Abstract
The ability of epidermal growth factor receptor (EGFR) to control cell fate is defined by its affinity for ligand. Current models suggest that ligand-binding heterogeneity arises from negative cooperativity in signaling receptor dimers, for which the asymmetry of the extracellular region of the Drosophila EGFR has recently provided a structural basis. However, no asymmetry is apparent in the isolated extracellular region of the human EGFR. Human EGFR also differs from the Drosophila EGFR in that negative cooperativity is found only in full-length receptors in cells. To gain structural insights into the human EGFR in situ, we developed an approach based on quantitative Förster resonance energy transfer (FRET) imaging, combined with Monte Carlo and molecular dynamics simulations, to probe receptor conformation in epithelial cells. We experimentally demonstrate a high-affinity ligand-binding human EGFR conformation consistent with the extracellular region aligned flat on the plasma membrane. We explored the relevance of this conformation to ligand-binding heterogeneity and found that the asymmetry of this structure shares key features with that of the Drosophila EGFR, suggesting that the structural basis for negative cooperativity is conserved from invertebrates to humans but that in human EGFR the extracellular region asymmetry requires interactions with the plasma membrane.
INTRODUCTION
The human epidermal growth factor receptor (hEGFR) is a classic receptor tyrosine kinase (26). The hEGFR was originally identified in A431 epithelial carcinoma cells (47) and consists of an extracellular growth factor-binding region, a single-pass transmembrane region, and a cytoplasmic domain that has tyrosine kinase activity (42).
The extracellular domain of hEGFR is comprised of four subdomains (I to IV). The unliganded receptor monomer is held in a closed conformation by an intramolecular tether formed by loops in subdomains I and III (17). In ligand-occupied receptor dimers, the intramolecular tether is broken, and the receptor is opened into an extended conformation which interacts with another monomer, forming a back-to-back dimer (19, 36). Ligand-induced receptor dimerization is thought to be the key stimulatory step, leading to allosteric transactivation of the two associated intracellular hEGFR kinases (42).
The EGFR family has diversified during evolution from one ligand/one receptor in Caenorhabditis elegans, to multiple ligands and one receptor in flies, and to a family of multiple ligands and four closely related receptors that form homo- and heterodimers in humans (10). The EGFR itself is highly conserved across species. Wild-type hEGFR and Drosophila EGFR (dEGFR) show extensive conservation, with 55% homology in the kinase domain and 41% homology in the ligand-binding portion of the extracellular domain (39).
An important unresolved question regarding the mechanism of hEGFR activation is the structural basis for the concave-up Scatchard plots exhibited by epidermal growth factor (EGF) (31, 43). These plots were initially interpreted as indicating the presence of two receptor populations: a small minority of high-affinity receptors with dissociation constants (KD) of <1 nM that mediate most signaling events and a majority of low-affinity receptors with KD of >1 nM (14, 18, 40, 55). These two populations were initially believed to be composed of tethered monomers and extended dimers; however, in this model stabilization of the extended dimer configuration by one ligand molecule would facilitate the binding of a second ligand to the remaining receptor in the dimer, resulting in positive cooperativity, inconsistent with concave-up Scatchard plots (25, 33, 37, 38).
Using global modeling of EGF-binding data as a function of receptor number, Macdonald and Pike (29) recently showed that EGF-binding heterogeneity could be accounted for by the presence of negative cooperativity in hEGFR dimers. However, negative cooperativity requires that the binding of ligand to one subunit of the hEGFR dimer decrease the affinity of the other subunit for ligand. This would, in turn, require the interactions between ligand and the two subunits of the hEGFR dimer to be asymmetric, inconsistent with the symmetric structural data found for hEGFR dimers bound to two ligands (19, 36). In a recent breakthrough, a series of crystallographic structures have revealed that, unlike hEGFR, dEGFR extracellular domain asymmetry is induced by the binding of the first ligand, which structurally restrains the unoccupied binding site, reducing the affinity for binding of the second ligand (2). Interestingly, concave-up Scatchard plots were observed in preparations of the isolated extracellular region of the dEGFR, in direct contrast with its human counterpart, in which concave-up plots are observed only when ligands bind to full-length receptors in cells. These differences suggest that other receptor regions, conformations, and/or unknown cellular components must be involved in the regulation of ligand affinity in hEGFR (25).
Using Förster resonance energy transfer (FRET), a method extremely sensitive to short intermolecular distances (<10 nm) (46), we previously obtained initial evidence for two types of EGF/hEGFR complexes, tilted and upright, that are associated to high-affinity and low-affinity EGF binding, respectively (49). Subsequently, with molecular dynamics (MD) simulations we showed that, with minor rearrangements, the hEGFR back-to-back dimer can be aligned almost flat on the cell membrane, leading to conformational changes which further stabilize the extracellular dimer (23). Alignment on the cell surface and the interactions between the hEGFR dimer and the upper leaflet of the membrane that follow break the pseudo-2-fold symmetry of the hEGFR extracellular region, resulting in a highly asymmetric hEGFR dimer structure.
In this study, we investigate the relevance of this asymmetric model of the hEGFR dimer to EGF-binding heterogeneity and negative cooperativity. We first verified the existence of this conformation in cells by extending a FRET assay based on fluorescence lifetime imaging microscopy (FLIM) (3) to include FRET titrations as a function of acceptor concentration together with a data analysis method based on Monte Carlo simulations. Unlike the FRET method we employed previously (49), this new assay allows full quantification of the distance of closest approach of receptor-bound ligands to the surface of the plasma membrane of adherent epithelial cells, the assessment of the variation associated to the distance measurement, and the rejection of potential sources of artifacts which can be inherent in standard FRET measurements from cells (e.g., photobleaching, nonradiative transfer, and nonuniform distributions of FRET donors and acceptors). Crucially, the new FRET data obtained from hEGFR that display high-affinity for EGF can be reconciled with the accepted crystallographic hEGFR ectodomain dimer structure only if receptors are aligned flat on the membrane. In addition, we have reanalyzed our previous MD simulations of doubly liganded hEGFR dimers aligned on a model membrane and also performed new simulations of both singly liganded and unliganded hEGFR dimers under the same conditions. This has allowed us to demonstrate that the asymmetry resulting from alignment on the membrane shares a number of key features with that observed recently in soluble dEGFR (2).
MATERIALS AND METHODS
Sample preparation.
A431 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) without phenol red and supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, and 1% penicillin-streptomycin. HeLa cells were cultured in minimal essential medium (MEM) with Earle's salts without phenol red and supplemented with 10% FBS, 2 mM glutamine, and 1% penicillin-streptomycin (all Invitrogen). Cells were incubated at 37°C in the presence of 5% CO2 in air. Cell samples deprived of serum for 16 h upon reaching 80% confluence were loaded with acceptor by adding 100 μl of a 5 μM concentration of dialkylcarbocyanine fluorescent probe C18DiD, C18DiI, C16DiI, or C12DiI in serum-free medium to cells before incubation at 37°C for 8 to 15 min. A total of 40 to 80% of cells incorporated membrane probe into their membranes depending upon incubation time. Cells were incubated in 200 nM monoclonal antibody (MAb) 2E9 in phosphate-buffered saline (PBS) at 4°C for 4 h to bind MAb 2E9 to EGFRs. If required, cells were incubated in 100 nM phorbol myristate acetate (PMA), diluted in PBS and containing 0.01% dimethyl sulfoxide (DMSO), for 30 min at 37°C or incubated with 10 mM methyl-β-cyclodextrin (MβCD) diluted in serum-free medium for 30 min at 37°C to deplete their membranes of cholesterol. For EGF labeling cells were incubated at 4°C for 30 to 60 min with 100 nM, 1 nM, or 0.5 nM murine EGF-Atto 488 (AttoTec) or EGF-Alexa Fluor 546 (Invitrogen) in PBS as required (dye conjugation performed by Cambridge Research Biochemicals). Cells were fixed by incubation with 3% paraformaldehyde (Electron Microscopy Sciences) for 15 min at 4°C and then for 15 min at room temperature. Loading of cells with Fluo-4 AM ester was achieved by incubating cells for 1 h at room temperature in 5 μM Fluo-4 in PBS with 0.02% Pluronic F-127 (both Invitrogen).
