Abstract
Coenzyme A (CoA) plays a central and essential role in all living organisms. The pathway leading to CoA biosynthesis has been considered an attractive target for developing new antimicrobial agents with novel mechanisms of action. By using an arabinose-regulated expression system, the essentiality of coaBC, a single gene encoding a bifunctional protein catalyzing two consecutive steps in the CoA pathway converting 4′-phosphopantothenate to 4′-phosphopantetheine, was confirmed in Escherichia coli. Utilizing this regulated coaBC strain, it was further demonstrated that E. coli can effectively metabolize pantethine to bypass the requirement for coaBC. Interestingly, pantethine cannot be used by Pseudomonas aeruginosa to obviate coaBC. Through reciprocal complementation studies in combination with biochemical characterization, it was demonstrated that the differential characteristics of pantethine utilization in these two microorganisms are due to the different substrate specificities associated with endogenous pantothenate kinase, the first enzyme in the CoA biosynthetic pathway encoded by coaA in E. coli and coaX in P. aeruginosa.
INTRODUCTION
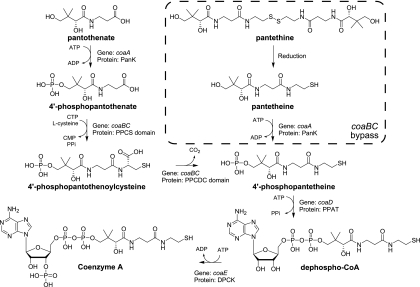
Coenzyme A (CoA) is a ubiquitous and essential cofactor in all living organisms, where it functions as a carrier for activated acyl groups in numerous central metabolic processes. Perhaps most notably, CoA provides the indispensable phosphopantetheine prosthetic group posttranslationally appended to acyl carrier proteins upon which fatty acids are biosynthesized; however, CoA participates in many other biochemical processes from the tricarboxylic acid (TCA) cycle to amino acid degradation (30). The five-step biochemical pathway for conversion of vitamin B5 pantothenate to CoA is conserved across all taxa and begins with formation of 4′-phosphopantothenate, catalyzed by pantothenate kinase (PanK). 4′-Phosphopantothenate is subsequently condensed with cysteine, which is in turn decarboxylated to form 4′-phosphopantetheine by the enzymes phosphopantothenoylcysteine synthetase (PPCS) and phosphopantothenoylcysteine decarboxylase (PPCDC), respectively. Finally, 4′-phosphopantetheine is converted to dephospho-CoA by the addition of an AMP moiety catalyzed by 4′-phosphopantetheine adenylyltransferase (PPAT) and then the 3′-hydroxyl of the AMP ribose is phosphorylated by dephospho-CoA kinase (DPCK) to form CoA (8) (Fig. 1).
Fig. 1.
The CoA biosynthetic pathway.
In bacteria, CoA plays a vital role in biogenesis of the cell envelope. Membrane lipid biogenesis utilizes fatty acid precursors that are biosynthesized using the iterative condensation of acetyl-CoA precursors (20). The peptidoglycan, an essential bacterial structure that maintains the mechanical integrity of the cell and participates in several key processes, including cell division and virulence (9), is biosynthesized from the primary precursor UDP-N-acetylglucosamine. GlmU, the final enzyme required to convert d-fructose-6-phosphate to UDP-N-acetylglucosamine, utilizes acetyl-CoA for the acetylation of glucosamine-1-phosphate (26). Furthermore, UDP-N-acetylglucosamine is a precursor to teichoic acid(s) in Gram-positive bacteria (19) and lipid A in Gram-negative bacteria (4), both of which are surface-exposed molecules that impact the permeability of the bacterial cell. Finally, the majority of Gram-positive pathogens, including Staphylococcus aureus, Enterococcus faecalis, and Streptococcus pneumoniae, utilize the mevalonate pathway for isoprenoid biosynthesis. In the mevalonate pathway, 3 units of acetyl-CoA are condensed to form isopentenyl pyrophosphate (IPP), the building block for the cell wall carrier lipid undecaprenyl pyrophosphate (UPP) (7).
Despite its significance in bacteria, the complete biosynthetic machinery for synthesis of CoA has been elucidated only relatively recently, culminating in identification of the Escherichia coli coaBC gene, which encodes a single bifunctional PPCS/PPCDC protein (43). The existence of coaBC as a single gene encoding one protein with 2 functional domains in bacteria is in stark contrast to the pattern seen with eukaryotes such as plants and mammals, where PPCS and PPCDC are individual enzymes encoded by separate coaB and coaC genes, respectively (1–3, 13, 28). Given the paucity of information concerning coaBC in bacteria and its potential as a target for antimicrobial intervention, we sought to investigate the effect on E. coli of disruption of coaBC. Interestingly, despite the validated essentiality of coaBC (13, 15) and reports that pantothenate is the most advanced precursor to CoA that has been characterized as being actively transported into bacteria (8, 30), we find that chemical complementation with pantethine renders coaBC nonessential in E. coli but not in Pseudomonas aeruginosa. This bypass mechanism is dependent on the presence of pantothenate kinase.
MATERIALS AND METHODS
General materials and procedures.
Standard DNA, molecular cloning, and microbiological procedures were performed as described previously (39). Plasmids, genomic DNA, PCR fragments run on agarose gels, and restriction digests were purified using a QIAprep spin miniprep kit, DNeasy blood and tissue kit, QIAquick gel extraction kit, and QIAquick PCR purification kit, respectively (Qiagen). Primers and ultramers were purchased from Integrated DNA Technologies. Phusion high-fidelity DNA polymerase, Quick ligase, and restriction enzymes were from NEB. Electroporation was performed on a MicroPulser Electroporator (Bio-Rad). One Shot TOP10 and PIR1 E. coli were from Invitrogen. DNA sequencing was performed by Beckman Coulter Genomics. LB, LB agar, and Pseudomonas isolation agar (PIA) were from Difco. NZYM media and sucrose were from MP Biomedicals. ATP, NADH, kanamycin, chloramphenicol, gentamicin, ampicillin, pantethine, CoA, dephospho-CoA, l-arabinose, lactate dehydrogenase, pyruvate kinase, and phosphoenolpyruvate (PEP) were from Sigma. Pantothenate, IPTG (isopropyl-β-d-thiogalactopyranoside), and imidazole were from Acros Organics. Carbenicillin was from Fisher Scientific, dithiothreitol (DTT) was from Promega, TCEP [tris(2-carboxyethyl)phosphine] was from Thermo Scientific, Complete protease inhibitor cocktail was from Roche, and cobalt Talon metal affinity resin was from Clontech. Fast protein liquid chromatography (FPLC) purification of proteins was performed using a HiLoad 16/60 Superdex 200 prep grade column, an ÄKTA FPLC system, and Unicorn 5.0 software (GE Healthcare).
Construction of coaBC- and coaD-regulated E. coli strains.
