Abstract
The HMG-box transcription factor LEF1 controls many developmentally regulated genes, including genes that activate expression of the T-cell antigen receptor alpha chain (TCR-alpha) in developing thymocytes. At least two distinct isoforms of LEF1 are expressed, resulting from variable inclusion of LEF1 exon 6; however, the expression pattern of these isoforms and mechanism of splicing regulation have not been explored. Here we demonstrate that inclusion of LEF1 exon 6 is increased during thymic development and in response to signaling in a cultured T-cell line in a manner which temporally correlates with increased expression of TCR-alpha. We further find that inclusion of exon 6 is dependent on the signal-induced increase in expression and binding of the splicing factor CELF2 to two intronic sequences flanking the regulated exon. Importantly, loss of exon 6 inclusion, through knockdown of CELF2 or direct block of the exon 6 splice site, results in reduced expression of TCR-alpha mRNA. Together, these data establish the mechanistic basis of LEF1 splicing regulation and demonstrate that LEF1 alternative splicing is a contributing determinant in the optimal expression of the TCR-alpha chain.
INTRODUCTION
A major question to arise from the sequencing of the human genome is how functional complexity is achieved from the mere 20,000 to 25,000 genes present in human cells (28). Of the many mechanisms eukaryotes use to regulate gene expression, alternative splicing has the unique feature of allowing multiple discrete proteins to be encoded by a single gene (28). This generation of protein diversity is accomplished through the differential inclusion or skipping of exons, or portions thereof, to generate distinct mRNAs. Importantly, upwards of 95% of human genes are alternatively spliced (30, 39). Therefore, regulation of splicing can be assumed to play a major role in shaping protein diversity and cellular function.
Interestingly, differential alternative splicing patterns are particularly prevalent in genes critical for neuronal and/or immune function (26). One notable example is the gene encoding lymphocyte enhancer factor 1 (LEF1). LEF1 is an HMG-box transcription factor that is widely expressed during embryonic development and then restricted to certain lymphocyte populations in adulthood (2, 38). LEF1 was first identified as a protein that drives expression of the T-cell antigen receptor alpha chain (TCR-alpha) through binding to the TCR-alpha enhancer (37, 40). Subsequent studies have further implicated LEF1 as a ubiquitous regulator of developmental programs triggered in response to Wnt signaling pathways (2).
The LEF1 gene is alternatively spliced to give rise to different LEF1 protein isoforms that have overlapping, but distinct, functions (2). In particular, skipping of the 84-nucleotide exon 6 results in a protein referred to as LEF1*, which lacks a portion of the context-dependent regulatory domain (CRD) (see Fig. 1A) (5). Transfection studies with cDNAs and reporter constructs have shown that the full CRD is required for maximal TCR-alpha enhancer activity. In contrast, LEF* retains the activation domain (AD) that mediates beta-catenin binding and Wnt-dependent transcription (2, 5, 11), and it lacks the binding site for HIC5, a repressor of beta-catenin-dependent function (10). Therefore, the alternative splicing of LEF1 exon 6 potentially allows for the uncoupling of the multiple activities of this important transcription factor. Surprisingly, however, there has been little investigation of the relative expression pattern of LEF* versus full-length LEF1 in normal tissues or whether acute changes in isoform expression actually alter transcription of endogenous target genes. Equally importantly, there is thus far no understanding of the molecular mechanisms that regulate LEF1 isoform choice in any cell type.
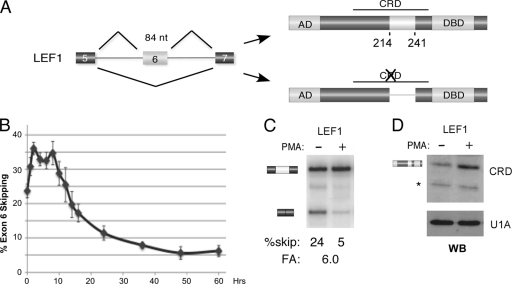
Fig. 1.
PMA activation of JSL1 cells induces expression of the version of LEF1 that includes exon 6. (A) Schematic of alternative splicing of the 84-nucleotide (nt) LEF1 exon 6 and the consequence of inclusion of this exon on the resulting protein domain structure. The beta-catenin-binding activation domain (AD), context-dependent regulatory domain (CRD), and DNA binding domain (DBD) are indicated. Skipping of exon 6 compromises the CRD by removing amino acids 214 to 241. (B) Percent skipping of LEF1 exon 6 graphed for cells grown from 0 to 60 hours in PMA. Data points are averaged from at least 4 independent experiments, with error bars indicating standard deviations. (C) Representative RT-PCR of the endogenous LEF1 gene, used for the graph in panel B, showing loss of exon 6 skipping (percent skip) in cells grown for 60 h in the presence (+) versus absence (−) of PMA. Fold activation (FA) is a measure of the increased inclusion of exon 6 upon PMA stimulation and was calculated from low-cycle RT-PCR as described in Materials and Methods. (D) Western blot (WB) of LEF1 protein in cells grown in the absence (−) or presence (+) of PMA for 60 h. The anti-LEF1 antibody is raised against an epitope in the CRD and thus recognizes only the full-length LEF1 protein. Anti-U1A is used as a loading control. The position of a nonspecific cross-reactive band is indicated by an asterisk to the left of the blot.
In general, alternative splicing is controlled by auxiliary (i.e., nonsplice site) elements located within variable exons and/or their flanking introns (15, 28). These cis-regulatory sequences typically bind to trans-acting proteins which, in turn, enhance or inhibit the inclusion of the exon into the final mRNA. One class of trans-acting proteins known to influence splicing is the CELF protein family (CUGBP and ETR-3 like factors), which typically bind to auxiliary sequences enriched in UG dipeptides (9). These proteins can have both stimulatory and repressive activities on exon choice. For example, binding of the protein CELF2 (also called CUGBP2, ETR-3, and Napor) to an intronic UG-rich element enhances inclusion of cardiac troponin T variable exon 5 (20), whereas binding of this protein to a sequence in the Tau gene inhibits the inclusion of exon 2 (23).
Here we show that alternative splicing of LEF1 exon 6 is regulated during pre-TCR signaling in thymic development and in response to activation of the JSL1 T-cell line and that this is driven by the activity of CELF2. Specifically, we find that CELF2 binds to evolutionarily conserved intronic sequences flanking either side of exon 6 that are both required for signal-induced inclusion of exon 6. During pre-TCR signaling in thymocytes and phorbol myristate acetate (PMA)-induced activation of JSL1 cells, expression of CELF2 increases, resulting in increased binding of CELF2 to the LEF1 regulatory sequences and enhanced inclusion of LEF1 exon 6. In contrast, knockdown of CELF2 reduces exon 6 inclusion. We further show that maximal transcription of the TCR-alpha locus, an essential downstream effect of pre-TCR signaling required for maturation of thymocytes, is dependent on CELF2-induced inclusion of LEF1 exon 6. Taken together, these data provide a comprehensive pathway from the mechanistic determinants to the functional consequences of LEF1 alternative splicing.
MATERIALS AND METHODS
Cell culture and reagents.
