Abstract
Transcriptional regulatory networks are fine-tuned systems that help microorganisms respond to changes in the environment and cell physiological state. We applied the comparative genomics approach implemented in the RegPredict Web server combined with SEED subsystem analysis and available information on known regulatory interactions for regulatory network reconstruction for the human pathogen Staphylococcus aureus and six related species from the family Staphylococcaceae. The resulting reference set of 46 transcription factor regulons contains more than 1,900 binding sites and 2,800 target genes involved in the central metabolism of carbohydrates, amino acids, and fatty acids; respiration; the stress response; metal homeostasis; drug and metal resistance; and virulence. The inferred regulatory network in S. aureus includes ∼320 regulatory interactions between 46 transcription factors and ∼550 candidate target genes comprising 20% of its genome. We predicted ∼170 novel interactions and 24 novel regulons for the control of the central metabolic pathways in S. aureus. The reconstructed regulons are largely variable in the Staphylococcaceae: only 20% of S. aureus regulatory interactions are conserved across all studied genomes. We used a large-scale gene expression data set for S. aureus to assess relationships between the inferred regulons and gene expression patterns. The predicted reference set of regulons is captured within the Staphylococcus collection in the RegPrecise database (http://regprecise.lbl.gov).
INTRODUCTION
Microorganisms are capable of adapting to different ecological niches, as facilitated by the versatility of the microbial physiology and the ability to rapidly respond to various environmental factors. Responses to environmental changes are implemented via alterations of gene expression that may be realized by different mechanisms at the transcriptional and translational levels. At the level of transcription, the main components of gene regulation are transcription factors (TFs) and TF binding sites (TFBSs). Under certain conditions, TFs specifically bind to their TFBSs and affect the expression of their target genes. TF regulons are defined as groups of genes under the direct control of a given TF. The regulatory interactions between an ensemble of various TFs and their multiple target genes in the cell form a complex system called a transcriptional regulatory network (TRN). TRN reconstruction is an important approach for an understanding of the cell physiology, the functional annotation of genes, metabolic reconstruction, and modeling.
To reconstruct a particular TRN, we need to specify which TFs bind to the regulatory regions of which genes and how this binding affects expression (e.g., whether a TF activates or represses the target genes). At present, there are various experimental techniques for studying transcriptional regulation, including traditional bottom-up genetic methods and relatively novel top-down approaches that utilize large sets of high-throughput gene expression and TF binding location data supplemented by an automated inference of TFBS motifs by computational techniques (39, 43). On the other hand, the current availability of a large number of complete microbial genomes has opened up an opportunity to perform a comparative genomic analysis of transcriptional regulatory elements and reconstruct the associated TRNs (14, 40). The comparative analysis of regulons linked to a particular TFBS motif in multiple phylogenetically related genomes helps to identify evolutionarily conserved regulatory sites and eliminate false-positive predictions. This analysis may be efficiently applied to the propagation of known regulons from model species to previously uncharacterized organisms as well as for the ab initio prediction of novel regulons (40). The combination of regulon reconstruction and other genome context techniques, such as the colocalization of genes on the chromosome and gene cooccurrence profiles, helps the functional annotation of genes and, often, helps predict functions for previously unknown genes (33). This integrated bottom-up approach was successively applied previously for the analysis of various regulatory systems in several groups of microorganisms (reviewed in reference 40).
The Gram-positive bacterium Staphylococcus aureus is an important human pathogen that has been intensively studied by a combination of postgenomic techniques (reviewed in references 17 and 49). Although regulatory mechanisms for genes involved in virulence, biofilm formation, and central metabolism in S. aureus were previously described in detail, our knowledge of the transcriptional regulation of many other metabolic pathways and biological processes is still incomplete. Previous automated computational analyses of S. aureus and Bacillus genomes have predicted multiple putative sets of coregulated genes associated with regulatory elements that have been conserved across multiple organisms (1). However, these putative regulons remained without a defined connection to specific TFs, which prevented an evaluation of their consistency. In this study, we used the availability of multiple complete genomes within the family Staphylococcaceae and accumulated experimental knowledge on transcriptional regulation in S. aureus to infer a reference set of 46 regulons using the comparative genomics approach. Seven representative microorganisms with complete genomes, including five pathogenic species (S. aureus N315, Staphylococcus capitis SK14, Staphylococcus epidermidis ATCC 12228, Staphylococcus haemolyticus JCSC1435, and Staphylococcus saprophyticus ATCC 15305) and two nonpathogenic bacteria (Staphylococcus carnosus TM300 and Macrococcus caseolyticus JCSC5402), were selected for this analysis. In this paper we report the first detailed reconstruction of the TRN in S. aureus and related species. We also perform a comparative analysis of this regulatory network with a large set of gene expression data.
MATERIALS AND METHODS
Seven complete genomes of the family Staphylococcaceae uploaded from the MicrobesOnline database (8) were analyzed in this study: Staphylococcus aureus N315, Staphylococcus capitis SK14, Staphylococcus epidermidis ATCC 12228, Staphylococcus carnosus TM300, Staphylococcus haemolyticus JCSC1435, Staphylococcus saprophyticus ATCC 15305, and Macrococcus caseolyticus JCSC5402.
Comparative genomics approaches were used to infer cis-acting regulatory elements (or TFBSs), build nucleotide frequency positional weight matrices (PWMs) for TFBS motifs, and reconstruct the corresponding regulons in the genomes of the family Staphylococcaceae (40). Two major components of this analysis are (i) the propagation of previously known regulons from model organisms (e.g., S. aureus and Bacillus subtilis) to others and (ii) the ab initio prediction of novel regulons. Here we used these two regulon reconstruction workflows implemented in the RegPredict Web server (http://regpredict.lbl.gov/) (30). Initially, the training set of regulatory regions containing either known or as-yet-unknown TFBSs was collected and used as an input for a suite of tools for regulatory motif identification and regulon reconstruction in microbial genomes. The expectation-maximization method implemented in the “Discover Profiles” program of the RegPredict server simultaneously optimizes the PWM description of a motif and the binding probabilities for its associated sites. The Discover Profiles program uses an iterative procedure of clustering all weak palindromic sequences in the training set of DNA fragments to identify a palindromic signal of a given length with the highest information content. One of the strong advantages of this algorithm for TFBS identification is that it accounts for a tendency of TFs in bacteria to bind to palindromic motifs. At the next stage, the constructed PWMs are used by the “Run Profile” computational algorithm to search for potential TFBSs in the analyzed genomes and find additional genes that share the same DNA motif within their regulatory regions.
The RegPredict server provides tools for both the genome-wide identification of TFBSs and comparison of gene sets in several genomes (30). The consistency check approach is based on the assumption that regulons have a tendency to be conserved between the genomes that contain orthologous TFs (40). The presence of the same TFBS upstream of orthologous genes is an indication that it is a true regulatory site, whereas TFBSs scattered at random in the genome are considered false positives. The simultaneous analysis of multiple genomes from the same taxonomic group allows one to make reliable predictions of TFBSs even with weak recognition rules. A gene was considered to be a candidate member of a regulon if it had a corresponding candidate TFBS in the upstream region in several genomes or it was included in the operon bearing such conserved TFBSs.
The comparative regulon analysis tools of the RegPredict server use gene orthologs from the MicrobesOnline database that were generated by a procedure based on the analysis of phylogenetic trees of protein domains (8). In addition, orthologs of TFs were validated by bidirectional genome-wide similarity searches using the Genome Explorer package (27). Functional annotations of the predicted regulon members and the associated metabolic subsystems were collected from the SEED database using recently developed Web services (9, 34). Initial data about target genes and TF binding sites for several regulons previously described for S. aureus (AgrA, CzrA, GlnR, LacR, MntR, Fur, Zur, and PerR) were extracted from the RegTransBase database (http://regtransbase.lbl.gov/), a database of regulatory interactions based on data in the literature (20). The reconstructed regulons, including TFs, their target genes and operons, and associated TFBSs, were uploaded to the RegPrecise database (http://regprecise.lbl.gov/) (29).
