Abstract
Endothelial cell activation and dysfunction underlie many vascular disorders, including atherosclerosis and inflammation. Here, we show that interleukin-4 (IL-4) markedly induced vascular cell adhesion molecule 1 (VCAM-1), both in cultured endothelial cells and in the intact endothelium in mice. Combined treatment with IL-4 and tumor necrosis factor alpha (TNF-α) resulted in further, sustained induction of VCAM-1 expression. IL-4-mediated induction of VCAM-1 and secondary monocyte adhesion was predominantly regulated by the transcription factor STAT6. Genome-wide survey of IL-4-mediated STAT6 binding from sequential chromatin-immunoprecipitation with deep sequencing (chromatin immunoprecipitation sequencing [ChIP-seq]) in endothelial cells revealed regions of transient and sustained transcription factor binding. Through the combination of DNA microarrays and ChIP-seq at the same time points, the majority of IL-4-responsive genes were shown to be STAT6 dependent and associated with direct STAT6 binding to their promoter. IL-4-mediated stable binding of STAT6 led to sustained target gene expression. Moreover, our strategy led to the identification of a novel functionally important STAT6 binding site within 16 kb upstream of the VCAM-1 gene. Taken together, these findings support a critical role for STAT6 in mediating IL-4 signal transduction in endothelial cells. Identification of a novel IL-4-mediated VCAM-1 enhancer may provide a foundation for targeted therapy in vascular disease.
INTRODUCTION
Endothelial cells are highly responsive to their extracellular milieu. Endothelial cell activation is a term used to describe the phenotypic response of endothelial cells to inflammatory mediators, including lipopolysaccharide, tumor necrosis factor alpha (TNF-α), and interleukin-1 (IL-1). The activation phenotype typically includes some combination of increased leukocyte adhesiveness, reduced barrier function, a shift in hemostatic balance toward the procoagulant side, and altered vasomotor tone. Many of these properties are mediated by changes in gene expression.
Vascular cell adhesion molecule 1 (VCAM-1) is a 110-kDa cell surface glycoprotein that is expressed in cytokine-activated endothelial cells. VCAM-1 is also expressed in other cell types, including smooth muscle cells and fibroblasts (8). The VCAM-1 promoter represents a potentially valuable tool for dissecting the molecular mechanisms of endothelial cell activation. Previous studies have implicated a role for NF-κB (17, 28, 29), GATA (28, 29, 44), Sp1 (34), activating protein 1 (2), interferon regulatory factor 1 (35), and SOX18 (13) in mediating inducible expression of VCAM-1. Several transcription factors have been shown to interfere with NF-κB-dependent expression of VCAM-1, including KLF4 and Oct1 (7, 12).
IL-4 is a 20-kDa pleiotropic cytokine expressed by T helper 2 (Th2) lymphocytes, eosinophils, basophils, and mast cells (reviewed in references 38 and 42). IL-4 has been shown to be necessary for stabilization of the Th2 phenotype and promotes the synthesis of IgE (reviewed in references 6 and 22). IL-4 has been implicated in the pathogenesis of atherosclerosis (reviewed in reference 21) and allergic asthma (reviewed in reference 6). Signaling of IL-4 in endothelial cells occurs via a heterodimeric IL-4 receptor (IL-4R), consisting of IL-4Rα and IL-13Rα subunits (36). Activation of the receptor results in Janus kinase 1/2 (JAK-1/2)-dependent tyrosine phosphorylation and subsequent dimerization of signal transducer and activation of transcription 6 (STAT6), which then translocates to the nucleus and binds to consensus sequences (TTCN3–4GAA) found within promoters of IL-4-regulated target genes (14, 27). Previous studies with endothelial cells have demonstrated that IL-4 induces the expression of CXCL-8, inducible nitric oxide synthase (iNOS) (15), urokinase-type plasminogen activator (u-PA) (46), vascular endothelial growth factor (VEGF) (15), P-selectin (20, 32, 47), monocyte chemoattractant protein 1 (MCP-1) (39), CCL26 (18), IL-6 (25), 15-lipoxygenase (24), and osteoprotegerin (41). In addition, previous studies have shown that IL-4 upregulates the expression of VCAM-1 in endothelial cells (4, 10, 14, 23, 26, 37, 40). In contrast, IL-4 does not lead to increased expression of intercellular adhesion molecule 1 (ICAM-1) (10, 43) and has a variable effect on E-selectin expression (3, 10, 15).
The mechanisms underlying IL-4-mediated induction of VCAM-1 are poorly understood. A previous analysis of the VCAM-1 promoter failed to reveal STAT6 binding sites (16). One study demonstrated that IL-4-dependent increase in VCAM-1 levels is mediated by stabilization of VCAM-1 mRNA (16). A role for reactive oxygen species has also been suggested (23). The goal of the present study was to delineate the molecular basis for IL-4-mediated induction of VCAM-1 expression in endothelial cells. Using a combination of chromatin immunoprecipitation sequencing (ChIP-seq) and functional promoter analyses, we show that IL-4 induction of VCAM-1 is mediated by a STAT6 binding site at kb −16 relative to the transcriptional start site.
MATERIALS AND METHODS
Mice.
All experiments were performed with 6- to 8-week-old male C57BL/6 mice (CLEA). Mouse IL-4 (PeproTech) or mouse TNF-α (PeproTech) was dissolved in phosphate-buffered saline (PBS) and injected intravenously (i.v.). All animal studies were approved by the University of Tokyo Institutional Animal Care and Use Committee.
Immunofluorescence studies.
Frozen tissue sections (10 μm) were fixed and incubated with a rat monoclonal anti-VCAM-1 antibody (1:50 dilution) (BD Pharmingen) and a goat polyclonal anti-ICAM-2 antibody (8 μg/ml) (R&D Systems) overnight at 4°C. Sections were washed 3 times in PBS and incubated with secondary antibody labeled with Alexa Fluor 594 (for VCAM-1) or Alexa Fluor 488 (for ICAM-2) (1:50 dilution) (Invitrogen) for 1 h at room temperature. The slides were then washed in PBS, mounted in ProLong Gold antifade reagent with DAPI (4′,6-diamidino-2-phenylindole) (Invitrogen), and examined by fluorescence microscopy.
Cell culture.
Human umbilical vein endothelial cells (HUVECs) were purchased from Lonza and cultured in EGM-2 medium containing 2% fetal bovine serum (FBS) (Lonza). Cells were used at passages 3 to 4. HUVECs were stimulated with human IL-4 (20 ng/ml; PeproTech) and/or human TNF-α (10 ng/ml; PeproTech). HEK-293 (ATCC CRL-1573) human embryonic kidney cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% FBS. Human U937 cells (JCRB-9021; Japan) were grown in RPMI 1640 medium supplemented with 10% FBS.
Generation of CA-STAT6 adenoviruses.
Human STAT6 was cloned by PCR using total RNA from HUVECs and STAT6-specific primers (sequences are shown in Table S1 in the supplemental material). Constitutively active STAT6 (CA-STAT6) was generated by mutating V547T548 to A547A548 in human STAT6 (5). The cDNA was subcloned into pIRES2-EGFP (Clontech) and then transferred into the pShuttle and Adeno-X DNA (Clontech) by using the Adeno-X adenoviral expression system (Clontech). All cloned constructs were confirmed by restriction enzyme digestions and automated DNA sequencing.
Quantitative real-time PCR.
Two micrograms of TRIzol-extracted RNA was reverse transcribed using SuperScript II enzyme and oligo(dT) primer as specified by Invitrogen. Real-time PCR was performed by using the SYBR green PCR reagent according to the manufacturer's instructions (Applied Biosystems). Primer pair DNA sequences are shown in Table S1 in the supplemental material.
Western blot analysis.
Endothelial cells were washed with ice-cold PBS, collected with a cell scraper, and lysed with radioimmunoprecipitation assay (RIPA) buffer as described previously (30). The membrane was blocked with Tris-buffered saline-Tween (TBS-T) containing 2% skim milk and incubated with primary antibody against total STAT6 (Santa Cruz Biotechnology), pSTAT6 (Cell Signaling), VCAM-1 (Santa Cruz Biotechnology), lamin A (Sigma), α-tubulin (Sigma), p65 NF-κB (Santa Cruz Biotechnology), or β-actin (Sigma).
Monocyte adhesion assays.
Monocyte adhesion to HUVECs was assayed as previously described (30). In brief, confluent HUVECs were infected with an adenovirus control (Ad-control) or adenovirus expressing constitutively active STAT6 (Ad-CA-STAT6) or transfected with a small interfering RNA (siRNA) control (si-control) or STAT6 siRNA (si-STAT6). Cells were then treated with 20 ng/ml IL-4 for 24 h. PKH-26 (Sigma)-labeled U937 cells were added to a HUVEC-seeded plate. Ninety minutes later, cells were washed and examined by fluorescent microscopy.