Growth and stimulation of the JSL1 cell line was done as described previously (24). The cells were cultured in RPMI 1640 medium plus 5% fetal calf serum at 37°C in 5% CO2. Stable cell lines containing vectors expressing the LEF1 minigenes, short hairpin RNA (shRNA), or microRNA miR-23b were created by transfecting 10 million cells with 10 μg of minigene plasmid by electroporation and grown under drug selection as described by Rothrock et al. (33). Transfections with morpholino oligomers were also done by electroporation; however, cells were directly harvested after 48. The morpholino oligomers used were as follows: CELF1, GTGGTCCAGGGTGCCGTTCATTTTC; polypyrimidine tract-binding protein (PTB), CTATATCTGGGACAATGCCGTCCAT; and LEF1 3′ splice site, ACCTTGCCTGAGGTCACAGAAGAAA. The CELF2 targeting shRNA vector was created by inserting the sequence CGCAGAGTAAAGGTTGTTGTT into the hairpin backbone described by Rao and Wilkinson (31). The miR-23b-expressing vector is pEP-has-mir-23b purchased from Cell Biolabs Inc. For stimulations, three independent clones of each minigene were either left untreated or treated with 20 ng/ml of phorbol myristate acetate (PMA) for the times indicated, after which cells were harvested and total RNA extracted using RNABee (Tel-Test).
Murine thymocytes.
Murine thymocytes were prepared as a single-cell suspension. To enrich for early progenitors, thymocytes expressing high levels of CD4 and CD8 were depleted using subsaturating amounts of anti-CD8α (53.6-7) and anti-CD4 (GK1.5), followed by removal of antibody (Ab)-coated cells with magnetic beads conjugated to goat anti-rat IgG (Qiagen). Cell preparations were stained with optimized Ab dilutions. Abs in the Lin cocktail include anti-B220 (RA3-6B2), anti-CD19 (1D3), anti-CD11b (M1/70), anti-Gr-1 (8C5), anti-CD11c (HL3), anti-NK1.1 (PK136), anti-Ter-119, anti-CD3 (2C11), anti-CD8 (53.6-7), anti-CD8β (53-5.8), anti-TCRβ (H57), and anti-TCR (GL-3). Additional Abs used included anti-Kit (2B8), anti-CD25 (PC61.5), anti-CD27(LG.7F9), anti-TCRβ (H57-597), anti-CD4 (GK1.5), and anti-CD8 (53.6-7). Abs were purchased from eBioscience or BD Pharmingen. For cell sorting, pooled thymocytes were first enriched for progenitors with the depletion described above, then stained, and sorted on a FACSAria (BD Biosciences). Some thymocytes were not enriched to cell sort for the immature intermediate CD8 single-positive (ISP), CD4+ CD8+ double-positive (DP), CD4+ single-positive (SP), and CD8+ SP populations. Dead cells were excluded through 4′,6-diamidino-2-phenylindole (DAPI) uptake. Doublets were excluded by using the following parameters: height versus width of forward scatter and height versus width of side scatter. Data were analyzed using FlowJo software (TreeStar).
RT-PCR.
Reverse transcription-PCR (RT-PCR) and analysis were carried out as previously described in detail (35). In brief, reverse transcription was done with a sequence-specific primer, followed by PCR done using a limited number of cycles (16 to 20 cycles) such that the signal detected is linear with respect to input RNA. Endogenous LEF1 was analyzed using the following primers: F (for forward), CCCATGCGGTCCATCCTCTCACC; R (for reverse), CTGGATGAGGGATGCCAGTTGTGTGG; murineF, CGTTGGACAGATCACCCCACCCATTGG; and murineR, GTCTTTTGGGCTCCTGCTCCTTTCTCTGTTCG. CELF2 splicing was analyzed using the following primers: F, CTGCAGATAGTGAAAAGTCAAACGCTGTGG; and R, GTGGCAGTGTTGAGCTGTTGCATCTGC. TCR-alpha and CELF2 mRNAs were quantified using the following primers: TCRa-F, CCTGACCCTGCCGTGTACCAGC; TCRa-R, GTCCATAGACCTCATGTCTAGCACAG; mTCRa-F, CTGCCTGTTCACCGACTTTGACTCCC; mTCRa-R, CAGTCAACGTGGCATCACAGGGAACG; CELF2-F, GATCACTCAGACCAACCAGACCCA; and CELF2-R, CCAAATGGAGAGAACATCACCCTG. Minigenes were analyzed using the vector-specific primers ACT and GE3R (sequence published in reference 33). Quantitation was done by densitometry using a Typhoon phosphorimager (Amersham Biosciences). Fold activation (FA) is a measure of the difference in splicing between resting and activated conditions and is calculated as (exon inclusion/exon exclusion)activated/(exon inclusion/exon exclusion)resting with the values for both exon inclusion and exon exclusion being percentages.
Minigenes.
LEF1 minigenes were synthesized by using PCR to isolate the indicated sequences from the endogenous LEF1 gene and to flank the sequences with an NdeI and BglII restriction site at the 5′ and 3′ end, respectively. These fragments were then inserted between beta-globin constitutive exons 1 and 2 in the SCglo parental minigene described previously (33). For the alt120A and alt120B constructs, an additional 120 nucleotides from beta-globin intron 3, or the reverse sequence, were inserted in the BglII site at the LEF1-beta-globin junction.
UV cross-linking.
Nuclear extract was purified from JSL1 cells using a standard protocol described previously (34). UV cross-linking was performed as described in reference 36.
Western blotting.
Western blotting was carried out as previously described (25). Antibodies for Western blots were as follows: LEF1 from Exalpha Biologicals, Inc. (Watertown, MA). Antibodies against CELF1, CELF2, MBNL (Muscleblind-like), and YB-1 were kind gifts from Tom Cooper (Baylor College of Medicine). Antibodies against U2AF35 and U2AF65 were provided by Tom Maniatis (Columbia), antibodies to U1A were provided by Iain Mattaj (EMBL), and antibodies to PTB were provided by Doug Black (UCLA).
RESULTS
LEF1 undergoes alternative splicing in response to cellular stimulation and thymic development.
An initial prediction that LEF1 isoform expression is subject to regulation in lymphocytes came from our previous study profiling global splicing changes upon stimulation of the human T-cell-derived JSL1 cell line (17). We first used our standard semiquantitative low-cycle reverse transcription-PCR (RT-PCR) assay to confirm whether inclusion of LEF1 exon 6 is indeed regulated in response to signaling pathways in JSL1 cells. We find that immediately following stimulation of JSL1 cells with the phorbol ester phorbol myristate acetate (PMA), LEF1 exon 6 undergoes a modest increase in skipping for the first few hours (∼25% to ∼35% skipping 2 to 8 h after PMA treatment; Fig. 1B). This increase in exon skipping is followed by a dramatic switch to exon inclusion starting 10 to 16 h after PMA addition, ultimately resulting in an almost total loss of exon skipping by 48 to 60 h after stimulation (Fig. 1B and C). Importantly, this reduction in exon skipping is reflected in a 3- to 4-fold increase in the full-length version of LEF1 protein, as demonstrated using an antibody that specifically recognizes an epitope encoded by the variable exon (Fig. 1D). We are unable to clearly detect expression of the smaller LEF1 isoform given the lack of specific antibodies and the minimal size difference between the two isoforms; however, the near loss of mRNA encoding the small isoform strongly suggests that this protein must ultimately decrease upon stimulation.