We performed analyses of the predicted regulons in the context of microarray expression data for Staphylococcus aureus. The entire data set was produced by using Affymetrix S. aureus GeneChips (10) and was generously provided by the Paul Dunman laboratory (University of Rochester, Rochester, NY). The data set consists of 850 arrays targeting a variety of experimental conditions and S. aureus strains (P. Dunman, personal communication). The expression data are available from Paul Dunman upon request. Affymetrix probe sequences were mapped to the S. aureus Mu50 genome found in the public SEED database (http://www.theseed.org) and were used to redefine the probe sets associated with specific genes. Expression data originating from CEL files were background corrected, normalized, and summarized by using robust multichip averaging (19). The resulting gene-based intensity values were used to calculate Pearson correlations (PCs) for pairs of genes over the entire set of 850 arrays. Pearson correlations were computed for all pairs of genes in a predicted regulon, as long as the predicted regulon had at least two members (44 of 46 regulons). The correlation of each regulon was summarized by using the mean, median, and 80th-percentile PCs for all pairs of genes in the regulon. Heat maps were generated based on gene pairwise PC values for selected regulons. The ordering of genes within the heat map was determined by using hierarchical clustering based on similarities calculated from PCs and using average linkages. Custom Perl and R scripts were used to perform data handling, normalization, statistical analyses, and visualization of regulons.
RESULTS AND DISCUSSION
Transcription factor repertoire in S. aureus.
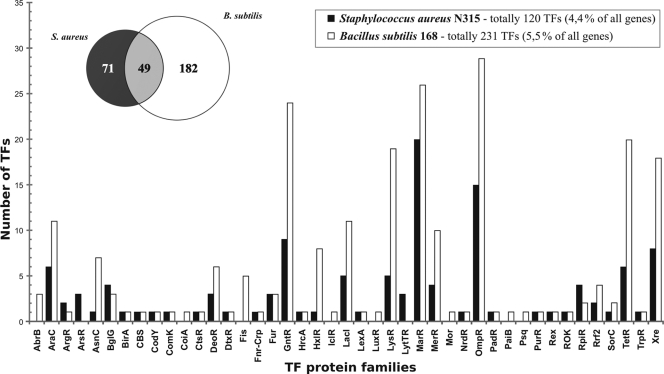
To analyze the TF repertoire in S. aureus, the complete set of known and predicted TFs was extracted from the DBD database (21) and compared with that of Bacillus subtilis strain 168 obtained from the DBTBS database (48) (see Table S1 in the supplemental material). The S. aureus genome appears to encode 120 predicted TFs, distributed in 37 protein families, which is approximately one-half of the B. subtilis repertoire, comprised of 231 TF genes in 40 families (Fig. 1); this is in agreement with an approximately 35% reduction in the total number of genes in the S. aureus genome. In addition to the reduction of family sizes, many TF protein families from B. subtilis (AbrB, CoiA, Fis, IclR, LuxR, Mor, PaiB, and Psq) have no representatives in S. aureus, whereas some TF families from S. aureus (ArsR and LytTR) are missing from B. subtilis. Protein similarity searches identified 49 TFs of S. aureus (40%) as orthologs of B. subtilis TFs. The remaining 71 TFs that are not conserved in B. subtilis include regulators of virulence (AgrA, SaeR, and multiple SarA-like proteins [2]), cell adhesion (IcaR and ArlR), drug and metal resistance (MepR, ArsR, and CzrA), the utilization of sugars (LacR, MalR, ScrR, RbsR, and GatR), and amino acids (ArcR and HutR). A significant reduction of the overall TF repertoire in S. aureus is accompanied by an increase in the number of TFs participating in pathogenesis processes.
Fig. 1.
Repertoire and major protein families of transcription factors in S. aureus and B. subtilis. The overlap between the TF repertoires in two organisms was calculated as the number of orthologous TFs.
A comparison of TF repertoires between seven representatives of the Staphylococcaceae with complete genomes revealed a core set of 39 universal regulators, an extensive set of partially conserved regulators (62 TFs), and a group of 15 regulators that are specific for S. aureus but absent in other species (see Table S1 in the supplemental material). The core set of TFs conserved in all studied species of the Staphylococcaceae is significantly enriched for regulators of the central metabolism of nitrogen and amino acids (ArgR, CodY, CymR, GlnR, and YerC), cofactors (BirA and PdxR), nucleotides (NrdR and PurR), fatty acids (FapR and WalR), and carbohydrates (CcpA, CcpN, CggR, FruR, and GntR) as well as metal homeostasis (Fur, MntR, and Zur), respiration (Rex and SrrA), stress and starvation responses (CtsR, HrcA, LexA, PerR, PhoP, and VraR), competence (ComK), and drug resistance (ArlR and MgrA). Three-quarters of these core TFs have orthologs in B. subtilis.
Inference of the reference regulon set in the Staphylococcaceae.
A comparative genomics approach for regulon inference, as implemented in the RegPredict Web server (30), was applied to the group of 7 related bacterial species from the family Staphylococcaceae. For the reconstruction of the regulatory network in the model species S. aureus N315, we first defined its repertoire of TFs and collected experimental knowledge about S. aureus TF regulons available from the literature (see Table S1 in the supplemental material). Next, depending on the availability of experimental data, we applied one of the following three computational workflows for regulon inference: (i) the expansion of regulons controlled by TFs in S. aureus that were previously characterized (21 TFs), (ii) the reconstruction of regulons for TFs whose orthologs were previously described for B. subtilis (15 TFs) and other related species (4 TFs), and (iii) the ab initio inference of regulons for previously uncharacterized TFs (6 TFs). In all three scenarios of regulon reconstruction, we used identification and multispecies comparisons of DNA target sequences in order to predict sets of target genes in each analyzed genome. A detailed description of the 46 TF regulons reconstructed by utilizing the three computational workflows is provided below and summarized in Table 1 and in Table S2 in the supplemental material. The reconstructed regulons are also captured in the Staphylococcus collection of regulons in the RegPrecise database (available at http://regprecise.lbl.gov) (29). In Table S3 in the supplemental material, we also provide detailed information on all TFBSs in S. aureus, including both previously described TFBSs and TFBSs first predicted in this work, as well as evidence codes for all TFBSs with references to previous experimental reports where the respective regulatory interactions have been confirmed.
Table 1.
