Abstract
Clostridium difficile expresses a number of cell wall proteins, including the abundant high-molecular-weight and low-molecular-weight S-layer proteins (SLPs). These proteins are generated by posttranslational cleavage of the precursor SlpA by the cysteine protease Cwp84. We compared the phenotypes of C. difficile strains containing insertional mutations in either cwp84 or its paralog cwp13 and complemented with plasmids expressing wild-type or mutant forms of their genes. We show that the presence of uncleaved SlpA in the cell wall of the cwp84 mutant results in aberrant retention of other cell wall proteins at the cell surface, as demonstrated by secretion of the proteins Cwp66 and Cwp2 into the growth medium. These phenotypes are restored by complementation with a plasmid expressing wild-type Cwp84 enzyme but not with one encoding a Cys116Ala substitution in the active site. The cwp13 mutant cleaved the SlpA precursor normally and had a wild-type-like colony phenotype. Both Cwp84 and Cwp13 are produced as proenzymes which are processed by cleavage to produce mature enzymes. In the case of Cwp84, this cleavage does not appear to be autocatalytic, whereas in Cwp13 autocatalysis was demonstrated as a Cys109Ala mutant did not undergo processing. Cwp13 appears to have a role in processing of Cwp84 but is not essential for Cwp84 activity. Cwp13 cleaves SlpA in the HMW SLP domain, which we suggest may reflect a role in cleavage and degradation of misfolded proteins at the cell surface.
INTRODUCTION
Clostridium difficile is a Gram-positive, spore-forming anaerobe that can cause serious gastrointestinal infections in humans and animals (8, 34). C. difficile infection (CDI) is most commonly seen in hospital environments and is associated with antimicrobial therapy that disrupts the normal microbiota (1). Clinical symptoms of disease range from mild or severe diarrhea to serious inflammatory conditions including pseudomembranous colitis (34). Although the elderly population still remains the largest at-risk group, CDI is increasingly being seen in younger patients and in patients in the community (16). The spores are the infectious form of the bacterium, as recently demonstrated in a mouse model of transmission (25).
Symptoms of disease are primarily caused by two secreted virulence factors, the toxins TcdA and TcdB. The mechanisms of action of these toxins have been well described, with both toxins exhibiting glucosyltransferase activity which inactivates Rho GTPases within host cells (21). This causes pleiotropic effects, including disruption of the actin cytoskeleton and tight junctions, induction of apoptosis, fluid accumulation, and destruction of the epithelium. Recent studies using C. difficile toxin knockout strains in the hamster model of infection have examined the essential nature of these toxins in disease (24, 27). Although the results of these studies were not in complete agreement, it appears that toxin A-negative strains were more virulent than toxin B-negative strains.
C. difficile has a cell wall typical of Gram-positive bacteria, comprising a cytoplasmic membrane and a thick peptidoglycan layer that may contain teichoic acids and other secondary cell wall polymers (31). In common with many bacteria, C. difficile expresses an S-layer, a two-dimensional proteinaceous array that coats the outer surface of the bacterium. The S-layer is composed primarily of two proteins, the high-molecular-weight S-layer protein (HMW SLP) and the low-molecular-weight (LMW) SLP (5). The SLPs can be removed from the cell by treatment with low pH glycine (5), which also removes other cell wall proteins (CWPs) present in relatively low amounts within the cell wall. The HMW SLP and the CWPs each contain three cell wall-binding motifs (Pfam 04122 [http://pfam.sanger.ac.uk/]) that appear to mediate noncovalent binding to the underlying cell wall by an uncharacterized mechanism. The majority of the CWPs have a second unique domain that in some cases specifies, or is predicted to specify, a function (11). Examples include Cwp66, a putative adhesin (41), CwpV, a phase-variable protein (12), and Cwp84, a cysteine protease (20).
The SLPs are present as a heterodimeric complex within the S-layer, and structural analysis has revealed the HMW SLP and LMW SLP interact through highly conserved domains present at the C terminus of the LMW SLP and the N-terminal domain of the HMW SLP (14). The remaining portions of the SLPs exhibit sequence divergence, in particular the LMW SLP, which shows immunological diversity between many C. difficile strains (4). The SLPs are derived from a precursor protein, SlpA, by proteolytic cleavage which removes the signal peptide, followed by a second cleavage resulting in the mature SLPs (5, 22). Recently, using both chemical and genetic techniques, the cysteine protease Cwp84 was shown to mediate cleavage of the mature SlpA precursor (9, 23). Cwp84 has also been implicated in degradation of extracellular matrix proteins such as fibronectin, laminin, and vitronectin (20). Neither chemical inhibition of Cwp84 (9) nor inactivation of the cwp84 gene (23) resulted in lethality, although severe growth defects were seen in both cases. These results indicate that correct processing of SlpA is important to retain healthy bacterial cells and suggest that perturbation of processing may affect the ability of bacteria to compete with other bacterial species in certain environments, for example, in the complex microbiota of the intestine. Interestingly, in the hamster model of acute infection, a C. difficile cwp84 mutant was not attenuated for virulence, which may be due to endogenous proteases within the hamster that could substitute for Cwp84 in cleavage of SlpA (23).
In addition to Cwp84, C. difficile 630 encodes a second, homologous protein termed Cwp13 (CD1751). cwp13 exhibits 70.4% nucleotide identity to cwp84, and the Cwp13 protein exhibits 63.2% amino acid identity to Cwp84. cwp13 is located outside the gene cluster that contains slpA, cwp84, and 10 other CWP genes. Cwp84 and Cwp13 are predicted to be members of the family of CA1 cysteine proteases (33). Proteases in this family (also termed papain proteases) possess a catalytic triad which in Cwp84 is proposed to comprise Cys116, His262, and Asn287 (37). We recently showed that Cwp84 containing the substitution Cys116Ala does not cleave SlpA in an Escherichia coli-based coexpression assay, confirming Cys116 as a catalytic residue (9). Papain peptidases are typically composed of an N-terminal signal peptide, a propeptide, and the catalytic domain (reviewed in reference 42). After removal of the signal peptide by a signal peptidase, the proenzyme undergoes self-cleavage, removing the proregion and generating the mature, active enzyme. The main function of the propeptide is to inhibit the activity of the proenzyme (15), which is potentially active in a manner independent of substrate binding due to the ionized state of the nucleophilic Cys. Furthermore, it has been proposed that the propeptide ensures the correct folding of the protein (6). There are, however, some papain peptidases that do not undergo autoprocessing, including human cathepsins X and C (29). Previous studies have indicated that Cwp84 undergoes autoprocessing based on two findings. First, an attempt to purify full-length Cwp84 from E. coli resulted in degradation of the recombinant protein. However, degradation was not prevented by treatment with cysteine protease inhibitors (37). Second, proteins of various sizes were purified from E. coli containing a plasmid designed to produce Cwp84 without its signal peptide (84 kDa) and treatment with dithiothreitol or trypsin resulted in purification of a single, active species of 61 kDa (20).
