Abstract
Toxin synthesis in Clostridium difficile increases as cells enter into stationary phase. We first compared the expression profiles of strain 630E during exponential growth and at the onset of stationary phase and showed that genes involved in sporulation, cellular division, and motility, as well as carbon and amino acid metabolism, were differentially expressed under these conditions. We inactivated the sigH gene, which encodes an alternative sigma factor involved in the transition to post-exponential phase in Bacillus subtilis. Then, we compared the expression profiles of strain 630E and the sigH mutant after 10 h of growth. About 60% of the genes that were differentially expressed between exponential and stationary phases, including genes involved in motility, sporulation, and metabolism, were regulated by SigH, which thus appears to be a key regulator of the transition phase in C. difficile. SigH positively controls several genes required for sporulation. Accordingly, sigH inactivation results in an asporogeneous phenotype. The spo0A and CD2492 genes, encoding the master regulator of sporulation and one of its associated kinases, and the spoIIA operon were transcribed from a SigH-dependent promoter. The expression of tcdA and tcdB, encoding the toxins, and of tcdR, encoding the sigma factor required for toxin production, increased in a sigH mutant. Finally, SigH regulates the expression of genes encoding surface-associated proteins, such as the Cwp66 adhesin, the S-layer precursor, and the flagellum components. Among the 286 genes positively regulated by SigH, about 40 transcriptional units presenting a SigH consensus in their promoter regions are good candidates for direct SigH targets.
INTRODUCTION
Clostridium difficile, a Gram-positive, anaerobic, spore-forming bacterium, is a major cause of nosocomial infections associated with antibiotic therapy. This enteropathogen can lead to antibiotic-associated diarrhea and pseudomembranous colitis, a potentially lethal disease. Transmission of C. difficile is mediated by contamination of the gut by spores (54). Spores also contribute to its survival during antibiotic therapy. The disruption of the colonic microflora by antimicrobial therapy precipitates C. difficile infection and colonization of the intestinal tract (54). Thus, spores germinate, vegetative forms multiply, and the major virulence factors, the two large toxins TcdA and TcdB, are produced. The toxin-encoding genes, tcdA and tcdB, are localized in the 19.6-kb pathogenicity locus with three other accessory genes, tcdR, tcdC, and tcdE. TcdR is an alternative sigma factor that directs transcription from the tcdA, tcdB, and tcdR promoters, while TcdC is an anti-sigma factor that negatively regulates TcdR-dependent transcription by destabilizing the TcdR-holoenzyme complex (33, 34). The tcdE gene encodes a holin-like protein that is required for the release of the toxins from the cell (R. Govind and B. Dupuy, unpublished results).
The spectrum of diseases caused by C. difficile is highly variable and depends on the level of toxin produced for the severe forms (1). This supports the hypothesis that regulation of toxin synthesis is a critical determinant of C. difficile pathogenicity. Toxin synthesis increases as cells enter into stationary phase, and many environmental factors influence their production. In the presence of phosphotransferase system (PTS) sugars, such as glucose, and of certain amino acids, like cysteine or proline, toxin production is inhibited (16, 30). Environmental stresses, such as alteration of the redox potential, high temperature, or limitation of biotin, also modulate toxin production. Three regulators involved in the control of toxin synthesis have already been identified: CcpA, mediating glucose-dependent repression (5), and two regulators controlling pre- or post-exponential events, CodY and Spo0A (14, 52).
To cope with nutrient limitations, bacterial cells have to change their metabolism and physiology when they enter into stationary phase. To adapt and survive, they can also express functions like competence, biofilm formation, or antibiotic production. Bacilli and clostridia also have the ability to undergo a cellular differentiation process leading to the formation of a spore that will germinate when growth-favoring conditions return. During transition from exponential to stationary growth phase, a complex regulatory network will determine which option, stationary phase or initiation of sporulation, is most suitable under the given conditions. The regulatory mechanisms involved in the transition phase have been extensively studied in Bacillus subtilis (38). In this bacterium, it has been shown that during the exponential growth phase, repressors such as AbrB, CodY, and SinR prevent the expression of functions that are only needed during stationary phase or sporulation (38, 43). Sporulation initiation in B. subtilis is controlled by the response regulator, Spo0A, and a phosphorelay involving five kinases (KinA to KinE), intermediary phosphorylated proteins (Spo0F and Spo0B), and several phosphatases (38, 45). Environmental and cellular stress signals are sensed by the sensor kinases that autophosphorylate. They transfer their phosphoryl group to Spo0F and then to the Spo0A regulator via the intermediary phosphorylation of Spo0B (38, 45). Phosphorylated Spo0A (Spo0A-P) activates or represses the expression of target genes by binding to a specific target site (TGNCGAA) in their promoter regions (35). If the Spo0A-P level reaches a critical threshold, Spo0A-P activates the transcription of genes required for sporulation, such as the spoIIAA-spoIIAB-sigF operon and the spoIIG and spoIIE genes (35, 38, 43). The alternative sigma factor, SigH, is also a key element in the control of transition phase and of the initiation of sporulation. The RNA polymerase containing SigH transcribes several genes that function during the transition state, including genes required for sporulation, like spo0A, spo0F, kinA, kinE, and spoIIAA. The consensus sequence of genes transcribed by SigH in B. subtilis is (A/G)NAGGA(A/T)3-N11-12-(A/G)NNGAAT (8).
Compared to B. subtilis, less is known about the complex regulatory network controlling post-exponential events, including the initiation of sporulation, in clostridia (37). The AbrB repressor is absent in the C. difficile genome, while CodY and a SinR-like repressor are present (41). Spo0A and SigH are present in all clostridia. In contrast, the phosphorelay is absent and the sporulation initiation pathway remains in clostridia as a two-component system with Spo0A and associated kinases which can phosphorylate Spo0A directly in the absence of Spo0F and Spo0B (37, 44, 56). A recent report showed that in C. difficile, Spo0A is phosphorylated by the CD1579 kinase in vitro. Moreover, inactivation of CD2492, a second kinase, reduces the sporulation capacity of C. difficile, while a spo0A mutant does not sporulate (52).
Whether the production of toxins in C. difficile is a stationary-phase event or is associated with sporulation remains to be clearly established. Among the regulators involved in the control of the transition from exponential growth to stationary phase, it has recently been shown that CodY represses toxin gene expression via direct binding to the tcdR promoter region (14). In addition, TcdA production decreases in spo0A and CD2492 mutants (52). Finally, the role of SigH in the stationary phase and in the initiation of sporulation, as well as in regulation of the toxin genes, has never been studied in clostridia.
In the present study, we analyzed the expression profile modifications between exponential growth and the onset of stationary phase in C. difficile and the role of SigH in these transcriptional changes. We then investigated the involvement of SigH in sporulation and virulence factor expression, including toxin production, in C. difficile.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The C. difficile strains and plasmids used in this study are presented in Table S1 in the supplemental material. C. difficile strains were grown anaerobically (10% H2, 10% CO2, 80% N2) in TY (16) or brain heart infusion (BHI) medium (Difco). Sporulation assays were performed with BHI supplemented with 5 g/liter yeast extract and 0.1% l-cysteine (BHIS). When necessary, cefoxitin (25 μg/ml), thiamphenicol (15 μg/ml), or erythromycin (2.5 μg/ml) was added to C. difficile cultures. Escherichia coli strains were grown in Luria-Bertani (LB) broth. When indicated, ampicillin (100 μg/ml) or chloramphenicol (15 μg/ml) was added to the culture medium. All routine plasmid constructions were carried out using standard procedures (40).
Construction of gene knockout mutants in C. difficile.
The ClosTron gene knockout system (23) was used to inactivate the sigH and spo0A genes, giving strains 630E sigH::erm (CDIP8) and 630E spo0A::erm (CDIP3), respectively (see Table S1 in the supplemental material). Primers to retarget the group II intron of pMTL007 to these genes (see Table S2 in the supplemental material) were designed with Targetron design software (Sigma-Aldrich). The PCR primer sets were used with the EBS universal primer and intron template DNA to generate by overlap extension PCR a 353-bp product for each gene that would facilitate intron retargeting. These PCR products were cloned into the HindIII and BsrGI restriction sites of pMTL007 (see Fig. S1 in the supplemental material), giving plasmids pMTL007::Cdi-spo0A178a and pMTL007::Cdi-sigH123s. DNA sequencing was performed to verify plasmid constructs, using the pMTL007-specific primers pMTL007-F and pMTL007-R (see Table S2 in the supplemental material). The derivative pMTL007 plasmids were transformed into E. coli HB101(RP4) and subsequently mated with C. difficile 630E (Δerm) (27). C. difficile transconjugants were selected by subculturing on BHI agar containing thiamphenicol (15 μg/ml) and cefoxitin (25 μg/ml) and then plated on BHI agar containing erythromycin (2.5 μg/ml). Chromosomal DNA of transconjugants was isolated as previously described (5). PCR using the ErmRAM primers confirmed that the Ermr phenotype was due to the splicing of the group I intron from the group II intron following integration (see Fig. S1 in the supplemental material). In order to verify the integration of the Ll.LtrB intron into the right gene targets, we performed PCR with two primers flanking the sigH (IMV358-IMV359) or spo0A (OBD530-OBD529) gene and, on one hand, a primer in sigH (IMV358) or spo0A (OBD530) and, on the other hand, with the intron primer EBSu (see Table S2 and Fig. S1 in the supplemental material).