Transcription factor regulons reconstructed in the Staphylococcaceae lineage
| TFa | TF locus tag | Genomeb |
TF regulon functional role | TF protein family | Workflowc | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S. aureus N315 | S. capitis SK14 | S. epidermidis ATCC 12228 | S. carnosus TM300 | S. haemolyticus JCSC1435 | S. saprophyticus ATCC 15305 | M. caseolyticus JCSC5402 | B. subtilis | |||||
| AgrA | SA1844 | RS | + | + | + | + | + | − | − | Regulation of quorum sensing | LytTR | A |
| ArcR | SA2424 | RS | + | + | + | + | − | − | − | Arginine catabolism | Crp | A |
| ArgR | SA1351 | + | + | + | + | + | + | + | RS | Arginine metabolism | ArgR | B |
| ArsR | SA1591 | + | + | + | − | + | + | + | RS | Arsenical resistance | ArsR | B |
| BglR* | SA0254 | + | − | − | − | − | − | − | + | Beta-glucoside utilization | GntR | C |
| BirA | SA1289 | + | + | + | + | + | + | + | RS | Biotin metabolism | BirA | B |
| BlaI | SAP012 | RS | + | + | − | + | − | − | − | Beta-lactam resistance | MarR | A |
| CcpA | SA1557 | RS | + | + | + | + | + | + | RS | Carbon catabolism | LacI | A |
| CggR | SA0726 | + | + | + | + | + | + | + | RS | Glycolysis | SorC | B |
| CodY | SA1098 | RS | + | + | + | + | + | + | RS | Amino acid metabolism | CodY | A |
| CtsR | SA0480 | RS | + | + | + | + | + | + | RS | Heat shock response | CtsR | A |
| CymR | SA1453 | RS | + | + | + | + | + | + | RS | Cysteine metabolism | Rrf2 | A |
| CzrA | SA1947 | RS | + | + | + | + | + | − | − | Zinc resistance | ArsR | A |
| FapR | SA1071 | + | + | + | + | + | + | + | RS | Fatty acid biosynthesis | DeoR | B |
| FruR | SA0653 | + | + | + | + | + | + | + | R | Fructose utilization | DeoR | B |
| Fur | SA1329 | RS | + | + | + | + | + | + | RS | Iron homeostasis | Fur | A |
| GlnR | SA1149 | + | + | + | + | + | + | + | RS | Nitrogen assimilation | MerR | B |
| GltC | SA0429 | + | + | + | + | + | + | − | RS | Glutamate synthase | LysR | B |
| GlvR | SA2115 | + | + | + | − | + | + | − | R | Maltose utilization | RpiR | B |
| GntR | SA2295 | + | + | + | + | + | + | + | RS | Gluconate utilization | GntR | B |
| HisR* | SA1723 | + | + | + | + | + | + | + | + | Histidine metabolism | TrpR | C |
| HrcA | SA1411 | RS | + | + | + | + | + | + | RS | Heat shock response | HrcA | A |
| HssR | SA2151 | RS | + | + | + | + | + | + | − | Heme efflux pump | OmpR | A |
| HutR* | SA2123 | + | + | − | + | − | + | + | − | Histidine utilization | LysR | C |
| IcaR | SA2458 | RS | + | − | + | − | + | − | − | Intercellular adhesin production | TetR | A |
| LacR | SA1998 | + | + | + | + | + | − | − | − | Lactose utilization | DeoR | B (S. xylosus) |
| LexA | SA1174 | RS | + | + | + | + | + | + | RS | DNA damage stress | LexA | A |
| MalR | SA1339 | + | − | − | − | + | + | + | − | Maltose utilization | LacI | B (S. xylosus) |
| ManR | SA2433 | + | + | + | + | − | − | − | R | Mannose utilization | BglG | B |
| MecI | SA0040 | RS | − | − | − | − | − | − | − | Methicillin-penicillin resistance | MarR | A |
| MepR | SA0322 | RS | − | − | + | + | + | − | − | Multidrug resistance | MarR | A |
| MntR | SA0590 | RS | + | + | + | + | + | + | RS | Manganese homeostasis | DtxR | A |
| MtlR | SA1961 | + | + | − | + | + | + | − | R | Mannitol utilization | BglG | B |
| MurR* | SA0187 | + | + | + | + | + | + | − | + | N-Acetylmuramate utilization | RpiR | C |
| NrdR | SA1509 | + | + | + | + | + | + | + | + | Deoxyribonucleotide biosynthesis | NrdR | C |
| NreC | SA2179 | RS | + | + | + | + | − | + | − | Nitrate and nitrite respiration | OmpR | A |
| PdxR* | SA0476 | + | + | + | + | + | + | + | − | Pyridoxine biosynthesis | GntR | C |
| PerR | SA1678 | RS | + | + | + | + | + | + | RS | Oxidative stress | Fur | A |
| PurR | SA0454 | + | + | + | + | + | + | + | RS | Purine biosynthesis | PurR | B |
| RbsR | SA0261 | + | − | − | + | + | + | + | + | Ribose utilization | LacI | B (L. casei) |
| Rex | SA1851 | RS | + | + | + | + | + | + | RS | Anaerobic metabolism | Rex | A |
| SaeR | SA0661 | RS | + | + | + | − | − | − | − | Virulence | OmpR | A |
| ScrR | SA1847 | + | + | + | − | + | + | + | − | Sucrose utilization | LacI | B (S. xylosus) |
| TreR | SA0434 | + | − | − | + | + | + | + | RS | Trehalose utilization | GntR | B |
| YtrA | SA1748 | + | + | + | + | + | + | − | R | Unknown | GntR | B |
| Zur | SA1383 | RS | + | + | + | + | + | + | RS | Zinc homeostasis | Fur | A |
Novel TF names introduced in this work are marked by asterisks.
R, TFs with previously characterized target genes in S. aureus and B. subtlis; RS, TFs with previously determined TFBS motifs; +, presence of TFs orthologs; −, absence of orthologs.
Workflow A, expansion and projection of a previously characterized S. aureus regulon; workflow B, projection of an orthologous regulon from B. subtilis or other species; workflow C, ab initio regulon inference.
(i) Projection and expansion of experimentally known S. aureus regulons.
In order to capture existing regulatory network information, project regulons to other species of the Staphylococcaceae, and expand possible targets of known TFs, we focused on the reconstruction of regulons for 21 TFs in S. aureus that were previously experimentally investigated (“workflow A”) (Table 1). For this group of TFs, we propagated the previously established regulatory interactions in S. aureus in other species of the Staphylococcaceae and predicted new regulon members by the comparative genomics approach. For each TF, we started from the construction of an initial positional weight matrix (PWM) using the training set of known TFBSs in S. aureus. All analyzed genomes that possess an orthologous TF were then scanned by the initial PWM profile, resulting in the identification of candidate TFBSs for genes that are orthologous to the previously known target genes in S. aureus. In the next step, a collection of upstream regions of genes with candidate TFBSs identified during the initial step for all Staphylococcaceae genomes was used to build a refined PWM profile for a studied TF regulon. Finally, the regulon was reconstructed by applying this refined PWM to the analyzed genomes and identifying clusters of orthologous coregulated operons, as defined previously (30). The inferred regulon was corroborated by taking into consideration individual scores and the overall conservation of TFBSs across the genomes as well as the genomic and functional context of regulated genes. The latter analysis was performed by using a combination of tools implemented in SEED (34) and MicrobesOnline (8).
The outcome was that 21 TF regulons previously described for S. aureus were propagated to six other species within the Staphylococcaceae lineage (see Table S2 in the supplemental material). Moreover, we report the significant expansion of nine previously characterized regulons in S. aureus by 114 novel regulatory interactions (Table 2). Three-quarters of the novel target operons in S. aureus were found to be under the regulation of two global regulators of carbon and amino acid metabolism. The catabolite control protein A (CcpA) regulon includes 46 newly discovered operons that are involved mostly in various sugar and amino acid utilization pathways, central carbohydrate metabolism, and the citric acid cycle. In addition, the CcpA regulon in S. aureus appears to control genes encoding virulence factors, such as toxin 1, immunoglobulin G binding protein, alpha-hemolysin, and beta-lactamase. A comparative analysis of the amino acid and nitrogen utilization regulon CodY revealed 39 novel target operons in S. aureus that are involved mainly in the amino acid biosynthesis and utilization pathways, the uptake of amino acids and ammonium, and the transport and hydrolysis of oligopeptides. The NAD+/NADH-responsive regulator Rex, controlling anaerobic respiration and fermentation, has an intermediate-sized regulon that was expanded by six newly found members in S. aureus, including alcohol dehydrogenase (adh2), the nitrate-nitrite antiporter (narK), and the cytochrome bd transport system (cydDC). The heat shock response regulon CtsR, previously known to control the hrcA-grpE-dnaKJ and groSL operons, was expanded by four novel target operons in S. aureus, including ctsR-SA0481-SA0482-clpC, clpB, and clpP, whose orthologs are known members of the CtsR regulon in B. subtilis. The hydrogen peroxide stress regulon PerR in S. aureus was expanded by two new candidate target operons involved in Fe-S assembly and heme biosynthesis, respectively. The cysteine metabolism regulon CymR reconstructed in species of the Staphylococcaceae includes seven new candidate target operons (four in S. aureus) encoding enzymes for inorganic sulfur assimilation and S-adenosylhomocysteine recycling. The iron-responsive regulon Fur in S. aureus was expanded by seven new possible target operons involved in iron storage, iron siderophore (staphylobactin) biosynthesis, ferrous iron transport, and the regulation of virulence. The reconstructed Zur regulon for zinc homeostasis in S. aureus includes four newly predicted target operons encoding putative components of zinc uptake systems and monooxygenase as well as a Zn-independent paralog of the ribosomal protein L33.
Table 2.
