Abstract
mTORC1 is a critical regulator of cell growth that integrates multiple signals and is deregulated in cancer. We previously reported that mTORC1 regulation by hypoxia involves Redd1 and the Tsc1/Tsc2 complex. Here we show that Redd1 induction by hypoxia is tissue dependent and that hypoxia signals are relayed to mTORC1 through different pathways in a tissue-specific manner. In the liver, Redd1 induction is restricted to the centrilobular area, and in primary hepatocytes, mTORC1 inhibition by hypoxia is independent of Redd1. Furthermore, Tsc1/Tsc2 and Arnt (Hif-1β) are similarly dispensable. Hypoxia signaling in hepatocytes involves Lkb1, AMP-activated protein kinase (AMPK), and raptor. Differences in signal relay extend beyond hypoxia and involve AMPK signaling. AMPK activation (using 5-aminoimidazole-4-carboxamide riboside [AICAR]) induces raptor phosphorylation and inhibits mTORC1 in both mouse embryo fibroblasts (MEFs) and hepatocytes, but whereas mTORC1 inhibition is Tsc1/Tsc2 dependent in MEFs, it is independent in hepatocytes. In liver cells, raptor phosphorylation is essential for both AMPK and hypoxia signaling. Thus, context-specific signals are required for raptor phosphorylation-induced mTORC1 inhibition. Our data illustrate a heretofore unappreciated topological complexity in mTORC1 regulation. Interestingly, topological differences in mTORC1 regulation by the tumor suppressor proteins Lkb1 and Tsc1/Tsc2 may underlie their tissue specificity of tumor suppressor action.
INTRODUCTION
Mammalian target of rapamycin complex 1 (mTORC1) is an important regulator of cell growth involved in tumor development (1). mTORC1 core components include the phosphoinositide kinase-related kinase mammalian target of rapamycin (mTOR) and an adaptor protein, regulatory-associated protein of mTOR (raptor) (22, 32, 41). Other mTORC1-interacting proteins are mammalian lethal with sec-thirteen protein 8 (mLST8) (33, 41), proline-rich Akt substrate of 40 kDa (PRAS40) (17, 45, 50, 61, 64, 66), and DEP domain TOR binding protein (DEPTOR) (46). The best characterized function of mTORC1 is in promoting translation initiation, and phosphorylation by mTORC1 of the eukaryotic initiation factor 4E (eIF4E) binding proteins (4E-BPs) releases eIF4E from their inhibition, thereby facilitating the assembly of a translation preinitiation complex at mRNA 5′ m7GTP moieties (58). mTORC1 also phosphorylates S6 kinase 1 (S6K1), priming it for further phosphorylation and activation (16), and S6K1 in turn phosphorylates, among others, eIF4B, which is then recruited into the translation preinitiation complex, and the small ribosomal subunit protein S6 (42).
mTORC1 is regulated by a variety of signals, including those reflecting the cellular energy state. In situations of low energy, cellular AMP levels rise, leading to the activation of the AMP-activated protein kinase (AMPK), a heterotrimeric protein kinase composed of a catalytic subunit (α) and two regulatory subunits (β and γ) (23). AMPK activation also requires phosphorylation by the tumor suppressor protein LKB1 (24), which forms a complex with sterile-20 related adaptor (STRAD) and mouse protein 25 (MO25), which maintain LKB1 in an active state (69). Once activated, AMPK phosphorylates tuberous sclerosis complex 2 (TSC2) (27), which forms a complex with tuberous sclerosis complex 1 (TSC1) protein, which acts as a GTPase-activating protein (GAP) toward Ras homologue enriched in brain (Rheb), a small G protein required for mTORC1 activation (3, 50). TSC2 phosphorylation by AMPK primes it for further phosphorylation by glycogen synthase kinase 3β (GSK3β) (26), which leads to Rheb-dependent inhibition of mTORC1.
Recently, a second mechanism whereby AMPK regulates mTORC1 was reported; AMPK was shown to directly phosphorylate raptor (20). However, the relative contribution of raptor phosphorylation to overall mTORC1 regulation by AMPK and the context in which raptor is implicated in mTORC1 regulation are not well understood.
mTORC1 activity is regulated by oxygen levels (63). mTORC1 regulation by oxygen occurs through a conserved pathway that is independent of energy signaling pathways (7, 38, 48). While anoxia or prolonged hypoxia would predictably lead to ATP depletion and activate AMPK (40), hypoxia per se does not activate AMPK (2). Furthermore, AMPK activity, like LKB1, is dispensable for hypoxia-induced mTORC1 inhibition (9).
mTORC1 regulation by hypoxia involves a 25-kDa protein, regulated in development and DNA damage response 1 (REDD1; also referred to as DDIT4). REDD1 is both necessary and sufficient for mTORC1 inhibition by hypoxia (9, 12, 15, 19, 52). In response to hypoxia, REDD1 expression is upregulated by hypoxia-inducible factor 1 (HIF-1) (55) and REDD1 overexpression is sufficient to inhibit mTORC1 (9, 12, 57). Importantly, REDD1-induced mTORC1 inhibition requires the TSC1/TSC2 complex (9, 12, 57), which is similarly required for mTORC1 regulation by hypoxia (9, 11, 30, 40). However, REDD1 has not been found to interact with TSC1/TSC2 (15, 65). TSC2 was previously reported to interact with 14-3-3 proteins (10, 56) and REDD1, which contains a putative 14-3-3 binding motif, was proposed to act by directly binding to and sequestering 14-3-3 proteins away from TSC2 (15). However, how REDD1 would sequester 14-3-3 proteins, which are very abundant, is unclear. In addition, the putative 14-3-3 binding motif does not conform to any 14-3-3 binding motif known, and REDD1 could not be docked onto 14-3-3β without steric clashes (65). Furthermore, residues typically implicated in 14-3-3 binding (S at position 0 and R at position −4) are not conserved and could be mutated without seemingly affecting REDD1 function (65). Thus, how REDD1 acts remains to be elucidated.
Multiple negative regulators of mTORC1 are bona fide tumor suppressor genes, including TSC1, TSC2, LKB1 as well as phosphatase and tensin homologue (PTEN). Germ line mutations in these genes result in syndromes with overlapping, but also distinct, features (8). Germ line mutations in the TSC1 and TSC2 genes lead to tuberous sclerosis complex, a syndrome characterized by hamartomas in multiple tissues (13), which in the skin is manifested by, among others, angiofibromas and periungual/ungual fibromas, and in these tumors, second-hit mutations are found in fibroblast-like cells (39). Germ line mutations in LKB1 result in Peutz-Jeghers, a syndrome characterized by the development of hamartomatous polyps in the gastrointestinal tract as well as mucocutaneous pigmentation abnormalities. In the sporadic setting, mutations in LKB1 are frequently observed in non-small cell lung cancer (51). While a role for LKB1 in human hepatocellular carcinoma (HCC) has not been definitively established (31), Lkb1 is an important tumor suppressor in the liver in the mouse, and Lkb1 heterozygous mice exhibit a high predisposition to develop HCC (44).
In this study, we show that hypoxia and AMPK engage different pathways to regulate mTORC1 in a tissue-specific manner. Whereas Redd1 is required for hypoxia-induced mTORC1 inhibition in thymocytes and intestinal crypts, as in mouse embryo fibroblasts (MEFs), Redd1 is dispensable for mTORC1 inhibition in hepatocytes. Furthermore, in hepatocytes, mTORC1 regulation by hypoxia is largely independent of Tsc1/Tsc2 as well as of the aryl hydrocarbon receptor nuclear translocator (Arnt, also called Hif-1β). In contrast, mTORC1 regulation by hypoxia in hepatocytes is Lkb1 and AMPK dependent. Interestingly, AMPK-induced mTORC1 inhibition is mediated through different pathways in different cell types and whereas Tsc1/Tsc2 is required for signaling in MEFs, it is largely dispensable in hepatocytes. Furthermore, while raptor is phosphorylated by AMPK in both cell types, Tsc1/Tsc2-deficient MEFs fail to downregulate mTORC1, indicating that raptor-induced mTORC1 inhibition is tissue specific. These data uncover a previously unknown topological complexity in mTORC1 regulation. Furthermore, the implication of TSC1/TSC2 in mTORC1 regulation by hypoxia in fibroblasts and LKB1 in hepatocytes provides a potential explanation for the tissue specificity of their tumor suppressor action.
MATERIALS AND METHODS
Reagents.
Antibodies against S6K1, S6, eukaryotic initiation factor 4E (eIF4E) binding protein 1 (4E-BP1), phosphorylated S6K1 (T389), phosphorylated S6 (S235/236), phosphorylated S6 (S240/244), Raptor, phosphorylated Raptor (S792), phosphorylated acetyl coenzyme A (acetyl-CoA) carboxylase (ACC) (S79), ACC, glutathione S-transferase (GST), and eIF4E were from Cell Signaling. ARNT antibodies were from BD Laboratories. Antibodies against Hif-1α, Tsc1, and REDD1 were from Bethyl. Anti-Tsc2 was from Santa Cruz, anti-cyclophilin B (Ppib) was from Abcam, and anti-β-galactosidase was from ICN. REDD1 polyclonal antibodies were a gift from Anke Klippel (53). 5-Aminoimidazole-4-carboxamide riboside (AICAR) and compound C were from Calbiochem. Tubulin, 4′,6′-diamidino-2-phenylindole (DAPI), dexamethasone, 3,3′,5-triiodo-l-thyonine (T3), insulin, 2-deoxyglucose (2DG), and type I collagen were from Sigma. Rapamycin was from LC Laboratories, S6 peptide was from Upstate, and P81 paper was from Whatman. Liver perfusion medium, liver digest medium, and medium 199 (M199) were from Invitrogen. m7GTP Sepharose was from GE Healthcare. VectaStain ABC kit was from Vector Laboratories, 3,3′-Diaminobenzidine (DAB) chromogen was from Covance, and nuclear fast red was from Poly Scientific. Restriction enzymes were from New England BioLabs, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was from Invitrogen, and Cell Titer Glo (CTG) was from Promega.
Sequencing.
Primers used for sequencing the gene trap insertion were as follows: 5′-CTCTGGGATCGTTTCTCGTC and 5′-GCAAGGCGATTAAGTTGGGT for the 5′ site and 5′-GAAGGGGTTGGGACTAACAGAA and 5′-TTAACAGCCCCTGGATCTTG for the 3′ site.