To complement the sigH mutant, the sigH gene with its promoter (−297 to +720 from the translational start site) was amplified by PCR. The PCR fragment was cloned into the XhoI and BamHI sites of pMTL84121 (24) to produce plasmid pDIA5932. Using the E. coli HB101(RP4) strain as donor, pDIA5932 was transferred by conjugation into the C. difficile 630E sigH::erm mutant, giving strain CDIP12 (see Table S1 in the supplemental material).
Sporulation assay.
Overnight cultures grown at 37°C in BHI were used to inoculate BHIS medium. After 72 h of growth, 1 ml of culture was divided into two samples. To determine the total number of CFU, the first sample was serially diluted and plated on BHI with 0.1% taurocholate (Sigma-Aldrich). Taurocholate is required for the germination of C. difficile spores (55). To determine the number of spores, the vegetative cells of the second sample were heat killed by incubation for 20 min at 65°C prior to plating on BHI with 0.1% taurocholate. The percentage of sporulation was determined as the ratio of the number of spores/ml and the total number of bacteria/ml times 100. The mean values for at least two independent experiments are presented.
Crude extract preparation and protein dot blot analysis.
To obtain C. difficile crude extracts, 10 ml of culture was centrifuged. Cells were washed with phosphate-buffered saline (PBS), resuspended in 1 ml of PBS, and disrupted in a FastPrep apparatus (2 times for 30 s at speed 6.5) in the presence of glass beads at 4°C. The soluble fractions were obtained by centrifugation to remove cells debris. For dot blot analysis, we directly spotted 200 ng of protein of each sample onto a nitrocellulose membrane (Hybond-C extra; Amersham Biosciences). The membranes were blocked with 5% (wt/vol) nonfat dried milk in Tris 50 mM, pH 7.5, containing 150 mM NaCl and then incubated overnight at 4°C with the primary anti-TcdA antibody. Following washing, membranes were incubated with the secondary antibody at 37°C for 60 min, and the TcdA protein was detected by using SuperSignal West Femto chemiluminescent substrate (Thermo Scientific) according to the manufacturer's instructions. Dot blots were quantified and normalized by Image Gauge (FujiFilm).
RNA extraction, quantitative RT-PCR analysis, and 5′ RACE.
We extracted total RNA from strains 630E, 630E sigH::erm, 630E spo0A::erm, and 630E sigH::erm complemented with sigH in trans grown in TY medium. Cells were harvested after 4 h or 10 h of culture by centrifugation for 2 min at 4°C. RNA extraction was performed as previously described (4), cDNA were synthesized from 5 μg of total RNA with 1 μl of hexamer oligonucleotide primers (5 μg/μl pdN6; Roche), 4 μl of deoxynucleoside triphosphates (dNTP; 10 mM each), and 6 μl of reverse transcription (RT) buffer in a final volume of 30 μl. Samples were heated for 5 min at 80°C. After a slow cooling, cDNAs were synthesized for 2 h at 42°C with avian myeloblastosis virus (AMV) reverse transcriptase (Promega). Real-time quantitative PCR was performed in a 20-μl reaction volume containing 10 ng or 10 pg (for 16S rRNA) of cDNAs, 12.75 μl of the SYBR PCR master mix (Applied Biosystems), and 400 nM gene-specific primers. Amplification and detection were performed as previously described (3). In each sample, the quantity of cDNAs of a gene was normalized to the quantity of cDNAs of the 16S rRNA gene. The relative change in gene expression was recorded as the ratio of normalized target concentrations (threshold cycle [ΔΔCT] method) (32). The transcriptional start sites of the spo0A, sigH, CD2492, spoIIAA, sigA2, dnaG, and sigG genes were determined using the 5′ rapid amplification of cDNA ends (RACE) system (Invitrogen) according to the manufacturer's recommendations. RNA preparations extracted from strains 630E or 630E sigH::erm were used as the template. The primers used for 5′ RACE analysis are listed in Table S2 in the supplemental material.
Microarray design for the C. difficile genome, DNA array hybridization, and data analysis.
The C. difficile strain 630 genome was obtained from the EMBL database. Probe design for the microarray was performed by using OligoArray 2.0 software (39). One or 2 oligonucleotides were designed for each 3,785 genes. We were unable to design oligonucleotides for 28 genes. Agilent produced the microarrays. Probes were replicated twice on the array to reach a final density of 14,224 probes per array. Five hundred thirty-six positive controls and 984 negative controls were also included. The description of the microarray design was submitted to the GEO database (accession number GPL10556).
Total RNA was extracted from cells of 4 independent cultures for each growth condition. RNA was labeled with either Cy3 or Cy5 fluorescent dye (GE Healthcare, Little Chalfont, United Kingdom) using a SuperScript Indirect cDNA labeling kit (Invitrogen) as previously described (4). The cDNAs were then mixed with Cy3 or Cy5 dyes, incubated for 1 h at room temperature in the dark, and purified on SNAP columns (Invitrogen). An amount of 200 pmol of Cy3- and Cy5-labeled cDNAs was mixed. Hybridization was performed for 17 h at 65°C according to the manufacturer's recommendations. Four differential hybridizations were performed for each experiment. The array was then washed successively with gene expression wash buffers 1 and 2 (Agilent). Scanning of arrays was performed with a GenePix Pro 6 dual-channel (635-nm and 532-nm) laser scanner (GenePix). All data were analyzed with R and Limma (Linear Model for Microarray data) software from the Bioconductor project (www.bioconductor.org). The background was corrected with the “Normexp” method (7), resulting in strictly positive values and reducing variability in the log ratios for genes with low levels of hybridization signal. Then, we normalized each slide with the Loess method (42). In order to identify differentially expressed genes, we used the Bayesian adjusted t statistics and performed the multiple testing correction of Benjamini and Hochberg (6) that is based on the false discovery rate. A gene was considered differentially expressed when the P value was <0.05.
Microarray data accession number.
The complete experimental data set was deposited in the Gene Expression Omnibus (GEO) database with the accession number GSE25521.
RESULTS AND DISCUSSION
Comparative analysis of gene expression profiles of strain 630E after 4 h and 10 h of growth.
We first performed growth kinetics of strain 630E in TY medium, and we observed that the transition between exponential growth phase and stationary phase occurred around 8 h of growth. Then, we analyzed by quantitative RT-PCR (qRT-PCR) the expression of transition-phase markers (spo0A, sigH, and spoIIAA) at different times of growth (4 h, 6 h, 8 h, 10 h, and 12 h). We showed that the expression of spo0A and sigH increased from 6 h to reach a plateau between 8 h and 10 h of growth, while the expression of spoIIAA occurred later, becoming enhanced after 8 h of growth. These patterns of transcription are in agreement with data obtained for Clostridium acetobutylicum (28). However, sigH expression is only slightly increased during transition phase in C. acetobutylicum (2), while in C. difficile, sigH expression is 15-fold induced at 10 h compared to its expression at 4 h.
To identify global changes in gene expression between exponential growth phase and stationary phase, we compared the gene expression profiles of strain 630E grown in TY medium for 4 h (exponential growth) and 10 h (early stationary phase). Seven hundred fifty-two genes were differentially expressed with a factor of ≥2. Three hundred twenty-five genes were expressed at higher levels and 427 were expressed at lower levels at 4 h than at 10 h (Table 1; also see Table S3 in the supplemental material). Genes involved in motility or in translation or encoding transporters and membrane proteins were expressed at higher levels during exponential growth than at the beginning of stationary phase, as reported for C. acetobutylicum (28). Conversely, several genes which participate in sporulation, cellular division, carbon metabolism, fermentation pathways, amino acid metabolism, and protein catabolism were induced at the onset of stationary phase (see Fig. S2 and S3 and Table S3 in the supplemental material). Actually, genes involved in the downstream part of glycolysis were induced in stationary phase in C. difficile, as already observed in C. acetobutylicum (2). In addition, the expression of genes involved in the biosynthesis of threonine, glycine, cysteine, aspartate, and glutamine, as well as genes encoding 17 putative proteases and peptidases or peptide (Opp, CD1404) and amino acid transporters (CD1259, CD1260, CD2693), increased after 10 h of growth (see Fig. S3 in the supplemental material). Finally, at the beginning of stationary phase, the expression of genes involved in the response to stress stimuli, including the oxidative stress response and the HrcA and CtsR regulons, were also induced (see Table S3 in the supplemental material) (2).
