Abstract
N-Acetylglucosamine (GlcNAc) is the most abundant carbon-nitrogen biocompound on earth and has been shown to be an important source of nutrients for both catabolic and anabolic purposes in Bacillus species. In this work we show that the GntR family regulator YvoA of Bacillus subtilis serves as a negative transcriptional regulator of GlcNAc catabolism gene expression. YvoA represses transcription by binding a 16-bp sequence upstream of nagP encoding the GlcNAc-specific EIIBC component of the sugar phosphotransferase system involved in GlcNAc transport and phosphorylation, as well as another very similar 16-bp sequence upstream of the nagAB-yvoA locus, wherein nagA codes for N-acetylglucosamine-6-phosphate deacetylase and nagB codes for the glucosamine-6-phosphate (GlcN-6-P) deaminase. In vitro experiments demonstrated that GlcN-6-P acts as an inhibitor of YvoA DNA-binding activity, as occurs for its Streptomyces ortholog, DasR. Interestingly, we observed that the expression of nag genes was still activated upon addition of GlcNAc in a ΔyvoA mutant background, suggesting the existence of an auxiliary transcriptional control instance. Initial computational prediction of the YvoA regulon showed a distribution of YvoA binding sites limited to nag genes and therefore suggests renaming YvoA to NagR, for N-acetylglucosamine utilization regulator. Whole-transcriptome studies showed significant repercussions of nagR deletion for several major B. subtilis regulators, probably indirectly due to an excess of the crucial molecules acetate, ammonia, and fructose-6-phosphate, resulting from complete hydrolysis of GlcNAc. We discuss a model deduced from NagR-mediated gene expression, which highlights clear connections with pathways for GlcNAc-containing polymer biosynthesis and adaptation to growth under oxygen limitation.
INTRODUCTION
N-Acetylglucosamine (GlcNAc) is a nitrogen-containing monosaccharide that constitutes a paramount building block ubiquitously found in the biosphere. GlcNAc-containing polymers, mainly chitin, chitosan, and peptidoglycan, are major constituents of arthropods' exoskeletons, filamentous fungi, and bacterial cell envelopes. Also, the matrix polysaccharides from biofilm of many bacteria consist of linear chains of GlcNAc residues in beta-(1,6) linkage. Considering partial deacetylation of polymeric N-acetylglucosamine, the substance is termed polysaccharide intercellular adhesin (PIA), although the abbreviation PNAG (poly-N-acetylglucosamine) also is found in the literature (20, 34).
As an amino-monosaccharide, GlcNAc represents a favorable nutrient source for a multitude of microorganisms to feed on, with several studies reporting its high position in the hierarchy of preferred carbon sources (18, 39). Distantly related bacteria, such as streptomycetes, firmicutes, and enterobacteriaceae, commonly use the phosphoenolpyruvate:phosphotransferase system (PTS) for uptake and phosphorylation of GlcNAc (1, 39, 42, 43, 61). In Bacillus, after uptake and concomitant phosphorylation with NagP as a probable PTS EIIBC component (48, 57), GlcNAc-6-phosphate (GlcNAc-6-P) is funneled into the glycolysis shunt after conversion to fructose-6-phosphate (Fru-6-P) by the consecutive action of the enzymes NagA (GlcNAc-6-P deacetylase) (64) and NagB (GlcN-6-P deaminase) (54, 63). On the other hand, when alternative carbon sources are available, GlcNAc is for the most part directed to peptidoglycan synthesis (39). GlcN-6-P is thereby converted into undecaprenol (UDP)-glucosamine, which is incorporated into the cell wall precursor lipid II in a multienzyme process (reviewed in reference 7). Since GlcNAc is exploited for both catabolic and anabolic purposes, its proper utilization should require rigorous and multilevel control, as documented in other model microorganisms, such as Streptomyces coelicolor and Escherichia coli (43, 45). Moreover, since complete hydrolysis of GlcNAc by the NagA and NagB enzymes generates acetate, ammonia (NH3), and Fru-6-P, we anticipate that GlcNAc utilization influences major biological processes, such as glycolysis, the tricarboxylic acid (TCA) cycle, respiration, nucleic acids, nitrogen, and fatty acid metabolism, as well as cell wall biosynthesis.
