Abstract
The transcription factor Nrf2 has emerged as a master regulator of cellular redox homeostasis. As an adaptive response to oxidative stress, Nrf2 activates the transcription of a battery of genes encoding antioxidants, detoxification enzymes, and xenobiotic transporters by binding the cis-antioxidant response element in the promoter regions of genes. The magnitude and duration of inducible Nrf2 signaling is delicately controlled at multiple levels by Keap1, which targets Nrf2 for redox-sensitive ubiquitin-mediated degradation in the cytoplasm and exports Nrf2 from the nucleus. However, it is not clear how Keap1 gains access to the nucleus. In this study, we show that Keap1 is constantly shuttling between the nucleus and the cytoplasm under physiological conditions. The nuclear import of Keap1 requires its C-terminal Kelch domain and is independent of Nrf1 and Nrf2. We have determined that importin α7, also known as karyopherin α6 (KPNA6), directly interacts with the Kelch domain of Keap1. Overexpression of KPNA6 facilitates Keap1 nuclear import and attenuates Nrf2 signaling, whereas knockdown of KPNA6 slows down Keap1 nuclear import and enhances the Nrf2-mediated adaptive response induced by oxidative stress. Furthermore, KPNA6 accelerates the clearance of Nrf2 protein from the nucleus during the postinduction phase, therefore promoting restoration of the Nrf2 protein to basal levels. These findings demonstrate that KPNA6-mediated Keap1 nuclear import plays an essential role in modulating the Nrf2-dependent antioxidant response and maintaining cellular redox homeostasis.
INTRODUCTION
Maintaining cellular redox homeostasis is required for proper functioning of the cell. Oxidative stress, characterized by excessive reactive oxygen species (ROS), is associated with the toxicity of many environmental insults and the pathogenesis of age-related diseases, such as cancer and neurodegenerative disorders (12, 14, 33, 34, 45). On the other hand, prolonged activation of cellular defense systems by genetic mutations in regulatory molecules is a common strategy adapted by cancer cells to promote malignant growth against deleterious microenvironments (7, 31). Mammalian intracellular redox homeostasis is maintained mainly through transcriptional control of a battery of antioxidant genes in response to oxidative stress. This requires that the transcription of those antioxidant genes is immediately induced in the presence of excess ROS and is quickly reduced back to basal levels once cells return to redox homeostasis. As a key component of such a control system, the antioxidant response element (ARE) is a conservative cis-acting element found in the promoter region of many genes encoding antioxidants and detoxification enzymes. The corresponding trans-acting factor for the ARE is a transcription factor named nuclear factor erythroid 2-related factor 2 (Nrf2) (28, 29, 44). The Nrf2-ARE system is responsible for both basal and inducible expression of many genes involved in the antioxidant response, such as NAD(P)H quinone oxidoreductase 1 (NQO1), heme oxygenase 1 (HO-1), glutathione S-transferase A1 (GSTA1, also known as GST-Ya in mice), and glutamate-cysteine ligase (also known as γ-glutamylcysteine synthetase, i.e., γGCS) modifier subunit (GCLM) and catalytic subunit (GCLC) (1, 24, 26, 42). The importance of the Nrf2-ARE system is evident from studies demonstrating that Nrf2 knockout mice are significantly sensitive to chemical toxicants and carcinogens (2, 32).
The current model of the molecular mechanism for Nrf2 activation is that oxidative stress modifies cysteine residues of Keap1, a component of an E3 ubiquitin ligase complex that targets Nrf2 for degradation, resulting in both compromised E3 activity and enhanced Nrf2 protein stability. The subsequent elevation in Nrf2 protein levels leads to Nrf2 nuclear accumulation, increased ARE-Nrf2 binding, and transactivation of its downstream target genes (5, 8, 15, 39, 46, 48, 49). In addition to the primary mode of regulation of its protein stability, Nrf2 is also regulated by Keap1 at the level of nucleocytoplasmic trafficking (13, 27, 37, 41). Keap1 contains a strong, leucine/isoleucine-rich nuclear export signal (NES) in its central linker region. Alanine substitution of the leucine/isoleucine residues within this NES retains Keap1 in the nucleus and leads to prolonged activation of Nrf2, suggesting that Keap1-mediated nuclear export of Nrf2 is a crucial step in Nrf2 repression (13, 27, 37, 41). However, how Keap1 gains access to the nucleus remains unclear.
Protein nuclear import is accomplished by the importin β superfamily proteins that either directly or indirectly, through adaptors such as the highly related importin α proteins, interact with their cargo proteins and escort them through nuclear pore complexes (4, 35). There are 22 putative members of the importin β family in mammals. The overall sequence similarity between various importin β family members is low and is restricted to the N-terminal domain in most cases. Only minor overlap occurs among the substrate specificity of the importin β family members (36). The importin α family has undergone considerable expansion throughout the course of evolution. Whereas the genome for yeast (Saccharomyces cerevisiae) contains a single importin α, the human genome contains six genes that fall into three phylogenetically distinct groups, the α1s, α2s, and α3s, which differ from one another in about 50% of their amino acids (9). Although the importin α family as a whole exhibits a broad functional redundancy, various importin α paralogs have been shown to uniquely bind and transport certain classical nuclear localization signal (cNLS) cargos. Furthermore, they exhibit unique temporal and spatial tissue-specific patterns of gene expression during different developmental stages (18, 38).
Early studies on the mechanism of importin-mediated nuclear transport have established the mechanistic principles that govern all nucleocytoplasmic trafficking pathways. Importin β can bind directly or indirectly to cargo proteins and mediate translocation. For the classical nuclear import, the interaction between cargo protein and importin β is mediated by the adaptor molecule importin α. In the best-characterized pathway, the first step of nuclear import occurs when importin α recognizes a cellular cargo protein via NLS. The cNLS for nuclear protein transport consists of either a single cluster (monopartite) or two clusters (bipartite) of basic amino acids (40). Monopartite cNLSs are exemplified by the simian virus 40 large T antigen NLS (126PKKKRRV132), and bipartite cNLSs are exemplified by the nucleoplasmin NLS (155KRPAA-TKKAGQAKKKK170). Structural studies have revealed that importin α is composed of a large curved domain consisting of 10 armadillo (ARM) motifs, which forms the cNLS-binding pocket, and a flexible N-terminal domain or the importin β-binding domain (IBB) (9). Both domains are required for binding to importin β and cargo dissociation (21). The ARM domain creates a long groove that can accommodate either two monopartite cNLS peptides or a single bipartite cNLS peptide (3). On the other hand, the IBB domain of importin α is believed to coordinate both the assembly and disassembly of the ternary cargo-importin α/β complex, primarily by controlling the access of cNLS peptides to the binding groove. When importin α is not bound to importin β, the autoinhibitory sequence within the IBB domain mimics cNLS and interacts with the NLS-binding pocket. Binding of importin β causes the removal of the autoinhibitory segment from the cNLS-binding site, thus converting importin α to a conformation that has a high affinity for cNLSs in cargo proteins (17).
Active nuclear import of cargo protein is initiated with the formation of the ternary cargo-importin α/β complex. Importin α provides the NLS-binding site for cargo proteins and interacts via its IBB domain with importin β. Importin β in turn interacts with components of the nuclear pore complex known as nucleoporins. Once in the nucleus, dissociation of the import complex is mediated by the small GTP Ran, which binds to importin β, displaces importin α, and releases the cargo protein. This autoinhibitory function of the IBB domain leads to the release of the cargo protein from importin α. After disassembly of the ternary cargo-importin α/β complex, the importins are recycled to the cytoplasm. It is hypothesized that the Ran GTP-importin β is returned directly to the cytoplasm, whereas importin α gets exported with the aid of an exportin, CAS (9, 23, 40).
In the present study, we identified an alpha karyopherin, KPNA6 (also known as importin α7), a nucleocytoplasmic transport adaptor, as a specific nuclear import adaptor protein for Keap1. Here, we show that Keap1 constitutively shuttles between the nucleus and the cytoplasm under basal conditions. The nuclear import of Keap1 is independent from either Nrf1 or Nrf2. Screening for Keap1-binding karyopherins led to the identification of KPNA6. KPNA6 interacts with the C-terminal Kelch domain of Keap1 and promotes its nuclear import. Knockdown of KPNA6 compromised Keap1 nuclear shuttling and inhibited Keap1-mediated ubiquitination of Nrf2, leading to enhanced Nrf2 activation upon exposure to oxidative stress. On the other hand, overexpression of KPNA6 facilitated Keap1 nuclear import and attenuated inducible Nrf2 signaling. These findings established that KPNA6 is a modulator of the cells' response to oxidative stress.
MATERIALS AND METHODS
Construction of recombinant DNA molecules.