Table 1.
Genes involved in sporulation controlled by SigH or differentially expressed at 4 h and 10 h of growth
| Locus tag | Gene namea | Functiona | Expression ratio in transcriptomeb |
Associated sigma factor(s) and/or Spo0A box(es)c | |
|---|---|---|---|---|---|
| sigH strain/630E | 4 h/10 h | ||||
| CD0057 | spo0H | RNA polymerase sigma-H factor | D | 0.19 | SigA*, Spo0A |
| CD1214 | spo0A | Stage 0 sporulation protein A | 0.20 | 0.07 | SigA*, SigH*, Spo0A |
| CD2492 | Histidine kinase associated to Spo0A | 0.35 | 0.55 | SigH* | |
| CD3670 | Putative selenocysteine lyase | 0.34 | 0.38 | ||
| CD3671 | spo0J | Spo0J, stage 0 sporulation protein J | 0.46 | — | |
| CD3672 | soj | Sporulation initiation inhibitor | 0.56 | — | SigH |
| CD3673 | Putative stage 0 sporulation protein, Spo0J-like | 0.21 | 0.28 | SigH | |
| CD0770 | spoIIAA | Anti-sigma F factor antagonist | 0.03 | 0.04 | SigH*, Spo0A |
| CD0771 | spoIIAB | Anti-sigma F factor | 0.03 | 0.04 | |
| CD0772 | sigF | RNA polymerase sigma-F factor | 0.05 | 0.08 | |
| CD2469 | spoIIP | Stage II sporulation protein P | 0.50 | 0.33 | |
| CD2470 | gpr | Spore endopeptidase | 0.48 | 0.51 | |
| CD2641 | Putative sporulation protein | 0.48 | 0.59 | ||
| CD2642 | sigG | RNA polymerase sigma-G factor | 0.20 | 0.09 | SigF* |
| CD2643 | sigE | RNA polymerase sigma-E factor | 0.17 | 0.18 | |
| CD2644 | spoIIGA | Sporulation sigma-E factor processing peptidase | 0.20 | 0.22 | |
| CD3490 | spoIIE | Phosphoprotein phosphatase | 0.03 | 0.02 | Spo0A |
| CD1192 | spoIIIAA | Stage III sporulation protein AA | 0.31 | 0.42 | |
| CD1193 | spoIIIAB | Stage III sporulation protein AB | 0.25 | 0.31 | |
| CD1194 | spoIIIAC | Stage III sporulation protein AC | 0.16 | 0.18 | |
| CD1195 | spoIIIAD | Stage III sporulation protein AD | 0.32 | 0.38 | |
| CD1196 | spoIIIAE | Stage III sporulation protein AE | 0.35 | 0.41 | |
| CD1197 | spiIIIAF | Stage III sporulation protein AF | 0.50 | 0.48 | |
| CD1198 | spoIIIAG | Stage III sporulation protein AG | 0.14 | 0.15 | |
| CD1199 | spoIIIAH | Stage III sporulation protein AH | 0.19 | 0.18 | |
| CD0126 | spoIIID | Stage III sporulation protein D | 0.07 | 0.12 | |
| CD2629 | spoIVA | Stage IV sporulation protein AA | 0.11 | 0.16 | |
| CD2656 | spoVD | Stage V sporulation protein D (penicillin-binding protein) | 0.15 | 0.18 | |
| CD2652 | spoVE | Cell division/stage V sporulation protein | 0.20 | 0.28 | |
| CD3516 | spoVG | Regulator required for spore cortex synthesis | 0.01 | 0.23 | SigH |
| CD1935 | spoVS | Stage V sporulation protein S | 0.03 | 0.08 | SigH |
| CD3499 | spoVT | Stage V sporulation protein T | — | 0.08 | |
| CD2967 | spoVFB | Dipicolinate synthase subunit B | — | 2.20 | |
| CD1230 | sigK | Fragment of RNA polymerase sigma-K factor (part 1) | — | 2.73 | |
| CD3563 | Putative spore cortex-lytic enzyme | 0.47 | 0.28 | ||
| CD3494 | Putative spore protein | 0.50 | 0.42 | ||
| CD0106 | cwlD | Germinaion N-acetylmuramoyl-l-alanine amidase | 0.31 | 0.31 | |
| CD3349 | bclA3 | Putative exosporium glycoprotein | 0.38 | 3.51 | Spo0A |
| CD3569 | Sporulation-specific protease | 0.33 | — | SigH, Spo0A | |
| CD1068 | Putative polysaccharide biosynthesis/sporulation protein | 0.39 | 0.4 | ||
| CD3541 | spmB | Spore maturation protein B | 0.43 | — | |
| CD2247 | cspBA | Serine peptidase | 0.50 | 0.43 | |
Gene names and functions correspond to those indicated in the MaGe database Clostriscope (https://www.genoscope.cns.fr).
A gene is considered differentially expressed when the P value is <0.05 using the statistical analysis described in Materials and Methods. The cutoff for biologically significant change was set at 2-fold. However, some genes had a fold change of less than 2-fold but were included because they appeared to be in the same transcription units with regulated genes for which the fold change was ≥2. Genes not differentially expressed between 4 h and 10 h of growth or between the 630E strain and the sigH mutant are indicated by a dash. D, deleted in the sigH mutant.
Using the B. subtilis SigH or Spo0A consensus, we searched for SigH-dependent promoters or Spo0A boxes. An asterisk indicates that we performed promoter cartography by 5′ RACE.
Changes in fermentation pathways from acidogenesis to solventogenesis occur at the onset of stationary phase in C. acetobutylicum (2). In the nonsolventogenic C. difficile, modifications in the expression of genes required for fermentation and energy metabolism were also observed between 4 h and 10 h of growth (see Fig. S2 in the supplemental material). Actually, the expression of the cat1 operon, involved in crotonyl-coenzyme A (CoA) production from succinate, and of two adhE genes (CD2966 and CD3006), encoding alcohol dehydrogenases, decreased after 10 h of growth. In contrast, the expression of the bcd2 operon, involved in acetyl-CoA to butyryl-CoA synthesis, and of genes encoding two phosphate butyryl-transferases (CD0112 and CD0715) and two butyrate kinases (CD0113 and CD2379) increased 5- to 10-fold at the onset of stationary phase. The genes involved in amino acid fermentations were not differentially expressed, with the exception of the ldhA gene and the hadA operon required for l-leucine reduction, which were induced after 10 h of growth (31). Finally, several genes involved in acetyl-CoA production from pyruvate, CO2, or CO or related to electron transfer systems, including genes encoding flavodoxins, ferredoxins, flavoproteins, oxidoreductases, ATP synthases, and hydrogenases, were also induced at the onset of stationary phase. All these results suggest a rerouting of energy metabolism at the end of exponential growth in C. difficile.
The large set of genes that are differentially expressed during exponential growth and at the beginning of stationary phase is required for a general adaptive response and survival-enhancing properties associated with entry into stationary phase. This also indicates the existence of complex regulatory networks modulating gene expression between these two growth phases, in agreement with the 29 genes encoding transcriptional regulators that are differentially expressed in these two conditions. Interestingly, the agrD gene, encoding an autoinducer prepeptide, was 2.5-fold induced at 10 h, suggesting that a system of quorum sensing could play a role at the end of exponential growth, as observed in several Gram-positive bacteria, including Clostridium perfringens and Clostridium botulinum (11, 36). Finally, the expression of genes encoding six sigma factors (sigA2, sigB, sigH, sigF, sigE, and sigG), including 4 out of 5 sigma factors involved in the sporulation process and one sigma factor sharing similarities with SigB, which is involved in the general stress response in Gram-positive bacteria, was induced at 10 h in our transcriptome analysis, in agreement with the major changes detected in the transcriptional patterns. Surprisingly, the expression of the first part of the sigK gene, which is inactivated by a sigK intervening (skin) element, as observed in B. subtilis (21), was downregulated at the onset of stationary phase.
Comparative analysis of gene expression profiles of strain 630E and the sigH mutant at the onset of stationary phase.
In B. subtilis, SigH plays a key role in the control of the transition phase and in the initiation of sporulation (8). The C. difficile SigH protein shares 63% identity with SigH of B. subtilis. To analyze the role of SigH in C. difficile, we inactivated the sigH gene using the ClosTron system (23). The group II intron of pMTL007 was retargeted to insert into the sigH gene in sense orientation immediately after the 123rd nucleotide in its coding sequence. Then, the derivative plasmid pMTL007::Cdi-sigH123s was transferred to C. difficile strain 630E (Erms) by conjugation and erythromycin-resistant transconjugants were isolated. To verify the insertion of the group II intron into sigH, PCRs were carried out using internal primers of sigH, the intron-specific primer EBSu, and the RAM-F and RAM-R primer pair from both wild-type and transconjugant DNA templates (Materials and Methods; also see Fig. S1 in the supplemental material). The results obtained confirmed the integration of the intron into sigH.