FRET pairs.
Two donor-acceptor pairs with different Förster radii (R0) were employed to allow optimal FRET-derived measurement of short and long distances and cross-checking of results. Atto 488 or Alexa Fluor 546 conjugated to the N terminus of murine EGF in a 1:1 stoichiometry were used as donors, and the acceptor common to both FRET pairs was the lipophilic dialkylcarbocyanine probe DiIC18(5) (DiD) that inserts itself in the plasma membrane with the chromophore lying on the outer leaflet. Dialkylcarbocyanine probes were selected because their absorption and emission dipoles lie parallel to the surface outer leaflet of the plasma membrane (4), and they are known not to flip between the outer and inner leaflets (53). R0 was calculated to be 5.6 nm for the EGF-Atto 488/DiD pair and 6.9 nm for the EGF-Alexa Fluor 546/DiD pair.
Confocal FLIM-FRET microscopy.
Data were acquired at room temperature using a purpose-built laser scanning confocal microscope with time-correlated single photon counting (TCSPC) electronics (SPC-730; Becker-Hickl GmbH) (3, 49) with the addition of a supercontinuum light source (Fianium SC450-4; 40 MHz repetition rate). EGF-Atto 488 was excited with 490-nm pulsed laser light, and fluorescence was detected between 505 to 530 nm using a fast photomultiplier tube (PMC-100; Becker-Hickl GmbH). EGF-Alexa Fluor 546 was excited with 545-nm pulsed laser light, and fluorescence was detected between 560 and 610 nm. Fluorescent intensity decays were best-fitted to a single exponential decay model where acceptor was absent and to a bi-exponential model when both donor and acceptor were present. Donor lifetimes for FRET efficiency calculations were obtained by taking the mean of the distribution of the fluorescence lifetimes of pixels. The occurrence of FRET results in a decrease in the fluorescence lifetime of the donor (τD) in cells loaded with acceptor (τDA). The FRET efficiency (EFRET) was calculated from fluorescence lifetime data using the following formula: EFRET = 1 − τDA/τD. Confocal images of acceptor from the same field of view were taken of DiD directly excited at 639 nm, and fluorescence was collected at >670 nm. Bleed-through between channels was negligible.
Monte Carlo modeling of the distance of closest approach.
The algorithm employed for the Monte Carlo modeling involves creating a random spatial distribution of donors and acceptors, determining a sequence in which pseudo-photons (excitons) are incident on donors, and then playing the Monte Carlo FRET scheme to see if and when the donors are excited and if and when each excited donor either fluoresces or transfers its energy to an acceptor (FRET).
Monte Carlo distribution of donors and acceptors.
Each sample was modeled as planes of donors and a parallel plane of acceptors, separated by a vertical distance d. At the concentrations of receptors being considered (50 to 1,000 per μm2), and with Förster radius R0, the typical separation of receptors is >4.5R0. The donors on each receptor can therefore be approximated as interacting with nearby acceptors independently of any other receptors. The simulation therefore modeled a donor (for receptor monomers) or a pair of donors (for receptor dimers) at a distance d above the midpoint of a square plane of acceptors of area 1/Σdon for monomers or 2/Σdon for receptor dimers (where Σdon = 500 μm−2 is the donor surface density). In the case of dimers the donors were placed at intervals of 180° around the circumference of a circle of radius 2 nm with a random orientation in the donor plane. The donors and acceptors were treated as occupying spherical volumes of a radius of 1 nm. The donors/donor multimers and acceptors were placed randomly in their respective planes with average concentrations Σdon and Σacc (fluorophores per unit area), respectively, and arranged such that there was no overlap between donors, acceptors, or one another. The acceptors were additionally excluded from a region of a radius of 1 nm in the acceptor plane under each donor to mimic the area excluded by the transmembrane receptor region. The acceptor distribution was created by generating a uniform hexagonally packed distribution of acceptors at the required concentration, filling a region slightly larger than that being simulated, and then applying 10 passes of position randomization. In each pass the acceptor positions were randomized one by one in a random order, with each position randomization consisting of allocating the acceptor the first randomly chosen position within the average acceptor separation which did not overlap any other acceptor or donor exclusion region. After all randomization steps, any fluorophores outside the simulation area were removed.
Monte Carlo exciton schedule.
The sequence in which excitors are incident on the donor is determined by the illumination properties of the laser. The illumination consists of pulses of duration tpulse repeated every trepeat with mean power (over many bursts), Plaser, and wavelength, λ. Eighty-four percent of the laser power is within a radius R84. The simulated receptors are all within the laser profile (we see fluorescence only from illuminated regions). The laser profile was assumed to be uniform within the simulated region with power per unit area: Flaser = (0.84 × Plaser)/πR842.
This gives the following equation for excitons per pulse in the simulation region: Nphot = λFlaserAsimulationtrepeat/hc, where Asimulation is the area of the simulation region, h is Planck's constant, and c is the speed of light. The absorption cross-section of each donor is Xdon, so the fraction of photons incident on a donor will be XdonΣdon, giving the following equation for photons incident on a donor per pulse: Nexcitons = XdonΣdonNphot. These excitons will be absorbed by the donor leading to FRET or direct fluorescence unless the donor is already excited when the exciton arrives. The schedule of excitons is created one pulse at a time, with a total of 500 pulses in each simulation. For each pulse, the number of photons is sampled from a Poisson distribution with mean Nphot. This gives the expected number of excitons for that pulse, from which the actual number of excitons is also sampled from a Poisson distribution. The time for each exciton is randomly selected from a uniform distribution for each pulse. Each exciton is assigned a random position in the simulation area, and the nearest donor to this position is chosen as the target.
Each simulation was repeated 2,000 to 6,000 times for a given set of simulation parameters, with a new fluorophore distribution and exciton schedule randomly generated for each such configuration. From these multiple configurations, the mean FRET efficiency was determined. Curves of FRET efficiency as a function of acceptor density were calculated for plane separations, d, from 0.5 to 15 nm in steps of 0.5 nm.
Calibration of acceptor density.
Two methods were used. Donor FLIM images and corresponding acceptor intensity images were acquired from samples of phosphatidylcholine monolayers dually labeled with DiI (donor) and DiD (acceptor). Acceptor photobleaching with 639-nm continuous-wave (CW) laser light was used to reduce the density of acceptors while keeping the donor density the same. The resulting variation in the efficiency of energy transfer between DiI and DiD as a function of DiD concentration was fitted to an analytical expression for zero distance of closest approach between donors and acceptors (52) to obtain a conversion factor between the measured acceptor intensity and the density of acceptors (7). Alternatively, the number and brightness (N&B) method (15) was used to estimate the brightness of individual acceptor molecules in the plasma membranes of A431 cells. The average acceptor brightness was then used to estimate the number of molecules contributing to each pixel in the acceptor images, which was converted to the acceptor density by dividing by the area of the microscope lateral point spread function (PSF; measured to be 243 ± 10 nm in diameter). The density of acceptors was calibrated in units of acceptors per R02. Both calibration methods return consistent results.
FRET ratio microscopy.
Fluorescence emission of EGF-Atto 488 excited by 488-nm light and the sensitized emission of C18DiD in the membranes of A431 cells were simultaneously imaged on a charge-coupled-device (CCD) chip (DV887; Andor) using an LD Plan-Neofluar, 0.6-numerical aperture (40×) objective (Carl Zeiss Inc.) and an image splitter (Optosplit III; Cairn Research). Fluo-4 fluorescence intensity was imaged as for EGF-Atto 488. Temperature was controlled by a bipolar temperature controller system (QE-1HC, TCM-1, and Cl-100 [Warner Instruments]).
