiments from frozen stocks of PA103 (frozen at mid-log phase; optical density at 540 nm of 0.8). PA103 was maintained in Vogel-Bonner minimal agar. Cultures were plated and grown overnight from frozen stock. Overnight plate cultures were then inoculated in tryptic soy broth supplemented with 1% glycerol and 100 mM sodium glutamate (TSB++) and grown by rotary shaking at 37°C to log phase (3).
Animal studies.
Male C57LB/6 mice (purchased from Jackson Laboratories) were acclimated at the University of Iowa Animal Care Facility and maintained according to all federal and institutional animal care guidelines and under a University of Iowa Institutional Animal Care and Use Committee (IACUC)-approved protocol. Mice were deeply anesthetized with ketamine (80 to 100 mg/kg of body weight, intraperitoneally [i.p.]) and xylazine (10 mg/kg, i.p.), and then the larynx was well visualized under a fiber optic light source before endotracheal intubation with a 3/400 24-gauge plastic catheter. Replication-deficient adenovirus (Ad5) alone or Adv-CaM (109 PFU in 50 μl of 10 mM Tris-HCl [pH 7.4], 150 mM NaCl, and 0.1% bovine serum albumin) was instilled intratracheally (i.t.) on day 1, after which animals were allowed to recover for 48 h. Following recovery, mice were deeply anesthetized again, followed by administration of P. aeruginosa (PA103; 107 CFU/mouse, i.t.) for 1 h. A tracheostomy was performed, and a metal 1.2-mm (internal diameter) tracheal cannula was inserted and tied firmly into place. An electrocardiograph tracing was monitored to ascertain any adverse effects of the ventilatory maneuvers. The mice were deeply anesthetized, paralyzed, and mechanically ventilated with a positive end expiratory pressure (PEEP) of 3, and a quasistatic volume pressure determination was performed by using a FlexiVent system (4). Lavage fluids were collected from mice to isolate surfactant, as described previously (20).
Immunoblot analysis.
Equal amounts of total protein in sample buffer were resolved by SDS-PAGE and transferred to nitrocellulose, and immunoreactive proteins were detected as described previously (4). The dilution factor for primary and secondary antibodies was 1:2,000. CCTα was purified to homogeneity from rat liver as described previously (4).
Coimmunoprecipitation.
Total cellular protein or the ubiquitination reaction mixture was precleared using protein A/G beads prior to incubation with primary antibodies (3). Beads were rinsed and processed prior to SDS-PAGE and immunoblotting as described previously (3). Cell lysates were also precleared using protein A/G beads prior to incubation with Skp1, Cullin1, Rbx1, or FBXL2 antibodies, and immunoprecipitates were pulled down by using protein A/G beads, washed, and eluted in glycine buffer, followed by centricon concentration.
Immunostaining.
Cells (2 × 105) were plated at 70% confluence on 35-mm MetTek glass-bottom culture dishes and infected with baculovirus-cameleon or transfected with cyan fluorescent protein (CFP)-FBXL2 or yellow fluorescent protein (YFP)-CCT. Immunofluorescent cell imaging was performed on a Zeiss LSM 510 confocal microscope, using a 458-nm, 488-nm, 514-nm, or 615-nm wavelength. All experiments were done with a Zeiss 63× oil differential interference contrast objective lens. Cells were washed with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde for 20 min, then exposed to 15% bovine serum albumin (BSA), 1:500 anti-Golgi 97 primary antibody, and 1:1,000 Alexa 633-labeled goat anti-mouse secondary antibody sequentially for immunostaining. In other studies, cells were incubated with a 1:2,000 dilution of Ca2+ Green dye for 30 min, followed by washing with PBS and fixing with 4% paraformaldehyde. A 458-nm wavelength was used to excite CFP, with fluorescence emission collected through a 475-to-500-nm filter. A 488-nm wavelength was used to excite Ca2+ Green dye, with fluorescence emission collected through a 505-to-530-nm filter. A 514-nm wavelength was used to excite YFP, with fluorescence emission collected through a 530-to-600-nm filter. A 613-nm wavelength was used to excite Alexa 633 dye, with fluorescence emission collected through a 633-nm filter.
PtdCho synthesis and CCT activity.
Phosphatidylcholine and CCT activities were assayed as described previously (3, 19, 38).
Construction of His-V5-tagged F-box proteins.
A series of F-box proteins were cloned using a cDNA library as a template for PCR amplification. The forward primer 5′-CACCATGGACCCGGCCGAG-3′ and the reverse primer 5′-TCTGGAGATGTAGGTGTATGTTCG-3′ were used to generate the FBXW1 fragment. The forward primer 5′-CACCATGGTTTTCTCAAACAATGATGAA-3′ and the reverse primer 5′-AAGAATGACACAGCACCTGC-3′ were used to generate the FBXL2 fragment. The resulting PCR products were purified, followed by one-step cloning into a pcDNA3.1D/V5-His vector. The PCR conditions were as follows: 98°C for 15 s and 35 cycles of 98°C for 15 s, 60°C for 15 s, and 72°C for 30 s.
Construction of CCTα, FBXL2, and CaM mutants.
A series of CCTα deletion mutants were constructed as described elsewhere (3, 4). FBXL2 deletion mutants were constructed as follows: pcDNA3.1D-FBXL2 was used as a template for PCR with primer pairs to clone FBXL2 fragments 67-423, 70-423, 80-423, 90-423, 101-423, 1-350, and 1-250. These fragments were then directionally cloned into the pcDNA3.1D/V5-His vector. NH2-terminal and carboxyl-terminal CaM deletion mutants were constructed using YFP-CaM full length as a template for PCR using primer pairs to clone CaM 1-80 and CaM 76-149 fragments. The forward primer has a BglII overhang, and the reverse primer has a SalI overhang. The PCR products were gel purified and subjected to BglII and SalII double digestion. The digested PCR products were gel purified again, followed by ligation into a linearized YFP vector.
FRET analysis.
Cells were plated and cotransfected with CFP-FBXL2 and YFP-CaM or YFP-CCTN40 plasmids as described. Interactions were detected at the single-cell level by using a combination laser-scanning microscope system (LSM510/ConfoCor2; Zeiss, Jena, Germany) as described. Fluorescence resonance energy transfer (FRET) efficiency was calculated as follows: EFRET = (1 − CFPbefore/CFPafter) × 100, where CFPbefore and CFPafter represent the CFP fluorescence levels measured before and after photobleaching, respectively. The efficiency of the fluorescence resonance energy transfer directly reflects the distance separating the donor and the acceptor.
In vitro ubiquitin conjugation assay.
The ubiquitination of CCTα was performed in a volume of 25 μl containing 50 mM Tris (pH 7.6), 5 mM MgCl2, 0.6 mM DTT, 2 mM ATP, 1.5 ng/μl E1 (Boston Biochem),10 ng/μl Ubc5, 10 ng/μl Ubc7, 1 μg/μl ubiquitin (Calbiochem), 1 μM ubiquitin aldehyde, and 4 to 16 μl of immunoprecipitated Cullin 1, Skp1, Rbx1, and FBXL2 from cells. Products (10%) were processed for CCTα immunoblotting. The rest of the reaction mixture was incubated with CCTα antibody, pulled down by using protein A/G beads, and then probed against ubiquitin antibody.
Protein interaction assays.
His-tagged FBXL2 transfectants were extracted in buffer A (50 mM Tris-HCl, 500 mM NaCl, 10 mM imidazole, 1% Triton X-100; pH 7.4). Aliquots (200 μl) of Talon beads (Clonetech) were then incubated with cell lysates at 4°C for 2 h. After incubation, the beads were slowly centrifuged and washed 3 times in buffer A to generate FBXL2 beads. GST-CCT or YFP-CaM transfectants were extracted in buffer B (50 mM Tris-HCl, 150 mM NaCl, 5 mM imidazole, 0.2% Triton X-100, 0.2% NP-40; pH 7.4). Forty microliters of FBXL2 beads was incubated with GST or YFP cell lysates at 4°C for 2 h prior to centrifugation and rinsing using buffer B. Proteins were eluted (100°C for 5 min) with sample buffer (80 μl) for immunoblotting. Cells transfected with His− FBXL2 mutants were also extracted in buffer B, and lysates were incubated with Talon beads (40 μl) at 4°C for 2 h and processed for subsequent immunoblotting. For CaM binding assays, CaM-Sepharose beads were incubated with V5-FBXL2-transfected cell lysates (50 μg), with or without Ca2+, at 4°C for 2 h as described. Beads and released products were processed for SDS-PAGE and immunoblotting as described elsewhere (3).
Mammalian two-hybrid assays.
CCTα-Gal4BD, CaM-Gal4AD, FBXL2-Gal4BD, and FBXL2-Gal4AD were constructed as described elsewhere (3, 4). CCTα/FBXL2, CaM/FBXL2, or CCT/CaM and a pFR-β-gal reporter vector were coelectroporated into cells per the manufacturers' instructions. At 24 h after transfection, cells were lysed and assayed for β-galactosidase activities. pM-53 and pVP16-T plasmids served as positive controls. pM3-VP16 and pVP16-CP plasmids served as negative controls.
In vivo micro-computed tomography (micro-CT) imaging.
A tracheotomy was performed following administration of ketamine-xylazine for anesthesia and a nonresponsive pedal reflex test. Bacteria were administered through the tracheotomy opening, and each mouse was connected to a ventilator.