In order to identify genes regulated by SigH, we compared the expression profiles of strain 630E and the sigH mutant at the onset of stationary phase (10 h). Four hundred seventy-six genes were differentially expressed between these two strains with a factor of ≥2: 190 genes were upregulated and 286 were downregulated in the sigH mutant compared to their expression in strain 630E (see Table S3 in the supplemental material). Of the genes regulated by SigH, 85% were differentially expressed between 4 h and 10 h of growth. We observed that SigH regulates numerous genes involved in various functions, such as motility, sporulation, cellular division, and protein catabolism, as well as carbon, amino acid, energy, and cell wall metabolism, virulence, and regulation. Thus, eight genes encoding peptidases or proteases and some genes involved in amino acid metabolism (a serine acetyl-transferase, an OAS-thiol-lyase, an aspartate aminotransferase, a glutamate synthase, a homoserine kinase, and a racemase) were induced via a SigH-dependent mechanism after 10 h of growth. Except for gapN, all genes involved in glycolysis that were regulated in a growth-dependent manner were negatively controlled by SigH (see Fig. S2 in the supplemental material). In addition, several genes implicated in fermentation and energy metabolism pathways were positively controlled by SigH (see Fig. S2 in the supplemental material). This includes the bcd2 operon that is involved in butyrate production, genes encoding a formate dehydrogenase, the bifunctional CO dehydrogenase/acetyl-CoA synthase, a hydrogenase (CD3313-CD3314), and an ATP synthase (CD2960-CD2954). Among the genes not differentially expressed during the growth phase, we observed that the expression of the grd operon, encoding the glycine reductase, a key enzyme of Stickland reactions, was 5- to 15-fold reduced in the sigH mutant compared to its expression in strain 630E (see Fig. S2 in the supplemental material). Finally, about 50 genes encoding proteins of unknown function were highly induced at the onset of stationary phase by a SigH-dependent mechanism, while most of the genes involved in the stress response were not regulated by SigH (see Table S3 in the supplemental material).
SigH positively controls the transcription of many genes involved in sporulation and cellular division.
In strain 630E, a total of 49 genes involved in sporulation and cellular division were induced at the onset of stationary phase, and most of these genes were expressed at lower levels in the sigH mutant than in strain 630E (Table 1). The spo0A and CD2492 genes, which encode the master regulator of the sporulation initiation and one of its associated kinases, respectively (52), were expressed at lower levels in the sigH mutant. Genes involved in chromosomal segregation during asymmetric division and prespore engulfment (spo0J, soj, and spoIIP) were positively controlled by SigH. Moreover, genes encoding sigma factors of sporulation SigF, SigE, and SigG and proteins controlling their activity (SpoIIAA, SpoIIAB, SpoIIE, and SpoIIGA) were expressed 5- to 30-fold less in the sigH mutant than in strain 630E. Finally, many genes involved in later stages of sporulation (spoIIIAA to spoIIIAH, spoIIID, spoIVA, spoVD, spoVE, spoVG, and spoVS) were also induced after 10 h of growth and positively regulated by SigH (Table 1).
Thus, as already observed in B. subtilis (8), SigH positively controls the expression of a large set of sporulation genes in C. difficile. To confirm the crucial role of SigH in the sporulation process, we compared the sporulation abilities of strain 630E, the sigH mutant, and a spo0A mutant inactivated with the ClosTron system and used as a control. After 72 h of incubation in BHIS medium, the percentage of sporulation in strain 630E was 9.3% ± 0.7% (mean ± standard deviation). Inactivation of the spo0A or sigH gene resulted in a complete inability of C. difficile to sporulate [(<2 × 10−4)% sporulation]. When we complemented the sigH mutant using a pMTL84121-sigH plasmid, which gave strain CDIP12, we obtained a sporulation efficiency of 17% ± 0.5%, indicating that the lack of sporulation is only due to the sigH gene disruption. Thus, in C. difficile, SigH is as essential as Spo0A for sporulation (52).
We also observed that 11 genes involved in cell division were induced at the onset of stationary phase and were less expressed in the sigH mutant (see Table S3 in the supplemental material). In B. subtilis, the formation of an asymmetric septum during sporulation is related to the control of several cellular division genes by sporulation factors (18, 26). The ftsZ gene, involved in the initiation of septum formation, and the minC, minD, and minE genes, required for proper placement of septum division, were expressed at 3- to 10-fold-lower levels in the sigH mutant. In B. subtilis, genes like ftsZ or minCD are also regulated by SigH (8). Using qRT-PCR, we confirmed the positive control of minC transcription by SigH. In addition, the expression of minC was restored in a sigH mutant complemented by pMTL84121-sigH. Finally, genes encoding DivIVA- and FtsQ-type proteins as well as FtsH, SepF, and MrdB were induced at the onset of stationary phase and positively controlled by SigH.
Modulation of PaLoc gene expression by SigH and growth phase.
In the VPI10463 strain, the expression of tcdA and tcdB increases as cells enter stationary phase (16). We tested whether the same regulation exists in strain 630E and the possible role of SigH in this regulation. The transcriptome analysis showed that the tcdA gene was expressed 4-fold more after 10 h than after 4 h of growth (see Table S3 in the supplemental material). Using qRT-PCR, we confirmed that tcdA expression increased 10-fold at the onset of stationary phase. Furthermore, tcdA is expressed 3.7- and 9-fold more in the sigH mutant than in strain 630E, as shown by transcriptome and qRT-PCR analysis, respectively (Fig. 1A; also see Table S3 in the supplemental material). In the sigH mutant complemented by pMTL84121-sigH, tcdA expression was restored to the level observed in strain 630E (Fig. 1A). So, tcdA expression is induced at the onset of stationary phase and is negatively controlled by SigH. We further measured the effect of SigH inactivation on TcdA protein synthesis. Crude extracts were obtained from the sigH mutant and strains 630E and CDIP12 grown in TY medium. TcdA production was assayed by dot blot assay using specific antibodies raised against TcdA (Fig. 1B). We observed an 8- and a 4-fold-increase in TcdA quantity after 10 h and 48 h of culture, respectively, in the sigH mutant compared to the levels in strain 630E. TcdA production decreased when the sigH gene was introduced in trans into the sigH mutant (Fig. 1B). The variation in the level of TcdA produced is in agreement with the increase of tcdA transcription in the sigH mutant.
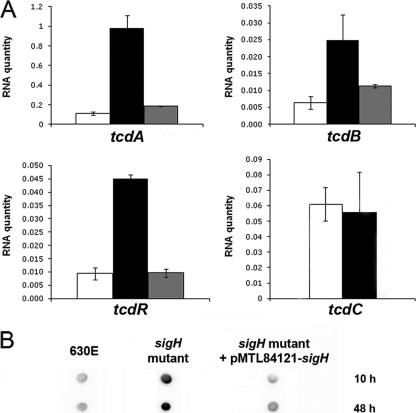
Fig. 1.
Effects of SigH inactivation on the expression of pathogenicity locus genes. (A) Expression of tcdA, tcdB, tcdR, and tcdC in strain 630E (white), in a sigH mutant (black), and in strain CDIP12 (sigH mutant complemented with pMTL84121-sigH) (gray) after 10 h of growth in TY medium. After reverse transcription, specific cDNAs were quantified by qRT-PCR, using the 16S rRNA gene for normalization and standard curve. The mean normalized quantity of RNA is indicated in arbitrary units. Error bars correspond to standard deviations from at least two biological replicates. (B) The TcdA level was estimated from crude extracts by protein dot blot analysis using specific monoclonal antibodies raised against TcdA. Crude extracts were obtained after 10 h and 48 h of growth in TY. The results shown are representative of at least three biological replicates of C. difficile crude extracts.
By qRT-PCR, we observed that the expression of tcdB and tcdR in strain 630E also increased 4- and 3-fold at the onset of stationary phase, respectively. In addition, tcdB and tcdR were expressed 5-fold more in the sigH mutant than in strain 630E (Fig. 1A). The levels of tcdB and tcdR expression were restored when the sigH gene was introduced in trans into the sigH mutant. In contrast, tcdC was transcribed similarly in strain 630E and in the sigH mutant (Fig. 1A). The absence of modulation of tcdC transcription in the sigH mutant compared to its expression in strain 630E indicates that the SigH-dependent control of tcdA, tcdB, and tcdR expression is not mediated by the regulation of TcdC synthesis. However, we cannot exclude the possibility that SigH might influence factors controlling TcdC stability or activity.