Molecular dynamics simulations.
Simulations of the doubly liganded EGFR extracellular region have been reported elsewhere (23). Briefly, simulations were performed with the NAMD program using the charmm 22 force field for the protein and the charmm 27 force field for a membrane composed of palmitoyloleoylphosphatidylcholine (POPC) lipids. An initial model of the liganded dimer was constructed from crystallographic structures 1IVO and 1NQL (Protein Data Bank) and simulated in water for 51 ns. The dimer was then relaxed on the membrane and simulated for over 70 ns. Snapshots from these simulations were used to produce figures. In the current study, we started from the asymmetric receptor dimer relaxed on the POPC membrane. To model the unliganded EGFR dimer, the ligands were removed, and the system was resolvated. The new system was simulated with NAMD, using the same options and conditions as before, for a total time of 50 ns. We also considered one possible singly liganded dimer by removing the upper ligand from the asymmetric receptor dimer on the membrane. The system was resolvated and simulated for a total of 60 ns. For all three simulations, we see an initial relaxation over 20 to 30 ns, followed by slower variation in quantities such as the root mean square deviation (RMSD) with respect to the initial structure. Given the size of the system, we cannot be certain that the trajectories have fully converged. Nevertheless, the structural features described and differences between the three cases are unambiguous.
RESULTS
A FRET-FLIM method to determine the distance of closest approach.
The distance of closest approach of the hEGFR ligand-binding site to the cell surface, which represents the minimum possible distance between the receptor-bound fluorescent donor probe on the EGF ligand and a lipid acceptor chromophore at the plasma membrane, can be determined from the variation of the efficiency of FRET (EFRET) between fluorescent donors and acceptors measured as a function of acceptor surface density. This FRET method has been previously used by several groups using cell-averaged steady-state intensity measurements (e.g., flow cytometry) (5, 7, 8). Here, we determine FRET efficiency by using time-resolved FLIM to image the shortening of the fluorescence decay time of donor-labeled EGF induced by energy transfer to nearby acceptors in adherent cells.
The A431 cell line was used for most of the experiments (except controls) because they express 1 × 106 to 3 × 106 receptors per cell, of which 30% are located at the cell surface (14, 16). We found that this number of cell surface receptors was optimal to obtain FLIM-FRET data with a good signal-to-background ratio from the small fraction of donor-labeled receptors relevant to high-affinity EGF binding. All binding was done at 4°C to block early temperature-dependent receptor signaling events, receptor internalization, and trafficking of EGF. EGF binding was followed by cell fixation with 3% paraformaldehyde to allow data collection at room temperature without ligand-induced receptor internalization. We note that room temperature is above the temperature at which a phase transition is known to occur in the plasma membrane of A431 cells (44).
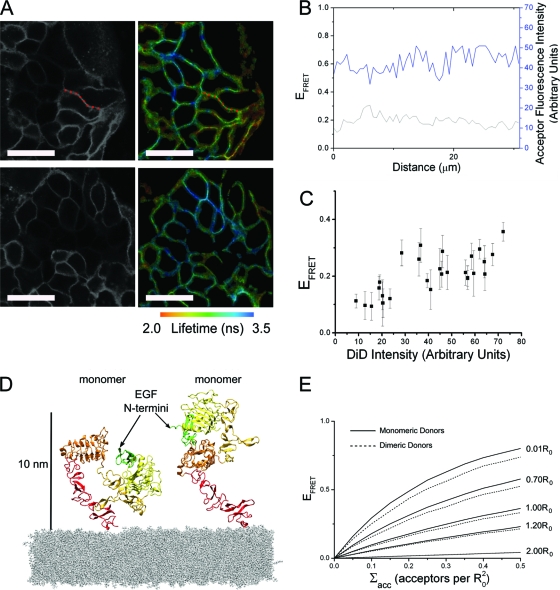
In the absence of lipid acceptor, we found little variation in donor-labeled EGF fluorescence intensity between A431 cells by confocal microscopy, indicating that receptor expression and EGF binding were homogeneous. Along the membrane of cells the acceptor intensity at optical resolution was also approximately constant, and the small variation observed did not correlate with variation in the measured FRET efficiency (Fig. 1 A and B). However, acceptor surface density varied between the membranes of different cells. We exploited this variation to plot FRET efficiency as a function of the acceptor density, pairing the mean EFRET value per cell with its associated mean acceptor density (Fig. 1C). Higher acceptor density values will result in larger numbers of acceptors available to donors. Consistent with this, higher acceptor densities are correlated with higher values of FRET efficiency. The acceptor density (Σacc) was calibrated as described in Materials and Methods in units of number of acceptors per R02, where R0 is the Förster radius of the donor/acceptor FRET pair. The error bars in Fig. 1C show the variation of FRET efficiency within each cell.
Fig. 1.
(A) Example FLIM maps and corresponding acceptor intensity images of A431 cells labeled with DiD and 1 nM EGF-Atto 488 (scale bar, 50 μm). (B) Plotting the calculated FRET efficiency and corresponding acceptor fluorescence intensity along a section of membrane indicated by the red dotted line in panel A shows small fluctuations with little cross-correlation. (C) The mean FRET efficiency from each area of homogenous DiD labeling in both example areas plotted as a function of mean DiD intensity. Error bars represent the standard deviation of the mean FRET efficiency. (D) The tethered and extended hEGFR structures place the N terminus of receptor-bound EGF at similar distances from the membrane. The figure is based on crystal structures 1NQL and 1IVO. At physiological pH, EGF is likely to bind to domain III of the tethered receptor rather than domain I as shown, in which case the distance to the membrane will be even closer to that of the extended receptor. (E) FRET efficiency as a function of acceptor density is shown for several donor-acceptor plane separations (normalized to the Förster radius, R0), produced by Monte Carlo simulations of monomeric or dimeric donors above a plane of randomly distributed acceptors.
We derived the theoretical dependence of EFRET on Σacc using the Monte Carlo techniques described in Corry et al. (12), producing simulations of monomeric or dimeric receptors with binding sites positioned at different distances above a plane of randomly distributed acceptors (see Materials and Methods for more details). The model did not need to distinguish between the tethered and extended conformations of monomers because their donor membrane distances are too close to be distinguishable by FRET (Fig. 1D). Sample results from the Monte Carlo calculations for monomers and dimers are shown in Fig. 1E.
A short ligand-membrane distance of closest approach is found in hEGFR.
We used two approaches to ascertain the vertical distance from the ligand-binding site of hEGFR to the cell surface as a function of ligand-binding affinity. First, receptors displaying high-affinity binding for EGF donor were distinguished from those of low affinity by the use of the monoclonal anti-EGFR antibody MAb 2E9, which is believed to block EGF binding to low-affinity receptor-binding sites without interfering with high-affinity binding (14). Second, cells were exposed to a subsaturating level of EGF donor, similarly biasing the population of occupied binding sites toward those that bind ligand with high affinity but removing the possible perturbation of antibody binding.
From the results of the fluorescence binding assays shown in Fig. 2A, we estimate that both treatment with 200 nM MAb 2E9 followed by 100 nM EGF and treatment with 1 nM EGF but no antibody will result in occupied receptor populations with approximately equal numbers of high- and low-affinity binding sites (for details, see the supplemental material). Figure 2A shows that 100 nM EGF binding is reduced by ∼75% in cells pretreated with MAb 2E9. When cells are also pretreated with phorbol 12-myristate 13-acetate (PMA), a treatment known to result in the complete loss of high-affinity EGF binding (13), MAb 2E9 reduces binding by ∼85%. We interpret the 10% additional receptor sites that are blocked by MAb 2E9 in cells treated with PMA to be an estimate of the fraction of high-affinity sites. Consistent with this, we note that an 86% blocking efficiency would be expected from 200 nM MAb 2E9 (which has an apparent KD of 32 nM [14]) if all receptors displayed low affinity.
