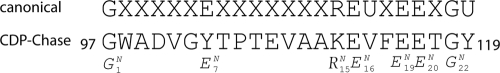Abstract
A Nudix enzyme from Bacillus cereus (NCBI RefSeq accession no. NP_831800) catalyzes the hydrolysis of CDP-choline to produce CMP and phosphocholine. Here, we show that in addition, the enzyme has a 3′→5′ RNA exonuclease activity. The structure of the free enzyme, determined to a 1.8-Å resolution, shows that the enzyme is an asymmetric dimer. Each monomer consists of two domains, an N-terminal helical domain and a C-terminal Nudix domain. The N-terminal domain is placed relative to the C-terminal domain such as to result in an overall asymmetric arrangement with two distinct catalytic sites: one with an “enclosed” Nudix pyrophosphatase site and the other with a more open, less-defined cavity. Residues that may be important for determining the asymmetry are conserved among a group of uncharacterized Nudix enzymes from Gram-positive bacteria. Our data support a model where CDP-choline hydrolysis is catalyzed by the enclosed Nudix site and RNA exonuclease activity is catalyzed by the open site. CDP-Chase is the first identified member of a novel Nudix family in which structural asymmetry has a profound effect on the recognition of substrates.
INTRODUCTION
Nudix hydrolases, divalent metal (Mg2+ or Mn2+)-requiring enzymes that hydrolyze nucleoside diphosphates linked to some other moiety, x, are present in viruses and prokaryotic and eukaryotic organisms of all levels of metabolic complexity (3, 24). All members of the Nudix family share the Nudix signature sequence GX5EX7REUXEEXGU, where X is any amino acid and U is a hydrophobic amino acid, typically isoleucine, leucine, or valine (Fig. 1) (3). The Nudix superfamily exhibits a great deal of functional diversity. In addition to helping control the concentrations of crucial metabolites (e.g., ADP-ribose [12, 19], UDP-glucose [46], and coenzyme A [4]), members of the superfamily also play key roles in biosynthetic pathways (e.g., folate biosynthesis [13]). It has been shown that some members of the Nudix family, i.e., Schizosaccharomyces pombe Dcp2 (31) and Escherichia coli and Bdellovibrio bacteriovorax RppH (7, 23), may also play a role in RNA degradation by decapping mRNA.
Fig. 1.
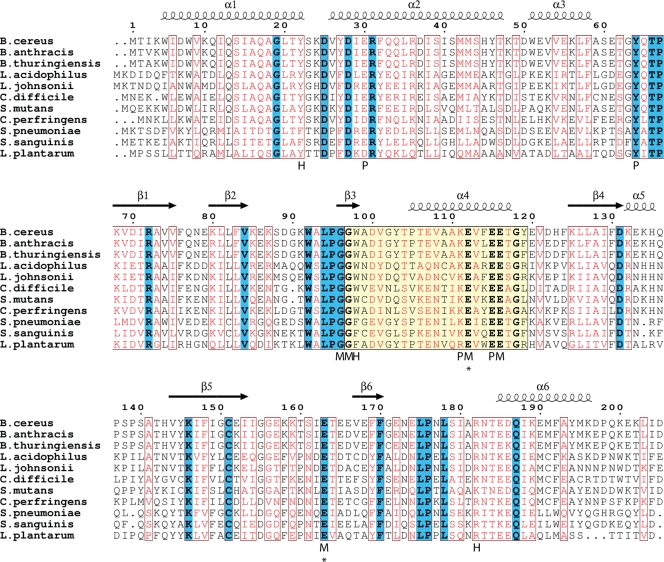
The Nudix signature sequence of B. cereus CDP-Chase. Conserved positions are shown in the canonical sequence. The nomenclature for these positions is shown below the CDP-Chase sequence in the form XjN, where X is the residue identity, N refers to the Nudix motif, and j refers to the placement in the Nudix signature sequence. Two exceptions to the canonical Nudix signature sequence are observed: E7N is a Tyr and R15N is a Lys in CDP-Chase. In the canonical sequence, X represents any amino acid and U represents a hydrophobic amino acid.
Although broad structural diversity is the basis for the functional diversity of members of the superfamily, all Nudix enzymes share a Nudix domain consisting of a mixed β-sheet of 4 strands, a 2-stranded antiparallel β-sheet, an α-helix to one side of the mixed β-sheet, and two α-helices on the other side of the same β-sheet. Nudix enzymes may include, in addition to the Nudix domain, other domains responsible for additional functionalities. For example, NadM-Nudix, a bifunctional enzyme involved in NAD biosynthesis and salvage pathways, contains two domains, an N-terminal Nudix domain and a C-terminal nicotinamide mononucleotide adenyltransferase domain (15). In other members of the superfamily, such as ADP-ribose pyrophosphatase (ADPRase), the addition of a second domain may also facilitate substrate recognition of the Nudix substrate (11). There are also examples in which the Nudix domain is catalytically inactive but binding of a Nudix substrate regulates the function of another domain. For example, the transcriptional regulator NrtR contains an N-terminal Nudix domain, as well as a C-terminal winged helix-turn-helix domain involved in DNA binding. Binding of ADP-ribose, 2′-phospho-ADP-ribose, or ADP by the Nudix domain relieves repression by the C-terminal domain of the prs-nadV operon which plays a role in NAD biosynthesis and salvage (14, 29). The Nudix domain may also serve as a scaffold, facilitating interactions with RNA or with other proteins in large complexes. Besides RppH and other decapping enzymes, such as Dcp2, which perform a Nudix reaction on the 5′ cap of mRNA, other enzymes that interact with RNA, such as human cleavage and polyadenylation specific factor-5 and cleavage factor Im-25, have a Nudix domain but do not perform a Nudix reaction on the RNA (6, 43). Due to this vast diversity in the Nudix superfamily, novel Nudix functions are still being discovered and few guidelines exist for the prediction of function directly from sequence information.
The anthrax pathogen Bacillus anthracis contains the highest number of Nudix genes of any sequenced organism (45). To take advantage of this extensive repertoire, the Nudix enzymes from the closely related but less pathogenic Bacillus cereus were cloned and expressed as a way of discovering enzymes that act on novel Nudix substrates. One of those enzymes is a CDP-choline pyrophosphatase (45) that we are calling CDP-Chase. The Nudix signature sequence of this enzyme is mutated from that of typical Nudix enzymes (Fig. 1). These changes affect a salt bridge present in the structures of other Nudix enzymes between residues E7N and R15N of the Nudix signature sequence (Fig. 1) (45). To gain insight into the structural and functional consequences of these differences, as well as into the relationship between sequence and specificity, we investigated the kinetics and determined the crystal structure of the B. cereus CDP-Chase. In an effort to relate enzymatic activity to function, we also determined the subcellular localization of CDP-Chase.
MATERIALS AND METHODS
Wild-type protein expression and purification.