Given the significant change in LEF1 splicing following stimulation of JSL1 cells and previous studies suggesting that the exon 6-included versus exon 6-skipped forms of LEF1 have distinct functions, we next wanted to determine whether regulation of LEF1 in JSL1 cells mimics a specific event in T-cell biology. On the basis of the fact that LEF1 has been linked to both TCR-alpha expression and cellular development, we focused our attention on thymic development of T cells. Thymic development is a highly regulated and ordered process in which T-cell precursors (early thymic precursors) must pass through a series of checkpoints to proceed to the next step in development. Each developmental step is defined by the presence of a distinct pattern of cell surface proteins. Using these well-defined cell surface markers and flow cytometry, we sorted total mouse thymocytes to isolate the CD4− CD8− double-negative (DN) DN2, DN3a, DN3b, and DN4 developmental populations, as well as the subsequent intermediate CD8 single-positive (ISP) and CD4+ CD8+ double-positive (DP) cells (see Fig. S1 in the supplemental material).
Interestingly, RT-PCR analysis of LEF1 mRNA in the isolated thymic populations reveals a biphasic change in LEF1 splicing between the DN3a and DP populations highly reminiscent of that observed in JSL1 cells (Fig. 2A). In the DN3a population of thymocytes, the beta chain of the T-cell receptor (TCR) has been rearranged and is expressed on the cell surface together with a surrogate version of the alpha chain, known as the pre-TCR-alpha chain (22). Signaling through this beta/prealpha heterodimer (pre-TCR signaling) initiates in the DN3a-to-DN3b transition. The similarity between the pattern of LEF1 splicing observed downstream of DN3a cells with that observed upon PMA treatment of JSL1 cells suggests that, at least in terms of LEF1 splicing, PMA stimulation of JSL1 cells is mimicking pre-TCR signaling in thymocytes. This is consistent with the fact that JSL1 cells express little to no detectable CD4 or CD8 (F. Heyd and K. W. Lynch, unpublished data).
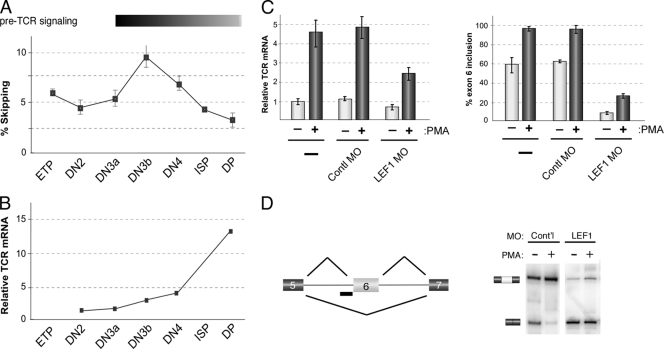
Fig. 2.
LEF1 splicing is regulated during thymic development coincident with TCR-alpha expression. (A) Percent skipping of LEF1 exon 6, as determined by low-cycle RT-PCR, in isolated thymic populations as indicated. The isolated thymic populations were early thymic precursors (ETP), CD4− CD8− double-negative (DN) DN2, DN3a, DN3b, and DN4 developmental populations, the subsequent intermediate CD8 single-positive (ISP), and CD4+ CD8+ double-positive (DP) cells. Thymocytes were sorted from 10 mice and pooled for analysis. The bar above the graph indicates normal timing of pre-TCR signaling and TCR-alpha expression in thymocyte development. (B) Total TCR-alpha mRNA, relative to the amount in DN2 set at 1, for subsequent thymic developmental states. For all data shown, similar results were obtained from an independent sort. (C) Graph of TCR-alpha mRNA (left) and LEF1 exon 6 inclusion (right) in cells transfected with a morpholino oligomer (MO) complementary to the 3′ splice site (3′ss) of LEF1 exon 6 to specifically block inclusion of this exon, grown under resting (without PMA) or stimulated (with PMA) conditions. Error bars indicate standard deviations from 2 or 3 independent experiments. Contl, control. (D) Schematic of experimental approach and representative RT-PCR gel showing switch in splicing induced by the 3′ss morpholino oligomer. Cont'l, control.
Arguably the most critical molecular event induced by pre-TCR signaling is the rearrangement and transcription of the actual TCR-alpha locus to yield expression of the mature alpha/beta TCR complex by the DP stage (22). Previous studies using reporter constructs have suggested that full-length LEF1 is a more potent enhancer of TCR-alpha transcription than LEF1* encoded by the exon 6-skipped mRNA (5, 11), though this has not been investigated with the endogenous TCR-alpha gene. Interestingly, we observe that increased TCR-alpha transcript levels between the DN3b and DP thymic populations correlate with decreased skipping of LEF1 exon 6 (Fig. 2B). These data suggest the intriguing possibility that the increased inclusion of LEF1 exon 6 is part of the mechanism by which TCR-alpha transcription is induced.
Consistent with the similarity between pre-TCR signaling in thymocytes and PMA stimulation of JSL1 cells, we observe an approximately 5-fold increase in TCR-alpha mRNA in JSL1 cells following treatment with PMA (Fig. 2C, left graph). To determine whether there is indeed a direct relationship between LEF1 splicing and transcription of the endogenous TCR-alpha gene, we took advantage of the ability to manipulate splicing in the JSL1 cells. Specifically, we used a morpholino oligomer complementary to the 3′ splice site of LEF1 exon 6 to directly block inclusion of this exon. As shown in Fig. 2D, transfection of the morpholino oligomer results in a dramatic loss of basal exon 6 inclusion in favor of the exon-skipped mRNA and significantly dampens the subsequent PMA-induced inclusion of exon 6 with little change in the total LEF1 mRNA expressed. Remarkably, this morpholino oligomer-induced loss of exon 6 results in a 40 to 50% decrease in the ability of PMA to induce TCR-alpha expression and even reduces basal TCR-alpha expression in resting cells (Fig. 2C, left graph). Therefore, we conclude that the alternative splicing of LEF1 exon 6 is an important contributing factor in the induced expression of the TCR-alpha chain.
Regulation of LEF1 exon 6 is conferred by elements in the flanking introns.
Having identified a functionally relevant change in LEF1 alternative splicing in thymocytes and JSL1 cells, we next set out to characterize the sequences and factors that determine the signal-responsive inclusion of LEF1 exon 6. We first identified the sequences within the LEF1 gene that confer signal-induced exon inclusion using chimeric minigenes. Inclusion of LEF1 exon 6 in minigene-derived mRNA was quantified by low-cycle RT-PCR using minigene-specific primers from total RNA harvested from cells stably transfected with the minigene. We express the signal responsiveness of exon 6 in the minigene as fold activation corresponding to the change in the ratio of exon-included to exon-skipped splicing product between resting and activated cells. This method allows for the direct comparison of the signal responsiveness of constructs in a manner that is separable from differences in the basal levels of exon inclusion (27).