In this study, we provide a compilation of in silico, in vitro, and in vivo evidence that defines YvoA (also named NagR; see below) as a main transcriptional repressor of genes involved in GlcNAc transport and utilization in Bacillus subtilis. A recent study revealed the crystal structure of YvoA (49), which belongs to the HutC subfamily of GntR-type transcriptional regulators (50). The ortholog of YvoA in streptomycetes, DasR (deficient in aerial hyphae and spore formation), has been analyzed in depth: next to its involvement in the control of GlcNAc utilization (52), DasR plays a pivotal role in the regulation of antibiotic synthesis and differentiation (51, 53). Here we aimed to shed light on further common or distinct features of these two regulators. We further intensively characterized the YvoA regulon by combining DNA microarrays and computational prediction of YvoA-binding sites to highlight direct and indirect connections to major regulators of B. subtilis. A model of YvoA-dependent pathways is presented and discussed.
MATERIALS AND METHODS
Materials and general methods.
Chemicals were purchased from Merck (Darmstadt, Germany), Roth (Karlsruhe, Germany), or Sigma (Munich, Germany) at the highest purity available. Enzymes for DNA restriction and modification were obtained from New England BioLabs (Frankfurt/Main, Germany), Roche (Mannheim, Germany) or PeqLab (Erlangen, Germany) and were used according to the manufacturers' recommendations. Isolation and manipulation of DNA were performed using standard techniques. Oligonucleotides were purchased from MWG-Biotech (Ebersberg, Germany) or biomers.net (Ulm, Germany) and are listed in Table 1. Sequencing was carried out according to the protocol provided by the manufacturer with an ABI Prism 310 genetic analyzer (Applied Biosystems, Weiterstadt, Germany), at MWG-Biotech (Ebersberg, Germany), or at GATC (Constance, Germany).
Table 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′ → 3′) |
|---|---|
| A_lacZ_fw | GCTCAGAATTCTTTCATTTTATCACTT |
| A_lacZ_rev | GTTGGATCCGTGCTGATTTTTCCGT |
| dre_cons_fw | AAACACCTCAaCTGGTCTAGAcCACTAGTCTGAAAA |
| dre_cons_rev | TTTTCAGACTAGTGgTCTAGACCAGtTGAGGTGTTT |
| dre_nagA_fw | AAACACCTCAGCTGGTCTAGATCACTAGTCTGAAAA |
| dre_nagA_rev | TTTTCAGACTAGTGATCTAGACCAGCTGAGGTGTTT |
| nagB_fw | TCAAGTCGCGAACACATGTTGTGACAT |
| nagB_rev | AGCTCCGGATTCAAGGTCTTAATGACGCG |
| P_lacZ_fw | AATAGAATTCACACGGACCTGGGAA |
| P_lacZ_rev | AATTGGATCCGCAGGCAGAACCG |
| yvnB_fw | GGAAACCGGGACGTCTTGTGCATCA |
| yvnB_rev | AGCCTCGAGCCCTTTTTAAGGATTG |
| yvoA_ov_fw | GGAACATGCTGACATATGAATATCAAT |
| yvoA_ov_rev | CATATCGTCCGGATCCAGACCAGT |
| yvoA_fw | CGACCCGGGTATGAATATCAATAAACAATCGCCT |
| yvoA_rev | ATGTATAGACGTCGCCTCTGTATACGGA |
Bacterial strains, genetic manipulations, and growth conditions.
Bacterial strains used in this study are listed in Table 2. Standard cloning procedures were applied using E. coli DH5α as a host. B. subtilis WH557 (6) served as the parental strain. It chromosomally harbors the tetR gene (encoding a tetracycline repressor), enabling repression of yvoA in derivative strains (see below). All plasmids constructed in this study were verified by sequencing. Integration of relevant plasmid portions by double homologous recombination into the genome was verified by PCR and sequencing of amplified products. Excision of lox-flanked resistance cassettes from B. subtilis chromosomes was achieved using the Cre recombinase expression plasmid pCrePA (46) or pRAB1 (29) as described previously (5). Briefly, strains subjected to Cre treatment were transformed with either of the two plasmids and initially incubated at 30°C (permissive for the plasmids' thermosensitive origins of replication) to allow cre expression. A temperature shift to 37°C with further incubation for 1 or 2 days resulted in plasmid loss. The results of site-specific recombination by Cre were checked by PCR, and the concomitant reinstated antibiotic sensitivity of respective strains was verified by an inability to grow on selective media. Cells were generally grown in LB medium unless stated otherwise. Media contained the following antibiotics, where appropriate: ampicillin (Ap) (100 mg/liter for E. coli), kanamycin (Km) (60 mg/liter for E. coli or 15 mg/liter for B. subtilis), and chloramphenicol (Cm) (25 mg/liter for E. coli or 5 mg/liter for B. subtilis). (Liquid) cultures were incubated (shaking) at 37°C unless stated otherwise.
