Abstract
Swarming colonies of independent Proteus mirabilis isolates recognize each other as foreign and do not merge together, whereas apposing swarms of clonal isolates merge with each other. Swarms of mutants with deletions in the ids gene cluster do not merge with their parent. Thus, ids genes are involved in the ability of P. mirabilis to distinguish self from nonself. Here we have characterized expression of the ids genes. We show that idsABCDEF genes are transcribed as an operon, and we define the promoter region upstream of idsA by deletion analysis. Expression of the ids operon increased in late logarithmic and early stationary phases and appeared to be bistable. Approaching swarms of nonself populations led to increased ids expression and increased the abundance of ids-expressing cells in the bimodal population. This information on ids gene expression provides a foundation for further understanding the molecular details of self-nonself discrimination in P. mirabilis.
INTRODUCTION
The gammaproteobacterium Proteus mirabilis is a leading cause of recurrent urinary tract infections and displays some extraordinary behaviors (15). P. mirabilis undergoes a morphologically distinct developmental cycle during growth on surfaces (reviewed in references 8, 15, and 18). Upon contact with a surface, flagellated rod-shaped swimmer cells (1 to 2 μm in length) differentiate into hyperflagellated, polynucleoid, elongated cells (10 to 80 μm in length). Colonies can swarm across surfaces rapidly, and the elongated cells, called swarmer cells, do not divide during migration. Expression of many virulence genes appears to be linked to swarmer cell differentiation, and there is a distinction between genes expressed during liquid growth and those expressed during surface-associated growth (1, 2, 5, 6, 16, 18). The migrating front of a swarm will periodically arrest movement; cells will dedifferentiate to short cells and consolidate through cell division. After another round of differentiation, swarmer cells will migrate forward once again. This behavior results in colonies with a bull's-eye-like pattern (reviewed in references 8 and 18).
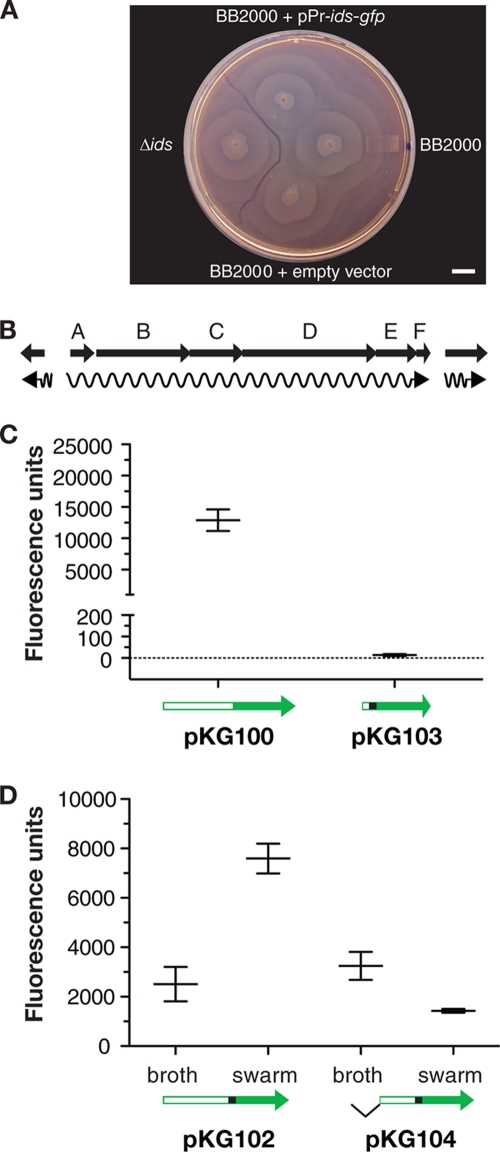
Of particular interest to us, a boundary will form between approaching swarms of different strains but not between two swarms of a single strain (Fig. 1A). The boundary formation demonstrates that P. mirabilis populations have an ability to distinguish self from nonself. We recently identified a gene cluster, idsABCDEF, involved in P. mirabilis self versus nonself recognition (10). Swarms of Ids mutants form a boundary with their parent. Two of the genes, idsD and idsE, encode functions for determining strain-specific identity. Of the remaining genes, idsB, idsC, and idsF are required for self versus nonself recognition but are not identity determinants. The idsA gene is not required for recognition, but it has been termed an ids gene because idsA is possibly cotranscribed with idsBCDEF. There is no available information on ids gene expression.