Plasmids expressing wild-type Keap1-CBD (chitin binding domain), a Keap1-NES mutant (L301A I304A L308A L310A), Keap1 domain deletion mutants, and hemagglutinin (HA)-Nrf2 proteins have been previously described (37, 47). To make retroviral vectors for stable cell line construction, full-length Keap1 wild type or the NES mutant were first cloned into BamHI/XcmI sites of pEGFP-N3 vector (Clontech), and cDNAs encoding the Keap1-GFP (green fluorescent protein) fusion proteins were further subcloned into BamHI/SalI sites of the pBabe-puro retroviral vector. GFP-tagged Keap1 truncations were generated by inserting PCR products for Keap1 truncations into the XhoI/BamHI sites of the pEGFP-C3 vector (Clontech). cDNAs for human KPNA6, KPNA5, KPNA4, KPNA2, KPNA1, IPO11, and IPO13 were purchased from Open Biosystems. Myc-tagged KPNA6 was generated by inserting PCR products for KPNA6 into the BamHI/EcoRI sites of the pcDNA3.1-Myc/His vector (Invitrogen). GST-tagged KPNA6-ΔIBB was generated by inserting the PCR product for KPNA6 (L108 to the C-terminal end) into the SmaI/XhoI sites of the pGEX-5X-3 vector (Pharmacia). Keap1 mutants containing alanine substitutions on positively charged clusters were generated by site-directed mutagenesis using the PCR-and-DpnI-based method.
Cell culture, transfection, RNA interference, and establishment of stable cell line.
NIH 3T3, HEK293T, and MDA-MB-231 cells were purchased from American Type Culture Collection (ATCC). Keap1−/− and Keap1+/+ mouse embryonic fibroblast (MEF) cells were generous gifts from Masayuki Yamamoto (Tohoku University, Japan). The Nrf1−/− Nrf2−/− MEF cells were kindly provided by Jefferson Chan (University of California, Irvine). Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) in the presence of 10% fetal bovine serum (FBS). Transfections of plasmid DNA were performed with Lipofectamine Plus reagent (Invitrogen) according to the manufacturer's instructions. Small interfering RNA (siRNA) against KPNA6 and scrambled control siRNA were purchased from Invitrogen. The sequence of the siRNA against human KPNA6 was CAG CCC UAC CUU GCC UUC UCC ACU U (Invitrogen catalog numbers 10620318 and 10620319). Transfection of siRNA was performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. A concentration of 80 pmol siRNA was used for a 35-mm dish of cells. For cotransfection of DNA and siRNA with Lipofectamine 2000, 40 pmol siRNA and 0.5 μg DNA were used for a 35-mm dish of cells. To establish stable cell lines harboring Keap1-CBD, the pBabe-puro retroviral vectors were transfected into SD-3443, a packaging cell line from ATCC. Viruses produced in the supernatant were collected and used to infect MEF cells in the presence of 3 μg/ml Polybrene. At 48 h postinfection, cells were put in selection medium containing 3 μg/ml puromycin. Stable cell lines were established once all the cells in the negative-control plate were killed. Stable cell lines were continuously grown in medium containing 3 μg/ml of puromycin.
Antibodies, immunoprecipitation, and immunoblot analysis.
The rabbit anti-Nrf2 (Santa Cruz), goat anti-Keap1 (Santa Cruz), rabbit anti-KPNA6 (Sigma), anti-lamin A (Santa Cruz), antitubulin (Santa Cruz), anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase; Santa Cruz), anti-NQO1 (Santa Cruz), anti-HO-1 (Santa Cruz), anti-Myc epitope (Santa Cruz), anti-HA epitope (Santa Cruz), mouse antiubiquitin (Sigma), and anti-CBD (New England BioLabs) were purchased from commercial sources. The rabbit anti-Keap1 antibody was generated in rabbits challenged with purified full-length His-tagged human Keap1 protein, followed by purification on a column with immobilized His-Keap1 protein (Pierce). To detect protein expression in total cell lysates, cells were lysed in sample buffer (50 mM Tris-HCl [pH 6.8], 2% SDS, 10% glycerol, 100 mM dithiothreitol [DTT], 0.1% bromophenol blue) 48 h following transfection. For immunoprecipitation assays, cells were lysed in radioimmunoprecipitation (RIPA) buffer (10 mM sodium phosphate [pH 8.0], 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS) containing 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride (PMSF), and a protease inhibitor cocktail (Sigma). Cell lysates were precleared with protein A beads and incubated with 2 μg of the affinity-purified antibody for 2 h at 4°C, followed by incubation at 4°C with protein A-agarose beads for 2 h. After six washes with RIPA buffer, immunoprecipitated complexes were eluted in sample buffer by boiling in water for 4 min, electrophoresed through SDS-polyacrylamide gels, and subjected to immunoblot analysis.
Immunofluorescence staining, live-cell imaging, and image analysis.
For immunofluorescence staining, NIH 3T3 cells were grown on glass coverslips in 35-mm plates. Cells were fixed in prechilled methanol at −20°C for 1 h and incubated at room temperature for 50 min with primary antibodies diluted 1:100 in phosphate-buffered saline (PBS) containing 10% FBS. The glass coverslips were washed in PBS and incubated at room temperature for 50 min with secondary antibody Alexa Fluor 488-conjugated anti-mouse IgG or Alexa Fluor 594-conjugated anti-rabbit IgG (Invitrogen) at 1:100 dilution in PBS containing 10% FBS and 1% Hoechst stain. The glass coverslips were washed in PBS and mounted with ProLong gold antifade reagent (Invitrogen) on glass slides. Both stained slides and living cells expressing GFP-tagged proteins were imaged using a 3i Marianas Ziess Observer Z1 system and Slidebook software (Intelligent Imaging Innovations). The images were exported from Slidebook as tiff files. Adobe Photoshop was used to construct the figures. Minimal alterations were performed on the digital images. Slidebook software was also used for image quantification analysis. The mean fluorescence intensities in the nucleus and cytoplasm were individually quantified within each cell, and a total of 20 cells were analyzed for each sample. The mean nucleus-to-cytoplasm ratio from 20 cells and statistically significant differences between experimental groups were evaluated with the Student t test. A P value of less than 0.05 was considered statistically significant.
Subcellular fractionation.
A nuclear extraction kit (catalog number 40010; Active Motif) was used to obtain nuclear and cytoplasmic subcellular fractions according to the manufacturer's instructions, with minor modifications. MDA-MB-231 cells in 60-mm dishes were trypsinized and collected. After two washes with PBS containing phosphatase inhibitors, cell pellets were resuspended in the 1× hypotonic buffer and incubated on ice for 15 min to allow cells to swell. To the swollen cells in lysis buffer, NP-40 was added to a final concentration of 0.2%, and then the cells were vortexed vigorously for 10 s, followed by immediate centrifugation for 1 min at 6,000 rpm. The pellets were collected in sample buffer as the nuclear extract. The supernatant was further purified by centrifugation at 14,000 rpm for 20 min and collected as the cytoplasmic extract.
Protein radiolabeling and in vitro binding assay.
Importin family proteins and truncated forms of Keap1 were radiolabeled with [35S]methionine using the in vitro TNT transcription/translation system (Promega). His-tagged Keap1 and GST-tagged KPNA6 proteins were expressed in E. coli Rosetta (DE3) LysS cells and purified with a Ni-nitrilotriacetic acid (NTA) agarose column (Qiagen) and glutathione Sepharose 4B matrix (Amersham Biosciences), respectively. For the in vitro binding assay, radiolabeled proteins and purified proteins were incubated in binding buffer (4.2 mM Na2HPO4, 2 mM KHPO4, 140 mM NaCl, 10 mM KCl, 0.2% bovine serum albumin [BSA], 0.02% Triton 100, 1 mM DTT) in the presence of Ni-NTA agarose beads or Sepharose beads at 4°C for 4 to 6 h. The beads were then washed six times with binding buffer without BSA. The proteins were eluted by boiling in SDS sample buffer, followed by SDS-PAGE and autoradiography analysis.
mRNA extraction and qRT-PCR.