Fig. 2.
(A) Comparison of the mean fluorescence intensity of EGF-Atto 647N measured in A431 cells after binding to saturating (100 nM) or subsaturating (1 nM) concentrations of EGF-Atto 647N, with or without prebinding of MAb 2E9. Also shown are the results of binding a saturating concentration of EGF after exposure to PMA. Each result was obtained from measurement of >500 cells. Error bars represent the standard errors of the means. (B) Mean fluorescence lifetime of EGF-Atto 488 bound to EGFR in A431 cell membranes after exposure to 0.5 nM, 1 nM, and 100 nM concentrations of EGF-Atto 488 obtained from 20, 27, and 54 cells, respectively. Error bars represent the standard deviations of the mean lifetimes.
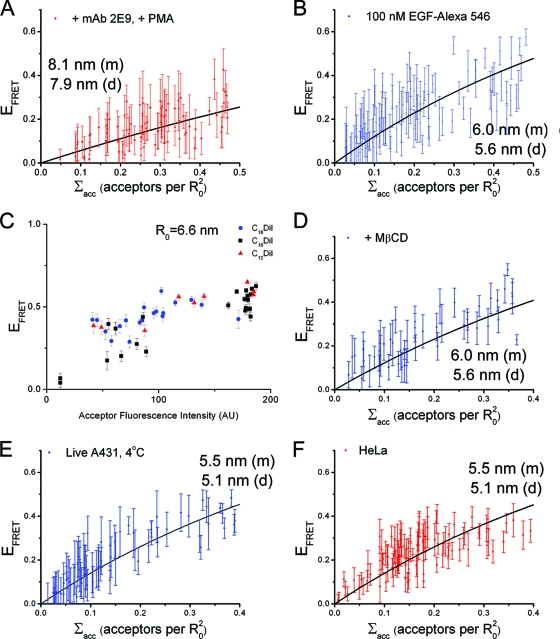
EGF-Atto 488 and the dialkylcarbocyanine membrane probe C18DiD were used as the FRET pair (Förster radius, R0, of 5.6 nm) in our FLIM-FRET experiments. Plots of FRET efficiency as a function of acceptor density, plus the results of fitting the model obtained from the Monte Carlo simulations to the data obtained from the two labeling approaches, are shown in Fig. 3A and B. The range of distances of closest approach obtained for cells treated with 200 nM MAb 2E9 plus 100 nM EGF-Atto 488 was 3.5 to 4.0 nm (the range indicates the extreme possibilities of all labeled receptors being monomers or dimers and reflects the effect of uncertainty in the oligomerization state of the donor-labeled receptor population) (Fig. 1E). For cells exposed to 1 nM EGF-Atto 488 the range obtained was 3.1 to 3.6 nm. The similarity between the distances determined from both measurements demonstrates that antibody binding does not perturb the FRET results. The distance of closest approach from cells exposed to 100 nM EGF-Atto 488 without MAb 2E9 was determined to be 6.9 to 7.1 nm. As a control, we measured donor fluorescence lifetimes in cells bound to EGF-Atto 488 in the absence of acceptor as a function of EGF concentration. We found no difference in donor fluorescence lifetime between 0.5 and 100 nM (Fig. 2B), indicating that the differences in FRET efficiency shown in Fig. 3A and B cannot be attributed to the different amount of donor-EGF binding, donor self-quenching, or cell autofluorescence.
Fig. 3.
Determination of the distance of closest approach of EGF binding sites to the cell surface. (A) Plots of FRET efficiency as a function of acceptor density measured in DiD-loaded A431 cells blocked with 200 nM MAb 2E9 before labeling hEGFRs with 100 nM EGF-Atto 488 (red) and without MAb 2E9 blocking (blue). (B) Results after labeling with 1 nM EGF-Atto 488. (C and D) Comparison of FRET data obtained from DiD-loaded A431 cells lightly fixed before or after labeling with 100 nM (C) and 1 nM (D) EGF-Atto 488. Each data point represents a stretch of cell membrane with homogeneous acceptor labeling, and error bars represent the standard deviations of the mean FRET efficiencies. The best fit of Monte Carlo simulation results to the data are shown and labeled with the corresponding distance of closest approach for monomeric (m) and dimeric (d) donors. The errors from uncertainty in the Monte Carlo model fitting were typically 1 to 1.5% of the calculated distance values.
It should be noted, however, that the FRET-reported distances are ensemble averages and will represent only actual vertical ligand-membrane separations in the case of monodisperse receptor conformations. We expect mixed populations of high-affinity and low-affinity binding sites; therefore, 3.0 to 4.0 nm likely represents an upper boundary for the actual short ligand-membrane distance because of the contribution of mixed-in long distances. Likewise, the distance of 6.9 to 7.1 nm will be a lower boundary of the long distance because of the contribution of mixed-in short distances.
We next compared results obtained in cells fixed before and after binding 100 nM or 1 nM EGF and found that changing the order in which EGF binding and fixation were carried out had no effect on the outcome of the experiment (Fig. 3C and D). The prefixation procedure has been shown to be sufficient to block subsequent conformational changes in membrane proteins (6), suggesting that hEGFR receptors might acquire different extracellular conformations that are associated with high- and low-EGF-binding affinity independent of ligand binding.
Abolishing high-affinity binding results in the loss of the short ligand-membrane distances.
To further investigate the association between high-affinity binding and short ligand-membrane distances, we examined the effect of PMA on the measured ligand-membrane distances. For these measurements we used a second FRET pair, EGF-Alexa Fluor 546/DiD, whose R0 (6.9 nm) is much closer to the longer distances observed and therefore more sensitive to changes around this distance. Cells were bound to 200 nM MAb 2E9 and then treated with PMA to abolish high-affinity binding, followed by receptor labeling with 100 nM EGF-Alexa Fluor 546. Figure 4A shows the resulting plot of FRET efficiency versus acceptor concentration. Under these conditions the range of distances of closest approach was 7.9 to 8.1 nm, which is significantly longer than the 3.5 to 4.0 nm observed when the PMA treatment was not applied between MAb 2E9 binding and EGF binding (Fig. 3A, red). Given that PMA treatment does not interfere with MAb 2E9 binding (Fig. 2A), we conclude that abolition of high-affinity EGF binding at the cell surface by PMA is accompanied by the loss of the receptor conformation that places EGF molecules at short distances from the membrane. We checked that the difference in ligand-membrane distance between PMA-treated and untreated cells was not due to the use of a different FRET pair by measuring the distance of closest approach in DiD-loaded A431 cells labeled with 100 nM donor EGF-Alexa Fluor 546 (Fig. 4B). As expected from the larger R0 of this FRET pair, the results show larger FRET efficiencies than acceptor-loaded cells treated with 100 nM donor EGF-Atto 488 (Fig. 3A, blue), but the returned distances of closest approach using EGF-Alexa Fluor 546 and EGF-Atto 488 as donors are consistent within the 10% error arising from the experimental calculation of the values of R0 (21). The distances in cells treated with PMA (Fig. 4A) are longer than those observed in wild-type cells under saturating ligand-binding conditions (without MAb 2E9 treatment) (Fig. 4B), which would be consistent with the latter having a small contribution from short distances associated with the presence of high-affinity binding.
Fig. 4.
