Abstract
Linker histones play essential roles in the chromatin structure of higher eukaryotes. While binding to the surface of nucleosomes is directed by an ∼80-amino-acid-residue globular domain, the structure and interactions of the lysine-rich ∼100-residue C-terminal domain (CTD), primarily responsible for the chromatin-condensing functions of linker histones, are poorly understood. By quantitatively analyzing binding of a set of H1 CTD deletion mutants to nucleosomes containing various lengths of linker DNA, we have identified interactions between distinct regions of the CTD and nucleosome linker DNA at least 21 bp from the edge of the nucleosome core. Importantly, partial CTD truncations caused increases in H1 binding affinity, suggesting that significant entropic costs are incurred upon binding due to CTD folding. van't Hoff entropy/enthalpy analysis and intramolecular fluorescent resonance energy transfer (FRET) studies indicate that the CTD undergoes substantial nucleosome-directed folding, in a manner that is distinct from that which occurs upon H1 binding to naked DNA. In addition to defining critical interactions between the H1 CTD and linker DNA, our data indicate that the H1 CTD is an intrinsically disordered domain and provide important insights into the biological function of this protein.
INTRODUCTION
Linker histones are essential proteins in higher eukaryotes that play multiple critical roles in eukaryotic chromatin, including defining nucleosome spacing (5, 14, 21), stabilizing folding and condensation of chromatin (7, 32, 37), and directly regulating gene expression (14, 32). In higher eukaryotes, these small, basic proteins are composed of three distinct domains: a well-conserved ∼80-amino-acid (aa)-residue globular domain, a long ∼100-residue C-terminal domain (CTD), and a short ∼25-residue N-terminal tail (41) (Fig. 1 A). While linker histone binding to the surface of nucleosomes is directed by the 80-aa globular domain (1, 6, 12), the chromatin-condensing functions of the protein are primarily provided by the highly basic CTD (2, 25). Indeed, the linker histones of some protists consist of only regions resembling the CTD (42). While the structure of the globular domain and its location within nucleosomes have been elucidated (29, 36), the specific interactions and structure of the CTD remain poorly defined.
Fig. 1.
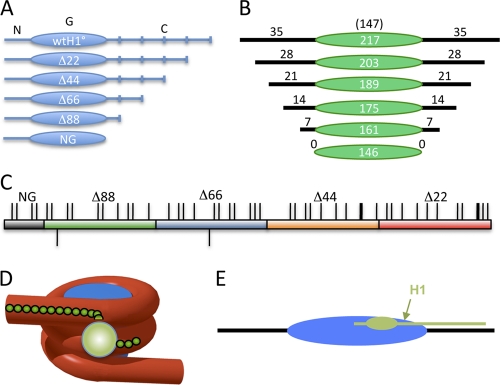
Schematic of the H1 C-terminal domain and model for the nucleosome-H1 complex. (A) Linker histone domains and deletion mutants used in this study. N, G, and C indicate the N-terminal, globular, and C-terminal domains of the protein. Deletion mutants are denoted according to the number of residues deleted from the C terminus of the protein. (B) Constructs used for nucleosome binding experiments. The green oval indicates the 147-bp nucleosome core DNA (not to scale). Black numbers and lines indicate base pairs in each linker region. (C) Schematic of the Xenopus laevis H1(0) CTD. Thin and thick ticks above the bar indicate lysines and arginines, respectively; ticks below indicate the two glutamic acid residues in the CTD. The red, orange, blue, green, and gray sections indicate the regions accumulatively deleted in Δ22, Δ44, Δ66, Δ88, and NG, respectively. Note that eight additional residues were deleted in NG compared to Δ88. (D and E) Models for H1 interaction with the nucleosome showing the globular domain binding at the nucleosome dyad (sphere or oval) and the CTD extending linearly along a linker DNA segment.
Previous analysis of the electrostatic properties of the C-terminal tail of linker histone H1 concluded that all basic residues are fully charged under physiological conditions (35). Moreover, analyses of the ion dependence of chromatin folding and the extent of DNA charge neutralization by H1 as predicted by Manning polyelectrolyte theory indicate that nearly all negatively charged residues in the CTD are involved in the neutralization of the linker DNA backbone in condensed chromatin (9). Thus, a simple model predicts a roughly linear association of the CTD with the linker DNA. However, recent work by Lu et al. provided evidence that two discontinuous regions of approximately 24 aa residues played distinct roles in array condensation and the organization of linker DNA (26). Interestingly, amino acid composition, and not specific sequence, appears to be important for these functions (25, 26). The CTD sequences of linker histones are not well conserved, yet these domains have remarkably similar amino acid compositions that are characteristic of intrinsically disordered proteins (15, 22). Such proteins do not exhibit a unique conformation in their native state but adopt a defined structure or ensemble of structures upon interaction with macromolecular partners (40). Indeed, the H1 CTD, while unstructured in solution, can adopt substantial α-helical structure in the presence of helix-stabilizing agents and other conditions thought to mimic the native chromatin environment (8, 31). Thus, it has been proposed that the CTD adopts secondary structure upon binding to DNA and nucleosome surfaces in chromatin to mediate close apposition of linker DNA and promote chromatin condensation. However, data regarding the structure(s) of the H1 CTD in a chromatin environment are not yet available.
In this work, we have used linker histone CTD deletion mutants to elucidate the contributions of specific regions within this domain to nucleosome binding. Analysis of binding to nucleosomes containing various lengths of linker DNA indicates that the CTD interacts with regions of linker DNA at least 21 bp from the edge of the nucleosome core. Surprisingly, truncation of the CTD does not result in a decrease in binding affinity, and van't Hoff enthalpy/entropy analysis and intramolecular fluorescent resonance energy transfer (FRET) studies indicate substantial folding of the CTD, which likely results in an entropic cost that offsets the enthalpic contribution of lysine-DNA interactions.
MATERIALS AND METHODS
Expression and purification of full-length H1 and the H1 CTD deletion mutants.