We initially engineered a minigene that contained LEF1 variable exon 6 and several hundred nucleotides of flanking intron inserted between exons of the beta-globin gene. Consistent with regulation conferred by cis-acting elements proximal to the variable exon, we find that inclusion of LEF1 exon 6 in this minigene is increased the same ∼5- to 6-fold following PMA stimulation as observed in the endogenous gene (Fig. 3A and B, LEF1MG370/160). We do observe a modest difference in the resting inclusion of the LEF1 variable exon in the minigene context (41%; Fig. 3C) relative to the endogenous gene (76%; Fig. 1B). This decrease in basal inclusion in the minigene context is likely due to the increased splice site strength of the competing beta-globin exons flanking the variable exon compared to the endogenous gene, although we cannot rule out loss of some regulatory element that controls the resting level of inclusion of the variable exon. Since we are focused here on the mechanisms controlling the signal-induced alternative splicing of LEF1, we have not further pursued this, or other, differences in basal exon inclusion in this present study. We also note that while the LEF1MG370/160 (MG for minigene) minigene recapitulates the signal-induced loss of exon skipping at 60 h after stimulation, we do not observe any significant short-term increase in exon skipping for this, or any other, LEF1 minigene tested (data not shown). Thus, we conclude that the transient increased skipping of exon 6 in the endogenous LEF1 gene is not regulated by exon-proximal sequences. As this transient repression of exon 6 is smaller than the subsequent increase in exon inclusion and the biologic implication of this is less clear, we have not further investigated the factors driving this regulation.
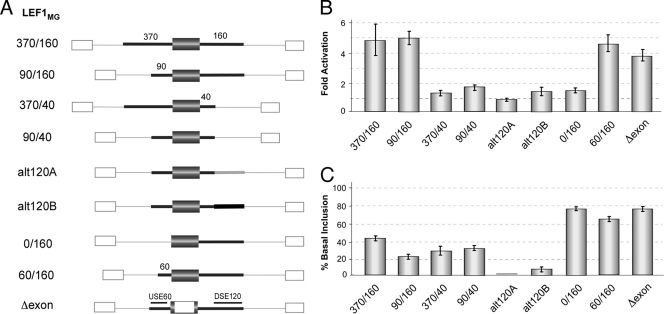
Fig. 3.
Intronic sequences flanking LEF1 exon 6 function as signal-induced enhancers of exon inclusion. (A) Schematic representations of the minigenes used. The gray box and thick lines correspond to exon 6 and flanking intron sequences from LEF1, respectively. The numbers above the lines give the approximate length of included LEF1 endogenous sequence. The white boxes and thin lines correspond to exon and intron sequences derived from the human beta-globin gene, respectively. The thick light gray and black lines in constructs alt120A and alt120B correspond to heterologous sequence used as filler. All constructs were expressed from a constitutive, heterologous promoter. (B) Graph of fold activation, as defined in the legend to Fig. 1, of exon 6 inclusion in each of the minigenes upon 60-h PMA stimulation of JSL1 cells. The values are the averages of 3 or 4 independent experiments with error bars indicating standard deviations. (C) Graph of the amount of exon 6 inclusion in each of the minigenes expressed in resting cells (basal inclusion). Values were calculated and graphed as for panel B.
Having confirmed that the LEF1MG370/160 minigene recapitulates the signal-induced enhancement of LEF1 variable exon inclusion, we next sought to identify the cis-acting sequences that determine this regulation. Deletion of ∼280 nucleotides of the upstream intron had no effect on signal-induced alternative splicing (Fig. 3A and B, LEF1MG90/160), whereas deletion of ∼120 nucleotides in the downstream intron virtually abolished all signal-induced inclusion of the LEF1 variable exon regardless of the length of the upstream intron (Fig. 3A and B, LEF1MG370/40 and LEF1MG90/40). The loss of regulation of the LEF1MG90/40 construct is not simply a consequence of shortening the downstream intron, as the addition of heterologous sequence of a similar length did not restore signal-induced regulation (Fig. 3A and B, alt120A and alt120B).
Surprisingly, while the downstream intron is necessary for the signal-induced regulation of exon 6, it is not sufficient. In particular, substitution of the upstream intron with respective sequence from a beta-globin intron results in a complete loss of signal-induced splicing (Fig. 3A and B, LEF1MG0/160). This substitution of the upstream intron also dramatically increases basal exon inclusion (Fig. 3C). However, the increase in basal inclusion is not the cause of the loss of signal responsiveness as returning 60 nucleotides of the endogenous LEF1 intron restores the responsiveness to cellular stimulation while maintaining strong basal inclusion (Fig. 3A to C, LEF1MG60/160). Finally, substitution of the sequences internal to LEF1 exon 6 itself also increases basal inclusion with only a modest decrease in the extent of inclusion induced by cellular activation (Fig. 3A to C, LEF1MGΔexon). Taken together, our minigene data highlight the 60 nucleotides immediately upstream of exon 6 and a region of 120 nucleotides in the intron downstream of exon 6 as together functioning as a signal-induced enhancer of exon inclusion. In subsequent studies, we designate the upstream 60 nucleotides as USE60 (upstream signal-induced enhancer 60 nucleotides) and the downstream 120 nucleotides as DSE120 (downstream signal-induced enhancer 120 nucleotides).
Interestingly, the USE60 and DSE120 display remarkable conservation across species. LEF1 exon 6 is highly conserved across mammals, with at least some evidence for alternative splicing even in armadillos, which is believed to have diverged from humans ∼120 million years ago (32). Alignment of the LEF1 variable exon and flanking intron from six divergent mammalian species reveals an expected ∼90% identity across the exon, but this high degree of identity continues throughout the entire USE60 before diverging rapidly upstream of the USE60 (Fig. 4). In contrast, alignment of the 60 nucleotides preceding the flanking constitutive LEF1 exons from these same species reveals only ∼50% identity (data not shown). Therefore, the ∼90% conservation in the USE60 is more than would be anticipated simply from the requirement for this region to contain the branch point sequence, polypyrimidine tract, and 3′AG, to direct splicing catalysis. Similarly, the intron downstream of LEF exon 6 is largely divergent between mammalian species with the exception of two blocks of sequence within the DSE120 that exhibit 85 to 90% identity (11/13 or 35/40, respectively; Fig. 4). Such a high degree of conservation within the intronic USE60 and DSE120 provide further evidence for a functional role of these sequence elements in controlling the accurate expression of LEF1.
Fig. 4.
Phylogenetic comparison reveals high conservation of sequences required for signal-responsive inclusion of LEF1 exon 6. Clustal W alignment of sequences surrounding LEF1 exon 6 with the corresponding sequences from armadillo (Dasypus novemcinctus) (aLEF1), dog (Canis lupus familiaris) (dLEF1), cow (Bos taurus) (cLEF1), rat (Rattus norvegicus) (rLEF1), mouse (Mus musculus) (mLEF1), and human (Homo sapiens) (LEF1). Nucleotides that are absolutely conserved are indicated by an asterisk. The elements are indicated by colors as follows: blue, USE60 (encompasses the 3′ splice site [3′ss]); red, exon 6; orange, 5′ splice site; green, DSE120 (highly conserved elements in DSE120 are in darker green).
CELF2 binds to both of the LEF1 regulatory sequences.
