Abstract
DMAP1 (DNMT1-associated protein 1) is a member of the TIP60-p400 complex that maintains embryonic stem (ES) cell pluripotency and a complex containing the somatic form of DNA methyltransferase 1 (DNMT1s). DMAP1 interacts with DNMT1s through a domain that is absent in Dnmt1V/V mice expressing just the oocyte form (DNMT1o). A Dmap1-null allele was generated to study the role of DMAP1 in development. Consistent with the phenotypes of loss of other members of the TIP60-p400 complex, Dmap1−/− mice died during preimplantation in both Dnmt1+/+ and Dnmt1V/V backgrounds. Unexpectedly, in the Dnmt1V/V background, Dmap1+/− parents produced mainly Dmap1+/− mice. Most Dmap1+/+ progeny died during midgestation, with loss of DNA methylation on imprinted genes, suggesting that DMAP1 influences maintenance methylation mediated by DNMT1o. In this regard, a DMAP1-DNMT1o complex was detected in ES cells when DNMT1o was stably expressed but not when transiently expressed, indicating a novel interaction between DMAP1 and DNMT1o. These results suggest that DMAP1-DNMT1s and DMAP1-DNMT1o interactions are essential for normal development and that DMAP1-DNMT1o complexes are not readily formed in the embryo. Therefore, DMAP1 mediates distinct preimplantation epigenetic reprogramming processes: TIP60-p400 nucleosome remodeling and DNMT1 maintenance methylation.
INTRODUCTION
A major biological role of chromatin is to control gene expression, through either transcriptional activation or repression. There are many types of chromatin, defined primarily by the organization and composition of nucleosomes, including specific posttranslational modifications of histones within nucleosomes. The significant changes in chromatin that occur during preimplantation development are of particular interest because of their likely association with epigenetic reprogramming, the inheritance of genomic imprints, and the generation of the embryo's stem (ES) cells. To gain insight into this process, we focused on understanding the role of the corepressor DNMT1-associated protein 1 (DMAP1). DMAP1 was initially identified as a protein associated with the N-terminal domain of DNMT1s and was shown to function as a transcriptional corepressor by interacting with histone deacetylase 2 (HDAC2) (26). Later, DMAP1 was found to be a component of the TIP60-p400 histone acetyltransferase complex (14, 28), which is critically important for the maintenance of ES cell pluripotency (8).
Genetic studies using Dnmt1 mutant mice (11, 12) and immunohistological studies of the intracellular locations of DNMT1 proteins (5, 12) indicate that DNMT1 catalyzes the maintenance of DNA methylation during preimplantation development. The maintenance of CpG methylation patterns is carried out by a combination of maternal (oocyte-derived) and zygotic (embryo-derived) DNMT1 proteins (5). The oocyte synthesizes two DNMT1 proteins: the somatic DNMT1s form of 1,621 amino acids (aa) whose N terminus interacts with DMAP1, and the oocyte form of 1,503 amino acids (DNMT1o) that lacks the known 118-amino-acid DMAP1 interaction domain (5, 18, 26). Both the DNMT1s and DNMT1o proteins coexist in the ooplasm of fully grown mouse oocytes (5). Oocyte-derived DNMT1s maintains methylation patterns during the first two embryonic S phases, whereas DNMT1o functions at the 4th embryonic S phase (4). After the 2-cell stage, zygotic DNMT1s is synthesized, which maintains methylation patterns during the remaining S phases of preimplantation (5, 23).
An important aspect of preimplantation maintenance methylation is its specificity for a subset of gamete-derived DNA methylation patterns. The level of genomic CpG methylation fluctuates during development, with a notable decline from the zygote to the blastocyst stages (3, 19). Differentially methylated domains (DMDs) of imprinted genes maintain (inherit) their gamete-derived methylation patterns during preimplantation development, despite the loss of the bulk of genomic methylation (17, 25). We have postulated that the selective maintenance of DMD methylation during this developmental window is part of a critically important reprogramming process mediated by activities of preimplantation DNMT1 proteins (1, 25). Thus, associations of DMAP1 with DNMT1s and the TIP60-p400 complex suggest that DMAP1 plays an important role in epigenetic reprogramming during preimplantation development. We approached this issue by generating a mouse line with a Dmap1-null allele and studying the effects of this allele in wild-type Dnmt1 and mutant Dnmt1V genetic backgrounds.
MATERIALS AND METHODS
Targeted deletion of Dmap1.
A targeting construct was made by inserting a loxP site into an SmaI restriction site in the first intron of Dmap1 and a Pgk-neor cassette at an engineered EagI site in the intron between exons 10 and 11. The cassette contained a Pgk-neor transcription unit flanked by FLP recombination target (FRT) sites and a 5′ loxP site (Fig. 1 A). Genotypes of Dmap1+/+, Dmap1floxed/+ and Dmap1+/− mice were confirmed by Southern blotting of tail DNA digested with either BglII or EcoRI and with the probes indicated (Fig. 1B and C). Dmap1+ and Dmap− alleles were also genotyped by PCR using oligonucleotides 1 (5′CCCCCTCCCTCAAATACTTC3′) and 2 (5′CAGCCATTGAGAGGAAAAGC3′) for the wild-type (WT) allele and oligonucleotides 3 (5′TCCTATCCGTGGGTCTTCAG3′) and 4 (5′GTCAACCCTCTCCTGTCGTC3′) for the null allele (Fig. 1D).
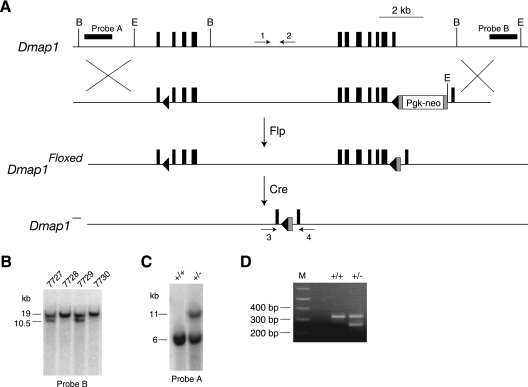
Fig. 1.