B. cereus CDP-Chase was cloned into a pET-24a vector as described previously (45). Plasmid DNA was used to transform E. coli BL21(DE3) cells. Cells were grown at 37°C in LB medium supplemented with 30 μg/ml kanamycin to an optical density of 0.8 at 600 nm, induced with 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), and grown at 15°C overnight. Cells were harvested via centrifugation, resuspended in 50 mM Tris-HCl, pH 7.5, 1 mM EDTA, and 0.1 mM dithiothreitol (DTT) (TED), and frozen at −80°C. Following thawing, cells were lysed by microfluidization and cell debris was removed by centrifugation. The lysate was fractionated by first bringing the solution to 30% saturation, discarding the pellet, and then subsequently increasing the saturation to 60% and 90% with (NH4)2SO4 and dissolving the precipitates in TED. The protein was further purified using hydrophobic interaction chromatography over a HiPrep 16/10 phenyl FF (high sub) column (GE Healthcare), dialyzed against TED, and subjected to anion exchange chromatography over a Source Q column (GE Healthcare). The eluted protein was passed over a HiPrep 26/60 Sephacryl S100 HR column (GE Healthcare) for size exclusion. Fractions of ≥95% purity were concentrated to 10 to 11 mg/ml and stored at −80°C.
Expression and purification of selenomethionine-derivatized protein.
The pET-24a vector containing the CDP-Chase gene was used to transform B834 (DE3) competent cells (Novagen). Cells were grown overnight at 37°C in 60 ml M9 medium supplemented with 5% LB medium and 60 μg/ml kanamycin, harvested, and resuspended in M9 medium containing 0.1 mg/ml selenomethionine (Sigma) and 60 μg/ml kanamycin. After growth to an optical density of 0.6, expression was induced with 0.1 mM IPTG, and growth was continued at 25°C overnight. Cells were harvested and lysed, and the protein was purified and stored as described for the wild-type protein.
Protein crystallization.
Crystals of selenomethionine-derivatized B. cereus CDP-Chase were grown by hanging-drop vapor diffusion with 1 ml of a reservoir consisting of 0.1 M Tris-HCl, pH 8.5, 0.2 to 0.3 M Li2SO4, and 26 to 29% polyethylene glycol 4000 (PEG 4000). One microliter of reservoir was added to 1 μl of 10 mg/ml CDP-Chase in TED buffer with 0.15 M NaCl to form the drop. Crystals grew at 20°C in 1 to 4 days. Wild-type protein crystals were grown in a similar fashion. Gd3+-derivatized crystals were prepared by soaking wild-type protein crystals in 5 mM GdCl3 for 5 days.
Diffraction data collection and processing.
Data from the SeMet crystals were collected at the SGX-CAT beamline of the Advanced Photon Source (APS) at the Se peak wavelength of 0.9789 Å (Table 1). Data from the Gd3+-soaked crystals were collected with a copper rotating anode RU-H3R generator (Rigaku) as the source of X rays with an R-AXIS IV image plate detector at the X-ray facility of the Department of Biophysics and Biophysical Chemistry of the Johns Hopkins University School of Medicine (Table 1). Indexing and data reduction were carried out using HKL2000 (26).
Table 1.
Data collection and refinement statistics for B. cereus CDP-Chase-SeMet and B. cereus CDP-Chase-Gd3+
| Parameter | Result for: |
|
|---|---|---|
| B. cereus CDP-Chase-SeMet | B. cereus CDP-Chase-Gd3+ | |
| PDB ID | 3Q1P | 3Q4I |
| Data collection | ||
| Space group | P212121 | P212121 |
| Unit cell | ||
| a, b, c (Å) | a = 60.5, b = 71.4, c = 111.2 | a = 61.0, b = 70.9, c = 111.8 |
| α, β, γ (°) | α = β = γ = 90° | α = β = γ = 90° |
| Resolution range (Å)a | 50.00–1.80 (1.86–1.80) | 50.00–2.50 (2.59–2.50) |
| No. of observed reflections | 641,542 | 75,841 |
| No. of unique reflections | 45,135 | 16,640 |
| Redundancy | 14.2 (12.4) | 4.6 (4.5) |
| Completeness (%) | 99.9 (99.5) | 95.5 (97.9) |
| Rsymb | 0.078 (0.561) | 0.083 (0.544) |
| Mean I/σI | 42.6 (3.2) | 31.2 (3.7) |
| Refinement statistics | ||
| Resolution range (Å) | 50.00–1.80 | 50.00–2.50 |
| R | 0.194 | 0.223 |
| Rfree (5% of data) | 0.238 | 0.288 |
| No. of atoms refined | ||
| Protein | 3274 | 3326 |
| Water | 395 | 65 |
| SO42− | 10 | 0 |
| Gd3+ | 0 | 3 |
| RMSD from ideal geometry | ||
| Bond lengths (Å) | 0.010 | 0.010 |
| Bond angles (°) | 1.1 | 1.2 |
| Mean B factors (Å2) | 26.2 | 51.6 |
Data in parentheses correspond to the outermost resolution shell.
Rsym = ΣhklΣ|I − <I>|/ΣhklΣjIj, where<I> is the mean intensity of j observations from a reflection hkl and its symmetry equivalents.
Structure determination and refinement.
Phases were determined by single-wavelength anomalous diffraction (SAD) using data collected on selenomethionine-derivatized crystals at the selenium peak. The program SOLVE (34, 35, 37–40) was used to determine the positions of all 8 anomalously diffracting selenium atoms. RESOLVE (32, 33, 36) was used for density modification and initial automatic model building. Model building was completed through iterative cycles of manual model building with O (17, 18) and refinement with Refmac5 in the CCP4i suite (5). No clear density was observed for residue 1 and residues 87 to 89 in either molecule of the dimer. Also, in the monomer designated monomer B, no density was observed for residues 160 to 163. These residues were omitted from the final model.
The structure of the Gd3+-derivatized protein was determined by direct refinement using the coordinates of the selenomethionine-derivatized protein. The initial model was adjusted manually using O (17, 18) and refined with Refmac5 (5). An anomalous map was used to determine the locations of the Gd3+ ions. Unlike the selenomethionine-derivatized protein, density was present for all residues except for residue 1. Refinement statistics for both datasets are given in Table 1.
Search of Nudix enzymes similar to B. cereus CDP-Chase.
The protein sequence of B. cereus CDP-Chase was used to search the database of nonredundant protein sequences using the BLASTp algorithm. Selected protein sequences were aligned with ClustalW (41). Pairwise alignments of the B. cereus CDP-Chase N terminus (residues 1 to 68) were performed in EMBOSS (28).
Enzymatic assays of CDP-choline hydrolysis.
Reactions were performed in 50-μl reaction mixtures containing 50 mM Tris, pH 8.4, 2 mM MgCl2 or MnCl2, CDP-choline, 1 unit of calf intestinal alkaline phosphatase (CIP), and 3.14 milliunits of B. cereus CDP-Chase (1 unit of enzyme hydrolyzes 1 μmol of CDP-choline per min). CDP-Chase converts the CIP-insensitive substrate CDP-choline to CIP-reactive substrates CMP and phosphocholine. CIP releases orthophosphate. The final concentration of the different substrates in the reaction mixture was 2 mM for the assays comparing wild-type and mutated enzymes. After incubation for 15 min at 37°C, reactions were quenched by the addition of 30 μl of 100 mM EDTA and inorganic orthophosphate was quantified by the method of Fiske and Subbarow (10) as modified by Ames and Dubin (2). For the kinetic assays, the concentration of CDP-choline ranged from 0 to 15 mM. For each substrate concentration, 350-μl reaction mixtures were prepared in the relative activity assay. The data were fit by nonlinear least squares to the Michaelis-Menten equation.