Shown are plots of FRET efficiency as a function of acceptor density measured in DiD-loaded A431 cells labeled with 100 nM EGF-Alexa Fluor 546 after pretreatment with 200 nM MAb 2E9 and PMA (A) or from untreated cells (B). (C) FRET efficiency plotted as a function of acceptor fluorescence intensity for A431 cells labeled with 100 nM EGF-Atto 488 after loading with C18, C16, and C12 DiI. FRET efficiency is shown as a function of acceptor density measured in DiD-loaded A431 cells labeled with 100 nM EGF-Alexa Fluor 546 after depletion of membrane cholesterol by MβCD (D), in live cells at 4°C (E), and in HeLa cells (F). Where appropriate the best fit of Monte Carlo simulation results to the data are shown and labeled with the corresponding distance of closest approach for monomeric (m) and dimeric (d) donors.
The short distance of closest approach is not an artifact of differential donor/acceptor partitioning or overexpression.
Three control experiments were performed to confirm that the FRET differences shown in Fig. 3 and Fig. 4A and B can be attributed to different ligand-membrane distances. First, we wished to eliminate the possibility that differences in FRET efficiency values observed for different fractions of high- and low-affinity EGF-binding sites could be due to differences in lateral colocalization of receptor-bound donors and lipid acceptors on submicrometer scales. We treated cells with EGF-Atto 488 and compared the dependence of EFRET as a function of Σacc for three different dialkylcarbocyanine lipid acceptor probes with different alkyl chain lengths that preferentially partition into the membrane gel phase or fluid phase or show no marked preference (C18, C12, or C16, respectively) (45). We did not detect any appreciable difference in the FRET results (Fig. 4C), so we conclude that it is unlikely that heterogeneous colocalization of receptors and acceptors on submicrometer scales can explain the observed differences in FRET efficiency. Second, we found that depletion of cholesterol from the membrane using methyl-β-cyclodextrin (MβCD), a procedure that releases raft-localized receptors from the inhibitory effects of the raft environment and preserves high-affinity EGF binding (38, 41), produced the same results as in untreated cells (Fig. 4D). Third, we measured the steady-state anisotropy of the probes, which indicates that donors and acceptors can sample a large number of orientations within the lifetime of the donor (anisotropies of 0.077 for EGF-Atto 488, 0.194 for EGF-Alexa Fluor 546, and 0.154 for DiD in lipid vesicles were measured). This rules out adverse effects from static relative orientations between donors and acceptors and confirms that the dynamic approximation of the orientation factor can be used to calculate donor/acceptor distances with errors of approximately ±10% of the calculated distance values (21). From these controls we conclude that the FRET data shown in Fig. 3 and Fig. 4 accurately report the distances at which the EGF-binding sites of hEGFR can be located from the cell membrane.
Next, we assessed the possibility of adverse effects of the fixation procedure on our findings by repeating the measurements in live A431 cells at 4°C (Fig. 4E). We found that measurements of the distance of closest approach in these cells were similar to those in fixed cells (Fig. 4B), indicating that fixation does not affect the results. Likewise, similar results were obtained from fixed HeLa cells (Fig. 4F), which express many fewer cell surface EGFRs (∼50,000) than A431, suggesting that short ligand-membrane distances are also present in cells that do not over express hEGFR (we would expect ∼8 nm and not 5.5 nm if short distances were not present). Taken together, our data demonstrate that the short vertical ligand-membrane distances derived from the observed high FRET states are not an artifact due to differential donor/acceptor partitioning or receptor overexpression.
A transient conformational change accompanies high-affinity receptor signaling.
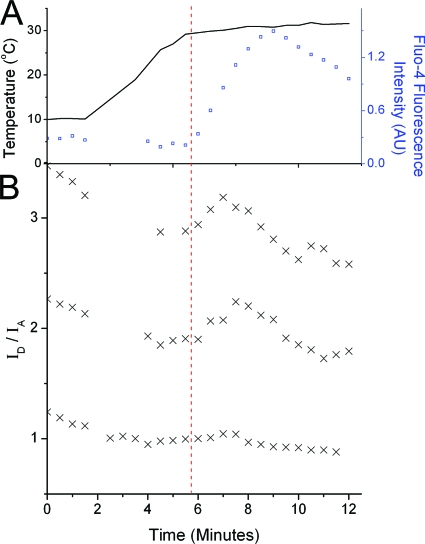
Given that receptors displaying high-affinity binding are mostly responsible for early signal transduction events (14), we investigated whether the hEGFR conformations bearing short ligand-membrane distances were relevant to signal transduction by following changes in the distance between the EGF-binding site and the membrane after ligand binding. For this we loaded live A431 cells with DiD acceptor, followed by incubation at 4°C with 1 nM EGF-Atto 488 for 1 h. We then raised the temperature to physiological values to allow the signal transduction events inhibited by the low temperature to progress and followed the ratio of donor and sensitized acceptor fluorescence intensity (ID/IA). The calcium indicator Fluo-4 was used to confirm that raising the temperature produced an increase in intracellular Ca2+ concentration, an EGFR-triggered second messenger event, demonstrating that the receptors were able to transduce the EGF signals. The profiles of temperature and Fluo-4 fluorescence intensity changes are shown in Fig. 5A. ID/IA ratios corrected for background and cross talk are inversely proportional to the FRET efficiency and were calculated from simultaneously acquired images of EGF-Atto 488 and DiD. This approach is less quantitative than the FLIM-FRET technique but allows real-time acquisition of images. Figure 5B shows the ID/IA ratio over time as the temperature is raised from 8 to 34°C for three example cells with various membrane acceptor densities. A slow decrease in the ID/IA ratio due to donor photobleaching is noticeable in addition to a transient increase that occurs as the calcium-competent signaling temperature is achieved. This transient increase corresponds to a transient decrease in FRET efficiency. These transients are unlikely to be caused by the direct effects of raising the temperature on the properties of the membrane as A431 cell membranes have heterogeneous lipid compositions that undergo a phase transition around 10 to 20°C (44), well below the temperature at which we observed the FRET transient. Also, in cells with very low acceptor concentration, the FRET transient is not detected (Fig. 5B, bottom). The observed FRET transient also appears to be too large to be caused by increased dimerization of receptors, which would cause a small change in FRET due to increased competition between donors for available FRET partners (Fig. 1E). We also discount receptor internalization as a cause of the FRET transient because this would lead to a permanent decrease in FRET efficiency (e.g., via separation of ligands from receptors or reduction of the surface density of DiD in endocytic vesicles). We therefore propose that it is caused by a transient decrease in the distance of closest approach.
Fig. 5.
EGFR ectodomain conformational changes that accompany signal transduction. (A) Raising the temperature of A431 cells labeled with 1 nM EGF-Atto 488 at 4°C results in an increase in free intracellular Ca2+ concentration (reported by an increase in Fluo-4 fluorescence intensity). (B) Example time courses of the FRET ratio ID/IA measured under the same experimental conditions. The upper two traces are taken from cells with high acceptor loading, and the bottom trace is from a cell displaying low acceptor loading. A transient increase in the FRET ratio (corresponding to a transient decrease in FRET efficiency) can be clearly seen above the slow decay in the FRET ratio caused by donor photobleaching. The red dashed line crossing both panels marks the onset of intracellular calcium release. AU, arbitrary units.
hEGFR alignment on the membrane leads to an extracellular conformation with similar asymmetry to the dEGFR.
We considered the implications of our FRET results for a model of the ectodomain dimer that combines the structure of the extended back-to-back dimer configuration of subdomains I to III with the structure of domain IV joined to the transmembrane domain and inserted in a lipid bilayer (Fig. 6A).