Respiratory paralysis was induced by administering 0.1 mg/kg pancuronium. To maintain sedation throughout the imaging process, 1.5% isoflurane was administered through the ventilator. A micro-CT II system (Siemens Pre-Clinical Solutions, TN) was kindly provided by Jessica C. Sieren and Geoffrey McLennan (University of Iowa) for in vivo scanning. Lung imaging during spontaneous respiration can affect image quality, and so a custom gated imaging process (intermittent isopressure breath hold) was used. This technique triggers image acquisition during a forced breath hold, with periods of hyperventilation between acquisitions. The micro-CT scanner setting for in vivo imaging was 60 kVp, 500 μA, and an exposure time of 500 ms. A total of 720 projections were acquired over 200°.
Statistical analysis.
Statistical comparisons were performed with the Prism program, version 4.03 (GraphPad Software, Inc., San Diego, CA) by using an analysis of variance or an unpaired t test, with a P value of <0.05 considered indicative of significance.
RESULTS
P. aeruginosa induces Ca2+ influx in MLE cells.
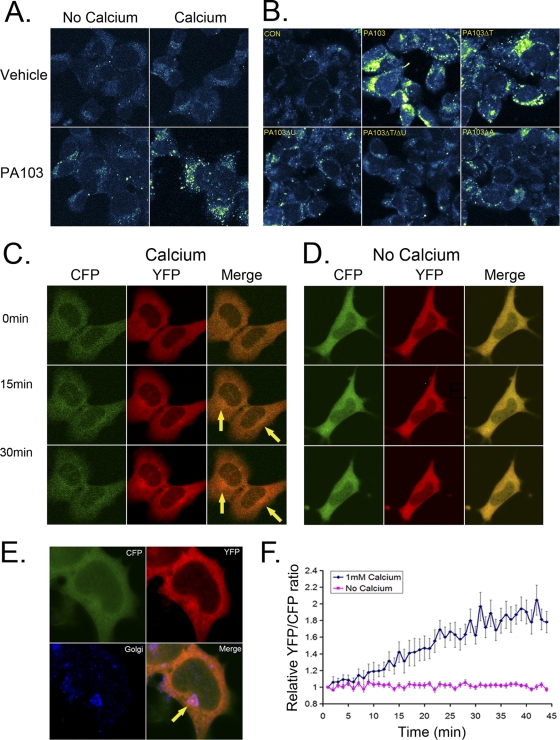
We assayed Ca2+ levels in lung epithelium infected with P. aeruginosa (PA103), a highly virulent pneumonia pathogen that induces CCTα degradation. In particular, P. aeruginosa encodes a type III secretion system (TTSS) that confers high-level virulence, allowing the bacterium to induce eukaryotic cell injury (27). This system contains bioeffector molecules, translocators, and ExsA, a transcriptional activator of the TTSS regulon. PA103 mutants devoid of the type III secretion apparatus (ExsA), or related toxins ExoT or ExoU, were thus tested. Cells cultured in the presence or absence of Ca2+ were incubated with Ca2+ Green after bacterial infection and processed for visualization by confocal microscopy. PA103-infected cells accumulated punctate cytosolic signals indicative of elevated Ca2+ levels when cultured in Ca2+-replete medium. PA103 infection failed to significantly increase Ca2+ levels in cells maintained in Ca2+-free medium, suggesting that the increased cellular Ca2+ originated from its influx, rather than from release of internal stores (Fig. 1 A). Cells infected with PA103 mutant strains defective in ExoU alone, in combination with an ExoT defect, or lacking ExsA failed to elicit a robust increase in elevated Ca2+ levels (Fig. 1B).
Fig. 1.
P. aeruginosa PA103 triggers Ca2+ influx in murine lung epithelia. (A and B) MLE cells were cultured with Ca2+ Green dye with or without PA103 (MOI, 10; 1 h) or with 1 mM Ca2+ followed by staining with Ca2+ Green dye (1:2,000) for 30 min; cells were then washed with PBS and fixed with 4% paraformaldehyde. A 488-nm wavelength was used to excite the Ca2+ Green dye, with visualization of fluorescence emission. In panel B, cells were also infected (MOI, 10; 1 h) with or without PA103 mutants defective in expression of either the TTSS gene (ExsA; ΔA), exotoxin U (ΔU), or ExoT (ΔT) or mutants harboring a double deletion for ExoU and ExoT (ΔT/ΔU). Cells were stained with Ca2+ Green dye, washed, and fixed, followed by visualization of fluorescence emission. (C and D) Cells were infected with baculovirus encoding cameleon prior to culture in medium replete with Ca2+ (1 mM) or depleted of Ca2+, followed by PA103 infection (MOI, 10; 1 h) prior to processing for visualization of fluorescent CFP and YFP signals. (E) Following PA103 infection, cells were washed, fixed, and immunostained using anti-Golgi 97 antibody (1:200) to visualize the Golgi complex. (F) Cells infected with baculovirus encoding cameleon and PA103 as described for panel C were processed for analysis of fluorescent CFP and YFP signals.
As a complementary approach, we performed cameleon FRET analysis to assess cellular Ca2+. We utilized a cameleon fluorescent indicator consisting of tandem fusions of CFP, CaM, the CaM binding peptide M13 (based on the calmodulin binding domain [CBD] of skeletal muscle myosin light chain kinase; residues 577 to 602, KRRWKKNFIAVSAANRFKKISSSGAL), and YFP (21). This construct senses Ca2+ binding to CaM, which is visualized in live cells; increased Ca2+ binding causes CaM to wrap around the M13 domain, thereby increasing FRET between the flanking CFP and YFP. Cells were infected with a baculovirus (BacMam; Invitrogen) expressing this construct and 24 h later were infected with PA103 prior to visualization by fluorescence confocal microscopy. A significant increase in YFP emission was seen only in PA103-infected cells (Fig. 1C, YFP). Interestingly, the highest signal intensity was detected in a small area close to the nucleus, near or within the Golgi complex (Fig. 1C, merge image, arrows). Consistent with data in Fig. 1A, the YFP emission did not change in cells kept in Ca2+-free medium (Fig. 1D, YFP). Immunostaining of cells with a Golgi marker confirmed that the intense YFP signal in PA103-infected cells originated near the Golgi complex, suggesting that PA103 infection might elevate Ca2+ levels within this organelle (Fig. 1E). To observe changes in the YFP/CFP emission ratio over time, cells were plated, infected with the cameleon construct, and 24 h later infected with PA103. YFP/CFP emission ratios increased for ∼30 min and then reached a plateau (Fig. 1F).
Identification of a ubiquitin E3 ligase for CCTα.
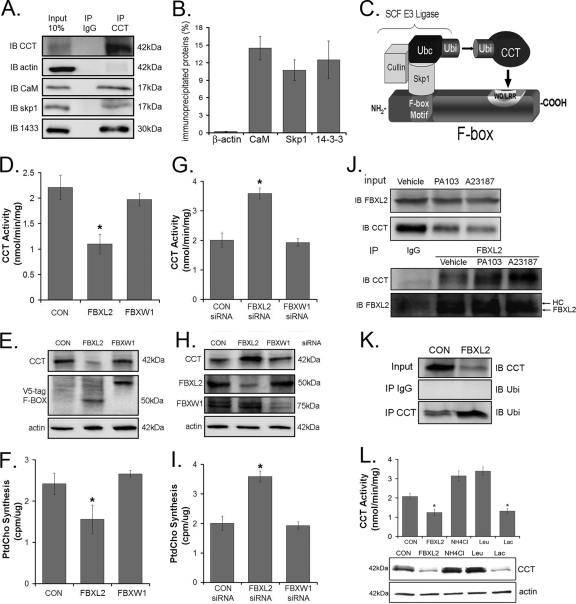
The enzyme CCTα is monoubiquitinated, degraded in lysosomes (4), and highly phosphorylated (35). We hypothesized that CCTα ubiquitination is catalyzed by an SCF complex, because this E3 ligase-like family targets phosphoproteins (12) and based on database analysis may contain subunits that are Ca2+ regulated. As confirmation, the SCF component Skp1 bound CCTα as determined by immunoblotting of individual components from immunoprecipitated CCTα from cells. 14-3-3 and CaM as positive controls bound CCTα, whereas β-actin served as a negative control (Fig. 2 A and B). Thus, the SCF complex is a potential E3 ubiquitin ligase for CCTα monoubiquitination. In the classic model, SCF ubiquitination of substrates occurs via the E2-conjugating enzyme Ubc (2, 17). In this model, the SCF complex uses the F-box motif to bind Skp1, whereas the leucine-rich/WD repeat motif is used for CCTα recognition (Fig. 2C). Here, the F-box protein serves as a critical linker to recruit the substrate within a close intermolecular distance to Ubc for enzyme ubiquitination. Our recent work identified a novel F-box protein, termed FBXL2, that appears to partake in lipid homeostasis (24). To assess functionality, FBXL2 and a specificity control, FBXW1, were cloned and expressed in lung epithelia (Fig. 2D to F). Only FBXL2 significantly decreased CCT activity, an effect mirrored in measurements of PtdCho synthesis (Fig. 2F). Levels of immunoreactive CCTα were reduced only after expression of FBXL2, indicative of selectivity of F-box proteins on regulating targets within the phospholipid pathway (Fig. 2E). To further test if FBXL2 is a putative E3 ubiquitin ligase component that targets CCTα for ubiquitination, cells were transfected with either FBXL2 siRNA, FBXW1 siRNA, or a control RNA for 48 h prior to functional assays (Fig. 2G to I). FBXL2 knockdown significantly increased CCTα activity, CCTα mass, and PtdCho synthesis. FBXW1 knockdown had no effect on these parameters. Together, these results demonstrate that both overexpressed and endogenous FBXL2 regulate phospholipid synthesis.