A new vector was created that allowed the one-step λ Red recombinase knockout method developed by Datsenko and Wanner (12) to be adapted to perform one-step incorporation of the PBAD promoter in front of any gene. The araC-PBAD region from the pBAD18 plasmid (17) was amplified by PCR using primers P1 and P2 (all primers and ultramers are listed in Table S1 in the supplemental material), and the majority of pKD4 (12) was amplified by PCR using primers P3 and P4. The PCR fragments were digested with ClaI/XhoI, ligated, and transformed into PIR1 chemically competent cells. The resulting plasmid, pKD4-PBAD, was used as a template in subsequent PBAD promoter integrations.
The PBAD promoter was integrated 15 bp upstream of the coaBC start codon by the use of the linear PCR product generated by amplification from a pKD4-PBAD template with the P5 and P6 ultramers, generating the BW25113PBADcoaBC strain (all strains are listed in Table S2 in the supplemental material). The PBAD promoter was integrated 25 bp upstream of the coaD start codon by the use of the linear PCR product generated by amplification from pKD4-PBAD template with the P7 and P8 ultramers, generating the BW25113PBADcoaD strain. Transformation and selection were performed as described previously (6). Briefly, overnight cultures of BW25113 cells harboring pKD46 (6) were diluted 100-fold into fresh LB containing 100 μg/ml ampicillin and 0.2% (wt/vol) arabinose, grown at 30°C until an optical density at 600 nm (OD600) of 0.5 was reached, washed twice with an equal volume of cold water, washed three times with 1 ml 10% glycerol, and finally resuspended in 10% glycerol using 1/250 of the initial culture volume. Competent cells (50 μl) were mixed with 100 to 300 ng (not exceeding 5 μl) of gel-purified PCR product to be inserted into the chromosome, electroporated in a 0.2-cm-gap cuvette using an EC2 setting (2.5 kV, 5 ms), recovered in 1 ml SOC medium (Invitrogen) containing 0.2% (wt/vol) arabinose at 37°C for 2 h, and finally plated on LB agar containing 30 μg/ml kanamycin and 1 mM arabinose. Colonies that grew the next day were replica plated to confirm loss of pKD46 and acquisition of arabinose-dependent growth. All integrants were confirmed by PCR and sequencing.
Deletion of panF and replacement of coaA from E. coli with coaX from P. aeruginosa in BW25113PBADcoaBC.
The panF gene was deleted, leaving only the first and last six codons, by using the linear PCR product generated by amplification from pKD3 (12) with the P9 and P10 ultramers, generating strain BW25113PBADcoaBCΔpanF::cat. For replacement of the coaA gene from E. coli (EccoaA), splicing by overlap extension PCR (21) was used to fuse a chloramphenicol resistance cassette downstream of the coaX gene from P. aeruginosa (PacoaX) to facilitate selection. In the first round of PCR, ultramer P11 and primer P12 were used to amplify PacoaX from P. aeruginosa genomic DNA, and primer P13 and ultramer P14 were used to amplify the cat gene from pKD3. In the second round of PCR, products from the first round were mixed and further amplified using P11 and P14. This second-round complete PCR product was used to replace the EccoaA gene from the start to the stop codon, generating strain BW25113PBADcoaBCΔcoaA::PacoaX. Transformation and selection were performed as described above except that, during final plating and selection, 25 μg/ml chloramphenicol was added. All integrants were confirmed by PCR and sequencing.
Deletion of coaBC in E. coli.
The EccoaBC gene was deleted, leaving only the first and last six codons, by using the linear PCR product generated by amplification from pKD4 with the P15 and P16 ultramers, generating strain BW25113ΔcoaBC::kan. Transformation and selection were performed as described above except that, during recovery and final plating, 1 mM pantethine was included and arabinose was omitted in the media. All integrants were confirmed by PCR and sequencing.
Deletion of coaBC in P. aeruginosa.
Marked deletions of the PacoaBC gene were constructed in PAO1 and the hyperpermeable Z61 mutant (5, 27, 51) (ATCC 35151) by the use of the pEX18Tc suicide vector (22), which utilizes a sacB-based selection method for resolving cointegrants to create double-crossover mutants. The PacoaBC gene between the first 10 codons and last 10 codons was replaced with the gentamicin resistance cassette. This replacement deletes the majority of coaBC while minimizing the chances of hitting neighboring genes. To accomplish this, splicing by overlap extension was used to fuse the flanking regions of the PacoaBC gene while introducing a midpoint PmlI restriction site. In the first round of PCR, 600 bp upstream of PacoaBC was amplified with the P17 and P18 primers and 600 bp downstream of PacoaBC was amplified with the P19 and P20 primers. In the second round of PCR, products from the first round were mixed and further amplified using P17 and P20. The PCR product from the second round was digested with BamHI/KpnI and ligated into a similarly digested pEX18Tc vector. This new plasmid was then digested with PmlI and ligated with the gentamicin cassette excised with SmaI from pUCGM (40). PCR was used to screen for plasmids in which the gentamicin cassette was ligated in the same orientation as the PacoaBC gene. The resulting pEX18TcΔPacoaBC::gent vector was mobilized into P. aeruginosa from S17-1 E. coli (41). Briefly, 500 μl of S17-1 E. coli harboring pEX18TcΔPacoaBC::gent grown overnight at 37°C in LB with 10 μg/ml gentamicin was mixed with 500 μl of PAO1 or Z61 P. aeruginosa grown in LB. Pelleted cells were spread as a nickel-sized plane of growth on LB agar, incubated at 37°C for 18 h, and then restreaked on PIA with either 100 μg/ml (for PAO1) or 10 μg/ml (for Z61) gentamicin to select for merodiploids. To resolve cointegrants to create double-crossover knockouts, single colonies were restreaked on PIA with 6% (wt/vol) sucrose, 1 mM pantethine, and either 100 μg/ml (for PAO1) or 10 μg/ml (for Z61) gentamicin to select for loss of a sacB-containing vector backbone.
These experiments were repeated after introducing the E. coli coaA gene on a replicative plasmid. EccoaA was cloned onto two versions of the pFlp2 plasmid (22). In both versions, the flp gene was removed, but for one version the sacB gene, which renders cells nonviable when grown on sucrose, was removed (pΔflp2ΔsacBEccoaA), whereas in the other version it was not (pΔflp2EccoaA). EccoaA was amplified from genomic DNA using the P21 and P22 primers. The pFlp2 plasmid was amplified using primer P23 and either primer P24 to include the sacB gene or primer P25 to omit the sacB gene. The vector and insert were digested with NcoI/XhoI, ligated, and transformed into TOP10 chemically competent cells. These plasmids were mobilized into both the wild-type PAO1 and membrane-permeable Z61 P. aeruginosa strains from S17-1 E. coli by the use of either 100 μg/ml (PAO1) or 10 μg/ml (Z61) carbenicillin for maintenance. Subsequently, the pEX18TcΔPacoaBC::gent suicide vector was introduced into these 4 backgrounds and cointegrants were selected per the protocol described above. Finally, to obtain full coaBC knockouts, the four strains—PAO1pEX18TcΔPacoaBC::gent/pΔflp2ΔsacBEccoaA, PAO1pEX18TcΔPacoaBC::gent/pΔflp2EccoaA, Z61pEX18TcΔPacoaBC::gent/pΔflp2ΔsacBEccoaA, and Z61pEX18TcΔPacoaBC::gent/pΔflp2EccoaA—were streaked onto media containing gentamicin (to select for retention of the PacoaBC deletion), 6% sucrose (to select for loss of pEX18Tc sacB containing backbone and pΔflp2EccoaA), and 1 mM pantethine (to complement loss of PacoaBC).