Total mRNA was extracted from cells using TRI Reagent (Sigma). Equal amounts of RNA were used for reverse transcription using a Transcriptor First-Strand cDNA synthesis kit (Roche). The following TaqMan probes from the universal probe library were used (Roche): Nrf2 (no. 70), NQO1 (no. 87), HO-1 (no. 25), GCLM (no. 18), KPNA6 (no. 26), and GAPDH (no. 25). The following primers were synthesized by Integrated DNA Technologies: Nrf2, forward (ACACGGTCCACAGCTCATC) and reverse (TGTCAATCAAATCCATGTCCTG); NQO1, forward (ATGTATGACAAAGGACCCTTCC) and reverse (TCCCTTGCAGAGAGTACATGG); HO-1, forward (AACTTTCAGAAGGGCCAGGT) and reverse (CTGGGCTCTCCTTGTTGC); GCLM, forward (GACAAAACACAGTTGGAACAGC) and reverse (CAGTCAAATCTGGTGGCATC); KPNA6, forward (GAGGAACCCCTGAGCAGAT) and reverse (AGCAAGTCACATAGGGGTTTG); and GAPDH, forward (CTGACTTCAACAGCGACACC) and reverse (TGCTGTAGCCAAATTCGTTGT). Quantitative real-time PCR (qRT-PCR) was performed on the LightCycler 480 system (Roche) as follows: one cycle of initial denaturation (95°C for 4 min), 45 cycles of amplification (95°C for 10 s and 60°C for 30 s), and a cooling period. The data presented are relative mRNA levels normalized to the level of GAPDH, and the value from the undifferentiated cells was set as 1. PCR assays were performed two times with duplicate samples, which were used to determine the means ± standard deviations. The Student t test was used to evaluate statistically significant differences between two samples.
Ubiquitination assay.
To detect ubiquitinated endogenous Nrf2, cells were exposed to 10 μM MG132 (Sigma) for 4 h. Cells were lysed by boiling in a buffer containing 2% SDS, 150 mM NaCl, 10 mM Tris-HCl, and 1 mM DTT. This rapid lysis procedure inactivated cellular ubiquitin hydrolases to preserve ubiquitin-Nrf2 conjugates present in cells prior to lysis. Protein-protein interactions, including the association of Nrf2 with Keap1, were also disrupted by this lysis procedure. For immunoprecipitation, these lysates were diluted 5-fold in buffer lacking SDS and incubated with an anti-Nrf2 antibody. Immunoprecipitated proteins were analyzed by immunoblotting with antibodies directed against ubiquitin.
Protein half-life measurement.
To measure the half-life of Nrf2, 50 μM cycloheximide (Sigma) was added to block protein synthesis in MDA-MB-231 cells. Total cell lysates were collected at different time points and subjected to immunoblot analysis with an anti-Nrf2 antibody. The relative intensities of the bands were quantified by using the ChemiDoc CRS gel documentation system and Quantity One software from Bio-Rad (Hercules, CA).
Fluorescence recovery after photobleaching (FRAP) microscopy experiment.
Keap1-GFP knock-in cells were cultured in Delta T glass-bottom culture dishes (Bioptechs, Inc.) using phenol red-free medium and were transfected with control siRNA or siRNA against KPNA6 as described above. Before imaging, cells were treated with 5 nM leptomycin (LMB) for predetermined times to ensure equal nuclear/cytosolic distribution of Keap1. The dishes were mounted in a Bioptechs Delta T stage insert to maintain the cells at 37°C and secured to the stage of an upright Zeiss LSM 510 meta confocal microscope (Carl Zeiss, Inc.). Experiments were performed using a 40×, numeric aperture 1.0, Zeiss W Plan-Apochromat “dipping” objective. For excitation of GFP, the 488-nm Ar laser was used, and a 505-nm long-pass filter was used to capture the emission. Pre- and postbleach images were captured at low laser powers (5%), and the photobleaching was done at 100% power (7.1 Å). Experiments were conducted using images of 512 by 512 pixels at 2-times zoom and an optical slice thickness of 8 μm. Each image acquisition required 393 ms, and no additional delay was added between image acquisitions. Nuclei were photobleached using regions of interest to target the 15 rapid photobleach exposures. An unbleached cell was included in each frame as a control. The percentage of fluorescence recovery was as described by Daelemans et al. (6).
RESULTS
Keap1 is constantly shuttling between the nucleus and the cytoplasm under basal conditions.
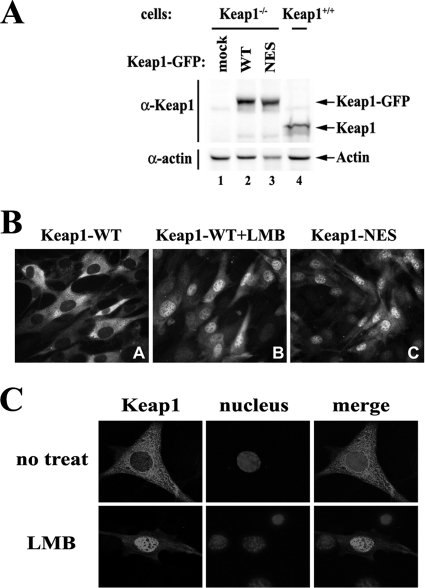
In order to test whether Keap1 shuttles between the nucleus and the cytoplasm in living cells, stable cell lines expressing GFP-tagged Keap1 wild type or NES mutant were constructed. The expression of Keap1-GFP fusion proteins and endogenous Keap1 was confirmed by Western blotting (Fig. 1 A). Blocking Keap1 nuclear export, either by leptomycin (LMB) or by mutating the NES, led to nuclear accumulation of Keap1-GFP fusion proteins in living cells (Fig. 1B). These data suggest that Keap1 is constitutively shuttling between the nucleus and the cytoplasm under basal conditions.
Fig. 1.
The nuclear import of Keap1 occurs at its physiological protein level and is not dependent on Nrf1 or Nrf2. (A) GFP-tagged Keap1 proteins were expressed at levels similar to the levels of endogenous Keap1 in the stable MEF cell lines. Keap1−/− MEF cells were infected with pBabe-puro retroviral vectors expressing GFP-tagged Keap1 wild type (WT) or a mutant containing mutations in the nuclear export signal (NES). After puromycin selection, cells were harvested for immunoblot analysis with antibodies against Keap1 and β-actin. (B) Nuclear accumulation of Keap1-GFP fusion proteins. Stable Keap1−/− MEF cells expressing Keap1-GFP were either left untreated or treated with 5 nM LMB for 4 h, followed by live-cell imaging of GFP fluorescence. (C) The nuclear import of Keap1 is not dependent on Nrf1 or Nrf2. Nrf1−/− Nrf2−/− MEF cells were transfected with the expression vector for Keap1. Cells were treated with 5 nM LMB for 2 h, followed by indirect immunofluorescence staining with a rabbit anti-Keap1 antibody.
Nuclear import of Keap1 is independent of Nrf1 or Nrf2.
It was assumed that Keap1 relies on Nrf2 for nuclear access, since no nuclear localization signal (NLS) has been identified in Keap1. We have previously shown that Keap1 is still able to travel into the nucleus in Nrf2−/− cells, suggesting the existence of an Nrf2-independent mechanism that allows Keap1 to localize in the nucleus (37), which is a prerequisite for Keap1-mediated Nrf2 nuclear export to occur. Another candidate that could potentially mediate Keap1 nuclear import is Nrf1, another member of the NF-E2 family. Nrf1 contains the conserved Keap1-binding motifs (DLG motif and ETGE motif) on its N-terminal Neh2 domain and is able to associate with Keap1, although Keap1 does not seem to regulate the transcriptional activity of Nrf1 (43, 50). When Nrf1−/− Nrf2−/− MEF cells were transfected with Keap-GFP, Keap1 still accumulated in the nucleus after LMB treatment (Fig. 1C), suggesting that Keap1 does not rely on Nrf1 or Nrf2 for its nuclear entry.
The C-terminal Kelch domain of Keap1 mediates its nuclear entry.
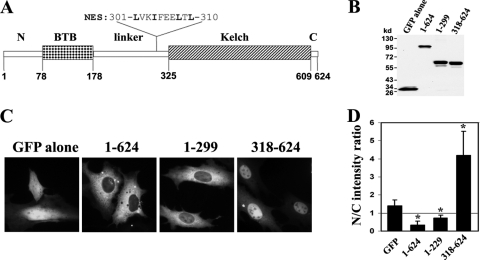
Human Keap1 protein is comprised of 624 amino acids that can be divided into an N-terminal BTB (bric a brac, tramtrack, broad complex) domain, a C-terminal Kelch domain, and a central linker region in between (Fig. 2 A). The hydrophobic NES is located at the C-terminal end of the linker region (amino acids 301 to 310) (Fig. 2A). To determine which part of Keap1 is responsible for its nuclear import, two deletion mutants that do not contain the NES were constructed (amino acids 1 to 299 and 318 to 624). The expression of Keap1-GFP fusion proteins was confirmed by immunoblot analysis (Fig. 2B). When GFP-tagged Keap1 truncations were expressed in living cells, it was apparent that the C-terminal Kelch domain was required and sufficient for Keap1 nuclear entry (Fig. 2C and D). Further deletion within the Kelch domain seemed impossible, because deletion of part of the Kelch domain renders the mutant proteins unstable. Therefore, we were unable to further define the motifs or residues that mediate Keap1 nuclear import.
Fig. 2.