We examine here in detail the phenotype of a C. difficile cwp84 mutant and show that some other CWPs are dependent on Cwp84 activity for correct anchoring to the cell wall. Using a plasmid-localized copy of the wild-type gene in a cwp84 background, we characterized the processing of cwp84. We have created a mutant in cwp13 and show that this has subtle effects of the processing of Cwp84. Finally, we have analyzed the role of Cwp84 and Cwp13 in the cleavage of SlpA and of Cwp13 in the processing of the phase-variable protein CwpV (12).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
E. coli strains Top10 (Invitrogen) and NovaBlue (Novagen) were used routinely for cloning and propagation of plasmids. E. coli strain CA434 was used for conjugating plasmids into C. difficile. E. coli strains were routinely grown in LB broth (2) or on LB agar supplemented with appropriate antibiotics: carbenicillin at 50 μg/ml for the selection of derivatives of pUC19 and chloramphenicol at 12.5 μg/ml for the selection of derivatives of pMTL007 or pMTL960. The C. difficile strains and plasmids used in the present study are described in Table 1. C. difficile was routinely grown on blood agar base (Oxoid) supplemented with 7% defibrinated horse blood (TCS Biosciences), on brain heart infusion (BHI) agar (Oxoid), or in BHI broth (Oxoid). Thiamphenicol at 15 μg/ml was used for selection of derivatives of pMTL007 or pMTL960 in C. difficile. Growth was carried out under anaerobic conditions (10% H2, 10% CO2, 80% N2) at 37°C. Plasmids were introduced by conjugation into C. difficile as described previously (32) using LB medium for washing the pellets of the overnight cultures of the donor strains and BHI instead of phosphate-buffered saline (PBS) for scraping the transconjugants off BHI plates.
Table 1.
Bacterial strains and plasmids used in this study
| Strain of plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| C. difficile strains | ||
| 630Δerm | Also named WT in this study | 19 |
| NF2184 | 630Δerm cwp84347a::erm; also named cwp84 mutant in this study | This study |
| NF2233 | 630Δerm cwp13833a::erm; also named cwp13 mutant in this study | This study |
| Plasmids | ||
| pMTL960 | E. coli-C. difficile shuttle vector | 32 |
| pCBR023 | Contains modified gusA gene under the control of the cwp2 promoter; pUC19 backbone | 12 |
| pMTL007C-E2-cwp84-347/348a | pMTL007C-E2 with the Ll.ltrB intron retargeted to insert after base 347 in the 630Δerm cwp84 ORF | 18 |
| pLRP024 | Contains the cwp84 gene under the control of the cwp2 promoter; pUC19 backbone | This study |
| pLRP025 | Contains a derivative of the cwp84 gene, with a mutation for generating the substitution Cys116Ala, under the control of the cwp2 promoter; pUC19 backbone | This study |
| pCwp84WT | Insert from pLRP024 cloned into pMTL960 | This study |
| pCwp84C116A | Insert from pLRP025 cloned into pMTL960 | This study |
| pMTL007C-E5-cwp13-833/834a | pMTL007C-E5 with the Ll.ltrB intron retargeted to insert after base 833 in the 630Δerm cwp13 ORF | 18 |
| pLRP033 | Contains the cwp13 gene under the control of the cwp2 promoter; pUC19 backbone | This study |
| pLRP035 | Insert from pLRP033 cloned into pMTL960 | This study |
| pCwp13WT | pMTL960 containing the cwp13 gene, with the RBS-ORF spacer region of cwp84 and a mutation for generating the substitution Val1Met, under the control of the cwp2 promoter | This study |
| pCwp13C109A | pMTL960 containing the cwp13 gene, with the RBS-ORF spacer region of cwp84 and two mutations for generating the substitutions Val1Met and Cys288Ala, under the control of the cwp2 promoter | This study |
| pCBR044 | pMTL960 containing the full-length cwpV gene from 630 under the control of cwp2 promoter | 12 |
Plasmid construction.
DNA manipulations were performed according to standard techniques (35). Primers used in the present study are detailed in Table 2. Inserts for cloning were amplified from genomic DNA, which was isolated as described previously (5), using KOD Hot Start DNA polymerase (Novagen), which was also used for inverse PCR. Standard PCR was performed using Taq DNA polymerase (Sigma). Restriction enzymes were provided by New England BioLabs. Ligations were carried out using Quick-Stick ligase (Bioline) according to the protocol given by the manufacturer. All recombinant DNA molecules that were generated in the present study were confirmed to be correct by sequencing (GATC Biotech).
Table 2.
Primers used in this study
| Primer | Sequence (5′–3′)a | Relevant characteristic | Source or reference |
|---|---|---|---|
| NF722 | ACGCGTTATATTGATAAAAATAATAATAGTGGG | ErmRam-F | 17 |
| NF723 | ACGCGTGCGACTCATAGAATTATTTCCTCCCG | ErmRam-R | |
| NF1063 | CGAAATTAGAAACTTGCGTTCAGTAAAC | EBS Universal | 17 |
| NF1163 | CCATAAAACTCTAGATGGAG | Fw primer used for screening of insertion of the Ll.ltrB intron after base 347 in the 630Δerm cwp84 ORF | This study |
| NF1165 | GTTACATTTGAACTCCCTG | Rv primer for NF1163 | This study |
| NF1225 | AGTCTTAATACAGCATGGTCTTTTTCAG | Fw primer used for introducing the Cys116Ala mutation in Cwp84 by inverse PCR from plasmid pLRP024 | 9 |
| NF1226 | TCCTTGATTTTTTGCTGGTGTAGTAAGAC | Rv primer for NF1225 | 9 |
| NF1250 | CCGGAATTCATACATATAAGGGGGTAAACATGAG | Fw primer for amplification of cwp84 | This study |
| NF1251 | CGCGGATCCCTAGTGGTGGTGGTGGTGGTGCTCGAGTTTTCCTAAAAGAGTATTTAG | Rv primer for NF1250 | This study |
| NF1462 | CCGGAATTCCTCAAAATAAGGGGGAGAAAGCGTG | Fw primer for amplification of cwp13 | This study |
| NF1463 | CGCGGATCCCTAGTGGTGGTGGTGGTGGTGCTCGAGTTTAGCACTTTTTAATTTTACTAACTCT | Rv primer for NF1462 | This study |
| NF1479 | TAAACATGAAAAAATTTACTTCAAAAAAAGTAACA | Fw primer used for replacing the RBS-ORF spacer region of cwp13 by that of cwp84 and introducing a Val1Met substitution | This study |
| NF1480 | CCCCCTTATTTTGAGGAATTCTTAT | Rv primer for NF1479 | This study |
| NF1487 | CTGTGTGTGTAAGTAATGACATAAATTCTAC | Fw primer for screening of insertion of the Ll.ltrB intron after base 833 in the 630Δerm cwp13 ORF | This study |
| NF1488 | GTTAAGAACATTACAGATTTTAGAGTTTCATCGCCTCTATTG | Rv primer for NF1487 | This study |
| NF1489 | GATTTAGGAATCGCATGGGATTTTG | Fw primer used for substituting Cys109 by Ala in Cwp13 | This study |
| NF1490 | CTCTTGATTTTTTATTGATGTCATAAGAC | Rv primer for NF1489 | This study |
Restriction endonuclease sites (EcoRI or BamHI) are italicized. Sequences encoding the Leu-Glu-His6 tag are underlined. Mutations introduced by primers are indicated in boldface and are described in detail in column 3.