Cloning, expression, and purification of E. coli and P. aeruginosa pantothenate kinase.
E. coli coaA was amplified using genomic DNA and primers P26 and P27. P. aeruginosa coaX was amplified using genomic DNA and primers P28 and P29. The resulting PCR fragments were digested with BamHI/SalI and ligated into a similarly digested pTrcHis2B vector (Invitrogen). To express the C-terminal His6-tagged protein, overnight cultures of TOP10 cells harboring expression plasmids were diluted 100-fold into 3 liters of fresh NZYM broth containing 50 μg/ml carbenicillin. Cultures were grown at 37°C until the OD600 reached 0.5 (approximately 3 h), at which point cultures were induced with 1 mM IPTG, transferred to 18°C, and grown for an additional 48 h.
For purification, 3-liter cultures were harvested by centrifugation and resuspended in 30 ml of ice-cold lysis buffer (50 mM Tris [pH 8.0], 250 mM NaCl, 2 mM MgCl2, 100 μg/ml lysozyme, 1 mM TCEP, 0.1 mg/ml DNase I, 1 Complete protease inhibitor cocktail tablet, 10% glycerol). For P. aeruginosa PanK purification, 20 mM imidazole was included in the lysis and wash (see below) buffers. Cells were passed four times through a French press (Thermo) at 1,100 lb/in2 and centrifuged at 12,000 × g for 30 min at 4°C, and the supernatant was loaded onto 10 ml of cobalt Talon resin that had been preequilibrated with wash buffer (50 mM Tris [pH 8.0], 250 mM NaCl, 2 mM MgCl2, 1 mM TCEP). The resin was washed with 20 column volumes of wash buffer, after which cobalt-bound His6 protein was eluted with 3 column volumes of elution buffer (50 mM Tris [pH 8.0], 250 mM NaCl, 2 mM MgCl2, 1 mM TCEP, 500 mM imidazole, 10% glycerol). Eluted fractions containing protein were concentrated to ∼2 ml using an Amicon Ultra 10,000-molecular-weight cutoff (10,000 MWCO) centrifugal device (Millipore) and then injected for FPLC size exclusion chromatography using a buffer consisting of 50 mM Tris [pH 8.0], 250 mM NaCl, 2 mM MgCl2, 1 mM TCEP, and 10% glycerol. Fractions (2.5 ml each) in the largest peak in the UV trace were analyzed for purity on a 4 to 15% Tris-HCl sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel (Bio-Rad), and those showing a single band at the correct molecular weight were pooled. Protein concentrations were determined at 595 nm using Bio-Rad protein reagent with bovine serum albumin (BSA) as the standard. Purified enzymes were stored in aliquots at −80°C.
Biochemical assays with pantothenate kinase.
Kinase activity was measured in vitro using a pyruvate kinase/lactate dehydrogenase-coupled assay in which the formation of ADP from the PanK reaction was coupled to the formation of pyruvate by pyruvate kinase, which was in turn coupled to the oxidation of NADH by lactate dehydrogenase. After optimizing PanK enzyme activity with regard to buffer, pH, salt, dimethyl sulfoxide (DMSO), magnesium, DTT, detergent, and BSA, optimal reaction conditions were established at 100 mM HEPES [pH 7.3], 50 mM KCl, 1 mM MgCl2, 0.01% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 20 μg/ml BSA, 1 mM DTT, 150 μM pantothenate or pantethine, 400 μM ATP, 500 μM PEP, 200 μM NADH, 20 U/ml pyruvate kinase, 20 U/ml lactate dehydrogenase, and 50 nM PanK. For E. coli PanK Km determinations, either ATP was fixed at 1 mM and pantothenate or pantethine was adjusted to between 0 and 1,000 μM or pantothenate or pantethine was fixed at 1 mM and ATP was adjusted to between 0 and 2,000 μM. For P. aeruginosa PanK Km determinations, either ATP was fixed at 16 mM and pantothenate was adjusted to between 0 and 40 μM or pantothenate was fixed at 80 μM and ATP was adjusted to between 0 and 16 mM. Reactions were monitored by measuring the decrease in fluorescence resulting from the coupled oxidation of NADH (excitation at 340 nm and emission at 460 nm) by the use of a SpectraMax Gemini XS microplate spectrofluorometer (Molecular Devices). To convert relative fluorescence units (RFU) to concentration values, the molar fluorescence coefficient of NADH was experimentally determined to be 14.81 RFU/μM.
Growth curves.
Overnight cultures of desired strains were centrifuged such that resuspension of cells in 1 ml of fresh LB yielded an OD600 of 1.0. These normalized cultures were then diluted 10,000-fold into fresh LB, and 100 μl was aliquoted into a 96-well U-bottom polystyrene plate (Falcon). Medium components (pantothenate, pantethine, dephospho-CoA, CoA, arabinose, and antibiotics for strain maintenance) were added as necessary either before aliquoting (when a single concentration was required for the assay) or to the first column of the plate followed by 2-fold serial dilutions made across the plate when titration of a component was necessary. Plates were incubated at 37°C with shaking, and OD600 readings were taken every minute using a SpectraMax Plus 384 plate reader. All experiments were performed at least 3 times to confirm the observed effects on growth.
RESULTS AND DISCUSSION
Construction and analysis of a coaBC-regulated E. coli strain.
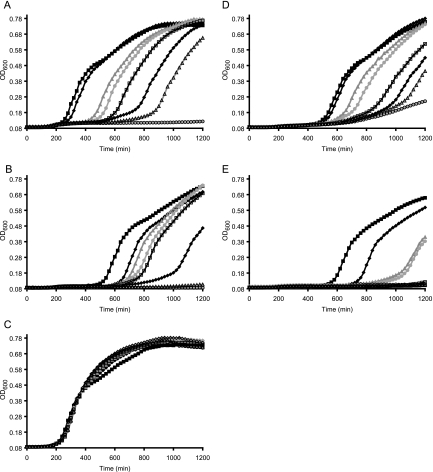
In order to investigate the essentiality of coaBC and the effects of its depletion on E. coli, a regulated strain was constructed in which expression of coaBC was placed under the control of the arabinose-inducible PBAD promoter. The growth of the resulting strain, BW25113PBADcoaBC, was dependent on the presence of arabinose, which is consistent with the reported essentiality of coaBC (13, 15). Furthermore, the lag time for detectable growth scaled with the concentration of arabinose in the growth media (Fig. 2A), with increased lag times observable below 62 μM arabinose. In fact, the midpoint for the exponential growth phase (OD600 = 0.35) shifted from 340 min for growth in the presence of 125 μM arabinose to 380, 540, 600, 680, 820, and 960 min for growth in the presence of 63, 16, 8, 4, 2, and 1 μM arabinose, respectively (Fig. 2A).