Generation of a mouse Dmap1− allele. (A) Schematic of targeted-mutagenesis scheme in mouse ES cells used to generate a mouse conditional allele (Dmap1floxed), which in turn was used to generate a mouse null allele (Dmap1−). The targeting plasmid contains a 5′ loxP site in the first intron and a 3′ loxP site in the last intron of Dmap1. loxP sites are shown by black triangles and exons by vertical black rectangles. Gray rectangles represent FRT sites. Probes used for Southern blots are indicated by horizontal black rectangles. A mouse line generated with a Pgk-neoR-containing heterozygous ES cell clone was crossed to a CAGGS-flp transgenic mouse line (gift from G. Homanics) to remove the Pgk-neoR fragment and generate a Dmap1floxed mouse line, which was then crossed to an EIIa-cre transgenic line (15) to remove the bulk of the Dmap1 gene between the 5′ and 3′ LoxP sites. BglII and EcoRI restriction sites are indicated by “B” and “E,” respectively. The positions of the oligonucleotides used to genotype mice are indicated by arrows. (B) Southern blot of EcoRI-digested genomic DNA from four offspring (no. 7727 to 7730) derived from a cross between a wild-type mouse and a heterozygous Dmap1floxed mouse. The 19-kb band is the wild-type allele, and the 10.5-kb band is the Dmap1floxed allele. (C) Southern blot of BglII-digested genomic DNA from a wild-type Dmap1+/+ mouse and the founder Dmap1+/− mouse derived from a cross between a Dmap1floxed/+ mouse and an EIIA-cre transgenic mouse. The 6-kb band is the wild-type allele, and the 11-kb band is the Dmap1− allele. (D) Genotypes of a wild-type mouse and a Dmap1+/− mouse identify a 320-nucleotide (nt) wild-type allele and a 250-nt Dmap− allele in PCR assays by using the oligonucleotide pairs shown in panel A. M, molecular size markers.
Mouse lines.
The Dmap1−, Dnmt1V, and TR2 + 3/Igmyc mouse lines were all maintained in an inbred 129/Sv genetic background. Dnmt1+/+ and Dnmt1V alleles were genotyped as previously described (6). TR2 + 3/Igmyc mice were identified by genotyping as previously described (25). All mouse experiments were approved and conformed to the standards of the Institutional Animal Care and Use Committee of the University of Pittsburgh.
Cell lines.
Dnmt1V/V ES cell lines were generated from blastocysts isolated from crosses between Dnmt1V/V mice (6). The R1 mouse ES cell line has been previously described (21). Primary mouse embryonic fibroblasts (MEFs) were established from embryonic day 14.5 (E14.5) embryos.
Expression plasmids.
Dmap1 cDNA amplified from mouse spleen total RNA was cloned in Topo blunt vector (Invitrogen), and the sequence was verified and subcloned into EF1-myc version B expression plasmid (Invitrogen) to stably express DMAP1-MYC fusion peptide in ES cells. An EF1-internal ribosome entry site (IRES)-hyg expression vector constructed by replacing the c-myc promoter in pIRES-hyg (Clontech) with PCR-amplified EF1α promoter from EF1-myc vector was used for expression of DNMT1s and DNMT1o proteins. Stable transfections were carried out by electroporation. Transient transfections of ES cells with Lipofectamine 2000 (Invitrogen) gave a transfection efficiency of ∼75%. Cell lysates were prepared 48 h after transfection.
DMAP1 immunoprecipitation.
Whole-cell lysates prepared using a nondenaturing cell lysis buffer (20 mM Tris HCl [pH 7.5], 137 mM NaCl, 10% glycerol, 1% Nonidet P-40 [NP-40], and 2 mM EDTA) containing protease inhibitor cocktail (Roche) were centrifuged at 12,000 rpm for 20 min at 4°C. The supernatants were transferred to fresh tubes and incubated overnight with anti-MYC antibody conjugated with agarose (Santa Cruz Biotechnology). Protein complexes were pulled down by centrifugation, washed three times with the lysis buffer, and denatured for analysis by immunoblotting.
Immunoblotting.
Protein lysates resolved on 5% to 15% gradient polyacrylamide gels were Western blotted and probed with the UPTC21 antibody that detects both DNMT1s and DNMT1o (23), anti-DMAP1 antibody (ab2848; Abcam), and anti-MYC antibody (Covance). Bands on Western blots were quantified with Image J software (http://rsbweb.nih.gov/ij/).
Embryo collection.
Blastocysts were obtained from a cross between Dmap1+/−, Dnmt1V/V parents. One-half of the DNA sample from each blastocyst was used to determine the Dmap1 genotype. The remaining DNA samples from Dmap1+/−, Dnmt1V/V blastocysts were collected into one pool of two blastocysts, and the remaining DNA samples from Dmap1+/+, Dnmt1V/V blastocysts were collected into a separate pool of two blastocysts.
Methylation analysis.
Samples of genomic DNA were treated with sodium bisulfite (EZ DNA methylation kit; Zymo Research) and amplified with primers specific to H19, Snurf/Snrpn, and Gtl2 differentially methylated domains (DMD) and the imprinted TR2 + 3/Igmyc transgene sequences (1, 25). Methylation levels of Snurf/Snrpn and Gtl2 DMDs were estimated with combined bisulfite restriction analysis (COBRA) assays (1). The level of methylation on H19/Igf2 DMD and the TR2 + 3/Igmyc transgene was determined by sequencing individual cloned DNA fragments amplified from the bisulfite-treated DNA (1, 25).
RESULTS
Homozygous Dmap1−/− embryos show a lethal embryonic phenotype.
To explore the function of the DMAP1 protein in embryonic development, we generated a mouse line with a null allele of the mouse Dmap1 gene. We first generated mice with a conditional allele of Dmap1 (Dmap1floxed) in which loxP sites were placed in the first and last introns of the Dmap1 gene. By crossing Dmap1floxed mice with mice carrying an EIIa-cre transgene (15), a mouse line carrying an allele lacking nine exons coding for amino acids 1 to 448 of the 468-aa DMAP1 protein was produced (Fig. 1). As shown in Table 1, homozygous Dmap1−/− neonatal mice from crosses between Dmap1+/− parents were not recovered, and heterozygous and wild-type mice were recovered at an approximate ratio of 2:1 (cross 1), indicating that homozygous Dmap1−/− mice died during embryonic development, whereas the development of heterozygous Dmap1+/− mice is normal. To define the age of embryonic lethality, we determined the genotypes of E10.5 embryos, E3.5 blastocysts, and 8-cell embryos from crosses between heterozygous Dmap1+/− parents. No homozygous mutant Dmap1−/− 8-cell embryos or blastocysts were identified (cross 1). We conclude that the DMAP1 protein is required for preimplantation development.