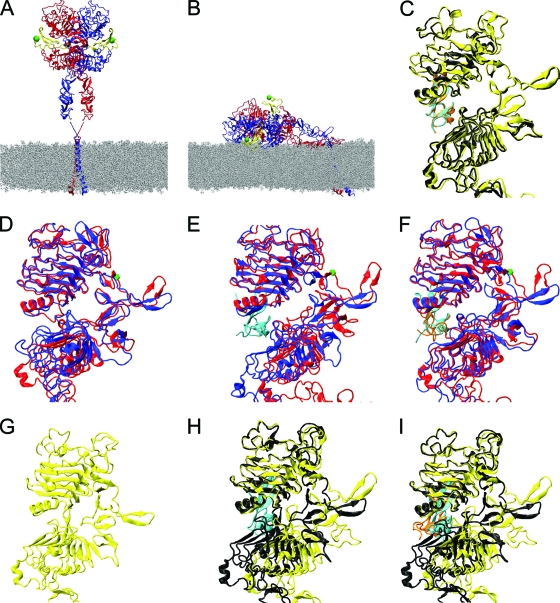
Fig. 6.
(A) The extended hEGFR ectodomain dimer with two bound ligands, modeled on crystallographic structures 1IVO and 1NQL and placed above modeled transmembrane helices in a POPC membrane. Receptor monomers are shown in red and blue ribbon representation, and both ligands are in yellow. Green spheres indicate the N termini of the ligands to which donor dyes are attached. (B) Endpoint of a molecular dynamics simulation of a doubly liganded, tilted ectodomain hEGFR dimer, relaxed on the membrane (23). Also shown are overlays of the left and right subunits of receptor dimers using domain I as a reference for doubly liganded soluble hEGFR (1IVO) (C), simulation of unliganded hEGFR relaxed on the membrane (D), simulation of singly liganded hEGFR relaxed on the membrane (E), simulation of doubly liganded hEGFR relaxed on the membrane (F), unliganded soluble dEGFR (3I2T) (G), singly liganded soluble dEGFR (3LTG) (H), and doubly liganded soluble dEGFR (3LTF) (I). The hEGFR residue Asp238 is highlighted by a green mark in panels D to F.
Comparison of the FRET-derived ligand-membrane distances with those predicted by the model in Fig. 6A shows that the distance of ∼8 nm found in receptors that do not display high-affinity binding (Fig. 4A) is consistent with the commonly assumed upright orientation. However, the short ligand-membrane distance of <4 nm associated with the occurrence of high-affinity binding (Fig. 3A, red, and B) is clearly inconsistent with the upright orientation, and these receptors must therefore adopt a different conformation. Although we find in our simulations that the extracellular domain is quite flexible at the level of subdomains, the only known large-scale rearrangement possible is the transformation between extended and tethered configurations. This involves a hinge motion between domains II and III and a bending of domain II but would not correspond with a large change in ligand-membrane separation (Fig. 1D). There is also no evidence for any flexibility between domains III and IV, with both extended and tethered structures giving a consistent view (17, 19, 36). To place ligand-binding sites within 4 nm of the cell surface, all four receptor domains would have to bend significantly toward the membrane. We note that the bulky ectodomain dimer is tethered to the membrane by only two transmembrane helices and could in principle adopt a range of tilted orientations around the recently described flexible hinge encompassing residues 615 to 621 (28); the upright conformation represents one extreme orientation (Fig. 6A), and the flat conformation represents the other (Fig. 6B). Given that the smallest cross-section of each hEGFR subunit in the dimer is ∼4 nm, we propose that only the flat conformation would place bound EGF molecules at separations consistent with a ligand-membrane distance of <4 nm.
Previously (23), we reported results for molecular dynamics simulations of the extracellular region of the hEGFR dimer placed on a membrane surface. We showed that the dimer remained stable but lost the approximate 2-fold symmetry of the crystal structure. The average dimerization interface area rose from 1,197 Å2 to 1,483 Å2, largely as a result of additional interactions N-terminal to the dimerization arm, while the two ligands buried 1,166 Å2 and 1,211 Å2 at distinct binding sites. Intriguingly, a similar asymmetry has recently been observed in the soluble ectodomain dimer of dEGFR (2), which leads to an increase in interface area per receptor from 1,131 Å2 to 1,698 Å2 and distinct ligand-binding sites.
Motivated by these structures of the dEGFR ectodomain dimer, we reanalyzed the results of our earlier simulation of the doubly liganded hEGFR ectodomain dimer aligned on a membrane (23). Starting from the energy-minimized endpoint of the MD simulation, we superimposed the two monomers using domain I as the reference (Fig. 6F); for comparison, the equivalent result for the solubilized extracellular domain is shown in (Fig. 6C). Fig. 6F shows the distortion of one dimerization arm reported earlier (23), which allows one monomer to be raised and, thus, the complete dimer to lie flat on the membrane. In addition, however, the two copies of domain II are seen to diverge around residue Asp238. The corresponding residue in dEGFR is precisely where a ligand-induced kink was identified (2). This region was also identified as the source of divergence of the ErbB1- and ErbB3-tethered structures (17), which have significantly different orientations of domain I and the N-terminal region of domain II.
To investigate extracellular conformational changes further, we carried out new MD simulations of the extracellular region of the unliganded extended hEGFR dimer aligned on the membrane (we note that although the soluble hEGFR extracellular region appears not to dimerize in the absence of ligand, there is evidence that it does predimerize in the cell membrane [9]). Starting from the asymmetric liganded dimer simulated previously (23), we removed both ligands and simulated the system for a further 50 ns. Minor subdomain reorientations were observed (of the order 10° to 20°), driven by the removal of the ligand connecting subdomains I and III, after which the structure appeared to be stable. The kink in domain II around residue Asp238 was reduced (Fig. 6D), and the interface area per receptor was observed to fall from the high value of the initial structure to a value of 1,315 Å2 (averaged over the second half of the trajectory), closer to the value for the symmetric dimer. Despite this, the ectodomain dimer retains significant asymmetry with a root mean square deviation (RMSD) between the monomers of 8.6 Å (averaged over snapshots taken from the second half of the simulation), compared to 4.3 Å observed for the doubly liganded dimer (Fig. 6F).
We also carried out a new MD simulation of a singly liganded hEGFR ectodomain dimer on the membrane, with the upper ligand removed and the resulting system simulated for 60 ns. The receptor interface area falls to a value of 1,363 Å2 (averaged over the second half of the trajectory), intermediate between the unliganded and doubly liganded results. However, the dimer is more asymmetric than in the other simulations (Fig. 6E), with an RMSD between the monomers of 12.8 Å (averaged over snapshots taken from the second half of the simulation). The average interface area between the remaining ligand and its receptor is 1,269 Å2, slightly higher than the areas observed for the doubly liganded dimer on the membrane, possibly indicating better ligand binding when only one ligand is present and driving the larger receptor asymmetry.
We conclude from the simulations that the presence of the membrane surface, together with the inherent flexibility of the ectodomain, leads to asymmetry in the dimer similar to that observed recently for dEGFR (2). Some asymmetry is always present but is particularly marked when a single ligand is bound.
DISCUSSION
In this report we show that treatment with MAb 2E9 and use of subsaturating concentrations of EGF, both of which enhance the contribution from high-affinity EGF-binding sites to the FRET-derived distance measurements, allows the detection of a hEGFR conformation in A431 cells in which EGF-binding sites are located <4 nm from the cell surface (Fig. 3A and 4B). Based on current models, in which the extended back-to-back EGF/hEGFR extracellular conformation is believed to represent the high-resolution structure of active hEGFR dimers, our FRET results provide strong evidence that receptors bearing high-affinity binding sites are extended receptor dimers with their extracellular domains aligned with the plasma membrane, while receptors bearing low-affinity sites display only a upright conformation. These results implicate two different receptor extracellular conformations in the occurrence of EGF-binding heterogeneity and concave-up Scatchard plots.