The C-terminal Kelch domain of Keap1 mediates its nuclear entry. (A) Schematic of conserved domains in human Keap1 protein. Keap1 contains an N-terminal BTB domain, a C-terminal Kelch domain, and a linker region in between the two domains. The nuclear export signal (NES) is located in the linker region as indicated. Amino acids in boldface were mutated for the NES mutant. (B) The expression of GFP-tagged Keap1 mutant proteins (amino acid numbers as indicated) was analyzed by immunoblotting with an anti-GFP antibody. (C) NIH 3T3 cells were transfected with expression vectors for GFP-tagged full-length or truncated Keap1. The GFP fluorescence was examined in living cells 48 h after transfection. (D) Fluorescence images were analyzed with Slidebook 4.2 software (Intelligent Imaging Innovations, Inc.). The mean fluorescence intensities in the nucleus and cytoplasm were individually quantified within each cell, and a total of 20 cells were analyzed for each sample. The mean nucleus-to-cytoplasm (N/C) ratios were calculated and plotted. The error bars indicate standard deviations (n = 20). Asterisks indicate significant differences from the results for GFP alone (P < 0.05).
KPNA6 interacts with the Kelch domain of Keap1.
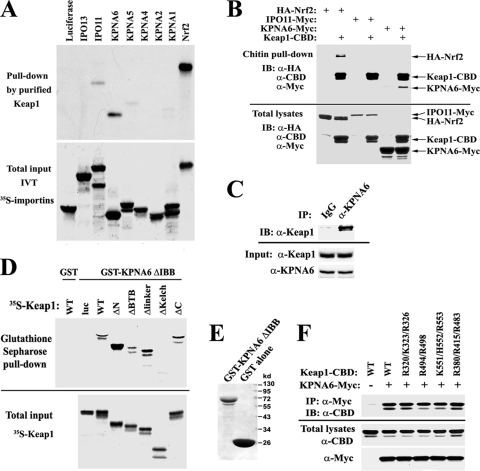
Since Keap1 does not rely on Nrf1 or Nrf2 for its nuclear entry, we hypothesized that Keap1 associates directly with one or more of the importin proteins that is/are immediately responsible for Keap1 nuclear entry. In an effort to identify such importin(s), an array of in vitro-translated [35S]methionine-labeled importin proteins was incubated with purified Keap1 proteins. Both KPNA6 and, to a lesser degree, IPO11, as well as Nrf2 (positive control), were pulled down by Keap1, while the negative-control luciferase was not (Fig. 3 A). To confirm the interaction in vivo, HEK293T cells were transfected with an expression vector for Myc-tagged IPO11, Myc-tagged KPNA6, or HA-tagged Nrf2 (positive control) along with or without an expression vector for CBD-tagged Keap1. KPNA6 but not IPO11 was pulled down by Keap1 (Fig. 3B). To further confirm the interaction between endogenous KPNA6 and Keap1 proteins, HEK293T cell lysates were collected in RIPA buffer containing 0.1% SDS, followed by immunoprecipitation with an anti-KPNA6 antibody or normal rabbit IgG (negative control). KPNA6-bound Keap1 was readily detected by immunoblot analysis with the anti-Keap1 antibody (Fig. 3C).
Fig. 3.
KPNA6 interacts with the Kelch domain of Keap1. (A) KPNA6 binds Keap1 in vitro. Purified His-tagged Keap1 proteins were incubated with the indicated in vitro-translated (IVT) 35S-labeled proteins of the importin family, followed by pulldown with Ni-NTA agarose beads. The Keap1-bound proteins were visualized by autoradiography. Nrf2 served as a positive control for Keap1 binding, and luciferase was a negative control. Human importins are labeled by their official names. Their corresponding common names are as follow: IPO13, importin 13; IPO11, importin 11; KPNA6, importin α7; KPNA5, importin α6; KPNA4, importin α3; KPNA2, importin α1 or α2; and KPNA1, importin α5. (B) KPNA6 binds Keap1 in vivo. HEK293T cells were transfected with an expression vector for HA-tagged Nrf2, Myc-tagged IPO11, or Myc-tagged KPNA6 with or without an expression vector for CBD (chitin binding domain)-tagged Keap1. Cell lysates were collected in RIPA buffer containing 0.1% SDS and were subjected to pulldown by chitin beads, followed by immunoblot (IB) analysis with anti-HA, anti-Myc, and anti-CBD antibodies. (C) Endogenous KPNA6 interacts with Keap1. HEK293T cell lysates were collected in RIPA buffer containing 0.1% SDS, followed by immunoprecipitation (IP) with a rabbit anti-KPNA6 antibody and immunoblotting with a goat anti-Keap1 antibody. (D) Kelch domain in Keap1 interacts with KPNA6. 35S-labeled Keap1 wild type (WT) and the indicated deletion mutants were incubated with purified GST alone or GST-tagged KPNA6 with its N-terminal importin β-binding domain deleted (ΔIBB). The KPNA6-bound Keap1 was pulled down by glutathione Sepharose beads and visualized by autoradiography. 35S-labeled luciferase (luc) and GST alone were included as negative controls. ΔN, N-terminal deletion mutant; ΔC, C-terminal deletion mutant. (E) Purified GST-tagged KPNA6-ΔIBB and GST alone were visualized by Coomassie stain. (F) The interaction with KPNA6 is not mediated by positively charged primary motifs. HEK293T cells were cotransfected with expression vectors for Myc-tagged KPNA6 and CBD-tagged Keap1 bearing alanine substitutions for the indicated positively charged residues. Cell lysates were subjected to immunoprecipitation with an anti-Myc antibody and immunoblotted with an anti-CBD antibody.
To map the interaction domains in Keap1 and KPNA6, a truncated form of KPNA6 was constructed with its N-terminal IBB domain (amino acids 1 to 107) deleted (KPNA6-ΔIBB). Both GST itself and GST-tagged KPNA6-ΔIBB were expressed and purified from bacteria (Fig. 3E). GST-KPNA6-ΔIBB retains the ability to bind 35S-labeled Keap1 in an in vitro binding assay, suggesting that the C-terminal ARM domain of KPNA6 mediates the interaction with Keap1 (Fig. 3D). Binding analysis using Keap1 proteins with deletions of each individual domain showed that the Kelch domain is required for the interaction with KPNA6 (Fig. 3D). As negative controls, GST alone did not pull down 35S-labeled Keap1, and GST-KPNA6 did not pull down 35S-labeled luciferase, demonstrating the specificity of the interactions (Fig. 3D). These data suggest that the Kelch domain (325 to 609) of Keap1 associates with the C-terminal ARM domain of KPNA6 (amino acids 108 to 536).
Attempts to further map the KPNA6-interacting sites within the Keap1 Kelch domain were futile. The Kelch domain of Keap1 forms a ring-like 6-bladed propeller structure (22), and therefore, truncations or deletions within the Kelch domain tend to disrupt the overall structural integrity. In the classical mode of interaction between importin α and its cargo substrate, the ARM domain binds to clusters of positively charged residues in the cargo protein. The Keap1 Kelch domain contains several positively charged clusters composed of multiple arginine (R), lysine (K), or histidine (H) residues. Alanine substitutions for positive residues in several clusters were constructed, and the resultant mutants were tested for their ability to associate with KPNA6. However, none of them showed loss of interaction (Fig. 3F), suggesting that either we did not find the right clusters or the interaction between KPNA6 and Keap1 may be different from the classical mode. Further, it is worth noting that mutations of the three key arginines (R380, R415, and R483) within the Nrf2 binding pocket of Keap1 had no effect on its interaction with KPNA6, suggesting that KPNA6 and Nrf2 bind different residues in the Kelch domain of Keap1 (Fig. 3F).
Overexpression of KPNA6 facilitates nuclear import of Keap1.
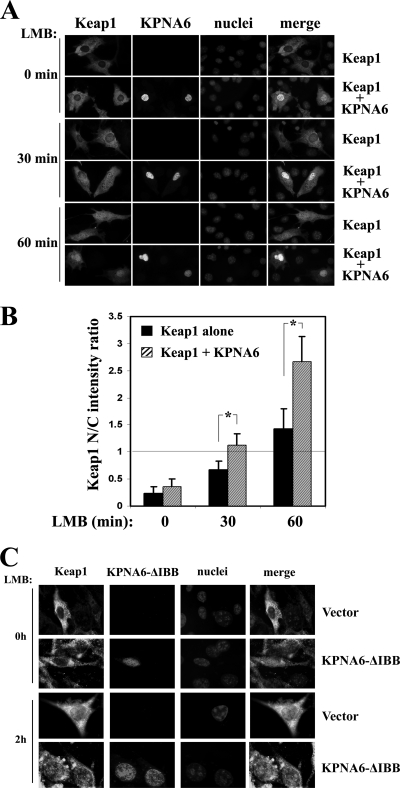
NIH 3T3 cells were transfected with an expression vector for Keap1, along with an empty vector or an expression vector for Myc-KPNA6. Cells were treated with 5 nM LMB for different time periods, fixed, and subjected to double indirect immunofluorescence staining with a rabbit anti-Keap1 antibody and a mouse anti-Myc antibody. Nuclear accumulation of Keap1 was significantly enhanced in cells overexpressing KPNA6 and treated with LMB at both the 30- and 60-min time points (Fig. 4 A and B). In contrast, Myc-KPNA6-ΔIBB had a decreased ability to mediate nuclear import of Keap1 (Fig. 4C), possibly due to loss of interaction between importin α and importin β so that cargo protein could not be transported into the nucleus. These data suggest that overexpression of full-length KPNA6 is sufficient to accelerate Keap1 nuclear import.