Table 1.
Genotypes of offspring from crosses of Dnmt1 and Dmap1 parents
| Cross | Parent genotype | Developmental stagea |
Dmap1 genotype |
Total no. of mice | ||
|---|---|---|---|---|---|---|
| +/+ | +/− | −/− | ||||
| 1 | Dmap1+/−, Dnmt1+/+ × Dmap1+/−, Dnmt1+/+ | Adult | 21 | 62 | 0 | 83 |
| E10.5 | 6 | 17 | 0 | 23 | ||
| Blastocyst | 9 | 20 | 0 | 29 | ||
| 8-cell | 10 | 17 | 0 | 27 | ||
| 2 | Dmap1+/−, Dnmt1V/V × Dmap1+/−, Dnmt1V/V | Adult | 4b | 74 | 0 | 78 |
| E9.5 | 5b | 63 | 0 | 68 | ||
| E8.5 | 1 | 8 | 0 | 9 | ||
| E7.5 | 3 | 21 | 0 | 24 | ||
| Blastocyst | 6 | 18 | 0 | 24 | ||
| 8-cell | 2 | 8 | 0 | 10 | ||
| 3 | Dmap1+/−, Dnmt1V/V × Dmap1+/+, Dnmt1V/V | Adult | 0b | 21 | 0 | 21 |
| 4 | Dmap1+/+, Dnmt1V/V × Dmap1+/−, Dnmt1V/V | Adult | 0b | 23 | 0 | 23 |
| 5 | Dmap1+/−, Dnmt1V/V × Dmap1+/+, Dnmt1+/+ | Adult | 7 | 9 | 0 | 16 |
| 6 | Dmap1+/+, Dnmt1+/+ × Dmap1+/−, Dnmt1V/V | Adult | 5 | 9 | 0 | 14 |
| 7 | Dmap1+/−, Dnmt1V/V × Dmap1+/+, Dnmt1V/+ | Adult | 0b | 9 | 0 | 9 |
| 8 | Dmap1+/+, Dnmt1V/+ × Dmap1+/−, Dnmt1V/V | Adult | 6 | 5 | 0 | 11 |
| 9 | Dmap1+/−, Dnmt1V/+ × Dmap1+/+, Dnmt1V/V | Adult | 4 | 7 | 0 | 11 |
| 10 | Dmap1+/+, Dnmt1V/V × Dmap1+/−, Dnmt1V/+ | Adult | 4 | 7 | 0 | 11 |
“Adult” indicates mice genotyped at 3 weeks after birth.
Significant deviation from expected ratio of Dmap1+/+ to Dmap1+/− offspring.
DMAP1 is required for development even in the absence of its known DNMT1 interaction domain.
The DMAP1 interaction domain was mapped to the first 118 aa of DNMT1s, and DNMT1o was suggested not to interact with DMAP1 (26). We previously showed that Dnmt1V/V mice that produce a mutant form of DNMT1s lacking this interaction domain (equivalent to DNMT1o) are viable and fertile (6), indicating that the DMAP1 interaction domain is not required for normal development. Here we were interested in knowing whether loss of DMAP1 would have no consequences in a Dnmt1V/V background. Heterozygous Dmap1+/− mice in a Dnmt1V/V background (Dmap1+/−, Dnmt1V/V) were crossed, and progeny were genotyped at multiple stages. As shown in Table 1 (cross 2), none of the progeny (examined as early as the 8-cell stage) were Dmap1−/−, indicating that the Dnmt1V/V background did not suppress or modify in any discernible way the lethality of homozygous Dmap1−/− mice. We conclude that DMAP1 serves an essential function in preimplantation that is independent of its specific interaction with DNMT1s.
Contrary to expectations, we also did not recover mice with the Dmap1+/+ genotype at the expected frequency from crosses between Dmap1+/−, Dnmt1V/V mice (Table 1, cross 2). When the progeny of these parents were genotyped at different stages of development, we observed that the Dmap1+/+ embryos died around 9.5 days of gestation. Because the progeny of the same genotype (Dmap1+/+, Dnmt1V/V) are viable when the parents are Dmap1+/+, Dnmt1V/V, we examined whether Dmap1+, Dnmt1V gametes from Dmap1+/−, Dnmt1V/V mice are compromised. When the genotypes of the opposite parents are Dmap1+/+, Dnmt1V/V, these crosses also resulted in only Dmap1+/− mice (Table 1, crosses 3 and 4), indicating that the Dmap1+, Dnmt1V gametes from Dmap1+/−, Dnmt1V/V parents are not equivalent to those from the Dmap1+/+, Dnmt1V/V mice in their ability to support the development of Dmap1+/+, Dnmt1V/V embryos. This observed transmission ratio distortion indicates that interactions between Dmap1 and Dnmt1 alleles are complex.