Fig. 2.
Arabinose-dependent growth of BW25113PBADcoaBC. (A) Growth curves of BW25113PBADcoaBC grown at 37°C in LB with 30 μg/ml kanamycin and various concentrations of arabinose: 125 μM (black filled squares), 63 μM (dark gray filled diamonds), 16 μM (medium gray filled triangles), 8 μM (light gray filled circles), 4 μM (black unfilled squares), 2 μM (dark gray unfilled diamonds), 1 μM (medium gray unfilled triangles), and 0 μM (no arabinose; light gray unfilled circles). (B) Same as described for panel A but with 1 mM pantothenate under all conditions. (C) Same as described for panel A but with 1 mM pantethine under all conditions. (D) Same as described for panel A but with 1 mM dephospho-CoA under all conditions. (E) Same as described for panel A but with 1 mM CoA under all conditions. For figure clarity, curves are illustrative of a single replicate, though experiments were performed at least 3 times.
Nutrient supplementation.
Given that CoA biosynthesis is a primary metabolic process leading to the formation of a small-molecule cofactor, it was of interest to determine whether supplementing the media with substrates, intermediates, and/or products of the pathway could rescue deficiencies in coaBC expression. Precedents for chemical complementation of PPCDC defects can be found in studies on Arabidopsis thaliana. This plant possesses two genes that encode PPCDC, HAL3A and HAL3B. While plants containing homozygous transfer DNA (T-DNA)-disrupted alleles of either hal3a-1 or hal3b are viable, a hal3a-1 hal3b double mutant is embryonically lethal, indicating a redundant and yet essential function for these genes. Interestingly, although hal3b individuals heterozygous for HAL3A behave similarly to wild-type plants, hal3a-1 individuals heterozygous for HAL3B were severely impaired for seedling establishment (38), a result that supports the notion that HAL3A plays a dominant role with respect to HAL3B, as suggested by differences in transcript levels (14). This severe defect could be rescued by supplementation with pantethine, restoring normal seedling establishment.
To investigate whether a similar phenomenon could occur in E. coli, the growth phenotype of BW25113PBADcoaBC as a function of arabinose concentration was measured in the presence of 1 mM pantothenate, pantethine, dephospho-CoA, or CoA. As expected, the addition of pantothenate, the primary precursor of the CoA pathway, was unable to rescue the arabi-nose-dependent growth phenotype of BW25113PBADcoaBC (Fig. 2B). This result is consistent with the fact that PPCS/PPCDC acts downstream of pantothenate formation in CoA biosynthesis and implies that increasing the concentration of the substrate cannot overcome a deficiency in coaBC expression. It was also noted that the lag time seen with all concentrations of arabinose increased when adding pantothenate, suggesting that E. coli is subject to additional stress imposed by the exogenous addition of this metabolite. Nonetheless, the apparent change in lag time reflects only the difference in growth conditions and does not alter the conclusion that pantothenate is unable to bypass the need for endogenous PPCS/PPCDC function. Next, addition of pantethine did deregulate arabinose-dependent growth in BW25113PBADcoaBC. As can be seen in Fig. 2C, in the presence of 1 mM pantethine, the lag times for growth were equivalent in the presence or absence of arabinose. Finally, addition of dephospho-CoA or CoA, the penultimate and ultimate products of the pathway, did not rescue the increased lag time for growth due to underexpression of coaBC (Fig. 2D and E). Although dephospho-CoA and CoA formation are downstream with respect to PPCS/PPCDC function, these highly charged phosphate-containing molecules are unable to enter cells. As was the case with pantothenate, an overall increase in lag time for the bacterial cultures regardless of the arabinose concentration was observed when either dephospho-CoA or CoA was added to the medium, likely due to a change in the overall growth conditions. Additionally, there was a slight amount of growth observed with 1 mM dephospho-CoA in the absence of arabinose (Fig. 2D), which could have been caused by the breakdown of dephospho-CoA to pantetheine over time or by the presence of a small amount of pantetheine or pantethine in the sample, as the purchased dephospho-CoA was only 90% pure. In summary, regardless of any general changes in growth that chemical supplementation may have caused, the primary phenotype of arabinose-dependent growth and increased lag time caused by regulating the expression of coaBC was rescued only in the presence of pantethine.
Pantethine is a dimeric thiol-oxidized version of the PPCS/PPCDC product that is missing the 4′-phosphate appended by PanK. The results showing that it can complement deficiencies in coaBC expression indicates that it can both enter cells and bypass the need for PPCS/PPCDC activity. To demonstrate that pantethine was not being converted to CoA by an alternative pathway, chemical complementation experiments were repeated in a strain in which coaD, which encodes the PPAT enzyme that catalyzes the step subsequent to PPCS/PPCDC formation in CoA biosynthesis, was placed under the control of the arabinose-inducible PBAD promoter. As with coaBC, coaD was confirmed to be essential, because growth of BW25113PBADcoaD was dependent on coaD expression, and the lag time for growth scaled with the concentration of arabinose in the media (see Fig. S1A in the supplemental material). As can be seen in Fig. S1B to E in the supplemental material, addition of 1 mM pantothenate, pantethine, dephospho-CoA, or CoA was unable to rescue the arabinose-regulated phenotype of BW25113PBADcoaD. Although slight differences in the overall growth could be observed when exogenous metabolites were added, especially at low concentrations of arabinose, addition of any of these nutrients did not change the fact that the lag time for growth was inversely proportional to the amount of arabinose present and that in the absence of arabinose, no growth was observed. This result confirms the idea that increasing the amount of upstream precursors such as pantothenate cannot overcome deficiencies in downstream enzyme activity and that dephospho-CoA and CoA were not entering the cells, since both were downstream of PPAT activity. More importantly, these data demonstrate that pantethine cannot bypass the expression of coaD, indicating that pantethine is ultimately converted to the PPAT substrate phosphopantetheine by a PPCS/PPCDC-independent mechanism.