H1(0) from Xenopus laevis (here referred to as H1) was expressed in bacterial cells using the plasmid pET3aH1(0)a (17). The coding sequences for H1 CTD deletion mutants were generated by PCR from this plasmid using the upstream primer TAGCCATATGGCAGAGAATTCAGCC and downstream primers GCTAGGATCCTTATTACTTGGATGCCCTCAC, GCGCGGATCCTTATTAAGGCTTTTTCTTAGCTAC, GCTAGGATCCTTATTAAGACTTTGCAGCTTTTTTG, GCTAGGATCCTTATTATCCTTTGGTCTGTTTGAG, and GCTAGGATCCTTATTAAGGCTTCTTTGCTGG to generate Δ22, Δ44, Δ66, Δ88, and NG, respectively (Fig. 1). These were inserted into the pET3a vector at the NdeI and BamHI restriction sites and transformed into BL21(DE3) bacterial cells (Novagen). Proteins were expressed and purified as described previously (17). The concentration of the purified protein stocks was determined by amino acid analysis (Molecular Structure Facility, UC Davis).
DNA fragments for nucleosome reconstitution.
DNA fragments for nucleosome reconstitution containing various lengths of linker DNA were generated by PCR using the 601 nucleosome positing sequence as a template (24) and appropriate primers (IDT Technologies) (sequences available upon request). The predicted nucleosome dyad axis is located at the center of each DNA fragment such that linker DNA is equally distributed on either side of the nucleosome. Primers (0.5 μg each) were radiolabeled before PCR by incubation with 18 μl [γ-32P]ATP and 10 units of polynucleotide kinase (Promega) according to standard methods. After precipitation, the primers were used for PCR with 0.1 μg of EcoRI/HindIII-digested CP10ZY plasmid, which contains the 601 nucleosome positioning sequence. After PCR, products were isolated from 8% polyacrylamide gels.
Nucleosome reconstitution.
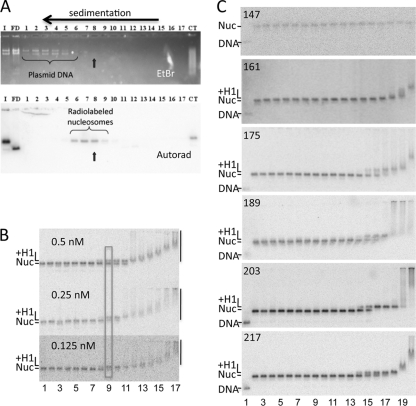
Reconstitution conditions were empirically optimized by independent adjustment of H3/H4 and H2A/H2B amounts to maximize generation of mononucleosome species. Typically, reconstitution conditions (200 μl) contained 2.8 μg of H3/H4 tetramer, 7 μg of BamHI-digested pBS plasmid, 175,000 cpm of a radiolabeled 601 DNA fragment, and 2.9 μg of H2A/H2B dimer in reconstitution buffer (10 mM Tris, pH 8.0, 1 mM EDTA, 5 mM dithiothreitol [DTT], and 2 M NaCl). Nucleosomes were reconstituted via standard salt dialysis (18). After reconstitution, nucleosomes were purified by sedimentation through 7 to 20% sucrose gradients (10 mM Tris-HCl, pH 8.0, and 1 mM EDTA [TE]) with ultracentrifugation at 34,000 × g for 18 h in a Beckman SW41 rotor at 4°C. Nucleosome fractions were collected in 0.6-ml siliconized tubes (Axygen) pretreated with bovine serum albumin (BSA) (0.3 mg/ml) in TE overnight at 4°C. BSA was added to peak fractions to 0.15 mg/ml to prevent dissociation (34). Fractions were analyzed by electrophoresis on nucleoprotein gels (0.7% agarose, 0.5× TBE [1× 90 mM Tris base, 90 mM boric acid, 2.5 mM EDTA]), stained with ethidium bromide, and autoradiographed to ensure that carrier DNA was separated from radiolabeled nucleosomes (see Fig. 2A).
Fig. 2.
H1 binding to nucleosomes with different lengths of linker DNA. (A) Purification of reconstituted nucleosomes. Nucleosomes were fractionated on 7 to 20% sucrose gradients to remove plasmid carrier DNA. Fractions 1 to 17 were run on a 0.7% agarose nucleoprotein gel. I, input sample after nucleosome reconstitution; FD, free DNA; CT, a nucleosome control in which samples were reconstituted with calf thymus DNA instead of plasmid as a carrier. Top and bottom panels show the ethidium bromide (EtBr)-stained gel and autoradiograph of the same gel after drying. Fraction 8 was chosen for H1 binding experiments. (B) Binding is driven by protein concentration. Three binding experiments with decreasing 217N nucleosome concentrations, as indicated, are shown. H1 concentrations in lanes 1 to 17 were 0, 0.03, 0.05, 0.1, 0.2, 0.5, 1, 2, 4, 8, 15, 30, 60, 100, 200, 300, and 400 nM, respectively. Bands corresponding to free nucleosome (Nuc), H1-nucleosome complex (+H1), and H1-induced nucleosome aggregates (vertical bars) are indicated. The approximate midpoint in the binding transition is indicated by the rectangle. (C) Representative gels showing H1 binding to 147N, 161N, 175N, 189N, 203N, and 217N nucleosomes as depicted in Fig. 1B. Lanes 1 and 2 show the naked DNA fragment and nucleosomes in the absence of H1. Nucleosomes in lanes 2 to 20 were incubated with H1 at final concentrations of 1, 2, 4, 8, 15, 30, 60, 120, and 240 pM and 0.48, 0.96, 1.9, 3.9, 7.7, 15, 30, 60 and 120 nM, respectively. The positions of naked DNA, nucleosomes, H1-bound nucleosomes, and H1-induced aggregates are indicated as in panel B.
Linker histone binding assay.