Genomics-based expansion of experimentally characterized regulons in S. aureus
| TF | Functional role | Total no. of targetsa | No. of known targetsb | No. of new targetsc | Reference(s) |
|---|---|---|---|---|---|
| CcpA | Carbon catabolism | 73 | 27 | 46 | 46 |
| CodY | Amino acid metabolism | 62 | 23 | 39 | 25, 38 |
| Rex | Anaerobic metabolism | 21 | 15 | 6 | 35 |
| LexA | DNA damage stress | 10 | 9 | 1 | 7 |
| CtsR | Heat shock response | 6 | 2 | 4 | 5 |
| PerR | Oxidative stress | 9 | 7 | 2 | 18 |
| CymR | Cysteine/sulfur metabolism | 12 | 8 | 4 | 50 |
| Fur | Iron homeostasis | 19 | 12 | 7 | 52 |
| Zur | Zinc homeostasis | 8 | 4 | 4 | 24 |
| Total | 220 | 107 | 113 |
Total number of operons constituting a reconstructed TF regulon in S. aureus.
Number of operons in the predicted regulon previously shown to be under the control of the corresponding TF in S. aureus.
Number of newly predicted target operons for the reconstructed regulon in S. aureus.
(ii) Reconstruction of regulons by projection from B. subtilis and other related species.
A slightly different computational workflow was utilized for the reconstruction of 19 regulons controlled by TFs with previously studied orthologs in B. subtilis and other members of the Firmicutes (“workflow B”) (Table 1). The following strategy was used for the reconstruction of these regulons in the Staphylococcaceae genomes: (i) the construction of an initial PWM using known TFBSs from the studied organism (e.g., B. subtilis), (ii) the finding of orthologs for a TF and its known target genes in the Staphylococcaceae, (iii) the scanning of the genomes using the initial PWM to identify candidate TFBSs upstream of the orthologs of known target genes, (iv) the building of a novel PWM using upstream regions of genes with identified candidate TFBSs, and (v) applying the Staphylococcus-specific PWM to obtain the final regulon reconstruction using the comparative approach (see above). Workflow B was used for the reconstruction of regulons for 15 TFs that have orthologs in B. subtilis that were previously experimentally investigated and for 4 TFs, namely, LacR, MalR, RbsR, and ScrR, that were studied for Staphylococcus xylosus or Lactobacillus casei (Table 1).
The comparative contents of 19 Staphylococcaceae regulons reconstructed using workflow B are shown in Table S2 in the supplemental material. All of these regulons are local regulons that control up to six target operons per genome. In most cases, the genes encoding these local TFs were found in close proximity to the regulated target genes. Ten regulons controlling individual catabolic pathways for the utilization of various carbohydrates, such as fructose (FruR), gluconate (GntR), lactose (LacR), maltose (MalR and GlvR), mannose (ManR), mannitol (MtlR), ribose (RbsR), sucrose (ScrR), and trehalose (TreR), are not uniformly distributed among the analyzed Staphylococcaceae genomes. Furthermore, the appearance of the corresponding TFs in the genomes correlates strictly with the presence of orthologs of their target regulated genes.
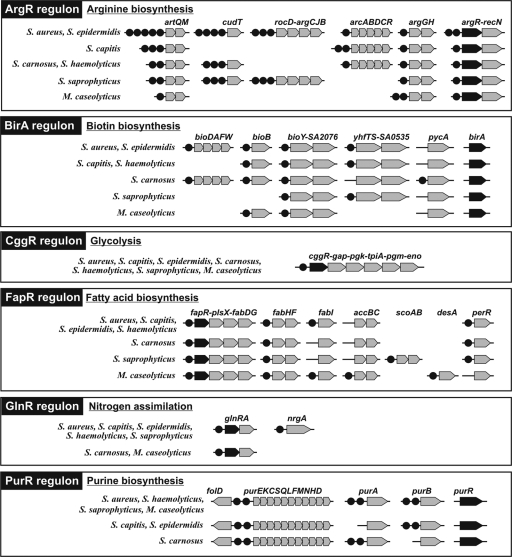
The six other regulons controlling genes involved in the essential biosynthetic pathways, such as the arginine (ArgR), biotin (BirA), fatty acid (FapR), glutamine (GlnR), and purine (PurR) regulons, and the central glycolytic pathway (CggR) are universally conserved in all studied members of the Staphylococcaceae (Fig. 2). Most of these regulons have a core set of coregulated genes conserved across all species, while the remaining genes in the regulon are not universally conserved. For instance, the arginine regulon ArgR includes a conserved core of operons in all members of the Staphylococcaceae (argGH, artQM, and argR-recN), whereas the remaining regulon members (cudT, rocD-argCJB, and arcABDCR) are present in a subset of species; the latter set of operons is always coregulated by ArgR. The purine biosynthesis regulon PurR includes two strongly conserved target operons (folD and purEKCSQLFMNHD), whereas two other predicted PurR-regulated genes (purA and purB) have missing PurR binding sites in some species of the Staphylococcaceae. The conserved core of the BirA regulon in the Staphylococcaceae includes all genes from the biotin biosynthesis and uptake subsystem (bioDAFW, bioB, and bioY) as well as the putative long-chain fatty acid coenzyme A (CoA) ligase and acetyl-CoA C-acetyltransferase (yhfTS). An additional candidate BirA-regulated gene encoding biotin-dependent pyruvate carboxylase (pycA) was identified uniquely in S. carnosus, but this regulatory interaction is not conserved in other species. The fatty acid biosynthesis regulon FapR contains two candidate target operons (fapR-plsX-fabDG and fabHF) with totally conserved FapR binding sites and a single gene (fabI) whose FapR-dependent regulation is not conserved in two species of the Staphylococcaceae. Another predicted member of the FapR regulon with a candidate binding site conserved in six Staphylococcus species encodes the peroxide-responsive regulator PerR, which is not directly involved in fatty acid metabolism. In addition, the FapR regulon has species-specific expansions to include some fatty acid metabolism genes, such as the acetyl-CoA carboxylase accBC and the fatty acid desaturase desA in M. caseolyticus and the 3-oxoacid CoA-transferase scoAB in S. saprophyticus.
Fig. 2.
Novel Staphylococcus regulons for essential biosynthetic and glycolytic pathways reconstructed by using the projection of known B. subtilis regulons. Genes from each regulon are shown by arrows; TF genes are in black. TF binding sites are shown by black circles.
(iii) Ab initio inference of regulons.
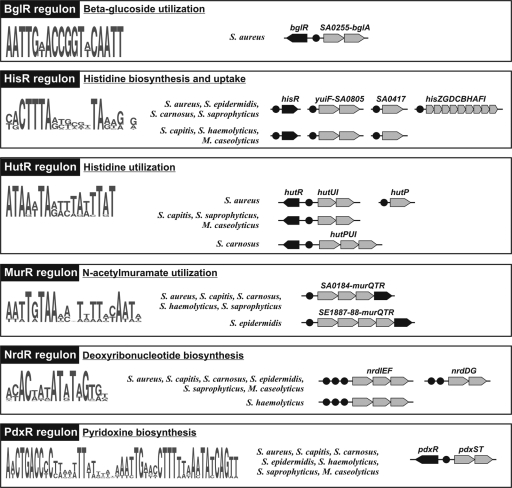
The identification of 6 novel regulons for previously uncharacterized TFs in the Staphylococcaceae was performed by a genome- and functional context-based approach, which utilizes sets of potentially coregulated genes from different input data (“workflow C”) (Table 1). Three regulons were inferred by the analysis of conserved gene neighborhoods, including a putative TF gene: the pyridoxine biosynthesis regulon PdxR*, the N-acetylmuramate utilization regulon MurR*, and the β-glucoside catabolism regulon BglR* (asterisks indicate a new TF name introduced in this study). Based on observations that functionally related genes tend to colocalize on the chromosome and that such a functional gene colocalization is often conserved across species (33), we analyzed the conserved loci containing TF genes and analyzed putative regulatory regions for the presence of conserved motifs. Three novel regulons for histidine metabolism (HisR* and HutR*) and deoxyribonucleotide biosynthesis (NrdR) were identified by the subsystem-based approach (41), when conserved motifs were found in upstream regions of different genes from the same metabolic subsystem (Fig. 3). Candidate TFs were then attributed to the regulons by using a combination of various genomic associations (i.e., colocalization, cooccurrence, and coregulation) of a putative TF gene with the predicted genes in the regulon.