Plasmids, transient transfections, and luciferase assays.
The construction of VCAM-1-luc (bp −1716 and +119) was previously described (29). The STAT6 binding regions (kbp −16, −11, and +15) were cloned by PCR with genomic DNA from HUVECs and specific primers containing SacI and XhoI sites (shown in Table S1 in the supplemental material). SacI- and XhoI-digested enhancer fragments were subcloned into SacI/XhoI-digested VCAM-1-luc. HUVECs were transiently transfected with plasmid DNA using FuGENE HD reagent (Roche Molecular Biomedicals), and luciferase activity was measured with the dual-luciferase assay kit (Promega) as previously described (31).
DNA microarrays.
HUVECs were transfected with either si-control or si-STAT6 (oligo1 or oligo2) for 48 h. Alternatively, HUVECs were infected with Ad-control or Ad-CA-STAT6. RNA was harvested and purified with TRIzol (Invitrogen). Preparation of cRNA and hybridization of probe arrays were performed according to the manufacturer's instructions (Affymetrix). Data were analyzed according to the MIAME rule.
ChIP.
HUVECs were cross-linked with 1 mM disuccinimidyl glutarate (Pierce) for 30 min at room temperature, washed once with ice-cold PBS, and cross-linked again using 1% formaldehyde for 10 min at room temperature. Cells were prepared for chromatin immunoprecipitation (ChIP) (data are available at http://www.lsbm.org/MCB_2146207). Antibodies against histone H3 lysine 4 trimethyl (H3K4me3) (ab8580; Abcam), histone H3 lysine 4 monomethyl (H3K4me1) (kindly provided by H. Kimura), p300 (05-257; Upstate), acetylated histone H4 (H4Ac) (06-866; Upstate), and STAT6 (sc-621; Santa Cruz) were added and immunoprecipitated with protein A/G-conjugated magnetic beads (Invitrogen). Prepared DNA was quantified by using Qubit (Invitrogen), and more than 10 ng of DNA was processed for ChIP-seq and ChIP-quantitative PCR (qPCR).
ChIP-seq.
All protocols for Illumina/Solexa sequence preparation, sequencing, and quality control are provided by Illumina. A brief summary of the technique and minor protocol modifications are available at http://www.lsbm.org/MCB_2146207.
Statistics.
Data are shown as means ± standard deviations (SD). P values were calculated by using the two-tailed unpaired Student's t test. A P value of <0.05 was considered significant.
Accession numbers.
Annotation of the probe numbers and targeted sequences are shown on the Affymetrix web page under accession no. GSE 28117. ChIP-seq data are available under accession no. SRA030735.1.
RESULTS
IL-4 induces VCAM-1 but not ICAM-1 expression in cultured endothelial cells.
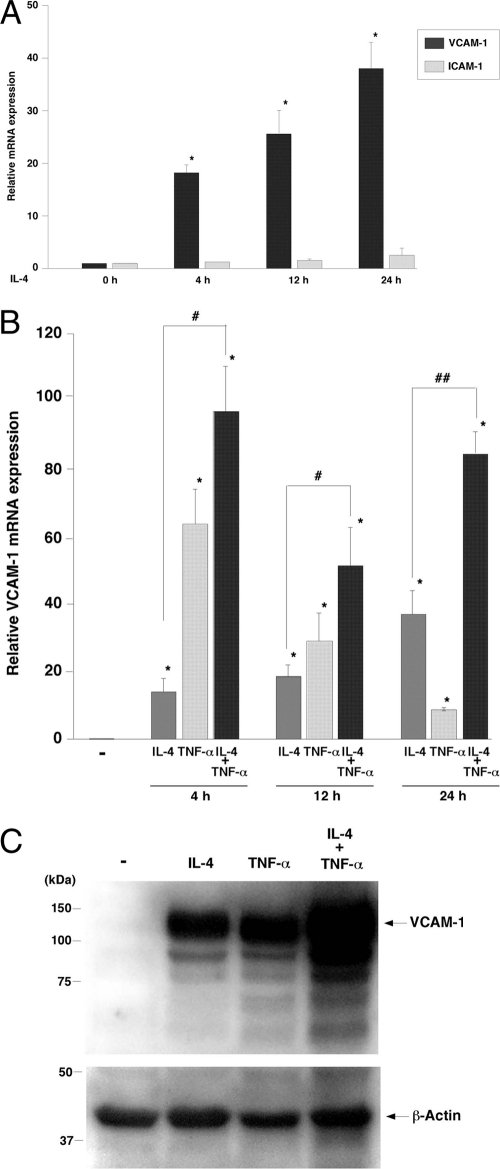
Previous studies have shown that IL-4 induces VCAM-1 expression in cultured endothelial cells (15). Consistent with these data, incubation of human umbilical vein endothelial cells (HUVECs) with 20 ng/ml IL-4 resulted in a time-dependent increase in IL-4 mRNA, as measured by real-time PCR, with peak levels (39.6-fold) occurring at 24 h (Fig. 1A). In contrast, IL-4 did not affect ICAM-1 mRNA expression. Compared with IL-4, TNF-α resulted in an earlier though far greater peak (64.5-fold at 4 h) in VCAM-1 mRNA levels (Fig. 1B). Combined treatment with IL-4 and TNF-α resulted in additive (at 4 and 12 h) and synergistic (at 24 h) induction of VCAM-1 expression (Fig. 1B). In Western blot analyses, treatment of HUVECs for 24 h with IL-4 or TNF-α resulted in comparable induction of VCAM-1 protein levels, while combined treatment with IL-4 and TNF-α resulted in further induction of VCAM-1 (Fig. 1C). Thus, IL-4 results in sustained induction of VCAM-1 expression in cultured endothelial cells.
Fig. 1.
IL-4-mediated induction of VCAM-1 expression in primary cultured endothelial cells. (A) Confluent HUVECs were serum starved for 16 h and treated with 20 ng/ml IL-4 for the indicated times. Total RNA was harvested and assayed by quantitative real-time PCR for VCAM-1 and ICAM-1 mRNA levels. Data are expressed relative to untreated cells as means ± standard deviations (n = 4). *, P < 0.001 compared with untreated control. (B) HUVECs were treated with IL-4 and/or TNF-α for the indicated times and then assayed for VCAM-1 mRNA levels using quantitative real-time PCR. Data are expressed relative to untreated cells as means ± standard deviation (n = 4). *, P < 0.01 compared with untreated cells; #, P < 0.01, and ##, P < 0.05, compared with IL-4-treated cells at each time point. (C) HUVECs were treated as described above for 24 h. Western blotting was performed using anti-VCAM-1 antibody. The membrane was stripped and reprobed with anti-β-actin antibody as a loading control. Shown are representative data from three independent experiments.
IL-4 induces VCAM-1 but not ICAM-1 expression in endothelial cells in vivo.
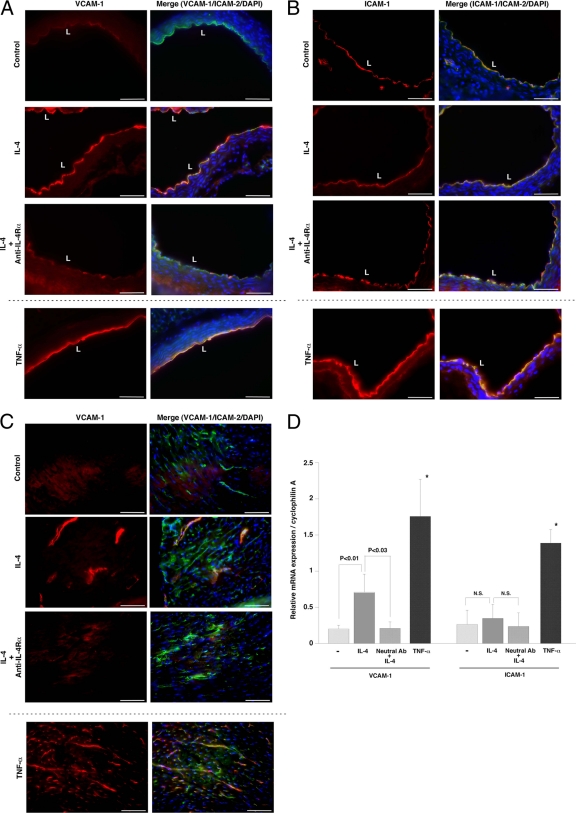
To determine whether IL-4 induces VCAM-1 expression in vivo, mice were injected intravenously (i.v.) with IL-4 (0.5 mg/kg) or an equal volume of saline (control). Aortas and hearts were harvested 6 h later, and cryosections were processed for immunostaining of VCAM-1 or ICAM-1 and the endothelial marker ICAM-2. Compared with the control, IL-4 treatment resulted in marked upregulation of VCAM-1 protein in the aorta, the majority of which colocalized with ICAM-2 in the endothelium (Fig. 2A). This effect was blocked by pretreatment with IL-4 receptor-α blocking antibody. TNF-α (0.1 mg/kg i.v.) injection resulted in a comparable induction of VCAM-1 protein expression in aortic endothelium. In contrast, TNF-α, but not IL-4, also resulted in induction of ICAM-1 in both vascular endothelial cells and smooth muscle cells of the aorta (Fig. 2B).