Radiolabeled 601 nucleosomes (∼300 cpm; ∼0.5 fmol) and H1 proteins were incubated in binding buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 50 mM NaCl, 150 ng/μl BSA, and 5% [vol/vol] glycerol). H1 stocks were prepared in dilution buffer (10 mM Tris-HCl, pH 8.0, 50 mM NaCl, 1 mM EDTA, 0.1 mg/ml BSA, and 10% [vol/vol] glycerol), and 1/10 volume (1.5 μl) was added to the binding reaction mixtures to achieve the concentrations listed in the figure legends. Binding reaction mixtures were incubated at 25°C for 30 min and then loaded directly onto running (30 V) 0.7% or 1.0% (for smaller CTD deletion mutants) agarose gels (0.5× TBE). Gels were run at 90 to 120 V for 2 to 3 h, dried, and autoradiographed. H1 binding was analyzed by plotting the concentration of linker histone in each sample against the fraction of histone H1-bound nucleosome, determined by dividing the intensity of the H1-nucleosome band by the sum of the unbound and H1-bound nucleosome bands. Quantification of images was accomplished with ImageQuant and GraphPad Prism 4 software. Dissociation constants were obtained from global least-squares fits determined as follows:
| (1) |
where Nuc is free nucleosome and Ka is an association constant.
van't Hoff enthalpy/entropy analysis.
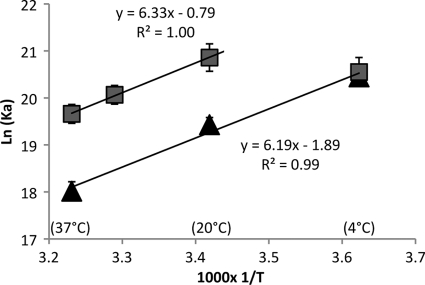
Binding analysis was performed as described above except that binding reactions and gel electrophoresis were carried out at the temperatures indicated below with full-length H1 or Δ66. All buffers and components were equilibrated at temperature for at least 4 h before the binding reactions were initiated. Dissociation constants for binding of either full-length (wild-type [WT]) H1 or Δ66 to N203 nucleosomes were determined at the temperatures indicated below (see Table 3), and then ln(1/Kd) was plotted versus 1/T and the slope (ΔH/R) and y intercept (ΔS/R) were determined. Enthalpy and entropy were calculated using R = 1.985 cal·K−1·mol−1.
Table 3.
van't Hoff entropy and enthalpy of linker histone binding to nucleosomesa
| H1 | Temp (°C) | ΔH/R (kcal/mol) | ΔS/R (mol) | ΔH (kcal/mol) | ΔS·T (kcal/mol) | Calc ΔG (kcal/mol) | Calc Kd (nM) | Avg Kd (nM) |
|---|---|---|---|---|---|---|---|---|
| WT | 20 | 6.19 | −1.89 | −12.3 | −1.10 | −11.2 | 4.2 | 4.0 |
| WT | 37 | 6.19 | −1.89 | −12.3 | −1.17 | −11.1 | 13.5 | 15 |
| Δ66 | 20 | 6.33 | −0.787 | −12.6 | −0.46 | −12.1 | 0.91 | 0.72 |
| Δ66 | 37 | 6.33 | −0.787 | −12.6 | −0.48 | −12.1 | 2.8 | 3.0 |
Dissociation constants for binding of either full-length (WT) or Δ66 to N203 nucleosomes were determined at the indicated temperatures and values for ΔH and ΔS·T were determined as described in Materials and Methods. ΔG and a corresponding Kd were calculated (Calc) from the apparent entropy and enthalpy for each temperature and compared to experimentally determined Kds, with an estimated average error of ±10% (see Fig. 3).
FRET analysis.
FRET analysis was performed with the H1(0) double mutant G101C/K195C. This protein was prepared, expressed, and purified as described above and reduced with 50 mM DTT for 1 h, DTT was removed, and the protein was purified by ion-exchange chromatography and quick-frozen (16). The protein was labeled with either maleimido-Cy3, maleimido-Cy5, or a 50/50 mix of both according to the manufacturer's instructions (Pierce), and excess reagent was removed by another round of chromatography. The labeled proteins were diluted to a concentration of 5 to 20 nM in 150 μl of H1 binding buffer and placed in a siliconized glass cuvette. Emission spectra were recorded with excitation at 515- and 610-nm wavelengths (see Fig. 4; data not shown) on a Horiba Jovin Yvon FluoroMax-4 spectrofluorometer with 2-nm slit widths in both excitation and emission channels. Spectra were recorded in the absence or presence of increasing amounts of 207-bp-gradient-purified mononucleosomes or the 207-bp DNA fragment alone (free DNA) as indicated in the figure legends. FRET efficiency was calculated as described in reference 28 using maximum peak heights and a value of εA(610) = 161,103 (Cy5), εA(515) = 6,078 (Cy5), εD(515) = 92,058 (Cy3), and d+ = 1.
Fig. 4.
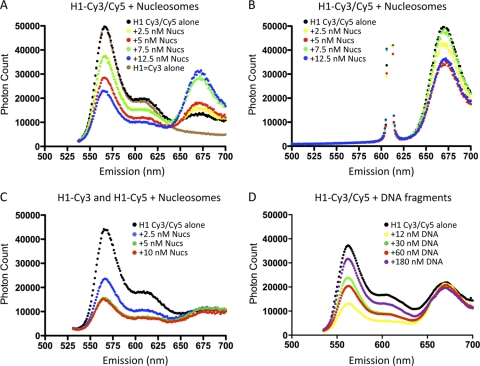
FRET analysis of H1 binding to nucleosomes indicates intramolecular folding of the CTD. (A) Emission spectra of H1 (10 nM) labeled at either end of the CTD as described in Materials and Methods in the absence of nucleosomes (black circles) or in the presence of 2.5, 5.0, 7.5, or 12.5 nM 217N nucleosomes, as indicated. The spectrum of H1 labeled exclusively with Cy3 is also shown (brown line). Excitation was at 515 nm. (B) As in panel A except with excitation at 610 nm. (C) Nucleosome-induced FRET is entirely intramolecular. H1 was labeled exclusively with either Cy3 or Cy5 and then mixed in a 1:1 ratio, and emission spectra were examined in the absence of nucleosomes (black circles) or in the presence of 2.5, 5, or 10 nM nucleosomes. (D) FRET of H1 labeled with both Cy3 and Cy5 (as in panel A) was determined in the presence of increasing amounts of 207-bp naked 601 DNA fragments with excitation at 515 nm. Corresponding emission spectra for excitation at 610 nm are not shown.