Preparation of RNA substrates for RNA exonuclease assay.
A 43-nucleotide RNA (5′-GGAAUCUCUCUCUCUCUCUAUGCUCUCUCUCUCUCUCUCUCUC-3′) was transcribed and purified as previously described (1, 23). To prepare the 5′-labeled substrate, phosphates on the 5′ end were removed by Antarctic phosphatase (New England BioLabs) according to the manufacturer's instructions and rephosphorylated with [γ-32P]ATP (Perkin Elmer) with a T4 polynucleotide kinase (New England BioLabs).
To prepare the 3′-labeled RNA substrate, phosphates were removed from the 5′ end with Antarctic phosphatase and the RNA was labeled using T4 RNA ligase (New England BioLabs) and [5′-32P]pCp (Perkin Elmer). The reaction mixture contained the manufacturer's supplied buffer, the phosphatase-treated RNA, 6 μl [5′-32P]pCp, and 10 μl T4 RNA ligase (New England BioLabs) in a final volume of 100 μl. The reaction was carried out for 18 h at 16°C. An Illustra G-25 spin column (GE Healthcare) was used to remove any unreacted [5′-32P]pCp. The RNA was phenol and chloroform extracted, ethanol precipitated, and resuspended in water.
RNA exonuclease assay.
Reactions were carried out in 50 mM Tris, pH 8.25, 5 mM MgCl2, 1 mM DTT, and 0.3 μg CDP-Chase with 40 nM 5′- or 3′-end-labeled RNA in a 100-μl total volume. For the RNase assay involving the mutated enzymes and the RNase assay in the presence of CDP-choline, 0.6 μg CDP-Chase was used. For the RNase competition assay, 0.3 μg CDP-Chase and 20 nM 5′-end-labeled RNA was used and the unlabeled RNA was varied from 0 to 180 nM. Ten-microliter samples were taken at various time points (see Fig. 4C) and quenched with 95% formamide, 25 mM EDTA, 0.02% bromophenol blue, and 0.02% cyanol blue. Reaction products were visualized on a 10% urea denaturing sequencing gel loaded with 2-μl samples. Gels were dried for 2 h, exposed overnight to a phosphorimager screen, and scanned with a Typhoon imager (GE Healthcare). Gel quantification was performed using ImageQuant (Molecular Dynamics) to monitor the disappearance of the top band, which corresponds to the 43-nucleotide RNA species.
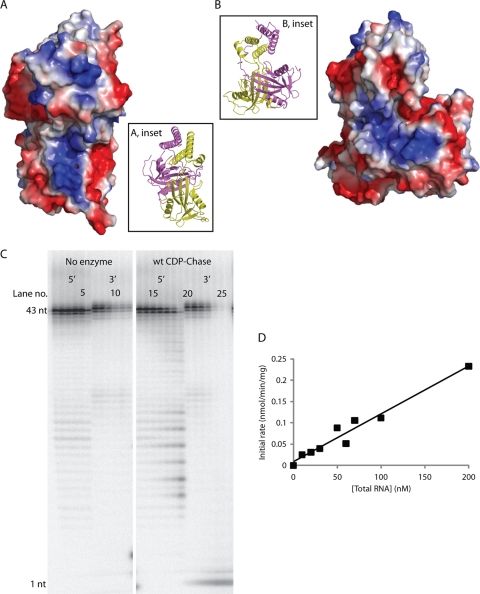
Fig. 4.
CDP-Chase has an RNA exonuclease activity. CDP-Chase contains two positively charged grooves, one on each monomer, which may facilitate RNA binding. (A) Electrostatic surface of CDP-Chase centered on the positively charged electrostatic groove of monomer A. (Inset) Ribbon diagram of protein in the same orientation as the electrostatic surface. The coloring of the protein backbone is the same as in Fig. 2A. (B) Electrostatic surface of CDP-Chase centered on the positively charged electrostatic groove of monomer B. (Inset) Ribbon diagram of protein in the same orientation as the electrostatic surface. (C) To identify any RNase activity by B. cereus CDP-Chase, an RNase assay was performed on RNA labeled with 32P-monophosphate on either the 5′ end or the 3′ end in the absence (lanes 1 to 6 for 5′-end-labeled RNA and lanes 7 to 12 for 3′-end-labeled RNA) or presence (lanes 13 to 19 for 5′-end-labeled RNA and lanes 20 to 26 for 3′-end-labeled RNA) of enzyme. In the assay with no enzyme, the two sets of six lanes represent time points of 0, 5, 10, 20, 30, and 60 min, from left to right. In the assay with enzyme (wt CDP-Chase), the two sets of seven lanes represent time points of 1, 2, 5, 10, 20, 30, and 60 min, from left to right. wt, wild type; nt, nucleotide. (D) Competition assay of RNA degradation by CDP-Chase. The data were fit to a linear model.
Molecular modeling of CDP-choline binding.
CDP-choline was docked into the active site of monomer B of the CDP-Chase structure containing Gd3+ using the program AUTODOCK (16, 25). In the best prediction, CDP-choline bound to a narrow channel with the phosphates located close to the metal binding site. Based on other Nudix structures containing bound substrates and Mg2+, the docked CDP-choline was adjusted manually and a Mg2+ was placed in a position similar to that of the Gd2 site, although slightly closer to coordinating residues to correct for the fact that the Gd3+ ionic radius is larger than that of Mg2+. Two water molecules were removed from the active site as they were situated too close to the fitted CDP-choline. The model was optimized by energy minimization of CDP-choline and residues within 4.5 Å of it using the Molecular Operating Environment (Chemical Computing Group, Inc.).
Immunoelectron microscopy.
Rat antisera were developed using a modification of the procedure by Larsson and Nilsson (21). Ten microliters of B. cereus CDP-Chase at 11.2 mg/ml were placed onto each of two 1-cm2 coupons of sterile nitrocellulose paper and allowed to dry. Each nitrocellulose coupon was then surgically implanted into the peritoneal cavity of a Sprague Dawley rat. The procedure was repeated on days 10, 20, and 30 with similarly prepared coupons. On day 40, rats were bled via heart puncture, and the sera were prepared and stored at −20°C.
B. cereus ATCC 14579 was grown overnight at 30°C in NB medium. Bacteria were pelleted, and cells were fixed and samples prepared according to the method of Tokuyasu (42). Briefly, cells were fixed in 4% formaldehyde in 0.1 M sodium cacodylate buffer with 3% sucrose and 3 mM CaCl2, cryopreserved in 2.3 M sucrose in 20% polyvinylpyrrolidone (Sigma), and frozen in liquid nitrogen. Ultrathin sections were cut with a Leica UCT microtome and placed on 200-mesh nickel Formvar-coated grids. Grids were floated overnight at 4°C, section side down, on drops of primary antibody diluted in phosphate-buffered saline (PBS) with 10% fetal bovine serum. Primary antibody was detected with 12-nm goat anti-rat secondary gold antibody (Jackson ImmunoResearch) diluted 1:20 in PBS for 1 h at room temperature. Final contrasting of the sections was done by incubation in 2% methyl cellulose (Sigma) and 0.3% uranyl acetate for 10 min at 4°C. All sections were viewed either on a Philips CM 120 transmission electron microscope at 80 kV equipped with a Gatan Orius SC 1000 digital camera or a Zeiss Libra 120 at 120 kV equipped with an Olympus Cantega camera.