Fig. 2.
Identification of an E3 ligase for CCTα. (A and B) CCTα was immunoprecipitated using 2 μg of CCTα antibody from 100 μg of MLE cell lysates and processed for CCT, β-actin, CaM, Skp1, and 14-3-3 immunoblotting (A), and bands were quantitated (B). (C) Cartoon illustrating a classic model of the SCF-E3 ligase complex substrate engagement. (D to F) Two micrograms of each plasmid encoding two F-box proteins was transfected into cells (1 × 106) by nucleofection. Twenty-four hours later, cells were lysed and assayed for CCT activity (D), immunoblotting (E), or phosphatidylcholine synthesis (F). (G to I) Cells (1 × 106) were transfected with 0.2 nmol of scrambled (CON) RNA, FBXL2 siRNA, or FBXW1 siRNA by nucleofection. Forty-eight hours later, cells were lysed and processed for CCT activity (G), immunoblotting (H), and phosphatidylcholine synthesis (I). (J) Cells were treated either with vehicle, PA103 (MOI, 10; 1 h), or A23187 (10 nM) for 4 h. Cells were lysed, and 10 μg of lysate was resolved by SDS-PAGE prior to FBXL2 or CCTα immunoblotting (upper panel). FBXL2 was immunoprecipitated in 200 μg of lysate and processed for CCTα or FBXL2 immunoblotting (lower panel). (K) Cells were transfected with either empty vector or FBXL2 plasmid and lysed; 10 μg of lysate was resolved by SDS-PAGE prior to CCTα immunoblotting (upper panel). CCTα was immunoprecipitated in 200 μg of lysate and processed for ubiquitin immunoblotting (lowest panel). (L) Cells (1 × 106) were transfected with 2 μg of FBXL2 and treated with vehicle, the lysosomal inhibitors leupeptin (1:1,000 dilution) or NH4Cl (10 mM), or the proteasomal inhibitor lactacystin (1:1,000 dilution). Twenty-four hours later, cells were lysed, and assays of CCT activity (upper) and CCTα mass (lower) were performed.
We next assessed binding of FBXL2 to CCTα in a cell model of Ca2+ activation. Cells were treated with PA103 or the Ca2+ ionophore A23187 and lysed, and FBXL2 was immunoprecipitated prior to CCTα immunoblotting. Both PA103 and A23187 promoted FBXL2-CCTα interaction (Fig. 2J). Cells were also transfected with FBXL2 and lysed, and CCTα was immunoprecipitated prior to ubiquitin immunoblotting. The results suggested that overexpression of FBXL2 increases levels of monoubiquitinated CCTα (Fig. 2K). Lastly, cells transfected with FBXL2 and simultaneously treated with lysosomal inhibitors (leupeptin or NH4Cl) or the proteasomal inhibitor lactacystin differentially regulated enzyme levels. Unlike lactacystin, both lysosomal inhibitors completely blocked the effects of FBXL2 and tended to increase CCT activity and mass (Fig. 2L), indicating that monoubiquitinated CCTα is degraded within the lysosomal pathway.
The FBXL2-CCTα interaction occurs within defined motifs.
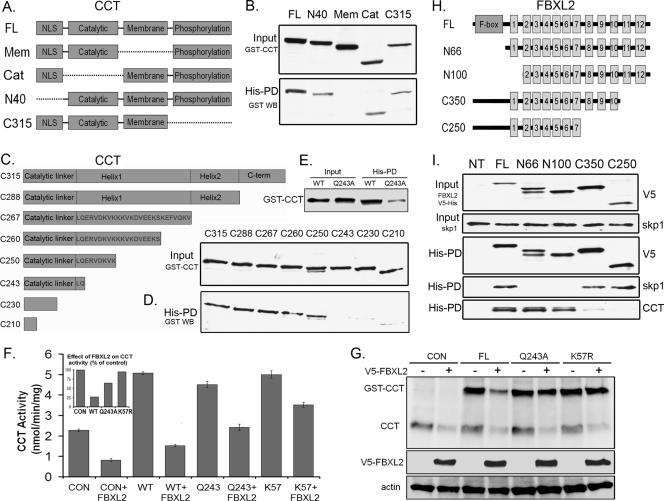
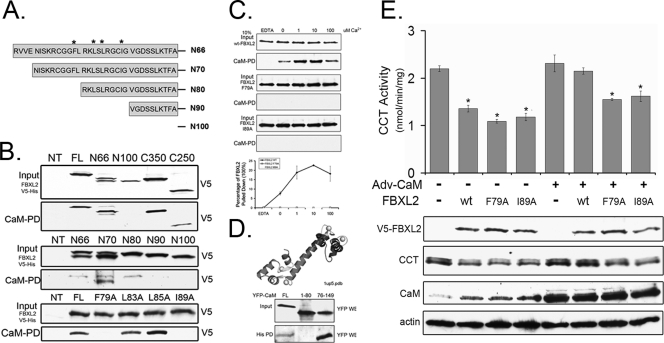
To map the FBXL2 binding sites within CCTα, GST-tagged CCTα lacking various functional domains was expressed separately in cells (Fig. 3 A). Immunoblotting confirmed that recombinant GST-tagged proteins were expressed (Fig. 3B, upper panel). These lysates were then applied to His-tagged FBXL2 prebound to Talon cobalt affinity beads, and samples were eluted and processed for GST immunoblotting. FBXL2 bound GST-tagged full-length CCTα (CCTαFL) and GST-CCTα mutants lacking either the catalytic core (Cat), NH2-terminal sequence (amino acids [aa] 1 to 40; N40), or the carboxyl terminus (C315); however, deletion of the CCTα membrane binding domain disrupted the FBXL2-CCTα association, indicating that FBXL2 binds CCTα within its membrane binding domain. When CCTα constructs progressively truncated at the carboxyl terminus of the membrane binding domain were tested (Fig. 3C), binding of FBXL2 to CCTα mutants (C243, C230, and C210) was significantly reduced (Fig. 3D). This region of CCTα (aa 242 to 250, LQERVDKVK) contains a highly conserved CaM binding IQ motif (3). Thus, FBXL2 also might utilize this molecular signature to target CCTα for ubiquitination by SCF. To assess this further, we introduced a point mutation at Q243, which disrupts the IQ motif (3). GST-CCTFL or GST-CCTQ243A plasmids were expressed in cells and also processed using FBXL2 pulldown assays prior to GST immunoblotting and functional assays. As shown in Fig. 3E, binding of FBXL2 to the CCTα Q243A mutant was significantly reduced, underscoring the importance of the IQ motif in the FBXL2-CCT interaction. This molecular site (CCTQ243) was critical for FBXL2 targeting, as overexpression of FBXL2 resulted in an ∼75% decrease in activity of both endogenous and overexpressed GST-CCTFL (Fig. 3F). However, in cells transfected with GST-CCTQ243A or a CCTα plasmid encoding a mutated ubiquitin acceptor site (CCTK57R) (4), enzyme activity was significantly higher (Fig. 3F). Partial resistance to actions of FBXL2 were also recapitulated in immunoblotting experiments. Here, both endogenous and overexpressed immunoreactive GST-CCTFL was significantly reduced after FBXL2 overexpression, but levels were better preserved after expression of the CCTα point mutants (Fig. 3G). Thus, Q243 may serve as a key dock site for access of FBXL2. Interestingly, this region also binds CaM (3), strongly suggesting that FBXL2 and CaM compete for occupancy with CCTα.
Fig. 3.
FBXL2 interaction with CCTα. (A) Map of GST-CCTα mutants. Dashed lines represent deleted motifs. (B) Five micrograms of plasmids encoding GST-tagged mutants (mapped in panel A) were expressed in MLE cells (3 × 106), and 24 h later cells were lysed and processed for GST immunoblotting (upper panel) or purification on His-V5-tagged FBXL2 complexed on cobalt affinity beads prior to GST immunoblotting (lower panel). (C) Map of CCTα mutants lacking motifs within its membrane binding domain. (D) The mutants in panel C were processed similar to those shown in panel B for GST immunoblotting of lysates (upper panel) or after affinity purification (lower panel). His-PD, His-tagged pulldown product. (E) Five micrograms of plasmids encoding either GST-CCT WT or GST-CCTQ243A was expressed in cells (3 × 106), and 24 h later cells were lysed and processed for GST immunoblotting after purification with His-V5-tagged FBXL2 cobalt affinity beads. (F and G) Cells were cotransfected with 3 μg of plasmids encoding FBXL2 and 2 μg of plasmids encoding either WT GST-CCT, GST-CCTQ243A, or GST-CCTK57R and then processed for CCT activity (F) or immunoblotting (G). The inset in panel F shows CCT activity as a percentage of control after FBXL2 expression. (H) Map of FBXL2 mutants. (I) His-V5-FBXL2 mutants were expressed in cells, and lysates were processed for V5 and Skp1 immunoblotting or purified on cobalt affinity beads (His-PD) prior to immunoblotting for V5, Skp1, and CCTα.