Genotyping.
Mouse tails were digested in 100 mM Tris (pH 8.0), 200 mM NaCl, 1 mM EDTA, and 0.2% SDS supplemented with 0.5 mg/ml proteinase K (Invitrogen), ethanol precipitated, and resuspended in Tris-EDTA (TE) buffer. Redd1 genotyping PCR was carried out as follows: 3 min at 95°C, 40 cycles (1 cycle consisting of 30 s at 94°C, 30 s at 55°C, and 2 min at 72°C), followed by 7 min at 72°C; primers 5′-CTCTGGGATCGTTTCTCGTC, 5′-TTAACAGCCCCTGGATCTTG, and 5′-CTGGTGAGGCCAAGTTTGTTTCC were used. Tsc1 genotyping PCR was carried out as follows: 2 min at 94°C and 35 cycles of 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C, followed by 72°C for 7 min, with primers 5′-AGGAGGCCTCTTCTGCTACC, 5′-CAGCTCCGACCATGAAGTG, and 5′-TGGGTCCTGACCTATCTCCTA. Lkb1 genotyping PCR was carried out as follows: 2 min at 94°C and 35 cycles of 25 s at 94°C, 20 s at 64°C, and 35 s at 72°C, followed by 7 min at 72°C. Primers 5′-GGGCTTCCACCTGGTGCCAGCCTGT, GAGATGGGTACCAGGAGTTGGGGCT, and 5′-TCTAACAATGCGCTCATCGTCATCCTCGGC were used for the wild-type and loxP alleles, and primers 5′-GGGCTTCCACCTGGTGCCAGCCTGT and 5′-GATGGAGGACCTCTTGGCCGGCTCA were used for the null allele. Arnt genotyping PCR was carried out as follows: 2 min at 94°C and 35 cycles of 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C, followed by 72°C for 7 min; primers 5′-TGCCAACATGTGCCACCATGT and 5′-GTGAGGCAGATTTCTTCCATGCTC were used for the wild-type and loxP alleles, and primers 5′-GTGAGGCAGATTTCTTCCATGCTC and 5′-ACGCACTACAACACCTGAGCTAA were used for the null allele.
Cell isolation, culture, and cell-based hypoxia experiments. (i) ES cells.
A total of 2 × 106 embryonic stem (ES) cells were plated on 0.1% gelatin (Sigma)-coated plates and grown in Dulbecco modified Eagle medium (DMEM) (Cellgro) supplemented with 10% fetal calf serum (FCS) (Gemini), 1% penicillin-streptomycin (P-S) (Sigma), 1× nonessential amino acids (Cellgro), 49 μM β-mercaptoethanol (Sigma), 1,000 units/ml leukocyte inhibitory factor (LIF) (Millipore), and 150 μg/ml G418 (Cellgro). The medium was changed daily, and cells were passaged every 2 days.
(ii) MEFs.
Embryos on embryonic day 13.5 (E13.5) were collected, and after the brain, liver, and heart were discarded, the embryos were minced in trypsin and incubated at 37°C for 15 min. DMEM supplemented with 10% FCS (HyClone) and 1% P-S was added, and cells were further disaggregated, plated, and grown until they reached confluence. Mouse embryo fibroblasts (MEFs) were maintained in DMEM supplemented with 10% FCS and 1% P-S. For hypoxia experiments, 0.8 × 106 to 1 × 106 MEFs were plated in alpha minimum essential medium (α-MEM) (Invitrogen) containing 4% FCS and 1% P-S in each well of a 6-well plate, and the following day, the cells were placed in a hypoxia chamber (Coy Laboratories) at 1% O2, 5% CO2, and 100% humidity.
(iii) Thymocytes.
Four- to 6-week-old mice were sacrificed using isofluorane. The thymus was carefully removed from each mouse, and the cells were dispersed with a pestle in a 1.5-ml tube in DMEM containing 10% FCS and 1% P-S. After dilution in additional medium, cells were counted using a 1:1 TURKS solution (3% glacial acetic acid, 0.1% crystal violet in H2O) and plated for experiments. The thymocytes were cultured in DMEM containing 10% FCS and 1% P-S and plated in T75 flasks at 5 × 106 cells/ml for hypoxia experiments. Hypoxia experiments were carried out in a hypoxia chamber at 1% O2, 5% CO2, and 100% humidity.
(iv) Hepatocytes.
The hepatocytes were isolated as described previously (25) with some modifications. Mice were sacrificed using isofluorane, the portal vein was canulated, and the liver was perfused using 15 to 20 ml prewarmed liver perfusion medium followed by 30 ml prewarmed liver digest medium. Subsequently, hepatocytes were released by gently shaking the liver in cold DMEM containing low glucose (Invitrogen) supplemented with 5% FCS and 1% P-S and were filtered through a 70-μm mesh. Cells were pelleted two times at 50 × g for 3 min each time, and an aliquot was diluted 1:1 in 0.4% trypan blue (Invitrogen) and used to calculate the cell number. For experiments, 1 × 106 to 2 × 106 cells (50 to 75% viability) were plated in collagen-coated plates for 2 to 4 h until they attached, and the medium was changed to M199 supplemented with 1 nM insulin, 100 nM dexamethasone, 100 nM T3, and 1% P-S (68). Replication-deficient recombinant adenovirus encoding Cre recombinase (Ad-Cre) (Ad5CMVCre) at a multiplicity of infection (MOI) of 20 was added, and after 2 days, the medium was changed to M199 (supplemented as above). Hypoxia experiments were carried out on day 3 in a hypoxia chamber at 1% O2, 5% CO2, and 100% humidity.
Huh7 cells (ATCC) were grown in DMEM (Invitrogen) with high glucose and supplemented with 10% FCS and 1% P-S. Cells were split the day before as to be 70 to 80% confluent for hypoxia experiments (1% O2, 5% CO2).
5-Aminoimidazole-4-carboxamide riboside (AICAR) was used at 0.5 mM and 1 mM in H2O for hepatocytes and MEFs, respectively. Where indicated, cells were pretreated with 10 μM compound C dissolved in dimethyl sulfoxide (DMSO) for 30 min prior to hypoxia or the administration of AICAR. 2DG was used at 50 mM in H2O.
Ad-Cre expansion and titer determination.
Recombinant replication-deficient Ad5CMVCre adenoviruses (Gene Transfer Vector Core, University of Iowa, IA) were expanded as described previously (36) with some modifications using human embryonic kidney (HEK) 293 cells. Adenoviruses were purified by discontinuous CsCl gradient ultracentrifugation and desalted in PD-10 columns (GE Healthcare). The titers of adenoviruses were determined by infecting Rosa26-LoxSTOPLox-LacZ MEFs (59) with serial dilutions and comparison to a control virus of known titer (PFU/ml). The cells were then stained with X-Gal solution, and the number of recombinant events, as indicated by blue cells, was counted and compared to the control virus to infer PFU/ml.
MEF immortalization with SV40 early region.
Phoenix cells were transfected with a pBabe-zeo vector containing the early region of simian virus 40 (SV40) (21) in OPTI-MEM using Lipofectamine Plus (Invitrogen) according to the manufacturer's recommendations. After 3 h, the medium was changed to DMEM with 10% FCS with no antibiotics, and following an overnight incubation, it was changed to DMEM containing 10% FCS and 1% P-S. Six hours after the medium was changed, the medium was collected, supplemented with FCS and Polybrene (Sigma) to a final concentration of 4 μg/ml, filtered through a 0.45-μm filter, and added to early passage Tsc1F/F or Redd1βgeo/βgeo and wild-type controls from littermate embryos. The infection was repeated two more times the next day. Cultures were grown to 80 to 90% confluence and selected in 100 μg/ml zeocin (Invitrogen). Following antibiotic selection, and once uninfected-cell cultures were not viable, polyclonal populations were expanded, and cell aliquots were frozen for later experiments.
Generation of Huh7 cells expressing various AMPKs.
Huh7 cells were cotransfected with the pEBG empty vector (EV) or a vector containing the AMP-activated protein kinase (AMPK) α dominant-negative (DN) mutants (T172A [TA] mutant or K45R[KR] mutant) (54) along with a pcDNA3 vector using Lipofectamine Plus reagents according to the manufacturer's instructions. Two days after transfection, the cells were selected with G418 (Cellgro) at 0.6 mg/ml for 2 weeks.
Generation of Huh7 cells expressing ectopic wild-type or mutant raptor.
Raptor AA mutant (S722A/S792A) was generated by site-directed mutagenesis of a human raptor cDNA in a pRK5 vector (laboratory database identification [ID] 694). After sequencing verification, AA mutant (and wild-type [WT]) cDNAs were digested with SalI and NotI, filled with Klenow fragment, and subcloned into a pBabe-hygro vector previously digested with SnaBI. The appropriate orientation of the insert was confirmed by PCR (ID 695 [WT] and 696 [AA]). Subsequently, Phoenix cells were transfected, and supernatant was collected for Huh7 cell transduction. Following selection with 0.2 mg/ml hygromycin (Invitrogen), endogenous raptor was knocked down using a previously validated (20) lentiviral pLKO.1 puromycin short hairpin RNA (shRNA) vector targeting the raptor 3′ untranslated region (3′ UTR) (Addgene) followed by further selection in 3 μg/ml puromycin.
Small interfering RNA (siRNA) transfections.
Huh7 cells were transfected with the corresponding oligonucleotides (9, 57) using DharmaFECT3 transfection reagent (Dharmacon) according to the manufacturer's instructions.
RNA extraction and Northern blotting.