To further investigate the effects of Dmap1 and Dnmt1 interactions during development, we conducted a series of crosses in which the Dnmt1V allele was replaced by the wild-type Dnmt1 allele. In the first set of experiments, Dmap1+/−, Dnmt1V/V mice were crossed with wild-type mice (Table 1, crosses 5 and 6). These crosses yielded both Dmap1+/+ and Dmap1+/− mice in the expected ratios, suggesting that in Dmap1+/−, Dnmt1V/V mice, both Dmap1− and Dmap1+ gametes are viable and postzygotic expression of a Dnmt1+ allele is sufficient to suppress the lethality in Dmap1+/+ mice obtained from Dmap1+/−, Dnmt1V/V parents. Second, we conducted reciprocal crosses in which one of the parents was Dmap1+/−, Dnmt1V/V and the other was Dmap1+/+, Dnmt1V/+ (Table 1, crosses 7 and 8). Dmap1+/+ mice were obtained when the Dnmt1+ allele was from the mother (cross 8) but not from the father (cross 7). When the Dmap1+/+ progeny from cross 8 were genotyped for Dnmt1 alleles, there was a 1:1 distribution of both Dnmt1+ and Dnmt1V alleles. This suggests that a zygotic Dnmt1+ allele is not required for normal development of the Dmap1+/+ progeny, and the presence of a Dnmt1+ allele in the mother seems to be sufficient to support development. In these mothers, both DNMT1o and DNMT1s proteins that are expressed in the germ line are deposited in the Dmap1+, Dnmt1V oocytes, and transmission of maternal DNMT1s is able to fully rescue the Dmap1+/+ genotype in the offspring. In conclusion, Dmap1+/+ offspring can be recovered at the expected frequencies from a homozygous Dnmt1V/V carrier if the offspring inherit a Dnmt1+ allele from the opposite parent or, alternatively, if the female parent is heterozygous Dnmt1V/+.
To more precisely define the developmental requirements for interactions between Dmap1 and Dnmt1 alleles, we crossed Dmap1+/−, Dnmt1V/+ mice with Dmap1+/+, Dnmt1V/V mice (Table 1, crosses 9 and 10). Unlike the previous crosses (crosses 7 and 8), both male and female parents had the same ability to produce Dmap1+/+ offspring at expected ratios. Moreover, when Dmap1+/+ progeny were genotyped further for the presence of Dnmt1 alleles, we observed an equal representation of both Dnmt1+ and Dnmt1V alleles among the Dmap1+/+ progeny, suggesting that, as was the case in cross 8, there was no absolute requirement for the continued presence of a Dnmt1+ allele for the survival of Dmap1+/+ progeny produced from these parents. A comparison between crosses 3 and 9 or 4 and 10 indicated that the Dmap1+, Dnmt1V gametes from Dmap1+/−, Dnmt1V/+ parents are capable of supporting development, whereas the gametes of the same genotype from Dmap1+/−, Dnmt1V/V parents were compromised. Taking the results shown in Table 1 together, we observed that Dmap1+, Dnmt1V gametes produced from the Dmap1+/−, Dnmt1V/V parent were defective in supporting the development of Dmap1+/+, Dnmt1V/V progeny. This defect can be corrected by the presence of a Dnmt1+ allele during gametogenesis or early embryogenesis. This conclusion is indicated in the schematic shown in Fig. 2.
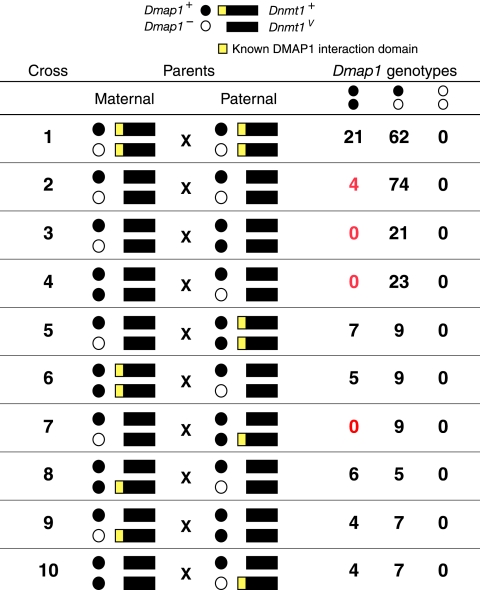
Fig. 2.
Summary of genotypes of adult offspring from crosses of Dmap1 and Dnmt1 parents. The data are derived from Table 1, and crosses 1 to 10 correspond to the crosses labeled in Table 1. The symbols correspond to different Dmap1 and Dnmt1 alleles (top). Statistically significant deviations from the expected number of homozygous Dmap1+/+ offspring are highlighted in red.
Imprinted DNA methylation in Dmap1+/+, Dnmt1V/V embryos.
Abnormalities in the inheritance of genomic imprints during preimplantation can lead to defects in development and death of postimplantation embryos (12). Therefore, we examined the methylation pattern of imprinted genes in progeny of Dmap+/−, Dnmt1V/V parents. We first compared the methylation of the endogenous H19/Igf2 DMD, which is normally methylated on the paternal allele, in Dmap1+/−, Dnmt1V/V and Dmap1+/+, Dnmt1V/V blastocysts, a time before the onset of Dmap1+/+, Dnmt1V/V lethality. There was a small, but detectable reduction in H19/Igf2 DMD methylation in Dmap1+/+, Dnmt1V/V blastocysts compared to Dmap1+/−, Dnmt1V/V blastocysts (Fig. 3A).
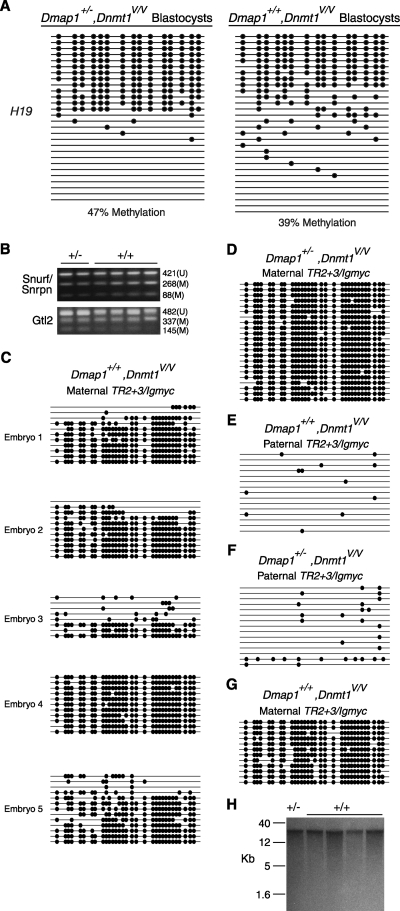
Fig. 3.