FBXL family proteins contain LRR for substrate targeting. In FBXL2, residues 1 to 66 contain a classic F-box domain that interacts with Skp1 (33). Residues 80 to 423 contain 12 LRRs that display extensive internal homology (Fig. 3H) (36). His-V5-tagged FBXL2 constructs lacking various LRRs were expressed in cells (Fig. 3I, upper panel), copurified on Talon cobalt affinity beads, and eluted prior to immunoblotting with CCTα and Skp1 antibodies. Full-length FBXL2 and FBXL2 mutants lacking either the F-box domain (N66) or the first LLR (N100) bound CCTα (Fig. 3I, bottom panel); however, deletion of the last five LLRs (C250) or last two LLRs (C350) markedly disrupted the FBXL2-CCTα association. Thus, CCTα binds FBXL2 within its last two LLR domains (aa 350 to 423). As a control, full-length FBXL2 and FBXL2 with deletion of LLRs (C250 and C350) all bound Skp1; however, deletion of the F-box domain (N66 and N100) markedly disrupted the FBXL2-Skp1 association (Fig. 3I, fourth panel from the top).
FBXL2 degradation of CCT is Ca2+ dependent and blocked by CaM.
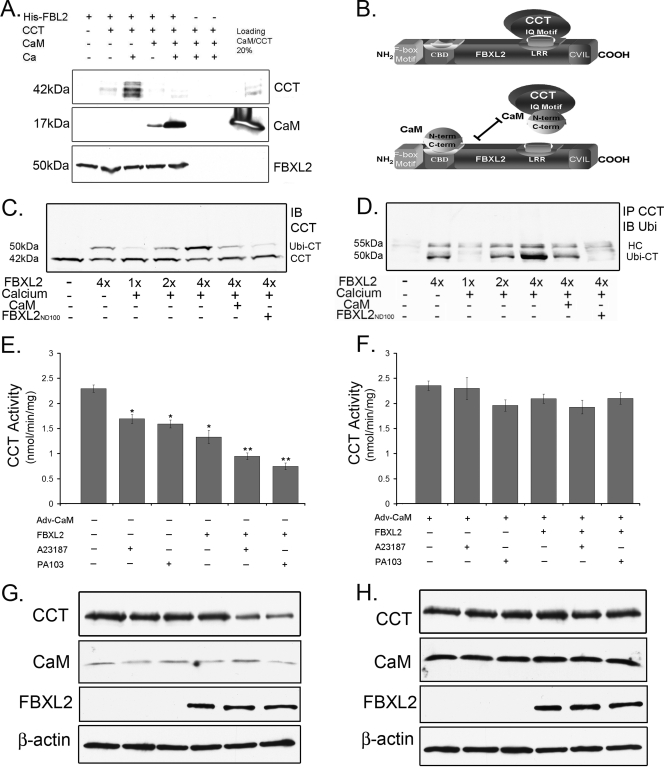
As shown above, both FBXL2 and CaM bind to the CCTα IQ motif, LQERVDKVK, which resides within the CCTα membrane binding domain. The data suggest CaM and FBXL2 compete for CCTα binding. This was tested in pull-down experiments in the presence or absence of Ca2+ in which FBXL2 was immobilized on beads and used as bait for CCTα and CaM (Fig. 4 A). Three negative controls were included: (i) FBXL2-agarose alone was assayed to control for CCTα and CaM contamination; (ii) CCTα and CaM with Ca2+ were run over empty Talon beads and eluted and proteins were resolved by SDS-PAGE followed by CCTα and CaM immunoblotting to ensure that associations were FBXL2 specific; and (iii) V5 immunoblotting was used as a loading control, to ensure that pulldown experiments contained equivalent amounts of FBXL2. The results indicated that not only does FBXL2 directly interact with both CCTα and CaM in a Ca2+-dependent manner but also that excess CaM disrupts the FBXL2 interaction with CCTα (Fig. 4A and B).
Fig. 4.
SCF (FBXL2) ubiquitination of CCTα is Ca2+ dependent and opposed by CaM. (A) FBXL2-agarose beads (used as bait) were generated and incubated with combinations of 1 μg purified CCTα or CaM with or without Ca2+. After washing of beads (150 mM NaCl, 0.1% Triton X-100), proteins were eluted and resolved by SDS-PAGE followed by CCT, CaM, and V5 immunoblotting. (B) Cartoon illustrating the proposed FBXL2 targeting mechanism. (C) In vitro ubiquitination of CCTα and immunoblotting. Purified CCTα was incubated with the full complement of immunoprecipitated Cullin 1, Skp1, Rbx1, and FBXL2 from cells, plus ubiquitin (∼8.5 kDa) and ATP. Reactions proceeded in the presence or absence of excess Ca2+, CaM, or the FBXL2N100 mutant. Reaction products were processed for CCTα immunoblotting. (D) Coimmunoprecipitation of monoubiquitinated CCTα. CCTα was immunoprecipitated from the in vitro ubiquitination reactions shown in panel C and processed for ubiquitin immunoblotting. (E to H) Cells (4 × 106) were infected with Adv-empty or Adv-CaM (MOI, 40) for 12 h prior to harvest and transfection with FBXL2 plasmid (5 μg) for an additional 24 h. Cells were then infected with PA103 (MOI, 10) for 1 h or treated with A23187 (10 nM) for 4 h prior to analysis of CCT activity (E and F) or immunoblotting (G and H).
To test whether FBXL2 is an authentic E3 ligase component that partakes in CCTα ubiquitination, we performed in vitro ubiquitination assays. Purified CCTα was incubated with the full complement of immunoprecipitated Cullin 1, Skp1, Rbx1, and FBXL2 from cells plus ubiquitin (∼8.5 kDa) and ATP. Reactions proceeded in the presence or absence of excess Ca2+, CaM, or the FBXL2N100 mutant. Products were processed for CCTα immunoblotting, which showed that purified conjugation enzymes generate an additional CCTα species of ∼50 kDa, a size consistent with monoubiquitinated CCTα (Fig. 4C). The proportion of this species significantly increased with addition of FBXL2, and levels of the 50-kDa species were reduced with inclusion of CaM. A portion of the reaction mixture was also incubated with CCTα antibody, pulled down by protein A/G beads, and then probed with ubiquitin antibody (Fig. 4D). Excess Ca2+ increased the intensity of the ∼50-kDa monoubiquitinated product, whereas both CaM and dominant negative FBXL2N100 significantly reduced levels of the monoubiquitinated form. The results in Fig. 4D recapitulate the data shown in Fig. 4C, showing a dominant band (∼50 kDa) just below the heavy chain (HC), strongly suggesting that FBXL2 serves as an E3 ligase subunit that within the SCF complex ubiquitinates CCTα.
We observed that PA103 infection and the Ca2+ ionophore A23187 each promote CCT degradation and impair its function via Ca2+ signals (Fig. 1) (38, 39). Cells infected with PA103 or treated with A23187 displayed only a modest reduction in CCTα activity when we employed a low level of stimulus (Fig. 4E). However, when cells were transfected with FBXL2, inhibitory effects of either A23187 or PA103 on CCT activity were more pronounced, coupled with reduced immunoreactive enzyme levels (Fig. 4E and G). When cells were preinfected with a replication-deficient adenovirus expressing CaM (Adv-CaM) and then transfected with FBXL2 with either stimulus, CaM effectively restored CCTα activity and CCTα protein levels (Fig. 4F and H). Together, these experiments strongly suggest CCTα is monoubiquitinated by FBXL2 in a Ca2+-regulated manner and degraded in a lysosomal pathway and that CaM stabilizes the enzyme from the ligase in vitro and in vivo. Further, abrogation of the FBXL2-CaM interaction regulates CCTα enzymatic behavior.
The FBXL2-CaM interaction occurs within defined motifs.
To map the CaM binding sites within FBXL2, first we performed a database analysis, which suggested that FBXL2 is a non-EF hand Ca2+ binding protein with a strong CaM binding domain at residues 66 to 100 (Fig. 5 A) (Calmodulin Target Database). To confirm this, cell lysates expressing V5-tagged full-length FBXL2 or FBXL2 mutants were purified on CaM-Sepharose beads, the beads were extensively rinsed using buffer containing 0.1% NP-40 and 0.1% Triton X-100, and products were eluted and processed for V5 immunoblotting (Fig. 5B, top panel). The results indicated that full-length FBXL2 and FBXL2 mutants (N66, C350, and C250) all bound CaM, with the exception of N100, which failed to bind CaM. Thus, CaM interacts with a putative CBD located within the first LLR domain (residues 66 to 100) of the ubiquitin ligase subunit. To further map this region, we similarly tested several FBXL2 mutants in CaM pulldown studies that were progressively truncated within this span of the NH2 terminus (residues 66 to 100). FBXL2 mutants (N66, N70, and N80) all bound CaM, with the exception of N90 and N100, indicating that CaM interacts with a putative CBD residing at residues 80 to 90 of FBXL2 (Fig. 5B, middle panel). Several bulky and hydrophobic residues within this region might be important for CaM interaction. Four FBXL2 variants in which amino acids (F79, L83, L85, and I89) were mutated to alanine were similarly tested in CaM pulldown assays. Indeed, FBXL2 mutants (F79A and I89A) lose the ability to bind CaM (Fig. 4B, lower panel).