Protein structure accession numbers.
The atomic coordinates and structure factors of the selenomethionine-derivatized B. cereus CDP-Chase and the Gd3+-derivatized B. cereus CDP-Chase have been deposited in the Protein Data Bank (PDB) with identification (ID) codes PDB ID 3Q1P and 3Q4I, respectively.
RESULTS
Structure of the free enzyme.
B. cereus CDP-Chase crystallizes as a dimer (Fig. 2A), in agreement with its elution in size exclusion chromatography (data not shown). Each monomer consists of two domains, an N-terminal domain (residues 1 to 58) and a C-terminal domain (residues 68 to 205), separated by a linker region (residues 59 to 67).
Fig. 2.
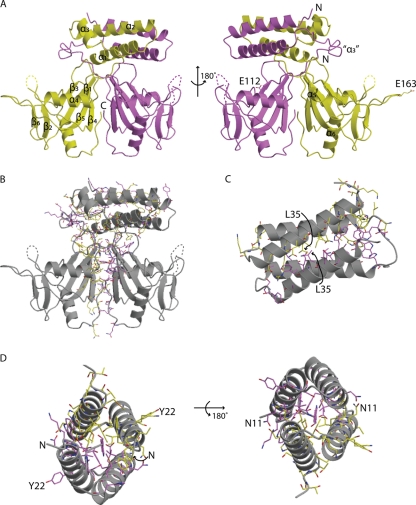
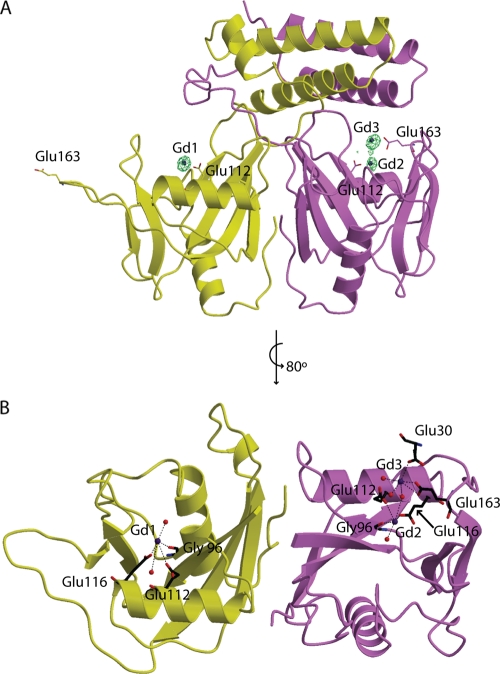
Crystal structure of B. cereus CDP-Chase. CDP-Chase assembles as an asymmetric dimer. (A) The A monomer is colored yellow, and the B monomer is colored magenta. Alternate views of the CDP-Chase structure are shown in with the secondary structural elements labeled. In the view on the right, E163 of monomer A and E112 of monomer B are shown as sticks. Helix α3 is a well-ordered helix in one monomer but is a disordered coil (designated “α3”) in the other monomer. (B) There is an extensive network of residues (shown as sticks with coloring corresponding to that in panel A) involved in hydrogen bonding networks and salt bridges, as well as hydrophobic contacts that stabilize the dimer interface. (C) The N-terminal helical bundle is a well-packed bundle. (D) At either end of the bundle, there is a network of stabilizing intermolecular interactions. Residues shown in sticks are residues involved in such interactions. In panels C and D, residues of the bundle are also labeled, to emphasize the symmetry of the N-terminal bundle.
The C-terminal domain has the fold found in Nudix hydrolases, consisting of two β-sheets (one antiparallel and one mixed) and two helices. The mixed β-sheet consists of β3 (residues 96 to 98), β1 (residues 68 to 75), β5 (residues 144 to 153), and β4 (residues 125 to 131). The antiparallel β-sheet consists of β2 (residues 80 to 84) and β6 (residues 167 to 170). Helix α4 (residues 105 to 117) of the Nudix signature sequence is located on one side of the mixed β-sheet, and helices α5 (residues 132 to 135) and α6 (residues 185 to 196) are located on the other. B. cereus CDP-Chase contains a modified Nudix signature sequence where position E7N is a tyrosine (Tyr 103) and position R15N is a lysine (Tyr111) (Fig. 1). The interaction of Tyr103 and Lys111 is similar to the interaction between E7N and R15N typically observed in the Nudix box. The two C-terminal domains are related by an approximately 2-fold axis forming a dimer stabilized by an interface that buries 2,030 Å2 surface area.
The secondary structure of the N-terminal domain differs between the two monomers of the dimer. Monomer A contains three helices: α1 (residues 5 to 22), α2 (residues 26 to 47), and α3 (residues 51 to 58). In monomer B, residues 51 to 58 fold as a loop instead of an α-helix. Helices α1 and α2 of each molecule form a tightly packed 4-helix bundle held together by hydrogen bonding networks and salt bridges at either end of the bundle, as well as hydrophobic packing in the center of the bundle. The dimer interface in the 4-helix bundle buries 1,770 Å2 surface area.
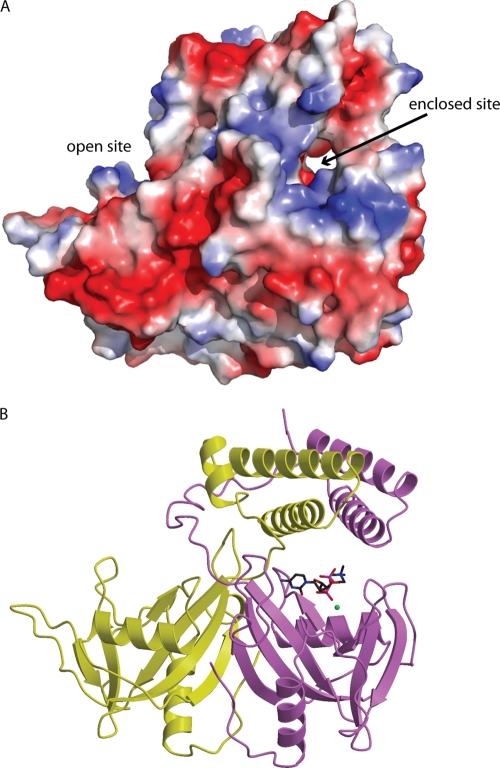
The asymmetry in the N-terminal domain results in differences in accessibility for the two Nudix catalytic sites. The catalytic site in monomer A is more accessible to solvent than that of monomer B. The positioning of the N-terminal domain bundle over the catalytic domain of monomer B results in the formation of a small channel that is ideally suited to accommodate a Nudix substrate (see Fig. 7A). This site will be referred to as the “enclosed” site, while the catalytic site of monomer A will be referred to as the “open” site.
Fig. 7.
Model of CDP-choline binding. (A) An electrostatic surface representation of B. cereus CDP-Chase showing the narrow channel comprising the enclosed site and the open cavity comprising the open site. (B) The energy-minimized complex of CDP-Chase and CDP-choline (sticks). The modeled Mg2+ is shown as a green sphere.