Despite its inability to cross the cytoplasmic membrane in E. coli, extracellular pantothenate can be used as a precursor to CoA biosynthesis due to its active uptake by the pantothenate permease encoded by panF (24, 45, 47). In order to determine whether the uptake of pantethine also was dependent on panF, knockouts were constructed in the BW25113PBADcoaBC background. As can be seen in Fig. S2 and S3 in the supplemental material, arabinose-dependent growth is deregulated after addition of pantethine in a manner similar to that seen in comparisons of the parent BW25113PBADcoaBC to the mutant BW25113PBADcoaBCΔpanF::cat. This deregulation is observable as a decrease in lag time for the different concentrations of arabinose. Furthermore, the degrees of deregulation seen with increasing concentrations of pantethine were similar for the two strains (see Fig. S2B to E and S3B to E in the supplemental material). These data indicate that the ability of pantethine to enter E. coli is not dependent on the presence of panF. This finding is consistent with studies in which an azido-pantetheine analog was capable of entering cells and being processed to ultimately label endogenous acyl carrier proteins in a panF-independent manner (34).
Constructing coaBC knockouts.
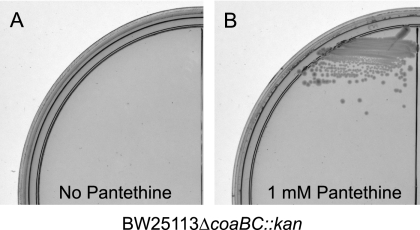
To rule out the possibility that residual PPCS/PPCDC activity in BW25113PBADcoaBC was responsible for cell viability due to unknown regulatory mechanisms triggered by addition of pantethine, a full deletion of coaBC was attempted. Indeed, when pantethine-supplemented media were used, mutants could be obtained with a full knockout in the coaBC gene. The BW25113ΔcoaBC::kan strain, whose identity was confirmed by sequencing, was not viable on media lacking exogenous pantethine (Fig. 3). Pantothenate, dephospho-CoA, and CoA could not be used as surrogates for restoring growth, and plating performed multiple times with 1012 cells did not yield any suppressors of the pantethine auxotrophy. Although these data are consistent with previous studies indicating that CoA cannot be transported across the bacterial cell envelope (23, 25, 42), they alter the notion that pantothenate is the most advanced precursor to CoA that can permeate cells (24, 42, 45).
Fig. 3.
BW25113ΔcoaBC::kan is auxotrophic for pantethine. (A) BW25113ΔcoaBC::kan grown on LB agar with 30 μg/ml kanamycin. (B) BW25113ΔcoaBC::kan grown on LB agar with 30 μg/ml kanamycin and 1 mM pantethine.
Biochemical experiments using PanK.
The use of pante-thine to chemically complement a coaBC deficiency in a coaD-dependent fashion suggests that E. coli is capable of converting this molecule to the PPCDC product phosphopantetheine. This transformation could be accomplished rather directly through reduction of the disulfide to the free thiol and phosphorylation of the 4′ hydroxyl (Fig. 1). Although the general reducing environment of the cytoplasm could suffice for accomplishing disulfide reduction, the phosphorylation event most likely would require a specific enzyme. It has been known for some time that specific enzyme preparations purified from cellular extracts could phosphorylate both pantothenate and pantethine (1, 31), and ultimately this activity was attributed to the protein involved in the first step in CoA biosynthesis, PanK. There are three types of bacterial PanKs that differ with respect to structure, substrate affinity, and feedback regulation. The type I PanKs are the only type modulated through feedback inhibition by CoA and are exemplified by the prototypical member encoded by coaA in E. coli (46). Type II PanKs are related to the eukaryotic isoforms of PanK and are exemplified by the product of the coaA gene in Staphylococcus aureus (29). Finally, type III PanKs are the most divergent of the three, have high Km values for their substrates, and are exemplified by the product of the coaX gene in Helicobacter pylori (10).
The coaA gene, encoding the type I E. coli PanK (EcPanK), was cloned into the C-terminal His6-tagged vector pTrcHis2B. Expression in TOP10 E. coli at 18°C with 1 mM IPTG induction yielded 30 to 40 mg/liter after purification to homogeneity using cobalt affinity and gel filtration chromatography in tandem. Initial activity titration assays performed using a pyruvate kinase/lactate dehydrogenase-coupled system demonstrated that EcPanK was active and readily phosphorylated both pantothenate and pantethine.
Once initial activities were established, a full set of Michaelis-Menten parameters for the kinase reaction were measured under optimized reaction conditions. The data in Table 1 indicate that the EcPanK Km values for ATP are within a 2-fold range whether using pantothenate or pantethine as substrate and that the Km for the natural pantothenate substrate is 2-fold greater than the Km for the surrogate pantethine. These data demonstrate that EcPanK can phosphorylate pantethine and that the specific activity for this conversion is 2-fold greater than that for the natural pantothenate substrate.
Table 1.
Kinetic properties of PanKa
| Enzyme | Substrate adjusted | Substrate held constant | Km (mM) | kcat (s−1) | kcat/Km (mM−1 s−1) |
|---|---|---|---|---|---|
| EcPanK | ATP | Pantothenate | 0.093 ± 0.008 | 0.77 ± 0.02 | 8.33 ± 0.75 |
| EcPanK | ATP | Pantethine | 0.15 ± 0.02 | 1.48 ± 0.06 | 9.79 ± 1.36 |
| EcPanK | Pantothenate | ATP | 0.66 ± 0.08 | 1.11 ± 0.07 | 1.68 ± 0.23 |
| EcPanK | Pantethine | ATP | 0.30 ± 0.04 | 1.05 ± 0.06 | 3.46 ± 0.50 |
| PaPanK | ATP | Pantothenate | 7.3 ± 0.9 | 0.090 ± 0.005 | 0.012 ± 0.002 |
| PaPanK | Pantothenate | ATP | 0.005 ± 0.001 | 0.22 ± 0.01 | 49.6 ± 10.2 |
± values represent standard errors calculated using GraFit 5 software.
The promiscuity of type I and II PanKs has been exploited in the use of pantothenamide antimetabolites that can act as substrates for PanK as well as for downstream enzymes. These molecules have been utilized as labels for carrier proteins in vivo (32) and as antimicrobial agents; however, there is still debate as to whether the mechanism of action for the pantothenamide class of antimetabolites operates by direct inhibition of CoA biosynthetic enzymes (11, 44), by inhibition of fatty acid biosynthesis through the accumulation of nonfunctional acyl carrier proteins (29, 49), or by inhibition of CoA- and acetyl-CoA-utilizing enzymes (42). Given this promiscuity, it was not surprising that E. coli PanK was demonstrated here to efficiently utilize pantethine as a substrate in biochemical assays.
As a parallel comparison, the type III PanK enzyme was also isolated by cloning and expressing the coaX gene from the clinically relevant pathogen Pseudomonas aeruginosa. Possessing a rather impermeable outer membrane and a diverse range of efflux pumps (35, 36), P. aeruginosa is inherently recalcitrant to antimicrobial intervention. It is of particular interest given that it has emerged in the clinic as a major cause of nosocomial infections in immunocompromised patients and is well known to be a cause of declining lung function in cystic fibrosis patients (16, 33).