Fig. 3.
Novel Staphylococcus regulons reconstructed using the ab initio regulon inference approach. Genes from each regulon are shown by arrows; TF genes are in black. TF binding sites are shown by black circles.
The HisR* regulon operates by a 20-bp palindromic DNA motif that appears upstream of the histidine biosynthesis his operon in four Staphylococcaceae genomes, whereas the other three studied genomes lack the his operon. Scanning of the genomes with the constructed PWM identified three additional genes that share a similar DNA motif. The inferred regulon includes a hypothetical TF from the TrpR repressor family (SA1723) and two hypothetical solute transporters (SA0804 and SA0417) that have absolutely conserved candidate binding sites corresponding to the novel motif. A similar DNA motif was identified for the his operon and orthologs of SA0804 (yuiF) and SA1723 (yerC) in Bacillus subtilis and related genomes (A. G. Vitreschak, unpublished observation), suggesting that YerC (HisR*) is a novel histidine-responsive transcriptional repressor for the histidine biosynthesis operon and the putative histidine uptake genes yuiF and SA0417 in the Bacillus-Staphylococcus group.
The second regulon inferred for the Staphylococcaceae by the subsystem-based approach, HutR*, operates by a conserved 17-bp palindromic DNA motif that coregulates the histidine utilization operon hutUI and the histidine uptake permease hutP. The inferred regulon also includes a hypothetical LysR family TF gene (SA2123), which is located just upstream of the hutUI operon in the divergent direction, and thus, it shares the same regulatory region with the target operon. Orthologs of SA2123 were identified only in those five Staphylococcaceae genomes that encode the histidine utilization genes. Based on these combined genomic context evidences, we tentatively proposed that SA2123 (HutR*) is a novel histidine utilization regulator in the Staphylococcaceae.
Analysis of the “ribonucleotide reduction” subsystem in the SEED database revealed two ribonucleotide reductase operons, nrdIEF and nrdDG, that are conserved in all studied Staphylococcaceae genomes, with the exception that the nrdDG operon is absent from S. haemolyticus. Conserved 16-bp palindromic sites have been identified in the candidate regulatory regions of all nrd genes (Fig. 3). In most cases, these candidate sites were found in a tandem arrangement with two or three sites per regulatory region. The inferred conserved motif for the nrd-associated candidate binding sites is highly similar to the sequences of NrdR repressor binding sites previously described for Escherichia coli (51) and Streptomyces coelicolor (16). We identified orthologs of NrdR in S. aureus (SA1509) and all other studied Staphylococcaceae genomes. Based on the above-described observations, we proposed that SA1509 is the NrdR repressor that binds to the candidate 16-nucleotide (nt) binding sites in the regulatory regions of nrd genes.
Comparison of site motifs for orthologous regulators in the Staphylococcaceae and B. subtilis.
Thirty-one analyzed TF regulons in the Staphylococcaceae have orthologous regulators in B. subtilis (Table 1). Orthologs of five TFs (BglR*, HisR*, MurR*, NrdR, and RbsR) in B. subtilis were not studied experimentally. Five other orthologous regulators (FruR, GlvR, ManR, MtlR, and YtrA) in B. subtilis were previously studied, but their TFBSs are still unknown. For these last two groups of orthologous TFs in S. aureus, we report, for the first time, the likely identity of their cognate TFBSs (see the corresponding regulon pages in the RegPrecise database [29]). For the other 21 regulons reconstructed for the Staphylococcaceae, the deduced TFBS motifs were compared to previously known motifs for orthologous regulators in B. subtilis by using the TBTBS database (48) (see Table S4 in the supplemental material). Four TFBS motifs (for ArsR, CggR, MntR, and PurR) in the Staphylococcaceae are substantially different from the respective motifs in B. subtilis. Only minor differences were detected in the consensus HrcA binding-site sequences that are inverted repeats; however, the distance between these two repeats is 2 bp longer in the Staphylococcaceae. Five other TFBS motifs (for ArgR, FapR, GltC, TreR, and Zur) in the Staphylococcaceae are moderately different (2 to 4 mismatches in the conserved positions) from the known motifs in B. subtilis. The remaining 11 TFs appear to have binding motifs that are well conserved or only slightly variable in comparison with the motifs of their previously characterized B. subtilis orthologs. Remarkably, among the latter group of TFs, there are many global regulators (CcpA, CodY, Fur, and Rex) and regulators that control 6 to 12 target operons (CtsR, CymR, LexA, and PerR). The latter observation suggests that the conservation of TFBS motifs might have a positive correlation with the regulon size.
Comparative analysis of predicted regulons and microarray expression data for S. aureus.
Microarrays are a standard method used to evaluate the global gene expression patterns of many organisms under a variety of experimental conditions. The amount of expression data for certain model organisms is rapidly increasing and becoming more readily available to the broader scientific community through large repositories such as the Gene Expression Omnibus (GEO) (3), ArrayExpress (36), and M3D (11). With the availability of a large number of data from microarray experiments for any one organism, it becomes possible to evaluate the tendency of any two genes in an organism to be expressed in a coordinated fashion under a wide variety of biological conditions. We have computed the Pearson correlations (PCs) between each pair of genes in the S. aureus genome based on the expression of each gene in 850 microarrays, kindly provided by the Paul Dunman laboratory, and we have evaluated the predicted regulons constructed in this work in the context of these data. In so doing, our goal was not to validate or refute any particular relationship between genes in the predicted regulons but rather to assess an overall consistency of the predicted regulons with a large-scale expression data set.
To facilitate comparative analyses, we calculated the means, medians, and 80th percentiles to summarize all pairwise PCs within each of the 44 predicted regulons containing 2 or more genes. For example, in a regulon containing 10 genes, there are 55 pairs of genes, and the PC is computed for each pair. Pairwise PC values are then summarized using the means, medians, and 80th percentiles as a measure of the overall correlation of genes within each regulon-defined set. The calculated summary PC values indicate how consistent the genes in a set are with respect to changes in gene expression over the entire set of 850 microarray experiments. In order to observe how genes linked by a particular TF are correlated in the context of global expression profile experiments and to establish a baseline for comparisons of summary PC values, we focused on the set of TFs and their targets that have been experimentally characterized. This subset included only those genes in the target set for which experimental evidence of interactions existed prior to this study (i.e., all genes in a regulon highlighted in blue in Table S2 in the supplemental material). The result is a set of TF-target gene relationships for 20 regulons that we considered to be a “gold standard.” The summary PC values for these regulons vary greatly (see Table S5 in the supplemental material), ranging from highly correlated regulons (mean PC of 0.87 for hssR) to less well-correlated regulons (mean PC of 0.20 for saeR). Comparisons of summary PC values derived from random sets of genes of the same size showed that 14 of the 20 (70%) gold-standard regulons have summary PC values that would not be expected to occur by chance (P ≤ 0.05). Gene sets with summary PC values as low as 0.36 were still statistically significant, which included the second largest regulon, ccpA. This indicates that summary PC values even for those regulons that contain only experimentally validated TF-target gene interactions can vary widely and can be found to be no better than what would be expected to occur by chance. The low summary PC values for known regulons can be explained by the intrinsic complexity of real TRNs caused by many cross talks between TFs (multiple-regulation, regulatory cascades).
Considering the entire set of predicted regulons described in this study, the patterns seen for the summary PC values mirror those of the gold standard, ranging from highly correlated gene sets (mean PC of 0.93 for murR) to less well-correlated gene sets (mean PC of 0.25 for codY). In this case, 33 of the 44 predicted regulons (75%) have summary PC values considered to be significantly better than what would occur by chance. Most of the regulons considered significantly correlated have summary mean PC values above 0.5, but significantly correlated gene sets range down to a summary mean PC value of 0.31 (ccpA). This indicates that while one should expect a reasonably high PC value for truly coregulated gene sets, this is not uniformly the case, as some regulons yield low PC values.