RESULTS
The ∼100-residue C-terminal domain typically represents about half the mass of metazoan linker histone proteins and is essential for chromatin folding under physiological conditions, yet the structure and interactions of this domain remain poorly understood. A simple model posits that this uniformly lysine-rich domain interacts in a linear fashion with the linker DNA (Fig. 1C, D, and E). In order to map interactions of the H1 CTD binding in nucleosomes, we analyzed binding of H1 to a set of nucleosomes with progressively shorter linker DNAs reconstituted with DNA fragments containing the 601 nucleosome positioning element in the center of the fragment (Fig. 1A and B). We verified that our gel-based assay reliably reported binding driven solely by protein concentration, since decreasing the nucleosome concentration did not alter the binding profile (Fig. 2 B; see Materials and Methods). Quantitative analysis of such gels indicated that H1 bound to a 217-bp nucleosome (217N) with an affinity of ∼5 nM (Table 1), consistent with other measures of H1 binding affinity (17, 39). Deletion of 7 bp from each linker region (14 bp total) to generate a 203-bp nucleosome (203N) did not significantly alter the binding affinity. However, deletion of 7 or 14 bp from each linker, to generate 189N and 175N nucleosomes, respectively (to 21- or 14-bp linker DNA on each side of the core region), resulted in a decrease in affinity for H1, with the two nucleosomes exhibiting dissociation constants of ∼8 nM and ∼10 nM, respectively (Table 1). Further deletion to generate nucleosomes with 161 bp of DNA (7 bp of linker DNA) resulted in a large decrease in affinity (Kd > 50 nM) such that H1 association did not result in unique species on the gel (Fig. 2C), indicative of nonspecific binding and consistent with prior delineation of the minimal length of DNA required to bind H1 in the chromatosome particle (33, 41). These results indicate that energetically significant interactions between the CTD and linker DNA can be detected beyond the edge of the chromatosome, as far as 21 to 28 bp from the edge of the core region.
Table 1.
Dissociation constants for binding of full-length and H1 CTD deletion mutants to nucleosomes containing different amounts of linker DNA
| DNA fragment size (bp) | Binding affinitya |
Linker DNA length (bp)b | |||||
|---|---|---|---|---|---|---|---|
| WT H1 (nM) | Δ22 (nM) | Δ44 (nM) | Δ66 (nM) | Δ88 (nM) | NG (nM) | ||
| 147 | >50 | >50 | >50 | >50 | >50 | >50 | 0 |
| 161 | >50 | >50 | >50 | >50 | >50 | >50 | 7 |
| 175 | 10.5 ± 3.0 | 6.3 ± 1.2 | 3.5 ± 0.8 | 2.4 ± 0.6 | 4.1 ± 1.0 | 12.5 ± 3.5 | 14 |
| 189 | 7.8 ± 1.8 | 3.8 ± 0.7 | 1.8 ± 0.2 | 0.9 ± 0.2 | 5.1 ± 1.2 | 15.1 ± 2.5 | 21 |
| 203 | 3.7 ± 0.3 | 1.3 ± 0.2 | 1.3 ± 0.2 | 0.6 ± 0.07 | ND | ND | 28 |
| 217 | 4.8 ± 0.9 | 1.1 ± 0.2 | 2.0 ± 1.0 | 2.1 ± 0.4 | ND | 15.0 ± 5.0 | 35 |
Binding affinities for full-length and the H1 CTD deletion mutants. Specific binding constants of >50 nM were not determined due to nonspecific association. ND, not determined.
Length of each linker DNA, on each side of the 147-bp nucleosome core.
We next tested a set of CTD truncation mutants for binding to the set of 601 nucleosomes (Fig. 1A). Interestingly, deletion of 22 amino acid residues from the end of the CTD (Δ22) resulted in a 2- to 3-fold increase in binding affinity to nucleosomes containing 217, 203, 189, or 175 bp of DNA (Table 1). No discrete binding products were detected with 161N and 147N nucleosomes, similar to results with full-length H1. Deletion of an additional 22 aa residues in Δ44 did not further alter binding to 217N or 203N nucleosomes but did cause a modest increase in binding affinity to 189N and 175N nucleosomes. These results are surprising, given that there are 11 and 20 positively charged residues deleted in Δ22 and Δ44, respectively (see Fig. 1C), which are believed to interact with the negatively charged DNA backbone (9).
We also tested deletions of 66 and 88 residues from the CTD as well as deletion of the entire CTD domain (Fig. 1A). These deletions result in progressive loss of 11, 10, and 4 lysines from the CTD, respectively, while only two acidic residues are lost (Fig. 1C). In most cases, H1 lacking the last 66 residues bound to the nucleosomes with affinities similar to those found for Δ44. For example, both proteins bound to 175N with comparable affinities of 3.5 and 2.4 nM, while truncation of 7 additional bp from each linker DNA to generate 161N caused a drastic loss of affinity for both proteins, indicating deletion of a region of strong interactions. We also note that Δ66 bound with a detectably higher affinity to 189N and 203N than did Δ44 (Table 1); thus, again, truncation of the CTD and loss of ∼10 positively charged residues that presumably interact with the DNA backbone actually led to an increase in binding affinity. Similar results were obtained with Δ88, although deletion of the entire CTD in NG resulted in a distinct loss of binding affinity (see below).