Fig. 5.
FBXL2 interaction with CaM. (A) Map of FBXL2 mutants. (B) The FBXL2 truncation mutants shown in Fig. 3H, shown in panel A here, or FBXL2 point mutants were expressed in MLE cells, and lysates were processed for V5 immunoblotting (top panel, each pair) or after incubation and elution with CaM-Sepharose beads (40 μl; lower panel, each pair). (C) Cells (4 × 106) were transfected with 5 μg of plasmids encoding WT-V5-FBXL2, V5-FBXL2-F79A, or V5-FBXL2-I89A. Twenty-four hours later, cells were harvested and 100 μg of cell lysate was incubated with CaM-Sepharose beads at different Ca2+concentrations. After extensive rinsing, elusion products were resolved by SDS-PAGE prior to V5 immunoblotting. Levels of V5 proteins were quantified using densitometry and graphed (lowest panel). Data represent two independent experiments. (D) Crystal structure of CaM, showing the NH2 domain (residues 1 to 80) and the carboxyl-terminal domain (residues 76 to 149). Four Ca2+ ions are bound to two homologous domains composed of two helix-loop-helix motifs, connected by a 5-residue flexible linker (black). YFP-tagged full-length (FL) CaM, CaM containing the first 80 residues (1 to 80), or a stretch of carboxyl-terminal 73 residues (76 to 149) was coexpressed in cells with His-V5-tagged FBXL2. Lysates from transfectants were resolved by SDS-PAGE prior to YFP immunoblotting (upper panel), or lysates were purified on Talon cobalt affinity beads prior to processing for YFP immunoblotting (lower panel). (E) Cells (4 × 106) were plated in 100-mm dishes for 24 h, infected with Ad-CaM or an empty vector (Ad-Con) at an MOI of 40 for 12 h, and cells were then harvested and transfected with 5 μg of plasmid encoding WT-FBXL2, FBXL2-F79A, or FBXL2-I89A for an additional 24 h. Cells were then assayed for CCT activity (upper panel) or immunoblotting (lower panel).
CaM binding proteins often interact with CaM in a Ca2+-dependent manner, and many Ca2+-dependent CaM recognition motifs are characterized by a basic amphipathic helix, moderate to high helical hydrophobic moment, and net positive charge (25). Consistent with this, the FBXL2 CBD is enriched with basic residues, and database analysis predicts a basic amphipathic helix within this region (data not shown). Thus, we hypothesized that FBXL2 is a Ca2+-dependent CaM binding protein. To confirm this, recombinant wild-type (WT) V5-FBXL2, FBXL2-F79A, and FBXL2-I89A proteins were incubated with CaM-agarose beads in the presence of increasing concentrations of Ca2+ (0 to 100 μM). Beads were rinsed as described above, and products were eluted and subjected to V5 immunoblotting. FBXL2 interacts with CaM in a Ca2+-dependent manner, whereas the mutants showed no interaction with CaM despite an excess of calcium (Fig. 5C). CaM also contains two homologous domains (NH2-terminal domain and C domain), each of which binds two Ca2+ ions. Interestingly, the C domain has a 10-fold-greater Ca2+ affinity than the NH2-terminal domain and is required for FBXL2 interaction (Fig. 5D). Importantly, when cells were transfected with WT FBXL2, FBXL2-F79A, or FBXL2-I89A, which do not bind CaM, all three FBXL2 forms effectively decreased CCTα activity and CCTα protein levels (Fig. 5E), suggesting that mutant FBXL2s are functional. However, when cells were preinfected with a replication-deficient adenovirus expressing CaM (Adv-CaM) and then transfected with these FBXL2 plasmids, CaM gene transfer was not able to totally rescue CCTα activity and CCTα protein levels when cells were transfected with FBXL2-F79A or FBXL2-I89A (Fig. 5E).
Intermolecular competition by FBXL2 and CaM for CCTα occurs within the Golgi complex.
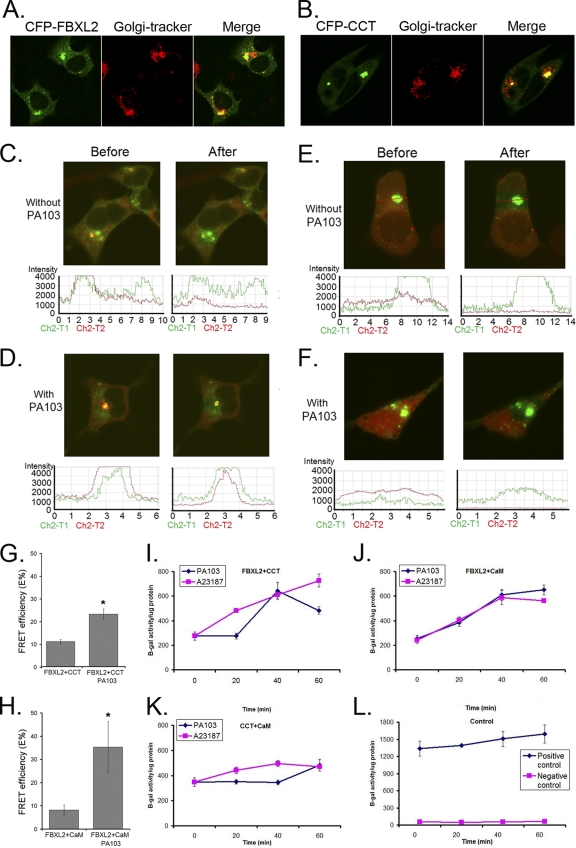
PA103 infection appears to increase Ca2+ concentrations in or around the Golgi complex (Fig. 1E). To investigate FBXL2 cellular localization, we monitored recombinant CFP-FBXL2. A strong CFP signal was detected in the perinuclear region and this colocalized with a Golgi marker (Fig. 6 A). Overexpressed recombinant CCTα was detected in the nucleus; in contrast, endogenous CCTα also localizes to the ER and the Golgi complex (3, 4, 26). To circumvent such an artifact, we constructed CFP-CCTN40, which lacks the nuclear localization signal. Indeed, CFP-CCTN40 primarily localizes within the Golgi complex of cells, making it a suitable target for FBXL2 (Fig. 6B).
Fig. 6.
P. aeruginosa degradation of CCTα involves molecular interactions of FBXL2 and CaM within the Golgi complex. (A and B) MLE cells (2 × 105) were transfected with CFP-FBXL2 or CFP-CCTα plasmids (1 μg) and immunostained with anti-Golgi complex (anti-Golgi 97) antibodies. (C to F) Cells were cotransfected with CFP-FBXL2/YFP-CCTα (C and D) or CFP-FBXL2/YFP-CaM (E and F) for 24 h. Cells were then infected with or without PA103 at an MO1 of 10 for 1 h. Cells were washed with PBS and fixed with 4% paraformaldehyde for 20 min and observed using a confocal microscope. The FBXL2-CCT or FBXL2-CaM interaction at the single-cell level was imaged using laser scanning microscopy before and after photobleaching. Shown in the upper sets of panels are single-cell images before and after acceptor photobleaching fluorescence with intensities of YFP and CFP (C to F). (Bottom) The same FRET in each panel was confirmed quantitatively and is shown graphically. FRET efficiencies (E%) were calculated and are graphed (G and H). (I to L) Mammalian two-hybrid assay. Cells (1 × 106) were cotransfected using electroporation with combinations of CCTα-Gal4BD, CaM-Gal4AD, FBXL2-Gal4BD, and FBXL2-Gal4AD plasmids as fusion proteins with a pFR-β-galactosidase reporter vector prior to assays for β-galactosidase activities.
Next, we examined if FBXL2 engages CCTα or CaM under a Ca2+ stimulus and whether this interaction is accentuated by PA103. CFP-FBXL2 and YFP-CCTN40 or YFP-CaM were cotransfected into cells prior to PA103 infection. Cells were analyzed by fixation followed by irreversible photobleaching using FRET (Fig. 6C to H). FRET is observed, indicating protein interaction between two partners, when the donor emission (CFP) signal increases after a nearby acceptor fluorophore (YFP) is inactivated by irreversible photobleaching. The emission fluorescence levels of both the donor CFP-FBXL2 and acceptor YFP-CCTα (Fig. 6C and D) or YFP-CaM (Fig. 6E and F) before and after acceptor photobleaching are shown in Fig. 6 (upper images and lower plots; the region of interest around the Golgi complex is marked with a red arrow). The data from three independent experiments and >12 randomly selected cells for each condition were analyzed, and FRET efficiency is presented in Fig. 6G and H). The results indicated that upon bleaching, there is decreased acceptor fluorescence (YPF) coupled with increased donor emission fluorescence (CFP), which is consistent with an FBLX2 interaction with either CaM or CCTα after PA103 infection in cells (Fig. 6D and F). In separate studies we tested the potential interaction of the CCT Q243A mutant with FBXL2 by FRET analysis, and the results indicated that these proteins do not interact in cells (data not shown). Together, these results suggest that PA103 infection induces molecular interactions between CaM, FBXL2, and CCTα that may regulate FBXL2-mediated CCTα degradation.