Although P. aeruginosa PanK (PaPanK) catalyzed robust phosphorylation of pantothenate with Michaelis-Menten parameters similar to those found for the type III PanK from H. pylori (10) (Table 1), it was unable to utilize pantethine as a substrate. This result held true despite exploration of several sets of conditions in which high concentrations of enzyme and/or substrate were used and large amounts of reducing agent were supplied to form the free thiol pantetheine. The inability of PaPanK to utilize pantethine is also consistent with the inability of H. pylori PanK to accept N-pentylpantothenamide as an alternative substrate (10).
Pantethine complementation dependence on PanK in vivo.
The inability of PaPanK to phosphorylate pantethine was further explored in vivo in P. aeruginosa through an attempt to create a coaBC knockout. Initially, deletion was performed in a wild-type PAO1 background; however, single integrants never resolved to double-crossover knockout mutants. Since Pseudomonas is known for possessing an especially impermeable outer membrane (5, 18), it is possible that pantethine is unable to penetrate PAO1. Therefore, the knockout experiments were repeated with the hyperpermeable Pseudomonas aeruginosa strain Z61 (ATCC 35151) (5, 27, 51). Despite the lower permeability threshold, coaBC knockout mutants were not obtained.
The result showing that PaPanK is unable to phosphorylate pantethine and therefore is unable to form the PPAT substrate phosphopantetheine suggested that perhaps the inability to delete coaBC in P. aeruginosa and thus the inability to generate a pantethine auxotroph was not due to the lack of cellular penetration of pantethine but rather due to an inability to metabolize the molecule. If this were indeed the case, then introducing the E. coli coaA gene (EccoaA) into P. aeruginosa should allow it to utilize pantethine to bypass the PPCS/PPCDC function, rendering the coaBC deletion viable. Conversely, if the coaA gene function in E. coli were paramount for incorporating pantethine metabolism into CoA, replacing it with the coaX gene from P. aeruginosa (PacoaX) should render the pantethine bypass of coaBC ineffective.
To generate the P. aeruginosa pantethine auxotroph, E. coli coaA was introduced into the wild-type PAO1 and membrane-permeable Z61 P. aeruginosa strains on either of two replicative plasmids. The pΔflp2EccoaA plasmid contains the sacB gene, which renders cells nonviable when grown on sucrose, whereas the pΔflp2ΔsacBEccoaA plasmid does not contain the sacB gene. The use of these parallel plasmids can demonstrate the essentiality of EccoaA in pante-thine complementation of coaBC deficiency. During the process of generating knockouts, single integrants of the pEX18TcΔPacoaBC::gent suicide vector are forced to resolve to double-crossover knockouts because the use of gentamicin selects for retention of the resistance cassette incorporated into the target insert and the use of sucrose selects for loss of plasmid backbone which contains the sacB gene. Concurrently, the use of sucrose in this selection also selects for loss of the pΔflp2EccoaA plasmid but would not select for loss of the pΔflp2ΔsacBEccoaA plasmid. If EcPanK were responsible for the ability to utilize pantethine, thus bypassing PPCD/PPCDC activity, only cells with pΔflp2ΔsacBEccoaA would be viable after selection on sucrose, since only they would retain a copy of EccoaA.
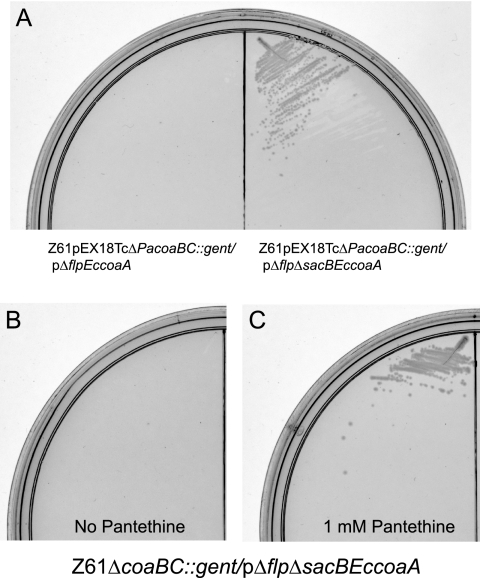
As evidenced in Fig. 4A, only Z61pEX18TcΔPacoaBC::gent/pΔflp2ΔsacBEccoaA bacteria were viable when selection for knockouts on gentamicin, sucrose, and pantethine was performed. The Z61pEX18TcΔPacoaBC::gent/pΔflp2EccoaA strain failed to grow, indicating that EccoaA is required for growth on pante-thine. Colonies from this selection were isolated and sequenced to confirm that they were double-crossover knockouts of coaBC. As can be seen in Fig. 4B and C, the resulting Z61ΔcoaBC::gent/pΔflp2ΔsacBEccoaA strain was viable only in the presence of pantethine. These results indicate that PanK phosphorylation of pantethine is necessary and sufficient for pantethine complementation of coaBC deficiency, even in P. aeruginosa, where the endogenous PanK encoded by coaX is unable to catalyze this phosphorylation. Interestingly, single integrants of the PAO1 strain containing either the pΔflp2EccoaA or pΔflp2ΔsacBEccoaA plasmid never resolved to double-crossover knockout mutants, even when pantethine was supplied at concentrations as high as 5 mM in the media. This indicates that the permeability barrier in PAO1 is too high to allow pantethine to enter.
Fig. 4.
EccoaA is required for pantethine bypass of coaBC in P. aeruginosa. (A) Z61pEX18TcΔPacoaBC::gent/pΔflp2EccoaA and Z61pEX18TcΔPacoaBC::gent/pΔflp2ΔsacBEccoaA plated on PIA with 10 μg/ml gentamicin, 10 μg/ml carbenicillin, 1 mM pantethine, and 6% (wt/vol) sucrose. (B) Z61ΔcoaBC::gent/pΔflp2ΔsacBEccoaA grown on LB agar with 10 μg/ml gentamicin. (C) Same as described for panel B except that the plates also contained 1 mM pantethine.
To further confirm these results, the reciprocal pantethine complementation experiment was conducted using E. coli. Using the BW25113PBADcoaBC background, we replaced coaA with the coaX gene from P. aeruginosa. The growth of the resulting strain, BW25113PBADcoaBCΔcoaA::PacoaX, was assayed using various concentrations of arabinose, with or without the addition of 1 mM pantethine. As shown in Fig. 5, unlike the parental BW25113PBADcoaBC strain, in which addition of pantethine bypasses arabinose dependence (Fig. 2C), in BW25113PBADcoaBCΔcoaA::PacoaX, addition of pantethine had no effect on the increased lag time observed with decreasing concentrations of arabinose. These results indicate that, when EccoaA is replaced with PacoaX, E. coli can no longer metabolize pantethine into phosphopantetheine and bypass deficiencies in coaBC. It also suggests that PanK is the only enzyme in E. coli that can phosphorylate pantethine.
Fig. 5.