RNA was extracted using either Trizol (Invitrogen) or RNA minikit (Qiagen) and resuspended in RNase-free H2O. Twenty micrograms of total RNA was resuspended in 0.5 volume of glyoxal sample loading buffer (Ambion), incubated at 65°C for 15 min, run on a 1.2% agarose–formaldehyde denaturing gel, transferred by capillary action in 10× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) to a nylon membrane (Amersham), cross-linked using UV light, and prehybridized at 68°C for 1 h in ExpressHyb solution. The membrane was hybridized with heat-denatured probes in ExpressHyb solution for 1 h at 68°C, washed, and exposed to film. Probes were generated using High Prime DNA labeling kit and [α-32P]dCTP according to the manufacturer's instructions. Templates were generated by PCR using the following primers: 5′-TTATCGATGAGCGTGGTGGTTATGC and 5′-GCGCGTACATCGGGCAAATAATATC for βgeo, 5′-CGGCTTCTGTGCGCCTTCAT and 5′-CTCCGGCCCGAAGCCACTGT for Redd1 (exon 2), 5′-ACTCCTCATACCTGGATGGGG and 5′-TTAACAGCCCCTGGATCTTGfor Redd1 (exon 3), 5′-GGATTATGTTGTCCCTGAGCCCA and 5′-AAGGCCTGAGGAGAAGCGACCT for Redd2, and 5′-TGCTCCTCCTGAGCGCAAGTACTC and 5′-CTCAGACCTGGGCCATTCAGAAAT for Actin.
qRT-PCR.
cDNA was synthesized from 1 to 4 μg of total RNA using random primers (Invitrogen) and Moloney murine leukemia virus reverse transcriptase (M-MLV-RT) (Invitrogen). Quantitative reverse transcription-PCR (qRT-PCR) was performed as described by S. Peña-Llopis et al. (submitted for publication) using Platinum SYBR green qPCR SuperMix-UDG (Invitrogen) in an Applied Biosystems 7500 real-time PCR system as follows: 2 min at 50°C, 5 min at 95°C, and 40 to 50 cycles of 15 s at 95°C and 35 s at 60°C. Reference amounts of the sequence to be amplified were included in each reaction plate as standards. Primers were 5′-CGGCTTCTGTGCGCCTTCAT and 5′-CAATCGCGCCTGGGACAGGC for Redd1, 5′-CGGCTTCTGTGCGCCTTCAT and 5′-GCAAGGCGATTAAGTTGGGT for Redd1βgeo, 5′-CCTGCTCATCAATCGTAACGAGG and 5′-CGACCCTCTTCTTTCATCTCC for Glut-1, 5′-GCCTGCCTCGGTTTACCCTTC and 5′-CCACCGTCCAACAACTCTGAAATG for Redd2, and 5′-CATTGTTACCAACTGGGACG and 5′-CAGAGGCATACAGGGACAG for Actin.
Western blotting.
Western blotting was performed as described by Vega-Rubin-de-Celis et al. (65).
m7GTP affinity chromatography.
Cells were lysed in immunoprecipitation (IP) buffer (50 mM Tris HCl at pH 7.5, 150 mM NaCl, and 0.5% Igepal CA-630 [USB]) containing phosphatase and protease inhibitors, and after the protein amounts were normalized, the samples were rocked with a 1:1 slurry of 7methyl-GTP Sepharose beads in IP buffer at 4°C for 2 h. The beads were pelleted, washed with IP buffer, and denatured by boiling in 1× loading buffer (2.23% SDS, 11.1% glycerol, 100 mM dithiothreitol [DTT] with bromophenol blue). Samples were subsequently resolved by SDS-PAGE and analyzed by Western blotting.
S6K1 assay.
Cells were lysed in IP buffer containing phosphatase inhibitors and protease inhibitors and after adjusting for protein concentration, samples were rocked with anti-S6K1 antibody for 2 h. Immune complexes were recovered using protein A-Sepharose, washed with IP buffer and subsequently with kinase buffer (100 mM Tris [pH 7.4], 25 mM morpholinepropanesulfonic acid [MOPS], 25 mM MgCl2, 2 mM EDTA). Immunoprecipitates were incubated with kinase buffer in the presence of S6 peptide, ATP, and [γ-32P]ATP (MP Biomedicals) for 15 min at 37°C. Reaction mixtures were spotted on P81 phosphocellulose paper, air dried, washed in 1% phosphoric acid, and evaluated following the addition of scintillation fluid in a Beckman LS 6500 scintillation counter.
ATP measurement assay using Cell Titer Glo.
A total of 1 × 104 wild-type SV40-transformed MEFs were plated in triplicate in α-MEM (Invitrogen) supplemented with 4% FCS and 1% P-S in 24-well plates. A total of 5 × 105 wild-type hepatocytes were plated in triplicate in collagen-coated 12-well plates in M199 medium (Invitrogen) supplemented with dexamethasone, insulin, and T3. The following day, cells were exposed to 1% oxygen, 5% CO2, and 100% humidity for 0, 3, 6, and 9 h. At 9 h, the plates were removed from the incubator, cells were washed with 1 ml of room temperature α-MEM medium (without additives), and 150 μl of a 1:1 mix of room temperature Cell Titer Glo (CTG) reagent (diluted 1:1 in 1% Triton X-100 in water) and α-MEM were added to the cells. After lysis for 10 min on a rocker, 140 μl was transferred to a 96-well, white-walled, clear-bottom, assay plate (Costar 3903; Corning Inc.) and read on a PolarStar Optima illuminometer (BMG Labtech, Durham, NC) with a 0.5-s interval. A well containing no cells was used as a blank control.
Mouse hypoxia experiments.
All mouse experiments were approved by the IACUC. Mice in individual boxes with wet food were placed in a hypoxia chamber with 18% O2 and 0% CO2 at 25°C, and O2 was slowly brought down to 6% at 3% decrements every 10 min.
Rapamycin treatment of mice.
Rapamycin in ethanol (5 mg/ml) was diluted to 0.1 mg/ml in a solution of 5% PEG400, 5% Tween 80, and 5% dextrose (pH 7.2). Mice were treated once intraperitoneally (i.p.) with rapamycin (0.5 mg of rapamycin per kg of body weight) or vehicle and sacrificed 3 h afterwards by isofluorane overdose.
X-Gal staining.
MEFs or hepatocytes were fixed in 0.5% glutaraldehyde for 10 min, washed three times in phosphate-buffered saline (PBS) for 5 min each time, incubated overnight in X-Gal solution, and washed three times in PBS. For mouse tissues, samples were fixed in 0.5% glutaraldehyde in PBS at 4°C for 8 h, washed three times in PBS, and placed in PBS containing 30% sucrose at 4°C overnight. The tissue samples were placed in optimum cutting temperature (OCT) medium and frozen on dry ice. Six-micron sections were cut at −20°C, postfixed in 0.5% glutaraldehyde for 5 min, and washed three times in PBS. The slides were incubated overnight in X-Gal staining solution (1 mg/ml X-Gal, 5 mM potassium ferricyanide/potassium ferrocyanide, 2 mM MgCl2) at 37°C, washed in PBS, counterstained with nuclear fast red for 1 min, and washed five times in water. The slides were dehydrated (95% ethanol for 1 min [two times], 100% ethanol for 1 min [three times], and xylene for 1 min [three times]) and mounted with Cryoseal 60 (Richard-Allen Scientific).
Immunohistochemistry (IHC).
Mouse tissue samples were fixed in 0.5% glutaraldehyde for 8 h, transferred to 30% sucrose in PBS overnight, and embedded in OCT medium. Six-micron sections were cut, placed on slides, and washed with PBS. For paraffin embedding, the tissue samples were fixed in formalin for 24 h, dehydrated, paraffinized in a Microm STP 120, and embedded in paraffin blocks. Five-micron sections were cut, placed on slides, deparaffinized in xylene for 2 min (three times), hydrated (1 min in 100% ethanol [three times], 1 min in 95% ethanol [two times], and 2 min in H2O). Antigen unmasking was performed with 10 mM sodium citrate buffer (pH 6.0) at 95 to 100°C for 10 min. The tissue samples were blocked with 5% normal goat serum in Tris-buffered saline containing 0.05% Tween 20 (TBS-T) for 1 h, incubated with primary antibody in blocking solution overnight, washed three times in TBS-T, incubated with anti-rabbit secondary antibody for 30 min and then with VectaStain ABC solution for 30 min, and developed with DAB solution for 2 min. Controls were treated as described above but omitting incubation with primary antibody. Slides were counterstained with hematoxylin.
Statistical analyses.
Statistical analyses were performed using SAS 9.1.3 Service Pack 3 (SAS Institute Inc., Cary, NC) and Microsoft Excel. qRT-PCR data were normalized, where indicated, to β-actin and reported as means and standard error (SE) along with the sample sizes. Data of S6K1 in vitro kinase assays were normalized to IgG control and reported as means and SE along with the sample sizes. Expression changes over time for Redd1βgeo, Redd1, Redd2, and Glut1 were estimated using simple linear regressions and were compared across the individual tissues using a pair-wise two-sample t test (two-sided). Multiple comparisons were not adjusted. A P value of 0.05 or less was considered statistically significant.
RESULTS
βgeo insertion disrupts Redd1 function.
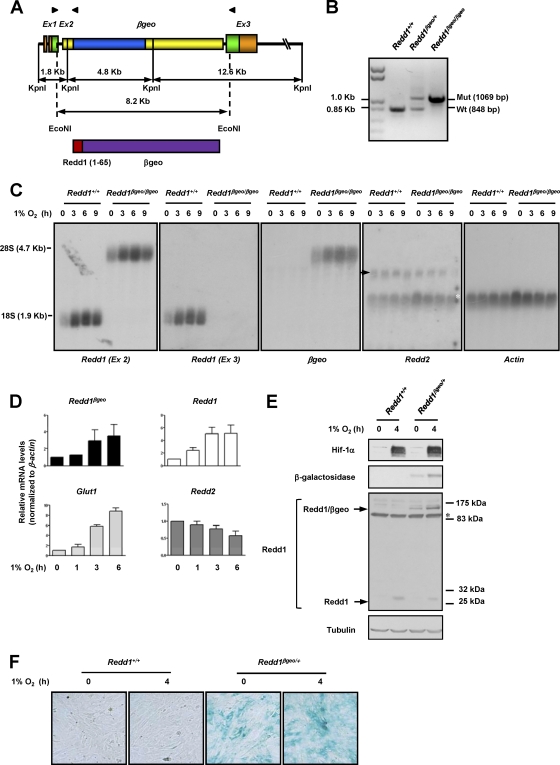
Because of its genomic structure, the murine Redd1 gene is particularly suitable for disruption by gene trap strategies. Among the 3 exons that constitute Redd1, exon 1 is noncoding, and exon 2 encodes the first 65 amino acids, which are poorly conserved and are dispensable for function (65). Thus, the insertion of a gene trap vector in either intron 1 or 2 precluding the expression of sequences downstream would result in a loss-of-function allele. In addition, since there are no other known genes in the region, it is unlikely that such an insertion would disrupt other functions. A search of a publicly available gene trap library (BayGenomics) identified two embryonic stem (ES) cell lines with an insertion of a βgeo cassette (β-galactosidase fused to the neomycin gene) in the Redd1 locus. Southern blotting and sequencing analyses showed that one ES cell line contained a single βgeo insertion in intron 2 of Redd1 (Fig. 1A and B; also data not shown). The βgeo coding sequence was in frame with exon 2, and thus, a chimeric protein containing the first 65 amino acids of Redd1 fused to the βgeo protein was predicted (Fig. 1A).