Fig. 1.
Proteus mirabilis territorial boundaries, the ids locus, and transcription from the idsA promoter. (A) Swarm plate of the wild-type strain BB2000, BB2000 idsABCDEF deletion mutant (Δids), and BB2000 with the idsA-gfp vector pKG100 (labeled pPr-ids-gfp) or with an empty vector. A visible boundary formed between swarms of the wild type and the deletion mutant. Swarms of the wild type merged regardless of the presence of pKG100. The bar shown is 1 cm. (B) Organization of the P. mirabilis idsABCDEF locus. The arrows indicate open reading frames. The region between idsA and the diverging open reading frame (ORF) is 1,899 bp. The distance between the idsA and idsB ORFs is 73 bp. The distance between idsF and the downstream ORF is 180 bp. (C) Fluorescence of P. mirabilis cells containing the idsA promoter-gfp expression vector pKG100 or pKG104, which contains the idsAB intergenic region with gfp fused to idsB. Cells were harvested at a culture density of 4 to 5. (D) P. mirabilis BB2000 containing the idsA-gfp vector pKG102, which contains 788 bp upstream of idsA, or pKG104, which contains only 435 bp upstream of idsA, were grown either in broth or on swarm-agar plates. Broth-grown cells were obtained at a culture density of 4 to 5. Swarm cells were obtained from agar plates after 24 h (at this time active swarming had ceased). Cells were suspended in PBS, fluorescence and optical density were measured, and fluorescence units were normalized to the optical density. Bars indicate the means with standard deviations. GFP fluorescence units were normalized to optical density and measured as described in the Materials and Methods.
Bacterial colonies can be considered either as populations of cells or as populations of individual members of a species. The ability to distinguish self from nonself is a behavior conserved in both cellular and organismal population dynamics. Territoriality, a social behavior, is conserved among many organisms and is often driven by competition for resources, competition for mates, or the need to segregate genetic pools (22). We view swarm colony boundary formation as an example of territoriality. Because we have begun to identify genetic determinants for the self-recognition required for this territoriality, we believe P. mirabilis can serve as a model to help understand the molecular mechanisms of recognition and territoriality and can provide insights about their evolution. To better develop this model, we initiated a study of ids gene expression. Here we show that idsABCDEF constitute an operon, that expression of this operon results in a bimodal pattern consistent with bistability, and that this expression positively correlates with increasing cell densities. We also provide evidence that ids gene expression is altered by an approaching swarm of a strain recognized as nonself.
MATERIALS AND METHODS
Bacterial strains and growth media.
We used Escherichia coli Top10 (Invitrogen), E. coli S17-1λpir (7), P. mirabilis BB2000 (3), P. mirabilis Δids (10), and P. mirabilis BB2000 constitutively expressing gfp (10). We used LB-Lennox (LB) broth supplemented with 1 mM MOPS (morpholinepropanesulfonic acid) buffer for growth of P. mirabilis swimmer cells. The media used for P. mirabilis colonial growth were either CM55 blood agar base (Remel Inc., Lenexa, KS) for swarm colony growth or low-swarm (LSW−) agar for isolation of single colonies (3). E. coli was grown in LB broth and on LB agar. Antibiotics were used as follows: kanamycin (Km) at 35 μg/ml; rifampin at 100 μg/ml; and chloramphenicol at 35 μg/ml for E. coli and 75 μg/ml for P. mirabilis. All media contained antibiotics appropriate for selection or maintenance of plasmids.
Plasmid constructions.