A variety of factors could influence the extent of the correlation of genes within a particular regulon, including the number of genes in the regulon, the number of different loci represented in the regulon, the autoregulation of the regulon (i.e., the TF that regulates the set is a target of self-regulation), and, possibly, the type of workflow used for regulon predictions. In order to understand which factors were most strongly and uniquely associated with the observed summary PC values across the 49 regulons, we conducted a multiple-regression analysis, predicting summary PC values (separate models for means, medians, and 80th percentiles) by the four factors mentioned above. We found that there was a significant (P ≤ 0.05) inverse relationship between the number of genes in a regulon and the observed summary PC values; namely, as the number of genes in a regulon increases, the summary PC value decreases after controlling for the other three factors. Likewise, there was a significant relationship between the operonal organization of the regulon (single locus/operon versus multiple loci/operons) and the summary PC value (single-locus summary PC values were higher). In contrast, there was not a significant relationship between the workflow used for regulon inference and the observed summary PC values. Additionally, there was little difference in the summary mean PC values considered to be significant for projected (16/19 [85%]) and ab initio (5/6 [83%]) methods (see Table S5 in the supplemental material). These two observations indicate that the ab initio prediction of regulons (workflow C) may perform as well as prediction methods for regulons that are derived, in part, from previously reported data for a TF (workflows A and B). Lastly, we note that the multiple-regression analysis showed no evidence that the autoregulation of an operon contributes significantly to the summary PC values for a regulon.
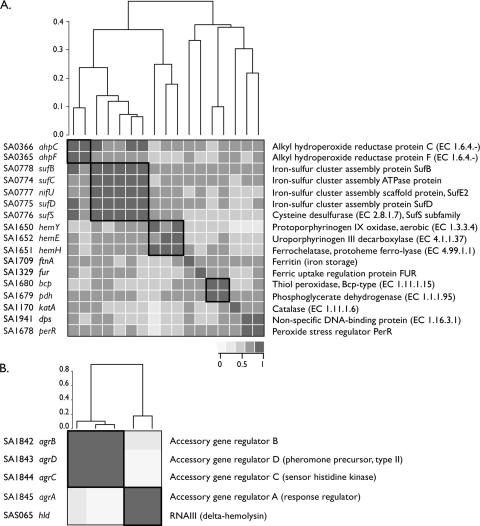
However, we have anecdotally observed that there are autoregulated regulons with a tendency for the TF to be well correlated with the rest of the genes in the regulon and those where the TF is not well correlated. For example, compare the regulons in Fig. 4A and B, which depict heat map representations of the pairwise PC for each gene in the regulons controlled by PerR and AgrA, respectively. It is clear that the regulator in each operon behaves very differently: perR expression is well correlated, whereas agrA expression is not well correlated with the rest of the regulon. This is also evident in the summary PC values for the regulons when the value for all members of the regulon is compared to the value obtained excluding the TF gene (means of 0.62 versus 0.62 for perR and means of 0.42 versus 0.52 for agrA) (see Table S5 in the supplemental material). When making the same comparison for autoregulated regulons with at least 4 loci and fewer than 20 loci, there is not a statistically significant difference in the summary PC values (0.62 with regulator versus 0.66 without regulator; P = 0.08). Despite the lack of a statistically supported trend, it is clear that in specific cases, the TF does not always have an expression pattern similar to those of its targets. The reasons for this could be the influence of other factors affecting the expression of the TF as well as a basal level of TF expression under all experimental conditions in the microarray data set.
Fig. 4.
Heat maps of Pearson correlations for the PerR and AgrA regulons. (A) Pairwise Pearson correlations of all predicted PerR regulon members depicted as a heat map. The shading of each box corresponds to the PC value between the gene pair according to the scale shown below the heat map. Hierarchical clustering was used to order the genes based on distances calculated by using the PC values, and the resulting dendrogram is shown above the heat map. The scale bar represents the calculated distance, with a value of 0.0 corresponding to a PC of 1 (perfectly correlated). The identity of each gene is given on the left, and the corresponding function is given on the right. Genes corresponding to operonal units are boxed in the heat map. (B) Pairwise Pearson correlations of all predicted AgrA regulon members depicted as a heat map. Boxes in the heat map highlight the organization of the regulon into two distinct clusters, which are separated by a distance of greater than 0.8 (corresponding to a PC of less than 0.2).
The analysis of PC values for individual regulons can lend confidence to the predicted regulatory interactions and allows one to infer the substructure within a regulon. Take, for example, the PerR regulon, which has been expanded by two operons (sufCDF-nifU and hemEHY) in this study. There is good correlation between these novel operons and experimentally known members of the regulon (Fig. 4A). This provides evidence that PerR, which senses oxidative stress and maintains metal homeostasis in S. aureus (18), also controls the final steps of protoheme biosynthesis and the formation of iron-sulfur clusters via the SufBCD-SufSE system (22, 42, 53). The heme biosynthesis pathway was identified as a member of the PerR regulon in B. subtilis (6). Finally, there is an expected substructure within the PerR regulon when genes in the same operon are highly correlated within the expression data (Fig. 4A).
The AgrA regulon serves as a striking example of a regulator being controlled by other factors. The agr locus has been extensively studied as one of the primary regulators that control the expression of virulence factors in S. aureus (31). AgrA is a cell density-dependent activator of the agr locus, which is composed of two divergent transcripts, RNAII and RNAIII. The activity of RNAIII controls the regulation of downstream targets of AgrA (that is, they are indirectly controlled by AgrA). The heat map of AgrA regulon members clearly shows two groups of genes, one composed of agrBDC and the other composed of agrA and RNAIII (represented by hld) (Fig. 4B). In fact, there is no apparent correlation between the transcription of agrA and that of agrBDC, despite these genes clearly being organized as an operon (agrBDCA), and the RNAII transcript, including all four genes (32). This suggests the presence of a promoter that drives the expression of agrA independent of the upstream genes. Evidence for a weak promoter upstream of agrA was seen in an early study of the agr locus (37). More recently, an analysis of the CodY regulon in S. aureus (25) demonstrated that CodY binds to the agr locus upstream of agrA within agrC and acts as a repressor of agr locus transcription. The presence of this additional binding site is fully consistent with the pattern of expression seen in Fig. 4B. The current evaluation of the expression of the agr locus represents the behavior of the locus under diverse conditions and in diverse strains, suggesting that the predominant form of agrA expression is independent of the rest of the agr RNAII locus. It should be noted that the 850 experiments used in this analysis include the array data used in the previously reported study of CodY (25) (representing S. aureus codY, agrA, and codY-agrA mutants), but the removal of the CodY-specific array data does not affect the observed expression pattern (data not shown).
Interconnectivity in the reconstructed S. aureus transcriptional regulatory network.
Potential cross talk between regulons can be identified through the identification of binding sites for multiple TFs in the upstream regions of the same operons. Among 271 target operons in the reconstructed TRN of S. aureus, 41 operons share two or more TFs, revealing an overlap between the respective regulons (see Table S2 in the supplemental material). The greatest extent of regulatory cross talk was found for the arcABDCR operon, which belongs to the CcpA, Rex, ArgR, and ArcR regulons. This observation suggests a complex regulation of the anaerobic arginine catabolic pathway by multiple cellular signals, including the availability of electron acceptors and carbon sources and the presence of arginine. The global regulons CcpA and CodY overlap with many local regulons jointly controlling the expression of 30 S. aureus operons involved in the metabolism of amino acids, nitrogen, and carbohydrates. For instance, the CcpA regulon overlaps with 8 out of 10 local regulons that control various sugar catabolic pathways (FruR, GlvR, GntR, MalR, ManR, RbsR, ScrR, and TreR), whereas the CodY regulon controls several operons from amino acid metabolism regulated by their specific TFs (ArgR, CymR, HisR, GlnR, and GltC). Two other noteworthy examples of partially overlapping regulons include the NreC and Rex regulons, controlling nitrate/nitrite respiration and the redox response, and the HrcA and CtsR regulons that control a heat shock response (5). The autoregulation of a TF is a common regulatory network motif in S. aureus: 33 of the 46 studied TFs were predicted to control their own expression. Multiple regulatory cascades between various TFs and feed-forward loop motifs can be detected in the reconstructed S. aureus TRN. For instance, the global regulator CcpA controls 10 local TFs that are mostly specific for various sugar catabolic subsystems. Other regulatory cascades identified in S. aureus include CodY for glnR, Rex for arcR and nreC, CtsR for hrcA, PerR for fur, and FapR for perR.