Methylation of imprinted DNA sequences in Dmap+/−, Dnmt1V/V and Dmap+/+, Dnmt1V/V embryos derived from Dmap1+/−, Dnmt1V/V mice. (A) H19/Igf2 DMD methylation in two pools of blastocysts. Each horizontal line is an H19/Igf2 DMD allele whose methylation was determined by bisulfite genomic sequencing (1). Filled circles represent methylated CpG dinucleotides, and the absence of circles indicates unmethylated CpGs. (B) COBRA of Snurf/Snrpn and Gtl2 DMD sequences from E9.5 embryos. Snurf/Snrpn PCR products were digested with BstUI and Gtl2 PCR products with TaqI. Fragments representing methylated (M) or unmethylated (U) DNA and their sizes in nucleotides are indicated. +/−, Dmap1+/−, Dnmt1V/V; +/+, Dmap1+/+, Dnmt1V/V. (C) Bisulfite sequencing of the maternal TR2 + 3/Igmyc transgene in five different Dmap1+/+, Dnmt1V/V E9.5 embryos from crosses between Dmap1+/−, Dnmt1V/V parents. (D) Methylation of the maternal TR2 + 3/Igmyc transgene in a Dmap1+/−, Dnmt1V/V embryo from a cross between Dmap1+/−, Dnmt1V/V parents. (E and F) Methylation of the paternal TR2 + 3/Igmyc transgene in a Dmap1+/+, Dnmt1V/V and a Dmap1+/−, Dnmt1V/V embryo, both from a cross between crosses between Dmap1+/−, Dnmt1V/V parents. (G) Methylation of the maternal TR2 + 3/Igmyc transgene in a Dmap1+/+, Dnmt1V/V E9.5 embryo from a cross between a Dmap1+/−, Dnmt1V/+ female and a Dmap1+/+, Dnmt1V/V male mouse. (H) Southern blot of HpaII-digested DNA hybridized with an IAP probe (1). DNA was obtained from one E9.5 Dmap1+/−, Dnmt1V/V embryo (D) and four E9.5 Dmap1+/+, Dnmt1V/V embryos (1 to 4 in panel C).
To further evaluate the integrity of DMD methylation in Dmap1+/+, Dnmt1V/V embryos, we compared the levels of maternal methylation of Snurf/Snrpn DMD and paternal methylation of Gtl2 DMD between Dmap1+/−, Dnmt1V/V and Dmap1+/+, Dnmt1V/V E9.5 embryos. As shown in Fig. 3B, both DMDs in all six embryos examined showed both methylated and unmethylated DMD alleles, with no obvious and consistent differences in Snurf/Snrpn and Gtl2 methylation between the two different embryonic genotypes. We conclude from this that for the DMDs tested here, allele-specific DMD methylation is largely intact.
Because the COBRA assay provides at best a semiquantitative measurement of methylation on specific DNA sequences, it is possible that significant differences in imprinted methylation between Dmap1+/−, Dnmt1V/V and Dmap1+/+, Dnmt1V/V embryos would not be detected. For instance, a partial loss of methylation from the normally methylated parental allele would not be easily detectable in the presence of the normally unmethylated, opposite parental allele. To better assess imprinted methylation, we examined methylation of the TR2 + 3/Igmyc transgene in E9.5 embryos. TR2 + 3/Igmyc is an imprinted transgene in which Igf2r DMD2 sequences were incorporated into the imprinted RSVIgmyc transgene (4, 24, 25). Importantly, the parental origin of TR2 + 3/Igmyc can be unambiguously determined in crosses between a TR2 + 3/Igmyc hemizygous carrier and a nontransgenic mouse, and in wild-type strain backgrounds, there are strict patterns of methylation, with the maternal TR2 + 3/Igmyc highly methylated and the paternal TR2 + 3/Igmyc unmethylated (4). As shown in Fig. 3C, four out of five Dmap1+/+, Dnmt1V/V E9.5 embryos from crosses between Dmap1+/−, Dnmt1V/V parents showed deficiencies in maternal TR2 + 3/Igmyc methylation. In comparison, Dmap1+/−, Dnmt1V/V E9.5 embryos from Dmap1+/−, Dnmt1V/V crosses showed complete methylation of maternal TR2 + 3/Igmyc alleles (Fig. 3D). Paternally inherited TR2 + 3/Igmyc alleles in both types of E9.5 embryos were unmeth-ylated (Fig. 3E and F). The maternal TR2 + 3/Igmyc allele was also completely methylated in a Dmap1+/+, Dnmt1V/V adult from a cross between a Dmap1+/−, Dnmt1V/+ female and a Dmap1+/+, Dnmt1V/V male (Fig. 3G); there is no evidence of a decrease in viability in Dmap1+/+, Dnmt1V/V offspring of this cross (cross 9) and its reciprocal (cross 10) (Table 1). No differences in methylation between Dmap1+/−, Dnmt1V/V and Dmap1+/+, Dnmt1V/V E9.5 embryos were seen at intracisternal A particle (IAP) retrotransposon loci (Fig. 3H). Collectively, the findings indicate that there are defects in imprinted DMD methylation in Dmap1+/+, Dnmt1V/V embryos. Such defects are reminiscent of the preimplantation imprinting defects observed in embryos derived from Dnmt1Δ1o/Δ1o female mice (12) and likely account for the lethality of Dmap1+/+, Dnmt1V/V mice derived from Dmap1+/−, Dnmt1V/V parents.
DNMT1s and DNMT1o proteins physically associate with DMAP1 with different affinities.
The ability of Dmap1+, Dnmt1V gametes to support embryonic development varies based on their parental genotypes. Gametes from Dmap1+/−, Dnmt1V/V parents are less capable of supporting development than those from Dmap1+/+, Dnmt1V/V parents. This suggests that DNMT1o might functionally interact with DMAP1 during gametogenesis. To measure physical associations between DMAP1 and DNMT1 proteins, we first expressed an epitope (MYC)-tagged version of DMAP1 in wild-type and Dnmt1V/V ES cell lines derived from Dnmt1V/V blastocysts. As shown in Fig. 4A, ES cell clones expressing exogenous DMAP1-MYC protein were isolated. To determine if DMAP1 interacts with DNMT1o, we immunoprecipitated DMAP1. As shown in Fig. 4B, both DNMT1s and DNMT1o formed complexes with DMAP1-MYC. However, the amount of DNMT1o protein associated with DMAP1 was approximately 5 times lesser than the amount of DNMT1s protein associated with DMAP1. To directly compare the association of DNMT1s and DNMT1o with DMAP1, we stably transfected a Dnmt1s cDNA construct into Dnmt1V/V ES cells expressing DMAP1-MYC. As shown in Fig. 4C, both DNMT1o and DNMT1s can be immunoprecipitated with DMAP1-MYC. Although the level of expression of DNMT1s is similar to the level of endogenous DNMT1o in these transfected cells, the ratio of coimmunoprecipitated DNMT1s to DNMT1o was approximately 5:1 (Fig. 4C). Collectively, these findings suggest that although the strongest physical association between DNMT1 and DMAP1 is through the previously described 118-amino-acid DMAP1 interaction domain (26), other regions of DNMT1 enable formation of a complex with DMAP1.