B. cereus CDP-Chase, a prototype for other asymmetric Nudix enzymes.
A search of the sequence databases revealed other putative Nudix hydrolases, all in Gram-positive bacteria, with N-terminal domains similar (≥50% sequence similarity) to that of B. cereus CDP-Chase (Fig. 3). Residues in the linker region between the N- and C-terminal domains (in particular Tyr64, Thr66, and Pro67) are highly conserved among these Nudix hydrolases. Many residues in the N-terminal domain are also conserved, particularly hydrophobic residues that may contribute to stabilization of helix-helix packing (i.e., Ile12 and Phe32, which may be replaced by a Leu or Met at position 12 and Tyr at position 32). β-Branched residues, such as the frequently observed Val9, are entropically favored in helix-helix oligomerization. Leu35 (sometimes replaced by Ile) may act in a similar manner. Asp25 and Glu30, located on the exterior of the bundle, are also conserved. Arg31, a residue involved in a stabilizing interaction at one end of the N-terminal bundle, is another conserved charged residue. Other conserved residues (Ser14, Ala16, Gln17, Ala18, Thr21, Gln33, Gln34, Ser39, Met42, Met43, and Ser44) may be important for overall helix stability, and the strictly conserved Gly19 may disrupt the continuation of α1, allowing a small loop (residues 23 to 25) before α2. Additional conserved residues are found in α3 (Val54, Glu55, Leu57, and Phe58). Similar helix bundles have been observed in the structure of a redesigned Rop (PDB ID 1F4N) (44), a pyogenecin immunity protein Spy2152 (PDB ID 2FU2), and a synthetic coiled-coil tetramer formed by hnRNPC (PDB ID 1TXP).
Fig. 3.
Family of CDP-Chase-like enzymes. A BLAST search reveals many putative Nudix hydrolases with a high degree of sequence similarity to CDP-Chase. Identical residues are in boldface and highlighted in blue, and residues with high similarity are shown in red. The Nudix signature sequence is highlighted in yellow. The secondary structural assignment above the sequences corresponds to that of monomer A of CDP-Chase. Below the sequences, residues are labeled according to their predicted interactions with CDP-choline: H marks residues that are within hydrogen bonding distance, M marks residues that either coordinate the catalytic Mg2+ or interact with these residues through solvent-mediated interactions, and P marks other residues that line the binding pocket of CDP-choline. Asterisks indicate residues that were mutated to study activity. B. thuringiensis, Bacillus thuringiensis; L. acidophilus, Lactobacillus acidophilus; L. johnsonii, Lactobacillus johnsonii; C. difficile, Clostridium difficile; S. mutans, Streptococcus mutans; C. perfringens, Clostridium perfringens; S. sanguinis, Streptococcus sanguinis; L. plantarum, Lactobacillus plantarum.
Kinetics of CDP-choline hydrolysis.
Catalysis of CDP-choline hydrolysis was characterized by measuring initial rates as a function of substrate concentration in the presence of Mg2+. These initial rates as a function of the CDP-choline concentration were fitted with a Michaelis constant (Km) of 1.48 ± 0.09 mM (mean ± standard deviation), a turnover rate (kcat) of 4.2 ± 0.3 s−1, and a Vmax of 10.6 ± 0.2 units/mg. This specific activity is similar to that of other Nudix hydrolases (13, 22).
Determination of RNase activity and directionality.
Close examination of the electrostatic surface of the CDP-Chase structure shows a negatively charged groove ideally suited to bind the sugar-phosphate backbone of a single-stranded nucleic acid such as RNA. The Nudix active site of monomer B is ideally situated to hydrolyze the RNA (Fig. 4A and B). We used either 5′- or 3′-end-radiolabeled RNA molecules as substrates and monitored the degradation of these RNA molecules in the absence and presence of the enzyme. Aliquots were removed from the reaction mixture at different times and quenched with excess EDTA, and products at each time point were visualized on a 10% urea denaturing sequencing gel (Fig. 4C). Over time, degradation products were observed for the 5′-end-labeled RNA, while only the appearance of the 3′-labeled nucleotide was observed for the 3′-end-labeled RNA. A small amount of degradation is observed in the absence of enzyme and is likely due to base-catalyzed hydrolysis of the RNA, since the incubation was done at pH 8.25. These results suggest that the enzyme acts as a 3′-to-5′ exonuclease.
Kinetic assay of RNA hydrolysis by wt CDP-Chase.
To further characterize the level of RNA hydrolysis, a competition assay was performed with a constant concentration of labeled RNA and varied amounts of unlabeled RNA. Here, the initial rate is defined as the rate of disappearance of the 43-nucleotide RNA species and was measured over a course of 2 to 5 min. Beyond this time, rates were no longer linear. The rates increased linearly with RNA concentration and did not level off in the range of substrate concentrations assayed (Fig. 4D). Assuming that Km is larger than 200 nM, the highest concentration of substrate used, and that Vmax is greater than 2.24 × 10−4 unit/mg protein, kcat must be greater than 5.52 × 10−3 min−1.
Structure of the metal-bound enzyme.
To gain insight into the mode of binding of the two substrates to CDP-Chase, we attempted to determine the structures of complexes of the enzyme with CDP-choline or RNA. Cocrystallization attempts with CDP-choline or RNA failed to produce structures with either substrate bound. Since catalytic metal ions have commonly been observed to bridge Nudix hydrolases to their substrates, wild-type protein crystals were soaked with Gd3+; this ion can mimic the binding of divalent metal cations and has more electrons than Mg2+, facilitating its detection in electron density maps. An anomalous map calculated with data obtained using CuKα radiation (1.54 Å) confirmed the location of the Gd3+ ions (Fig. 5). The overall fold of the metal-bound enzyme is similar to that of the native enzyme. The presence of Gd3+ stabilizes the loops containing residues 87 to 89, as well as residues 160 to 163 of monomer B. These residues were modeled into the density. Interestingly, the asymmetry of the N-terminal domains is reflected in the metal binding sites of the two monomers. Both monomers have a Gd3+ binding site coordinated by Gly96, Glu112, and Glu116 and two water molecules (Fig. 5, Gd1 and Gd2). The Gd1 and Gd2 coordination resembles the coordination of the catalytic metal observed in other Nudix enzymes (11, 13, 19). In monomer B, a second Gd3+ ion, Gd3, is coordinated by the side-chain carboxylic acid oxygens of Glu163 and a residue in the N-terminal domain, Glu30, which is also a highly conserved residue in this putative CDP-Chase family (Fig. 5 and 3). Gd3 is also coordinated to three water molecules, one of which bridges Gd2 to Gd3. This site does not exist in monomer A.
Fig. 5.
Gd3+-bound CDP-Chase structure. In both panels, the coloring of the protein monomers is similar to that in Fig. 2. (A) The structure of the enzyme with Gd3+ (blue spheres) is shown with an anomalous difference map contoured at 5σ (green mesh). Residues mutated are drawn in sticks. (B) The coordination of these Gd3+ ions is shown. (For simplicity, the N-terminal domain was omitted in this panel).
Mutational studies of active-site residues.