Further confirmation of the FBXL2 interaction with either CaM or CCTα after PA103 infection was obtained from a mammalian two-hybrid assay (Fig. 6I to L). In this experiment, plasmids expressing CCTα and FBXL2 were fused to the Gal4 binding domain (CCTα-Gal4BD and FBXL2-Gal4BD, respectively), and CaM and FBXL2 were fused to the Gal4 acceptor domain (CaM-Gal4AD and FBXL2-Gal4AD, respectively). Cells were coelectroporated with a reporter (pFR-β-gal) together with CCTα/FBXL2, CaM/FBXL2, or CCT/CaM; 24 h after transfection cells were infected with PA103 prior to harvest and assays for β-galactosidase activities with appropriate control plasmids. A23187 or PA103 significantly increased β-galactosidase activities in cells transfected with CCTα/FBXL2 or CaM/FBXL2, but not in cells transfected with CCT/CaM or the positive- or negative-control plasmids. Despite the ability of A23187 and PA103 to stimulate Ca2+ influx, the CCTα and CaM interaction was not altered, consistent with Ca2+-independent binding between CCTα and CaM (3). Thus, during PA103 infection the interaction of FBXL2 with either CCTα or CaM is enhanced within the Golgi compartment, and this interaction is suppressed by CaM.
Adenoviral CaM gene transfer ameliorates P. aeruginosa-induced lung injury.
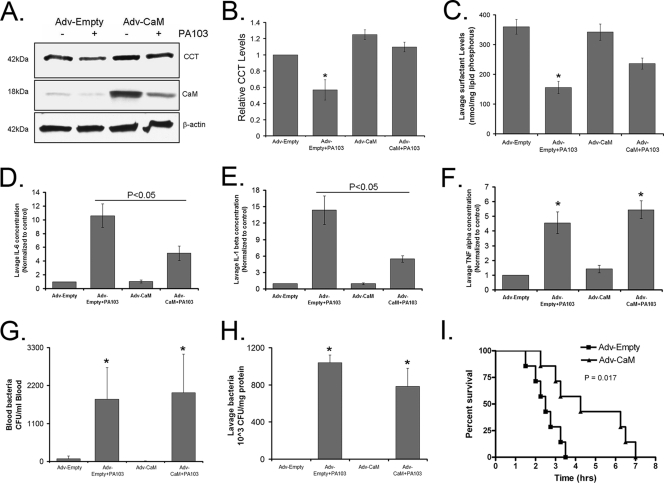
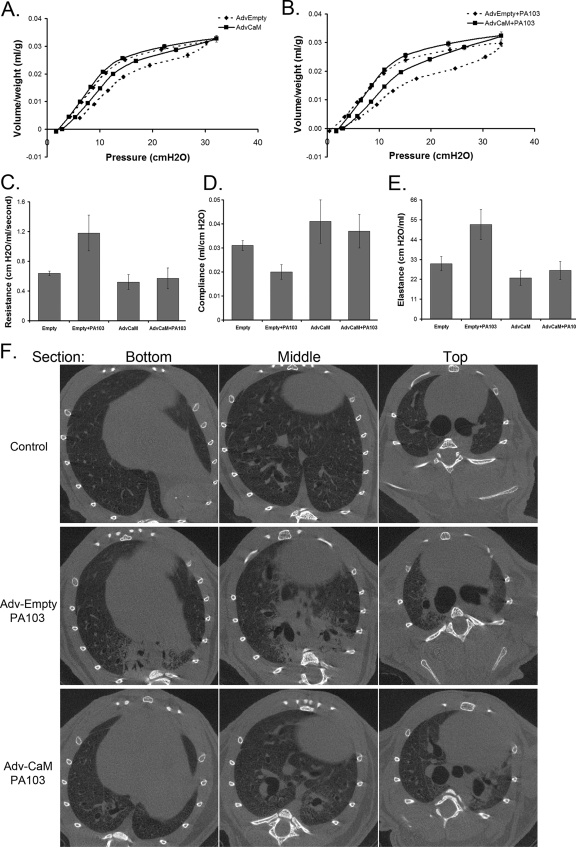
We next evaluated the biologic significance of these molecular interactions in an animal model of pneumonia. We hypothesized that during P. aeruginosa infection adenoviral CaM gene transfer would counteract actions of the E3 ligase FBXL2, thereby preserving CCT and surfactant levels. In mice, the PA103 pathogen decreased CCTα levels (Fig. 7A and B), thereby reducing surfactant phosphatidylcholine (Fig. 7C) needed to stabilize lung function (39). Mice infected with PA103 also exhibited increased inflammatory markers (Fig. 7D to F) with high bacterial loads (Fig. 7G and H). Mice given Adv-CaM had an ∼20-fold induction of immunoreactive CaM compared to control mice and restored CCTα levels and partially restored surfactant levels after PA103 infection (Fig. 7A to C). Although CaM did not alter circulating or lavage bacteria (Fig. 7G and H), Adv-CaM significantly attenuated PA103 increases in interleukin-6 (lL-6) and IL-1β (Fig. 7D and E) and increased survival of PA103-infected mice (Fig. 7I). Pneumonia also resulted in impaired lung mechanics, as evidenced by decreased lung compliance (Δvolume/pressure) and increased elastance (lung stiffness) (Fig. 8). Adv-CaM gene transfer had limited effects on pressure-volume relationships in control mice (Fig. 8A), but after PA103 infection there was improved lung compliance (Fig. 8B and D); CaM blocked PA103-induced increases in resistance (Fig. 8C) and tissue elastance (Fig. 8E). Mice were also deeply anesthetized and paralyzed prior to live imaging of lungs by micro-CT scanning after administration of PA103 (Fig. 8F). Control mice receiving an empty vector after PA103 infection showed significant areas of basilar consolidation within lung parenchyma (Fig. 8, middle row). In some areas, there was resolution of these abnormalities in mice overexpressing CaM (Fig. 8, middle panels, bottom two rows). Collectively, these observations indicate that pulmonary expression of CaM can partially improve inflammation or preserve lung mechanics in the setting of P. aeruginosa infection.
Fig. 7.
Adenoviral CaM gene transfer lessens the severity of P. aeruginosa-induced lung inflammation and injury. C57BL/6J mice were administered i.t. with Adv-empty or Adv-CaM (109 PFU/mouse) for 48 h, and 4 mice/group were inoculated with PA103 (1 × 107 CFU/mouse). Mice were euthanized after 1 h. Lungs were lavaged with saline, harvested, and then homogenized; blood was also withdrawn using cardiac puncture. Lung CCTα, CaM, and β-actin were assayed by immunoblotting (A), and the results were analyzed and quantified using ImageJ software (B). (C) Lavage surfactant lipids were extracted and resolved using thin-layer chromatography, and surfactant phosphatidylcholine mass was then assayed. (D to F) An aliquot of lavage samples was centrifuged, and 0.5% fatty acid-free BSA plus EDTA-free protease inhibitor cocktail was added to the supernatant. Forty microliters of supernatant was assayed for cytokines by using antibodies to IL-1β, IL-6, and tumor necrosis factor (TNF) alpha for immunoblotting, and the results were analyzed and quantified using ImageJ software. (G and H) Blood and whole-lavage diluents were plated on TSB agar plates to determine CFU. (I) Replication-deficient Ad5 alone or Adv-CaM (109 PFU/mouse) was instilled i.t. on day 1, after which animals were allowed to recover for 48 h. Following recovery, mice were deeply anesthetized, and 7 mice/group were inoculated with 1 × 107 CFU PA103. Mice were carefully monitored over time; moribund, preterminal animals were immediately euthanized and recorded as deceased. Kaplan-Meier survival curves were generated using Prism software. *, P < 0.05 versus control.
Fig. 8.
Adenoviral CaM gene transfer improves lung mechanics after P. aeruginosa infection. (A and B) Replication-deficient adenovirus (AdvEmpty) alone or Adv-CaM (109 PFU/mouse) was instilled i.t. on day 1, after which animals were allowed to recover for 48 h. Following recovery, mice were deeply anesthetized, followed by infection with P. aeruginosa PA103 (107 CFU/mouse, i.t.) for 1 h. Animals were then mechanically ventilated, and pressure-volume loops were measured (B). (C to E) Lung resistance, compliance, and elastance (lung stiffness). Each group contained five to six mice. (F to H) In separate studies, mice were given adenovirus and PA103 as described above and processed for micro-CT scanning to visualize lung infiltrates.
DISCUSSION
The plethora of various ubiquitin E3 ligases suggests highly significant and diverse roles, and yet the functions and molecular interplay of the majority of SCF-based E3 ligases remain unknown (2, 6, 28). These studies are the first demonstrating the ability of SCF-based E3 ligases to monoubiquitinate a substrate, that this activity requires FBXL2, and that CaM is a SCF complex antagonist (Fig. 9). FBXL2 functions as a substrate recognition subunit of a prototypical ubiquitin E3 ligase that is Ca2+ regulated (Fig. 9B), but the ubiquitinating activity of the SCF (FBXL2) complex is significantly attenuated by CaM (Fig. 9C). FBXL2, via specific molecular determinants, competes with CaM to target a key lipogenic enzyme, CCTα, which is involved in the biosynthesis of a crucial structural component of animal membranes and of lung surfactant. These biochemical findings were extended to a murine model of pneumonia, in which CaM gene transfer stabilized CCTα levels, lessened inflammation, and improved pulmonary mechanics under conditions when ubiquitin ligase activity was activated by Ca2+. CaM's ability to oppose FBXL2 for substrate binding represents a potentially important model for ubiquitin-mediated proteolysis, as it opens the door for strategies directed at development of small-molecule F-box antagonists that could ultimately lessen the severity of inflammatory lung injury in patients with bacterial pneumonia.