pCwp84WT and derivatives were constructed as follows. cwp84 was amplified by using the primers NF1250 and NF1251 to introduce a C-terminal His6 tag. The fragment was cloned into the pCBR023 backbone, replacing the gusA gene (12) using EcoRI and BamHI sites, to generate plasmid pLRP024. Inverse PCR was carried out on pLRP024 by using the primers NF1225 and NF1226 as described previously (14) to obtain plasmid pLRP025, which contains a mutation substituting Cys116 in Cwp84 by Ala. The Pcwp2-cwp84WT and Pcwp2-cwp84C116A fragments were subcloned into pMTL960 using the Acc65I and BamHI sites to obtain the plasmids pCwp84WT and pCwp84C116A, respectively.
pCwp13WT and derivatives were constructed as follows. cwp13 was amplified by using the primers NF1462 and NF1463 to introduce a C-terminal His6 tag. Plasmid pLRP033 was obtained in the same way as pLRP024 and pLRP025. The Pcwp2-cwp13 fragment was subcloned into pMTL960 as detailed above to obtain plasmid pLRP035. Inverse PCR was carried out using pLRP035 and primers NF1479 and NF1480 to replace the ribosomal binding site (RBS), RBS-open reading frame (ORF) spacer and Val1 encoding GTG of cwp13 with the RBS, RBS-ORF, and Met1 encoding ATG of cwp84, obtaining plasmid pCwp13WT. Plasmid pCwp13C109A was generated by inverse PCR from pCwp13WT using the primers NF1489 and NF1490.
Construction of mutants in C. difficile 630Δerm.
The ClosTron gene knockout system (17) was used to obtain insertional mutants in cwp84 (CD2787) and cwp13 (CD1751) genes. cwp84 and cwp13 were analyzed by the Intron Design Tool (USG, University of Nottingham, Nottingham, United Kingdom) to identify insertion sites for the Ll.ltrB intron. The Ll.ltrB intron was inserted after base 347 in cwp84 and after base 833 in cwp13, both in the antisense orientation, using the retargeted plasmids pMTL007C-E2-cwp84-347/348a and pMTL007C-E5-cwp13-833/834a. The plasmids were synthesized (DNA2.0, Menlo Park, CA) and transformed into E. coli CA434 prior to conjugation into C. difficile 630Δerm. The following protocol for generating and selecting mutants was kindly provided by Lisa Dawson (London School of Hygiene and Tropical Hygiene, London, United Kingdom). First, 1 ml of an overnight culture of the donor strain was harvested by centrifugation at 1,700 × g for 5 min. The pellet was washed once in LB broth and then resuspended in 200 μl of a culture of the recipient strain in exponential phase of growth. Then, 20-μl drops of the bacterial suspension were spotted onto Brazier's agar (Bioconnections) supplemented with 4% egg yolk (Bioconnections) and 1% defibrinated horse blood (TCS Biosciences), followed by incubation overnight. Bacterial growth was scraped off the plates with 600 μl of PBS and were spread onto Brazier's agar supplemented with egg yolk, blood, 15 μg of thiamphenicol/ml, and cefoxitin-cycloserine supplements (Bioconnections). Transconjugants were allowed to grow for 2 to 3 days before single colonies were purified twice on the same medium. Pure transconjugants were resuspended in 200 μl of PBS, and 100-μl samples of undiluted bacterial suspension and of 1:100, 1:1,000, and 1:10,000 dilutions were spread onto Brazier's agar supplemented with egg yolk, blood, and 5 μg of erythromycin/ml. Erythromycin-resistant colonies were streaked twice onto the same medium. For PCR screening of the insertion of the intron within the target gene, genomic DNA was extracted and used as a template for the following PCRs (see Fig. S1 in the supplemental material). (i) Splicing of the group I intron from the group II intron was confirmed by using the primers NF722 and NF723. (ii) Integration of the Ll.ltrB intron was confirmed by using primers flanking the insertion site. The primers NF1163 and NF1165 were used in the case of the cwp84 mutant, and NF1487 and NF1488 were used in the case of the cwp13 mutant. The cwp13 gene in the cwp84 mutant and the cwp84 gene in the cwp13 mutant were also analyzed to confirm insertion of the Ll.ltrB intron in the specific target gene in each case. (iii) Integration in the antisense orientation was confirmed by using the intron-specific primer EBS Universal (17) and one of the two flanking primers.
Protein analysis.
Cell wall proteins were extracted from C. difficile cells using the low-pH glycine extraction method described previously (4). Culture supernatants were concentrated using Amicon 10-kDa cutoff centrifugal filter units (Millipore) according to the instructions given by the manufacturer. SDS-PAGE was carried out as described previously (13). Acrylamide in the resolving gel was used at 10% unless otherwise specified. For immunoblot analysis, proteins were transferred to Immobilon-PVDF membranes (Millipore) using a three-buffer semidry method according to the instructions provided by the manufacturer. The rabbit antibodies to HMW SLP and LMW SLPs were described previously (14). Antisera against Cwp84, Cwp66-Cter, and Cwp2 were raised in mice. Anti-Cwp84, anti-Cwp66, anti-Cwp2, anti-HMW SLP, and anti-LMW SLP were used at the dilutions 1:4,000, 1:20,000, 1:50,000, 1:100,000, and 1:200,000, respectively. Primary antibodies were detected by using horseradish peroxidase (HRP)-conjugated goat anti-rabbit antibody at 1:2,000 or rabbit anti-mouse antibody at 1:1,000 (Dako) and the SuperSignal West Pico chemiluminescent substrate (Thermo Scientific Pierce). His6 tag was detected using HRP-conjugated anti-histidine tag (Sigma).
N-terminal sequences were determined by Edman degradation at the Protein and Nucleic Acid Chemistry Facility of the University of Cambridge, Cambridge, United Kingdom. Samples were prepared according to the instructions given on http://www.bioc.cam.ac.uk/pnac/proteinsequencing.html. For a better resolution of the 77-kDa Cwp84 band and the 78-kDa and 84-kDa Cwp13 bands, proteins were separated using bis-Tris gels (http://openwetware.org/wiki/Sauer:bis-Tris_SDS-PAGE,_the_very_best) with 8% acrylamide.
RESULTS
Characterization of a C. difficile cwp84 mutant.
To examine the role of Cwp84 in cell wall biogenesis, we generated an insertionally inactivated mutant of cwp84 in C. difficile 630Δerm (WT) using targeted mutagenesis (17). Several independent mutants were analyzed and all exhibited the same genotype and phenotype. In the cwp84 mutant (Fig. 1A), the full-length SlpA precursor was the predominant band in the cell wall extract, and the mature HMW and LMW SLPs were not present, confirming the phenotype observed previously (23). In order to complement the mutation and confirm in C. difficile that Cys116 in Cwp84 is a catalytically active residue as described previously in E. coli (9), we used the shuttle plasmid pMTL960 to introduce into the cwp84 mutant strain either the wild-type cwp84 gene (pCwp84WT) or a derivative containing a Cys116Ala mutation (pCwp84C116A). In these plasmids the cwp84 gene was placed under the control of Pcwp2, a promoter we used previously to obtain constitutive expression of cell wall proteins in C. difficile (12). Both Cwp84 proteins incorporated a C-terminal His6 tag to aid identification.