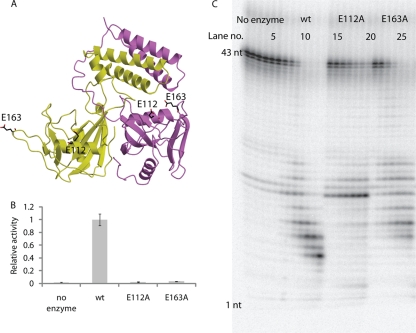
To further characterize both enzymatic activities, we performed single-site mutations of residues Glu112 and Glu163 to alanine. Glu112 is located in the Nudix signature sequence (residue E16N in Fig. 1), while Glu163 is located outside the Nudix box in a loop between β5 and β6 (Fig. 6A). Glu112 coordinates a Gd3+ ion in both monomers, while Glu163 is a ligand of Gd3 in monomer B (Fig. 5B), suggesting that these residues may be important in binding catalytic metals that bridge the enzyme to the substrate and/or in interacting with water molecules in the active site, which may form hydrogen bonds to the substrate. Activity assays measuring CDP-choline hydrolysis showed that both mutations have activities comparable to that of a no-enzyme control (Fig. 6B). The E112A mutation severely impairs RNA exonuclease activity, while the E163A mutation decreases RNA exonuclease activity only moderately (Fig. 6C).
Fig. 6.
Mutagenesis of E112 and E163. (A) Locations of mutated residues. (B) Level of CDP-choline hydrolysis plotted relative to that of the wild-type protein. Error bars show standard deviations over three independent measurements. (C) An RNA exonuclease assay was performed using 5′-32P-labeled RNA with no enzyme (lanes 1 to 6), wild-type CDP-Chase (lanes 7 to 13), CDP-Chase E112A (lanes 14 to 20), and CDP-Chase E163A (lanes 21 to 27). Lanes correspond to the same time points described for Fig. 4C.
Molecular modeling of CDP-choline binding.
Using the Gd3+-bound structure of CDP-Chase, we modeled the binding of CDP-choline to monomer B. The N-terminal helical bundle of CDP-Chase restricts the area near the metal binding sites in monomer B to a narrow channel (the enclosed site), preventing binding of the larger RNA substrate in this site (Fig. 7A). This suggests that RNA may bind to the open site in monomer A for catalysis or that a large movement of the N-terminal bundle takes place to accommodate RNA in monomer B. This narrow channel, however, is large enough to contain a diphosphate-choline moiety such as that present in CDP-choline (Fig. 7A and B). This binding site is similar to those of other Nudix hydrolases and can accommodate coordination of the substrate to Mg2+. Binding of CDP-choline in this region can be stabilized by several hydrogen bonds to the protein. In this model, the choline head group occupies a location similar to that of Gd3 in the metal-bound structure, suggesting that Gd3 is not a true physiological binding site for a catalytic metal ion but, rather, a positively charged placeholder in the absence of CDP-choline. All residues that form the CDP-choline binding pocket are highly conserved (Fig. 3). The open site in monomer A may be able to accommodate CDP-choline, but in comparison to the enclosed site in monomer B, fewer interactions would be available to stabilize the substrate. This model provides a rationale for the biochemical data that show that both E112 and E163 are important for catalysis of CDP-choline hydrolysis. The observation that E112 is important for RNA exonuclease activity and that E163 is less crucial may be explained by the use of the open site in monomer A for RNA binding and catalysis of exonuclease activity; E163 is in a loop pointed away from the active-site cleft, suggesting that it is less important for binding of RNA and for the exonuclease activity.
RNA exonuclease activity in the presence of CDP-choline.
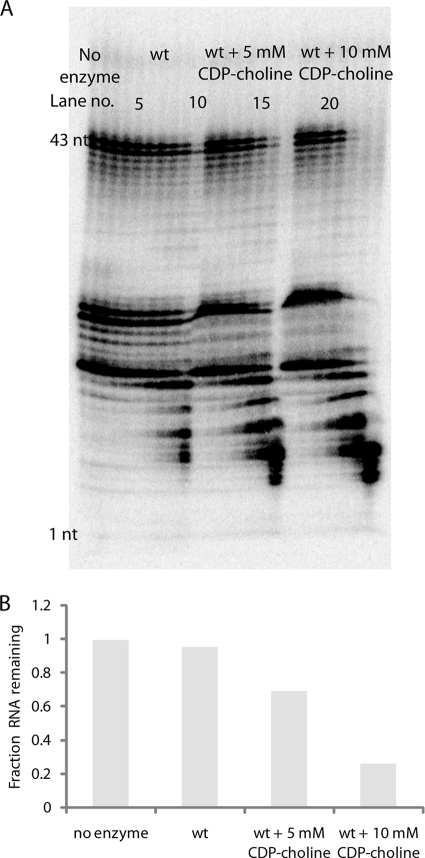
We assayed RNA exonuclease activity in the presence of excess CDP-choline to test the hypothesis that CDP-choline and RNA use different sites for catalysis (Fig. 8). Surprisingly, the presence of CDP-choline not only did not hinder exonuclease activity but increased the exonuclease activity. This result confirms that the two substrates do not use the same active site but also suggests that binding of CDP-choline modulates binding of RNA in such a way as to accelerate catalysis. As the CDP-choline concentration is increased from 5 mM to 10 mM, the RNA exonuclease activity is further accelerated.
Fig. 8.
RNA exonuclease activity in the presence of CDP-choline. (A) An RNA exonuclease assay was performed using 5′-32P-labeled RNA with no enzyme (lanes 1 to 3) and with 6.28 milliunits of wt CDP-Chase (lanes 4 to 24) either in the absence (lanes 4 to 10) or the presence of 5 mM (lanes 11 to 17) or 10 mM (lanes 18 to 24) CDP-choline. Results at the 0-, 20-, and 60-min time points are shown for the no-enzyme control, and results at the 1-, 2-, 5-, 10-, 20-, 30-, and 60-min time points are shown for assays with enzyme. (B) The fraction of 43-nucleotide RNA remaining at 20 min is plotted for the no-enzyme control, the wt CDP-Chase, and the wt CDP-Chase in the presence of 5 mM and 10 mM CDP-choline.
Immunoelectron microscopy of B. cereus ATCC 14579.
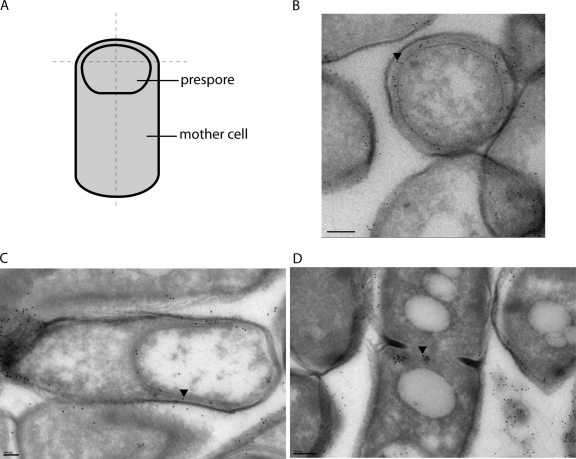
We determined the subcellular localization of CDP-Chase by raising an antibody against the enzyme, visualizing these primary antibodies with a gold particle-containing secondary antibody and using transmission electron microscopy (TEM) to image thin sections of cells (Fig. 9A and B). Our protocol for growing B. cereus resulted in the formation of sporangia, as well as vegetative cells. High cell density and nutrient starvation can trigger the beginning of the sporulation cycle. An asymmetric cell division results in a daughter cell, the prespore, which is then engulfed by the mother cell (Fig. 9A) and eventually matures into the spore that is released into the environment upon mother cell lysis (8, 30). Two different slices of stage III (engulfment) sporangia are depicted in Fig. 9B and C. In both, labeling of the B. cereus CDP-Chase is both close to the inner leaflet of the prespore membrane and between the prespore membrane and the mother cell membrane. Vegetative cells also contained B. cereus CDP-Chase molecules located at the periphery of the cell, but staining was more diffuse (data not shown). Interestingly, B. cereus CDP-Chase was also found at the division septum of mitotic cells (Fig. 9D).