Fig. 4.
Overexpression of KPNA6 facilitates nuclear import of Keap1. (A) NIH 3T3 cells were transfected with an expression vector for Keap1 with or without a vector for Myc-tagged KPNA6. Cells were treated with 5 nM LMB for the indicated times. After fixation in methanol, indirect immunofluorescence staining was done with rabbit anti-Keap1 and mouse anti-Myc antibodies. (B) The fluorescence images of Keap1 were analyzed as described for Fig. 2D. (C) Keap1-GFP knock-in cells were transfected with empty vector or an expression vector for Myc-KPNA6-ΔIBB. Cells were treated with 5 nM LMB for the indicated times. After fixation in methanol, indirect immunofluorescence staining was done with mouse anti-GFP and rabbit anti-Myc antibodies.
Overexpression of KPNA6 attenuates the inducible Nrf2 signaling in response to oxidative stress.
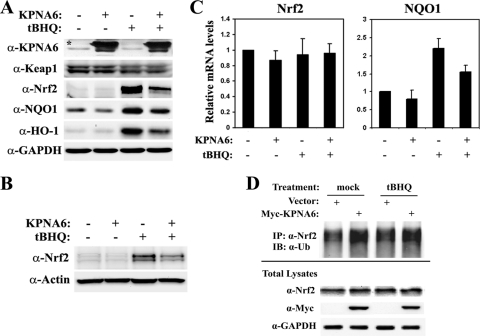
To test whether a change in Keap1 nuclear-shuttling dynamics caused by KPNA6 overexpression has any function in modulating the Nrf2-dependent antioxidant response, HEK293T cells were transfected with either an empty vector or an expression vector for KPNA6. Cells were then either left untreated or treated with tert-butylhydroquinone (tBHQ), a classical Nrf2 activator. The protein levels of Nrf2 and its downstream target genes, encoding NQO1 and HO-1, were significantly decreased in the presence of overexpressed KPNA6 (Fig. 5 A). Keap1 protein levels were not changed by KPNA6 overexpression (Fig. 5A) but were slightly decreased upon tBHQ treatment, which is consistent with a previous report (49). The blunted Nrf2 activation in the presence of overexpressed KPNA6 was further confirmed in MDA-MB-231 cells (Fig. 5B), arguing that the observed effects are not restricted to specific cell lines. qRT-PCR analysis showed that KPNA6 did not change Nrf2 mRNA levels (Fig. 5C), implying that KPNA6 modulates Nrf2 at the protein level by changing Keap1 nucleocytoplasmic shuttling dynamics. In order to assess the effect of KPNA6 overexpression on Nrf2 ubiquitination and degradation, MDA-MB-231 cells transfected with control vector or Myc-KPNA6 were treated with tBHQ and a proteasome inhibitor, MG132, before cells were lysed. Endogenous Nrf2 proteins were immunoprecipitated, and ubiquitination was detected by immunoblot analysis with an antiubiquitin antibody. In the presence of KPNA6, ubiquitination of Nrf2 increased under both basal and induced conditions, suggesting that overexpression of KPNA6 could repress the Nrf2-dependent antioxidant response by promoting Nrf2 ubiquitination and degradation (Fig. 5D).
Fig. 5.
Overexpression of KPNA6 attenuates the inducible Nrf2 signaling in response to oxidative stress. (A) KPNA6 decreases the inducible Nrf2 protein level and the expression of its downstream detoxification genes without altering Keap1 protein levels. HEK293T cells were transfected with either an empty vector or an expression vector for Myc-tagged KPNA6. Cells were either left untreated or treated with 50 μM tBHQ overnight before being harvested and used for immunoblot analysis with the indicated antibodies. The asterisk indicates endogenous KPNA6. (B) The same experiment in MDA-MB-231 cells gives similar results. (C) KPNA6 reduces the transcription of Nrf2 target genes without changing Nrf2 mRNA levels. HEK293T cells were transfected and treated as described above. Cells were harvested for mRNA extraction followed by qRT-PCR. Error bars indicate standard deviations of the results of two independent experiments. (D) MDA-MB-231 cells were transfected with an empty vector or an expression vector for Myc-KPNA6 as described in the text. At 48 h after transfection, cells were cotreated with 100 μM tBHQ and 10 μM MG132 for 4 h and harvested in denaturing conditions. Cell lysates were diluted and subjected to immunoprecipitation with an anti-Nrf2 antibody, followed by immunoblotting with an antiubiquitin antibody.
Knockdown of KPNA6 inhibits nuclear import of Keap1.
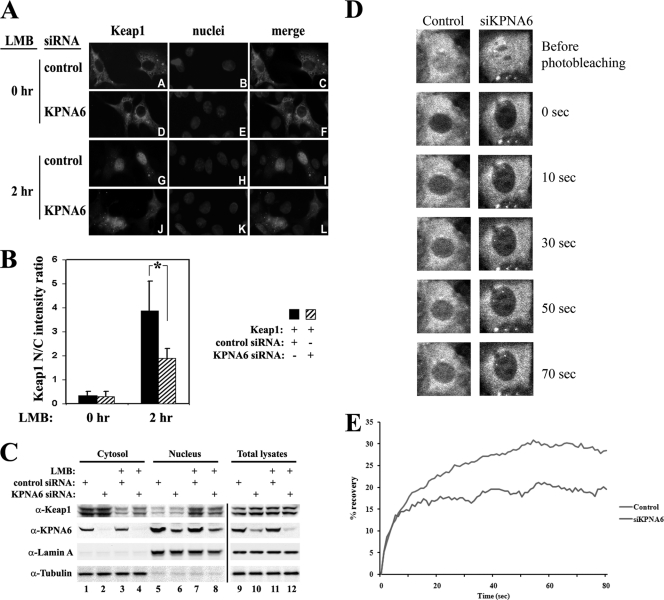
So far, we have demonstrated that overexpression of KPNA6 is sufficient to repress Nrf2 activity through accelerating Keap1 nucleocytoplasmic shuttling and the export of Nrf2 from the nucleus (37). Next, to determine if KPNA6 is necessary for Keap1 nuclear entry and Nrf2 repression, MDA-MB-231 cells were cotransfected with either scrambled control siRNA or KPNA6 siRNA, along with an expression vector for Keap1. The nuclear import of Keap1 was significantly hindered in the presence of KPNA6 siRNA, as shown by immunofluorescence staining of Keap1 at 2 h after LMB treatment (Fig. 6 A and B). The requirement of KPNA6 for endogenous Keap1 nuclear entry was further demonstrated by subcellular fractionation analysis. MDA-MB-231 cells were transfected with either scrambled control siRNA or KPNA6 siRNA. Cells were harvested and fractionated into cytosolic and nuclear fractions. The nuclear accumulation of endogenous Keap1 at 2 h after LMB treatment was decreased when KPNA6 was knocked down (Fig. 6C, compare lane 3 to 4 and lane 7 to 8). There was still a considerable amount of Keap1 switching from the cytosol to the nucleus after LMB treatment even in the presence of KPNA6 siRNA (Fig. 6C, compare lane 2 to 4 and lane 6 to 8), which was probably due to either the existence of a KPNA6-independent mechanism or insufficient knockdown of KPNA6. Additionally, fluorescence recovery after photobleaching (FRAP) experiments were carried out in Keap1-null cells expressing the GFP-tagged Keap1. Knockdown of KPNA6 was sufficient to decrease nuclear accumulation of Keap1 compared to the control group (Fig. 6D and E), confirming our results shown in Fig. 6A. Collectively, these results demonstrate that KPNA6 is required for efficient Keap1 nuclear entry.
Fig. 6.