Surprisingly, the effects of SigH or Spo0A inactivation on toxin production differ. While a spo0A mutation reduces the amount of TcdA produced (52), a sigH mutation stimulates TcdA production via the control of its transcription. However, neither a Spo0A-binding motif nor a SigH promoter is present upstream of tcdA, tcdB, and tcdR, indicating an indirect, still-uncharacterized effect of these two proteins on toxin gene expression.
Control of surface-associated proteins and flagellum synthesis by SigH.
Surface-associated proteins and factors, which play a role in cell adherence and could be involved in intestinal colonization, have been identified in C. difficile (12, 57). Several genes encoding surface-associated proteins and adhesion factors were differentially expressed between 4 h and 10 h of growth and/or controlled by SigH (see Table S3 in the supplemental material). The expression of CD2789 (cwp66), encoding a cell surface protein with adhesive properties (53), was induced at the onset of stationary phase and positively regulated by SigH. The slpA gene, encoding the S-layer precursor (19), was upregulated in the sigH mutant. The high-molecular-weight S-layer protein (HMW-SLP) binds to epithelial cells and plays a role in adherence (9). The C. difficile genome contains 28 genes encoding proteins with domains similar to the HMW-SLP (41). Some of them are located on the cell surface (57). CD0514, a gene controlled by phase variation that encodes the cell wall protein CwpV (17), was upregulated in the sigH mutant, as observed for slpA. The expression of some other genes, CD2713, CD1803 and CD2784, encoding SLP-like proteins, dropped at the onset of stationary phase and was not controlled by SigH. In addition, two genes encoding SLP-like proteins (CD2518 and CD0440) were expressed more at the onset of stationary phase and were positively regulated by SigH, similarly to CD2789. We confirmed by qRT-PCR 4-, 82-, and 500-fold decreases in transcription in the sigH mutant for the cwp66, CD0440, and CD2518 genes, respectively. Their transcription was restored when the sigH mutant was complemented with pMTL84121-sigH. Interestingly, we also observed that the expression of CD3145, encoding a protein that may bind to extracellular matrix components of eukaryotic cells, was higher after 4 h of growth and in the sigH mutant. So, the expression of several genes encoding surface-associated proteins considered putative accessory virulence factors is growth phase dependent and/or regulated by SigH.
The role of flagellum in adherence has also been shown in C. difficile (49). Most of the genes of the flagellum locus (43 genes) were downregulated at the onset of stationary phase and upregulated in a sigH mutant (see Table S3 in the supplemental material). The levels of expression of the fliC and fliD genes, encoding the flagellin and the flagellum cap protein, were 6.4 and 8.3-fold higher, respectively, in the sigH mutant in the transcriptome and we confirmed by qRT-PCR a 13-fold increase of the fliC mRNA in this mutant. Furthermore, the expression of CD0240, encoding a glycosyl-transferase enzyme involved in flagellin glycosylation (51), was also 4-fold increased in a sigH mutant. However, we failed to detect an increased motility of the sigH mutant under the conditions used (BHI plates or tubes containing 0.3% agar) (data not shown). In C. acetobutylicum, Spo0A negatively controls a large set of genes involved in motility (50), suggesting that both SigH and Spo0A repress motility-associated genes at the onset of the stationary phase in clostridia, as observed in B. subtilis (8, 20). In B. subtilis, the effect of SigH and Spo0A on flagellum synthesis is mediated by SinR. Interestingly, the expression of the C. difficile sinR-like gene, CD2214, was 16-fold higher in a sigH mutant than in strain 630E, while the expression of the sigD gene, encoding the SigD sigma factor of late flagellum genes, hardly increased (1.5-fold). In C. difficile, SinR might be involved in the SigH-dependent control of flagellin synthesis.
Transcriptional control of sigH, spo0A, CD2492, and spoIIAA by SigH and Spo0A.
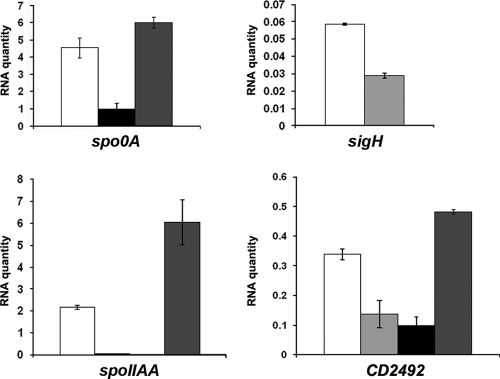
To test whether SigH and Spo0A mutually regulate their transcription, we performed qRT-PCR using RNAs extracted from strain 630E, the sigH or spo0A mutant, and CDIP12 grown for 10 h in TY medium (Fig. 2). We also tested the transcriptional regulation of CD2492, encoding the Spo0A-associated kinase and the spoIIAA operon, by SigH and Spo0A. In the spo0A mutant, we observed that the transcript levels of sigH, CD2492, and spoIIAA were 2-, 3-, and 40-fold lower, respectively, than in strain 630E. In the sigH mutant, the spo0A, CD2492, and spoIIAA transcript levels were 4.5-, 3-, and 200-fold lower, respectively, than in strain 630E (Fig. 2). Using antibodies raised against Spo0A of B. subtilis, we also showed that the quantity of Spo0A decreased in a sigH mutant (data not shown). In all cases, when we complemented the sigH mutant using a pMTL84121-sigH plasmid, we restored the expression of these genes to the level observed in strain 630E (Fig. 2). Thus, SigH positively controls spo0A, CD2492, and spoIIAA transcription, in agreement with our transcriptome data (Table 1), while Spo0A activates the expression of sigH, CD2492, and spoIIAA. In C. acetobutylicum, Spo0A also positively controls the expression of the spoIIAA operon, of the cac3319 and cac0903 genes, encoding Spo0A-associated histidine kinases, and of cac0437, encoding a protein that dephosphorylates Spo0A-P (44, 50).
Fig. 2.
qRT-PCR analysis of the role of SigH and Spo0A in the control of spo0A, sigH, CD2492, and spoIIAA gene expression. Total RNAs were extracted from C. difficile strain 630E (white), the spo0A mutant (pale gray), the sigH mutant (black), and strain CDIP12 (sigH mutant complemented with pMTL84121-sigH) (dark gray) grown in TY medium. After reverse transcription, specific cDNAs were quantified by qRT-PCR, using the 16S rRNA gene for normalization and standard curve. Error bars correspond to standard deviations from at least two biological replicates.
We further characterized the sigH, spo0A, CD2492, and spoIIAA transcription initiation sites by the 5′ RACE technique and looked for the presence of a Spo0A binding motif using the consensus sequence TGNCGAA of B. subtilis (35). We identified a transcription initiation site 32 bases upstream of the sigH gene start codon. The deduced −35 (TTGAAA) and −10 (TATAAT) boxes correspond to the consensus sequence of a SigA-dependent promoter. A Spo0A box is present 67 bases upstream of the transcription initiation site (Fig. 3A), in agreement with the positive control of sigH transcription by Spo0A. Transcription of spo0A is initiated 65 bases upstream of its start codon. A sequence sharing similarities with the consensus of SigH-type promoters of B. subtilis (GGAGGATAT-N11-GTCGAAT) is present upstream of this transcriptional start site (Fig. 3B). This is in accordance with the positive control of spo0A transcription by SigH. As found in B. subtilis (48), a Spo0A binding site overlaps the −10 box of this promoter, suggesting a repression of its own transcription by Spo0A. We noted that spo0A transcription was still detectable in the sigH mutant (Fig. 2), suggesting the existence of a second promoter. So, we performed a 5′ RACE experiment using RNAs extracted from the sigH mutant. We identified a second transcription initiation site 139 bases upstream of the start codon. A consensus sequence (TGGTAT-N17-TAAAAT) probably recognized by SigA is present (Fig. 3B).
Fig. 3.
Promoter regions of sigH (A), spo0A (B), spoIIAA (C), and CD2492 (D) genes. The transcriptional start sites are indicated by gray backgrounds and “+1.” The −35 and −10 boxes corresponding to SigA or the conserved sequences of SigH-dependent promoters are underlined. The Spo0A binding sites are boxed, while the translational start sites are indicated in bold.
The transcription initiation site of CD2492 was mapped 35 bases upstream of the start codon. We found a SigH-dependent promoter (GAAGGAATT-N12-ATCGTAT), in agreement with its positive control by SigH (Fig. 3D). For the spoIIAA operon, the transcription initiation site was located 39 bases upstream of the start codon. A consensus sequence recognized by SigH (GAAGGAATA-N11-ATAGAAT) and a Spo0A box located 63 bases upstream of the transcription initiation site are present (Fig. 3C). This is in accordance with the positive regulation of spoIIAA transcription by both Spo0A and SigH (Fig. 2).