Fig. 9.
Subcellular localization of B. cereus CDP-Chase. (A) Depiction of a stage III sporangium. Dashed lines indicate manner of cell slicing. (B) A stage III sporangium treated with primary antibody against CDP-Chase and visualized with gold-conjugated secondary antibody (black dots). This cell is sliced in a manner similar to the horizontal dashed line in panel A. (C) A stage III sporangium sliced in an orientation similar to the vertical dashed line in panel A. (D) A dividing vegetative cell. Black arrowheads were placed in B, C, and D to indicate the locations of single gold particles as a guide for visualization. Bars, 200 nm (B and D) and 100 nm (C).
DISCUSSION
The two activities of CDP-Chase are mediated by active-site asymmetry.
The structure of B. cereus CDP-Chase shows that the two identical monomers fold as an asymmetric dimer with two different catalytic sites: an enclosed Nudix site (monomer B) and a large open cavity (monomer A) (Fig. 7A), suggesting that each monomer is tailored to a specific function. The enzyme is capable of performing a Nudix reaction on CDP-choline as well as a non-Nudix exonuclease reaction on RNA in the 3′-to-5′ direction. Several other proteins, such as the activated form of the receptor tyrosine kinase ErbB4, as well as an NADPH-bound form of the dinucleotide-binding protein HSCARG, have also been found to fold as asymmetric dimers (20, 27). The most obvious effect of the N-terminal domain asymmetry is to change the accessibility of the active sites in the dimer. Our model of CDP-choline binding supports the hypothesis that the asymmetry is required for recognition of CDP-choline by the enclosed site and that a different mode of binding for the other substrate is likely to be operational in the open site. In the enclosed site, the presence of the N-terminal domain would hinder RNA binding. In the other site, the N-terminal domain does not block binding of RNA and the C-terminal Nudix domain may bind RNA in a manner similar to that of CPSF5 and the cleavage factor Im-25 (6, 43). Our structural and enzymatic data not only support the use of two distinct active sites for CDP-choline and RNA but also suggest that binding of CDP-choline in the enclosed site increases exonuclease activity in the open site.
Physiological function of CDP-Chase.
The immunoelectron microscopy showed that CDP-Chase is expressed near the cell membrane of B. cereus in the vegetative state of the cell and between the prespore and mother cell membranes during sporulation. The enzyme may regulate phosphocholine and CDP-choline concentrations during cell wall polysaccharide biosynthesis, cell division, and/or sporulation. Both teichoic acids (TA) and lipoteichoic acids (LTA) in the cell wall require the incorporation of phosphocholine groups donated by CDP-choline (9). Furthermore, phosphocholine expression at the cell surface is required for a number of cell functions in addition to incorporation into TA and LTA in Gram-positive bacteria (9). In Streptococcus pneumoniae, for example, phosphocholine expression is required for interaction with the host cell membrane during infection (47). Phosphocholine is also required for the separation of daughter cells after mitosis. Both mitosis and sporulation involve a division septum (Fig. 9A). In B. cereus, CDP-Chase was found to be localized at the site of formation of this septum.
A possible physiological role for the RNA exonuclease activity is degradation of RNA after commitment to sporulation. Synthesis of mRNA does stop at the beginning of sporulation and decreases afterwards (30). Since CDP-Chase is already induced in this stage, it may provide one of the means for RNA degradation.
CDP-Chase is a member of a novel Nudix family.
Interestingly, we found enzymes similar to this unconventional asymmetric Nudix enzyme present in other Gram-positive bacteria (Fig. 3). The high degree of sequence similarity between these proteins (≥50% in the N-terminal domain and ≥25% over the entire protein) suggests that this asymmetry is a conserved feature of the members of this family and is crucial to their function, as well as to substrate recognition. This subfamily's exclusive presence in Gram-positive bacteria and its localization in B. cereus at the periphery of the vegetative cell membrane, as well as between the membranes of the prespore and mother cell in sporangia, strongly suggest that the function of these enzymes is to regulate phosphocholine and CDP-choline concentrations at and beyond the bacterial cell membrane. As cell surface and cell wall expression of phosphocholine is critical for the survival of many Gram-positive bacteria, this family of enzymes may serve as a promising target for attenuating the infectivity of these pathogens.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (grants GM066895 and NS061827 to L.M.A.) and by a National Science Foundation graduate research fellowship to K.C.D. K.C.D. was also supported by NIH training grant T32 GM 008403 to the Program in Molecular Biophysics at Johns Hopkins University. Use of the Advanced Photon Source at Argonne National Laboratory was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. DE-AC02-06CH11357.
Use of the Lilly Research Laboratories Collaborative Access Team (LRL-CAT) beamline at Sector 31 of the Advanced Photon Source was provided by Eli Lilly Company, which operates the facility. We also thank the beamline staff at NSLS X6A for their assistance in data collection, as well as Jon Lorsch, Sarah Mitchell, Sarah Walker, and Jagpreet Nanda for their assistance with the RNA exonuclease assays and the transcription and labeling of the RNA substrates, Carol Cooke for preparation of immunoelectron microscopy samples and collection of images, and Susan Welkos and Adam Driks for their assistance in interpretation of the immunoelectron microscopy images.
Footnotes
Published ahead of print on 29 April 2011.