In analyzing the USE60 and DSE120, we also noted that these sequence elements were enriched in TG and CTG nucleotides. This is particularly conspicuous in the most highly conserved block of 40 nucleotides in the DSE120. Such a sequence pattern bears the hallmarks of the optimal binding site for the CELF and MBNL (Muscleblind-like) families of splicing regulatory proteins (9, 12). To investigate whether any CELF or MBNL proteins function as regulators of LEF1 splicing, we first used a UV cross-linking assay to determine in an unbiased fashion what proteins bind to the LEF1 regulatory sequences. UV cross-linking of radiolabeled DSE120 RNA with nuclear extract from JSL1 cells, followed by degradation of the RNA and resolution by SDS-PAGE, revealed almost exclusive association of DSE120 with a single protein species migrating at approximately 50 kDa (Fig. 5A). Consistent with a functional role of this protein in signal-induced splicing, the efficiency of this protein-RNA cross-link was greater in extracts from stimulated JSL1 cells than in extracts from resting cells, despite similar concentrations of total protein in the two extracts (Fig. 5A, ∼3-fold increase in binding between extracts from resting and stimulated cells relative to control bands). UV cross-linking using USE60 RNA gave notably similar results to the DSE120, though some additional RNA-binding proteins were evident, including one constitutive protein migrating immediately above the stimulation-induced 50-kDa protein (Fig. 5B). In contrast, RNA corresponding to ∼100 nucleotides from an unrelated intron displayed a markedly distinct protein binding pattern in the UV cross-linking assay (Fig. 5C), demonstrating at least some degree of specificity for the proteins bound to DSE120 and USE60.
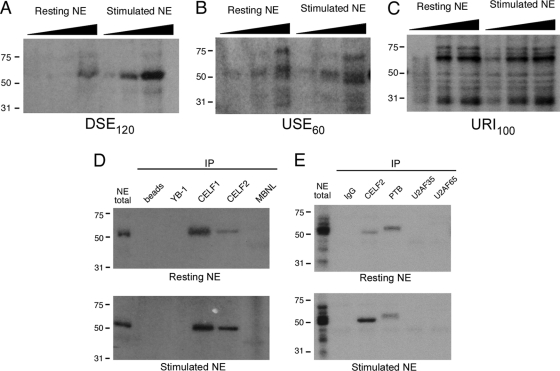
Fig. 5.
Both intronic regions controlling LEF1 exon 6 inclusion bind to the splicing factor CELF2 in a PMA-inducible manner. (A) UV cross-linking with increasing amounts of nuclear extracts (NE) prepared from resting or stimulated (+PMA) JSL1 cells with body-labeled DSE120 RNA, followed by RNase digestion and resolution by SDS-PAGE. The extract concentrations used were 1.5, 5, and 15 μg total protein and are indicated by the height of the black triangle above the lane. The migration positions of molecular mass markers (in kilodaltons) are shown to the left of the gels. (B) Same as panel A but with USE60 RNA. The top band of the apparent doublet at ∼50 kDa corresponds to PTB, while the bottom band is CELF2 (see panel E). (C) Same as panel A but with unrelated intron (URI100) RNA. (D) Immunoprecipitation (IP) of UV cross-linking reactions from panel A (DSE120, after RNase treatment) with antibodies as shown. Antibody to CELF1 cross-react with CELF2 but not vice versa. (E) Same as panel D but following cross-linking with USE60.
Many of the CELF family members are approximately 50 kDa in size, whereas the MBNL family members typically migrate in the range of 30 to 40 kDa. However, as several splicing factors are also of similar sizes, we utilized antibodies to more conclusively determine the identity of the protein cross-linked to the USE60 and DSE120. As shown in Fig. 5D, antibodies specific to CELF1 and CELF2 both efficiently precipitated a protein cross-linked to DSE120 which comigrates with the major cross-linked species in total nuclear extract. The protein precipitated by the anti-CELF1 antibody increases only slightly if at all between the resting and stimulated cell extracts, whereas there is an 2- to 3-fold enrichment in the anti-CELF2 reactive protein in the stimulated cell extracts, consistent with the increase in cross-linking observed in total extract. No cross-linked protein is precipitated by beads alone or by antibodies specific for YB-1, another 50-kDa splicing factor that binds to CA-rich sequences (16). A slight signal is detected with antibodies specific for MBNL, but this comigrates with a faint band observed below the 50-kDa species in long exposures of total extract, and thus cannot account for the 50-kDa protein itself.
Similar to the results obtained with DSE120, the antibody specific for CELF2 precipitates a protein cross-linked to USE60 which exhibits preferential abundance in extracts from stimulated cells. In contrast, the ubiquitous cross-linked protein migrating above 50 kDa appears to be the splicing factor polypyrimidine tract-binding protein (PTB), known to preferentially bind to UCUU sequences (7). Interestingly, although the USE60 overlaps the polypyrimidine tract of the intron upstream of exon 6, cross-linking of USE60 to the core splicing factors that recognize the polypyrimidine tract in the spliceosome (U2AF65 and U2AF35) is observed only upon overexposure of the gel (data not shown). Taken together, the cross-linking data we show here strongly suggest that CELF2 is the primary signal-responsive protein bound to both the DSE120 and USE60, while CELF1 and PTB appear to bind in a signal-independent manner to the DSE120 and USE60, respectively.
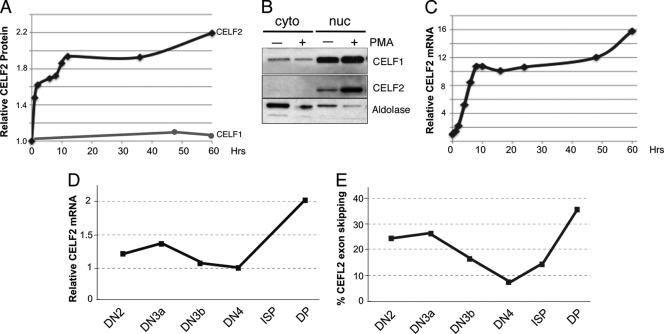
Importantly, the increased binding of CELF2 to the LEF1 regulatory elements reflects an increase in the expression of CELF2 protein that is triggered by PMA stimulation of the JSL1 cells, reaching a plateau at ∼10 h after stimulation (Fig. 6A and B). This increase in CELF2 protein correlates with a notable increase in expression of the CELF2 mRNA (Fig. 6C), suggesting that the upregulation of CELF2 protein is a direct result of increased transcription. We also detect changes in the CELF2 transcript levels during thymic development (Fig. 6D). This increase in CELF2 transcript is followed by an autoinhibitory splicing event in CELF2 pre-mRNA, also observed in JSL1 cells (17), which is indicative of increased CELF2 protein levels (Fig. 6E) (8). Therefore, we conclude that both PMA stimulation of JSL1 cells and pre-TCR signaling in thymocytes result in an increase in CELF2 expression, which in turn drives the increase in CELF2 binding to the LEF1 regulatory sequences.
Fig. 6.
Expression of CELF2 in JSL1 cells and thymocytes. (A) Quantification of total CELF2 and CELF1 protein in JSL1 cells following stimulation with PMA. Protein level was determined by Western blotting and normalized to actin. (B) Representative Western blot of CELF2 and CELF1 as used for panel A at 48 h following stimulation. Cells were fractionated upon lysis and blotted for adolase as a measure of the purity of cytoplasmic (cyto) and nuclear (nuc) fractions. Results indicate that while expression of CELF2 changes upon stimulation, subcellular localization of this protein is not altered. (C) Quantification of total CELF2 mRNA in JSL1 cells following stimulation with PMA. mRNA was determined by RT-PCR and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). (D) Quantification of CELF2 mRNA in murine thymic populations. Analysis was done as described above for panel C. (E) Quantification of percent exon skipping in CELF2 mRNA in the indicated murine thymic populations, showing a sharp rise in skipping between DN4 and DP, where the increase in mRNA and presumably protein is also observed.