Similar to B. subtilis (38, 43), there is reciprocal control of the synthesis of SigH and Spo0A in C. difficile. However, the positive control of sigH transcription by Spo0A in B. subtilis is indirect, with a negative effect of Spo0A-P on the synthesis of AbrB, which in turn represses sigH expression (46, 47). In the absence of the AbrB repressor in C. difficile, the Spo0A box that is located upstream of the SigA-dependent promoter of sigH (Fig. 3) strongly suggests direct positive control of sigH transcription by Spo0A. The transcription of spo0A itself is initiated from two promoters recognized by the RNA polymerase containing SigA or SigH. We further showed that the increase of spo0A expression at 10 h was abolished in a sigH mutant (data not shown). So, SigH contributes to the induction of spo0A expression at the onset of the stationary phase, as in B. subtilis (10). A second level of control of Spo0A activity probably occurs via the regulation of the synthesis of the CD2492 kinase by SigH. In contrast, the synthesis of the other putative Spo0A-associated kinases, including CD1579, which is able to phosphorylate Spo0A in vitro, and CD1492 (52) is not controlled by the growth phase or by SigH under our experimental conditions (Table 1). The possible regulation of the expression of CD1579 and CD1492 by Spo0A or by other regulators remains to be established.
Identification of potential direct target genes of SigH.
In the transcriptome analysis, 286 genes are positively regulated by SigH. To identify genes directly controlled by SigH, we searched for the presence of the consensus sequence of B. subtilis SigH-dependent promoters (A/G)NAGGA(A/T)3-N11-12-(A/G)NNGAAT in the 200-bp region upstream of start codons of C. difficile genes using the GenoList web server (http://genodb.pasteur.fr/cgi-bin/WebObjects/GenoList) and allowing two mismatches. About 40 genes and operons are positively regulated by SigH and contain a SigH-like consensus sequence in their promoter regions, suggesting their direct regulation by SigH (Table 2). In B. subtilis, there are about 50 transcriptional units directly controlled by SigH (8). Surprisingly, only eight of them are in common with C. difficile: spo0A, spoIIAA, spoVG, spoVS, ftsZ, minC, glgC, and CD0142/yvyD. In addition, genes encoding Spo0A kinases in B. subtilis (kinA and kinE) and C. difficile (CD2492) are also transcribed by SigH in these two bacteria. Most of the common genes that are direct SigH targets are involved in sporulation or cell division, confirming the crucial role of this sigma factor in the early steps of the sporulation process in both microorganisms. Furthermore, in C. difficile, the soj-spo0J-CD3670 operon and the CD3673 gene, encoding a Spo0J-like protein, are probably transcribed by SigH (Table 2). The effects of SigH on the other sporulation genes are probably indirect and mediated at least partly by SigF and/or Spo0A. For example, a Spo0A box is present upstream of spoIIE in C. difficile, as in B. subtilis (58). Moreover, we mapped a SigF-type promoter upstream of the sigG gene (see Fig. S4 in the supplemental material), which was expressed 5-fold less in a sigH mutant (Table 1). In contrast, some other genes that are direct SigF targets in B. subtilis, such as spoIIR and spoIVB, are not induced in C. difficile after 10 h of growth compared to their expression at 4 h and are not controlled by SigH in the transcriptome. These differences could be due either to low expression after 10 h of growth or to a different circuit of control of these genes in C. difficile than in B. subtilis.
Table 2.
Genes or operon positively controlled by SigH and with a SigH consensus sequence in their promoter region
| Genea | Promoter recognized by SigHb | Expression ratio in sigH mutant/strain 630E | Function |
|---|---|---|---|
| spo0A* | taGGAGGAATAtaattttggagtGTCGAATat | 0.2 | Stage 0 sporulation protein A |
| CD2492 | aaGAAGGAATTataatgtcgagtATCGTATaa | 0.35 | Histidine kinase associated to Spo0A |
| soj | atACAGGAATTgatgctaagtatAATGAAAta | 0.56 | Sporulation initiation inhibitor |
| spoIIAA* | ttGAAGGAATAaaaat-ataattATAGAATtg | 0.03 | Anti-sigma F factor antagonist |
| spoVS* | atAAAGGTTTTcttaaaacgattATAGAAGta | 0.03 | Stage V sporulation protein S |
| spoVG* | aaAGAGGATATccctagttgttcATAGAATta | 0.01 | Regulator required for spore cortex synthesis |
| CD3673 | atAAAGGAATTattaaagtagatGCAGAAAta | 0.21 | Putative stage 0 sporulation protein, Spo0J-like |
| CD3569 | aaGAAGGGTTTagtgaagatgatATAGAAAag | 0.33 | Sporulation-specific protease |
| ftsZ* | aaAAAGGAAAAtttac-gtttttGTGGAATat | 0.34 | Cell division protein FtsZ |
| minC* | taAAAGGGTTTaaagc-gtatttGAAGAATat | 0.09 | Cell division regulator |
| CD2624 | taAAAGGGTTTaaagc-gtatttGAAGAATat | 0.33 | Conserved hypothetical protein |
| sigA2 | aaGGAGGATATtgctg-ttagaaGTAGAATaa | 0.05 | Transcription |
| CD0142* | aaAGAGGATTAtgaga-gttcgtGTAGAATat | 0.14 | Putative RNA-binding protein |
| CD0838 | taCAAGGATTTtaaag-ataaatATAGAAAtt | 0.26 | DNA-binding protein |
| gapN | ttATAGGtAAAcgtttatgaaaaAAAGAAAtg | 0.48 | Glyceraldehyde-3-P dehydrogenase |
| glgC* | agGAAGGATATggaaa-ttaaagGTCGAATta | 0.19 | Glucose-1-P adenylyltransferase |
| fdhF | taAGAGGAATTgtgag-aaaattGTTGAATtt | 0.31 | Formate dehydrogenase |
| cobT | atAAAGGAAATgaaca-taaaatGTAGAATaa | 0.35 | Nicotinate-nucleotide-dimethylbenzimidazole |
| CD3456 | agGGAGGAATTatttt-atgaaaAAAGAATtt | 0.49 | 5-Formyltetrahydrofolate cycloligase |
| CD2738 | agGGAGGGATAatattgcaagagAGAGAAGaa | 0.49 | Putative cytosine permease |
| CD0760 | gtTTAGGATAAataaattttattATGGAAAaa | 0.4 | Putative Ca2+/Na+ antiporter |
| CD2789 | caAGAGGAgAAgttattaagtgcAATGAATgc | 0.39 | Putative adhesin Cwp66 |
| CD0440 | atAAAGGAAAAtattc-ttttatGTAGAATta | 0.25 | Putative cell wall-binding protein |
| CD2518 | aaGGAGGAAAAgaaaa-tttaatGTAGAATta | 0.14 | Cell surface protein |
| lplA | ttAAAGGAgTTtttat-attattGTCGAATta | 0.34 | Lipoate protein-ligase |
| CD3458 | ttAAAGGAAATtgtaggtagtttATCGAATtg | 0.07 | Membrane protein |
| CD2800 | ctATAGGAAcTgttaaatctaaaAAAGAAGaa | 0.06 | Membrane protein |
| CD2295 | taGGAGGGATTttatg-gattttGGGGAATtt | 0.28 | Membrane protein |
| CD1590 | aaAAAGGAAAAcgtctaaagtatTGGAATTac | 0.44 | Membrane protein |
| CD0865 | taACAGGATTTatgtaggtgtttATAGAAAta | 0.08 | Putative ADP-ribose binding protein |
| CD2447 | taGAAGGAATTttgctataacatGTAGAAAtt | 0.06 | Putative histidine triad (HIT) protein |
| CD3654 | aaAGAGGAAGAacgtatagttaaGGAGAATat | 0.24 | Putative DNA replication protein |
| CD0022 | aaAGAGGACAAttacctccagatGTAGAAAta | 0.04 | Elongation factor G (EF-G) |
| CD2137 (frr) | atAAAGGTATTtgagcttacaacAGAGAATat | 0.4 | Ribosome-recycling factor |
| CD1941 | aaGTAGGTATAtataacaaggaaACAGAACcc | 0.05 | Unknown function |
| CD1543A | atAAAGGAAAAacctcttttaatGTAGAAAct | 0.07 | Unknown function |
| CD1264 | aaAGAGGAAAAatcatt-ttaatGTAGAATaa | 0.08 | Unknown function |
| CD1061 | ccAGAGGATATgaaagagttaatATCGAAAtt | 0.25 | Unknown function |
| CD1317 | taAAAGGAAAGacaggat-aaatATAGAAAtt | 0.42 | Unknown function |
| CD1622 | atAAAGGGTTTagagggcataatATAGAATaa | 0.46 | Unknown function |
The transcriptional start site was identified for underlined genes. We used the search pattern program of the GenoList Web server (http://genodb.pasteur.fr/cgi-bin/WebObjects/GenoList) to detect the presence of the consensus sequence of B. subtilis SigH-dependent promoters (A/G)NAGGA(A/T)3-N11-12-(A/G)NNGAAT in the 200-bp region upstream of C. difficile genes, allowing two mismatches. An asterisk indicates genes or operons with a SigH promoter in B. subtilis (9).