Our idsA promoter reporter vector pKG100 was constructed by digesting pidsBBΔABC (10) with NheI and HincII and ligating the ids promoter-containing digestion product to pPROBE-GFP-AT digested with SpeI and EcoRV (14). The resulting pKG100 construct contains idsA at bp −788 to −1 fused to gfp. To construct the promoterless gfp control vector, pKG101, gfp was PCR amplified from pPROBE-GFP-AT by using a primer complementary to the first nine codons of gfp and an NheI restriction site tail and a primer complementary to the last nine codons of gfp with an AgeI restriction site tail. The PCR product was digested with NheI and AgeI and ligated to similarly digested pBBR1-NheI (10) to yield pKG101. The idsA-gfp fusion vector pKG102 was constructed from a PCR product consisting of the 788-bp region upstream of idsA and the first three codons of idsA amplified from pidsBB (10). The PCR primers were designed to have NheI and SacI restriction site tails. After restriction enzyme treatment, the PCR product was ligated to NheI- and SacI-digested pKG101 to form pKG102, which contained gfp fused in frame with the first nine codons of idsA and the idsA upstream promoter region. To test whether the DNA between the idsA and idsB coding regions exhibited detectable promoter activity, we generated pKG103, which contained the complete 72-bp intergenic region and the first 3 codons of idsB fused in frame with gfp from pKG101. As described above, the PCR product was generated using primers to the desired region with NheI and SacI restriction site tails. The PCR product was ligated into NheI-SacI-digested pKG101 to form pKG103. We constructed pKG104, a pKG100 deletion derivative by removal of bp −788 to −435 with respect to the idsA translation start site via excision of the 353-bp NheI-XbaI fragment.
In all cases, plasmids were isolated from ligation mixtures by transformation of E. coli Top10. Transformants were selected on Km-agar plates. Constructs were confirmed by DNA sequencing of the inserted region (SeqWright DNA Technology Services, Houston, TX). Plasmids were moved to E. coli S17-1λpir by transformation and then moved from E. coli S17-1λpir to P. mirabilis BB2000 by conjugation. Transconjugants were selected on LSW− plates containing Km and rifampin.
Reverse transcription-PCR (RT-PCR).
Wild-type P. mirabilis strain BB2000 was grown to late logarithmic phase in LB broth. Cells were harvested and stored at −20°C. After RNA extraction, contaminating DNA was digested with RQ DNase (Promega Co., Madison, WI) and removed by using an RNeasy kit and the RNA Clean-Up protocol (Qiagen Inc., Valencia, CA). cDNA was generated by using a SuperScript III first-strand synthesis kit (Invitrogen Co., Carlsbad, CA) with either random hexamer or oligo(dT) primers. Reaction mixtures in which reverse transcriptase was omitted served as controls for DNA contamination. PCR was performed by using Taq polymerase (Invitrogen Co.) and gene-specific primers. Samples were isolated after 35 cycles and examined by agarose gel electrophoresis.
Imaging swarming P. mirabilis colonies.
CM55 agar plates were inoculated from stationary-phase cultures with an inoculation needle at a spacing of approximately 2 cm. Images were captured on an Optio W10 digital camera (Pentax Imaging Company, Westminster, CO) after overnight incubation at 37°C. False-colored microscope images were prepared by overlaying the fluorescence channel as green and the phase channel as red in Adobe Photoshop CS5 (Adobe Systems, San Jose, CA). Image contrast was equally increased across the entire raw image, and images were then cropped to size. Microscopy was performed as described previously (10).
Fluorescence measurements.
For liquid cultures, cells were grown aerobically in LB-Lennox broth at 37°C. Cells were harvested by centrifugation, washed, and then suspended in phosphate-buffered saline (PBS). For cells isolated from swarms, P. mirabilis was permitted to form swarms on CM55 blood agar media overnight at 37°C as described above. Cells were scraped from colonies with a wooden dowel and suspended in 500 μl PBS. The cell suspension was centrifuged, and the cell pellet was washed with PBS and finally suspended in 500 μl PBS. Samples (200 μl) were dispensed into wells of a sterile Costar 96-well plate (Corning Inc., Lowell, MA). The optical density (OD) at 595 nm (for culture density) and fluorescence at 485/535 nm in each well were measured by using a Tecan GENios Pro-Basic microplate reader (Tecan Group Ltd., Männedorf, Switzerland). Fluorescence measurements of an entire swarm were performed in Costar 6-well plates. All swarm well data consist of at least three experiments. Each well contained 5 ml of CM55 agar. Where indicated, we placed a sterile 0.22-μm filter across the middle of the well. Agar-containing wells were inoculated as described above. After an overnight incubation at 37°C, absorbance and fluorescence in wells were measured as described above. Background fluorescence (uninoculated medium) was subtracted, and the data are presented as fluorescence values normalized to absorbance.
Microscopy.