Knockdown of KPNA6 inhibits nuclear import of Keap1. (A) NIH 3T3 cells were cotransfected with either scrambled control siRNA or KPNA6 siRNA, along with an expression vector for Keap1. At 48 h after transfection, cells were treated with 5 nM LMB for the indicated times. After fixation in methanol, indirect immunofluorescence staining was done using a rabbit anti-Keap1 antibody. (B) The fluorescence images of Keap1 were analyzed as described for Fig. 2D. (C) MDA-MB-231 cells were transfected with either scrambled control siRNA or siRNA against KPNA6. At 48 h after transfection, cells were either left untreated or treated with 5 nM LMB for 2 h. Cells were harvested and fractionated into cytosolic and nuclear fractions using a nuclear extraction kit. Both fractions and total lysates were analyzed by immunoblotting with the indicated antibodies. Lamin A and tubulin served as markers for the nuclear and cytosolic fractions, respectively. (D) Representative postbleach images of Keap1-GFP in control and KPNA6 knockdown groups at indicated time points during a 2-min time course. Keap1-GFP knock-in cells were transfected with either control siRNA or siRNA against KPNA6 (siKPNA6). Cells were treated with 5 nM LMB for predetermined times to ensure equal nuclear/cytosolic distribution of Keap1 before being subjected to FRAP. (E) Quantitative analyses of FRAP results demonstrate that the nuclear import of Keap1 is slower in KPNA6 knockdown cells than in the control group. The fluorescence intensities of control and KPNA6 knockdown cells were obtained by briefly photobleaching the nucleus and monitoring the recovery of fluorescence within an interval of 393 ms. The intensity of fluorescence at each time point after photobleaching was calculated and plotted.
KPNA6 modulates the antioxidant response by promoting ubiquitination and degradation of Nrf2.
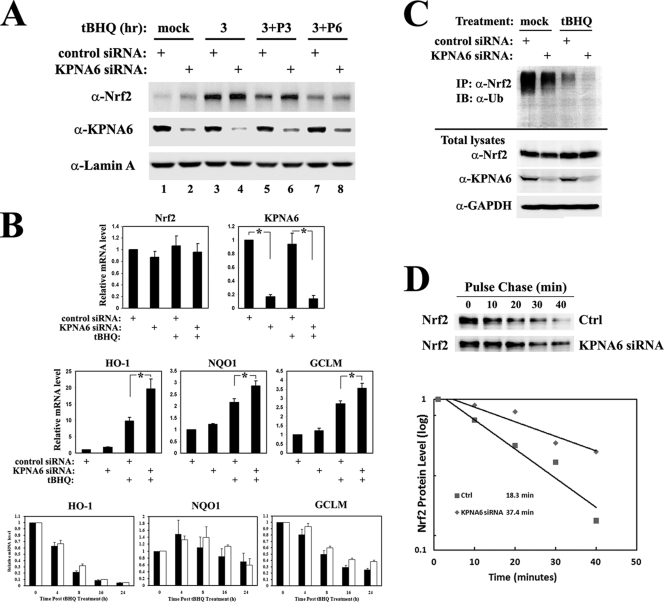
To test whether KPNA6 is required for efficient repression of nuclear Nrf2 protein levels during the induction and postinduction phases of the antioxidant response, MDA-MB-231 cells were transfected with KPNA6 siRNA, followed by a 3-h pulse treatment with tBHQ. After the removal of tBHQ, cells were further incubated in normal medium for 3 h (Fig. 7 A, 3+P3) or for 6 h (Fig. 7A, 3+P6). Nuclear fractions were extracted for immunoblot analysis. The increase of endogenous nuclear Nrf2 protein levels was readily detectable in both the induction and early postinduction phases when KPNA6 was knocked down (Fig. 7A, compare lane 3 to 4 and lane 5 to 6). The downstream effect of the elevated nuclear Nrf2 proteins was confirmed using qRT-PCR analysis (Fig. 7B). The inducible transcription of Nrf2 target genes, encoding NQO1, HO-1 and GCLM, was significantly enhanced when KPNA6 was efficiently knocked down (Fig. 7B, middle panel). The recovery time course of Nrf2 target gene expression was also examined, and the duration of target gene activation was extended when KPNA6 was knocked down in cells (Fig. 7B, lower panel). Nrf2 mRNA levels did not change upon knockdown of KPNA6 (Fig. 7B, upper panel), arguing that the observed increase in nuclear Nrf2 protein levels is probably due to a decrease in nuclear import of Keap1, thus impairing Nrf2 export and its subsequent ubiquitination and degradation in the cytoplasm. Next, MDA-MB-231 cells transfected with control siRNA or KPNA6 siRNA were treated with tBHQ and MG132 for 4 h before cells were lysed. Endogenous Nrf2 proteins were immunoprecipitated, and ubiquitination was detected by immunoblotting with an antiubiquitin antibody. Ubiquitination of Nrf2 decreased in the absence of KPNA6 under both basal and induced conditions (Fig. 7C), and the half-life of Nrf2 after the removal of tBHQ doubled in KPNA6 knockout cells (Fig. 7D). Collectively, these data indicate that KPNA6-mediated Keap1 nuclear shuttling is required for postinduction repression of the Nrf2-dependent antioxidant response by promoting Nrf2 ubiquitination and degradation.
Fig. 7.
KPNA6 modulates the antioxidant response by promoting ubiquitination and degradation of Nrf2. (A) KPNA6 is required for efficient repression of nuclear Nrf2 protein levels during the postinduction phase of the antioxidant response. MDA-MB-231 cells were transfected with either scrambled control siRNA or siRNA against KPNA6. At 48 h posttransfection, cells were either left untreated (mock) or treated with 100 μM tBHQ for 3 h (3). After removal of tBHQ by washing, cells were further incubated in normal medium for 3 h (3+P3, where “P” stands for postinduction) or for 6 h (3+P6). Nuclear extracts of the cells were analyzed by immunoblotting with anti-Nrf2, anti-KPNA6, and anti-lamin A antibodies. (B) KPNA6 represses the transcription of Nrf2 downstream target genes through posttranscriptional regulation of Nrf2. MDA-MB-231 cells were transfected with either control siRNA (■) or siRNA against KPNA6 (□). Cells were either untreated or treated with 50 μM tBHQ for either 16 h (upper and middle panels) or the indicated time period (lower panel) before being harvested and used for qRT-PCR analysis. Error bars indicate standard deviations of the results from three independent experiments. Asterisks indicate significant differences between two samples. (C) KPNA6 promotes polyubiquitination of Nrf2. MDA-MB-231 cells were transfected with siRNA as described above. At 48 h after transfection, cells were cotreated with 100 μM tBHQ and 10 μM MG132 for 4 h and harvested in denaturing conditions. Cell lysates were diluted and subjected to immunoprecipitation with an anti-Nrf2 antibody, followed by immunoblotting with an antiubiquitin antibody (α-Ub). (D) Half-life of Nrf2 protein is extended in the absence of KPNA6. MDA-MB-231 cells were transfected with siRNA as described above. At 48 h after transfection, cells were treated with 100 μM tBHQ for 4 h. Cells were then washed once with PBS and incubated with fresh medium containing 50 μM cycloheximide for the indicated times. Cell lysates were analyzed by immunoblotting with an anti-Nrf2 antibody. The relative intensities of the Nrf2 bands were quantified and plotted on a semilog scale. The calculated half-life of Nrf2 in each group is shown.
DISCUSSION
How Keap1 is transported into the nucleus is somewhat controversial: Keap1 was initially considered a cytosolic protein. It was thought that the major role of Keap1 was to physically bind to Nrf2 in the cytoplasm, prevent nuclear accumulation of Nrf2, and block trans activation of ARE-regulated genes (10, 11). However, accumulating evidence shows that Keap1 is a nuclear-cytoplasmic shuttling protein equipped with a nuclear export signal (NES) that confers nuclear-cytoplasmic shuttling of the Nrf2-Keap1 complex (13, 27, 37, 41). In this study, we observed that Keap1 shuttles between the cytoplasm and the nucleus, since nuclear export of Keap1 can be blocked, either by leptomycin (LMB) or by NES mutations. Furthermore, nuclear import of Keap1 is independent of Nrf1 or Nrf2 (Fig. 1B and C). Stable cell lines derived from Keap1−/− cells that stably express comparable levels of GFP-tagged wild-type Keap1 or the Keap1-NES mutant were used to eliminate the possible artifacts from Keap1 overexpression (Fig. 1A). Therefore, our data provide further evidence that Keap1 translocates into the nucleus and modulates Nrf2 signaling.
Although evidence in favor of Keap1 as a nuclear-cytoplasmic shuttling protein continues to grow, the mechanism of nuclear import of Keap1 and its function inside the nucleus remains disputable. In silico analysis revealed that Keap1 does not contain a cNLS. The results of domain mapping confirmed that the Kelch domain of Keap1 is indispensable for its nuclear localization (13, 30), which is in agreement with our domain deletion analyses (Fig. 2C and D). To explain how Keap1 translocates into the nucleus, one hypothesis suggests that Nrf2 carries Keap1 into the nucleus, since Nrf2 possesses two putative NLSs and Keap1 does not (41). Our data indicate that Keap1 still accumulates in the nucleus in Nrf1−/− Nrf2−/− MEF cells after LMB treatment (Fig. 1C), suggesting that Keap1 does not rely on Nrf1 or Nrf2 for its nuclear entry. Another group hypothesized that a small nuclear protein, prothymosin α (PTMα), mediates nuclear import of the Keap1-Cul3-Rbx1 complex, leading to ubiquitination and degradation of Nrf2 inside the nucleus (30). However, how PTMα itself shuttles between the nucleus and the cytosol and mediates Keap1 nuclear import is still unclear.