Fig. 2.
IL-4-mediated induction of VCAM-1 expression in the intact endothelium. Mice were pretreated with saline (control) or blocking antibody against IL-4Rα and then injected i.v. with control saline, 0.5 mg/kg IL-4, or 0.1 mg/kg TNF-α. Tissues were harvested 6 h later, and cryosections of the aorta (A and B) and heart (C) were collected and stained with anti-VCAM-1 (A and C) or anti-ICAM-1 (B) antibody (left panels, red), ICAM-2 (right panels, green), and DAPI (right panels, blue). Merged images are shown in the right panels. Bar, 50 μm. L, lumen. (D) Total RNA was harvested from hearts and assayed for VCAM-1 or ICAM-1 mRNA levels by quantitative real-time PCR. Ab, antibody. The results show the means and standard deviations of expression levels relative to cyclophilin A from at least four independent sets of mice. *, P < 0.001 compared with VCAM-1 or ICAM-1 expression levels with saline treatment. N.S., nonsignificant.
In the heart, IL-4 treatment resulted in increased VCAM-1 protein expression in the endothelial lining of medium-size blood vessels. In contrast, TNF-α induced VCAM-1 expression both in capillaries and in larger vessels (Fig. 2C). IL-4 treatment had no effect on ICAM-1 levels in the heart (data not shown). To quantitate the effect of systemic IL-4 and TNF-α on VCAM-1 expression, mouse hearts were harvested for RNA and processed for real-time PCR. As shown in Fig. 2D, IL-4 and TNF-α treatment resulted in significant (3.3-fold and 8.5-fold, respectively) increases in VCAM-1 mRNA levels. The effect of IL-4 was blocked (98%) by pretreatment with IL-4 receptor blocking antibody. TNF-α, but not IL-4, induced ICAM-1 mRNA expression in the heart (Fig. 2D). Collectively, these findings suggest that IL-4 results in vascular-bed-specific induction of VCAM-1 in the intact endothelium.
STAT6 is required for IL-4-mediated induction of VCAM-1 in cultured endothelial cells.
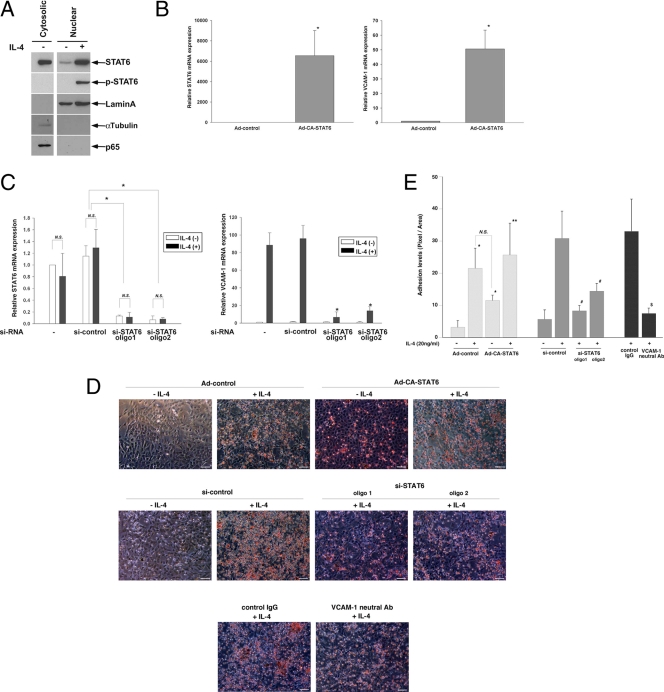
Previous studies have shown that IL-4 activates STAT6 in endothelial cells (36, 40). Moreover, STAT6 has been implicated in IL-4-mediated induction of P-selectin and eotaxin-3 in endothelial cells (18, 20, 32). As shown in Fig. 3A, incubation of HUVECs with 20 ng/ml IL-4 resulted in tyrosine phosphorylation of STAT6 and translocation of STAT6 from the cytosol to the nucleus (at 60 min). In contrast to TNF-α or thrombin, IL-4 failed to promote nuclear translocation of p65 NF-κB (at 60 min).
Fig. 3.
Role of STAT6 in IL-4-mediated induction of VCAM-1 and monocyte adhesion in primary cultured endothelial cells. (A) HUVECs were treated in the presence or absence of 20 ng/ml IL-4 and then processed for cytosolic and nuclear fractions. Western blot analysis was carried out using antibodies against total STAT6, phospho-STAT6, nuclear lamin A, cytosolic α-tubulin, and p65 NF-κB. (B) HUVECs were treated with Ad-control or Ad-CA-STAT6. STAT6 (left) and VCAM-1 (right) mRNA levels were measured by quantitative real-time PCR. The results show the means and standard deviations of expression levels relative to Ad-control, derived from three independent experiments. *, P < 0.001 compared with Ad-control. (C) HUVECs were transfected with si-control or two independent siRNAs against STAT6 (oligo1 or oligo2), serum starved, and then treated with 20 ng/ml IL-4 for 24 h. STAT6 (left) and VCAM-1 (right) mRNA levels were measured by quantitative real-time PCR. The results show the means and standard deviations of expression levels relative to si-control in the absence of IL-4 treatment, derived from at least four independent experiments. *, P < 0.01 compared with si-control-treated cells in the presence of IL-4. N.S., nonsignificant. (D and E) U937 monocytic cell adhesion assays were carried out as described in Materials and Methods. HUVECs were infected with Ad-control or Ad-CA-STAT6 or transfected with si-control or si-STAT6, preincubated with control IgG or neutralizing antibody against VCAM-1, treated with 20 ng/ml IL-4 for 24 h, and then washed and incubated with U937 monocytes. The results are representative of four independent optical images from three independent experiments. Bar, 50 μm. Adhesion levels were quantitated (E). *, P < 0.01, compared with Ad-control minus IL-4; **, P = 0.045 compared with Ad-CA-STAT6 minus IL-4; #, P < 0.01 compared with si-control plus IL-4; and $, P < 0.01 compared with control IgG.
To determine whether STAT6 induces VCAM-1 expression, HUVECs were infected with adenovirus expressing constitutively active STAT6 (Ad-CA-STAT6). In the absence of cytokine stimulation, Ad-CA-STAT6 resulted in a 52-fold induction of VCAM-1 mRNA levels (Fig. 3B). In contrast, CA-STAT6 had no effect on ICAM-1 mRNA expression (data not shown; see Table S2 in the supplemental material). To determine whether endogenous STAT6 plays a role in mediating basal and/or inducible expression of VCAM-1, HUVECs were transfected with two independent siRNAs against STAT6 (oligo1 and oligo2). Each si-STAT6 resulted in >85% reduction of STAT6 and VCAM-1 mRNA in cells treated in the absence or presence of IL-4 (Fig. 3C). Taken together, these findings suggest that VCAM-1 is a STAT6-responsive gene and that IL-4 induces VCAM-1 expression via a STAT6-dependent mechanism.
IL-4-mediated STAT6 activation augments monocyte adhesion to cultured endothelial cells.
To determine whether STAT6-mediated induction of VCAM-1 has functional consequences in IL-4-treated endothelial cells, we carried out cell adhesion assays using HUVECs and U937 monocyte cells. IL-4 treatment (20 ng/ml for 24 h) of Ad-control-transfected HUVECs resulted in 7.1-fold increased monocyte adhesion (Fig. 3D and E). In the absence of IL-4 treatment, Ad-CA-STAT6 resulted in a 3.7-fold increase in cell adhesion. Treatment of CA-STAT6-expressing HUVECs with IL-4 resulted in further induction of adhesion, but the absolute levels were comparable with those observed in IL-4-treated Ad-control-infected cells. IL-4 treatment of si-control-treated HUVECs resulted in 5.5-fold increased monocyte adhesion (Fig. 3D and E). This effect was attenuated 73.4% and 55.4% by si-STAT6 oligo1 and oligo2, respectively (Fig. 3D and E). Treatment of HUVECs with neutralizing antibody against VCAM-1 resulted in a comparable reduction (74.6%) of IL-4-mediated monocyte adhesion (Fig. 3D and E). Together, these data suggest that STAT6 plays a critical role in IL-4-mediated VCAM-1-dependent monocyte adhesion to endothelial cells.