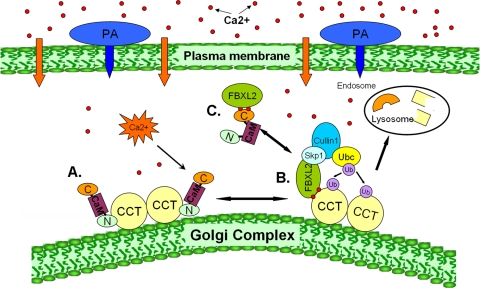
Fig. 9.
Molecular interplay between FBXL2, its substrate CCTα, and CaM controls phosphatidylcholine (PC) synthesis. (A) CCTα is a dimeric lipogenic enzyme normally bound and stabilized by CaM that is activated by membrane binding (e.g., to the Golgi complex). (B) P. aeruginosa increases intracellular Ca2+ (red dots) that recruits FBXL2 to membrane-bound CCTα (lower right); the activated SCF E3 ligase complex (Skp1-Cullin 1-FBXL2) binds and ubiquitinates CCTα, the rate-limiting enzyme required for phosphatidylcholine synthesis. The monoubiquitinated CCTα is targeted for disposal via the endosome-lysosome pathway. (C) Excess CaM competitively rescues CCTα from ubiquitination by directly binding to the FBXL2 E3 ligase via its carboxyl-terminal domain.
A central phenomenon that emerged from our studies is that elevations in intracellular Ca2+ are an initial event that regulates the behavior of the E3 ligase proteolytic apparatus. Our data provide the first direct evidence that P. aeruginosa activates Ca2+ flow in the lung and that this occurs from an influx of extracellular Ca2+ rather than from internal stores. Although Ca2+ concentrations in alveolar fluid are relatively high (∼2 mM), lung epithelial cells maintain tight control of Ca2+ levels with very low intracellular Ca2+ concentrations ([Ca2+]i), at nanomolar to micromolar levels. Elevated [Ca2+]i induced by PA103 triggered an increase in FRET signals in a perinuclear zone that was consistent with [Ca2+]i increases within the Golgi compartment, and each of the binding partners either colocalized or interacted within this organelle. Positive FRET signals within the nucleus were not observed, indicating either very low or undetectable Ca2+ concentrations within the nucleus or cytosolic expression of cameleon protein. Cells costained with the ER stain Tracker red indicated that the positive FRET signal does not localize within this organelle (data not shown). Thus, the Golgi complex appears to be a subcellular site for the molecular interaction of FBXL2 with CCTα and CaM, regulating ubiquitination during [Ca2+]i induction prior to lysosomal degradation. Although we identified that ESCRT I (the endosomal sorting complex required for transport) assists in lysosomal targeting of CCTα (4), its ubiquitination within the Golgi complex by FBXL2 suggests that other sorting elements, such as GGA (Golgi-localized, gamma ear-containing, Arf binding) proteins, through their GAT domain might also be components of the molecular machinery that aids in enzyme sorting (22). Interestingly, P. aeruginosa exotoxins that degrade CCTα (11) also sort through the Golgi apparatus, suggesting that this compartment is an active site for enzyme modification, sorting, and destabilization.
Our RNAi and expression studies showed that so far only FBXL2 has been identified as functionally relevant (Fig. 2). Both endogenous and overexpressed FBXL2 bound CCTα under physiologic conditions when Ca2+ increased, and this association occurred via specific domains (Fig. 3). CCT activity was assayed after cellular expression of full-length FBXL2 and two additional constructs that would be predicted to disrupt the SCF-ligase complex, resulting in impaired ligase activity. An FBXL2 mutant devoid of the F-box domain (N66) lacked the ability to bind Skp1, and a mutant (C350) where the last two LLRs were removed resulted in an inability to bind substrate and led to a dominant negative effect. It is unlikely that the C350 construct was misfolded, because like full-length CFP-FBXL2, a CFP-FBXL2-C350 mutant also localized to the Golgi complex (data not shown). As a whole, FBXL2 may be one bona fide E3 ligase component that destabilizes CCTα via substrate interaction, using its last two LLR domains (aa 350 to 423). The results do not exclude the possibility that other F-box proteins shuttling within SCF complexes also ubiquitinate CCTα.
The observation that FBXL2 binds CCTα within its canonical IQ motif (LQERVDKVK; residues 243 to 250) located in helix 1 of CCTα was unexpected, as this region was also mapped as a site for CaM binding (3). CaM, however, interacts with this motif irrespective of Ca2+ availability (3), whereas FBXL2 interacts with this motif in a Ca2+-dependent manner (Fig. 4A). Although FBXL2 has no sequence similarity with CaM, protein database analysis suggests that FBXL2 resembles a non-EF hand Ca2+ binding protein. Thus, constitutive levels of CCTα may be governed by CaM, but during agonist-induced increases in [Ca2+]i, stoichiometric levels of CaM versus FBXL2 in lung epithelia might control CCTα life span. CCTα is also heavily phosphorylated, and F-box proteins with a carboxyl-terminal LRR motif (e.g., FBXL1) target phosphoproteins (8). However, our data (Fig. 3) and our prior work (4) suggest that the CCTα carboxyl-terminal phosphorylation domain is not required either for CCTα ubiquitination or for FBXL2 targeting.
Importantly, FBXL2 and CaM interact; both partners compete for binding to a substrate within specific molecular determinates, and these interactions are functionally relevant in vivo. FBXL2 employs its first LRR motif (specifically, amino acids 79 to 89) to bind CaM (Fig. 5B), and CaM engages FBXL2 via its carboxyl terminus (Fig. 5D). FBXL2, through a calcium-dependent binding domain (79FLRKLSLRGCI89), binds CaM. There are three major groups of calcium-dependent CaM binding domains: 1-10, 1-12, and 1-14 motifs, in which two bulky hydrophobic residues are spaced by 8, 10, or 12 amino acids. The 1-10 motif, for example, can be represented by (FILVW)xxxxxxxx(FILVW). In some cases, additional anchoring residues are present in the middle of the 1-10 motif: (FILVW)xxxx(FAILVW)xxxx(FILVW). In the FBXL2-CaM binding domain, there are 9 residues between two bulky residues (F79 and I89) and 2 anchoring residues (L83 and L85). Single substitution of bulky residues completely disrupted the motif; however, mutation of the anchoring residues did not affect the FBXL2-CaM interaction. Other work is needed to determine whether these sites alter CaM binding affinities.
The complexity of CCTα posttranslational regulation is underscored by its ubiquitination and lysosomal degradation by SCF E3 ligases, as shown here, but also by its destabilization by calcium-activated neutral proteinases (calpains) (3). Our results are not unlike sensitivities of other substrates that are cleaved and targeted by multiple proteolytic systems. For example, IkBα and tau proteolysis levels are mediated by both the calpain and ubiquitin proteolytic pathways (10, 37). Calpains, lysosomes, and the proteasome also regulate myofibrillar protein turnover (9). Calpains execute limited proteolysis of their substrates but play critical roles in presenting substrates for proteasomal or lysosomal elimination (5). CCTα turnover is entirely consistent with the multiple modes of regulation seen with structural proteins, given that the kinetics of enzyme degradation by calpains and the ubiquitin-lysosome system in lung epithelial cells also differ significantly. Calcium ionophores activate calpains after long-term exposure to degrade CCTα (38). After transient exposure, as employed here, calpain activity remains unaltered (data not shown), making it unlikely that these proteinases contribute to CCTα degradation. SCF complexes might also regulate CCTα monoubiquitination, multiubiquitination, or even polyubiquitination (18), depending on whether different E2 enzymes are involved and the appropriate in vivo context is present, similar to other regulatory proteins (e.g., p53) or hormone receptors that are channeled via different degradative pathways (13, 31, 32).
CaM gene transfer in mice significantly ameliorated adverse effects of P. aeruginosa on CCTα protein stability, inflammation, and mechanical properties and extended survival, underscoring the biologic relevance of its actions within the ubiquitination pathway. Our data are in line with CaM inhibition reducing phospholipid synthesis and impairing lung growth (23, 34) and the ability of agents that induce [Ca2+]i to lessen lung injury (30). Mechanistically, CaM may have other beneficial effects. As an abundant cellular protein that chelates four Ca2+ molecules, gene transfer sufficiently induced CaM levels ∼20-fold that could act as a sink, sequestering [Ca2+]i after PA103 infection. Alternatively, CaM could regulate Ca2+ influx, or it could itself be subject to ubiquitination by FBXL2. CaM localized to the Golgi complex with FBXL2 (Fig. 6) and PA103 reduced immunoreactive levels (Fig. 7A), suggesting a role for CaM as a decoy ubiquitin substrate. CaM ubiquitination is also Ca2+ dependent; however, in other systems this modification appears to be executed by a ubiquityl-CaM synthetase (15). FBXL2 also localizes to the plasma membrane to target some P4-type ATPases involved in lipid import (24). Thus, FBXL2 appears to be an E3 ligase component of fundamental importance, and its opposition by CaM is a pivotal regulatory mechanism for maintenance of cellular homeostasis.