Binding of full-length H1 to nucleosomes with 161 or 146 bp of DNA did not occur with measurable affinities in our assays. As mentioned above, these nucleosomes contain less than 168 bp of DNA, which has been described for H1-containing “chromatosome” particles (33). In order to examine the chromatosome- and subchromatosome-sized nucleosomes more closely, we generated a second set of nucleosomes in which the linker DNAs differed by 1 bp in length within each linker region (Table 2). Interestingly, full-length H1 bound with measurable affinity only to 167N, 171N, and 175N nucleosomes, containing a minimum of 10 bp of linker DNA on each side of the nucleosome core; deletion of only 1 bp or more from each linker DNA to generate 165N nucleosomes caused a drastic loss of affinity (Table 2). Similar results were observed for Δ22 and Δ44. Consistent with previous trends, Δ66 bound with about 2-fold-higher affinity to 167N nucleosomes than did full-length H1 and with weak but measurable affinity to 163N nucleosomes with only 8 bp of linker DNA. Importantly, Δ88 bound with high affinity to 163N nucleosomes with 8 bp of linker DNA but ∼10-fold more weakly to 161N nucleosomes with only 1-bp-shorter linkers. These results are consistent with recent modeling showing that a stretch of basic residues just beyond the H1 globular domain (and present within Δ88) interacts with the DNA backbone ∼8 bp from the edge of the core region (36). In support, this region is deleted in NG, which consistently exhibits a 4- to 5-fold weaker affinity for binding all nucleosomes tested.
Table 2.
Dissociation constants for binding of full-length and H1 CTD deletion mutants to nucleosomes with linker DNAs differing by 1 bp in length
| DNA fragment size (bp) | Binding affinitya |
Linker DNA length (bp)b | |||||
|---|---|---|---|---|---|---|---|
| WT H1 (nM) | Δ22 (nM) | Δ44 (nM) | Δ66 (nM) | Δ88 (nM) | NG (nM) | ||
| 161 | >50 | >50 | >50 | >50 | >50 | >50 | 7 |
| 163 | >50 | >50 | >50 | 43.7 ± 7.0 | 4.6 ± 1.6 | 16.7 ± 2.9 | 8 |
| 165 | >50 | >50 | >50 | 3.4 ± 0.5 | 4.7 ± 1.4 | 19.3 ± 6.3 | 9 |
| 167 | 9.7 ± 1.3 | 8.4 ± 1.4 | 6.7 ± 1.2 | 4.2 ± 0.8 | 0.6 ± 0.1 | 8.3 ± 2.7 | 10 |
| 171 | 9.7 ± 1.0 | 7.4 ± 1.2 | 4.6 ± 0.6 | 4.3 ± 0.8 | 6.5 ± 0.8 | ND | 12 |
| 175 | 10.5 ± 3.0 | 6.3 ± 1.2 | 3.5 ± 0.8 | 2.4 ± 0.6 | 4.1 ± 1.0 | 12.5 ± 3.5 | 14 |
Binding affinities for full-length and H1 CTD deletion mutants. Specific binding constants of >50 nM were not determined due to nonspecific association. ND, not determined.
Length of each linker DNA, on each side of the 147-bp nucleosome core.
The above data suggest that there are energetically significant interactions between the H1 CTD and linker DNA well beyond the edge of the chromatosome and that despite numerous lysine-DNA interactions, truncations of the CTD have a net positive impact on overall binding affinity. To further investigate this phenomenon, we measured van't Hoff enthalpy and entropy contributions to binding affinity by determining how specific dissociation constants varied with temperature. Full-length H1 exhibited an apparent linear temperature dependence of ln(1/Kd) and 1/T between 4°C and 37°C for binding to 203N nucleosomes (Fig. 3). Analysis of these data indicates that binding of the full-length protein is enthalpically driven with a small net negative entropic contribution, which amounts to about 1.2 kcal/mol at 37°C (Table 3). Interestingly, Δ66 exhibits an apparent linear temperature dependence between 25°C and 37°C but is nonlinear in the 25-to-−4°C range, indicating that binding is largely entropy driven at low temperatures. Investigation of binding at an intermediate temperature point (31°C) supports that the plot is linear in the 25-to-37°C range, with a slope almost identical to that observed for the WT protein (Fig. 3). Thus, binding of Δ66 involves a favorable enthalpic component similar to that of full-length H1 in the physiological temperature range. However, binding of Δ66 involves a more favorable entropic component than binding of full-length H1, partly accounting for the higher binding affinity of this protein (Table 3). These data indicate that removal of the terminal 66 residues from the CTD significantly reduces the entropic cost incurred upon H1 binding to the nucleosome.
Fig. 3.
van't Hoff enthalpy and entropy contributions to binding affinity. Nucleosomes were reconstituted on the 203-bp fragment and binding to full-length H1 (gray squares) and Δ66 (black triangles) tested at various temperatures and ln(Ka) [or/ln(1/Kd)] plotted versus 1,000 × 1/T. Linear least-squares fits were carried out for all data points for full-length H1, while only the data in the range of 20 to 37°C were fitted for Δ66.
A significant folding or ordering of a disordered CTD might explain the entropic cost observed upon binding. To determine whether this domain undergoes such folding, we determined end-to-end distance across the CTD by FRET. The G101C K195C double mutant of H1 was modified with Cy3-maleimide and Cy5-maleimide; then, emission spectra were measured in the absence or presence of 207N nucleosomes with excitation at 515 or 610 nm (Fig. 4 A and B). Cy5 fluorescence emission (λmax, ∼660 nm) was low when H1-Cy3/Cy5 was irradiated at 515 nm in the absence of nucleosomes, consistent with an extended domain (Fig. 4A, compare brown and black lines). The calculated FRET efficiency of the free double-labeled protein (∼0.1) combined with the R0 (Förster distance) for the Cy3/Cy5 FRET pair of 5.4 nm indicates an approximate distance between fluorophores of 8.5 nm, consistent with the predicted end-to-end distance for an ∼100-aa residue disordered domain of ∼9.0 nm (38). Upon addition of nucleosomes, emission from Cy3 (λmax, ∼560 nm) is significantly reduced and emission from Cy5 is greatly increased, indicating a substantial increase in FRET efficiency. Samples prepared with lower nucleosome:H1 ratios yielded FRET efficiencies of 0.32 and 0.44 at 2.5 and 5.0 nM nucleosomes, respectively, while at nucleosome:H1 ratios near 1:1, where native nucleosome binding is maximal, FRET efficiency peaked (∼0.8 at 12.5 nM nucleosomes), indicating a significant reduction of the mean distance between the ends of the CTD, from ∼9.0 nm in the free protein to ∼5.2 nm when bound to nucleosomes. We note that at low nucleosome:H1 ratios, some quenching of Cy3 is observed (Fig. 4A, compare red and green lines), likely due to native and nonnative binding modes for H1 (see below). These data are consistent with significant folding of the CTD upon nucleosome binding.