The observation of two liganded hEGFR conformations coexisting at the cell surface does not necessarily mean that we have identified two independent classes of high-affinity and low-affinity receptors. Our results represent snapshots of dynamic receptor populations, and it is likely that the high degree of flexibility at the base of the hEGFR extracellular region adjacent to the plasma membrane (28) allows receptors to sample a range of extracellular orientations. Interestingly, it has recently been shown that deletion or release of the Cys571-Cys593 disulfide bridge in close proximity to the flexible linker has a marked impact on EGF binding, either abrogating or causing an extreme enhancement of negative cooperativity, respectively (1). Within the context of the usual dynamic behavior attributed to cell surface hEGFRs, the flat orientation could represent a previously unconsidered intermediate species where interactions with the membrane produce additional conformational adjustments in ectodomain dimers that promote high-affinity ligand binding. Our observation of a conformational transient in hEGFR extracellular domains, consistent with a rocking motion that occurs in signaling-competent cells, provides some evidence of the relevance of extracellular axial rotations to hEGFR signaling.
Alvarado et al. argued that the observed asymmetry in doubly liganded dEGFR explains the negative cooperativity seen in soluble dEGFR ectodomains and provides a structural basis for high- and low-affinity sites (2). They suggest that the same is not observed in the soluble hEGFR extracellular region because intracellular interactions are required to stabilize the asymmetric singly liganded dimers. In contrast, our MD simulations show that the alignment of hEGFR ectodomains on a membrane surface is sufficient to introduce an asymmetry to receptor dimers very similar to that observed in soluble dEGFR ectodomains, suggesting that, with the aid of the membrane, negative cooperativity could be achieved via asymmetry in hEGFR.
However, the mechanism that regulates extracellular changes in orientation of hEGFR dimers with respect to the membrane it is not yet known. It is tempting to speculate that the flat configuration may be stabilized via interactions between hEGFR and other membrane components, including other membrane proteins, such as sugar-mediated interactions with gangliosides in the upper leaflet of the membrane (24, 44), consistent with prior observations that high-affinity EGF binding to EGFR is not seen outside cell membranes (22) and that altering the glycosylation of EGFRs in cells produces altered equilibrium binding kinetics (51). Yet intracellular interactions must also be involved in promoting the flat configuration, despite the apparent loose coupling between the extracellular and intracellular regions (28), because hEGFR mutants with deleted intracellular regions do not show negative cooperativity (25, 30) but have extracellular regions that are presumably competent to interact with other membrane components.
More generally, our MD simulations illustrate an ability of hEGFR extracellular subdomains to reorient themselves according to the presence or absence of ligand and according to the environment, consistent with a recent report (54). We speculate that such flexibility, as well as allowing the generation of asymmetry with the additional interaction with a membrane, may also be important if members of the mammalian EGFR family are to have the ability to form several heterodimers with other members of the family. There are as yet no crystal structures of ectodomain heterodimers, but formation of different interfaces may involve rearrangements similar to those seen here. It is also possible that the apparent preferential blocking of low-affinity binding sites by MAb 2E9 could arise from an inability to bind to tilted asymmetric ectodomain dimers due to disruption of its epitope by conformational changes.
We propose that the flat conformation of hEGFR found in A431 cells should also be at the surface of any other cell line, independent of receptor expression levels, as we obtained indirect evidence of short distances in HeLa cells. In addition, A431 cells display properties relevant to the formation of EGF/hEGFR complexes identical to those of other cell lines (9, 11, 14, 20, 41). Differences introduced by receptor overexpression reside in the proportions of unliganded dimers (35), the dimerization frequency in the cell periphery (9), interactions with other receptors (50), and the degree of nuclear localization of hEGFR (27).
EGF binding was performed at low temperature, and therefore we cannot rule out that this has an effect on our results. However, published data show that ligand-receptor complexes form normally at the cell surface at 4°C (34, 40, 48) and that neither receptor dimerization nor the initial stages of signal transduction prior to internalization are inhibited by the low temperature (32, 49). We believe that any effect is likely to be to arrest EGF/receptor complexes at conformations representative of an early state in signal transduction.
To conclude, our combined experimental and computational study has revealed an asymmetric conformation of human EGFR in contact with the plasma membrane that shares key features with that in Drosophila EGFR, suggesting that the structural basis for negative cooperativity is conserved from invertebrates to humans.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge funding from the United Kingdom Biotechnology and Biological Sciences Research Council (grant number BB/G006911/1).
We thank S. Webb and C. Ballestrem for reading the manuscript and B. Corry for providing the ExiFret code that was modified for use in this paper.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 28 March 2011.
REFERENCES
- 1. Adak S., Deandrade D., Pike L. J. 2011. The tethering arm of the EGF receptor is required for negative cooperativity and signal transduction. J. Biol. Chem. 286:1545–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alvarado D., Klein D. E., Lemmon M. A. 2010. Structural basis for negative cooperativity in growth factor binding to an EGF receptor. Cell 142:568–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Askari J. A., et al. 2010. Focal adhesions are sites of integrin extension. J. Cell Biol. 188:891–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Axelrod D. 1979. Carbocyanine dye orientation in red cell membrane studied by microscopic fluorescence polarization. Biophys. J. 26:557–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bagossi P., Horváth G., Vereb G., Szöllösi J., Tözsér J. 2005. Molecular modeling of nearly full-length ErbB2 receptor. Biophys. J. 88:1354–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barisas B. 1999. Dynamics of molecules involved in antigen presentation: effects of fixation. Mol. Immunol. 36:701–708 [DOI] [PubMed] [Google Scholar]
- 7. Carraway K. L., Koland J. G., Cerione R. A. 1990. Location of the epidermal growth factor binding site on the EGF receptor. A resonance energy transfer study. Biochemistry 29:8741–8747 [DOI] [PubMed] [Google Scholar]
- 8. Chigaev A., Buranda T., Dwyer D. C., Prossnitz E. R., Sklar L. A. 2003. FRET detection of cellular α4-integrin conformational activation. Biophys. J. 85:3951–3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chung I., et al. 2010. Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature 464:783–787 [DOI] [PubMed] [Google Scholar]
- 10. Citri A., Skaria K. B., Yarden Y. 2003. The deaf and the dumb: the biology of ErbB-2 and ErbB-3. Exp. Cell Res. 284:54–65 [DOI] [PubMed] [Google Scholar]
- 11. Cochet C., et al. 1988. Demonstration of epidermal growth factor-induced receptor dimerization in living cells using a chemical covalent cross-linking agent. J. Biol. Chem. 263:3290–3295 [PubMed] [Google Scholar]
- 12. Corry B., Jayatilaka D., Rigby P. 2005. A flexible approach to the calculation of resonance energy transfer efficiency between multiple donors and acceptors in complex geometries. Biophys. J. 89:3822–3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Davis R. J. 1988. Independent mechanisms account for the regulation by protein kinase C of the epidermal growth factor receptor affinity and tyrosine-protein kinase activity. J. Biol. Chem. 263:9462–9469 [PubMed] [Google Scholar]