Table 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics or sequencea | Reference or source |
|---|---|---|
| Strains | ||
| E. coli DH5α | recA1 endA1 gyrA96 thi relA1 hsdR17(rK− mK+) supE44 φ80dlacZΔ ΔlacU169 | 23 |
| E. coli FT1/pLysS | BL21(DE3) Δ(ptsHI crr)/pLysS, Kmr | 44 |
| B. subtilis WH557 | trpC2 lacA::pt17-tetR-lox66-aphAIII-lox71 | 6 |
| B. subtilis WH558 | trpC2 lacA::pt17-tetR-lox66-aphAIII-lox71 amyE::>InsTetG+2Cm>-lacZ | 6 |
| B. subtilis FT1 | trpC2 lacA::pt17-tetR-lox72 | This study |
| B. subtilis FT2 | trpC2 lacA::pt17-tetR-lox72 amyE::drenagA-nagA′-lacZ | This study |
| B. subtilis FT3 | trpC2 lacA::pt17-tetR-lox72 amyE::drenagP-nagP′-lacZ | This study |
| B. subtilis FT10 | trpC2 lacA::pt17-tetR-lox66-aphAIII-lox71 >InsTetG+2Cm>-yvoA | This study |
| B. subtilis FT11 | trpC2 lacA::pt17-tetR-lox72 >InsTetG+2Cm>-yvoA | This study |
| B. subtilis FT12 | trpC2 lacA::pt17-tetR-lox72 >InsTetG+2Cm>-yvoA amyE::drenagA-nagA′-lacZ | This study |
| B. subtilis FT13 | trpC2 lacA::pt17-tetR-lox72 >InsTetG+2Cm>-yvoA amyE::drenagP-nagP′-lacZ | This study |
| B. subtilis FT20 | trpC2 lacA::pt17-tetR-lox66-aphAIII-lox71 yvoA::cat | This study |
| B. subtilis FT21 | trpC2 lacA::pt17-tetR-lox72 yvoA::cat | This study |
| B. subtilis FT22 | trpC2 lacA::pt17-tetR-lox72 yvoA::cat amyE::drenagA-nagA′-lacZ | This study |
| B. subtilis FT23 | trpC2 lacA::pt17-tetR-lox72 yvoA::cat amyE::drenagP-nagP′-lacZ | This study |
| Plasmids | ||
| pAC7 | Vector for integration into amyE of B. subtilis, pBR322 derivative, Apr Cmr | 66 |
| pAC7-nagA | drenagA-nagA′-lacZ, Apr Cmr, pAC7 derivative | This study |
| pAC7-nagP | drenagP-nagP′-lacZ, Apr Cmr, pAC7 derivative | This study |
| pCrePA | PpagA-cre, thermosensitive, expression of cre, Apr Ermr | 46 |
| pRAB1 | PpagA-cre, thermosensitive, expression of cre, Apr Cmr | 29 |
| pWH1935-2Cm | InsTetG+2Cm, pUC19 derivative, Apr Cmr | 6 |
| pWH1935-2CmNA | >InsTetG+2Cm->yvoA, pWH1935-2Cm derivative | This study |
| pWH1935-2CmNB | yvoA::cat, pWH1935-2Cm derivative | This study |
| pET-3c | Apr, PT7 | Novagen |
| pET-3c-yvoA | PT7-yvoA, pET-3c derivative | This study |
Angle brackets denote the promoter direction in InsTetG+1a or InsTetG+2Cm. Abbreviations: Ap, ampicillin; Cm, chloramphenicol; Km, kanamycin; superscript r, resistant.
Phylogeny analysis.