CELF2 binding to the intronic regulatory sequences is functionally required for LEF1 splicing.
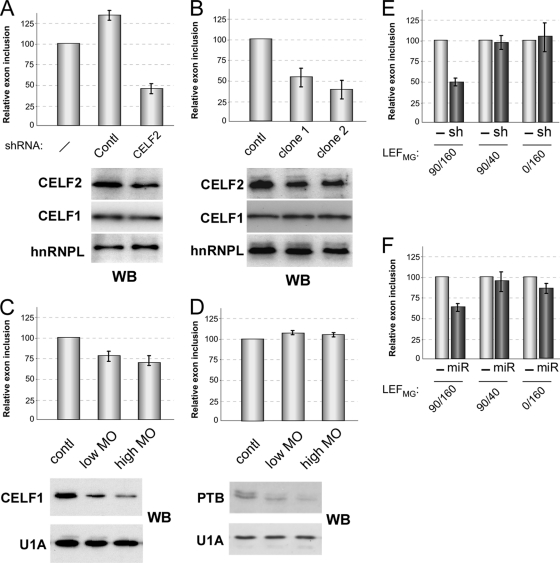
To determine the functional relevance of CELF2, CELF1, or PTB binding to the regulation of LEF1 exon 6, we knocked down these proteins in our JSL1 cell line and assayed splicing of the endogenous LEF1 gene by RT-PCR. Stable expression of an shRNA directed against CELF2 decreased expression of this protein by about 50% in resting cells and correspondingly resulted in a 2- to 3-fold decrease in exon 6 inclusion relative to untransfected cells or those expressing an unrelated shRNA (Fig. 7A). A similar trend was observed in activated cells as shown below.
Fig. 7.
Knockdown of CELF2 results in loss of exon 6 inclusion in a manner dependent on the presence of the DSE120 and USE60. (A) Percent inclusion of exon 6 in endogenous LEF1 gene in resting JSL1 cells expressing a shRNA against CELF2, an unrelated splicing protein PSF (control [contl]), or no shRNA (−). The values are the averages of 3 or 4 independent experiments normalized to the control values, with error bars indicating standard deviations. Western blots (WB) below the graph show the effect of shRNAs on CELF2 and CELF1 relative to hnRNPL loading control. (B) Same as panel A for 2 independent clones stably expressing miR-23b. (C) Same as panel A for cells transfected with a translation-blocking morpholino oligomer (MO) directed against the AUG of CELF1. (D) Same as panel A for cells transfected with a translation-blocking morpholino oligomer directed against the AUG of PTB. (E) Relative percent inclusion of exon 6 in the indicated minigenes cotransfected into resting JSL1 cells with the shRNA against CELF2 as in panel A. (F) Same as panel E but with miR-23b-expressing cells.
As a complementary method to decrease CELF protein expression, we also generated stable clones that ubiquitously expressed miR-23b. miR-23b was recently shown to be an endogenous regulator of CELF1 and CELF2 in heart (18), though it does not appear to be expressed at significant levels in our JSL1 cells (M. J. Mallory and K. W. Lynch, unpublished data). Consistent with the shRNA results, vector-driven expression of miR-23b resulted in a 50 to 60% reduction in the inclusion of LEF1 exon 6 in two independent stable clones (Fig. 7B). Although miR-23b can repress expression of both CELF1 and CELF2, we reason that the effect on LEF1 splicing is due primarily to the loss of CELF2, as miR-23b had no detectable effect on the expression of CELF1 in the JSL1 cells (Fig. 7B, Western blots). Moreover, specific knockdown of CELF1 has a much more modest effect on LEF1 splicing than observed with either miR-23b or the CELF2-directed shRNA (Fig. 7C versus Fig. 7A and B). Finally, depletion of PTB had no discernible influence on the inclusion of exon 6, further highlighting the specificity of the results with CELF2 (Fig. 7D).
The above functional data, together with the binding data in Fig. 5, suggest a model in which binding of CELF2 to the DSE120 and/or USE60 enhances inclusion of LEF1 exon 6. To more conclusively link the function of CELF2 to its binding of the DSE120 and USE60 elements, we next tested the responsiveness of representative minigenes to CELF2 knockdown. As anticipated, the minigene LEF1MG90/160, which contains both the USE60 and DSE120, showed a marked decrease in exon 6 inclusion in cells expressing the CELF2 shRNA (Fig. 7E) or miR-23b (Fig. 7F). In contrast, splicing of LEF1MG90/40, which lacks the DSE, was unaffected by CELF2 knockdown (Fig. 7E and F). Similarly, removal of the USE also reduced the sensitivity of the resulting minigene to CELF2 knockdown (Fig. 7E and F, LEF1MG0/160). Therefore, both of the CELF2 binding sites are required for the activity of CELF2 in promoting exon inclusion.
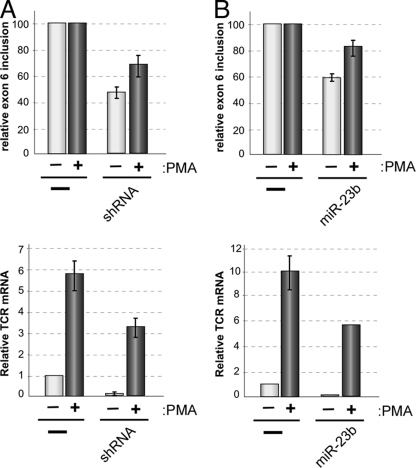
Taken together, our data demonstrate that binding of CELF2 to the USE and DSE elements flanking LEF1 exon 6 promotes inclusion of this exon in the final spliced transcript and imply that the increased binding of CELF2 to the USE and DSE which we observe under stimulated conditions (Fig. 5) drives the signal-induced inclusion of exon 6. To make the full link between CELF2 regulation of LEF1 and TCR-alpha expression, we sought to determine whether reducing the expression of CELF2 inhibited the stimulation-induced transcription of TCR-alpha that we have already shown to be dependent on LEF1 exon 6 inclusion (Fig. 2). As anticipated, knockdown of CELF2 by expression of either the CELF2 shRNA or miR-23b reduced inclusion of LEF1 exon 6 in PMA-stimulated cells similar to that observed in the resting cells (Fig. 8A and B, top graphs). Strikingly, in these cells, we also observe a marked decrease in TCR-alpha expression relative to the corresponding control cells, under both resting and stimulated conditions (Fig. 8A and B, bottom graphs). The decrease in TCR-alpha expression in the shRNA- or miR-23b-containing cells is highly similar to our results with the LEF1 splice-blocking morpholino oligomer (Fig. 2); therefore, we conclude that CELF2-mediated inclusion of LEF1 exon 6 plays an important functional role in contributing to the signal-induced expression of the TCR-alpha chain.
Fig. 8.
Depletion of CELF2 to reduce LEF1 exon 6 inclusion inhibits signal-induced TCR-alpha expression. (A) Graph of LEF1 exon 6 inclusion (top) and TCR-alpha mRNA (bottom) in cells depleted of CELF2 using the shRNA as described in the legend to Fig. 6. Error bars indicate standard deviations from 2 or 3 independent experiments in cells grown for 48 h in the absence (−) or presence (+) of PMA. Values are shown relative to the JSL1 control. (B) Same as panel A for cells expressing miR-23b.