Importantly, a mixture of Cy3-only and Cy5-only labeled H1s did not exhibit significant increases in FRET efficiencies upon nucleosome binding (Fig. 4C), indicating that the observed FRET signal observed with the double-labeled protein in the presence of nucleosomes must be due to intramolecular energy transfer. We also note that binding of H1-Cy3/Cy5 to naked DNA also resulted in significant FRET, but unlike nucleosomes, FRET was maximal at low DNA:H1 ratios (∼0.9), with efficiencies decreasing to ∼0.5 with increasing concentrations of DNA fragment (Fig. 4D). Binding at low DNA:H1 ratios resulted in greater quenching of Cy3 fluorescence than was observed with nucleosomes. Also in contrast to the result with nucleosomes, similar high FRET efficiencies were observed upon DNA fragment binding to a combination of H1-Cy3 and H1-Cy5 individually labeled proteins (data not shown), indicating that H1 FRET with naked DNA was at least in part due to intermolecular energy transfer. These results are consistent with H1 binding cooperatively to naked DNA to form large “tram track” structures, with close packing of H1 molecules (10) at low DNA:H1 ratios and a switch to a more singular, noncooperative binding mode at high ratios (11), and are consistent with the notion of binding to linear DNA in a fundamentally different manner than nucleosomes (19).
DISCUSSION
While important for stabilizing native chromatin and essential for life in higher organisms, the structures and interactions of linker histones in chromatin are not well understood. Our data define critical interactions between the H1 CTD and linker DNA and show that this domain undergoes substantial folding upon nucleosome binding. These data support current proposals that the CTD is an intrinsically disordered domain that adopts a defined structure(s) upon binding in chromatin and provide important insights into the biological function of H1.
Early nuclease protection studies provided an indication that at least a portion of the H1 CTD interacts with linker DNA (41), while later analysis of the electrostatic mechanism of chromatin folding led to the conclusion that nearly all lysines within the H1 CTD were involved in DNA charge neutralization in condensed chromatin, likely within the linker DNA region (9). In addition, while binding of the globular domain occurs in the DNA minor groove at the dyad axis, the actual sites of interaction of the CTD have not been determined. The changes in H1 binding affinity that we find upon truncation of the linker DNA indicate that interactions between the CTD and linker DNA can be detected some 21 to 28 bp from the edge of the core region. Moreover, these results do not support a simple linear association model for CTD interaction with the linker DNA, as sequential deletion of the linker DNA does not lead to a gradual reduction in H1 affinity. Thus, H1 CTD-DNA interactions occur well outside the classically defined chromatosome region and, assuming such interactions are symmetrically disposed, would involve a significant fraction of linker DNA for chromatin with an average nucleosome repeat spacing of ∼200 bp. For example, the H1 employed in the current study [H1(0) from Xenopus laevis] is associated with chromatin with an average nucleosome repeat spacing of ∼192 bp (43) and ∼45 bp of linker DNA. Thus, CTD contacts to 21 to 28 bp of linker DNA on only one or both sides of the core would involve approximately half or all, respectively, of the linker DNA in this chromatin.
Recent hydroxyl radical footprinting suggests that binding of H1 to nucleosomes results in significant organization of the linker DNA (36), perhaps into a stem-like structure protruding away from the nucleosome along the dyad axis (4). Formation of this structure is in part due to binding of H1 globular domain to the widened DNA minor groove at the nucleosome dyad and to the first helical turn of DNA in one linker region (36). In addition, the model envisions that a basic patch of residues located within the CTD immediately adjacent to the globular domain is critical for this organization, interacting with the second linker segment (references 32 and 36 and data not shown). Multiple aspects of our data support this view. First, we find that Δ88, which contains this patch, consistently binds with ∼5-fold-higher affinity compared to NG, which lacks the entire CTD. In addition, Δ88 exhibits a significant drop in affinity when the linker DNA is reduced below ∼8 bp on each side of the core, whereas binding to nucleosomes containing longer linkers occurs with similar high affinities (Table 2). Moreover, Δ88 binds to N167 with the highest affinity recorded in our assays, 0.6 nM, suggesting that this length of linker DNA provides optimal interaction with the basic patch but does not involve energetically costly close apposition of the two linker DNAs. Interestingly, we find increasing the length of linker by even 1 bp (N169) results in an ∼8-fold decrease in affinity. These data support a model where the basic patch interacts with a region approximately 8 to 10 bp from the edge of the nucleosome core and, in conjunction with the globular domain, organizes a structure in which the two linkers are in close proximity about 1 DNA helical turn from the edge of the nucleosome core region (13, 36).
Given the high concentration of positively charged residues in the CTD (∼40/100 residues), and evidence that the vast majority of these residues contact the polyanionic backbone of DNA (17), one would expect a significant energetic contribution to binding free energy provided by lysines throughout this domain. However, we find that deletions of portions of the CTD actually result in an increase in binding affinity and that H1 binding to nucleosomes involves an entropic cost that is reduced upon deletion of a significant fraction of the CTD (Fig. 3). Correspondingly, it is likely that nucleosomes with shortened linkers alter the entropy/enthalpy balance, resulting in the differences in binding constants observed in our experiments. We propose that binding of full-length H1 involves an entropic cost that offsets the enthalpic contribution from interactions of positively charged lysine residues with the DNA backbone. These results are consistent with biophysical analyses showing that CTD peptides are unstructured in solution but can acquire secondary structure in helix-stabilizing solvents or in the presence of nucleic acids (8, 30, 31). In support of these results, our FRET experiments indicate that the CTD exhibits an extended structure in solution with an average end-to-end distance expected for an unstructured domain of similar size (38). Importantly, H1 binding to nucleosomes elicits a drastic increase in FRET, suggesting a folding of the CTD (Fig. 5). These results support models in which the CTD is predicted to behave as an intrinsically disordered domain based on the amino acid content and the chromatin condensation properties of CTD mutants (15).