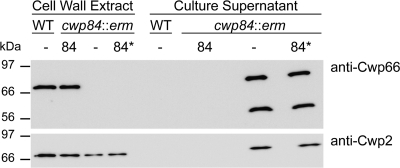
Fig. 1.
Phenotype and complementation of a cwp84 mutant of C. difficile 630Δerm. (A) SDS-PAGE analysis of cell wall proteins and culture supernatants. Bacteria containing plasmids (see below) were grown in BHI broth. Cell wall proteins were extracted, and culture supernatants were concentrated from bacterial cultures at late exponential phase (OD600 = 0.5). Plasmids: –, pMTL960; 84, pCwp84WT; 84*, pCwp84C116A. (B) Colony morphologies. Serial dilutions (100 μl) of overnight cultures were plated on BHI agar supplemented with 15 μg of thiamphenicol/ml and grown for 2 days, and representative colonies were photographed.
The plasmid-encoded wild-type cwp84 gene completely restored cleavage of SlpA in the cwp84 mutant strain, as judged by the appearance of the mature SLPs in the cell wall extracts (Fig. 1A), whereas the strain harboring the cwp84(C116A) allele was indistinguishable from the cwp84 mutant. Analysis of the culture supernatants showed that the cwp84 mutant harboring either the vector pMTL960 or pCwp84C116A secreted large amounts of unprocessed SlpA into the culture medium, in contrast to the cwp84 mutant complemented with pCwp84WT or the WT strain, where the secretion of SlpA was barely detectable.
Previously, it was noted that deletion of cwp84 resulted in a small-colony phenotype (23). As shown in Fig. 1B, cwp84 mutant colonies lack the rough, irregular edges typical of C. difficile and instead are shiny and have a defined edge and a raised center. Complementation of the cwp84 mutant with pCwp84WT restored the colony morphology seen with C. difficile 630Δerm, whereas the small-colony morphology was retained in the cwp84 mutant carrying pCwp84C116A.
Deletion of cwp84 results in instability of the cell wall.
Since the colony morphology of the cwp84 mutant was so different from the WT strain, we investigated whether there were any other changes to the cell wall composition of this strain. Cell wall extracts and supernatants were prepared from WT and cwp84 mutant cultures containing plasmids pCwp84WT or pCwp84C116A and analyzed by Western blotting. Using antibody prepared against the C-terminal domain of Cwp66, a cell wall protein described as an adhesin (41), a band of ∼70 kDa can be seen in the cell wall extracts of 630Δerm (WT) (Fig. 2, anti-Cwp66). In contrast, this band was absent in the cwp84 mutant but present when it harbored pCwp84WT but not pCwp84C116A. Analysis of the culture supernatants of the cwp84 mutant containing the vector pMTL960 or the pCwp84C116A plasmid revealed two bands, one corresponding to the full-length Cwp66 and a second of ∼45 kDa which is presumably a degradation product (Fig. 2, anti-Cwp66). These results indicate that the absence of active Cwp84 in the cell wall, as well as the consequential lack of mature HMW and LMW SLPs, results in a defect in retention of Cwp66. We repeated these experiments using an antibody against Cwp2, a cell wall protein that is conserved in a majority of C. difficile strains (4). A similar result was seen with Cwp2, although the effect was not as dramatic (Fig. 2, anti-Cwp2). Cwp2 was present in the cell wall extracts of 630Δerm and the cwp84 mutant and also in the supernatant of the cwp84 mutant. The presence of plasmid pCwp84WT, but not pCwp84C116A, complemented the phenotype of shedding of Cwp2 to the supernatant in the cwp84 mutant.
Fig. 2.
Cell wall protein localization is defective in a cwp84 mutant. C. difficile 630Δerm (WT) and the cwp84 mutant harboring plasmids (see below) were grown in BHI broth to late exponential phase (OD600 = 0.5). Cell wall extracts and culture supernatants were prepared and analyzed by SDS-PAGE, followed by Western blotting with antibodies against Cwp66 and Cwp2. Plasmids: –, pMTL960; 84, pCwp84WT; 84*, pCwp84C116A.
Processing of Cwp84.
Previous studies have indicated that, when expressed in E. coli, Cwp84 may undergo posttranslational processing (20). Our recent work in C. difficile using biotinylated inhibitors of cysteine proteases in combination with streptavidin pulldown experiments identified two species of Cwp84, migrating with apparent masses of 84 and 77 kDa (9). We therefore investigated processing of Cwp84 in C. difficile using an antibody raised against recombinant (E. coli-produced) Cwp84. In the WT strain, Cwp84 is visible in the cell wall as a single species of 77 kDa (Fig. 3A, anti-Cwp84). We infer this to be the active form of the enzyme. When WT cwp84 is expressed from plasmid pCwp84WT in the cwp84 mutant, an additional band at 84 kDa is visible (Fig. 3A, anti-Cwp84). Interestingly, this 84-kDa band is not visible when Cwp84C116A is expressed from the plasmid. Most probably, this protein is the 84-kDa species we previously identified as Cwp84 (9). The 77- and 84-kDa bands are also visible when probed with an anti-histidine tag antibody, which recognizes the C-terminal His6 tag present on these proteins (Fig. 3A, anti-His), indicating that the two forms of Cwp84 must differ at their N termini. In order to determine whether the 77-kDa band, which is also detected by Coomassie blue staining (Fig. 1A), is a processed form of Cwp84, we determined its N-terminal sequence. The N-terminal sequence of the 77-kDa band from the cwp84 mutant carrying pCwp84WT was determined to be SSVAY. Unfortunately, the 84-kDa protein could not be sequenced due to insufficient protein on the gel. The sequence SSVAY is located 92 residues downstream of the N-terminal Met residue and 61 residues downstream of the predicted site of cleavage by a signal peptidase (Fig. 3B). These results strongly suggest that the mature form of Cwp84 present on the cell wall results from two proteolytic processing events; a signal peptidase removes the signal peptide generating the 84-kDa proenzyme, followed by a second cleavage to generate the 77-kDa mature active enzyme. The 77-kDa band is also observed in the cwp84 mutant expressing the Cwp84C116A protein, indicating that the cleavage event generating the mature protease is unlikely to be due to self-cleavage. In the supernatant of the cwp84 mutant overexpressing the WT or mutant Cwp84, several fragments of Cwp84 were detected which are presumed to be degradation products.
Fig. 3.
Processing of Cwp84. (A) Detection of Cwp84 in the cwp84 mutant overexpressing either WT or the Cys116Ala protein. C. difficile 630Δerm (WT) and the cwp84 mutant harboring plasmids as indicated were grown in BHI broth to late exponential phase (OD600 = 0.5). Cell wall extracts and culture supernatants were prepared and analyzed by SDS-PAGE, followed by Western blotting with antibodies against Cwp84 and the C-terminal His6 tag. Plasmids: –, pMTL960; 84, pCwp84WT; 84*, pCwp84C116A. The upper band (84 kDa) recognized by these antibodies in the cwp84 mutant carrying pCwp84WT is indicated (◀). The N-terminal sequence of the 77-kDa protein corresponding to the lower band was determined as SSVAY. (B) Line diagram of domain structure of Cwp84 and location of the signal peptide and propeptide domains. At the top is the domain structure of Cwp84 showing the location of the signal peptide (black rectangle), as predicted by SignalP, and the cysteine protease domain (white box), as predicted by Pfam (PF00112). The three cell wall binding motifs (PF04122) are shaded in gray. Below is the N-terminal amino acid sequence of Cwp84 showing the sites of cleavage to release the signal peptide (▾) and the mature protein (▿).