Microscopy of swarms was performed as described previously (10). Cells grown in broth were harvested, washed with PBS, mounted onto glass slides, and imaged immediately. We measured fluorescence intensity by using MetaMorph 6.3r2 (Molecular Devices, Sunnyvale, CA), and subsequent analysis was performed with Microsoft Excel 2004 (Microsoft Co., Redmond, WA) and GraphPad Prism 5.0b (GraphPad Software, Inc., La Jolla, CA). For analysis of single cells from broth cultures, the Integrated Morphometry Analysis function of MetaMorph was used to identify and measure the physical dimensions of individual cells in phase images. The average, minimum, and maximum intensities of each identified cell from fluorescence images were then measured. The slide background fluorescence intensity was subtracted from the total cellular intensity. The average intensities of individual cells were sorted to study the variation in idsA-gfp expression from cell to cell. For image analysis in swarms, we used the Linescan function in MetaMorph to measure the average and maximum intensities of pixels from one point to another across the image, specifically from a region inside the swarm of the reporter strain, across the boundary or merger region, and into the swarm of the apposing strain. Each recorded pixel intensity consisted of the mean, or maximum, intensity of a 1-pixel-wide by 20-pixel-tall slice, which corresponded to a 0.65-μm by 13-μm slice through a swarm. We report the averages from multiple images.
RESULTS
The idsABCDEF genes constitute an operon.
The organization of idsABCDEF and the flanking genes is shown in Fig. 1B. Our previous studies showed that idsB, idsC, idsD, idsE, and idsF were involved in self versus nonself recognition, but idsA mutants did not have a self versus nonself recognition defect (10). This taken together with the facts that the adjacent stop and start codons overlap in the five other genes, idsB, idsC, idsD, idsE, and idsF, and that there is a 72-bp noncoding region between idsA and idsB suggest that idsA might be transcribed independently from the other genes (10, 17). We analyzed mRNA extracted from P. mirabilis wild-type strain BB2000 by reverse transcription-PCR (RT-PCR) with primers that spanned adjacent genes. Our results showed that idsABCDEF, but not the genes flanking idsABCDEF, were on a single transcript. That is, primers spanning intergenic regions between each gene in the ids cluster yielded PCR products of the predicted sizes. There was no detectable product with primers spanning the region between idsA and the gene upstream of it or idsF and the gene downstream, nor was there any product in reaction mixtures in which reverse transcriptase was omitted (data not shown).
It is nevertheless conceivable that there is a transcription start site in the region upstream of idsA and a secondary site in the intergenic region between idsA and idsB. To address this possibility, we constructed a transcriptional reporter plasmid containing the 788-bp region between idsA and the adjacent upstream gene fused to gfp (pKG100). We also constructed a plasmid containing the idsA-idsB intergenic region (72 bp) and the first 9 bp of idsB fused to gfp (pKG103). We introduced these plasmids into our wild-type strain P. mirabilis BB2000 and measured green fluorescent protein (GFP) expression. The plasmid containing the region upstream of idsA (pKG100) directed significant GFP expression, whereas pKG103 with gfp fused to the idsA-idsB intergenic region did not direct appreciable GFP expression (Fig. 1C). We conclude that expression of all of the ids genes is entirely dependent on a promoter or promoters upstream of idsA.
To examine the promoter region more closely, we constructed two plasmids, one with the complete 788-bp region upstream and the first nine base pairs of idsA-gfp (pKG102) and the other with only the proximal 435-bp region upstream and the first nine base pairs of idsA (pKG104). The GFP expression directed by each plasmid in P. mirabilis BB2000 was measured (Fig. 1D). Cells grown in broth (i.e., swimmer cells) showed similar levels of idsA-gfp expression from either promoter construct. In swarms, higher levels of idsA-gfp expression occurred with the complete idsA upstream region than with the −435 to +9 region (Fig. 1D). These data suggest the possibility of surface-contact-dependent expression of the P. mirabilis ids operon, but further work is required to address this question. Regardless, it appears that the minimal element for idsA transcription resides within a region extending 435 bp upstream of the idsA translation start site.
Bimodal and density-correlated expression of the ids operon.