The −10 and −35 boxes are indicated by uppercase letters.
Among the genes that are potentially direct targets of SigH, we found genes involved in metabolism and translation and encoding membrane and surface-associated proteins or proteins of unknown function. The glgCDAP-CD0886 operon, involved in a glycogen-type polymer synthesis and degradation (glgP-CD0886), is probably transcribed from a SigH-dependent promoter, as observed in B. subtilis (8). In C. acetobutylicum, there is no SigH-dependent promoter upstream of the glgCDA operon, which is involved in granulose production, while a promoter recognized by SigH is present upstream of glgP (2, 28). This suggests a diversity of control of the glg system among clostridia. In C. difficile, there are two copies of the sigA gene (CD1455/sigA1 and CD1498/sigA2). The sigA1 gene forms an operon with dnaG, as observed in B. subtilis. Interestingly, sigA2 transcription increased at the onset of stationary phase and was strongly dependent on SigH (see Table S3 in the supplemental material). We confirmed by qRT-PCR a 180-fold decrease of sigA2 transcription in a sigH mutant and the restoration of its expression in a sigH mutant complemented with pMTL84121-sigH. We further demonstrated the presence of a unique SigH-dependent promoter upstream of sigA2, while we mapped a SigA-dependent promoter upstream of the dnaG-sigA1 operon (see Fig. S4 in the supplemental material). In B. subtilis, the dnaG-sigA operon is transcribed by several promoters, two of them being SigH dependent (8), while in C. difficile, one copy of sigA is transcribed by SigH and the other by SigA. Finally, it is worth noting that among the genes encoding surface-associated proteins with HMW-SLP-type domains that are induced at the onset of stationary phase in a SigH-dependent manner, three genes (cwp66 [CD2789], CD0440, and CD2518) are probably transcribed by SigH (Table 2).
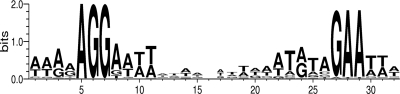
The alignment of all the probable SigH-dependent promoters listed in Table 2 allows the proposal of a consensus for the promoters transcribed by the RNA polymerase associated with SigH in C. difficile (Fig. 4). This consensus includes two well-conserved motifs, AGG and GAA, that are separated by 18 to 19 bases and is rather similar to the promoter sequences of the SigH regulon of B. subtilis (8).
Fig. 4.
Consensus of SigH-dependent promoters in C. difficile. Using an alignment of all the proposed direct targets of SigH listed in Table 2, the sequence logo was created on the WebLogo website (http://weblogo.berkeley.edu). The height of the letters is proportional to their frequency.
Conclusion.
We showed that about 20% of C. difficile genes are differentially expressed during the exponential growth phase and the onset of stationary phase. About 60% of these genes are regulated directly or indirectly by SigH, demonstrating the crucial role of this sigma factor during the post-exponential growth phase in C. difficile. However, other regulators, such as CodY or Spo0A, also contribute to the transcriptional changes observed during the transition phase. Indeed, among the genes whose expression varies depending on the growth phase, 80 (10.5%) are controlled by CodY in the transcriptome (13), and a putative Spo0A box is located upstream of 70 genes (9.5%). The regulatory network modulating gene expression at the end of exponential growth is probably more complex, since we showed that 29 additional regulatory genes were induced or repressed at the onset of stationary phase in the transcriptome.
Among the genes regulated by SigH, we found genes involved in sporulation, cellular division, motility, metabolism (carbon and energy, amino acids, and protein catabolism), and virulence, as well as genes encoding membrane and surface-associated proteins. Only a small number of genes activated by SigH in clostridia are found in B. subtilis, as proposed for Spo0A (35). This may be due to the fact that clostridia are anaerobes, leading to major differences in the physiology of these bacteria, and the fact that initiation of sporulation appears to occur via a different pathway with the lack of the phosphorelay (44).
Actually, the global impact of SigH on transcription in C. difficile largely exceeds the 40 genes or operons probably directly transcribed by SigH (Table 2). Most of the other genes, including those negatively controlled, are indirectly regulated by SigH via other regulators or changes in the pools of signaling molecules. We demonstrated that the regulation of SigH and Spo0A are intertwined, with SigH modulating spo0A transcription and Spo0A activating sigH expression. The identification of the Spo0A target genes would be helpful to better understand the global regulatory role of SigH during the transition phase. Part of the indirect effects of SigH may also be due to the alteration of the association of alternate sigma factors, such as SigB or SigD, with core RNA polymerase in the presence or absence of SigH. Competition between sigma factors for the core RNA polymerase has been observed within the σ70 family or between σS and σN in E. coli (15) and between SigA and SigH, SigE, or SigK in B. subtilis (25, 29). Finally, among the 14 genes encoding transcriptional regulators, including CggR and SinR, whose expression is modulated by SigH, only CD0838, encoding a DNA binding protein of unknown function, seems to be directly transcribed by SigH (Table 2). How this regulator may contribute to the control of gene expression by SigH will be interesting to explore in future investigations. Moreover, SinR, a repressor of genes expressed in stationary phase in B. subtilis, is also a good candidate for participation in the global regulatory effect of SigH.
Finally, the sigH mutant is unable to sporulate but still produces toxins, demonstrating that toxin synthesis is not associated with sporulation, contrary to the production of enterotoxin in C. perfringens (22). Toxin production in C. difficile is therefore clearly a stationary-phase event. The mechanism of control of toxin synthesis by SigH remains to be established. Several hypotheses could be suggested. This effect could be due to decreased competition between SigH and TcdR for the core RNA polymerase binding at the onset of stationary phase in the sigH mutant, to control of TcdC activity, or to a regulatory cascade involving other transcriptional factors. Further work will be required to analyze the molecular mechanisms of SigH-dependent control of toxin production.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Fujita for the gift of the anti-Spo0A antibody and N. Minton and J. Heap for providing the pMTL007 and pMTL84121 plasmids. We also acknowledge M. Thibonnier for helpful discussions.
This work was supported by funds from the Institut Pasteur. L. Saujet has a fellowship from the University Paris 7.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 13 May 2011.