REFERENCES
- 1. Acker M. G., Kolitz S. E., Mitchell S. F., Nanda J. S., Lorsch J. R. 2007. Reconstitution of yeast translation initiation. Methods Enzymol. 430:111–145 [DOI] [PubMed] [Google Scholar]
- 2. Ames B. N., Dubin D. T. 1960. The role of polyamines in the neutralization of bacteriophage deoxyribonucleic acid. J. Biol. Chem. 235:769–775 [PubMed] [Google Scholar]
- 3. Bessman M. J., Frick D. N., O'Handley S. F. 1996. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J. Biol. Chem. 271:25059–25062 [DOI] [PubMed] [Google Scholar]
- 4. Cartwright J. L., Gasmi L., Spiller D. G., McLennan A. G. 2000. The Saccharomyces cerevisiae PCD1 gene encodes a peroxisomal nudix hydrolase active toward coenzyme A and its derivatives. J. Biol. Chem. 275:32925–32930 [DOI] [PubMed] [Google Scholar]
- 5. Collaborative Computational Project, Number 4 1994. The CCP4 Suite—programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 [DOI] [PubMed] [Google Scholar]
- 6. Coseno M., et al. 2008. Crystal structure of the 25 kDa subunit of human cleavage factor Im. Nucleic Acids Res. 36:3474–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deana A., Celesnik H., Belasco J. G. 2008. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature 451:355–358 [DOI] [PubMed] [Google Scholar]
- 8. Errington J. 2003. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 1:117–126 [DOI] [PubMed] [Google Scholar]
- 9. Fischer W. 2000. Phosphocholine of pneumococcal teichoic acids: role in bacterial physiology and pneumococcal infection. Res. Microbiol. 151:421–427 [DOI] [PubMed] [Google Scholar]
- 10. Fiske C. H., Subbarow Y. 1925. The colorimetric determination of phosphorus. J. Biol. Chem. 66:375–400 [Google Scholar]
- 11. Gabelli S. B., Bianchet M. A., Bessman M. J., Amzel L. M. 2001. The structure of ADP-ribose pyrophosphatase reveals the structural basis for the versatility of the Nudix family. Nat. Struct. Biol. 8:467–472 [DOI] [PubMed] [Google Scholar]
- 12. Gabelli S. B., et al. 2002. Mechanism of the Escherichia coli ADP-ribose pyrophosphatase, a Nudix hydrolase. Biochemistry 41:9279–9285 [DOI] [PubMed] [Google Scholar]
- 13. Gabelli S. B., et al. 2007. Structure and function of the E. coli dihydroneopterin triphosphate pyrophosphatase: a Nudix enzyme involved in folate biosynthesis. Structure 15:1014–1022 [DOI] [PubMed] [Google Scholar]
- 14. Huang N., et al. 2009. Structure and function of an ADP-ribose-dependent transcriptional regulator of NAD metabolism. Structure 17:939–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang N., et al. 2008. Bifunctional NMN adenylyltransferase/ADP-ribose pyrophosphatase: structure and function in bacterial NAD metabolism. Structure 16:196–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huey R., Morris G. M., Olson A. J., Goodsell D. S. 2007. A semiempirical free energy force field with charge-based desolvation. J. Comput. Chem. 28:1145–1152 [DOI] [PubMed] [Google Scholar]
- 17. Jones T. A., Kjeldgaard M. 1997. Electron-density map interpretation. Methods Enzymol. 277:173–208 [DOI] [PubMed] [Google Scholar]
- 18. Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47(Pt. 2):110–119 [DOI] [PubMed] [Google Scholar]
- 19. Kang L. W., Gabelli S. B., Cunningham J. E., O'Handley S. F., Amzel L. M. 2003. Structure and mechanism of MT-ADPRase, a nudix hydrolase from Mycobacterium tuberculosis. Structure 11:1015–1023 [DOI] [PubMed] [Google Scholar]
- 20. Lamb H. K., Stammers D. K., Hawkins A. R. 2008. Dinucleotide-sensing proteins: linking signaling networks and regulating transcription. Sci. Signal. 1:pe38. [DOI] [PubMed] [Google Scholar]
- 21. Larsson A., Nilsson B. O. 1988. Immunization with nanogram quantities of nitrocellulose-bound antigen, electroblotted from sodium dodecyl sulphate-polyacrylamide gels. Scand. J. Immunol. 27:305–309 [DOI] [PubMed] [Google Scholar]
- 22. Legler P. M., Massiah M. A., Mildvan A. S. 2002. Mutational, kinetic, and NMR studies of the mechanism of E. coli GDP-mannose mannosyl hydrolase, an unusual Nudix enzyme. Biochemistry 41:10834–10848 [DOI] [PubMed] [Google Scholar]
- 23. Messing S. A., et al. 2009. Structure and biological function of the RNA pyrophosphohydrolase BdRppH from Bdellovibrio bacteriovorus. Structure 17:472–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mildvan A. S., et al. 2005. Structures and mechanisms of Nudix hydrolases. Arch. Biochem. Biophys. 433:129–143 [DOI] [PubMed] [Google Scholar]
- 25. Morris G. M., et al. 1998. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comp. Chem. 19:1639–1662 [Google Scholar]
- 26. Otwinowski Z., Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Macromol. Crystallogr. A 276:307–326 [DOI] [PubMed] [Google Scholar]
- 27. Qiu C., et al. 2008. Mechanism of activation and inhibition of the HER4/ErbB4 kinase. Structure 16:460–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rice P., Longden I., Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276–277 [DOI] [PubMed] [Google Scholar]
- 29. Rodionov D. A., et al. 2008. Transcriptional regulation of NAD metabolism in bacteria: NrtR family of Nudix-related regulators. Nucleic Acids Res. 36:2047–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schaeffer P. 1969. Sporulation and production of antibiotics, exoenzymes, and exotoxins. Bacteriol. Rev. 33:48–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. She M., et al. 2008. Structural basis of dcp2 recognition and activation by dcp1. Mol. Cell 29:337–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Terwilliger T. C. 2003. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. D Biol. Crystallogr. 59:38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Terwilliger T. C. 2003. Automated side-chain model building and sequence assignment by template matching. Acta Crystallogr. D Biol. Crystallogr. 59:45–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Terwilliger T. C. 1994. MAD phasing: Bayesian estimates of F(A). Acta Crystallogr. D Biol. Crystallogr. 50:11–16 [DOI] [PubMed] [Google Scholar]
- 35. Terwilliger T. C. 1994. MAD phasing: treatment of dispersive differences as isomorphous replacement information. Acta Crystallogr. D Biol. Crystallogr. 50:17–23 [DOI] [PubMed] [Google Scholar]
- 36. Terwilliger T. C. 2000. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56:965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Terwilliger T. C., Berendzen J. 1997. Bayesian correlated MAD phasing. Acta Crystallogr. D Biol. Crystallogr. 53:571–579 [DOI] [PubMed] [Google Scholar]
- 38. Terwilliger T. C., Eisenberg D. 1987. Isomorphous replacement: effects of errors on the phase probability-distribution. Acta Crystallogr. A 43:6–13 [Google Scholar]
- 39. Terwilliger T. C., Eisenberg D. 1983. Unbiased 3-dimensional refinement of heavy-atom parameters by correlation of origin-removed Patterson functions. Acta Crystallogr. A 39:813–817 [Google Scholar]
- 40. Terwilliger T. C., Kim S. H., Eisenberg D. 1987. Generalized method of determining heavy-atom positions using the difference Patterson function. Acta Crystallogr. A 43:1–5 [Google Scholar]
- 41. Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tokuyasu K. T. 1973. Technique for ultracryotomy of cell suspensions and tissues. J. Cell Biol. 57:551–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tresaugues L., et al. 2008. The crystal structure of human cleavage and polyadenylation specific factor-5 reveals a dimeric Nudix protein with a conserved catalytic site. Proteins 73:1047–1052 [DOI] [PubMed] [Google Scholar]
- 44. Willis M. A., Bishop B., Regan L., Brunger A. T. 2000. Dramatic structural and thermodynamic consequences of repacking a protein's hydrophobic core. Structure 8:1319–1328 [DOI] [PubMed] [Google Scholar]
- 45. Xu W., Dunn C. A., Jones C. R., D'Souza G., Bessman M. J. 2004. The 26 Nudix hydrolases of Bacillus cereus, a close relative of Bacillus anthracis. J. Biol. Chem. 279:24861–24865 [DOI] [PubMed] [Google Scholar]
- 46. Yagi T., et al. 2003. Cloning, expression and characterization of a mammalian Nudix hydrolase-like enzyme that cleaves the pyrophosphate bond of UDP-glucose. Biochem. J. 370:409–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang J. R., Idanpaan-Heikkila I., Fischer W., Tuomanen E. I. 1999. Pneumococcal licD2 gene is involved in phosphorylcholine metabolism. Mol. Microbiol. 31:1477–1488 [DOI] [PubMed] [Google Scholar]