Examination of the edge of an advancing swarm of P. mirabilis containing pKG100 revealed a mixture of bright and dark individual cells (Fig. 2A). Cells at the advancing edge of a swarm tended to move in small packs; both bright and dark cells were often in an individual pack. In contrast, dark cells were only rarely observed in swarms of P. mirabilis constitutively expressing gfp; a relatively consistent expression level of fluorescence was observed across the population (Fig. 2B). The two modes of fluorescence in the P. mirabilis (pKG100) swarms, bright and dark, suggest that in advancing swarms of a single strain, only a subset of cells expresses the ids genes and therefore is capable of sensing self. As such, when two separate swarms of a single strain approach, only a subset of cells in either of the swarms is capable of sensing self and presumably initiating a merger with the apposing swarm.
Fig. 2.
Migrating fronts of P. mirabilis swarms. (A) Bimodal expression of the pKG100 idsA-gfp fusion in cells of P. mirabilis at the outer edge of a swarm. The phase image is shown on the left, the fluorescence image is in the center, and the false-colored image is on the right and shows the agar background in red, fluorescence bright cells in green, and dark cells in black. Asterisks mark examples of dark cells, and arrowheads indicate swarm tracks. (B) Control. Outer swarm edge of BB2000 cells carrying a chromosomally integrated constitutive gfp (10). Asterisks mark dim cells (dark cells were not evident), and arrowheads indicate swarm tracks. The marker bars shown are 50 μm.
For several reasons, it proved difficult to obtain quantitative information on the bimodal distribution of bright and dark cells in swarming populations. Cells that are not in the leading edge of the swarm are irregularly shaped, cells move in packs and are closely aligned along each other, and the developmental stage at the edge of a swarm is variable (consolidation rings versus advancing swarms). Therefore, we sought to determine whether swimmer cells grown in broth exhibited two modes of fluorescence expression. In fact, broth-grown cells did exhibit a bimodal pattern of ids gene expression (Fig. 3A). The relative abundance of dark and bright cells changed as culture density increased. At optical densities below 0.8, dark cells predominated, while at those above 0.8, bright cells were dominant (Fig. 3A and B). This shift toward a higher percentage of idsA-gfp-expressing cells at densities above 0.8 was reflected in overall culture fluorescence measurements. When normalized to optical density, culture fluorescence decreased after inoculation until the optical density reached above 0.4 to 0.5, and then the fluorescence per unit of cell mass increased (Fig. 3C). A similar relationship between culture density and gfp expression was observed when we examined cells containing pKG102, with idsA fused in frame to gfp. Furthermore, expression of gfp in cells containing pKG104 with the idsA −435 to +9 region-gfp showed bimodal and cell density-related expression patterns similar to those described for constructs with the complete 788-bp idsA upstream region (data not shown). As a control, we show that cells with plasmid-borne constitutive gfp fluorescence do not exhibit bimodality (see Fig. S1 in the supplemental material). Thus, our evidence supports the view that ids gene expression is bimodal and cell density dependent in broth-grown swimmer cells or in swarmer cells on an agar surface.
Fig. 3.
The idsA promoter shows a bimodal and culture density-dependent pattern of expression. (A) Histograms of individual cell fluorescence intensity in an LB broth culture at different cell densities (OD at 600 nm). We measured fluorescence of 135 individual cells at an OD of 0.13, 255 cells at an OD of 0.60, 419 cells at an OD of 1.63, and 728 cells at an OD of 4.10. Cells with fluorescence levels below 100 are dark or very dim. (B) Percentage of LB broth-grown P. mirabilis BB2000 (pKG100) cells expressing idsA-gfp in relation to culture density. (C) Expression of idsA as a function of culture density in LB broth. Fluorescence units are normalized to the optical density.
Influence of an approaching swarm on ids expression.