Fig. 1.
Insertion of a βgeo cassette in intron 2 of Redd1 results in a hypoxia-inducible chimeric protein that reports on Redd1 and disrupts Redd1 function. (A) (Top) Diagram depicting the genomic structure of Redd1βgeo. The genomic structure is not drawn to scale. Coding sequences are depicted in green and blue. Arrowheads illustrate the positions of primers used in panel B (see Materials and Methods for primer sequences). Ex, exon. (Bottom) Diagram of the predicted protein with amino acids 1 to 65 of Redd1. (B) PCR analysis of genomic DNA from samples of the indicated genotypes. Mut, mutant; Wt, wild type. (C) Northern blot analyses using the indicated probes on extracts from MEFs of the two different genotypes exposed to hypoxia for the indicated number of hours (the arrow indicates Redd2, and the white asterisk indicates residual actin signal). (D) qRT-PCR analysis of Redd1βgeo/+ MEFs exposed to hypoxia for the indicated number of hours (data are normalized to β-actin and represent means plus standard errors [SE] [error bars]; n = 2). (E) Western blots of MEFs of the two genotypes exposed to hypoxia for the indicated number of hours (the asterisk indicates cross-reacting protein). (F) X-Gal staining of MEFs of the two genotypes and exposed to hypoxia for the indicated number of hours.
To further characterize Redd1βgeo, mouse embryo fibroblasts (MEFs) were generated from Redd1βgeo/βgeo embryos, which were recovered at Mendelian ratios. While in wild-type MEFs, Northern blot analyses with a Redd1 exon 2 probe identified one hypoxia-inducible band of the expected molecular size for Redd1, this band was absent in Redd1βgeo/βgeo MEFs (Fig. 1C). In Redd1βgeo/βgeo MEFs, the exon 2 probe identified a single band with a substantially higher molecular weight, but similarly induced by hypoxia, which was also recognized by a βgeo probe, and of a size consistent with a Redd1βgeo fusion transcript (4.21 kb) (Fig. 1C). Northern blot analyses with an exon 3 probe showed a band corresponding to Redd1 in Redd1+/+ MEFs, but no bands in Redd1βgeo/βgeo MEFs, indicating that the insertion of βgeo precluded the expression of exon 3 (Fig. 1C). Of note, disruption of Redd1 did not result in a compensatory upregulation of the paralogue Redd2 (also referred to as DNA damage-inducible transcript 4-like) (Fig. 1C), which in contrast to Redd1, was also not induced by hypoxia (Fig. 1C and D).
To quantitatively evaluate the degree to which Redd1βgeo reported on Redd1 induction by hypoxia, qRT-PCR analyses were performed in Redd1βgeo/+ MEFs using a shared primer hybridizing to both Redd1 and Redd1βgeo cDNAs. The kinetics and magnitude of Redd1βgeo upregulation in response to hypoxia very closely resembled those for Redd1, even compared to Glut1, a gene similarly induced by Hif-1 in response to hypoxia (Fig. 1D).
Redd1βgeo gave rise to a chimeric protein that was induced by hypoxia and was recognized by antibodies against both N-terminal sequences of Redd1 as well as β-galactosidase (Fig. 1E) and which could also be monitored by using a β-galactosidase substrate (Fig. 1F).
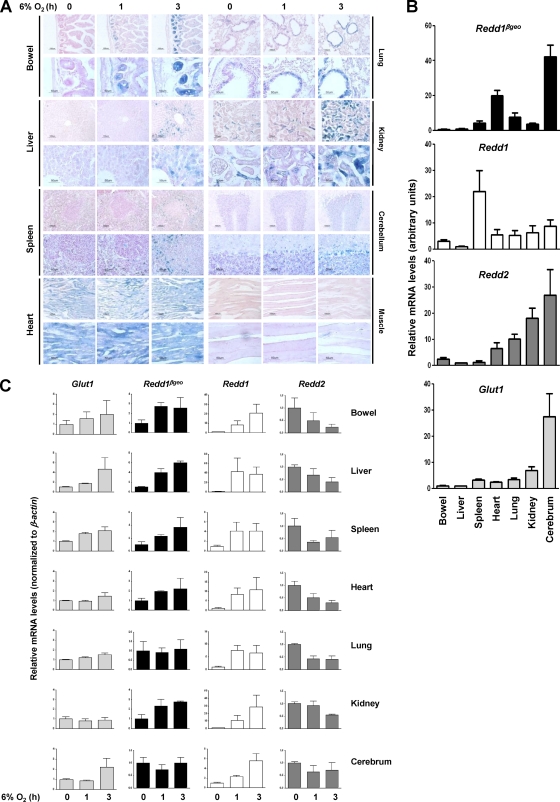
Temporal and cell-type-dependent upregulation of Redd1 by hypoxia in the mouse.
Exposure of Redd1βgeo/+ reporter mice to hypoxia revealed the upregulation of Redd1 in a manner that was dependent on both the cell type and the duration of hypoxia (Fig. 2A). In the small bowel, Redd1 was prominently induced in cells in the crypts; and in the lungs, Redd1 induction was observed primarily in bronchial epithelial cells. In both organs, there was already appreciable Redd1 induction after 1 h of hypoxia. In the liver, Redd1 induction was modest and restricted to zone 3, which is the most hypoxic at baseline and corresponds to the innermost zone of the hepatic lobule furthest from the arterial blood supply. In the kidney, Redd1 was induced in scattered tubular cells particularly in the collecting ducts. In the spleen, Redd1 induction was observed in the red pulp and in histiocytes, but not in the white pulp. In the cerebellum, Redd1 induction was prominent in Purkinje cells. Incidentally, in most organs examined, Redd1 appeared to be induced by hypoxia in endothelial cells. While differing in baseline X-Gal staining, no induction was appreciable in either cardiac or striated muscle.
Fig. 2.
Characterization of the pattern of Redd1 induction by hypoxia in the mouse using a Redd1βgeo reporter that faithfully reports on Redd1 regulation by hypoxia. (A) Two different magnifications of tissue sections from Redd1βgeo/+ mice exposed to 6% oxygen for the indicated periods of time and evaluated for β-galactosidase activity and counterstained with nuclear fast red. (B) qRT-PCR analysis of equal amounts of cDNA from Redd1βgeo/+ mouse tissues (data are means plus SE; n = 2 to 4). (C) qRT-PCR analysis from tissues of Redd1βgeo/+ mice exposed to hypoxia for the indicated periods of time (data are normalized to expression values in the bowel and represent means plus SE; n = 2 to 4).
Redd1βgeo faithfully reports on Redd1 regulation by hypoxia but not at baseline.
Substantial β-galactosidase activity was observed in some tissues at baseline (Fig. 2A). This activity was secondary to Redd1βgeo expression, as minimal activity was found across tissues in Redd1+/+ mice of comparable ages (data not shown). To determine how well Redd1βgeo reported on Redd1 expression in tissues, qRT-PCR analyses were performed in Redd1βgeo/+ mice using a shared primer. As the relative expression of no housekeeping gene is likely to be identical across tissues, but regardless of whether expression levels were normalized or not, the baseline pattern of Redd1 and Redd1βgeo in tissues was found to be very different (Fig. 2B; also data not shown). Redd1 expression was highest in the spleen, whereas Redd1βgeo expression was highest in the cerebrum. Multiple studies have shown that Redd1 expression in the brain is low by comparison to other tissues (6, 19, 55). Thus, Redd1βgeo does not report on baseline Redd1 expression across tissues.
In contrast, similar qRT-PCR analyses showed that there was a very close correlation between Redd1βgeo and Redd1 expression in response to hypoxia in tissues (Fig. 2C). Despite the fact that Redd1βgeo was induced to a lesser extent than Redd1, statistical analyses comparing the rates of induction of Redd1βgeo and Redd1 across individual tissues showed them to be (i) indistinguishable from each other and (ii) different from Redd2 or even Glut1.
Redd1 is induced in response to hypoxia in intestinal crypts and is involved in mTORC1 regulation.
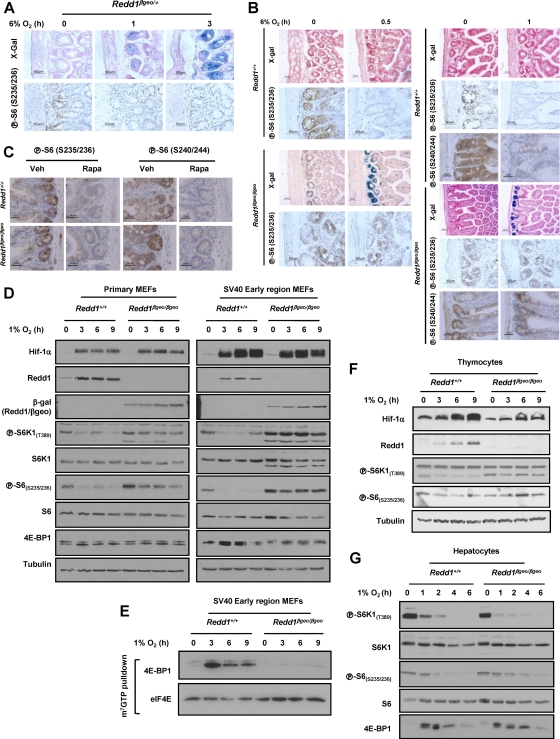
Having established that as in MEFs, in Redd1βgeo/βgeo mouse tissues, Redd1 exon 3 sequences were not expressed (data not shown), we sought to evaluate the effects of Redd1 disruption in hypoxia-induced mTORC1 inhibition. Western blot studies were limited by substantial variability in baseline mTORC1 activity in tissues in mice (not sufficiently corrected even after fasting for 24 h) and the inability to evaluate specific cell types within a tissue. For these reasons, immunohistochemistry studies were performed. We focused on intestinal crypts, a compartment where a robust induction of Redd1 by hypoxia was observed, and in which baseline mTORC1 activity was consistently high (Fig. 3A, B, and C). Hypoxia led to a downregulation in mTORC1 activity in intestinal crypts of wild-type mice, and this response was not appreciably different in Redd1βgeo/+ mice (Fig. 3A and data not shown). In Redd1βgeo/βgeo mice, a trend toward less mTORC1 inhibition was observed, suggesting that Redd1 plays a role in hypoxia-induced mTORC1 inhibition in intestinal crypts (Fig. 3B). Similar results were obtained with two different anti-phospho-S6 antibodies (Fig. 3B), and in keeping with the notion that the signal reflects mTORC1 activity, it was abolished by treatment of the mice with rapamycin (Fig. 3C).