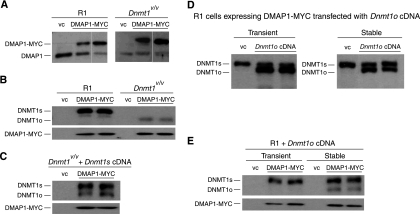
Fig. 4.
Physical interactions between DMAP1 and DNMT1 proteins. (A) Stable expression of DMAP1-MYC in WT R1 and Dnmt1V/V ES clones. Both endogenous DMAP1 (Mr, 52.9) and the exogenous DMAP1-MYC (Mr, 55.9) proteins were detected by anti-DMAP1 antibody. vc, ES cell clone with stably integrated empty expression vector. (B) DMAP1 is present in a complex with both DNMT1 and DNMT1o. Immunoprecipitates from clones stably expressing DMAP1-MYC were probed with UPTC21 to detect DNMT1o (Mr, 165,000) and DNMT1s (Mr, 190,000) proteins (top) and with anti-DMAP1 antibody (bottom). (C) DMAP1 binds preferentially to DNMT1s. Lysates from Dnmt1V/V ES cell clones stably expressing DMAP1-MYC DNMT1s proteins were immunoprecipitated with anti-MYC, resolved on SDS-PAGE gels, and probed with UPTC21 and anti-DMAP1 antibodies. (D) Levels of DNMT1o in R1 cells after transient transfection (left panel) and stable transfection (right panel) with DMAP1-cDNA expression plasmid. (E) Detection of DMAP1-DNMT1o complex in ES cells stably expressing DNMT1o. Immunopreciptates from the cell types shown in panel D were probed with UPTC21 and anti-DMAP1 antibodies.
To further investigate the nature of the interaction between DMAP1 and DNMT1o, we compared the levels of DMAP1-DNMT1o complexes formed in R1 ES cells expressing DNMT1o for a short time (transiently transfected) to those in R1 cells expressing DNMT1o for a long time (stably transfected). We detected higher levels of DNMT1o in transiently transfected cells than in stably transfected cells (Fig. 4D). However, DMAP1-DNMT1o complexes were detected in only the stably transfected cells (Fig. 4E). These findings are consistent with the notion that physical interactions between DMAP1 and DNMT1o proteins did not readily occur.
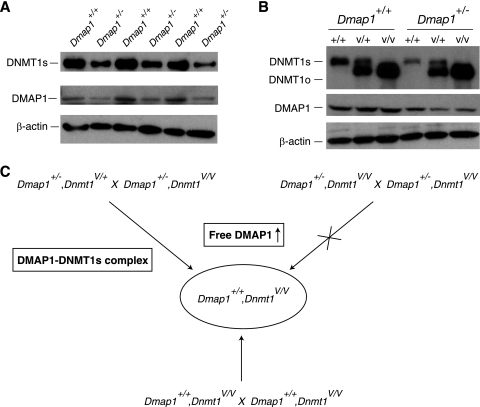
In addition to ES cells, we examined the relationship between DMAP1 and DNMT1 protein levels in somatic cells. As shown in Fig. 5A, in primary mouse embryonic fibroblasts (MEFs) obtained from wild-type and Dmap1+/− heterozygous embryos, the concentration of DMAP1 protein was proportional to the number of wild-type alleles, such that wild-type MEFs have roughly twice the concentration of DMAP1 as Dmap1+/− MEFs. Moreover, the level of DNMT1s protein in Dmap1+/+ MEFs was approximately twice that seen in Dmap1+/− MEFs. The same positive correlation between DMAP1 and DNMT1 protein levels was evident in Dnmt1+/+ spleens, but not in Dnmt1V/V spleens (Fig. 5B). These observations suggest that the concentration of DNMT1s depends on the concentration of DMAP1, possibly because DNMT1s is stabilized in the DMAP1-DNMT1s complexes through its known DMAP1 interaction domain. The absence of correlation between DMAP1 and DNMT1o concentrations in spleen cells may be due to the absence of requirement for stabilization through the DMAP1-DNMT1o interaction or simply to an innately stable DNMT1o protein.
Fig. 5.
Relationships between Dmap1 and Dnmt1 in mutant mice. (A) Immunoblots showing expression of DMAP1 and DNMT1s proteins in three Dmap+/+, Dnmt1+/+ and three Dmap1+/−, Dnmt1+/+ mouse embryonic fibroblast (MEF) lines. The concentration of β-actin protein was measured to ensure that approximately equal amounts of cellular protein were loaded in each lane. (B) Immunoblots showing expression of DMAP1 and DNMT1s proteins in spleens from six different mice. Genotypes are indicated at the top, with Dnmt1 genotypes listed below Dmap1 genotypes. +/+, Dnmt1+/+; V/+, Dnmt1V/+; V/V, Dnmt1V/V. The concentration of the β-actin protein was measured to normalize the amount of protein loaded in each lane. (C) Effects of parental Dmap1 and Dnmt1 genetic backgrounds on outcome of Dmap1+/+, Dnmt1V/V offspring. Three different crosses are shown, all of which result in Dmap1+/+, Dnmt1V/V embryos. Despite identical genotypes, Dmap1+/+, Dnmt1V/V embryos derived from crosses between Dmap1+/−, Dnmt1V/V mice are rarely recovered.