Genome-wide survey of IL-4-regulated genes in endothelial cells.
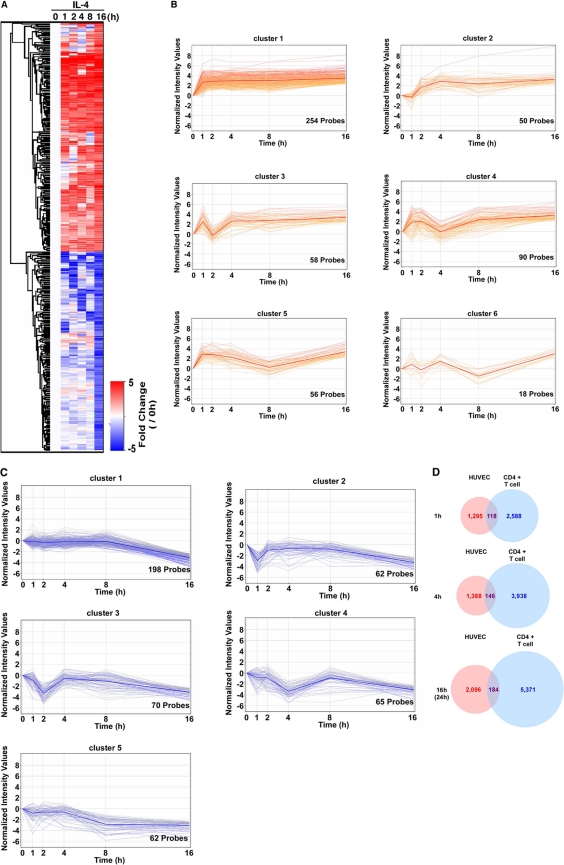
IL-4 resulted in the sustained induction of VCAM-1. Our next goal was to determine the extent to which IL-4 induced other genes in the endothelial cells. To that end, we carried out whole-genome expression arrays in HUVECs treated in the absence (0 h) or presence of IL-4 (1, 2, 4, 8, and 16 h). IL-4-regulated genes were collected, as those that were up- or downregulated more than 2-fold via IL-4 treatment at 16 h. Using these criteria, a total of 983 gene probes were identified (Fig. 4A). A total of 526 of these were upregulated at 16 h (Fig. 4A, red), and 457 were downregulated at 16 h (Fig. 4A, blue). The IL-4-induced genes were further divided into 6 clusters according to temporal changes in mRNA expression: (i) induced at all time points, (ii) induced after 2 h, (iii) transiently reduced at 2 h, (iv) transiently reduced at 4 h, (v) transiently reduced at 8 h, and (vi) altered in a fluctuating pattern (Fig. 4B). (The full probe list is shown in Table S2 in the supplemental material.) Those genes that were downregulated at 16 h were divided into five temporal patterns: (i) progressively downregulated, (ii) downregulated at 1 h, (iii) downregulated at 2 h, (iv) downregulated at 4 h, and (v) downregulated at 8 h (Fig. 4C). (The full probe list is shown in Table S2 in the supplemental material.)
Fig. 4.
Genome-wide analysis of IL-4-mediated up- or downregulated genes in primary cultured endothelial cells. (A) Heat map representation of IL-4-regulated genes. The color intensity indicates the expression level (upregulated in red and downregulated in blue relative to the median in white). A total of 983 probes demonstrating >5-fold upregulation or downregulation in the presence of IL-4 for 16 h were clustered at indicated time points. (B) Kinetics of IL-4-induced genes (526 probes) using Gene Spring software (Agilent). The bold line indicates the median of all probes in each cluster. (C) Kinetics of IL-4-repressed genes (457 probes) using Gene Spring software. The bold line indicates the median of all probes in each cluster. (D) Venn diagram showing overlap of IL-4-induced genes in HUVECs and CD4+ T cells. Whole probes were selected based on >2-fold induction after IL-4 treatment at 1, 4, and 16 h (HUVECs) or 1, 4, and 24 h (T cells).
In a recent study, genome-wide information was reported for IL-4-regulated genes in CD4+ T cells (9). Thus, we had the opportunity to compare expression patterns in IL-4-treated endothelial cells and T lymphocytes. Only ≈10% of the IL-4-inducible gene probes in HUVECs were also upregulated in CD4+ T cells at similar time points (Fig. 4D). For example, suppressor of cytokine signaling 1 (SOCS1) was commonly upregulated in both cell types. (The full list of commonly induced gene probes is shown in Table S3 in the supplemental material.) However, VCAM-1 was specifically induced in endothelial cells, whereas T-cell receptor was selectively upregulated in T cells. Taken together, these findings suggest that IL-4 signaling in endothelial cells results in temporally diverse cell-type-specific changes in gene expression.
Genome-wide survey of IL-4-mediated STAT6 binding in endothelial cells.
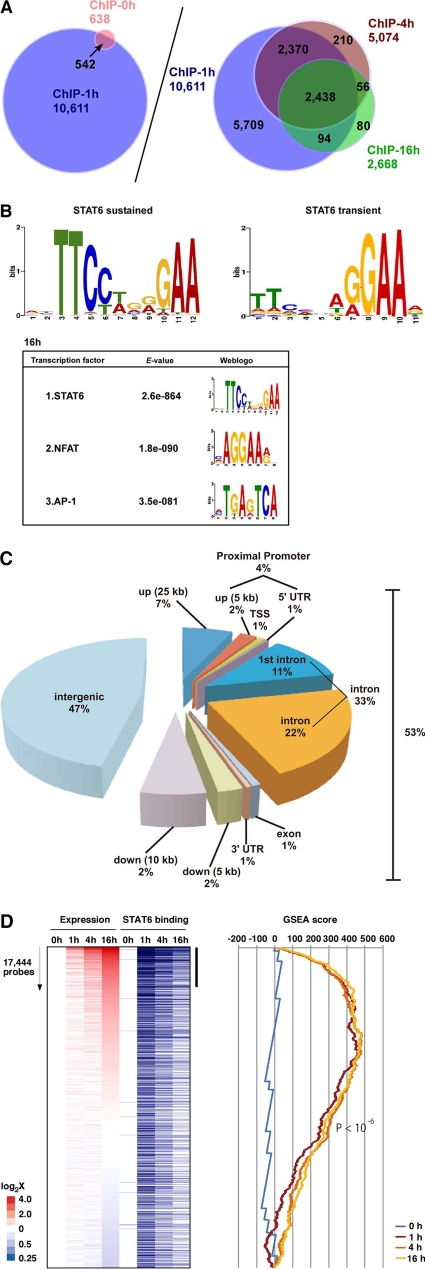
We next wished to determine whether IL-4-regulated gene expression correlated with STAT6 binding. To identify genes directly bound by STAT6 in IL-4-treated endothelial cells, we carried out chromatin immunoprecipitation (ChIP) with STAT6 antibody in HUVECs treated in the absence (0 h) or presence of IL-4 (1, 4, and 16 h), followed by ChIP-seq analysis. Nonimmunoprecipitated DNA (input DNA) was used as a negative control to define nonspecific binding. Totals of 12,741,937 (0 h), 13,828,587 (1 h), 14,150,991 (4 h), and 14,816,881 (16 h) nonredundant reads were aligned onto the human genome. In IL-4-treated HUVECs, we identified a total of 10,611 STAT6-occupied peaks (Fig. 5A). A total of 542 of these were present in untreated HUVECs and thus likely represent weak, basal STAT6 binding and/or false-positive binding (Fig. 5A, left). All of the remaining binding regions were induced at 1 h. Half of these bindings (5,709/10,069 regions) were transient (average signal ratio, 15.4), occurring at 1 h but not 4 or 16 h (Fig. 5A, right), whereas the rest were sustained at 4 or 16 h (average signal ratio, 59.7) (calculation data not shown).
Fig. 5.
Genome-wide ChIP-seq analysis of IL-4-mediated STAT6 binding in primary cultured endothelial cells. (A) ChIP-seq analysis was used to survey genome-wide STAT6 binding in HUVECs treated with IL-4 for 0, 1, 4 and 16 h. (B, upper) De novo search for STAT6 binding recognition sequence based on sustained or transient binding in HUVEC. (Lower) Second and third enriched motifs calculated by the MEME method. The E-value indicates the probability of de novo enriched sequences obtained from ChIP-seq. (C) Distribution of STAT6 binding sites in the proximal promoter (within 5 kb upstream of the 5′ untranscribed region [UTR] from the transcriptional start site [TSS]), exon, intron, and intergenic regions (defined by regions 25 kb upstream and 10 kb downstream from the TSS). (D) IL-4-responsive genes (based on 17,444 probes [representing ∼8,500 genes] in DNA microarrays) at 1, 4, and 16 h, sorted according to the induction ratio at 16 h (left). Genes are aligned with results of ChIP-seq (blue bars indicate STAT6 binding). The vertical black bar indicates a group of highly induced IL-4-responsive genes at 16 h that are enriched in STAT6 binding. A graphic representation of the GSEA enrichment score is shown on the right.