Fig. 3.
Evaluation of the role of Redd1 in mTORC1 regulation by hypoxia in the mouse. (A) Evaluation of small bowel tissue sections from Redd1βgeo/+ mice exposed to 6% oxygen for the indicated periods of time and assayed for β-galactosidase activity and S6 phosphorylation. (B) Immunohistochemistry (IHC) and β-galactosidase studies of small bowel sections from mice of the stated genotypes exposed to 6% oxygen for the indicated periods of time. (C) IHC of small bowel sections from mice treated with rapamycin (Rapa) or control mice given only vehicle (Veh). (D) Western blots of MEFs of the two genotypes (either primary or transformed with the SV40 early region) and exposed to hypoxia for the indicated number of hours. (E) m7GTP affinity chromatography experiments from cells exposed to hypoxia for the indicated number of hours. (F and G) Western blots of primary thymocytes (F) and hepatocytes (G) from mice of the different genotypes and exposed to hypoxia, ex vivo, for the indicated number of hours.
Redd1 is required for hypoxia-induced mTORC1 inhibition in MEFs and thymocytes but is dispensable in hepatocytes.
We next evaluated the role of Redd1 in mTORC1 regulation by hypoxia in various cell types ex vivo, where greater control over experimental conditions could be achieved. Redd1βgeo/βgeo MEFs were impaired in their downregulation of S6K1 phosphorylation, and this resulted in abnormally increased phosphorylation of S6 (Fig. 3D). These studies were limited, however, by the modest downregulation of mTORC1 activity by hypoxia in primary MEFs. Because tumor cell lines typically exhibit a more robust inhibition of mTORC1 by hypoxia (9), MEFs were transformed using the early region of the SV40 tumor virus (21), which is sufficient for oncogenic transformation of murine fibroblasts. In transformed MEFs, hypoxia resulted in a more robust inhibition of mTORC1 (Fig. 3D), and loss of Redd1 markedly impaired this response (Fig. 3D). In addition, whereas in wild-type MEFs, hypoxia led to a marked increase in 4E-BP1 binding to eIF4E, this was not the case in Redd1βgeo/βgeo MEFs (Fig. 3E). In thymocytes, Redd1 disruption similarly impaired mTORC1 inhibition by hypoxia, but as in MEFs, there was mild inhibition of mTORC1 (Fig. 3F and data not shown). In contrast to thymocytes and primary MEFs, the inhibition of mTORC1 by hypoxia in primary hepatocytes was quite profound (Fig. 3G). Unexpectedly, however, mTORC1 inhibition by hypoxia in hepatocytes was independent of Redd1 (Fig. 3G).
mTORC1 inhibition by hypoxia in hepatocytes is Arnt independent.
In Redd1βgeo/+ reporter mice, we had observed no detectable Redd1 induction by hypoxia in the majority of hepatocytes in zones 1 and 2. Similarly, primary hepatocytes in culture failed to upregulate Redd1 in response to hypoxia (Fig. 4A). In fact, Redd1 levels were actually downregulated in response to hypoxia (Fig. 4A). A similar response was observed for Redd1/βgeo (Fig. 4B and C). Redd1 induction by hypoxia is mediated by Hif-1 (55), but whereas in MEFs, Hif-1α was profoundly and persistently upregulated by hypoxia (Fig. 1E and 3D), this was not the case in hepatocytes (Fig. 4C).
Fig. 4.
Arnt-independent regulation of mTORC1 by hypoxia in hepatocytes. (A) Western blot analysis of wild-type hepatocytes exposed to hypoxia for the indicated number of hours. Ppib, cyclophilin B. (B and C) X-Gal staining (B) and Western blot (C) of hepatocytes of the three different genotypes exposed to hypoxia for the indicated number of hours. (D) Wester blot of Ad-Cre-transduced ArntF/F and Arnt+/+ hepatocytes exposed to hypoxia for the indicated number of hours. (E) Western blots of wild-type hepatocytes, treated with Ad-Cre or not treated with Ad-Cre (−), and exposed to hypoxia for the indicated number of hours.
To unequivocally address the role of Hif in hypoxia signaling in hepatocytes, we generated hepatocytes deficient for the Hif shared β subunit, the aryl hydrocarbon receptor nuclear translocator (Arnt, also referred to as Hif-1β). Primary hepatocytes harvested from ArntfloxP/floxP (henceforth ArntF/F) or from wild-type littermate controls, were transduced ex vivo with a replication-deficient recombinant adenovirus encoding Cre recombinase (Ad-Cre) and exposed to hypoxia (Fig. 4D). Incidentally, previous experiments had determined that Ad-Cre did not affect mTORC1 regulation by hypoxia (Fig. 4E and data not shown). Ad-Cre transduction of ArntF/F hepatocytes led to a substantial downregulation in Arnt protein levels (Fig. 4D). However, Arnt disruption did not affect mTORC1 inhibition by hypoxia (Fig. 4D). Thus, like Redd1, Arnt is also dispensable for hypoxia-induced mTORC1 inhibition in hepatocytes.
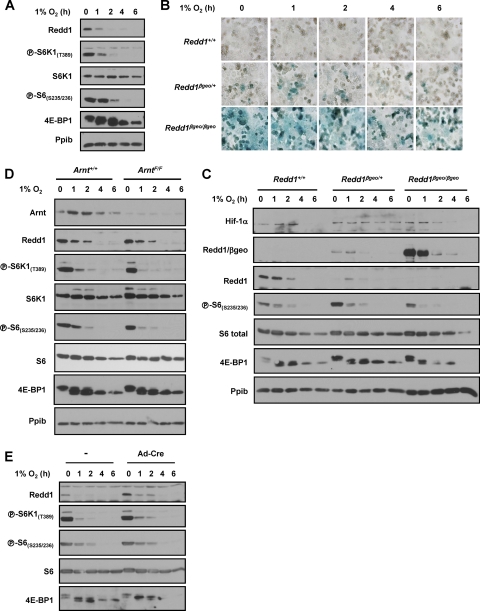
Tsc1/Tsc2 is required for mTORC1 regulation by hypoxia in MEFs but not hepatocytes.
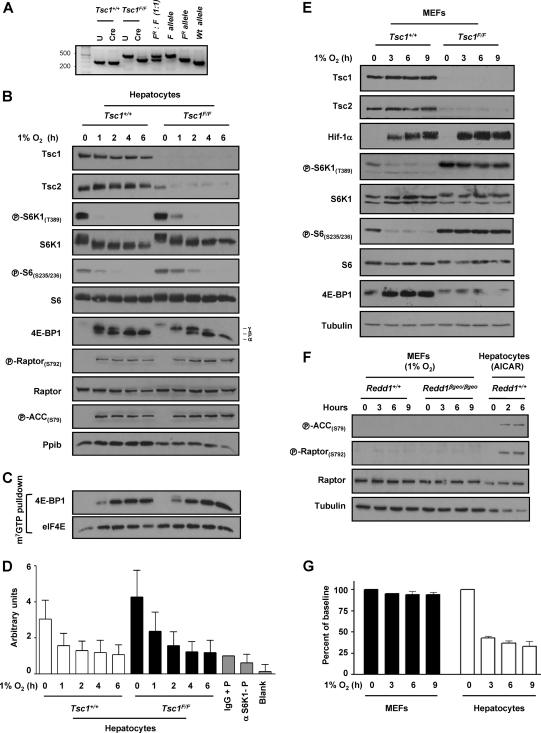
Next, we examined the role of the Tsc1/Tsc2 complex, which is implicated in hypoxia signaling in MEFs downstream of Redd1 (9, 11, 30, 40). Primary hepatocytes harvested from Tsc1floxP/floxP (Tsc1F/F) mice (37) or wild-type littermate controls, were transduced ex vivo with Ad-Cre. Ad-Cre transduction resulted in very efficient recombination of loxP sites (Fig. 5A) with consequent downregulation in the Tsc1 protein (and subsequently also the Tsc2 protein) (Fig. 5B). While loss of Tsc1 resulted in a slight delay in mTORC1 inhibition by hypoxia, mTORC1 was profoundly inhibited by hypoxia in Tsc1-deficient hepatocytes (Fig. 5B to D). In Tsc1-deficient hepatocytes, hypoxia markedly inhibited S6K1 phosphorylation (Fig. 5B) and its activity (as determined by S6 phosphorylation and through in vitro kinase assays [Fig. 5D]). In addition, 4E-BP1 phosphorylation was substantially inhibited by hypoxia in Tsc1-deficient hepatocytes, and this was accompanied by a marked increase in 4E-BP1 binding to eIF4E (Fig. 5C). These data are in marked contrast to MEFs, in which loss of Tsc1 profoundly disrupts mTORC1 inhibition by hypoxia (compare Fig. 5B with Fig. 5E). While MEFs and hepatocytes are cultured in different growth media, even when MEFs were cultured in hepatocyte medium, Tsc1/Tsc2 was essential for hypoxia-induced mTORC1 inhibition (data not shown). Thus, unlike MEFs, where hypoxia-induced mTORC1 inhibition is dependent on both Redd1 and the Tsc1/Tsc2 complex, in hepatocytes, mTORC1 inhibition by hypoxia is independent of both Redd1 and the Tsc1/Tsc2 complex.
Fig. 5.