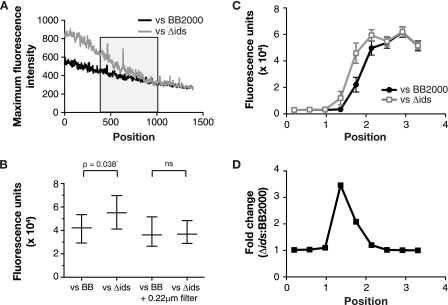
We next sought to determine whether an approaching P. mirabilis swarm, either self or nonself, influenced expression of the ids operon by analyzing cellular fluorescence in the wild-type strain BB200 carrying the idsA-gfp expression vector pKG102 at swarm edges that interface with either BB2000 (self) or an ids deletion mutant (Δids) derived from BB2000 (nonself). In a first analysis, we measured the fluorescence intensity of the brightest pixel in slices of 13 μM by 0.65 μm that extended across the region where two swarms either merged or formed a boundary (Fig. 4A). As expected, when strain BB2000 with the idsA-gfp vector pKG102 approached self (BB2000 with the backbone plasmid), the brightest pixel in a slice decreased progressively. A similar pattern was observed when the gfp-tagged BB2000 approached nonself (the ids deletion strain), except that fluorescence was higher in every slice through the boundary and receded into the denser regions of the swarm of gfp-tagged cells. This higher level of fluorescence suggested that the approach of and interaction with nonself somehow induced high-level idsA expression in at least a subset of cells. This induction may result from contact with a nonself cell that has migrated across the boundary, which we know occurs from our previous work (10) and for which there is evidence shown in Fig. 3A; we observed some slices with a bright pixel in the boundary and on the nonself side of the boundary.
Fig. 4.
Effects of social recognition on ids expression. (A) Maximum fluorescence intensity averaged across swarm boundaries. Boundaries are between P. mirabilis BB2000 containing the idsA-gfp vector pKG102 and either BB2000 or the ids deletion mutant containing the vector control pKG101 (to allow antibiotic maintenance of the plasmids in all bacteria). We measured fluorescence in 0.65-μm by 13-μm intervals. The “0” position is in the swarm of BB2000 (pKG102) and the “1500” position is in the swarm of the vector-containing bacteria. The black box indicates the location where swarms merged or where the boundary formed. Background fluorescence in dark colonies is about 200 units. (B) Total fluorescence of P. mirabilis BB2000 (pKG102). The swarms were approaching either BB2000 containing the vector control pKG101 or the ids deletion strain carrying pKG101. Where indicated, a 0.22-μm filter divided the approaching swarms. The data are normalized to the optical density. The boxes show ranges, and the horizontal lines denote the means. There was a significant difference without the filter and no significant difference (ns) with the filter. (C) Fluorescence of P. mirabilis BB2000 (pKG102) calculated as described for panel B. Fluorescence, normalized to the optical density, was measured at discrete points along the swarm and was plotted such that the x axis measures the distance from one edge of a well to the other. The swarms met between 1 cm and 2.2 cm from the edge of the well. Markers denote means, and bars show standard errors of the means. (D) Ratios (Δids/BB2000) of the data shown in panel C, plotted as a function of the distance from the edge of the well.
We next wanted to obtain additional evidence that migration toward a swarm recognized as nonself enhanced expression of the ids operon and to ask whether this phenomenon might require contact between cells from the two advancing swarms. To do this, we used six-well plates and a fluorescence plate reader. Wells contained agar on which swarms of gfp-tagged BB2000 and swarms of either BB2000 or the ids deletion derivative moved toward each other. In some wells, we placed a piece of a 0.22-μm filter standing on the edge into the agar between the two swarms. The filters allowed diffusion of small soluble molecules but did not allow cell passage. Consistent with a previous report that cell-cell contact is required for boundary formation (4), boundaries formed between self and nonself swarms in wells without a filter but not between swarms divided by a filter. Without a dividing filter, the fluorescence was significantly higher in wells with gfp-tagged BB2000 and the nonself ids deletion mutant than in wells with gfp-tagged BB2000 and BB2000 (Fig. 4B). The increased fluorescence due to interactions with nonself cells occurred primarily at the boundary, specifically at the swarm edge that directly interacted with the apposing swarm (Fig. 4C); here there was a 3-fold increase in fluorescence when nonself cells were present (Fig. 4D). These findings support the hypothesis that migration toward a swarm recognized as nonself enhances expression of the ids operon. Furthermore, in wells with a dividing filter, self or nonself did not differently influence the fluorescence (Fig. 4B). These findings indicate that cell-cell contact or a nondiffusible factor is not only required for boundary formation but is also required for the nonself stimulation of ids expression.