The interaction between the PerR and FapR regulons was first proposed in this study based on the identification of a candidate FapR binding site upstream of perR. We assessed this regulatory cross talk prediction by using microarray expression data. The heat map of the FapR regulon is consistent with the prediction that perR is a member of the regulon, although perR is an outlier with respect to other genes of the FapR regulon (see Fig. S1A in the supplemental material). This observation is consistent with perR being well correlated with members of the PerR regulon (see above) (Fig. 4A). However, the observation that the perR expression level in S. aureus does not significantly increase in response to H2O2 exposure, in contrast to most of the PerR regulon (18), indicates that there must be a more complex regulation of perR.
The nature of the interaction between these two regulons suggests a possible link between the cell oxidative stress response and fatty acid homeostasis, both of which are critical to cell survival: a perR deletion was shown previously to attenuate the virulence of S. aureus (18), and fatty acid biosynthesis remains an attractive target for antibiotic development (54). Many macromolecules are affected during oxidative stress (including DNA, proteins, and lipids), which was shown previously to slow down growth (13, 47). One of the primary defenses of S. aureus in response to peroxide-induced stress is the activation of the PerR regulon, which promotes the removal of H2O2 and damaged cellular components (18). PerR senses peroxide stress by interacting with H2O2, which leads to irreversible changes in the conformation of PerR. These changes cause the permanent release of PerR from DNA and result in the upregulation of the genes in the regulon (reviewed in reference 15). Thus, there are apparently two conditions that must be met in order to repress the activated PerR regulon: new PerR must be synthesized, and H2O2 levels must be reduced. However, in the regulatory cascade predicted in this study, FapR potentially imposes a third restriction by repressing the transcription of perR: FapR would be able to bind the upstream region of perR in the absence of PerR occupancy, since the binding sites for FapR and PerR upstream of perR are predicted to partially overlap (see Fig. S1B in the supplemental material). The FapR regulon in B. subtilis has been extensively characterized, revealing that FapR suppresses the expressions of many of the type II fatty acid synthesis genes by sensing intracellular pools of malonyl-CoA (44, 45) and/or malonyl-acyl carrier protein (ACP) (26). As these pools accumulate, FapR is released from DNA, allowing the increased transcription of genes for fatty acid synthesis in response to the cellular demand (26). Global transcription profiles of B. subtilis (28) and S. aureus (4) under conditions of peroxide-induced oxidative stress show that genes in the FapR regulon are downregulated, and in S. aureus, this coincides with a temporary growth arrest. Under such conditions, it is plausible that malonyl-CoA/ACP pools would be altered, affecting the occupancy of FapR regulatory sites. Upon encountering oxidative stress, PerR would be released from the perR binding site, but FapR would be available initially to occupy the vacancy. It was shown previously that AccBC activity is linked to macromolecular synthesis and the growth rate (23, 54), and this would lead to increased levels of malonyl-CoA/ACP pools in response to more favorable growth conditions (i.e., the alleviation of oxidative stress). Such a regulatory circuit would ensure that the timing of perR expression coincided with cellular processes indicative of the growth state and that excess PerR would not be produced in a situation where PerR interacts with high levels of peroxide and is rendered unable to bind cognate DNA sites. Despite the speculative nature of this interpretation, it could be tested by the analysis of perR expression in a fapR mutant.
Conclusions.
Transcriptional regulation in the family Staphylococcaceae has been previously studied through various experimental approaches with S. aureus. With the sequencing of multiple complete genomes, the further development of comparative genomic methods, and the accumulation of a large amount of experimentally derived data on transcriptional regulation, the integrative approach for the reconstruction of TRNs has become possible and was applied in this study to reconstruct TRN in S. aureus. The inferred network includes 46 TFs connected to ∼550 target genes that are organized into ∼270 candidate operons using ∼400 candidate TFBSs. To our knowledge, this is the largest TRN of S. aureus that has been assembled to date, comprising 20% of its genome. A total of 147 (46%) of 317 regulatory interactions in this network are based on experimental studies of 21 TFs in S. aureus, whereas the remaining 170 interactions are novel, which were predicted by utilizing the comparative genomics approach. These predictions await future experimental validation and include 6 novel TF regulons that control the carbohydrate utilization pathways (BglR* and MurR*), the histidine biosynthesis and utilization pathways (HisR* and HutR*), the pyridoxine biosynthesis pathway (PdxR*), and ribonucleotide reductases (NrdR).
The bottom-up genomic approach for regulon reconstruction employed here is complementary to alternative top-down approaches to TRN reconstruction using high-throughput gene expression data (e.g., the context likelihood of relatedness algorithm [12] and the cMonkey algorithm based on the integrated biclustering of heterogeneous genome-wide data sets [39]), but it also has a number of important advantages over context likelihood of relatedness (CLR)-type approaches. Although microarray data provide us with a potentially very rich source of information, the regulatory network inference from these data is a challenging and as-yet-unresolved task. Among many complications, the observed patterns of gene coexpression are obscured by regulatory cascades and other complex interactions within the TRN that may not be easily resolved. In other words, relationships between regulons (that are well defined at the genomic level) and so-called modulons (deduced from microarray data) are fairly complicated. We believe that a systematic comparison of regulons reconstructed from genomic data with microarray data would substantially contribute to our understanding of regulatory interactions in the cells. To this end, we performed a comparative analysis of the reconstructed regulons reported in this study with data from 850 microarray experiments with S. aureus. This analysis contributes to confidence in the predicted regulatory interactions, helps to formulate testable hypotheses for the identified substructure within regulons (such as the hypothesis that FapR controls perR [see above]), and can yield new insights into overall regulatory patterns of genes under diverse conditions.
Along with a number of advantages, the comparative genomics approach for TRN reconstruction has several limitations. First, this approach allows the confident prediction of regulatory interactions that are conserved between related microbial species, but it often fails with respect to the prediction of species-specific regulatory interactions that are not conserved in closely related microbial genomes. Second, the genome context analysis has a limited ability to assign a novel TF to a candidate TFBS motif for novel inferred regulons. This approach is useful primarily to assign TFs to local transcriptional regulons, when a TF-encoding gene is colocalized with the inferred genes in the regulon. However, the assignment of TFs to novel global regulons inferred by either the bottom-up genomic approach applied in this work or the top-down high-throughput expression data-based algorithms remains a challenging task.
Supplementary Material
ACKNOWLEDGMENTS
We thank Paul Dunman for providing the S. aureus microarray data set and Ross Overbeek for assistance with the RAST/SEED system.
This work was supported by the National Science Foundation under award DBI-0850546 (M.D.) and by the Office of Science (BER), U.S. Department of Energy, under contract DE-SC0004999 (D.A.R.). The Lawrence Berkeley National Laboratory is funded by the U.S. Department of Energy Genomics GTL program (grant DE-AC02-05CH11231). Additional funding was provided by the Russian Academy of Sciences (program Molecular and Cellular Biology) and the Russian Foundation for Basic Research (10-04-01768).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 29 April 2011.