ACKNOWLEDGMENTS
We thank David Price and Robert Piper for critical review of the manuscript and helpful suggestions. We thank Michael Feldkamp for analysis of the calmodulin crystal structure. We specially thank Jessica C. Sieren and Geoffrey McLennan for their help on the micro-CT scan study.
This material is based upon work supported, in part, by the Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development, Biomedical Laboratory Research and Development. This work was supported by a Merit Review Award from the Department of Veterans Affairs and NIH R01 grants HL081784, HL096376, HL097376, and HL098174 (to R.K.M.).
The contents of this report do not represent the views of the Department of Veterans Affairs or the U.S. Government.
Footnotes
Published ahead of print on 22 February 2011.
REFERENCES
- 1. Brandt N. R., Caswell A. H., Brandt T., Brew K., Mellgren R. L. 1992. Mapping of the calpain proteolysis products of the junctional foot protein of the skeletal muscle triad junction. J. Membr. Biol. 127:35–47 [DOI] [PubMed] [Google Scholar]
- 2. Cenciarelli C., et al. 1999. Identification of a family of human F-box proteins. Curr. Biol. 9:1177–1179 [DOI] [PubMed] [Google Scholar]
- 3. Chen B. B., Mallampalli R. K. 2007. Calmodulin binds and stabilizes the regulatory enzyme, CTP: phosphocholine cytidylyltransferase. J. Biol. Chem. 282:33494–33506 [DOI] [PubMed] [Google Scholar]
- 4. Chen B. B., Mallampalli R. K. 2010. Masking of a nuclear signal motif by monoubiquitination leads to mislocalization and degradation of the regulatory enzyme cytidylyltransferase. Mol. Cell. Biol. 29:3062–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Costelli P., et al. 2005. Ca2+-dependent proteolysis in muscle wasting. Int. J. Biochem. Cell Biol. 37:2134–2146 [DOI] [PubMed] [Google Scholar]
- 6. Dardente H., Mendoza J., Fustin J. M., Challet E., Hazlerigg D. G. 2008. Implication of the F-box protein FBXL21 in circadian pacemaker function in mammals. PLoS One 3:e3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. d'Azzo A., Bongiovanni A., Nastasi T. 2005. E3 ubiquitin ligases as regulators of membrane protein trafficking and degradation. Traffic 6:429–441 [DOI] [PubMed] [Google Scholar]
- 8. Frescas D., Pagano M. 2008. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat. Rev. Cancer 8:438–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goll D. E., Neti G., Mares S. W., Thompson V. F. 2008. Myofibrillar protein turnover: the proteasome and the calpains. J. Anim. Sci. 86:E19–E35 [DOI] [PubMed] [Google Scholar]
- 10. Han Y., Weinman S., Boldogh I., Walker R. K., Brasier A. R. 1999. Tumor necrosis factor-alpha-inducible IκBα proteolysis mediated by cytosolic m-calpain. A mechanism parallel to the ubiquitin-proteasome pathway for nuclear factor-κb activation. J. Biol. Chem. 274:787–794 [DOI] [PubMed] [Google Scholar]
- 11. Henderson F. C., Miakotina O. L., Mallampalli R. K. 2006. Proapoptotic effects of P. aeruginosa involve inhibition of surfactant phosphatidylcholine synthesis. J. Lipid Res. 47:2314–2324 [DOI] [PubMed] [Google Scholar]
- 12. Ho M. S., Tsai P. I., Chien C. T. 2006. F-box proteins: the key to protein degradation. J. Biomed. Sci. 13:181–191 [DOI] [PubMed] [Google Scholar]
- 13. Huang F., Kirkpatrick D., Jiang X., Gygi S., Sorkin A. 2006. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol. Cell 21:737–748 [DOI] [PubMed] [Google Scholar]
- 14. Ilyin G. P., Rialland M., Glaise D., Guguen-Guillouzo C. 1999. Identification of a novel Skp2-like mammalian protein containing F-box and leucine-rich repeats. FEBS Lett. 459:75–79 [DOI] [PubMed] [Google Scholar]
- 15. Jennissen H. P., et al. 1992. Ca2+-dependent ubiquitination of calmodulin in yeast. FEBS Lett. 296:51–56 [DOI] [PubMed] [Google Scholar]
- 16. Johnson G. V., Greenwood J. A., Costello A. C., Troncoso J. C. 1991. The regulatory role of calmodulin in the proteolysis of individual neurofilament proteins by calpain. Neurochem. Res. 16:869–873 [DOI] [PubMed] [Google Scholar]
- 17. Kipreos E. T., Pagano M. 2000. The F-box protein family. Genome Biol. 1:REVIEWS300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mallampalli R. K., Ryan A. J., Salome R. G., Jackowski S. 2000. Tumor necrosis factor-alpha inhibits expression of CTP:phosphocholine cytidylyltransferase. J. Biol. Chem. 275:9699–9708 [DOI] [PubMed] [Google Scholar]
- 19. McCoy D. M., Fisher K., Ryan A. J., Mallampalli R. K. 2006. Transcriptional regulation of lung cytidylyltransferase in developing transgenic mice. Am. J. Respir. Cell Mol. Biol. 35:394–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miakotina O. L., McCoy D. M., Shi L., Look D. C., Mallampalli R. K. 2007. Human adenovirus modulates surfactant phospholipid trafficking. Traffic 8:1765–1777 [DOI] [PubMed] [Google Scholar]
- 21. Miyawaki A., et al. 1997. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388:882–887 [DOI] [PubMed] [Google Scholar]
- 22. Pelham H. R. 2004. Membrane traffic: GGAs sort ubiquitin. Curr. Biol. 14:R357–R359 [DOI] [PubMed] [Google Scholar]
- 23. Rasouli M., Trischuk T. C., Lehner R. 2004. Calmodulin antagonist W-7 inhibits de novo synthesis of cholesterol and suppresses secretion of de novo synthesized and preformed lipids from cultured hepatocytes. Biochim. Biophys. Acta 1682:92–101 [DOI] [PubMed] [Google Scholar]
- 24. Ray N. B., et al. 2010. Dynamic regulation of cardiolipin by the lipid pump ATP8b1 determines the severity of lung injury in experimental pneumonia. Nat. Med. 16:1120–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rhoads A. R., Friedberg F. 1997. Sequence motifs for calmodulin recognition. FASEB J. 11:331–340 [DOI] [PubMed] [Google Scholar]
- 26. Ryan A. J., et al. 2008. 15-Deoxy-δ12,14-prostaglandin J2 impairs phosphatidylcholine synthesis and induces nuclear accumulation of thiol-modified cytidylyltransferase. J. Biol. Chem. 283:24628–24640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sato H., et al. 2003. The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J. 22:2959–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Skaar J. R., Pagan J. K., Pagano M. 2009. SnapShot: F box proteins I. Cell 137:1160–1161 [DOI] [PubMed] [Google Scholar]
- 29. Sun L., Chen Z. J. 2004. The novel functions of ubiquitination in signaling. Curr. Opin. Cell Biol. 16:119–126 [DOI] [PubMed] [Google Scholar]
- 30. Toyofuku T., Koyama S., Kobayashi T., Kusama S., Ueda G. 1989. Effects of polycations on pulmonary vascular permeability in conscious sheep. J. Clin. Invest. 83:2063–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Varghese B., et al. 2008. Polyubiquitination of prolactin receptor stimulates its internalization, postinternalization sorting, and degradation via the lysosomal pathway. Mol. Cell. Biol. 28:5275–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Walrafen P., et al. 2005. Both proteasomes and lysosomes degrade the activated erythropoietin receptor. Blood 105:600–608 [DOI] [PubMed] [Google Scholar]
- 33. Wang C., et al. 2005. Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol. Cell 18:425–434 [DOI] [PubMed] [Google Scholar]
- 34. Wang J., Campos B., Kaetzel M. A., Dedman J. R. 1996. Expression of a calmodulin inhibitor peptide in progenitor alveolar type II cells disrupts lung development. Am. J. Physiol. 271:L245–L250 [DOI] [PubMed] [Google Scholar]
- 35. Wang Y., Kent C. 1995. Effects of altered phosphorylation sites on the properties of CTP:phosphocholine cytidylyltransferase. J. Biol. Chem. 270:17843–17849 [DOI] [PubMed] [Google Scholar]
- 36. Winston J. T., Koepp D. M., Zhu C., Elledge S. J., Harper J. W. 1999. A family of mammalian F-box proteins. Curr. Biol. 9:1180–1182 [DOI] [PubMed] [Google Scholar]
- 37. Zhang J. Y., et al. 2009. Inhibition of autophagy causes tau proteolysis by activating calpain in rat brain. J. Alzheimers Dis. 16:39–47 [DOI] [PubMed] [Google Scholar]
- 38. Zhou J., Ryan A. J., Medh J., Mallampalli R. K. 2003. Oxidized lipoproteins inhibit surfactant phosphatidylcholine synthesis via calpain-mediated cleavage of CTP:phosphocholine cytidylyltransferase. J. Biol. Chem. 278:37032–37040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou J., et al. 2006. Adenoviral gene transfer of a mutant surfactant enzyme ameliorates Pseudomonas-induced lung injury. Gene Ther. 13:974–985 [DOI] [PubMed] [Google Scholar]