DISCUSSION
During our investigations into the roles of DNMT1 proteins and their interactions with the DMAP1 protein, we identified two Dmap1 lethal genotypes, Dmap1−/− embryos from crosses between Dmap1+/− heterozygous parents, and Dmap1+/+, Dnmt1V/V embryos from crosses between Dnmt1+/−, Dnmt1V/V parents. Dmap1−/− embryos die prior to the 8-cell stage of preimplantation development, whereas most Dmap1+/+, Dnmt1V/V embryos die around midgestation. We conclude that the cause of embryonic death in Dmap1−/− embryos is different from the cause of death in Dmap1+/+, Dnmt1V/V embryos, indicating two distinct and essential DMAP1-mediated processes.
The early preimplantation lethality of Dmap1−/− embryos is most likely due to inactivation of the TIP60-p400 complex, which functions in DNA repair and transcriptional regulation (28, 29). An RNA interference (RNAi) screen performed in mouse ES cells showed that reduction of the expression of individual components of the complex, including DMAP1, caused a loss of characteristic ES cell morphology and activation of genes associated with cell differentiation (8). Specifically, downregulation of DMAP1 expression induced by the stable expression of a Dmap1 short hairpin RNA (shRNA) induced a flattening of the normal ES colony morphology and loss of expression of genes associated with ES cell pluripotency. Genes encoding different components of the TIP60-p400 complex are expressed not only in ES cells, but also in early mouse embryos, and embryos lacking Tip60 or Trrap, two components of the complex, die before implantation (9, 10). We illustrated that the lack of another component of the complex, Dmap1, also led to preimplantation lethality as early as the 8-cell stage, possibly by the severe disruption to transcriptional control in preimplantation stages. In contrast to Dmap1−/− embryos, Dmap1+/+, Dnmt1V/V embryos perish because of epigenetic defects, as shown by the decreased methylation of DMDs of both maternally and paternally imprinted genes (Fig. 3 and 4). Although we cannot formally exclude a role of the TIP60-p400 complex in DMD methylation, the lack of DNMT1 in the purified TIP60-p400 complex (2, 7) indicates that DMAP1 interacts with DNMT1 in a distinct complex. Thus, DMAP1-dependent functions of the TIP60-p400 complex appear to be independent of the DMAP1 function associated with DNMT1 proteins.
How might maintenance methylation be disrupted in Dmap1+/+, Dnmt1V/V embryos but not in Dmap1+/−, Dnmt1V/V embryos derived from Dmap1+/−, Dnmt1V/V parents? Based on the genetic evidence, we propose that in the absence of DMAP1-DNMT1s complex, a transgenerational change in the level of DNMT1s-free DMAP1 (free DMAP1) is lethal in embryogenesis. In support of this hypothesis, we show three crosses producing Dmap1+/+, Dnmt1V/V embryos (Fig. 5C). The expected numbers of Dmap1+/+, Dnmt1V/V offspring were recovered in two crosses—one in which the parents were both Dmap1+/+ and the other in which one of the parents carried a wild-type Dnmt1+ allele. In the case of the Dmap+/+ parents, there is no change in the level of free DMAP1, whereas in the case of the cross in which one parent carries a Dnmt1+ allele, the inheritance of DMAP1-DNMT1s complex from this parent enables the embryo to survive the transgenerational change in the level of free DMAP1. In crosses between Dmap1+/−, Dnmt1V/V mice, there is no DNMT1s-DMAP1 complex and the Dmap1+/+, Dnmt1V/V embryos experience a sudden increase in free DMAP1 and do not survive. A plausible mechanism whereby free DMAP1 could lead to embryonic death is through interference with the normal preimplantation functions of DMAP1-DNMT1s and DMAP1-DNMT1o complexes. For example, free DMAP1, which is slow in forming a complex with DNMT1o, as suggested by Fig. 4, would interfere with the resident DMAP1-DNMT1o complex, possibly by occupying the sites of maintenance methylation and blocking access of preformed DMAP1-DNMT1o complex to them. If these sites include DMDs of imprinted genes, DMD methylation would be irreversibly lost, and this would show up as a loss of DMD methylation later in embryogenesis. Alternatively, free DMAP1 may have an indirect effect on DNMT1o maintenance methyltransferase activity. In this regard, free DMAP1 might influence TIP60-p400 activity in preimplantation, which in turn would lead to abnormalities in chromatin and effects on DNMT1o activity.
Associations of DMAP1 with both DNMT1s and DNMT1o suggest the possibility that the interaction with DMAP1 is needed for a DNMT1 protein's enzymatic function. In addition to the 4-fold-lower avidity of DNMT1o for DMAP1 seen in ES cells (Fig. 4), Dnmt1V/V mice have a roughly 4-fold-higher level of DNMT1o protein compared to the level of DNMT1s in the wild-type mouse (6). Thus, the concentrations of DMAP1-DNMT1 complexes would be the same in wild-type and Dnmt1V/V mice and would predict that all or most DMAP1 protein is in complex with either DNMT1s or DNMT1o. Additional support for this notion is the direct correlation between the Dmap1 allelic dose or the cellular DMAP1 concentration and the level of DNMT1s in MEFs and in spleens from mice with different Dmap1 and Dnmt1 genotypes (Fig. 5). Thus, neither DNMT1s nor DNMT1o would be expected to function independently of an interaction with DMAP1 protein. This notion could explain the observation that reduction of DMAP1 increases gene expression, possibly because of a reduction in DNA methylation (22, 26).
In addition to DMAP1, what other proteins comprise a DMAP1-DNMT1 complex, and which of these is required for enzymatic function of DNMT1 in cells? Rountree et al. (26) showed that the noncatalytic amino terminus of DNMT1s interacts with histone deacetylase 2 (HDAC2) as well as with DMAP1. They identified the interaction between DNMT1s and DMAP1 in a 2-hybrid screen using the first 125 amino acids of DNMT1s as the bait protein. Their approach to identifying DMAP1 suggested that DMAP1 interacts with DNMT1s via a domain not present in the oocyte-derived DNMT1o protein. DNMT1s and DMAP1 exist as a complex during the entirety of S phase, whereas HDAC2 is associated with the DNMT1-DMAP1 complex during late S phase. In the same study, DMAP1 was shown to have intrinsic repressive activity and was also associated with the corepressor TSG101 (26, 27). Subsequent reports added further support to the notion that DMAP1 is a corepressor (13, 16, 20). Regardless of the precise transcriptional function of DMAP1, it is clear that DMAP1 and DNMT1s directly physically interact via a DNMT1s-specific N-terminal domain. This direct interaction presumably accounts for the higher avidity of the DMAP1-DNMT1s interaction (approximately a 4-fold difference in avidity based on the results of immunoprecipitation experiments). Whether DMAP1 directly interacts with DNMT1o or whether the interaction is due simply to coexistence within a complex needs to be investigated further.