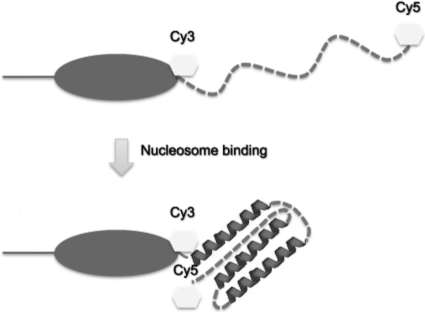
Fig. 5.
Model for CTD folding upon nucleosome binding. FRET experiments (this work) indicate a significant reduction in the end-to-end distance across the CTD, potentially with induced α-helical and β-sheet structures (8, 30, 31). However, little is known regarding the actual structure of the folded CTD.
H1 exhibits multiple binding modes and rapid equilibration in live nuclei (23, 27). Previous work has demonstrated that H1 binds strongly and cooperatively to naked DNA, to produce “tram track” structures with H1 binding sites approximately 30 bp apart (11). Moreover, H1 binds in a distinct fashion to nucleosomes in vitro, with a single linker histone binding preferentially and noncooperatively to nucleosomes over naked DNA fragments (19, 36). Concordantly, we find distinct H1 binding behavior with nucleosomes and naked DNA, with binding to nucleosomes yielding exclusively intramolecular FRET, while binding to naked DNA induces both intra- and intermolecular FRET. Moreover, the magnitude of the intramolecular FRET in each case suggests that a different extent of CTD folding occurs upon nucleosome or naked DNA binding. It is likely that both binding modes are relevant in vivo and may account for the multiple modes observed in fluorescence recovery after photobleaching (FRAP) experiments in live cells (27). First, binding affinities in vitro for both modes differ by only a fewfold (19). Second, we observe both modes under conditions of slight H1 excess, such as occurs in Xenopus erythrocyte nuclei and other tissues (3, 41). Third, experiments by Hendzel and colleagues (20) have demonstrated that the CTD is a primary determinant of H1 binding in chromatin in vivo. The contribution of each binding mode to the overall structure of chromatin remains to be elucidated.
One function of an intrinsically disordered CTD might be to “tune” the overall affinity of linker histone binding, allowing charge neutralization in extended regions of linker DNA and associated enthalpic contributions to binding with compensation via entropic costs to attain affinities appropriate for function. If the CTD were a classically ordered domain, the affinity of the protein for binding nucleosomes in chromatin might be too high to allow the appropriate rapid rates of exchange of the protein (27). A nonmutually exclusive alternative is that the disorder-to-order transition allows multiple structures to be adopted upon binding, providing efficient charge neutralization in different chromatin secondary structures (15). Recent work by Lu and Hansen has shown that only two regions within the CTD are primarily involved in stabilizing higher-order chromatin structure (26). Thus, the relatively strict conservation of the length and amino acid residue content of this domain may be related to the fact that H1 interacts with multiple surfaces and numerous other factors in chromatin. Indeed, we find that with respect to binding mononucleosomes, specific regions apparently contribute more to binding. In this context, it will be interesting to determine affinities of the CTD deletion mutants for binding within condensed nucleosome arrays.
ACKNOWLEDGMENTS
We thank Mark Dumont and Prahnesh Akshayalingham for technical assistance with fluorescence experiments.
This work was supported by NIH grant GM52426.
Footnotes
Published ahead of print on 4 April 2011.
REFERENCES
- 1. Allan J., Hartman P. G., Crane-Robinson C., Aviles F. X. 1980. The structure of histone H1 and its location in chromatin. Nature 288:675–679 [DOI] [PubMed] [Google Scholar]
- 2. Allan J., Mitchell T., Harborne N., Bohm L., Crane-Robinson C. 1986. Roles of H1 domains in determining higher order chromatin structure and H1 location. J. Mol. Biol. 187:591–601 [DOI] [PubMed] [Google Scholar]
- 3. Bates D. L., Thomas J. O. 1981. Histones H1 and H5: one or two molecules per nucleosome? Nucleic Acids Res. 2:5883–5894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bednar J., et al. 1998. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc. Natl. Acad. Sci. U. S. A. 95:14173–14178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blank T. A., Becker P. B. 1995. Electrostatic mechanism of nucleosome spacing. J. Mol. Biol. 252:305–313 [DOI] [PubMed] [Google Scholar]
- 6. Brown D. T., Izard T., Misteli T. 2006. Mapping the interaction surface of linker histone H1(0) with the nucleosome of native chromatin in vivo. Nat. Struct. Mol. Biol. 13:250–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carruthers L. M., Bednar J., Woodcock C. L., Hansen J. C. 1998. Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays: mechanistic ramifications for higher-order chromatin folding. Biochemistry 37:14776–14787 [DOI] [PubMed] [Google Scholar]
- 8. Clark D. J., Hill C. S., Martin S. R., Thomas J. O. 1988. Alpha-helix in the carboxy-terminal domains of histones H1 and H5. EMBO J. 7:69–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clark D. J., Kimura T. 1990. Electrostatic mechanism of chromatin folding. J. Mol. Biol. 211:883–896 [DOI] [PubMed] [Google Scholar]
- 10. Clark D. J., Thomas J. O. 1988. Differences in the binding of H1 variants to DNA. Cooperativity and linker-length related distribution. Eur. J. Biochem. 178:225–233 [DOI] [PubMed] [Google Scholar]
- 11. Clark D. J., Thomas J. O. 1986. Salt-dependent co-operative interaction of histone H1 with linear DNA. J. Mol. Biol. 187:569–580 [DOI] [PubMed] [Google Scholar]
- 12. Crane-Robinson C. 1997. Where is the globular domain of linker histone located on the nucleosome? Trends Biochem. Sci. 22:75–77 [DOI] [PubMed] [Google Scholar]