Tsc1 is dispensable for mTORC1 inhibition by hypoxia in hepatocytes but not in MEFs. (A) PCR of Tsc1+/+ and Tsc1F/F primary hepatocytes either untreated (U) or infected with Ad-Cre (Cre). A defined mixture of recombined and unrecombined DNA was used as a control. The positions of molecular size markers (in base pairs) are shown to the left. (B) Western blot of primary hepatocytes from mice of the indicated genotypes following Ad-Cre infection ex vivo and exposed to hypoxia for the indicated number of hours. (C) m7GTP affinity chromatography from the same extracts as those used in panel B. (D) S6K1 in vitro kinase assays of primary hepatocytes of the stated genotypes following Ad-Cre and exposed to hypoxia for the indicated number of hours. Controls include rabbit IgG immunoprecipitates incubated with peptide (IgG + P), anti-S6K1 immunoprecipitates in the absence of peptide (α S6K1-P), and a blank sample. Data are normalized to IgG control and represent means plus SE (n = 6). (E) Western blots of SV40 early region-transformed MEFs and Ad-Cre-treated MEFs of the indicated genotypes. (F) Western blot analysis of MEFs of the stated genotypes exposed to hypoxia for the indicated number of hours. Redd1+/+ hepatocytes treated with AICAR are shown as a positive control. (G) ATP levels in MEFs and hepatocytes exposed to hypoxia for the indicated number of hours. Data are normalized to baseline levels and represent means plus SE (n = 6 to 18).
AMPK is activated by hypoxia in hepatocytes and inhibits mTORC1 in a Tsc1/Tsc2-independent manner.
While 1% O2 does not activate AMPK in MEFs (Fig. 5F), given the already observed differences between MEFs and hepatocytes with respect to Tsc1/Tsc2, we examined whether AMPK was activated by hypoxia in hepatocytes. As determined by the phosphorylation of its canonical target, acetyl coenzyme A (acetyl-CoA) carboxylase (ACC), AMPK was rapidly and robustly activated by hypoxia in hepatocytes (Fig. 5B). Given these differences, we sought to determine whether 1% O2 differentially affected ATP levels in MEFs and hepatocytes; whereas ATP levels were minimally affected in MEFs, 1% O2 substantially downregulates ATP levels in hepatocytes (Fig. 5G).
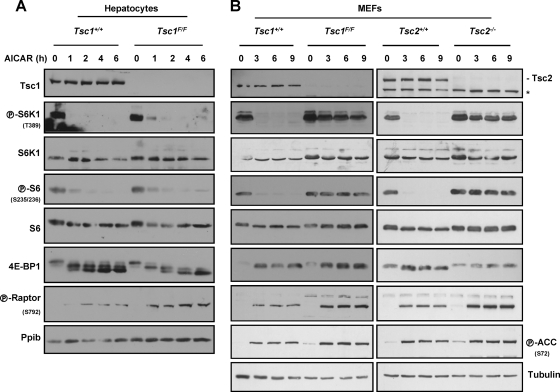
Because Tsc1/Tsc2 is largely dispensable for hypoxia-induced mTORC1 inhibition in hepatocytes (Fig. 5B), should AMPK be involved in the relay of hypoxia signals in hepatocytes, AMPK-induced mTORC1 inhibition would be expected to be independent of Tsc1/Tsc2. To determine whether AMPK could inhibit mTORC1 independently of Tsc1/Tsc2 in hepatocytes, we used the AMPK agonist, AICAR (5-aminoimidazole-4-carboxamide riboside), which is converted in cells to an AMP analogue that binds to and activates AMPK. Importantly, AMPK is necessary for AICAR-induced mTORC1 inhibition (29). AICAR profoundly inhibited mTORC1 even in Tsc1-deficient hepatocytes (Fig. 6A). AMPK has been recently shown to phosphorylate raptor (20), and AMPK activation by AICAR led to raptor phosphorylation (Fig. 6A). Thus, in hepatocytes, mTORC1 inhibition by AMPK is Tsc1/Tsc2 independent and may involve raptor.
Fig. 6.
Tsc1/Tsc2-dependent inhibition of mTORC1 by AMPK in MEFs, but not hepatocytes. (A) Western blots of primary hepatocytes of the stated genotypes treated with Ad-Cre and exposed to AICAR for the indicated number of hours. (B) Western blots of Tsc1+/+ or Tsc1F/F MEFs treated with Ad-Cre or immortalized Tsc2+/+ or Tsc2−/− MEFs and exposed to AICAR for the indicated number of hours. The asterisk indicates a cross-reactive band.
AMPK-induced mTORC1 inhibition in MEFs is Tsc1/Tsc2 dependent.
In keeping with the idea that AMPK is not activated by hypoxia in MEFs, raptor phosphorylation was not appreciably induced following hypoxia in this cell type (Fig. 4F). However, AMPK activation with AICAR did lead, as expected, to raptor phosphorylation in MEFs (Fig. 6B). Interestingly, however, despite a marked increase in raptor phosphorylation, MEFs deficient for Tsc1 or Tsc2 failed to inhibit mTORC1 in response to AICAR (Fig. 6B). Thus, in contrast to hepatocytes in which mTORC1 inhibition by AMPK was Tsc1/Tsc2 independent (Fig. 6A), in MEFs, signaling by AMPK required Tsc1/Tsc2 (Fig. 6B). This was the case despite the fact that in Tsc1/Tsc2-deficient MEFs, raptor was phosphorylated to an even greater extent in response to AICAR than in wild-type MEFs. Thus, raptor phosphorylation per se is not sufficient to inhibit mTORC1, and cell-type-dependent factors are required for raptor-induced mTORC1 inhibition.
In summary, whereas in MEFs, hypoxia did not activate AMPK (Fig. 5F) and AMPK activation by AICAR inhibited mTORC1 in a Tsc1/Tsc2-dependent manner (Fig. 6B), AMPK was activated by hypoxia in hepatocytes (Fig. 5B), and AMPK inhibited mTORC1 in a Tsc1/Tsc2-independent manner in this cell type (Fig. 6A).
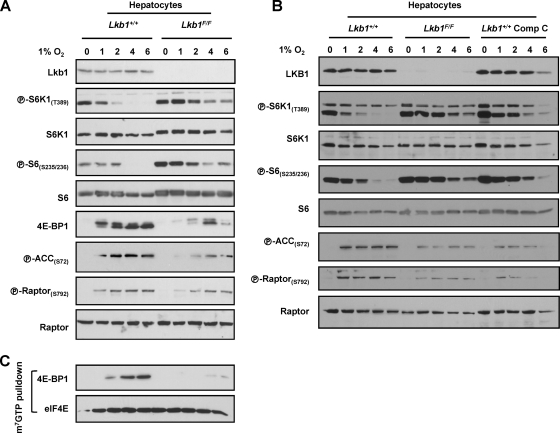
LKB1 is required for hypoxia-induced mTORC1 inhibition in primary hepatocytes.
To examine the role of AMPK in hypoxia-induced mTORC1 inhibition in liver cells, we utilized both genetic and pharmacologic approaches. AMPK activation requires phosphorylation by LKB1 (24). Ad-Cre treatment of Lkb1F/F hepatocytes resulted in a marked downregulation in Lkb1 protein levels (Fig. 7A and B). However, Lkb1 disruption dampened, but did not abolish, AMPK activation (Fig. 7A and B). The simplest explanation for these data is that despite the substantial downregulation in Lkb1 protein, residual Lkb1 function remained. Even in this setting, mTORC1 inhibition by hypoxia was clearly impaired by a reduction in Lkb1 levels (Fig. 7A and B). In Lkb1-depleted hepatocytes, S6K1 and S6 remained phosphorylated for longer periods of time, and the levels of 4E-BP1 bound to eIF4E were substantially reduced (Fig. 7C). The requirement for Lkb1 in hypoxia signaling in hepatocytes stands in contrast to other cell types, such as MEFs or HeLa cells (which are functionally deficient for LKB1) (62), in which mTORC1 inhibition by hypoxia is LKB1 independent (9, 14, 18, 28, 43). Treatment of hepatocytes with the AMPK inhibitor compound C also delayed mTORC1 inhibition by hypoxia (Fig. 7B), although as for most kinase inhibitors, compound C does not exclusively inhibit AMPK (4).
Fig. 7.
Lkb1 and AMPK are required for mTORC1 inhibition by hypoxia in hepatocytes. (A to C) Western blots (A and B) and m7GTP affinity chromatography (C) of Ad-Cre-treated Lkb1+/+ and Lkb1F/F hepatocytes exposed to hypoxia for the indicated number of hours. Where indicated, Lkb1+/+ hepatocytes were pretreated for 0.5 h with the AMPK inhibitor compound C (Comp C).
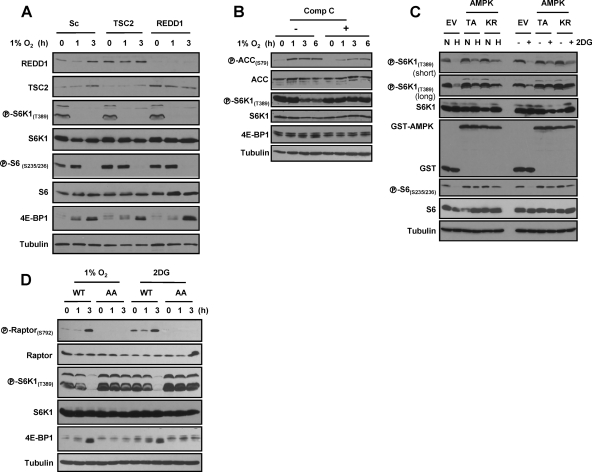
AMPK-dependent inhibition of mTORC1 in HCC cells.
To further evaluate the role of AMPK in hypoxia signaling in hepatocytes, we turned to a more experimentally tractable system, Huh7 hepatocellular carcinoma (HCC) cells. As in primary hepatocytes, hypoxia signaling in Huh7 cells was independent of both REDD1 and TSC2 (Fig. 8A). Also in keeping with the findings in primary cells, exposure to hypoxia led to the activation of AMPK (Fig. 8B), and AMPK inhibition with compound C blocked the effects (Fig. 8B). To further address the role of AMPK, Huh7 cells were generated stably expressing two AMPK dominant-negative (DN) mutants (a kinase-dead K45R [KR] mutant and a nonactivatable T172A [TA] mutant). In contrast to cells transduced with an empty vector, cells expressing AMPK DN mutants failed to inhibit mTORC1 in response to 2-deoxyglucose (2DG), and a similar effect was observed in response to hypoxia (Fig. 8C). These results further substantiate the importance of AMPK in hypoxia signaling in liver cells and extend our previous results in primary hepatocytes.