Characterization of Cwp13, a Cwp84 paralog.
C. difficile 630 contains a second gene, cwp13, that encodes a very similar protein to Cwp84 (see the introduction). It was of interest to investigate whether Cwp13 contained cysteine protease activity and, if so, what the substrate(s) of this protease might be. Using insertional mutagenesis, we constructed a mutant of cwp13 in strain 630Δerm and analyzed the phenotype of this strain. We first investigated whether the cwp13 mutant had any defect in processing of SlpA. In contrast to the cwp84 mutant, the cwp13 mutant appeared to process SlpA normally, as seen by the appearance of the HMW and LMW SLPs in the cell wall (Fig. 4A, Coomassie blue stain), and it retained the large-colony morphology of the parental strain (data not shown). The WT cwp13 gene and a derivative encoding a mutation at the predicted catalytic cysteine (Cys109Ala substitution) were introduced into the C. difficile cwp13 mutant using the plasmids pCwp13WT and pCwp13C109A, respectively. Cell wall extracts and culture supernatants from these strains showed very similar profiles with the HMW SLP and LMW SLP clearly visible, indicating cleavage had occurred mediated by Cwp84 (Fig. 4A, Coomassie). Interestingly, wild-type Cwp13 migrated at 78 kDa, whereas the Cys109Ala mutant protein migrated at 84 kDa. Since the Cwp13 proteins introduced on these plasmids contain a C-terminal His6 tag, the processing of this protein could be assessed by using Western blotting. (Fig. 4A, anti-His). Both proteins were recognized by the anti-His tag antibody showing that the proteins differed at their N termini. The N-terminal sequences of these two proteins were identified as APTSY for the 78-kDa fragment of Cwp13 and DNSNT for the 84-kDa fragment of Cwp13C109A. These results show that the signal peptide of Cwp13 consists of the first 30 amino acids, as predicted by SignalP. They also reveal that Cwp13 undergoes autocleavage between Thr84 and Ala85, but this processing does not occur in the Cwp13C109A protein (see Fig. 4B).
Fig. 4.
Characterization of a cwp13 mutant and processing of Cwp13. (A) Analysis of the effects of an insertional mutation in cwp13 by SDS-PAGE and Western blotting. C. difficile 630Δerm (WT) and the cwp13 mutant harboring plasmids as indicated were grown in BHI broth supplemented with 15 μg of thiamphenicol/ml to late exponential phase (OD600 = 0.5). Cell wall extracts and culture supernatants were prepared and analyzed by Coomassie blue staining (top) and Western blotting with antibodies against the C-terminal His6 tag (6% acrylamide), Cwp84 (6% acrylamide) and LMW SLP. Plasmids: –, pMTL960; 13, pCwp13WT; 13*, pCwp13C109A. The 77-kDa Cwp13WT and 84-kDa Cwp13C109A proteins detected by Coomassie blue staining and the anti-His6 tag are indicated (◁ and ◀), and their N-terminal sequences were determined as DNSNT and APTSY, respectively. (B) Domain structure of Cwp13 and location of the signal peptide and propeptide domains. At the top is domain structure of Cwp13 showing the location of the signal peptide (black rectangle) and the cysteine protease domain (white box), as predicted by Pfam (PF00112). The cell wall anchoring domains (PF04122) are shaded in gray. Below is the N-terminal amino acid sequence of Cwp13 showing the sites of cleavage to release the signal peptide (▾) and the mature protein (▿). (C) Domain structure of SlpA. The vertical bar indicates the cleavage site of Cwp84 to produce the mature HMW SLP and LMW SLP. The black triangle (▴) shows the approximate site of cleavage, within a cell wall binding domain, by Cwp13 to generate a 47-kDa N-terminal product.
Another interesting observation was the detection of unprocessed Cwp84. In the cwp13 mutant harboring either pMTL960 or pCwp13C109A, both the proenzyme migrating at 84 kDa and the mature 77-kDa species were observed. In contrast, in the cell wall fraction of the 630Δerm parental strain harboring the vector pMTL960 and in the cwp13 mutant harboring pCwp13WT only the processed form of Cwp84, migrating at 77 kDa, was seen. The Cwp84 preprotein was also observed in the supernatant of the cwp13 mutant harboring the pCwp13C109A plasmid (Fig. 4A, anti-His), but not in supernatants from the other strains. These results indicate that Cwp13 may have a role in cleaving and/or processing Cwp84.
Finally, we analyzed in greater detail the effects of the cwp13 mutation on processing of SlpA using an antibody recognizing the LMW SLP. Mature LMW SLP (34 kDa) was seen in the cell wall in all strains, with no full-length SlpA detected, in agreement with the stained gel (Fig. 4A, anti-LMW). In the culture supernatants of 630Δerm carrying pMTL960 or in the cwp13 mutant carrying pCwp13WT, a band of ∼47 kDa is seen. However, in the cwp13 mutant carrying either pMTL960 or pCwp13C109A this band is decreased in intensity and full-length SlpA is visible. This 47-kDa protein was investigated further (see below).
Cwp13 cleaves SlpA and does not complement the cwp84 mutant.
In order to better understand whether Cwp13, which shares 63.2% sequence identity with Cwp84, plays any role in the processing of SlpA we introduced plasmids pCwp13WT and pCwp13C109A in the cwp84 mutant by conjugation. Cell wall extracts and culture supernatants of these strains were analyzed by SDS-PAGE, followed by either Coomassie staining or Western blotting (Fig. 5). Corresponding samples of cwp84 mutant overexpressing Cwp84WT and Cwp84C116A were prepared alongside for comparison. Wild-type Cwp13 was not detected on the stained gel, but, interestingly, a small amount of the Cwp13C109A derivative was detected (Fig. 5, Coomassie), which was confirmed by Western blotting (data not shown). However, the size of Cwp13C109A when overexpressed in the cwp84 mutant is 78 kDa instead of the expected 84 kDa that was observed in the cwp13 mutant background (Fig. 4, Coomassie). This indicates that recombinant Cwp13C109A is processed from 84 to 77 kDa in this strain, presumably by the chromosomally encoded cwp13.
Fig. 5.
Cwp13 cleaves SlpA but does not complement the cwp84 mutant. C. difficile cwp84 mutant harboring different plasmids was grown in BHI broth supplemented with thiamphenicol (15 μg/ml) till late exponential phase (OD600 = 0.5). Cell wall extracts and culture supernatants were prepared and analyzed by Coomassie blue staining (top) and Western blotting with antibody against the LMW SLP (middle and bottom). The bottom panel shows the 34-kDa LMW SLP species on an overexposed Western blot. Plasmids: –, pMTL960; 84, pCwp84WT; 84*, pCwp84C116A; 13, pCwp13WT; 13*, pCwp13C109A. Open arrow, 77-kDa Cwp84; solid arrow, 78-kDa Cwp13C109A processed by endogenous Cwp13; ◀, 47-kDa fragment of SlpA.