Previous studies have demonstrated that STAT proteins bind to a TTCN3–4GAA consensus element (14). A recent report using ChIP-seq in T cells showed that while STAT4 bound to a GAA palindrome with a 3-nucleotide spacer, STAT6 preferentially bound to a GAA palindrome separated by a 4-bp spacer (TTCN4GAA) (45). To determine and compare the binding element(s) in our genome-wide analysis between 1 h and 16 h of IL-4 treatment, we searched the consensus motifs of STAT6-bound regions. Similar to the results in T lymphocytes, our data revealed enrichment for the TTCN4GAA STAT6 binding site at 16 h (Fig. 5B). Transient STAT6 binding yielded a weaker enrichment score, typically at the 5′-TTC-3′ region in the consensus element. Finally, STAT6 binding at 16 h but not 1 h was associated with coenrichment in other transcription factor binding sites, including NFAT and AP-1 (Fig. 5B).
To determine the location of STAT6 binding sites with IL-4 treatment for 16 h, we divided the human genome into 5 regions relative to the transcriptional start site (TSS) of the genes. As shown in Fig. 5C, 53% of STAT6 binding sites were located between −25 kbp and +10 kbp of the genes. Of these binding sites, 4% were in the proximal promoter region (between −5,000 and +1), while 33% and 1% were located in the introns and exons, respectively. The remaining 47% of STAT6 binding sites were localized to intergenic regions.
Binding of STAT6 to DNA does not necessarily correlate with changes in gene expression. To determine the association between genome-wide STAT6 binding and target gene expression, we compared the results of ChIP-seq with those of DNA microarrays of control and IL-4-treated endothelial cells. We selected microarray gene set probes that exhibited significant expression, as defined by >100 average difference in control HUVECs or HUVECs treated with IL-4 for 1, 4, or 16 h. A total of 17,444 probes (∼8,500 genes) met these criteria and were sorted by the log-fold induction/reduction ratio of the expression levels in HUVECs. IL-4 treatment for 1, 4, and 16 h showed a highly significant enrichment score (P < 10−6) (Fig. 5D, right). The correlation between IL-4-mediated gene induction and enriched STAT6 binding within the proximal (within 20 kb upstream and downstream of the TSS) was greatest at 16 h of treatment, compared with 1 h and 4 h (Fig. 5D, left). Interestingly, the majority of genes whose expression peaked at 16 h demonstrated STAT6 binding as early as 1 h (see Fig. S1 in the supplemental material). In contrast to IL-4-inducible genes, those genes whose expression was inhibited by IL-4 failed to reveal significant correlation with proximal STAT6 binding (see Fig. S1).
Identification of STAT6-dependent genes in endothelial cells.
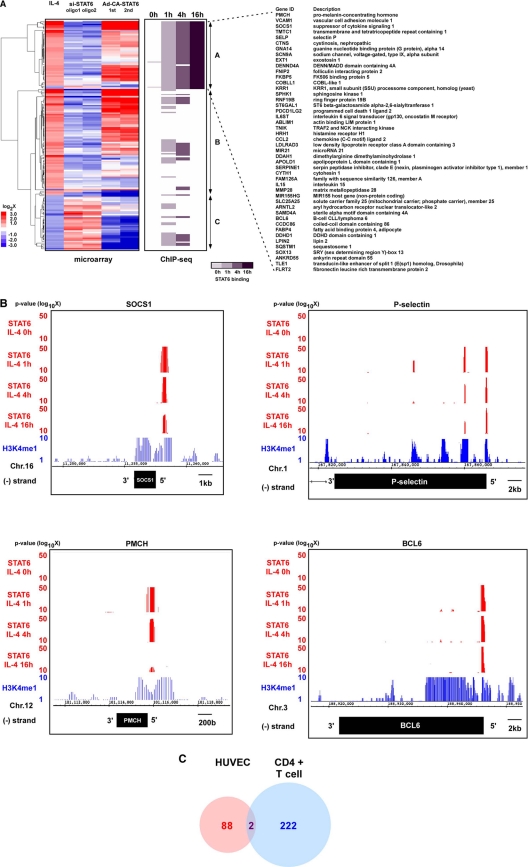
Subsequently, to determine which IL-4-responsive genes were dependent on STAT6, we carried out duplicate DNA microarrays of control-siRNA- or si-STAT6-transfected HUVECs treated in the absence or presence of IL-4 or HUVECs infected with Ad-CA-STAT6. (The full array data are shown in Table S2 in the supplemental material.) Three gene clusters were identified: (i) IL-4-induced genes stably bound by STAT6 (1, 4, and 16 h) (Fig. 6A, upper one-third, group A), (ii) IL-4-induced genes not stably bound by STAT6 (Fig. 6A, middle one-third, group B), and (iii) IL-4-repressed genes (Fig. 6A, lower one-third, group C). STAT6 knockdown inhibited the effect of IL-4 on 79% of its target genes. The majority of IL-4-inducible STAT6-dependent genes were activated by Ad-CA-STAT6, while many IL-4-STAT6-repressible genes were downregulated by Ad-CA-STAT6 (Fig. 6A). Interestingly, few of the genes that were downregulated by IL-4 demonstrated stable or even transient binding of STAT6. Representative ChIP-seq data from group A are shown for SOCS1, P-selectin, pro-melanin-concentrating hormone (PMCH), and BCL6 (Fig. 6B). In each case, the STAT6 binding region was adjacent to a histone H3 lysine 4 monomethyl (H3K4me1)-positive region (Fig. 6B, blue lines), suggesting that STAT6 binding is associated with transcriptional activation in group A of Fig. 6A. Finally, of those genes that were induced by IL-4 in T cells and HUVECs in a STAT6-dependent manner (at 4 h where data for comparison are available), only 2 were common to both cell types (SOCS1 and elongation factor for PolII [ELL]) (Fig. 6C). (The full gene list is shown in Table S4 in the supplemental material.) Taken together, these data suggest that the majority of IL-4-responsive genes are transcriptionally dependent on STAT6. Approximately half of the IL-4-inducible genes are associated with sustained STAT6 binding on the promoter in endothelial cells. Although the IL-4-STAT6 signaling axis is common to endothelial and T cells, the two lineages demonstrate cell-type-specific transcriptional responses.
Fig. 6.
Genome-wide analysis of STAT6-regulated genes in primary cultured endothelial cells. (A) Combined representation of microarrays and ChIP-seq results. Array genes were selected by the criteria with more than 2-fold up-or downregulated via IL-4 and were aligned with the results from ChIP-seq (middle column). (B) Representative ChIP-seq data (genome browser view) from SOCS1, P-selectin, PMCH, and BCL6. STAT6 binding signals are shown in red. H3K4me1 signals are shown in blue. Chr.16, chromosome 16. (C) Venn diagram showing the STAT6-dependent IL-4-inducible genes in HUVECs and CD4+T cells. Each number indicates the gene volume, which was induced via IL-4 and bound by STAT6 within the region from kb −20 upstream promoter, TSS, 5′ UTR, or 1st intron.
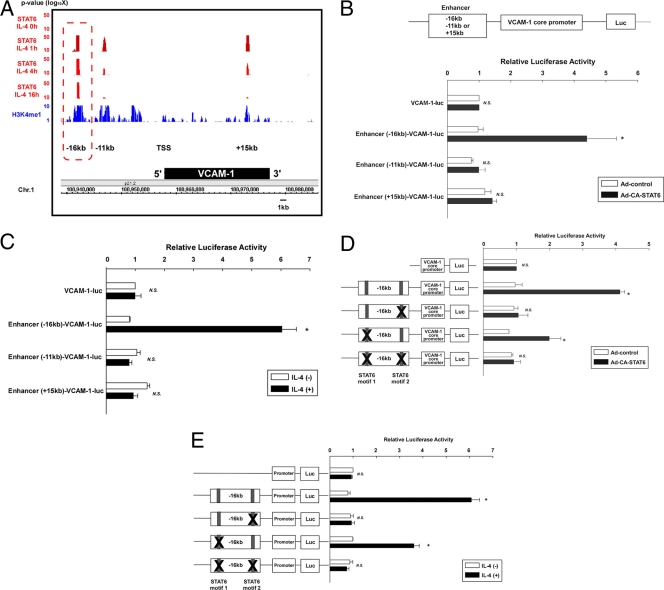
IL-4-mediated induction of VCAM-1 involves the inducible binding of STAT6 to a distal kb −16 enhancer in the VCAM-1 gene.