Fig. 8.
AMPK and raptor phosphorylation are essential for mTORC1 inhibition by hypoxia in liver cells. (A) Western blot of Huh7 cells transfected with the indicated siRNAs (or a scrambled control [Sc]) and exposed to hypoxia for the indicated number of hours. (B) Western blot of Huh7 cells pretreated (+) with compound C (Comp C) and exposed to hypoxia for the indicated number of hours. (C) Western blot of Huh7 cells stably transduced with AMPK dominant-negative (DN) mutants (T172A [TA] or K45R [KR] mutant) or an empty vector (EV) control and exposed to hypoxia (1% O2) (H) or 2-deoxyglucose (2DG) (normoxia [N]). (D) Western blots of Huh7 cells in which endogenous raptor has been replaced by a phosphorylation-deficient raptorS722A/S792A (AA) or by a wild-type raptor control (WT).
Raptor phosphorylation is required for hypoxia and AMPK signaling in HCC cells.
Next, we examined the role of raptor in hypoxia-induced mTORC1 inhibition. For these experiments, endogenous raptor was replaced by a phosphorylation-defective raptor mutant (S722A/S792A) or, as a control, wild-type raptor; Huh7 cells were stably transduced with retroviruses expressing wild-type or mutant raptor cDNAs, and subsequently, endogenous raptor was depleted using a previously validated shRNA targeting the 3′ UTR (20). raptorS722A/S792A did not appear to interfere with raptor function, and baseline mTORC1 activity was indistinguishable from mTORC1 activity in the controls (Fig. 8D). However, whereas in cells expressing wild-type raptor, raptor phosphorylation was induced by hypoxia, raptor phosphorylation did not occur in cells expressing raptorS722A/S792A (Fig. 8D). raptorS722A/S792A-expressing cells failed to normally downregulate mTORC1 in response to 2DG and hypoxia (Fig. 8D). Taken together, these data show that AMPK and raptor are critical for mTORC1 regulation by hypoxia in liver cells.
DISCUSSION
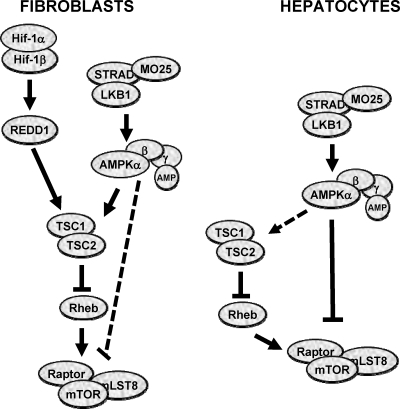
In this study, we show that hypoxia and AMPK signals are relayed to mTORC1 through different pathways in a tissue-dependent manner (Fig. 9). Whereas the Tsc1/Tsc2 complex plays a critical role in mTORC1 regulation in fibroblasts, this complex is largely dispensable in hypoxia and energy signaling in hepatocytes, which rely on Lkb1. To our knowledge, this is the first report of tissue-specific regulation by two different tumor suppressors of a validated cancer target.
Fig. 9.
Tissue-specific regulation of mTORC1 by hypoxia and AMPK. Dominant pathways are indicated by continuous black lines.
Using a novel Redd1βgeo strain we show that Redd1 upregulation by hypoxia is cell type specific. Notably, the pattern of Redd1 upregulation by hypoxia bore some resemblance to that of Hif-1α upregulation (60). However, as there were also some differences, other transcription factors besides Hif-1α may be involved in Redd1 regulation by hypoxia (7). In contrast, Redd2 was not induced by hypoxia. Importantly, while Redd1βgeo faithfully reported on Redd1 induction by hypoxia, Redd1βgeo failed to accurately report on baseline Redd1 expression levels, and we speculate that the reason for this discrepancy is that whereas in response to hypoxia, Redd1 is upregulated primarily through a transcriptional mechanism, at baseline, Redd1 levels are governed by additional mechanisms such as those controlling mRNA stability. In fact, consistent with this notion, the 3′ UTR of Redd1, which is absent in Redd1βgeo, is the target of a microRNA, miR-221 (47).
We show that Redd1 plays an important role in hypoxia signaling not only in MEFs but also in thymocytes and possibly intestinal crypts. Interestingly, by comparison to primary MEFs, MEFs transformed with the early region of the tumor virus SV40, which disrupts, among others, the tumor suppressor proteins p53 and retinoblastoma, exhibited a much more robust Redd1- and Tsc1/Tsc2-dependent inhibition of mTORC1. While this represents one specific instance, it may be that cellular transformation results in a greater dependency on oxygen to sustain a heighted demand for mTORC1 activity and cell growth.
In hepatocytes, by contrast, hypoxia signals are transduced to mTORC1 through a pathway that is independent of Redd1, the Tsc1/Tsc2 complex, and Hif-1β. Hypoxia-induced mTORC1 inhibition in hepatocytes involved LKB1, AMPK, and raptor. In contrast, Lkb1 is dispensable for hypoxia signaling in MEFs and other cell types (9, 14, 18, 28, 43). mTORC1 inhibition by hypoxia in liver cells was disrupted by pharmacological inhibition of AMPK, AMPK dominant-negative mutants, and a raptor mutant defective in AMPK phosphorylation. In hepatocytes, 1% oxygen led to a precipitous drop in ATP levels, which is likely to explain the engagement of this pathway. This contrasts with MEFs, which appeared to adapt and were able to sustain near normal ATP levels in 1% O2. This difference may be due, at least in part, to the differential engagement of Hif-1 in MEFs and hepatocytes. Why Hif-1 does not play a more prominent role in hepatocytes is not known. However, a potential explanation may be found in the devastating consequences of sustained Hif upregulation in this cell type. As we showed recently, constitutive Hif activation in hepatocytes is sufficient to block mitochondrial respiration, resulting in hypoketotic hypoglycemic death within days (35).
The differences in mTORC1 regulation between MEFs and hepatocytes extend beyond hypoxia. AMPK signaling in MEFs led to mTORC1 inhibition in a manner that was dependent on the Tsc1/Tsc2 complex. Tsc1/Tsc2-deficient MEFs failed to inhibit mTORC1 despite pronounced raptor phosphorylation. Thus, raptor phosphorylation alone is not sufficient to inactivate mTORC1. These data are in contrast to a previous report using somewhat higher concentrations of AICAR (20). In contrast, raptor phosphorylation is critical for mTORC1 regulation by AMPK in liver cells. These data underscore a previously unappreciated complexity in both hypoxia and AMPK signaling and illustrate context/tissue-specific differences in signal relay to mTORC1 (Fig. 9).
The differential, cell-type-dependent regulation of mTORC1 by LKB1 and the TSC1/TSC2 complex may contribute to explain the tissue specificity of their tumor suppressor action. It remains unresolved why germ line mutations of a given tumor suppressor gene result in tumors in some tissues but not in others. With respect to sporadic tumors, it is poorly understood why a specific gene is mutated in a particular histological tumor type but not in another. These differences may be explained by differences in the rates with which a specific gene is mutated across cell types. Mutation frequencies in a particular cell type may vary according to carcinogen exposures, local chromatin state, or tissue-specific transcription rates. In addition, differences in the frequencies with which a specific tumor suppressor gene is mutated across different histological tumor types may reflect cell-type-specific tumor suppressor functions. In support of this notion, we show that mTORC1, a critical regulator of tumor development, is differentially regulated in response to hypoxia by the tumor suppressors Tsc1/Tsc2 and Lkb1 in a tissue-specific manner.
ACKNOWLEDGMENTS
We are grateful to A. Klippel for REDD1 polyclonal antibody, D. J. Kwiatkowski for Tsc1F/F mice, F. J. Gonzalez for ArntF/F mice, N. Bardeesy for Lkb1F/F mice, A. D. Nguyen and R. A. DeBose-Boyd for advice on hepatocyte processing and culture, and to members of the Brugarolas lab for discussions.
S.V.-R.-D.-C. was supported in part by a fellowship from Fundacion Caja Madrid. This work was supported by the following grants to J.B.: K08NS051843, RO1CA129387, Basil O'Connor scholar award (5FY06582), and a V scholar award. J.B. is a Virginia Murchison Linthicum Scholar in medical research at UT Southwestern.
The content is solely the responsibility of the authors and does not represent official views from any of the granting agencies. There are no conflicts of interest.
Footnotes
Published ahead of print on 7 March 2011.
REFERENCES
- 1. Abraham R. T., Eng C. H. 2008. Mammalian target of rapamycin as a therapeutic target in oncology. Expert Opin. Ther. Targets 12:209–222 [DOI] [PubMed] [Google Scholar]
- 2. Arsham A. M., Howell J. J., Simon M. C. 2003. A novel hypoxia-inducible factor-independent hypoxic response regulating mammalian target of rapamycin and its targets. J. Biol. Chem. 278:29655–29660 [DOI] [PubMed] [Google Scholar]
- 3. Avruch J., et al. 2009. Amino acid regulation of TOR complex 1. Am. J. Physiol. Endocrinol. Metab. 296:E592–E602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bain J., et al. 2007. The selectivity of protein kinase inhibitors: a further update. Biochem. J. 408:297–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reference deleted.