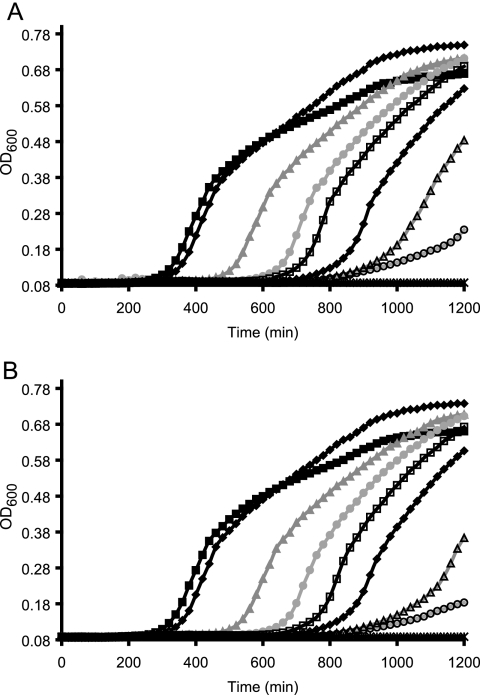
Arabinose-dependent growth of BW25113PBADcoaBCΔcoaA::PacoaX cannot be bypassed with pantethine. (A) Growth curves of BW25113PBADcoaBCΔcoaA::PacoaX grown at 37°C in LB with 30 μg/ml kanamycin, 25 μg/ml chloramphenicol, and various concentrations of arabinose: 500 μM (black filled squares), 63 μM (dark gray filled diamonds), 31 μM (medium gray filled triangles), 16 μM (light gray filled circles), 8 μM (black unfilled squares), 4 μM (dark gray unfilled diamonds), 2 μM (medium gray unfilled triangles), 1 μM (light gray unfilled circles), and 0 μM (no arabinose; dark gray black stars). (B) Same as described for panel A but with 1 mM pantethine under all conditions. For figure clarity, curves are illustrative of a single replicate, though experiments were performed at least 3 times.
The results of this study differ somewhat from findings demonstrating pantethine rescue of pantothenate-kinase-associated neurodegeneration (PKAN) in Drosophila studies. The neurodegenerative hereditary disease Hallervorden-Spatz syndrome, caused by mutations in human PANK2 (50), has been successfully recapitulated in Drosophila through analogous mutations in dPANK2/fbl (37, 48). The mutant flies have significantly reduced levels of CoA, resulting in impaired mitochondrial function, increased oxidative damage of proteins, defective locomotor abilities, and decreased life span. All of these phenotypes could be rescued to some degree by supplementation of food with pantethine (37), indicating that pantethine could be utilized as a substrate for generating CoA. Interestingly, these results suggest that in Drosophila, pantethine can be utilized in a PanK-independent manner, which is in contrast to the findings of this study, where pantethine utilization was found to be absolutely dependent on PanK activity. Although an alternate kinase or pathway for pantethine metabolism in Drosophila has yet to be found, ultimately, pantethine is most likely still converted to the intermediate 4′-phosphopantetheine, since pantethine could rescue decreased cell counts only in dPPCS-depleted cells and not in dPPAT-depleted cells (37). This is consistent with the results of this study, which demonstrate that pantethine supplementation could bypass arabinose-regulated growth of only a coaBC strain (BW25113PBADcoaBC) and not a coaD strain (BW25113PBADcoaD).
Supplementary Material
ACKNOWLEDGMENTS
We thank K. Poole (Queen's University) and H. Schweizer (University of Colorado) for strains and plasmids. We thank A. Jones, X. Shen, and C. Dean (Novartis) for helpful discussions.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 6 May 2011.
REFERENCES
- 1. Abiko Y. 1967. Investigations on pantothenic acid and its related compounds. IX. Biochemical studies. 4. Separation and substrate specificity of pantothenate kinase and phosphopantothenoylcysteine synthetase. J. Biochem. 61:290–299 [DOI] [PubMed] [Google Scholar]
- 2. Abiko Y. 1967. Investigations on pantothenic acid and its related compounds. X. Biochemical studies. 5. Purification and substrate specificity of phosphopantothenoylcysteine decarboxylase from rat liver. J. Biochem. 61:300–308 [DOI] [PubMed] [Google Scholar]
- 3. Abiko Y., Tomikawa M., Shimizu M. 1968. Further studies on phosphopantothenoylcysteine synthetase. J. Biochem. 64:115–117 [DOI] [PubMed] [Google Scholar]
- 4. Anderson M. S., Bulawa C. E., Raetz C. R. 1985. The biosynthesis of gram-negative endotoxin. Formation of lipid A precursors from UDP-GlcNAc in extracts of Escherichia coli. J. Biol. Chem. 260:15536–15541 [PubMed] [Google Scholar]
- 5. Angus B. L., Carey A. M., Caron D. A., Kropinski A. M., Hancock R. E. 1982. Outer membrane permeability in Pseudomonas aeruginosa: comparison of a wild-type with an antibiotic-supersusceptible mutant. Antimicrob. Agents Chemother. 21:299–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baba T., et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Balibar C. J., Shen X., Tao J. 2009. The mevalonate pathway of Staphylococcus aureus. J. Bacteriol. 191:851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Begley T. P., Kinsland C., Strauss E. 2001. The biosynthesis of coenzyme A in bacteria. Vitam. Horm. 61:157–171 [DOI] [PubMed] [Google Scholar]
- 9. Beveridge T. J. 1981. Ultrastructure, chemistry, and function of the bacterial wall. Int. Rev. Cytol. 72:229–317 [DOI] [PubMed] [Google Scholar]
- 10. Brand L. A., Strauss E. 2005. Characterization of a new pantothenate kinase isoform from Helicobacter pylori. J. Biol. Chem. 280:20185–20188 [DOI] [PubMed] [Google Scholar]
- 11. Choudhry A. E., et al. 2003. Inhibitors of pantothenate kinase: novel antibiotics for staphylococcal infections. Antimicrob. Agents Chemother. 47:2051–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Daugherty M., et al. 2002. Complete reconstitution of the human coenzyme A biosynthetic pathway via comparative genomics. J. Biol. Chem. 277:21431–21439 [DOI] [PubMed] [Google Scholar]
- 14. Espinosa-Ruiz A., Belles J. M., Serrano R., Culianez-MacIa F. A. 1999. Arabidopsis thaliana AtHAL3: a flavoprotein related to salt and osmotic tolerance and plant growth. Plant J. 20:529–539 [DOI] [PubMed] [Google Scholar]
- 15. Gerdes S. Y., et al. 2002. From genetic footprinting to antimicrobial drug targets: examples in cofactor biosynthetic pathways. J. Bacteriol. 184:4555–4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Govan J. R., Deretic V. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guzman L. M., Belin D., Carson M. J., Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hancock R. E., Nikaido H. 1978. Outer membranes of gram-negative bacteria. XIX. Isolation from Pseudomonas aeruginosa PAO1 and use in reconstitution and definition of the permeability barrier. J. Bacteriol. 136:381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harrington C. R., Baddiley J. 1985. Biosynthesis of wall teichoic acids in Staphylococcus aureus H, Micrococcus varians and Bacillus subtilis W23. Involvement of lipid intermediates containing the disaccharide N-acetylmannosaminyl N-acetylglucosamine. Eur. J. Biochem. 153:639–645 [DOI] [PubMed] [Google Scholar]