DISCUSSION
An important step in developing an understanding of how the P. mirabilis ids genes specify self-identity involves learning about ids expression. Here we show that the ids genes are transcribed as a single unit. This includes idsA, which itself is not a required identity determinant (10). Transcription of the ids operon depends on a promoter upstream of idsA, and the minimal promoter element resides within the adjacent 435 bp upstream of the predicted idsA translational start site. There may be additional regulatory sequences further upstream that are required for surface stimulation of ids gene expression (Fig. 1D). The question of whether ids transcription is stimulated by contact with an agar surface is interesting and deserves further study. We note that surface contact induces P. mirabilis swimmer cells to differentiate into swarmer cells and that this differentiation involves the master motility regulator FlhDC (9, 11).
When we examined ids gene expression at a cellular level by using P. mirabilis containing an idsA-gfp plasmid and fluorescence microscopy, there was obvious bimodal expression of ids promoter activity, which continued to be present after several passages (data not shown), suggesting that there might be a bistable switch controlling ids transcription (Fig. 2 and 3). This was true when we examined cells from broth-grown populations or cells swarming on agar plates. It remains to be determined if the readout from our plasmid-carried idsA-gfp construct reflects expression from the native chromosomal ids operon, but we have no reason to believe it does not.
Our data also indicate that ids expression increases when broth cultures grow past a critical density (Fig. 3), and visual analysis suggests that there might be higher ids expression in more dense areas of the swarms (data not shown). How the apparent cell density-dependent and bistable expression of the ids operon is involved in self versus nonself discrimination is not clear. In considering these aspects of ids regulation, it is important to remember that there are likely other as-yet-unidentified genes involved in recognition (10). It is also important to remember that all of our data on bistability and cell density-dependent expression come from studies of the idsA promoter on a plasmid we have introduced into P. mirabilis. Finally, it is prudent to keep in mind that in addition to the formation of the boundaries between isogenic strains, which we have studied here, there can be proticine-dependent killing within the boundaries that form between two different clinical isolates of P. mirabilis (19). We have dissected only one part of a complex set of behaviors occurring when swarms of two independent P. mirabilis isolates approach each other.
Given the limitations described in the previous paragraph, there are nevertheless some interesting implications that can be drawn from our experiments. First, our evidence is consistent with the idea that there are more cells expressing the ids genes in the interior regions of a swarm where the cells are at high density in multiple layers than at the periphery of a swarm where cells exist as small monolayered packs. Perhaps bistable expression of ids genes in some way enables alternative responses in cells at the advancing edge of a swarm. This idea is congruous with previously described functions of bistable switches; although not commonly encountered, they are known to regulate a variety of developmental processes, including Bacillus subtilis sporulation, competence, and motility (12, 13, 20). Bistability is thought to aid in population bet-hedging against uncertain environments (21). One uncertainty for an advancing swarm is whether it will encounter another advancing swarm of foreigners that can compete for resources and probably has a killing capability through proticine production or whether it will encounter fresh nutritional resources.
We present evidence that ids-expressing cells can traverse an established boundary and comingle with cells in a foreign population (Fig. 4A). We also present evidence that there is a higher level of ids expression in cells approaching a foreign swarm than in cells approaching a swarm recognized as self (Fig. 4). Enhancement of ids expression by a foreign swarm also appears to be dependent upon cell-cell contact or contact with a location that has been traversed by a cell or cells from a foreign swarm (Fig. 4B). In fact, P. mirabilis leaves visible tracks on the surface (Fig. 2). The enhanced ids expression in cells at the edge of a swarm boundary with a foreign swarm might be a response to foreigners that have traversed the boundary.
Though we believe that this analysis is an important step in developing a comprehensive view of how the ids genes serve as a self versus nonself discrimination system and that it provides important information that will need to be integrated into a molecular model for self versus nonself discrimination in P. mirabilis, it also raises many more questions than it answers. Is the observed surface stimulation of ids gene expression important, and what is the mechanism of stimulation? Is the increase in ids gene expression at high cell densities important for boundary formation? What is the mechanistic basis of the apparently bistable ids expression switch, and is this critical for self versus nonself swarm discrimination? Is cell-cell contact really required for development of boundaries, and if so, how is the information of self versus nonself transferred from one cell to the other?
Supplementary Material
ACKNOWLEDGMENTS
K.A.G. was supported by NIH training grant AI55396. The research was partially supported by USPHS grant GM-59026 to E.P.G.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 6 May 2011.