The VCAM-1 gene was present in the group A into the cluster (Fig. 6A), suggesting that it is a direct STAT6 target gene in IL-4-treated endothelial cells. ChIP-seq revealed three distinct STAT6 binding regions in the VCAM-1 locus (kb −16, −11, and +15, relative to the TSS) (Fig. 7A). Sustained STAT6 binding at 16 h was specifically enriched at the kb −16 region (Fig. 7A). To determine a functional role for the STAT6 binding sites, we carried out luciferase reporter assays in HUVECs with constructs containing a small (minimal) fragment of the human VCAM-1 promoter (between bp −287 and +119) coupled to the STAT6 binding region from kb −16 (244 bp), kb −11 (280 bp), and kb +15 (225 bp) (data not shown). As shown in Fig. 7B, expression of CA-STAT6 failed to induce activity of the minimal VCAM-1 promoter or the minimal promoter coupled to the kb −11 or +15 STAT6 binding region, but significantly increased (4.3-fold) reporter gene expression of the construct containing the core promoter linked to the kb −16 STAT6 binding region [Enhancer (-16)-VCAM-1-luc] (Fig. 7B). Finally, to determine whether IL-4-mediated induction of VCAM-1 requires the kb −16 STAT6 binding region, we carried out reporter analysis with HUVECs treated in the absence or presence of IL-4. IL-4 induced the expression of Enhancer (−16 kb)-VCAM-1-luc (>6-fold) but failed to activate the minimal VCAM-1 core promoter or the promoter coupled to the kb −11 or +15 binding region (Fig. 7C).
Fig. 7.
Identification of the STAT6-bound VCAM-1 enhancer. (A) ChIP-seq data in the VCAM-1 locus with the genome browser view. STAT6 binding signals are shown in red. H3K4me1 signals are shown in blue. The box (dashed red line) indicates the H3K4me1-positive region associated with stable binding of STAT6. Chr.1, chromosome 1. (B, upper) Schematic representation of the luciferase constructs. (Lower) HUVECs were transiently transfected with Enhancer-VCAM-1-luc, serum starved for 16 h, incubated with Ad-control or Ad-CA-STAT6 for 24 h, and then assayed for luciferase activity. The results show the mean ± standard deviations of luciferase light units (relative to VCAM-1-luc- and Ad-control-transfected cells) obtained in triplicate from at least 3 independent experiments. *, P < 0.01 compared with Ad-control-transfected cells. N.S., nonsignificant. (C) HUVECs were transiently transfected with VCAM-1-luc or VCAM-1-luc containing the enhancer, serum starved for 16 h, and incubated with IL-4 for 24 h. The results show the mean ± standard deviation of luciferase light units (relative to VCAM-1-luc in the absence of IL-4 treatment) obtained in triplicate from at least 3 independent experiments. *, P < 0.01 compared without IL-4 treatment. (D) HUVECs were transiently transfected with VCAM-1-luc, wild-type Enhancer (−16-kb)-VCAM-1-luc, or (Enhancer −16-kb)-VCAM-1-luc containing a point mutation of one or both STAT6 binding elements. Cells were assayed as described above. The results show the mean ± standard deviation of luciferase light units (relative to VCAM-1-luc- and Ad-control-transfected cells) obtained in triplicate from at least 3 independent experiments. *, P < 0.01, compared with Ad-control-transfected cells. (E) HUVECs were transiently transfected with plasmids, serum starved, and incubated with IL-4 for 24 h. The results show the mean ± standard deviation of luciferase light units (relative to VCAM-1-luc-transfected cells in the absence of IL-4 treatment) obtained in triplicate from at least 3 independent experiments. *, P < 0.05 compared with IL-4-nontreated cells.
The kb −16 enhancer region contains two STAT6 consensus binding motifs (Fig. 7D). To determine whether one or both of these elements are important for STAT6-mediated VCAM-1 promoter activation, we generated Enhancer (−16 kb)-VCAM-1-luc containing single or double point mutations (TTCN4GAA to TATN4GAA) of the STAT6 binding sites. As shown in Fig. 7D, CA-STAT6-mediated induction of promoter activity was abolished by mutation of the 3′ STAT6 binding site (STAT6 motif 2) or mutation of both sites and was partially inhibited (51.2%) by mutation of the 5′ STAT6 binding site (STAT6 motif 1). Consistent with the results from Ad-CA-STAT6-infected cells, IL-4-mediated activation of Enhancer (−16 kb)-VCAM-1-luc was abolished by mutation of STAT6 motif 2. In contrast, mutation of STAT6 motif 1 had partial (42.9%) blocking on IL-4-mediated VCAM-1 promoter activation (Fig. 7E). Taken together, these findings suggest that IL-4 increases expression of VCAM-1 in endothelial cells by inducing STAT6 binding to a single STAT6 binding element at kb −16.
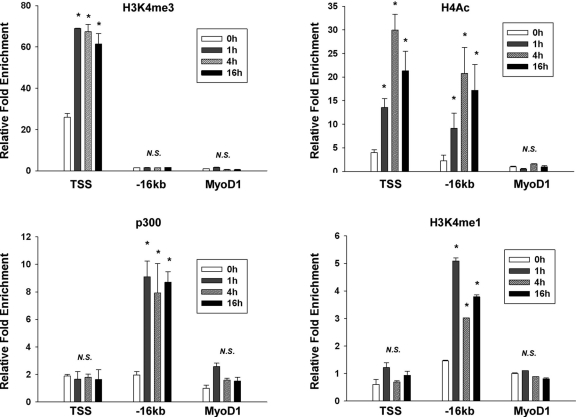
VCAM-1 enhancer is epigenetically activated with IL-4 treatment.
Having established the functional consequences of IL-4-inducible binding of STAT6 to the kb −16 STAT-binding element, we next wished to determine the effect of IL-4 on the epigenetic status of the VCAM-1 locus. To that end, we performed ChIP using antibodies against p300, acetylated histone H4 (H4Ac), H3K4me1, and H3K4me3 in the absence (0 h) or presence (1, 4, and 16 h) of IL-4. The precipitated genome fragments were subjected to the ChIP-qPCR. The MyoD1 promoter region was used as a negative control, since the promoter is silent in control and IL-4-treated endothelial cells. As shown in Fig. 8, IL-4 resulted in enriched binding of H4Ac and H3K4me3 in the proximal promoter region (TSS), consistent with increased capacity for transcriptional activation of the gene. Importantly, IL-4 induced stable binding of p300, H4Ac, and H3K4me1, but not H3K4me3, in the 16-kb region. Taken together, these findings suggest that the endogenous 16-kb STAT6 binding site functions as an IL-4-responsive enhancer. Thus, IL-4 treatment promotes an active chromatin configuration at the kb −16 STAT6 binding region of VCAM-1 in endothelial cells.
Fig. 8.
IL-4-mediated epigenetic modification of the kb −16 VCAM-1 enhancer. HUVECs were treated in the absence (0 h) or presence (1, 4, and 16 h) of IL-4, formalin fixed, and processed for immunoprecipitation of H4Ac, H3K4me3, p300, and H3K4me1. Precipitated genomic DNA was subjected to quantitative real-time PCR using primers specific to the VCAM-1 TSS, the kb −16 VCAM-1 enhancer, and the MyoD1 promoter. Shown is the ChIP enrichment level (mean ± standard deviation) relative to the ChIP-PCR value from the MyoD1 promoter without IL-4 (negative control). *, P < 0.05 compared to the same condition without IL-4 treatment. N.S., nonsignificant.
DISCUSSION
Endothelial cell activation describes the phenotypic response of endothelial cells to an inflammatory stimulus. When excessive, sustained, and/or uncoupled from local control mechanisms, endothelial cell activation may lead to dysfunction and vascular disease. VCAM-1 is involved in firm adhesion of leukocytes to the apical surface of endothelial cells through interactions with very late antigen 4 (VLA4), which is expressed primarily in lymphocytes and monocytes (reviewed in reference 33). Cross-linking VCAM-1 on the endothelial cell leads to increased cytosolic free calcium, activation of Rac1, and stimulation of reactive oxygen species (33). A pathogenic role for VCAM-1 has been implicated in atherosclerosis (reviewed in references 11 and 33). IL-4 has previously been shown to induce the expression of VCAM-1 in endothelial cells and to play a role in atherosclerotic lesion development. Thus, an understanding of how IL-4 regulates VCAM-1 expression may provide insights into the therapeutic potential of the IL-4-VCAM-1 signaling axis.