Here, we provide evidence that Keap1 itself is able to enter the nucleus with the help of an importin α protein, KPNA6. Interaction between Keap1 and KPNA6 was demonstrated not only in vitro but also in vivo with endogenous KPNA6 (Fig. 3A to D). Overexpression of KPNA6 significantly enhanced the nuclear accumulation of Keap1 after LMB treatment (Fig. 4A and B). Nrf2, NQO1, and HO-1 were diminished in the presence of KPNA6 (Fig. 5A and B), without alteration of Keap1 protein levels. Also, qRT-PCR analysis showed that KPNA6 changed the mRNA expression of Nrf2 target genes but not that of Nrf2 itself (Fig. 5C), supporting the notion that KPNA6 modulates Nrf2 at the protein level by changing Keap1's nucleocytoplasmic shuttling dynamics. On the other hand, knockdown of KPNA6 attenuates the nuclear import of Keap1 (Fig. 6A and B). There was less nuclear accumulation of Keap1 when MDA-MB-231 cells were transfected with KPNA6 siRNA and in the presence of LMB (Fig. 6C, compare lane 3 to 4, and lane 7 to 8). FRAP assays carried out in Keap1-null cells expressing GFP-tagged Keap1 also demonstrated that knockdown of KPNA6 was sufficient to decrease the nuclear accumulation of Keap1 (Fig. 6D and E). In addition, knockdown of KPNA6 promoted accumulation of endogenous nuclear Nrf2 protein levels during the induction and postinduction phases (Fig. 7A, compare lane 3 to 4 and lane 5 to 6). NQO1, HO-1, and GCLM were also significantly enhanced when KPNA6 was efficiently knocked down (Fig. 7B), while the ubiquitination and degradation of Nrf2 decreased in the absence of KPNA6 (Fig. 7C and D).
However, further attempts to map the KPNA6-interacting sites within the Keap1 Kelch domain were unsuccessful. Truncations or deletions within the Kelch domain disrupted the overall integrity of the ring-like 6-bladed propeller structure, thus providing no useful information. We also made several alanine (A) substitutions of positively charged arginine (R), lysine (K), or histidine (H) clusters within the Kelch domain, which should be responsible for the interaction between cargo protein and importin α in the classical nuclear import scenario, yet none of them showed loss of interaction (Fig. 3F). These data suggest that the interaction between KPNA6 and Keap1 may be different from the classical mode. Although the evidence to date has been biased toward classical monopartite or bipartite nuclear localization signals, many noncanonical NLSs that do not match the classical NLSs have been defined. Kosugi et al. have reported three classes of noncanonical monopartite NLSs that bind directly to importin α in the minor groove (3, 20), which also supports our hypothesis that Keap1 may interact with KPNA6 via a mechanism other than cNLS.
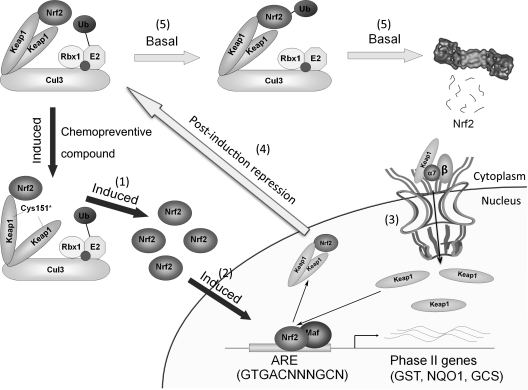
Keap1, a substrate adaptor protein for the Cul3-containing E3 ubiquitin ligase, negatively controls the activity of Nrf2 at the protein level. Nrf2 contains two binding sites for Keap1 in its Neh2 domain, a weak binding site (DLG) and a strong binding site (ETGE). This two-site substrate recognition model is also known as the “hinge-latch model.” The high-affinity ETGE motif functions as the “hinge,” and the lower-affinity DLG motif functions as the “latch.” Under basal conditions, the Keap1-homodimer recognizes and binds both motifs, positioning Nrf2 in the correct orientation for polyubiquitination and degradation to maintain low basal levels of Nrf2. In response to chemopreventive compounds, cysteine residues in Keap1 become modified, which may alter the structural confirmation and “unlatch” the weak binding DLG motif from Keap1, resulting in the stabilization and activation of Nrf2. (16, 25). Many cellular proteins, such as p62 and p21WAF1, are able to unlatch the weak interaction between Nrf2 and Keap1, resulting in activation of the Nrf2-mediated response (19, 37). This unlatch event in response to an activation signal should also expose the Kelch domain, which contains the NLS. Based on our work presented in this study, exposure of the NLS in Keap1 results in its nuclear translocation. Currently, the precise mechanism by which nuclear-cytoplasmic shuttling of Nrf2 or Keap1 is controlled in response to redox conditions remains uncertain. In this study, we found that kinetically, Nrf2 translocates into the nucleus faster than Keap1. Conceivably, the series of events occur in response to an activating signal, as follows. (i) Unlatching of the binding between Nrf2 and Keap1: Nrf2 activators cause a conformational change in the Keap1-Cul3-E3 ubiquitin ligase by acting on certain cysteine residues in Keap1. (ii) Nrf2 nuclear translocation: a decrease in Nrf2 degradation results in nuclear translocation of free Nrf2 and activation of the Nrf2's downstream target genes. (iii) Keap1 nuclear translocation: unlatching of the Kelch domain in Keap1 from an Nrf2 protein also exposes an NLS to KPNA6, and Keap1 enters the nucleus with the assistance of KPNA6/importin β. (iv) Turning off of the signal: Keap1 removes Nrf2 from the ARE of downstream target genes and transports the Keap1-Nrf2 complex out of the nucleus, using the strong NES in Keap1. (v) Once in the cytoplasm, the dimer is recruited to the ubiquitination and degradation machinery, leading to the degradation of Nrf2 and restoring low basal levels of Nrf2 (Fig. 8).
Fig. 8.
Schematic model of Nrf2 regulation by Keap1. Keap1 is a key regulator of the Nrf2-signaling pathway and serves as a molecular switch to turn the Nrf2-mediated antioxidant response on and off. (1) Oxidative stress or chemopreventive compounds cause a conformational change in the Keap1-Cul3-E3 ubiquitin ligase by acting on specific cysteine residues in Keap1. (2) Nuclear translocation of Nrf2: a decrease in Nrf2 degradation results in nuclear translocation of free Nrf2 and activation of Nrf2 target genes. (3) Nuclear translocation of Keap1: unlatching of the Kelch domain in Keap1 from Nrf2 also exposes an NLS to KPNA6, and Keap1 enters the nucleus with the assistance of KPNA6/importin β. (4) Turning off of the signal: Keap1 removes Nrf2 from the ARE of downstream genes. The Keap1-Nrf2 complex is then transported out of the nucleus by the strong NES in Keap1. (5) In the cytosol, the Nrf2-Keap1 complex associates with the Cul3-Rbx1 core ubiquitin machinery, leading to degradation of Nrf2.
In conclusion, we report a direct interaction between Keap1 and KPNA6 and have determined that Keap1 is transported into the nucleus by KPNA6. Moreover, we mapped the interaction domains to the Kelch domain of Keap1 and the N-terminal domain of KPNA6. By controlling the nuclear-cytoplasmic shuttling of Keap1, KPNA6 promotes the ubiquitination and degradation of Nrf2 and, thus, negatively regulates the protein level of Nrf2 and the transcription of its downstream target genes. Our data further support our previous work indicating that Keap1 is a postinduction repressor of Nrf2.
ACKNOWLEDGMENTS
This work is supported by the NIEHS, grant 1RO1ES015010-01, and the ACS, grant RSG-07-154-01.
We thank Doug Cromey, manager of Confocal Microscope Facility (AHSC Imaging facility), for technical support for the FRAP experiments.
Footnotes
Published ahead of print on 7 March 2011.