DISCUSSION
Recent profiling of RNA isoform expression in various cell types has indicated widespread changes in alternative splicing in response to cellular stimulation (1, 14, 17, 21). However, the pathways and proteins that mediate signal-induced alternative splicing, as well as the functional consequences of such regulation, remain poorly understood. In particular, there is virtually no concrete understanding as to whether any of the myriad alternative splicing events that have been identified in the immune system directly influence normal cellular responses. Here we describe a comprehensive pathway from the mechanistic determinants of LEF1 splicing regulation to a physiologic role for LEF1 alternative splicing. Specifically we demonstrate that manipulating the splicing pattern of LEF1 directly impacts expression of its downstream target gene TCR-alpha, we show that the endogenous splicing of LEF1 is regulated during T-cell development in a manner that correlates with altered TCR-alpha expression, and we uncover essential cis and trans regulators of LEF1 splicing.
CELF2 as a signal-responsive splicing regulator in T cells.
Although there has been long-standing evidence for multiple isoforms of LEF1, there has been no investigation as to the sequences or proteins that determine isoform expression. In this study, we identify two evolutionarily conserved intronic sequences flanking the LEF1 exon 6 that control the inclusion of this exon. Each of these regulatory elements binds the splicing regulatory protein CELF2. CELF2 expression and binding to LEF1 pre-mRNA increase in response to signals that promote exon 6 inclusion, whereas knockdown of CELF2 causes decreased inclusion of LEF1 exon 6. Previous studies have demonstrated CEFL2 as a critical regulator of splicing in the brain and during muscle development; however, to our knowledge, there has been no prior evidence of a role of CELF2 in determining splicing patterns in lymphocytes. Therefore, this work extends our knowledge of the tissue distribution and biologic impact of CELF2.
With regard to the mechanism by which cell signaling leads to changes in LEF1 splicing, we demonstrate that CELF2 expression increases in response to cellular stimulation, ultimately resulting in increased binding of CELF2 to the LEF1 intronic enhancer elements and increased inclusion of LEF1 exon 6. The signal-induced increase in CELF2 protein expression correlates with a dramatic increase in mRNA expression, suggesting that in the case of T-cell signaling the expression of CELF2 is controlled at the level of transcription. We note that this does not preclude other or additional levels of regulation of CELF2 expression, such as miRNA-based control as has been shown recently in cardiomyocytes (18). However, the absence of detectable endogenous expression of the CELF2-controlling miR-23a or miR-23b in either resting or stimulated JSL1 cells suggests that at least these miRNAs are not controlling CELF2 expression in T cells.
In the one example for which the details of CUGPB2 enhancement of exon inclusion has been studied, binding of CELF2 to a site downstream of cardiac troponin T exon 5 promotes recruitment of the U2 snRNP component of the spliceosome to the upstream splice site (13). Although some aspects of this mechanism may be relevant for LEF1, a distinction is that regulation of LEF1 requires the cooperative activity of both the DSE120 and USE60, as neither element alone is sufficient to confer signal-responsive inclusion. This suggests either that the dual sequences increase the affinity of CELF2 association and/or that binding of CELF2 to the two sites remodels the substrate in a way that promotes association of spliceosome components with the exon. There are currently no well-described models in which binding of a protein to either side of an exon induces exon inclusion. However, a recent bioinformatic study identified cooccurring intronic elements that promote inclusion of an intervening exon (19), suggesting that the configuration of regulatory elements that drive LEF1 exon 6 enhancement is not an isolated instance of such a mechanism.
Physiologic consequence of LEF1 alternative splicing.
Perhaps the most important conclusion from our work herein is the existence and functional relevance of LEF1 alternative splicing during thymic development. Specifically, we demonstrate an increase in exon 6 inclusion, leading to increased relative expression of full-length LEF1, precisely at the stage of thymic development in which the TCR-alpha enhancer is most active. Expression of TCR-alpha is the most critical checkpoint in the transition from DN to DP cells, and defects in expression of this gene result in a complete absence of active mature T cells. The peptide region encoded by LEF1 exon 6 promotes association of the LEF1 protein with cofactors Aly and Ets to form the enhanceosome complex which activates TCR-alpha expression (3, 4, 11). Consistent with this, cotransfection studies with reporter constructs have shown that LEF1* is a significantly less potent activator of the TCR-alpha enhancer than full-length LEF1 is (2, 5, 11). However, there has been no determination as to whether a shift in the isoform expression of the endogenous LEF1 gene truly impacts expression of downstream target genes, including the endogenous TCR-alpha gene.
We show here a direct causal relationship between LEF1 alternative splicing and expression of its most essential target gene, TCR-alpha, as specifically blocking inclusion of exon 6 reduces TCR-alpha expression. Knockout studies in mice demonstrate that LEF1 activity in thymocytes is at least partially redundant with its family member T-cell factor 1 (TCF1) during development (29); thus, we do not conclude that the regulation of LEF1 alternative splicing is absolutely required for TCR-alpha expression. We also note that we do not see a drop in TCR-alpha expression during the brief increase in LEF1 exon skipping initially induced by PMA. This could be due to either a threshold effect, in which the increase is not sufficiently large to shift transcription, and/or the transient nature of the increase, in which the change is sufficiently brief that it does not impact steady-state levels of LEF1 protein. Nevertheless, our data with the splice site morpholino oligomer clearly demonstrate that an acute and prolonged change in LEF1 splicing is sufficient to alter expression of TCR-alpha. Therefore, we favor a model in which the change in splicing of LEF1 during pre-TCR signaling redirects the activity of this protein toward the TCR-alpha enhancer to maximize TCR-alpha expression during a critical time in development.
Although outside the scope of our current study, given the essential role of Wnt signaling in cellular proliferation and development, modulation of LEF1 exon 6 inclusion may also influence other checkpoints in thymocyte development. Specifically, the loss of exon 6 skipping during thymic development may reduce the beta-catenin-dependent activities of LEF1 on Wnt-responsive genes by increasing its interaction with HIC5. Moreover, both CELF2 and LEF1 have been shown to play essential roles in the embryonic development of heart and other neuromuscular tissues (2, 6, 18, 20). Whether LEF1 splicing is regulated in these other tissues, and whether this is controlled by CELF2, remains to be determined. However, the possibility for broad control of cellular development through alternative splicing of LEF1 is an important implication of our studies and suggests exciting directions for further investigation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tom Cooper (Baylor College of Medicine) for the generous gift of antibodies and Avinash Bhandoola (University of Pennsylvania) for providing resources for thymocyte isolation. We also thank Tom Maniatis (Columbia University), Doug Black (UCLA), and Iain Mattaj (EMBL) for the gifts of antibodies to U2AF, PTB, and U1A, respectively, that they have provided in the past and were used in this study.
This work was funded by NIH grant R01 GM084034 to K.W.L.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 28 March 2011.