We have shown that IL-4 induces VCAM-1 mRNA expression in cultured endothelial cells and in the intact endothelium of mice. The in vitro findings are consistent with previously published data, while the observation that systemic administration of IL-4 induces VCAM-1 expression in vivo is novel. We cannot rule out the possibility that IL-4-mediated upregulation of VCAM-1 in the endothelium occurs indirectly via IL-4 signaling in another cell type. This limitation notwithstanding, the data provide strong support for VCAM-1 as an IL-4-responsive gene in endothelial cells.
The mechanisms underlying IL-4-mediated induction of VCAM-1 have remained elusive. Previous studies have failed to identify STAT6 binding sites on the VCAM-1 promoter. In the present study, several lines of evidence point to STAT6 as a key regulator of IL-4-inducible VCAM-1 expression. First, IL-4 treatment resulted in increased phosphorylation and nuclear translocation of STAT6. Second, constitutively active STAT6 induced the expression of VCAM-1. Third, IL-4-mediated induction of VCAM-1 was inhibited by si-STAT6. Fourth, ChIP-seq revealed IL-4-inducible binding of STAT6 to several regions of the VCAM-1 gene (at kb −16, −11, and +15). Fifth, in transient transfection assays, a STAT6 binding site 16 kb upstream of the transcriptional start site was found to mediate IL-4 induction of VCAM-1. (This region alone contains consensus STAT6 binding sequences [data are available at http://www.lsbm.org/MCB_2146207].) Finally, IL-4 induced accessible epigenetic marks in the 16-kb region (H3K4me1, H4Ac, and p300), consistent with an enhancing role of the STAT6 binding site at the level of the endogenous gene. Thus, contrary to previous reports, IL-4 does indeed induce VCAM-1 expression by a STAT6-dependent mechanism.
In contrast to the findings with VCAM-1, the IL-4-STAT6 signaling pathway failed to induce ICAM-1 expression in vitro and in vivo. These data add further evidence for the differential regulation of VCAM-1 and ICAM-1. For example, TNF-α and thrombin induce the expression of VCAM-1 via a GATA- and NF-κB-dependent mechanism, whereas induction of ICAM-1 occurs through NF-κB alone (28). We have shown that histone deacetylase (HDAC) inhibitors attenuate TNF-α-mediated induction of VCAM-1, but not ICAM-1 (19). Finally, FOXO1 has been implicated in VEGF stimulation of VCAM-1 alone (1). Taken together, these findings suggest that VCAM-1 is governed by a more complex repertoire of signaling pathways and transcription factors.
In addition to providing insights into the transcriptional regulation of VCAM-1, the genome-wide approaches used in the present study are the first to reveal the kinetics of IL-4-regulated genes and the time-dependent landscape of transcription factor binding of STAT6 in control and IL-4-treated endothelial cells. By combining DNA microarrays and ChIP-seq, we demonstrated that the majority (79%) of IL-4-responsive genes are STAT6 dependent, as defined by a reversal of the IL-4 effect in STAT6-deficient cells. In addition, IL-4 induction of many of these genes was associated with direct STAT6 binding to their promoter. Importantly, there was little overlap in IL-4-inducible STAT6-dependent genes between endothelial cells and T cells. Collectively, these findings strongly support a predominant cell-type-specific role for STAT6 in mediating IL-4 signal transduction in endothelial cells.
ChIP-seq with STAT6 demonstrated that more than 10,000 independent regions were rapidly occupied by STAT6 at 1 h. In many cases, binding was transient (absent at 4 h and 16 h). However, most genes that were induced at 16 h were already bound by STAT6 at an earlier time point. Thus, in some cases, STAT6 binding may be spurious, while in other cases, STAT6 binding may alter the chromatin microenvironment in such a way as to prime the gene for subsequent IL-4 activation. The enrichment calculation from our ChIP-seq data revealed that stable, but not transient binding of STAT6 occurred at the TTCN4GAA consensus sequence. Interestingly, stably (at 16 h) but not transiently (at 1 h) bound STAT6 genes demonstrated coenrichment in other transcription factor binding motifs, including NFAT and AP-1 (Fig. 5B). Thus, the stability of STAT6 binding may depend not only on the sequence of the STAT6 consensus motif, but also on an association of STAT6 with other transcription factors.
The use of ChIP-seq has proven useful in unveiling the histone code on a genome-wide scale. For example, a previous report on T cells demonstrated that STAT6 had a predominant role in antagonizing repressive marks on the genome (45). In the present study, we have shown that STAT6 binding to several target genes occurs at sites where epigenetic marks are permissive for transcriptional activation. Further studies are required to determine the extent to which STAT6 binding in endothelial cells causes changes in DNA methylation and the histone code.
In summary, we have employed complementary genome-wide approaches to delineate IL-4-responsive genes whose expression is associated with inducible binding of STAT6 in endothelial cells. The emerging picture is one in which IL-4 promotes rapid and sustained binding of STAT6 to a broad set of target genes. Importantly, our strategy led to the identification of a novel functional kb −16 STAT6 binding site in the VCAM-1 gene. Given the critical role of VCAM-1 in endothelial cell activation and dysfunction, the discovery of new transcriptional control elements may provide a foundation for targeted therapies in vascular disease.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Leading-Edge Research Promotion Fund from Japan Society for the Promotion of Science (to T.M.), in part supported by a Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science, and Technology in Japan (to T.M.), and in part supported by Mochida Memorial and Sankyo Science Foundation in Japan (to T.M.). This study was also in part supported by NIH grants HL082927 and HL076540 (to W.C.A.).
We are grateful to H. Kimura (Osaka University, Japan) for providing the monoclonal antibody against H3K4me1. We thank Akashi Izumi and Mai Miura (RCAST in the University of Tokyo) and Yuki Takagi (Tomy Digital Biology, Inc., Japan) for technical assistance.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 4 April 2011.
REFERENCES
- 1. Abid M. R., et al. 2006. A novel class of vascular endothelial growth factor-responsive genes that require forkhead activity for expression. J. Biol. Chem. 281:35544–35553 [DOI] [PubMed] [Google Scholar]
- 2. Ahmad M., Theofanidis P., Medford R. M. 1998. Role of activating protein-1 in the regulation of the vascular cell adhesion molecule-1 gene expression by tumor necrosis factor-alpha. J. Biol. Chem. 273:4616–4621 [DOI] [PubMed] [Google Scholar]
- 3. Bennett B. L., Cruz R., Lacson R. G., Manning A. M. 1997. Interleukin-4 suppression of tumor necrosis factor alpha-stimulated E-selectin gene transcription is mediated by STAT6 antagonism of NF-kappaB. J. Biol. Chem. 272:10212–10219 [DOI] [PubMed] [Google Scholar]
- 4. Bochner B. S., Klunk D. A., Sterbinsky S. A., Coffman R. L., Schleimer R. P. 1995. IL-13 selectively induces vascular cell adhesion molecule-1 expression in human endothelial cells. J. Immunol. 154:799–803 [PubMed] [Google Scholar]
- 5. Bruns H. A., Schindler U., Kaplan M. H. 2003. Expression of a constitutively active Stat6 in vivo alters lymphocyte homeostasis with distinct effects in T and B cells. J. Immunol. 170:3478–3487 [DOI] [PubMed] [Google Scholar]
- 6. Chapoval S., Dasgupta P., Dorsey N. J., Keegan A. D. Regulation of the T helper cell type 2 (Th2)/T regulatory cell (Treg) balance by IL-4 and STAT6. J. Leukoc. Biol. 87:1011–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de la Paz N. G., Simeonidis S., Leo C., Rose D. W., Collins T. 2007. Regulation of NF-kappaB-dependent gene expression by the POU domain transcription factor Oct-1. J. Biol. Chem. 282:8424–8434 [DOI] [PubMed] [Google Scholar]
- 8. Doucet C., et al. 1998. Interleukin (IL) 4 and IL-13 act on human lung fibroblasts. Implication in asthma. J. Clin. Invest. 101:2129–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Elo L. L., et al. 2010. Genome-wide profiling of interleukin-4 and STAT6 transcription factor regulation of human Th2 cell programming. Immunity 32:852–862 [DOI] [PubMed] [Google Scholar]