REFERENCES
- 1. Akerlund T., Svenungsson B., Lagergren A., Burman L. G. 2006. Correlation of disease severity with fecal toxin levels in patients with Clostridium difficile-associated diarrhea and distribution of PCR ribotypes and toxin yields in vitro of corresponding isolates. J. Clin. Microbiol. 44:353–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alsaker K. V., Papoutsakis E. T. 2005. Transcriptional program of early sporulation and stationary-phase events in Clostridium acetobutylicum. J. Bacteriol. 187:7103–7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Andre G., et al. 2008. S-box and T-box riboswitches and antisense RNA control a sulfur metabolic operon of Clostridium acetobutylicum. Nucleic Acids Res. 36:5955–5969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andre G., et al. 2010. Global regulation of gene expression in response to cysteine availability in Clostridium perfringens. BMC Microbiol. 10:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Antunes A., Martin-Verstraete I., Dupuy B. 2011. CcpA mediated repression of Clostridium difficile toxin gene expression. Mol. Microbiol. 79:882–899 [DOI] [PubMed] [Google Scholar]
- 6. Benjamini Y., Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57:289–300 [Google Scholar]
- 7. Breitling R., Armengaud P., Amtmann A., Herzyk P. 2004. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 573:83–92 [DOI] [PubMed] [Google Scholar]
- 8. Britton R. A., et al. 2002. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J. Bacteriol. 184:4881–4890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Calabi E., Calabi F., Phillips A. D., Fairweather N. F. 2002. Binding of Clostridium difficile surface layer proteins to gastrointestinal tissues. Infect. Immun. 70:5770–5778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chibazakura T., Kawamura F., Asai K., Takahashi H. 1995. Effects of spo0 mutations on spo0A promoter switching at the initiation of sporulation in Bacillus subtilis. J. Bacteriol. 177:4520–4523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cooksley C. M., et al. 2010. Regulation of neurotoxin production and sporulation by a putative agrBD signaling system in proteolytic Clostridium botulinum. Appl. Environ. Microbiol. 76:4448–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deneve C., Janoir C., Poilane I., Fantinato C., Collignon A. 2009. New trends in Clostridium difficile virulence and pathogenesis. Int. J. Antimicrob. Agents 33:24–28 [DOI] [PubMed] [Google Scholar]
- 13. Dineen S. S., McBride S. M., Sonenshein A. L. 2010. Integration of metabolism and virulence by Clostridium difficile CodY. J. Bacteriol. 192:5350–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dineen S. S., Villapakkam A. C., Nordman J. T., Sonenshein A. L. 2007. Repression of Clostridium difficile toxin gene expression by CodY. Mol. Microbiol. 66:206–219 [DOI] [PubMed] [Google Scholar]
- 15. Dong T., Yu R., Schellhorn H. 2011. Antagonistic regulation of motility and transcriptome expression by RpoN and RpoS in Escherichia coli. Mol. Microbiol. 79:375–386 [DOI] [PubMed] [Google Scholar]
- 16. Dupuy B., Sonenshein A. L. 1998. Regulated transcription of Clostridium difficile toxin genes. Mol. Microbiol. 27:107–120 [DOI] [PubMed] [Google Scholar]
- 17. Emerson J. E., et al. 2009. A novel genetic switch controls phase variable expression of CwpV, a Clostridium difficile cell wall protein. Mol. Microbiol. 74:541–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Errington J. 2003. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 1:117–126 [DOI] [PubMed] [Google Scholar]
- 19. Fagan R. P., et al. 2009. Structural insights into the molecular organization of the S-layer from Clostridium difficile. Mol. Microbiol. 71:1308–1322 [DOI] [PubMed] [Google Scholar]
- 20. Fawcett P., Eichenberger P., Losick R., Youngman P. 2000. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 97:8063–8068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Haraldsen J. D., Sonenshein A. L. 2003. Efficient sporulation in Clostridium difficile requires disruption of the sigmaK gene. Mol. Microbiol. 48:811–821 [DOI] [PubMed] [Google Scholar]
- 22. Harry K. H., Zhou R., Kroos L., Melville S. B. 2009. Sporulation and enterotoxin (CPE) synthesis are controlled by the sporulation-specific sigma factors SigE and SigK in Clostridium perfringens. J. Bacteriol. 191:2728–2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Heap J. T., Pennington O. J., Cartman S. T., Carter G. P., Minton N. P. 2007. The ClosTron: a universal gene knock-out system for the genus Clostridium. J. Microbiol. Methods 70:452–464 [DOI] [PubMed] [Google Scholar]
- 24. Heap J. T., Pennington O. J., Cartman S. T., Minton N. P. 2009. A modular system for Clostridium shuttle plasmids. J. Microbiol. Methods 78:79–85 [DOI] [PubMed] [Google Scholar]
- 25. Hicks K. A., Grossman A. D. 1996. Altering the level and regulation of the major sigma subunit of RNA polymerase affects gene expression and development in Bacillus subtilis. Mol. Microbiol. 20:201–212 [DOI] [PubMed] [Google Scholar]
- 26. Hilbert D. W., Piggot P. J. 2004. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol. Mol. Biol. Rev. 68:234–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hussain H. A., Roberts A. P., Mullany P. 2005. Generation of an erythromycin-sensitive derivative of Clostridium difficile strain 630 (630Deltaerm) and demonstration that the conjugative transposon Tn916DeltaE enters the genome of this strain at multiple sites. J. Med. Microbiol. 54:137–141 [DOI] [PubMed] [Google Scholar]
- 28. Jones S. W., et al. 2008. The transcriptional program underlying the physiology of clostridial sporulation. Genome Biol. 9:R114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ju J., Mitchell T., Peters H., III, Haldenwang W. G. 1999. Sigma factor displacement from RNA polymerase during Bacillus subtilis sporulation. J. Bacteriol. 181:4969–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Karlsson S., Lindberg A., Norin E., Burman L. G., Akerlund T. 2000. Toxins, butyric acid, and other short-chain fatty acids are coordinately expressed and down-regulated by cysteine in Clostridium difficile. Infect. Immun. 68:5881–5888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim J., Darley D., Selmer T., Buckel W. 2006. Characterization of (R)-2-hydroxyisocaproate dehydrogenase and a family III coenzyme A transferase involved in reduction of l-leucine to isocaproate by Clostridium difficile. Appl. Environ. Microbiol. 72:6062–6069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 33. Mani N., Dupuy B. 2001. Regulation of toxin synthesis in Clostridium difficile by an alternative RNA polymerase sigma factor. Proc. Natl. Acad. Sci. U. S. A. 98:5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Matamouros S., England P., Dupuy B. 2007. Clostridium difficile toxin expression is inhibited by the novel regulator TcdC. Mol. Microbiol. 64:1274–1288 [DOI] [PubMed] [Google Scholar]
- 35. Molle V., et al. 2003. The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 50:1683–1701 [DOI] [PubMed] [Google Scholar]
- 36. Ohtani K., et al. 2009. Virulence gene regulation by the agr system in Clostridium perfringens. J. Bacteriol. 191:3919–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Paredes C. J., Alsaker K. V., Papoutsakis E. T. 2005. A comparative genomic view of clostridial sporulation and physiology. Nat. Rev. Microbiol. 3:969–978 [DOI] [PubMed] [Google Scholar]
- 38. Phillips Z. E., Strauch M. A. 2002. Bacillus subtilis sporulation and stationary phase gene expression. Cell. Mol. Life Sci. 59:392–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rouillard J. M., Zuker M., Gulari E. 2003. OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. 31:3057–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 41. Sebaihia M., et al. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38:779–786 [DOI] [PubMed] [Google Scholar]
- 42. Smyth G. K., Speed T. 2003. Normalization of cDNA microarray data. Methods 31:265–273 [DOI] [PubMed] [Google Scholar]
- 43. Sonenshein A. L. 2000. Control of sporulation initiation in Bacillus subtilis. Curr. Opin. Microbiol. 3:561–566 [DOI] [PubMed] [Google Scholar]
- 44. Steiner E., et al. 2011. Multiple orphan histidine kinases interact directly with Spo0A to control the initiation of endospore formation in Clostridium acetobutylicum. Mol. Microbiol. 80:641–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stephenson K., Hoch J. A. 2002. Evolution of signalling in the sporulation phosphorelay. Mol. Microbiol. 46:297–304 [DOI] [PubMed] [Google Scholar]
- 46. Strauch M., Webb V., Spiegelman G., Hoch J. A. 1990. The SpoOA protein of Bacillus subtilis is a repressor of the abrB gene. Proc. Natl. Acad. Sci. U. S. A. 87:1801–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Strauch M. A. 1995. Delineation of AbrB-binding sites on the Bacillus subtilis spo0H, kinB, ftsAZ, and pbpE promoters and use of a derived homology to identify a previously unsuspected binding site in the bsuB1 methylase promoter. J. Bacteriol. 177:6999–7002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Strauch M. A., Trach K. A., Day J., Hoch J. A. 1992. Spo0A activates and represses its own synthesis by binding at its dual promoters. Biochimie 74:619–626 [DOI] [PubMed] [Google Scholar]
- 49. Tasteyre A., et al. 2001. Molecular characterization of fliD gene encoding flagellar cap and its expression among Clostridium difficile isolates from different serogroups. J. Clin. Microbiol. 39:1178–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tomas C. A., et al. 2003. DNA array-based transcriptional analysis of asporogenous, nonsolventogenic Clostridium acetobutylicum strains SKO1 and M5. J. Bacteriol. 185:4539–4547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Twine S. M., et al. 2009. Motility and flagellar glycosylation in Clostridium difficile. J. Bacteriol. 191:7050–7062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Underwood S., et al. 2009. Characterization of the sporulation initiation pathway of Clostridium difficile and its role in toxin production. J. Bacteriol. 191:7296–7305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Waligora A. J., et al. 2001. Characterization of a cell surface protein of Clostridium difficile with adhesive properties. Infect. Immun. 69:2144–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Walters B. A., Roberts R., Stafford R., Seneviratne E. 1983. Relapse of antibiotic associated colitis: endogenous persistence of Clostridium difficile during vancomycin therapy. Gut 24:206–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wilson K. H., Kennedy M. J., Fekety F. R. 1982. Use of sodium taurocholate to enhance spore recovery on a medium selective for Clostridium difficile. J. Clin. Microbiol. 15:443–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Worner K., Szurmant H., Chiang C., Hoch J. A. 2006. Phosphorylation and functional analysis of the sporulation initiation factor Spo0A from Clostridium botulinum. Mol. Microbiol. 59:1000–1012 [DOI] [PubMed] [Google Scholar]
- 57. Wright A., et al. 2005. Proteomic analysis of cell surface proteins from Clostridium difficile. Proteomics 5:2443–2452 [DOI] [PubMed] [Google Scholar]
- 58. York K., et al. 1992. Spo0A controls the sigma A-dependent activation of Bacillus subtilis sporulation-specific transcription unit spoIIE. J. Bacteriol. 174:2648–2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.