We then investigated cleavage of SlpA by visualizing the LMW SLP using antibody. Interestingly, a small quantity of the LMW SLP was detected both on the cell wall (Fig. 5, anti-LMW) and in the culture supernatant (Fig. 5, anti-LMW SLP) of the cwp84 mutant overexpressing Cwp13WT but not Cwp13C109A. This result shows that Cwp13 is weakly active on SlpA and that Cys109 is a catalytic residue for this cleavage. However, this cleavage does not result in restoration of the WT colony morphology (data not shown) or in complete SlpA processing. When the Western blot with the anti-LMW SLP is overexposed (Fig. 5, bottom), very faint bands corresponding the LMW SLP are detected on the cell wall and culture supernatants of cwp84 mutant containing pMTL960 or overexpressing Cwp84C116A or Cwp13C109A. This could be due to the activity of Cwp13 expressed from the genomic copy of cwp13.
Surprisingly, a protein of ∼47 kDa is detected by anti-LMW SLP predominantly in the culture supernatant of the cwp84 mutant overexpressing Cwp13WT (Fig. 5, anti-LMW). The same protein is present but to a less extent in the culture supernatants of the cwp84 mutant containing pMTL960 or pCwp84C116A and to an even lesser extent in the case of pCwp13C109A. Most probably, that protein is the same one that was detected previously (Fig. 4A, anti-LMW SLP). The N-terminal sequence of the 47-kDa protein was determined to be ATTGT, which is located at the N terminus of the LMW SLP. Since the LMW SLP is only 34 kDa, cleavage must occur within the cell wall binding domains of the SlpA precursor (see Fig. 4C). Thus, either Cwp13 recognizes a second cleavage site in SlpA or activates another protease that catalyzes this cleavage. This cleavage occurs most efficiently in the absence of active Cwp84, i.e., in the presence of full-length SlpA. We propose a model that summarizes our findings on the maturation and activities of Cwp13 and Cwp84 (see Fig. 7).
Fig. 7.
Model for processing and activities of Cwp84 and Cwp13. SlpA, Cwp84, and Cwp13 are produced as preproteins containing signal peptides that are removed during processing by the sec system (step a). The propeptides of Cwp84 and Cwp13 are removed (step b), either by autocatalysis in the case of Cwp13 or by an unknown activity together with Cwp13 activity in the case of Cwp84, to form the active enzyme species that are incorporated into the S-layer (step c). Mature Cwp84 cleaves the SlpA precursor (step d), which results in the formation of the H/L complex (step e). Misfolded proteins are recognized by Cwp13 and are cleaved in their cell wall binding domains to prevent incorporation into the S-layer, resulting in detachment from the cell and deposition into the growth medium (step f). S-L, S-layer; PG, peptidoglycan; Mem, membrane; Cyt, cytoplasm.
Cwp84 and Cwp13 are conserved across multiple strains of C. difficile.
The available complete or incomplete genome sequences of C. difficile strains 630, R20291, 196, BI1, CF5, M210, M68, 855, QCD32g58, and VPI10463 were searched for the cwp13 and cwp84 genes. A gene coding for a protein with very high sequence identity (≥95%) to Cwp13 from C. difficile 630 was found in all genomes searched, in contrast to a previous report suggesting cwp13 to be absent from the genome of strain R20291 (23). The predicted catalytic dyads within Cwp84 and Cwp13, which are Cys109 His256 and Cys116 His263, respectively, are fully conserved in all of these strains (data not shown). Furthermore, the site in Cwp84 at which the proenzyme is cleaved to produce the active enzyme (Lys91-Ser92 in strain 630) is conserved across Cwp84 from all of the strains. Notably, the 7-amino-acid sequence in Cwp84 containing the cleavage site and residues immediately upstream (Leu86 to Ser92) is absent in all Cwp13 sequences.
Cwp13 does not cleave CwpV.
CwpV is the largest member of the family of cell wall proteins and is expressed in a phase variable manner (12). CwpV is detected in ca. 5% of cells in a C. difficile population under laboratory conditions. CwpV is processed: two fragments of 40 and 120 kDa, corresponding to the N-terminal cell wall anchoring domain and the C-terminal repeats domain, respectively, are detected on the cell wall of cwpV-expressing cells.
It was shown that Cwp84 is not responsible for cleavage of CwpV (23). We therefore investigated whether Cwp13 catalyzes the cleavage of CwpV by introducing plasmid pCBR044, which directs the constitutive expression of CwpV, into the cwp84 and cwp13 mutants by conjugation. As Fig. 6 shows, both domains of CwpV were detected in the wild-type and both mutant backgrounds, confirming that neither Cwp84 nor Cwp13 catalyzes the cleavage of CwpV.
Fig. 6.
Investigation of cleavage of CwpV by Cwp84 and Cwp13. C. difficile strains containing either pMTL960 vector or pCBR044 carrying cwpV+ were grown overnight in BHI broth supplemented with thiamphenicol (15 μg/ml). Cell wall proteins were extracted and analyzed by SDS-PAGE. Plasmids: –, pMTL960; V, pCBR044 CwpV+.
DISCUSSION
A role for Cwp84 in cleavage of the S-layer protein precursor SlpA was demonstrated by using both chemical biology (9) and traditional genetic approaches (23). We have extended these studies here by using C. difficile strains containing gene knockouts in cwp84 or cwp13 coupled with plasmid-based expression of WT or mutant versions of the enzymes. These studies allowed us to confirm the phenotype of a cwp84 knockout and to demonstrate that the conserved Cys116 residue is a required catalytic residue. Importantly, complementation of the cwp84 mutant restored both SlpA cleavage and the WT colony phenotype, demonstrating that neither defect is due to second site mutations within the genome.
An unexpected consequence of the cwp84 mutation is defective localization of other CWPs. Analysis of the culture supernatants by Western blotting revealed detachment of Cwp66 and Cwp2 from the cell wall of the cwp84 mutant. It is possible that other members of the CWP family are affected in a similar way. Lack of cleavage of the S-layer also has a dramatic effect on colony morphology. The reasons why certain cell wall proteins do not fully associate with the cell wall and why the cwp84 mutant displays such a characteristic colony morphology are likely to lie in the altered nature of the S-layer rather than being a direct effect of the lack of Cwp84 cysteine protease activity. Clearly, full-length, uncleaved SlpA is tolerated on the cell wall, but it would appear that the S-layer is incorrectly formed. Our previous structural studies showed SlpA to be an elongated molecule with the HMW SLP and LMW SLP interacting through domains located on either side of the cleavage site (14). The lack of cleavage almost certainly prevents correct interaction between the two SLPs leading to an abnormal S-layer. The mechanism of anchoring of the S-layer proteins and the CWPs to the underlying cell wall is currently unknown. However, it is probable that a common mechanism exists for the S-layer proteins and the extended family of CWPs since they all contain three Pfam 04122 cell wall binding domains. Our finding that some CWPs are shed from the cell wall in the absence of an incorrectly formed S-layer suggests a complex mechanism of attachment, perhaps involving interactions between the CWPs and the SLP subunits in addition to putative interactions with underlying cell wall polymers.