REFERENCES
- 1. An P., Grabowski P. J. 2007. Exon silencing by UAGG motifs in response to neuronal excitation. PLoS Biol. 5:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arce L., Yokoyama N. N., Waterman M. L. 2006. Diversity of LEF/TCF action in development and disease. Oncogene 25:7492–7504 [DOI] [PubMed] [Google Scholar]
- 3. Balmelle N., Zamarreno N., Krangel M. S., Hernandez-Munain C. 2004. Developmental activation of the TCR alpha enhancer requires functional collaboration among proteins bound inside and outside the core enhancer. J. Immunol. 173:5054–5063 [DOI] [PubMed] [Google Scholar]
- 4. Bruhn L., Munnerlyn A., Grosschedl R. 1997. ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRalpha enhancer function. Genes Dev. 11:640–653 [DOI] [PubMed] [Google Scholar]
- 5. Carlsson P., Waterman M. L., Jones K. A. 1993. The hLEF/TCF-1 alpha HMG protein contains a context-dependent transcriptional activation domain that induces the TCR alpha enhancer in T cells. Genes Dev. 7:2418–2430 [DOI] [PubMed] [Google Scholar]
- 6. Chen X., et al. 2006. The beta-catenin/T-cell factor/lymphocyte enhancer factor signaling pathway is required for normal and stress-induced cardiac hypertrophy. Mol. Cell. Biol. 26:4462–4473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chou M. Y., Underwood J. G., Nikolic J., Luu M. H., Black D. L. 2000. Multisite RNA binding and release of polypyrimidine tract binding protein during the regulation of c-src neural-specific splicing. Mol. Cell 5:949–957 [DOI] [PubMed] [Google Scholar]
- 8. Dembowski J. A., Grabowski P. J. 2009. The CUGBP2 splicing factor regulates an ensemble of branchpoints from perimeter binding sites with implications for autoregulation. PLoS Genet. 5:e1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Faustino N. A., Cooper T. A. 2005. Identification of putative new splicing targets for ETR-3 using sequences identified by systematic evolution of ligands by exponential enrichment. Mol. Cell. Biol. 25:879–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ghogomu S. M., van Venrooy S., Ritthaler M., Wedlich D., Gradl D. 2006. HIC-5 is a novel repressor of lymphoid enhancer factor/T-cell factor-driven transcription. J. Biol. Chem. 281:1755–1764 [DOI] [PubMed] [Google Scholar]
- 11. Giese K., Kingsley C., Kirshner J. R., Grosschedl R. 1995. Assembly and function of a TCR alpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 9:995–1008 [DOI] [PubMed] [Google Scholar]
- 12. Goers E. S., Voelker R. B., Gates D. P., Berglund J. A. 2008. RNA binding specificity of Drosophila muscleblind. Biochemistry 47:7284–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goo Y. H., Cooper T. A. 2009. CUGBP2 directly interacts with U2 17S snRNP components and promotes U2 snRNA binding to cardiac troponin T pre-mRNA. Nucleic Acids Res. 37:4275–4286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hartmann B., et al. 2009. Global analysis of alternative splicing regulation by insulin and wingless signaling in Drosophila cells. Genome Biol. 10:R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. House A. E., Lynch K. W. 2008. Regulation of alternative splicing: more than just the ABCs. J. Biol. Chem. 283:1217–1221 [DOI] [PubMed] [Google Scholar]
- 16. Hui J., Stangl K., Lane W. S., Bindereif A. 2003. HnRNP L stimulates splicing of the eNOS gene by binding to variable-length CA repeats. Nat. Struct. Biol. 10:33–37 [DOI] [PubMed] [Google Scholar]
- 17. Ip J. Y., et al. 2007. Global analysis of alternative splicing during T-cell activation. RNA 13:563–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kalsotra A., Wang K., Li P. F., Cooper T. A. 2010. MicroRNAs coordinate an alternative splicing network during mouse postnatal heart development. Genes Dev. 24:653–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ke S., Chasin L. A. 2010. Intronic motif pairs cooperate across exons to promote pre-mRNA splicing. Genome Biol. 11:R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ladd A. N., Charlet N., Cooper T. A. 2001. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol. Cell. Biol. 21:1285–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee J. A., et al. 2007. Depolarization and CaM kinase IV modulate NMDA receptor splicing through two essential RNA elements. PLoS Biol. 5:e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leiden J. M., Thompson C. B. 1994. Transcriptional regulation of T-cell genes during T-cell development. Curr. Opin. Immunol. 6:231–237 [DOI] [PubMed] [Google Scholar]
- 23. Leroy O., et al. 2006. ETR-3 represses Tau exons 2/3 inclusion, a splicing event abnormally enhanced in myotonic dystrophy type I. J. Neurosci. Res. 84:852–859 [DOI] [PubMed] [Google Scholar]
- 24. Lynch K. W., Weiss A. 2000. A model system for the activation-induced alternative splicing of CD45 implicates protein kinase C and Ras. Mol. Cell. Biol. 20:70–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Melton A. A., Jackson J., Wang J., Lynch K. W. 2007. Combinatorial control of signal-induced exon repression by hnRNP L and PSF. Mol. Cell. Biol. 27:6972–6984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Modrek B., Resch A., Grasso C., Lee C. 2001. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 29:2850–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Motta-Mena L. B., Heyd F., Lynch K. W. 2010. Context-dependent regulatory mechanism of the splicing factor hnRNP L. Mol. Cell 29:223–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nilsen T. W., Graveley B. R. 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463:457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Okamura R. M., et al. 1998. Redundant regulation of T cell differentiation and TCRalpha gene expression by the transcription factors LEF-1 and TCF-1. Immunity 8:11–20 [DOI] [PubMed] [Google Scholar]
- 30. Pan Q., Shai O., Lee L. J., Frey B. J., Blencowe B. J. 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40:1413–1415 [DOI] [PubMed] [Google Scholar]
- 31. Rao M. K., Wilkinson M. F. 2006. Tissue-specific and cell type-specific RNA interference in vivo. Nat. Protoc. 1:1494–1501 [DOI] [PubMed] [Google Scholar]
- 32. Rosenbloom K., et al. 2008. Phylogenomic resources at the UCSC Genome Browser. Methods Mol. Biol. 422:133–144 [DOI] [PubMed] [Google Scholar]
- 33. Rothrock C., Cannon B., Hahm B., Lynch K. W. 2003. A conserved signal-responsive sequence mediates activation-induced alternative splicing of CD45. Mol. Cell 12:1317–1324 [DOI] [PubMed] [Google Scholar]
- 34. Rothrock C. R., House A. E., Lynch K. W. 2005. HnRNP L represses exon splicing via a regulated exonic splicing silencer. EMBO J. 24:2792–2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tong A., Nguyen J., Lynch K. W. 2005. Differential expression of CD45 isoforms is controlled by the combined activity of basal and inducible splicing-regulatory elements in each of the variable exons. J. Biol. Chem. 280:38297–38304 [DOI] [PubMed] [Google Scholar]
- 36. Topp J. D., Jackson J., Melton A. A., Lynch K. W. 2008. A cell-based screen for splicing regulators identifies hnRNP LL as a distinct signal-induced repressor of CD45 variable exon 4. RNA 14:2038–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Travis A., Amsterdam A., Belanger C., Grosschedl R. 1991. LEF-1, a gene encoding a lymphoid-specific [sic] with an HMG domain, regulates T-cell receptor alpha enhancer function. Genes Dev. 5:880–894(Erratum, 5:1113.) [DOI] [PubMed] [Google Scholar]
- 38. van Genderen C., et al. 1994. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice. Genes Dev. 8:2691–2703 [DOI] [PubMed] [Google Scholar]
- 39. Wang E. T., et al. 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456:470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Waterman M. L., Fischer W. H., Jones K. A. 1991. A thymus-specific member of the HMG protein family regulates the human T cell receptor C alpha enhancer. Genes Dev. 5:656–669 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.