REFERENCES
- 1. Alam J., et al. 1999. Nrf2, a Cap′n'Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J. Biol. Chem. 274:26071–26078 [DOI] [PubMed] [Google Scholar]
- 2. Chan K., Han X. D., Kan Y. W. 2001. An important function of Nrf2 in combating oxidative stress: detoxification of acetaminophen. Proc. Natl. Acad. Sci. U. S. A. 98:4611–4616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Conti E., Uy M., Leighton L., Blobel G., Kuriyan J. 1998. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell 94:193–204 [DOI] [PubMed] [Google Scholar]
- 4. Cook A., Bono F., Jinek M., Conti E. 2007. Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 76:647–671 [DOI] [PubMed] [Google Scholar]
- 5. Cullinan S. B., Gordan J. D., Jin J., Harper J. W., Diehl J. A. 2004. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell. Biol. 24:8477–8486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Daelemans D., Costes S. V., Lockett S., Pavlakis G. N. 2005. Kinetic and molecular analysis of nuclear export factor CRM1 association with its cargo in vivo. Mol. Cell. Biol. 25:728–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Engel R. H., Evens A. M. 2006. Oxidative stress and apoptosis: a new treatment paradigm in cancer. Front. Biosci. 11:300–312 [DOI] [PubMed] [Google Scholar]
- 8. Furukawa M., Xiong Y. 2005. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol. Cell. Biol. 25:162–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goldfarb D. S., Corbett A. H., Mason D. A., Harreman M. T., Adam S. A. 2004. Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell Biol. 14:505–514 [DOI] [PubMed] [Google Scholar]
- 10. Itoh K., et al. 1999. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 13:76–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kang M. I., Kobayashi A., Wakabayashi N., Kim S. G., Yamamoto M. 2004. Scaffolding of Keap1 to the actin cytoskeleton controls the function of Nrf2 as key regulator of cytoprotective phase 2 genes. Proc. Natl. Acad. Sci. U. S. A. 101:2046–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kanwar J. R. 2005. Anti-inflammatory immunotherapy for multiple sclerosis/experimental autoimmune encephalomyelitis (EAE) disease. Curr. Med. Chem. 12:2947–2962 [DOI] [PubMed] [Google Scholar]
- 13. Karapetian R. N., et al. 2005. Nuclear oncoprotein prothymosin alpha is a partner of Keap1: implications for expression of oxidative stress-protecting genes. Mol. Cell. Biol. 25:1089–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Klaunig J. E., Kamendulis L. M. 2004. The role of oxidative stress in carcinogenesis. Annu. Rev. Pharmacol. Toxicol. 44:239–267 [DOI] [PubMed] [Google Scholar]
- 15. Kobayashi A., et al. 2004. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 24:7130–7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kobayashi A., et al. 2006. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol. Cell. Biol. 26:221–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kobe B. 1999. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nat. Struct. Biol. 6:388–397 [DOI] [PubMed] [Google Scholar]
- 18. Kohler M., et al. 2002. Differential expression of classical nuclear transport factors during cellular proliferation and differentiation. Cell. Physiol. Biochem. 12:335–344 [DOI] [PubMed] [Google Scholar]
- 19. Komatsu M., et al. 2010. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 12:213–223 [DOI] [PubMed] [Google Scholar]
- 20. Kosugi S., et al. 2009. Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J. Biol. Chem. 284:478–485 [DOI] [PubMed] [Google Scholar]
- 21. Lange A., et al. 2007. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J. Biol. Chem. 282:5101–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li X., Zhang D. D., Hannink M., Beamer L. J. 2004. Crystal structure of the Kelch domain of human Keap1. J. Biol. Chem. 279:54750–54758 [DOI] [PubMed] [Google Scholar]
- 23. Liu S. M., Liu W. M. 2007. Recent developments in the understanding of nuclear protein import. Protein Pept. Lett. 14:723–733 [DOI] [PubMed] [Google Scholar]
- 24. McMahon M., et al. 2001. The Cap′n'Collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 61:3299–3307 [PubMed] [Google Scholar]
- 25. McMahon M., Thomas N., Itoh K., Yamamoto M., Hayes J. D. 2006. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “tethering” mechanism: a two-site interaction model for the Nrf2-Keap1 complex. J. Biol. Chem. 281:24756–24768 [DOI] [PubMed] [Google Scholar]
- 26. Moinova H. R., Mulcahy R. T. 1999. Up-regulation of the human gamma-glutamylcysteine synthetase regulatory subunit gene involves binding of Nrf-2 to an electrophile responsive element. Biochem. Biophys. Res. Commun. 261:661–668 [DOI] [PubMed] [Google Scholar]
- 27. Nguyen T., Sherratt P. J., Nioi P., Yang C. S., Pickett C. B. 2005. Nrf2 controls constitutive and inducible expression of ARE-driven genes through a dynamic pathway involving nucleocytoplasmic shuttling by Keap1. J. Biol. Chem. 280:32485–32492 [DOI] [PubMed] [Google Scholar]
- 28. Nguyen T., Sherratt P. J., Pickett C. B. 2003. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu. Rev. Pharmacol. Toxicol. 43:233–260 [DOI] [PubMed] [Google Scholar]
- 29. Nioi P., McMahon M., Itoh K., Yamamoto M., Hayes J. D. 2003. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochem. J. 374:337–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Niture S. K., Jaiswal A. K. 2009. Prothymosin-alpha mediates nuclear import of the INrf2/Cul3 Rbx1 complex to degrade nuclear Nrf2. J. Biol. Chem. 284:13856–13868 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31. Pennington J. D., et al. 2005. Redox-sensitive signaling factors as a novel molecular target for cancer therapy. Drug Resist. Updat. 8:322–330 [DOI] [PubMed] [Google Scholar]
- 32. Ramos-Gomez M., et al. 2001. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 98:3410–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ruef J., et al. 1999. Oxidative stress and atherosclerosis: its relationship to growth factors, thrombus formation and therapeutic approaches. Thromb. Haemost. 82(Suppl. 1):32–37 [PubMed] [Google Scholar]
- 34. Simonian N. A., Coyle J. T. 1996. Oxidative stress in neurodegenerative diseases. Annu. Rev. Pharmacol. Toxicol. 36:83–106 [DOI] [PubMed] [Google Scholar]
- 35. Stewart M. 2007. Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 8:195–208 [DOI] [PubMed] [Google Scholar]
- 36. Strom A. C., Weis K. 2001. Importin-beta-like nuclear transport receptors. Genome Biol. 2:REVIEWS3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sun Z., Zhang S., Chan J. Y., Zhang D. D. 2007. Keap1 controls postinduction repression of the Nrf2-mediated antioxidant response by escorting nuclear export of Nrf2. Mol. Cell. Biol. 27:6334–6349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Taniguchi Y., et al. 2006. Generation of medaka gene knockout models by target-selected mutagenesis. Genome Biol. 7:R116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tong K. I., et al. 2007. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol. Cell. Biol. 27:7511–7521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Toscano C. D., Prabhu V. V., Langenbach R., Becker K. G., Bosetti F. 2007. Differential gene expression patterns in cyclooxygenase-1 and cyclooxygenase-2 deficient mouse brain. Genome Biol. 8:R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Velichkova M., Hasson T. 2005. Keap1 regulates the oxidation-sensitive shuttling of Nrf2 into and out of the nucleus via a Crm1-dependent nuclear export mechanism. Mol. Cell. Biol. 25:4501–4513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Venugopal R., Jaiswal A. K. 1996. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc. Natl. Acad. Sci. U. S. A. 93:14960–14965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang W., Chan J. Y. 2006. Nrf1 is targeted to the endoplasmic reticulum membrane by an N-terminal transmembrane domain. Inhibition of nuclear translocation and transacting function. J. Biol. Chem. 281:19676–19687 [DOI] [PubMed] [Google Scholar]
- 44. Wasserman W. W., Fahl W. E. 1997. Functional antioxidant responsive elements. Proc. Natl. Acad. Sci. U. S. A. 94:5361–5366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yang C. S., Landau J. M., Huang M. T., Newmark H. L. 2001. Inhibition of carcinogenesis by dietary polyphenolic compounds. Annu. Rev. Nutr. 21:381–406 [DOI] [PubMed] [Google Scholar]
- 46. Zhang D. D. 2006. Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab. Rev. 38:769–789 [DOI] [PubMed] [Google Scholar]
- 47. Zhang D. D., Hannink M. 2003. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell Biol. 23:8137–8151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang D. D., Lo S. C., Cross J. V., Templeton D. J., Hannink M. 2004. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 24:10941–10953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang D. D., et al. 2005. Ubiquitination of Keap1, a BTB-Kelch substrate adaptor protein for Cul3, targets Keap1 for degradation by a proteasome-independent pathway. J. Biol. Chem. 280:30091–30099 [DOI] [PubMed] [Google Scholar]
- 50. Zhang Y., Crouch D. H., Yamamoto M., Hayes J. D. 2006. Negative regulation of the Nrf1 transcription factor by its N-terminal domain is independent of Keap1: Nrf1, but not Nrf2, is targeted to the endoplasmic reticulum. Biochem. J. 399:373–385 [DOI] [PMC free article] [PubMed] [Google Scholar]