We show that Cwp84 undergoes posttranslational processing to form the active enzyme. After removal of the signal peptide, the 84-kDa proenzyme is processed to an active 77-kDa species. The N terminus of this active protein was localized at Ser92; hence, the protein contains a 61-residue propeptide after removal of the signal peptide. Consistent with this are results from our previous study using biotinylated compounds that inhibited SlpA cleavage; two bands of 84 and 77 kDa were labeled from a cell wall extract and were identified by proteomic techniques as derived from Cwp84 (9). The sequence of Cwp84 is highly conserved across C. difficile strains (26, 37), and it is likely that the Cwp84 species detected in 17 C. difficile isolates from CDI patients (30) is the active form of the enzyme. Surprisingly, mutation of the Cys116 catalytic residue to Ala did not prevent processing of the 84-kDa proenzyme to the 77-kDa species, indicating that processing may not be autocatalytic or that an alternative mechanism for processing may exist (Fig. 7). In fact the Cwp84C116A protein was found exclusively in the 77-kDa form with no evidence of a 84-kDa species.
Cwp13 is highly related to Cwp84 in primary sequence, but our experiments show it has distinct activities. Cwp13 shows very weak activity in cleavage of the SlpA precursor to produce the HMW and LMW SLPs. However, Cwp13 does contain proteolytic activity, as shown by autoprocessing and by cleavage of SlpA at a sequence distinct from the cleavage site recognized by Cwp84. Cwp13 is localized to the cell wall so lack of activity against SlpA must be due either to poor recognition of the substrate or absence from the immediate environment of SlpA as it translocates through the membrane and is localized in the cell wall.
Our data also show that Cwp13 is synthesized as a proenzyme that is processed to produce the active enzyme. However, in contrast to Cwp84, this processing does appear to be autocatalytic as it is not observed in a cwp13 mutant expressing Cwp13C109A. As stated above, Cwp84 does not appear to undergo auto-processing. However, our data using a cwp13 mutant do point to a role for Cwp13 in processing Cwp84. Western blotting showed that the 84-kDa proenzyme form of Cwp84 was processed efficiently to the active 77-kDa species only in wild-type cells and in the cwp13 mutant when complemented with WT Cwp13 but not with Cwp13C109A. Where cwp13 activity was absent, both the proenzyme and the mature enzyme forms of Cwp84 were seen, indicating impaired processing of Cwp84. This suggests that Cwp13 has a role in cleavage of Cwp84 but that in the absence of Cwp13 cleavage of Cwp84 can still occur, albeit less efficiently.
The S-layer proteins are the major proteinaceous cell wall components and appear to be essential, as demonstrated by an apparent inability to create an slpA gene knockout (our unpublished data; Nigel Minton and Julian Rood, unpublished data). It might therefore be advantageous for the cell to have more than one mechanism to process Cwp84 to ensure correct processing of SlpA. It is possible that cleavage of Cwp84 may proceed via an unconventional self-processing route that is unrelated to its papain-like activity (3, 10). Alternatively, there could be a second protease that mediates cleavage of Cwp84.
Our results suggest that Cwp13 may have roles other than processing of Cwp84. In culture supernatants of a cwp13 mutant, accumulation of the mature form of SlpA was seen, whereas in the mutant complemented with active Cwp13, a 47-kDa cleavage product of SlpA was seen. This protein was shown to be derived from the LMW SLP and a portion of the HMW SLP, indicating cleavage was within the HMW SLP. SlpA is naturally expressed at very high levels, and excess protein would need to be efficiently removed from the cell wall to allow correct localization and assembly of the HMW and LMW SLPs to form the S-layer.
It is possible that Cwp13 might have a more general role in the cleavage of misfolded proteins secreted to the cell wall (see Fig. 7). Gram-negative bacteria produce a number of periplasmic chaperones that prevent misfolding of periplasmic and outer membrane proteins, ensuring safe transit of proteins and avoiding attack by proteases that recognize unfolded proteins with the consequential activation of the stress response (38). SurA and Skp are two such chaperones, and DegP is a chaperone-protease complex and member of the large HtrA family (7, 39, 40). In B. subtilis several chaperones and proteases have been identified that are located external to the plasma membrane, possibly within the space between the membrane and the thick peptidoglycan layer, termed the “Gram-positive periplasmic space” (28). These proteins include homologs of DegP and the lipoprotein PrsA, a putative cis-trans peptidyl proyl trans-isomerase (reviewed in reference 36). It is likely that all Gram-positive bacteria, including the clostridia, have chaperones and proteases located external to the plasma membrane since folding of proteins secreted through the sec pathway and correct assembly of protein complexes must occur immediately after traversal of the plasma membrane. Whatever the natural substrate(s) of Cwp13, its activity must be regulated either spatially or kinetically to avoid unwanted cleavage of the HMW SLP which is essential for correct formation of the S-layer. Notably, Cwp13 cleaves full-length SlpA within the region of the HMW SLP containing the three cell wall binding domains that mediate attachment to the underlying cell wall. Cleavage in this region would disrupt attachment to the cell wall, and the protein would be released from the cell. This then would provide a mechanism for the release of incorrectly folded SlpA precursor from the cell wall.
Another possibility is that Cwp13 may function to digest extracellular substrates, such as extracellular proteins. Cwp84 has been shown to possess degradative activity toward extracellular matrix proteins fibronectin, laminin, and vitronectin (20). Perhaps Cwp13 also provides similar activities to aid in destruction of host tissues, colonization of the host, and dissemination of infection, as suggested previously (20).
In conclusion, we show that C. difficile contains two active cysteine proteases in the cell wall with distinct functions. Both are produced as inactive proenzymes that are processed to active enzymes, however, only Cwp13 appears to undergo autocatalytic cleavage. These proteins both function in assembly of the S-layer, but the activity of Cwp13 seems to be dispensable, whereas that of Cwp84 is essential. Our finding that incorrect assembly of the S-layer has profound consequences for the localization of other CWPs is significant since it points to important interactions between the S-layer proteins and the diverse CWPs that together are major components of the cell wall. The possible activity of Cwp13 as a protease involved the regulation of protein misfolding is currently under investigation.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by a MRC Discipline Hopping grant to E.W.T. and N.F.F. (grant G0701834), a Wellcome Trust VIP award to L.D.L.R., a BBSRC studentship to S.E.W., and a BBSRC David Phillips Fellowship to E.W.T. (BB/D02014X/1).
We thank Lisa Dawson for advice on mutagenesis; Mike Weldon for N-terminal sequencing; John Heap and Nigel Minton for Clostron technology; and Cate Reynolds, Robert Fagan, and Tam Dang for their constructive advice and constant support.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 29 April 2011.