REFERENCES
- 1. Alkema W. B., Lenhard B., Wasserman W. W. 2004. Regulog analysis. Detection of conserved regulatory networks across bacteria: application to Staphylococcus aureus. Genome Res. 14:1362–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ballal A., Manna A. C. 2009. Expression of the sarA family of genes in different strains of Staphylococcus aureus. Microbiology 155:2342–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barrett T., et al. 2011. NCBI GEO: archive for functional genomics data sets—10 years on. Nucleic Acids Res. 39:D1005–D1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chang W., Small D. A., Toghrol F., Bentley W. E. 2006. Global transcriptome analysis of Staphylococcus aureus response to hydrogen peroxide. J. Bacteriol. 188:1648–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chastanet A., Fert J., Msadek T. 2003. Comparative genomics reveal novel heat shock regulatory mechanisms in Staphylococcus aureus and other Gram-positive bacteria. Mol. Microbiol. 47:1061–1073 [DOI] [PubMed] [Google Scholar]
- 6. Chen L., Keramati L., Helmann J. D. 1995. Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc. Natl. Acad. Sci. U. S. A. 92:8190–8194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cirz R. T., et al. 2007. Complete and SOS-mediated response of Staphylococcus aureus to the antibiotic ciprofloxacin. J. Bacteriol. 189:531–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dehal P. S., et al. 2010. MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res. 38:D396–D400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Disz T., et al. 2010. Accessing the SEED genome databases via Web services API: tools for programmers. BMC Bioinformatics 11:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dunman P. M., et al. 2004. Uses of Staphylococcus aureus GeneChips in genotyping and genetic composition analysis. J. Clin. Microbiol. 42:4275–4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Faith J. J., et al. 2008. Many Microbe Microarrays Database: uniformly normalized Affymetrix compendia with structured experimental metadata. Nucleic Acids Res. 36:D866–D870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Faith J. J., et al. 2007. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 5:e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Farr S. B., Kogoma T. 1991. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol. Rev. 55:561–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gelfand M. S. 2006. Evolution of transcriptional regulatory networks in microbial genomes. Curr. Opin. Struct. Biol. 16:420–429 [DOI] [PubMed] [Google Scholar]
- 15. Giedroc D. P. 2009. Hydrogen peroxide sensing in Bacillus subtilis: it is all about the (metallo)regulator. Mol. Microbiol. 73:1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grinberg I., et al. 2009. Functional analysis of the Streptomyces coelicolor NrdR ATP-cone domain: role in nucleotide binding, oligomerization, and DNA interactions. J. Bacteriol. 191:1169–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hecker M., Reder A., Fuchs S., Pagels M., Engelmann S. 2009. Physiological proteomics and stress/starvation responses in Bacillus subtilis and Staphylococcus aureus. Res. Microbiol. 160:245–258 [DOI] [PubMed] [Google Scholar]
- 18. Horsburgh M. J., Clements M. O., Crossley H., Ingham E., Foster S. J. 2001. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect. Immun. 69:3744–3754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Irizarry R. A., et al. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kazakov A. E., et al. 2007. RegTransBase—a database of regulatory sequences and interactions in a wide range of prokaryotic genomes. Nucleic Acids Res. 35:D407–D412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kummerfeld S. K., Teichmann S. A. 2006. DBD: a transcription factor prediction database. Nucleic Acids Res. 34:D74–D81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Layer G., et al. 2007. SufE transfers sulfur from SufS to SufB for iron-sulfur cluster assembly. J. Biol. Chem. 282:13342–13350 [DOI] [PubMed] [Google Scholar]
- 23. Li S. J., Cronan J. E., Jr 1993. Growth rate regulation of Escherichia coli acetyl coenzyme A carboxylase, which catalyzes the first committed step of lipid biosynthesis. J. Bacteriol. 175:332–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lindsay J. A., Foster S. J. 2001. zur: a Zn(2+)-responsive regulatory element of Staphylococcus aureus. Microbiology 147:1259–1266 [DOI] [PubMed] [Google Scholar]
- 25. Majerczyk C. D., et al. 2010. Direct targets of CodY in Staphylococcus aureus. J. Bacteriol. 192:2861–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martinez M. A., et al. 2010. A novel role of malonyl-ACP in lipid homeostasis. Biochemistry 49:3161–3167 [DOI] [PubMed] [Google Scholar]
- 27. Mironov A. A., Vinokurova N. P., Gel'fand M. S. 2000. Software for analyzing bacterial genomes. Mol. Biol. (Mosk.) 34:253–262 [PubMed] [Google Scholar]
- 28. Mostertz J., Scharf C., Hecker M., Homuth G. 2004. Transcriptome and proteome analysis of Bacillus subtilis gene expression in response to superoxide and peroxide stress. Microbiology 150:497–512 [DOI] [PubMed] [Google Scholar]
- 29. Novichkov P. S., et al. 2010. RegPrecise: a database of curated genomic inferences of transcriptional regulatory interactions in prokaryotes. Nucleic Acids Res. 38:D111–D118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Novichkov P. S., et al. 2010. RegPredict: an integrated system for regulon inference in prokaryotes by comparative genomics approach. Nucleic Acids Res. 38:W299–W307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Novick R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429–1449 [DOI] [PubMed] [Google Scholar]
- 32. Novick R. P., et al. 1995. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Genet. 248:446–458 [DOI] [PubMed] [Google Scholar]
- 33. Osterman A., Overbeek R. 2003. Missing genes in metabolic pathways: a comparative genomics approach. Curr. Opin. Chem. Biol. 7:238–251 [DOI] [PubMed] [Google Scholar]
- 34. Overbeek R., et al. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33:5691–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pagels M., et al. 2010. Redox sensing by a Rex-family repressor is involved in the regulation of anaerobic gene expression in Staphylococcus aureus. Mol. Microbiol. 76:1142–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Parkinson H., et al. 2011. ArrayExpress update—an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res. 39:D1002–D1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Peng H. L., Novick R. P., Kreiswirth B., Kornblum J., Schlievert P. 1988. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 170:4365–4372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pohl K., et al. 2009. CodY in Staphylococcus aureus: a regulatory link between metabolism and virulence gene expression. J. Bacteriol. 191:2953–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Reiss D. J., Baliga N. S., Bonneau R. 2006. Integrated biclustering of heterogeneous genome-wide datasets for the inference of global regulatory networks. BMC Bioinformatics 7:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rodionov D. A. 2007. Comparative genomic reconstruction of transcriptional regulatory networks in bacteria. Chem. Rev. 107:3467–3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rodionov D. A., et al. 2010. Genomic encyclopedia of sugar utilization pathways in the Shewanella genus. BMC Genomics 11:494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Saini A., Mapolelo D. T., Chahal H. K., Johnson M. K., Outten F. W. 2010. SufD and SufC ATPase activity are required for iron acquisition during in vivo Fe-S cluster formation on SufB. Biochemistry 49:9402–9412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schmid A. K., Pan M., Sharma K., Baliga N. S. 2011. Two transcription factors are necessary for iron homeostasis in a salt-dwelling archaeon. Nucleic Acids Res. 39:2519–2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schujman G. E., et al. 2006. Structural basis of lipid biosynthesis regulation in Gram-positive bacteria. EMBO J. 25:4074–4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schujman G. E., Paoletti L., Grossman A. D., de Mendoza D. 2003. FapR, a bacterial transcription factor involved in global regulation of membrane lipid biosynthesis. Dev. Cell 4:663–672 [DOI] [PubMed] [Google Scholar]
- 46. Seidl K., et al. 2009. Effect of a glucose impulse on the CcpA regulon in Staphylococcus aureus. BMC Microbiol. 9:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Semchyshyn H., Bagnyukova T., Storey K., Lushchak V. 2005. Hydrogen peroxide increases the activities of soxRS regulon enzymes and the levels of oxidized proteins and lipids in Escherichia coli. Cell Biol. Int. 29:898–902 [DOI] [PubMed] [Google Scholar]
- 48. Sierro N., Makita Y., de Hoon M., Nakai K. 2008. DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res. 36:D93–D96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Somerville G. A., Proctor R. A. 2009. At the crossroads of bacterial metabolism and virulence factor synthesis in staphylococci. Microbiol. Mol. Biol. Rev. 73:233–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Soutourina O., et al. 2009. CymR, the master regulator of cysteine metabolism in Staphylococcus aureus, controls host sulphur source utilization and plays a role in biofilm formation. Mol. Microbiol. 73:194–211 [DOI] [PubMed] [Google Scholar]
- 51. Torrents E., et al. 2007. NrdR controls differential expression of the Escherichia coli ribonucleotide reductase genes. J. Bacteriol. 189:5012–5021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Torres V. J., et al. 2010. Staphylococcus aureus Fur regulates the expression of virulence factors that contribute to the pathogenesis of pneumonia. Infect. Immun. 78:1618–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wollers S., et al. 2010. Iron-sulfur (Fe-S) cluster assembly: the SufBCD complex is a new type of Fe-S scaffold with a flavin redox cofactor. J. Biol. Chem. 285:23331–23341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang Y. M., Rock C. O. 2009. Transcriptional regulation in bacterial membrane lipid synthesis. J. Lipid Res. 50(Suppl.):S115–S119 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.