- 10. Fukushi J., Ono M., Morikawa W., Iwamoto Y., Kuwano M. 2000. The activity of soluble VCAM-1 in angiogenesis stimulated by IL-4 and IL-13. J. Immunol. 165:2818–2823 [DOI] [PubMed] [Google Scholar]
- 11. Galkina E., Ley K. 2007. Vascular adhesion molecules in atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 27:2292–2301 [DOI] [PubMed] [Google Scholar]
- 12. Hamik A., et al. 2007. Kruppel-like factor 4 regulates endothelial inflammation. J. Biol. Chem. 282:13769–13779 [DOI] [PubMed] [Google Scholar]
- 13. Hosking B. M., Wang S. C., Downes M., Koopman P., Muscat G. E. 2004. The VCAM-1 gene that encodes the vascular cell adhesion molecule is a target of the Sry-related high mobility group box gene, Sox18. J. Biol. Chem. 279:5314–5322 [DOI] [PubMed] [Google Scholar]
- 14. Hou J., et al. 1994. An interleukin-4-induced transcription factor: IL-4 Stat. Science 265:1701–1706 [DOI] [PubMed] [Google Scholar]
- 15. Huang H., Lavoie-Lamoureux A., Moran K., Lavoie J. P. 2007. IL-4 stimulates the expression of CXCL-8, E-selectin, VEGF, and inducible nitric oxide synthase mRNA by equine pulmonary artery endothelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 292:L1147–L1154 [DOI] [PubMed] [Google Scholar]
- 16. Iademarco M. F., Barks J. L., Dean D. C. 1995. Regulation of vascular cell adhesion molecule-1 expression by IL-4 and TNF-alpha in cultured endothelial cells. J. Clin. Invest. 95:264–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iademarco M. F., McQuillan J. J., Rosen G. D., Dean D. C. 1992. Characterization of the promoter for vascular cell adhesion molecule-1 (VCAM-1). J. Biol. Chem. 267:16323–16329 [PubMed] [Google Scholar]
- 18. Inomata M., Into T., Nakashima M., Noguchi T., Matsushita K. 2009. IL-4 alters expression patterns of storage components of vascular endothelial cell-specific granules through STAT6- and SOCS-1-dependent mechanisms. Mol. Immunol. 46:2080–2089 [DOI] [PubMed] [Google Scholar]
- 19. Inoue K., et al. 2006. Histone deacetylase inhibitor reduces monocyte adhesion to endothelium through the suppression of vascular cell adhesion molecule-1 expression. Arterioscler. Thromb. Vasc. Biol. 26:2652–2659 [DOI] [PubMed] [Google Scholar]
- 20. Khew-Goodall Y., Wadham C., Stein B. N., Gamble J. R., Vadas M. A. 1999. Stat6 activation is essential for interleukin-4 induction of P-selectin transcription in human umbilical vein endothelial cells. Arterioscler. Thromb. Vasc. Biol. 19:1421–1429 [DOI] [PubMed] [Google Scholar]
- 21. Kleemann R., Zadelaar S., Kooistra T. 2008. Cytokines and atherosclerosis: a comprehensive review of studies in mice. Cardiovasc. Res. 79:360–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kuperman D. A., Schleimer R. P. 2008. Interleukin-4, interleukin-13, signal transducer and activator of transcription factor 6, and allergic asthma. Curr. Mol. Med. 8:384–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee Y. W., Kuhn H., Hennig B., Neish A. S., Toborek M. 2001. IL-4-induced oxidative stress upregulates VCAM-1 gene expression in human endothelial cells. J. Mol. Cell Cardiol. 33:83–94 [DOI] [PubMed] [Google Scholar]
- 24. Lee Y. W., et al. 2001. Interleukin 4 induces transcription of the 15-lipoxygenase I gene in human endothelial cells. J. Lipid Res. 42:783–791 [PubMed] [Google Scholar]
- 25. Lee Y. W., Lee W. H., Kim P. H. 2010. Oxidative mechanisms of IL-4-induced IL-6 expression in vascular endothelium. Cytokine 49:73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Masinovsky B., Urdal D., Gallatin W. M. 1990. IL-4 acts synergistically with IL-1 beta to promote lymphocyte adhesion to microvascular endothelium by induction of vascular cell adhesion molecule-1. J. Immunol. 145:2886–2895 [PubMed] [Google Scholar]
- 27. Mikita T., Campbell D., Wu P., Williamson K., Schindler U. 1996. Requirements for interleukin-4-induced gene expression and functional characterization of Stat6. Mol. Cell. Biol. 16:5811–5820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Minami T., et al. 2003. Thrombin stimulation of vascular adhesion molecule-1 in endothelial cells is mediated by protein kinase C (PKC)-delta-NF-kappa B and PKC-zeta-GATA signaling pathways. J. Biol. Chem. 278:6976–6984 [DOI] [PubMed] [Google Scholar]
- 29. Minami T., Aird W. C. 2001. Thrombin stimulation of the vascular cell adhesion molecule-1 promoter in endothelial cells is mediated by tandem nuclear factor-kappa B and GATA motifs. J. Biol. Chem. 276:47632–47641 [DOI] [PubMed] [Google Scholar]
- 30. Minami T., Miura M., Aird W. C., Kodama T. 2006. Thrombin-induced autoinhibitory factor, Down syndrome critical region-1, attenuates NFAT-dependent vascular cell adhesion molecule-1 expression and inflammation in the endothelium. J. Biol. Chem. 281:20503–20520 [DOI] [PubMed] [Google Scholar]
- 31. Minami T., Rosenberg R. D., Aird W. C. 2001. Transforming growth factor-beta 1-mediated inhibition of the flk-1/KDR gene is mediated by a 5′-untranslated region palindromic GATA site. J. Biol. Chem. 276:5395–5402 [DOI] [PubMed] [Google Scholar]
- 32. Miyazaki Y., Satoh T., Nishioka K., Yokozeki H. 2006. STAT-6-mediated control of P-selectin by substance P and interleukin-4 in human dermal endothelial cells. Am. J. Pathol. 169:697–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Muller W. A. 2009. Mechanisms of transendothelial migration of leukocytes. Circ. Res. 105:223–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Neish A. S., Khachigian L. M., Park A., Baichwal V. R., Collins T. 1995. Sp1 is a component of the cytokine-inducible enhancer in the promoter of vascular cell adhesion molecule-1. J. Biol. Chem. 270:28903–28909 [DOI] [PubMed] [Google Scholar]
- 35. Neish A. S., et al. 1995. Endothelial interferon regulatory factor 1 cooperates with NF-kappa B as a transcriptional activator of vascular cell adhesion molecule 1. Mol. Cell. Biol. 15:2558–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Palmer-Crocker R. L., Hughes C. C., Pober J. S. 1996. IL-4 and IL-13 activate the JAK2 tyrosine kinase and Stat6 in cultured human vascular endothelial cells through a common pathway that does not involve the gamma c chain. J. Clin. Invest. 98:604–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Palmer-Crocker R. L., Pober J. S. 1995. IL-4 induction of VCAM-1 on endothelial cells involves activation of a protein tyrosine kinase. J. Immunol. 154:2838–2845 [PubMed] [Google Scholar]
- 38. Paul W. E. 1991. Interleukin-4: a prototypic immunoregulatory lymphokine. Blood 77:1859–1870 [PubMed] [Google Scholar]
- 39. Rollins B. J., Pober J. S. 1991. Interleukin-4 induces the synthesis and secretion of MCP-1/JE by human endothelial cells. Am. J. Pathol. 138:1315–1319 [PMC free article] [PubMed] [Google Scholar]
- 40. Schnyder B., et al. 2002. Phytochemical inhibition of interleukin-4-activated Stat6 and expression of VCAM-1. Biochem. Biophys. Res. Commun. 292:841–847 [DOI] [PubMed] [Google Scholar]
- 41. Stein N. C., et al. 2008. Interleukin-4 and interleukin-13 stimulate the osteoclast inhibitor osteoprotegerin by human endothelial cells through the STAT6 pathway. J. Bone Miner. Res. 23:750–758 [DOI] [PubMed] [Google Scholar]
- 42. Street N. E., Mosmann T. R. 1990. IL4 and IL5: the role of two multifunctional cytokines and their place in the network of cytokine interactions. Biotherapy 2:347–362 [DOI] [PubMed] [Google Scholar]
- 43. Thornhill M. H., Haskard D. O. 1990. IL-4 regulates endothelial cell activation by IL-1, tumor necrosis factor, or IFN-gamma. J. Immunol. 145:865–872 [PubMed] [Google Scholar]
- 44. Umetani M., et al. 2001. Function of GATA transcription factors in induction of endothelial vascular cell adhesion molecule-1 by tumor necrosis factor-alpha. Arterioscler. Thromb. Vasc. Biol. 21:917–922 [DOI] [PubMed] [Google Scholar]
- 45. Wei L., et al. 2010. Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity 32:840–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wojta J., et al. 1993. Interleukin-4 stimulates expression of urokinase-type-plasminogen activator in cultured human foreskin microvascular endothelial cells. Blood 81:3285–3292 [PubMed] [Google Scholar]
- 47. Yao L., Pan J., Setiadi H., Patel K. D., McEver R. P. 1996. Interleukin 4 or oncostatin M induces a prolonged increase in P-selectin mRNA and protein in human endothelial cells. J. Exp. Med. 184:81–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.