REFERENCES
- 1. Bartlett J. G. 2007. Clostridium difficile: old and new observations. J. Clin. Gastroenterol. 41(Suppl. 1):S24–S29 [Google Scholar]
- 2. Bertani G. 2004. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 186:595–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brannigan J. A., et al. 1995. A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature 378:416–419 [DOI] [PubMed] [Google Scholar]
- 4. Calabi E., Fairweather N. 2002. Patterns of sequence conservation in the S-layer proteins and related sequences in Clostridium difficile. J. Bacteriol. 184:3886–3897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Calabi E., et al. 2001. Molecular characterization of the surface layer proteins from Clostridium difficile. Mol. Microbiol. 40:1187–1199 [DOI] [PubMed] [Google Scholar]
- 6. Cappetta M., Roth I., Diaz A., Tort J., Roche L. 2002. Role of the prosegment of Fasciola hepatica cathepsin L1 in folding of the catalytic domain. Biol. Chem. 383:1215–1221 [DOI] [PubMed] [Google Scholar]
- 7. Chen R., Henning U. 1996. A periplasmic protein (Skp) of Escherichia coli selectively binds a class of outer membrane proteins. Mol. Microbiol. 19:1287–1294 [DOI] [PubMed] [Google Scholar]
- 8. Cloud J., Kelly C. P. 2007. Update on Clostridium difficile associated disease. Curr. Opin. Gastroenterol. 23:4–9 [DOI] [PubMed] [Google Scholar]
- 9. Dang T., et al. 2010. Chemical probes of surface layer biogenesis in Clostridium difficile. ACS Chem. Biol. 5:279–285 [DOI] [PubMed] [Google Scholar]
- 10. Dautin N., Barnard T. J., Anderson D. E., Bernstein H. D. 2007. Cleavage of a bacterial autotransporter by an evolutionarily convergent autocatalytic mechanism. EMBO J. 26:1942–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Emerson J., Fairweather N. F. 2009. Surface structures of Clostridium difficile and other clostridia, p. 157–167 In Bruggemann H., Gottschalk G. (ed.), Clostridia: molecular biology in the post-genomic era. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 12. Emerson J., et al. 2009. A novel genetic switch controls phase variable expression of CwpV, a Clostridium difficile cell wall protein. Mol. Microbiol. 74:541–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fagan R., Fairweather N. 2010. Dissecting the cell surface, p. 117–134 In Mullany P., Roberts A. P. (ed.), Methods Mol. Biol, 2010/07/03 ed., vol. 646 Humana Press, Inc., New York, NY: [DOI] [PubMed] [Google Scholar]
- 14. Fagan R. P., et al. 2009. Structural insights into the molecular organization of the S-layer from Clostridium difficile. Mol. Microbiol. 71:1308–1322 [DOI] [PubMed] [Google Scholar]
- 15. Fox T., de Miguel E., Mort J. S., Storer A. C. 1992. Potent slow-binding inhibition of cathepsin B by its propeptide. Biochemistry 31:12571–12576 [DOI] [PubMed] [Google Scholar]
- 16. Freeman J., et al. 2010. The changing epidemiology of Clostridium difficile infections. Clin. Microbiol. Rev. 23:529–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Heap J. T., Pennington O. J., Cartman S. T., Carter G. P., Minton N. P. 2007. The ClosTron: a universal gene knockout system for the genus Clostridium. J. Microbiol. Methods 70:452–464 [DOI] [PubMed] [Google Scholar]
- 18. Heap J. T., Pennington O. J., Cartman S. T., Minton N. P. 2009. A modular system for Clostridium shuttle plasmids. J. Microbiol. Methods 78:79–85 [DOI] [PubMed] [Google Scholar]
- 19. Hussain H. A., Roberts A. P., Mullany P. 2005. Generation of an erythromycin-sensitive derivative of Clostridium difficile strain 630 (630Δerm) and demonstration that the conjugative transposon Tn916E enters the genome of this strain at multiple sites. J. Med. Microbiol. 54:137–141 [DOI] [PubMed] [Google Scholar]
- 20. Janoir C., Pechine S., Grosdidier C., Collignon A. 2007. Cwp84, a surface-associated protein of Clostridium difficile, is a cysteine protease with degrading activity on extracellular matrix proteins. J. Bacteriol. 189:7174–7180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Just I., Gerhard R. 2004. Large clostridial cytotoxins. Rev. Physiol. Biochem. Pharmacol. 152:23–47 [DOI] [PubMed] [Google Scholar]
- 22. Karjalainen T., et al. 2001. Molecular and genomic analysis of genes encoding surface-anchored proteins from Clostridium difficile. Infect. Immun. 69:3442–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kirby J. M., et al. 2009. Cwp84, a surface-associated cysteine protease, plays a role in the maturation of the surface layer of Clostridium difficile. J. Biol. Chem. 284:34666–34673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kuehne S. A., et al. 2010. The role of toxin A and toxin B in Clostridium difficile infection. Nature 467:711–713 [DOI] [PubMed] [Google Scholar]
- 25. Lawley T. D., et al. 2009. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect. Immun. 77:3661–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lemee L., et al. 2005. Multilocus sequence analysis and comparative evolution of virulence-associated genes and housekeeping genes of Clostridium difficile. Microbiology 151:3171–3180 [DOI] [PubMed] [Google Scholar]
- 27. Lyras D., et al. 2009. Toxin B is essential for virulence of Clostridium difficile. Nature 458:1176–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Matias V. R., Beveridge T. J. 2005. Cryo-electron microscopy reveals native polymeric cell wall structure in Bacillus subtilis 168 and the existence of a periplasmic space. Mol. Microbiol. 56:240–251 [DOI] [PubMed] [Google Scholar]
- 29. Nagler D. K., et al. 1999. Human cathepsin X: a cysteine protease with unique carboxypeptidase activity. Biochemistry 38:12648–12654 [DOI] [PubMed] [Google Scholar]
- 30. Pechine S., et al. 2005. Immunological properties of surface proteins of Clostridium difficile. J. Med. Microbiol. 54:193–196 [DOI] [PubMed] [Google Scholar]
- 31. Poxton I. R., Cartmill T. D. 1982. Immunochemistry of the cell-surface carbohydrate antigens of Clostridium difficile. J. Gen. Microbiol. 128:1365–1370 [DOI] [PubMed] [Google Scholar]
- 32. Purdy D., et al. 2002. Conjugative transfer of clostridial shuttle vectors from Escherichia coli to Clostridium difficile through circumvention of the restriction barrier. Mol. Microbiol. 46:439–452 [DOI] [PubMed] [Google Scholar]
- 33. Rawlings N. D., Barrett A. J., Bateman A. 2010. MEROPS: the peptidase database. Nucleic Acids Res. 38:D227–D233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rupnik M., Wilcox M. H., Gerding D. N. 2009. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 7:526–536 [DOI] [PubMed] [Google Scholar]
- 35. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor, NY [Google Scholar]
- 36. Sarvas M., Harwood C. R., Bron S., van Dijl J. M. 2004. Post-translocational folding of secretory proteins in Gram-positive bacteria. Biochim. Biophys. Acta 1694:311–327 [DOI] [PubMed] [Google Scholar]
- 37. Savariau-Lacomme M. P., Lebarbier C., Karjalainen T., Collignon A., Janoir C. 2003. Transcription and analysis of polymorphism in a cluster of genes encoding surface-associated proteins of Clostridium difficile. J. Bacteriol. 185:4461–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sklar J. G., Wu T., Kahne D., Silhavy T. J. 2007. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 21:2473–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Strauch K. L., Johnson K., Beckwith J. 1989. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J. Bacteriol. 171:2689–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tormo A., Almiron M., Kolter R. 1990. surA, an Escherichia coli gene essential for survival in stationary phase. J. Bacteriol. 172:4339–4347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Waligora A. J., et al. 2001. Characterization of a cell surface protein of Clostridium difficile with adhesive properties. Infect. Immun. 69:2144–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wiederanders B. 2003. Structure-function relationships in class CA1 cysteine peptidase propeptides. Acta Biochim. Pol. 50:691–713 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.