- 13. Fan L., Roberts V. A. 2006. Complex of linker histone H5 with the nucleosome and its implications for chromatin packing. Proc. Natl. Acad. Sci. U. S. A. 103:8384–8389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fan Y., et al. 2005. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell 123:1199–1212 [DOI] [PubMed] [Google Scholar]
- 15. Hansen J. C., Lu X., Ross E. D., Woody R. W. 2006. Intrinsic protein disorder, amino acid composition, and histone terminal domains. J. Biol. Chem. 281:1853–1856 [DOI] [PubMed] [Google Scholar]
- 16. Hayes J. J. 1996. Site-directed cleavage of DNA by a linker histone—Fe(II) EDTA conjugate: localization of a globular domain binding site within a nucleosome. Biochemistry 35:11931–11937 [DOI] [PubMed] [Google Scholar]
- 17. Hayes J. J., Kaplan R., Ura K., Pruss D., Wolffe A. 1996. A putative DNA binding surface in the globular domain of a linker histone is not essential for specific binding to the nucleosome. J. Biol. Chem. 271:25817–25822 [DOI] [PubMed] [Google Scholar]
- 18. Hayes J. J., Lee K. M. 1997. In vitro reconstitution and analysis of mononucleosomes containing defined DNAs and proteins. Methods 12:2–9 [DOI] [PubMed] [Google Scholar]
- 19. Hayes J. J., Wolffe A. P. 1993. Preferential and asymmetric interaction of linker histones with 5S DNA in the nucleosome. Proc. Natl. Acad. Sci. U. S. A. 90:6415–6419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hendzel M. J., Lever M. A., Crawford E., Th'ng J. P. 2004. The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J. Biol. Chem. 279:20028–20034 [DOI] [PubMed] [Google Scholar]
- 21. Ito T., Bulger M., Kobayashi R., Kadonaga J. T. 1996. Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Mol. Cell. Biol. 16:3112–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kasinsky H. E., Lewis J. D., Dacks J. B., Ausio J. 2001. Origin of H1 linker histones. FASEB J. 15:34–42 [DOI] [PubMed] [Google Scholar]
- 23. Lever M. A., Th'ng J. P., Sun X., Hendzel M. J. 2000. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature 408:873–876 [DOI] [PubMed] [Google Scholar]
- 24. Lowary P. T., Widom J. 1998. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276:19–42 [DOI] [PubMed] [Google Scholar]
- 25. Lu X., Hamkalo B., Parseghian M. H., Hansen J. C. 2009. Chromatin condensing functions of the linker histone C-terminal domain are mediated by specific amino acid composition and intrinsic protein disorder. Biochemistry 48:164–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lu X., Hansen J. C. 2004. Identification of specific functional subdomains within the linker histone H10 C-terminal domain. J. Biol. Chem. 279:8701–8707 [DOI] [PubMed] [Google Scholar]
- 27. Misteli T., Gunjan A., Hock R., Bustin M., Brown D. T. 2000. Dynamic binding of histone H1 to chromatin in living cells. Nature 408:877–881 [DOI] [PubMed] [Google Scholar]
- 28. Poirier M. G., Oh E., Tims H. S., Widom J. 2009. Dynamics and function of compact nucleosome arrays. Nat. Struct. Mol. Biol. 16:938–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ramakrishnan V., Finch J. T., Graziano V., Lee P. L., Sweet R. M. 1993. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature 362:219–224 [DOI] [PubMed] [Google Scholar]
- 30. Roque A., Ponte I., Arrondo J. L., Suau P. 2008. Phosphorylation of the carboxy-terminal domain of histone H1: effects on secondary structure and DNA condensation. Nucleic Acids Res. 36:4719–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Roque A., Ponte I., Suau P. 2009. Role of charge neutralization in the folding of the carboxy-terminal domain of histone H1. J. Phys. Chem. B 113:12061–12066 [DOI] [PubMed] [Google Scholar]
- 32. Shen X., Yu L., Weir J. W., Gorovsky M. A. 1995. Linker histones are not essential and affect chromosome condensation in vivo. Cell 82:47–56 [DOI] [PubMed] [Google Scholar]
- 33. Simpson R. T. 1978. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry 17:5524–5531 [DOI] [PubMed] [Google Scholar]
- 34. Studitsky V. M., Clark D. J., Felsenfeld G. 1995. Overcoming a nucleosomal barrier to transcription. Cell 83:19–27 [DOI] [PubMed] [Google Scholar]
- 35. Subirana J. A. 1990. Analysis of the charge distribution in the C-terminal region of histone H1 as related to its interaction with DNA. Biopolymers 29:1351–1357 [DOI] [PubMed] [Google Scholar]
- 36. Syed S. H., et al. 2010. Single-base resolution mapping of H1-nucleosome interactions and 3D organization of the nucleosome. Proc. Natl. Acad. Sci. U. S. A. 107:9620–9625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thoma F., Koller T., Klug A. 1979. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J. Cell Biol. 83:403–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tran H. T., Pappu R. V. 2006. Toward an accurate theoretical framework for describing ensembles for proteins under strongly denaturing conditions. Biophys. J. 91:1868–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ura K., Nightingale K., Wolffe A. P. 1996. Differential association of HMG1 and linker histones B4 and H1 with dinucleosomal DNA: structural transitions and transcriptional repression. EMBO J. 15:4959–4969 [PMC free article] [PubMed] [Google Scholar]
- 40. Uversky V. N. 2010. Seven lessons from one IDP structural analysis. Structure 18:1069–1071 [DOI] [PubMed] [Google Scholar]
- 41. van Holde K. E. 1989. Chromatin. Springer Verlag, New York, NY [Google Scholar]
- 42. Wu M., Allis C. D., Richman R., Cook R. G., Gorovsky M. A. 1986. An intervening sequence in an unusual histone H1 gene of Tetrahymena thermophila. Proc. Natl. Acad. Sci. U. S. A. 83:8674–8678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Young D., Carroll D. 1983. Regular arrangement of nucleosomes on 5S rRNA genes in Xenopus laevis. Mol. Cell. Biol. 3:720–730 [DOI] [PMC free article] [PubMed] [Google Scholar]