- 14. Defize L. H., et al. 1989. Signal transduction by epidermal growth factor occurs through the subclass of high affinity receptors. J. Cell Biol. 109:2495–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Digman M. A., Dalal R., Horwitz A. F., Gratton E. 2008. Mapping the number of molecules and brightness in the laser scanning microscope. Biophys. J. 94:2320–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fabricant R. N., De Larco J. E., Todaro G. J. 1977. Nerve growth factor receptors on human melanoma cells in culture. Proc. Natl. Acad. Sci. U. S. A. 74:565–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ferguson K. M., et al. 2003. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol. Cell 11:507–517 [DOI] [PubMed] [Google Scholar]
- 18. Friedman B., et al. 1984. Tumor promoters block tyrosine-specific phosphorylation of the epidermal growth factor receptor. Proc. Natl. Acad. Sci. U. S. A. 81:3034–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Garrett T. P. J., et al. 2002. Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor α. Cell 110:763–773 [DOI] [PubMed] [Google Scholar]
- 20. Gusev K., et al. 2003. The store-operated calcium entry pathways in human carcinoma A431 cells: functional properties and activation mechanisms. J. Gen. Physiol. 122:81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Haas E., Katchalski-katzir E., Steinberg I. Z. 1978. Effect of the orientation of donor and acceptor on the probability of energy transfer involving electronic transitions of mixed polarization. Biochemistry 17:5064–5070 [DOI] [PubMed] [Google Scholar]
- 22. Jones J., Akita R. W., Sliwkowski M. X. 1999. Binding specificities and affinities of egf domains for ErbB receptors. FEBS Lett. 447:227–231 [DOI] [PubMed] [Google Scholar]
- 23. Kästner J., Loeffler H. H., Roberts S. K., Martin-Fernandez M. L., Winn M. D. 2009. Ectodomain orientation, conformational plasticity and oligomerization of ErbB1 receptors investigated by molecular dynamics. J. Struct. Biol. 167:117–128 [DOI] [PubMed] [Google Scholar]
- 24. Kawashima N., Yoon S.-J., Itoh K., Nakayama K. 2009. Tyrosine kinase activity of epidermal growth factor receptor is regulated by GM3 binding through carbohydrate to carbohydrate interactions. J. Biol. Chem. 284:6147–6155 [DOI] [PubMed] [Google Scholar]
- 25. Klein P., Mattoon D., Lemmon M. A., Schlessinger J. 2004. A structure-based model for ligand binding and dimerization of EGF receptors. Proc. Natl. Acad. Sci. U. S. A. 101:929–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lemmon M. A., Schlessinger J. 2010. Cell signaling by receptor tyrosine kinases. Cell 141:1117–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lin S. Y., et al. 2001. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat. Cell Biol. 3:802–808 [DOI] [PubMed] [Google Scholar]
- 28. Lu C., et al. 2010. Structural evidence for loose linkage between ligand binding and kinase activation in the epidermal growth factor receptor. Mol. Cell. Biol. 30:5432–5443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Macdonald J. L., Pike L. J. 2008. Heterogeneity in EGF-binding affinities arises from negative cooperativity in an aggregating system. Proc. Natl. Acad. Sci. U. S. A. 105:112–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Macdonald-Obermann J. L., Pike L. J. 2009. The intracellular juxtamembrane domain of the epidermal growth factor (EGF) receptor is responsible for the allosteric regulation of EGF binding. J. Biol. Chem. 284:13570–13576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Magun B. E., Matrisian L. M., Bowden G. T. 1980. Epidermal growth factor. Ability of tumor promoter to alter its degradation, receptor affinity and receptor number. J. Biol. Chem. 255:6373–6381 [PubMed] [Google Scholar]
- 32. Martin-Fernandez M., Clarke D. T., Tobin M. J., Jones S. V., Jones G. R. 2002. Preformed oligomeric epidermal growth factor receptors undergo an ectodomain structure change during signaling. Biophys. J. 82:2415–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mattoon D., Klein P., Lemmon M. A., Lax I., Schlessinger J. 2004. The tethered configuration of the EGF receptor extracellular domain exerts only a limited control of receptor function. Proc. Natl. Acad. Sci. U. S. A. 101:923–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McCune B. K., Earp H. S. 1989. The epidermal growth factor receptor tyrosine kinase in liver epithelial cells. The effect of ligand-dependent changes in cellular location. J. Biol. Chem. 264:15501–15507 [PubMed] [Google Scholar]
- 35. Nagy P., Claus J., Jovin T. M., Arndt-Jovin D. J. 2010. Distribution of resting and ligand-bound ErbB1 and ErbB2 receptor tyrosine kinases in living cells using number and brightness analysis. Proc. Natl. Acad. Sci. U. S. A. 107:16524–16529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ogiso H., et al. 2002. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell 110:775–787 [DOI] [PubMed] [Google Scholar]
- 37. Ozcan F., Klein P., Lemmon M. A., Lax I., Schlessinger J. 2006. On the nature of low- and high-affinity EGF receptors on living cells. Proc. Natl. Acad. Sci. U. S. A. 103:5735–5740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pike L. J., Casey L. 2002. Cholesterol levels modulate EGF receptor-mediated signaling by altering receptor function and trafficking. Biochemistry 41:10315–10322 [DOI] [PubMed] [Google Scholar]
- 39. Read R. D., Cavenee W. K., Furnari F. B., Thomas J. B. 2009. A Drosophila model for EGFR-Ras and PI3K-dependent human glioma. PLoS Genet. 5:e1000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rees A. R., Gregoriou M., Johnson P., Garland P. B. 1984. High affinity epidermal growth factor receptors of A431 cells have restricted lateral diffusion the surface. EMBO J. 3:1843–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ringerike T., Blystad F. D., Levy F. O., Madshus I. H., Stang E. 2002. Cholesterol is important in control of EGF receptor kinase activity but EGF receptors are not concentrated in caveolae. J. Cell Sci. 115:1331–1340 [DOI] [PubMed] [Google Scholar]
- 42. Schlessinger J. 2002. Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell 110:669–672 [DOI] [PubMed] [Google Scholar]
- 43. Shoyab M., De Larco J. E., Todaro G. J. 1979. Biologically active phorbol esters alter affinity of epidermal growth factor membrane receptors. Nature 279:387–391 [DOI] [PubMed] [Google Scholar]
- 44. Song W., Welti R., Hafner-Strauss S., Rintoul D. A. 1993. Synthesis and characterization of N-parinaroyl analogs of ganglioside GM3 and de-N-acetyl GM3. Interactions with the EGF receptor kinase. Biochemistry 32:8602–8607 [DOI] [PubMed] [Google Scholar]
- 45. Spink C. H., Yeager M. D., Feigenson G. W. 1990. Partitioning behavior of indocarbocyanine probes between coexisting gel and fluid phases in model membranes. Biochim. Biophys. Acta 1023:25–33 [DOI] [PubMed] [Google Scholar]
- 46. Stryer L., Haugland R. P. 1967. Energy transfer: a spectroscopic ruler. Proc. Natl. Acad. Sci. U. S. A. 58:719–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ullrich A., et al. 1984. Human epidermal growth factor receptor cDNA sequence and aberrant expression of the amplified gene in A431 epidermoid carcinoma cells. Nature 309:418–425 [DOI] [PubMed] [Google Scholar]
- 48. van Belzen N., et al. 1988. Direct visualization and quantitative analysis of epidermal growth factor-induced receptor clustering. J. Cell. Physiol. 134:413–420 [DOI] [PubMed] [Google Scholar]
- 49. Webb S. E. D., et al. 2008. Single-molecule imaging and fluorescence lifetime imaging microscopy show different structures for high- and low-affinity epidermal growth factor receptors in A431 cells. Biophys. J. 94:803–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Weihua Z., et al. 2008. Survival of cancer cells is maintained by EGFR independent of its kinase activity. Cancer Cell 13:385–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Whitson K. B., et al. 2005. Functional effects of glycosylation at Asn-579 of the epidermal growth factor receptor. Biochemistry 44:14920–14931 [DOI] [PubMed] [Google Scholar]
- 52. Wolber P., Hudson B. 1979. An analytic solution to the Förster energy transfer problem in two dimensions. Biophys. J. 28:197–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wolf D. E. 1985. Determination of the sidedness of carbocyanine dye labeling of membranes. Biochemistry 24:582–586 [DOI] [PubMed] [Google Scholar]
- 54. Zhang Z., Wriggers W. 2011. Polymorphism of the epidermal growth factor receptor extracellular ligand binding domain: the dimer interface depends on domain stabilization. Biochemistry 50:2144–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zidovetzki R., Johnson D. A., Arndt-Jovin D. J., Jovin T. M. 1991. Rotational mobility of high-affinity epidermal growth factor receptors on the surface of living A431 cells. Biochemistry 30:6162–6166 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.