ACKNOWLEDGMENTS
We thank Judy Yanowitz for helpful comments during preparation of the manuscript.
This research was supported by a grant from the National Institutes of Health.
Footnotes
Published ahead of print on 7 March 2011.
REFERENCES
- 1. Borowczyk E., Mohan K. N., D'Aiuto L., Cirio M. C., Chaillet J. R. 2009. Identification of a region of the DNMT1 methyltransferase that regulates the maintenance of genomic imprints. Proc. Natl. Acad. Sci. U. S. A. 106:20806–20811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cai Y., et al. 2003. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J. Biol. Chem. 278:42733–42736 [DOI] [PubMed] [Google Scholar]
- 3. Chaillet J. R., Vogt T. F., Beier D. R., Leder P. 1991. Parental-specific methylation of an imprinted transgene is established during gametogenesis and progressively changes during embryogenesis. Cell 66:77–83 [DOI] [PubMed] [Google Scholar]
- 4. Cirio M. C., et al. 2008. DNA methyltransferase 1o functions during preimplantation development to preclude a profound level of epigenetic variation. Dev. Biol. 324:139–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cirio M. C., et al. 2008. Preimplantation expression of the somatic form of Dnmt1 suggests a role in the inheritance of genomic imprints. BMC Dev. Biol. 8:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ding F., Chaillet J. R. 2002. In vivo stabilization of the Dnmt1 (cytosine-5)-methyltransferase protein. Proc. Natl. Acad. Sci. U. S. A. 99:14861–14866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Doyon Y., Selleck W., Lane W. S., Tan S., Côté J. 2004. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell. Biol. 24:1884–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fazzio T. G., Huff J. T., Panning B. 2008. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell 134:162–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gorrini C., et al. 2007. Tip60 is a haplo-insufficient tumor suppressor required for an oncogene-induced DNA damage response. Nature 448:1063–1067 [DOI] [PubMed] [Google Scholar]
- 10. Herceg Z., et al. 2001. Disruption of TRRAP causes early embryonic lethality and defects in cell cycle progression. Nat. Genet. 29:206–211 [DOI] [PubMed] [Google Scholar]
- 11. Hirasawa R., et al. 2008. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 22:1607–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Howell C. Y., et al. 2001. Genomic imprinting disrupted by a maternal-effect mutation in the Dnmt1 gene. Cell 104:829–838 [DOI] [PubMed] [Google Scholar]
- 13. Kang B. G., et al. 2007. Corepressor MMTR/DMAP1 is involved in both histone deacetylase 1- and TFIIH-mediated transcriptional repression. Mol. Cell. Biol. 27:3578–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Koizumi T., et al. 2010. Depletion of Dnmt1-associated protein 1 triggers DNA damage and compromises the proliferative capacity of hematopoietic stem cells. Int. J. Hematol. 91:611–619 [DOI] [PubMed] [Google Scholar]
- 15. Lakso M., et al. 1996. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. U. S. A. 93:5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee G. E., Kim J. H., Taylor M., Muller M. T. 2010. DNA methyltransferase 1 associate protein (DMAP1) is a co-repressor that stimulates DNA methylation globally and locally at sites of double strand break repair. J. Biol. Chem. 285:37630–37640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li E. 2002. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 3:662–673 [DOI] [PubMed] [Google Scholar]
- 18. Mertineit C., et al. 1998. Sex-specific exons control DNA methyltransferase activity in mammalian germ cells. Development 125:889–897 [DOI] [PubMed] [Google Scholar]
- 19. Monk M., Boubelik M., Lehnert S. 1987. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development 99:371–382 [DOI] [PubMed] [Google Scholar]
- 20. Muromoto R., et al. 2004. Physical and functional interactions between Daxx and DNA methyltransferase 1-associated protein, DMAP1. J. Immunol. 172:2985–2993 [DOI] [PubMed] [Google Scholar]
- 21. Nagy A., Rossant J., Nagy R., Abramour-Newerly W., Roder J. C. 1993. Derivation of completely cell-culture derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A. 90:8424–8428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Negishi M., et al. 2007. Bmi1 cooperates with Dnmt1-associated protein 1 in gene silencing. Biochem. Biophys. Res. Commun. 353:992–998 [DOI] [PubMed] [Google Scholar]
- 23. Ratnam S., et al. 2002. Dynamics of Dnmt1 methyltransferase expression and intracellular localization during oogenesis and preimplantation development. Dev. Biol. 245:304–314 [DOI] [PubMed] [Google Scholar]
- 24. Reinhart B., Eljanne M., Chaillet J. R. 2002. Shared role for differentially methylated domains of imprinted genes. Mol. Cell. Biol. 22:2089–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Reinhart B., Paoloni-Giacobino A., Chaillet J. R. 2006. Specific differentially methylated domain sequences direct the maintenance of methylation at imprinted genes. Mol. Cell. Biol. 26:8347–8356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rountree M. R., Bachman K. E., Baylin S. B. 2000. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 25:269–277 [DOI] [PubMed] [Google Scholar]
- 27. Ruland J., et al. 2001. p53 accumulation, defective cell proliferation, and early embryonic lethality in mice lacking tsg101. Proc. Natl. Acad. Sci. U. S. A. 98:1859–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sapountzi V., Logan I. R., Robson C. N. 2006. Cellular functions of TIP60. Int. J. Biochem. Cell Biol. 38:1496–1509 [DOI] [PubMed] [Google Scholar]
- 29. Squatrito M., Gorrini C., Amati B. 2006. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 16:433–442 [DOI] [PubMed] [Google Scholar]