- 20. Heath R. J., White S. W., Rock C. O. 2001. Lipid biosynthesis as a target for antibacterial agents. Prog. Lipid Res. 40:467–497 [DOI] [PubMed] [Google Scholar]
- 21. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59 [DOI] [PubMed] [Google Scholar]
- 22. Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., Schweizer H. P. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86 [DOI] [PubMed] [Google Scholar]
- 23. Jackowski S. 1996. Biosynthesis of pantothenic acid and coenzyme A, p. 687–694 In Neidhardt F. C., Curtiss R., Gross C. A., Ingraham J. L., Lin E. C. C., Low K. B., Magasanik B., Reznikoff W. S., Riley M., Schaechter M., Umbarger H. E. (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol. 1. American Society for Microbiology, Washington, DC [Google Scholar]
- 24. Jackowski S., Alix J. H. 1990. Cloning, sequence, and expression of the pantothenate permease (panF) gene of Escherichia coli. J. Bacteriol. 172:3842–3848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jackowski S., Rock C. O. 1984. Metabolism of 4′-phosphopantetheine in Escherichia coli. J. Bacteriol. 158:115–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kotnik M., Anderluh P. S., Prezelj A. 2007. Development of novel inhibitors targeting intracellular steps of peptidoglycan biosynthesis. Curr. Pharm. Des. 13:2283–2309 [DOI] [PubMed] [Google Scholar]
- 27. Kropinski A. M., Kuzio J., Angus B. L., Hancock R. E. 1982. Chemical and chromatographic analysis of lipopolysaccharide from an antibiotic-supersusceptible mutant of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 21:310–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kupke T., Hernandez-Acosta P., Culianez-Macia F. A. 2003. 4′-Phosphopantetheine and coenzyme A biosynthesis in plants. J. Biol. Chem. 278:38229–38237 [DOI] [PubMed] [Google Scholar]
- 29. Leonardi R., et al. 2005. A pantothenate kinase from Staphylococcus aureus refractory to feedback regulation by coenzyme A. J. Biol. Chem. 280:3314–3322 [DOI] [PubMed] [Google Scholar]
- 30. Leonardi R., Zhang Y. M., Rock C. O., Jackowski S. 2005. Coenzyme A: back in action. Prog. Lipid Res. 44:125–153 [DOI] [PubMed] [Google Scholar]
- 31. Levintow L., Novelli G. 1954. The synthesis of coenzyme A from pantetheine: preparation and properties of pantetheine kinase. J. Biol. Chem. 207:761–765 [PubMed] [Google Scholar]
- 32. Meier J. L., Mercer A. C., Rivera H. J., Burkart M. D. 2006. Synthesis and evaluation of bioorthogonal pantetheine analogues for in vivo protein modification. J. Am. Chem. Soc. 128:12174–12184 [DOI] [PubMed] [Google Scholar]
- 33. Mendelson M. H., et al. 1994. Pseudomonas aeruginosa bacteremia in patients with AIDS. Clin. Infect. Dis. 18:886–895 [DOI] [PubMed] [Google Scholar]
- 34. Mercer A. C., Meier J. L., Torpey J. W., Burkart M. D. 2009. In vivo modification of native carrier protein domains. Chembiochem 10:1091–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Poole K. 2005. Efflux-mediated antimicrobial resistance. J. Antimicrob. Chemother. 56:20–51 [DOI] [PubMed] [Google Scholar]
- 36. Poole K. 2004. Efflux-mediated multiresistance in Gram-negative bacteria. Clin. Microbiol. Infect. 10:12–26 [DOI] [PubMed] [Google Scholar]
- 37. Rana A., et al. 2010. Pantethine rescues a Drosophila model for pantothenate kinase-associated neurodegeneration. Proc. Natl. Acad. Sci. U. S. A. 107:6988–6993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rubio S., et al. 2006. An Arabidopsis mutant impaired in coenzyme A biosynthesis is sugar dependent for seedling establishment. Plant Physiol. 140:830–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 40. Schweizer H. D. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. Biotechniques 15:831–834 [PubMed] [Google Scholar]
- 41. Simon R., Priefer U., Puehler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Nat. Biotechnol. 1:784–791 [Google Scholar]
- 42. Strauss E., Begley T. P. 2002. The antibiotic activity of N-pentylpantothenamide results from its conversion to ethyldethia-coenzyme A, a coenzyme A antimetabolite. J. Biol. Chem. 277:48205–48209 [DOI] [PubMed] [Google Scholar]
- 43. Strauss E., Kinsland C., Ge Y., McLafferty F. W., Begley T. P. 2001. Phosphopantothenoylcysteine synthetase from Escherichia coli. Identification and characterization of the last unidentified coenzyme A biosynthetic enzyme in bacteria. J. Biol. Chem. 276:13513–13516 [DOI] [PubMed] [Google Scholar]
- 44. Thomas J., Cronan J. E. 2010. Antibacterial activity of N-pentylpantothenamide is due to inhibition of coenzyme A synthesis. Antimicrob. Agents Chemother. 54:1374–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vallari D. S., Rock C. O. 1985. Isolation and characterization of Escherichia coli pantothenate permease (panF) mutants. J. Bacteriol. 164:136–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vallari D. S., Rock C. O. 1987. Isolation and characterization of temperature-sensitive pantothenate kinase (coaA) mutants of Escherichia coli. J. Bacteriol. 169:5795–5800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vallari D. S., Rock C. O. 1985. Pantothenate transport in Escherichia coli. J. Bacteriol. 162:1156–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu Z., Li C., Lv S., Zhou B. 2009. Pantothenate kinase-associated neurodegeneration: insights from a Drosophila model. Hum. Mol. Genet. 18:3659–3672 [DOI] [PubMed] [Google Scholar]
- 49. Zhang Y. M., et al. 2004. Acyl carrier protein is a cellular target for the antibacterial action of the pantothenamide class of pantothenate antimetabolites. J. Biol. Chem. 279:50969–50975 [DOI] [PubMed] [Google Scholar]
- 50. Zhou B., et al. 2001. A novel pantothenate kinase gene (PANK2) is defective in Hallervorden-Spatz syndrome. Nat. Genet. 28:345–349 [DOI] [PubMed] [Google Scholar]
- 51. Zimmermann W. 1979. Penetration through the gram-negative cell wall: a co-determinant of the efficacy of beta-lactam antibiotics. Int. J. Clin. Pharmacol. Biopharm. 17:131–134 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.