- 6. Brafman A., et al. 2004. Inhibition of oxygen-induced retinopathy in RTP801-deficient mice. Invest. Ophthalmol. Vis. Sci. 45:3796–3805 [DOI] [PubMed] [Google Scholar]
- 7. Brugarolas J. 2010. mTORC1 signaling and hypoxia, p. 75–97 In Polunovsky V. A. A., Houghton P. J. J. (ed.), mTOR pathway and mTOR inhibitors in cancer therapy. Humana Press, Totawa, NJ [Google Scholar]
- 8. Brugarolas J., Kaelin W. G., Jr 2004. Dysregulation of HIF and VEGF is a unifying feature of the familial hamartoma syndromes. Cancer Cell 6:7–10 [DOI] [PubMed] [Google Scholar]
- 9. Brugarolas J., et al. 2004. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 18:2893–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cai S. L., et al. 2006. Activity of TSC2 is inhibited by AKT-mediated phosphorylation and membrane partitioning. J. Cell Biol. 173:279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Connolly E., Braunstein S., Formenti S., Schneider R. J. 2006. Hypoxia inhibits protein synthesis through a 4E-BP1 and elongation factor 2 kinase pathway controlled by mTOR and uncoupled in breast cancer cells. Mol. Cell. Biol. 26:3955–3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Corradetti M. N., Inoki K., Guan K. L. 2005. The stress-inducted proteins RTP801 and RTP801L are negative regulators of the mammalian target of rapamycin pathway. J. Biol. Chem. 280:9769–9772 [DOI] [PubMed] [Google Scholar]
- 13. Crino P. B., Nathanson K. L., Henske E. P. 2006. The tuberous sclerosis complex. N. Engl. J. Med. 355:1345–1356 [DOI] [PubMed] [Google Scholar]
- 14. Dayan F., et al. 2009. Activation of HIF-1alpha in exponentially growing cells via hypoxic stimulation is independent of the Akt/mTOR pathway. J. Cell. Physiol. 218:167–174 [DOI] [PubMed] [Google Scholar]
- 15. DeYoung M. P., Horak P., Sofer A., Sgroi D., Ellisen L. W. 2008. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev. 22:239–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dufner A., Thomas G. 1999. Ribosomal S6 kinase signaling and the control of translation. Exp. Cell Res. 253:100–109 [DOI] [PubMed] [Google Scholar]
- 17. Fonseca B. D., Smith E. M., Lee V. H., MacKintosh C., Proud C. G. 2007. PRAS40 is a target for mammalian target of rapamycin complex 1 and is required for signaling downstream of this complex. J. Biol. Chem. 282:24514–24524 [DOI] [PubMed] [Google Scholar]
- 18. Garcia-Maceira P., Mateo J. 2009. Silibinin inhibits hypoxia-inducible factor-1alpha and mTOR/p70S6K/4E-BP1 signalling pathway in human cervical and hepatoma cancer cells: implications for anticancer therapy. Oncogene 28:313–324 [DOI] [PubMed] [Google Scholar]
- 19. Gery S., et al. 2007. RTP801 is a novel retinoic acid-responsive gene associated with myeloid differentiation. Exp. Hematol. 35:572–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gwinn D. M., et al. 2008. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 30:214–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hahn W. C., et al. 2002. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol. Cell. Biol. 22:2111–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hara K., et al. 2002. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110:177–189 [DOI] [PubMed] [Google Scholar]
- 23. Hardie D. G. 2007. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 8:774–785 [DOI] [PubMed] [Google Scholar]
- 24. Hawley S. A., et al. 2003. Complexes between the LKB1 tumor suppressor, STRADalpha/beta and MO25alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. J. Biol. 2:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Horton J. D., Shimano H., Hamilton R. L., Brown M. S., Goldstein J. L. 1999. Disruption of LDL receptor gene in transgenic SREBP-1a mice unmasks hyperlipidemia resulting from production of lipid-rich VLDL. J. Clin. Invest. 103:1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Inoki K., et al. 2006. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell 126:955–968 [DOI] [PubMed] [Google Scholar]
- 27. Inoki K., Zhu T., Guan K. L. 2003. TSC2 mediates cellular energy response to control cell growth and survival. Cell 115:577–590 [DOI] [PubMed] [Google Scholar]
- 28. Jin H. O., et al. 2007. Hypoxic condition- and high cell density-induced expression of Redd1 is regulated by activation of hypoxia-inducible factor-1alpha and Sp1 through the phosphatidylinositol 3-kinase/Akt signaling pathway. Cell Signal. 19:1393–1403 [DOI] [PubMed] [Google Scholar]
- 29. Kalender A., et al. 2010. Metformin, independent of AMPK, inhibits mTORC1 in a rag GTPase-dependent manner. Cell Metab. 11:390–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kaper F., Dornhoefer N., Giaccia A. J. 2006. Mutations in the PI3K/PTEN/TSC2 pathway contribute to mammalian target of rapamycin activity and increased translation under hypoxic conditions. Cancer Res. 66:1561–1569 [DOI] [PubMed] [Google Scholar]
- 31. Kim C. J., et al. 2004. Genetic analysis of the LKB1/STK11 gene in hepatocellular carcinomas. Eur. J. Cancer 40:136–141 [DOI] [PubMed] [Google Scholar]
- 32. Kim D. H., et al. 2002. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110:163–175 [DOI] [PubMed] [Google Scholar]
- 33. Kim D. H., et al. 2003. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell 11:895–904 [DOI] [PubMed] [Google Scholar]
- 34.Reference deleted.
- 35. Kucejova B., et al. 10 January 2011. Uncoupling hypoxia signaling from oxygen sensing in the liver results in hypoketotic hypoglycemic death. Oncogene doi:10.1038/onc.2010.587. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kuhn R., Schwenk F., Aguet M., Rajewsky K. 1995. Inducible gene targeting in mice. Science 269:1427–1429 [DOI] [PubMed] [Google Scholar]
- 37. Kwiatkowski D. J., et al. 2002. A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum. Mol. Genet. 11:525–534 [DOI] [PubMed] [Google Scholar]
- 38. Lee S. J., Feldman R., O'Farrell P. H. 2008. An RNA interference screen identifies a novel regulator of target of rapamycin that mediates hypoxia suppression of translation in Drosophila S2 cells. Mol. Biol. Cell 19:4051–4061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li S., et al. 2008. Mesenchymal-epithelial interactions involving epiregulin in tuberous sclerosis complex hamartomas. Proc. Natl. Acad. Sci. U. S. A. 105:3539–3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu L., et al. 2006. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol. Cell 21:521–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Loewith R., et al. 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10:457–468 [DOI] [PubMed] [Google Scholar]
- 42. Ma X. M., Blenis J. 2009. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 10:307–318 [DOI] [PubMed] [Google Scholar]
- 43. Magagnin M. G., et al. 2008. The mTOR target 4E-BP1 contributes to differential protein expression during normoxia and hypoxia through changes in mRNA translation efficiency. Proteomics 8:1019–1028 [DOI] [PubMed] [Google Scholar]
- 44. Nakau M., et al. 2002. Hepatocellular carcinoma caused by loss of heterozygosity in Lkb1 gene knockout mice. Cancer Res. 62:4549–4553 [PubMed] [Google Scholar]
- 45. Oshiro N., et al. 2007. The proline-rich Akt substrate of 40 kDa (PRAS40) is a physiological substrate of mammalian target of rapamycin complex 1. J. Biol. Chem. 282:20329–20339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Peterson T. R., et al. 2009. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 137:873–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pineau P., et al. 2010. miR-221 overexpression contributes to liver tumorigenesis. Proc. Natl. Acad. Sci. U. S. A. 107:264–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Reiling J. H., Hafen E. 2004. The hypoxia-induced paralogs Scylla and Charybdis inhibit growth by down-regulating S6K activity upstream of TSC in Drosophila. Genes Dev. 18:2879–2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reference deleted.
- 50. Sancak Y., et al. 2007. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol. Cell 25:903–915 [DOI] [PubMed] [Google Scholar]
- 51. Sanchez-Cespedes M., et al. 2002. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Res. 62:3659–3662 [PubMed] [Google Scholar]
- 52. Schneider A., Younis R. H., Gutkind J. S. 2008. Hypoxia-induced energy stress inhibits the mTOR pathway by activating an AMPK/REDD1 signaling axis in head and neck squamous cell carcinoma. Neoplasia 10:1295–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schwarzer R., et al. 2005. REDD1 integrates hypoxia-mediated survival signaling downstream of phosphatidylinositol 3-kinase. Oncogene 24:1138–1149 [DOI] [PubMed] [Google Scholar]
- 54. Shaw R. J., et al. 2004. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell 6:91–99 [DOI] [PubMed] [Google Scholar]
- 55. Shoshani T., et al. 2002. Identification of a novel hypoxia-inducible factor 1-responsive gene, RTP801, involved in apoptosis. Mol. Cell. Biol. 22:2283–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shumway S. D., Li Y., Xiong Y. 2003. 14-3-3beta binds to and negatively regulates the tuberous sclerosis complex 2 (TSC2) tumor suppressor gene product, tuberin. J. Biol. Chem. 278:2089–2092 [DOI] [PubMed] [Google Scholar]
- 57. Sofer A., Lei K., Johannessen C. M., Ellisen L. W. 2005. Regulation of mTOR and cell growth in response to energy stress by REDD1. Mol. Cell. Biol. 25:5834–5845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sonenberg N., Hinnebusch A. G. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Soriano P. 1999. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21:70–71 [DOI] [PubMed] [Google Scholar]
- 60. Stroka D. M., et al. 2001. HIF-1 is expressed in normoxic tissue and displays an organ-specific regulation under systemic hypoxia. FASEB J. 15:2445–2453 [DOI] [PubMed] [Google Scholar]
- 61. Thedieck K., et al. 2007. PRAS40 and PRR5-like protein are new mTOR interactors that regulate apoptosis. PLoS One 2:e1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tiainen M., Ylikorkala A., Makela T. P. 1999. Growth suppression by Lkb1 is mediated by a G(1) cell cycle arrest. Proc. Natl. Acad. Sci. U. S. A. 96:9248–9251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tinton S. A., Buc-Calderon P. M. 1999. Hypoxia increases the association of 4E-binding protein 1 with the initiation factor 4E in isolated rat hepatocytes. FEBS Lett. 446:55–59 [DOI] [PubMed] [Google Scholar]
- 64. Vander Haar E., Lee S. I., Bandhakavi S., Griffin T. J., Kim D. H. 2007. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat. Cell Biol. 9:316–323 [DOI] [PubMed] [Google Scholar]
- 65. Vega-Rubin-de-Celis S., et al. 2010. Structural analysis and functional implications of the negative mTORC1 regulator REDD1. Biochemistry 49:2491–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang L., Harris T. E., Roth R. A., Lawrence J. C., Jr 2007. PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. J. Biol. Chem. 282:20036–20044 [DOI] [PubMed] [Google Scholar]
- 67.Reference deleted.
- 68. Yabe D., Komuro R., Liang G., Goldstein J. L., Brown M. S. 2003. Liver-specific mRNA for Insig-2 down-regulated by insulin: implications for fatty acid synthesis. Proc. Natl. Acad. Sci. U. S. A. 100:3155–3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zeqiraj E., Filippi B. M., Deak M., Alessi D. R., van Aalten D. M. 2009. Structure of the LKB1-STRAD-MO25 complex reveals an allosteric mechanism of kinase activation. Science 326:1707–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]