REFERENCES
- 1. Allison C., Coleman N., Jones P. L., Hughes C. 1992. Ability of Proteus mirabilis to invade human urothelial cells is coupled to motility and swarming differentiation. Infect. Immun. 60:4740–4746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allison C., Emody L., Coleman N., Hughes C. 1994. The role of swarm cell differentiation and multicellular migration in the uropathogenicity of Proteus mirabilis. J. Infect. Dis. 169:1155–1158 [DOI] [PubMed] [Google Scholar]
- 3. Belas R., Erskine D., Flaherty D. 1991. Transposon mutagenesis in Proteus mirabilis. J. Bacteriol. 173:6289–6293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Budding A. E., Ingham C. J., Bitter W., Vandenbroucke-Grauls C. M., Schneeberger P. M. 2009. The Dienes phenomenon: competition and territoriality in swarming Proteus mirabilis. J. Bacteriol. 191:3892–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burall L. S., et al. 2004. Proteus mirabilis genes that contribute to pathogenesis of urinary tract infection: identification of 25 signature-tagged mutants attenuated at least 100-fold. Infect. Immun. 72:2922–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Coker C., Poore C. A., Li X., Mobley H. L. 2000. Pathogenesis of Proteus mirabilis urinary tract infection. Microbes Infect. 2:1497–1505 [DOI] [PubMed] [Google Scholar]
- 7. de Lorenzo V., Cases I., Herrero M., Timmis K. N. 1993. Early and late responses of TOL promoters to pathway inducers: identification of postexponential promoters in Pseudomonas putida with lacZ-tet bicistronic reporters. J. Bacteriol. 175:6902–6907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fraser G. M., Hughes C. 1999. Swarming motility. Curr. Opin. Microbiol. 2:630–635 [DOI] [PubMed] [Google Scholar]
- 9. Furness R. B., Fraser G. M., Hay N. A., Hughes C. 1997. Negative feedback from a Proteus class II flagellum export defect to the flhDC master operon controlling cell division and flagellum assembly. J. Bacteriol. 179:5585–5588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gibbs K. A., Urbanowski M. L., Greenberg E. P. 2008. Genetic determinants of self identity and social recognition in bacteria. Science 321:256–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hay N. A., Tipper D. J., Gygi D., Hughes C. 1997. A nonswarming mutant of Proteus mirabilis lacks the Lrp global transcriptional regulator. J. Bacteriol. 179:4741–4746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kearns D. B., Losick R. 2005. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev. 19:3083–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Maamar H., Dubnau D. 2005. Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol. Microbiol. 56:615–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miller W. G., Leveau J. H., Lindow S. E. 2000. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol. Plant Microbe Interact. 13:1243–1250 [DOI] [PubMed] [Google Scholar]
- 15. Mobley H. L., Belas R. 1995. Swarming and pathogenicity of Proteus mirabilis in the urinary tract. Trends Microbiol. 3:280–284 [DOI] [PubMed] [Google Scholar]
- 16. Pearson M. M., Rasko D. A., Smith S. N., Mobley H. L. 2010. Transcriptome of swarming Proteus mirabilis. Infect. Immun. 78:2834–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pearson M. M., et al. 2008. Complete genome sequence of uropathogenic Proteus mirabilis, a master of both adherence and motility. J. Bacteriol. 190:4027–4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rather P. N. 2005. Swarmer cell differentiation in Proteus mirabilis. Environ. Microbiol. 7:1065–1073 [DOI] [PubMed] [Google Scholar]
- 19. Senior B. W. 1977. Typing of Proteus strains by proticine production and sensitivity. J. Med. Microbiol. 10:7–17 [DOI] [PubMed] [Google Scholar]
- 20. Veening J. W., Hamoen L. W., Kuipers O. P. 2005. Phosphatases modulate the bistable sporulation gene expression pattern in Bacillus subtilis. Mol. Microbiol. 56:1481–1494 [DOI] [PubMed] [Google Scholar]
- 21. Veening J. W., et al. 2008. Bet-hedging and epigenetic inheritance in bacterial cell development. Proc. Natl. Acad. Sci. U. S. A. 105:4393–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wilson E. O. 2000. Sociobiology: the new synthesis, 25th anniversary ed. Belknap Press of the Harvard University Press, Cambridge, MA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.