GntR/HutC subfamily regulators of B. subtilis 168 were used as a training set to generate phylogenetic trees performed on the Phylogeny.fr platform (16) using the “One click” mode, which provides ready-to-use pipeline chaining programs: MUSCLE for multiple alignment, Gblocks for automatic alignment curation, PhyML for tree building, and TreeDyn for tree drawing.
Computational prediction of YvoA-binding sites.
YvoA-binding sites upstream of nagP (drenagP; ATTGGTATAGATCACT) and upstream of the nagAB-yvoA locus (drenagA; GCTGGTCTAGATCACT) of B. subtilis were used to search similar sequences upstream of nagP and nagA orthologues in Bacillus species using the PREDetector software program (24). This list of 18 dre sites (see Table S1 in the supplemental material) was used to generate a position weight matrix (named NagR 2011) for NagR (YvoA) regulon prediction in Bacillus species.
Production and purification of YvoA in E. coli.
The plasmid pET-3c-yvoA (see Table 2) was constructed for overexpression of YvoA by cloning yvoA amplified with the primers yvoA_ov_fw and yvoA_ov_rev into pET-3c (Novagen) via NdeI/BamHI. Native YvoA was purified using a protocol and the same conditions established for purification of TetR (19). Briefly, E. coli FT1/pLysS (44) was transformed with pET-3c-yvoA, and T7-polymerase-dependent yvoA transcription was induced by isopropyl-β-d-thiogalactopyranoside (IPTG). After ultracentrifugation of crude cell extract, the supernatant was subjected to cation exchange chromatography and subsequent size fractioning. The concentration was determined by measuring the fluorescence of the solution at λ = 280 nm.
EMSA.
To detect binding of purified YvoA to DNA, electrophoretic mobility shift assays (EMSA) were conducted as follows. First, the complementary oligonucleotides dre_nagA_fw and dre_nagA_rev or dre_cons_fw and dre_cons_rev, respectively, were hybridized. To this end, equimolar amounts of each of two single-stranded oligonucleotides were mixed, heated at 96°C for 5 min, and allowed to cool to room temperature within 2 h, yielding the double-stranded 36-bp fragments drenagA and drecons. YvoA was added to the DNA fragments at indicated amounts and incubated for 10 min at room temperature in complex buffer (50 mM Tris-HCl [pH 7.5], 20 mM NaCl, 1 mM EDTA, and 1 mM dithiothreitol [DTT]). Assumed inducers of YvoA were added to final concentrations of 100 mM where applicable. Reaction mixtures were subjected to electrophoresis on a 10% polyacrylamide gel at 50 V in TBE buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA; pH 8.3 to 8.5), and DNA was detected by ethidium bromide staining.
Construction of B. subtilis strains bearing chromosomally inactivated or repressed yvoA.
A 296-bp fragment containing the 3′ part of the nagB gene (upstream of yvoA) including its stop codon was obtained using the primers nagB_fw and nagB_rev. This fragment was cloned into pWH1935-2Cm (6) via PciI/BspEI to yield pWH1935-2CmN. A 493-bp fragment containing a 3′ portion of the yvnB gene (downstream of yvoA) was obtained with primers yvnB_fw and yvnB_rev, digested with the enzymes AatII and XhoI, and inserted into the likewise restricted pWH1935-2CmN, yielding plasmid pWH1935-2CmNB. B. subtilis WH557 (6), which served as a parental strain (yvoA positive control), was transformed with the AhdI-linearized plasmid pWH1935-2CmNB by a standard procedure (27). The resulting strain was designated FT20 (ΔyvoA). Treatment of strain FT20 with the Cre recombinase gave rise to the marker-free strain FT21. The Km resistance gene was also removed by the same procedure from the parental strain B. subtilis WH557 to yield strain FT1.
In order to provide an yvoA repression mutant in addition to the yvoA knockout strain, we placed yvoA under the control of the TetR-controlled promoter Pxyl/tet (21). For this purpose, a 712-bp PCR product comprising almost the entire yvoA gene (obtained with primers yvoA_fw and yvoA_rev) was inserted into pWH1935-2CmN via AatII/XmaI. The resulting plasmid, pWH1935-2CmNA, was AhdI linearized and used to transform B. subtilis WH557. The resulting strain was designated FT10 (Pxyl/tet-yvoA), and the further steps required to obtain the marker-free strain FT11 were identical to those conducted to generate FT20 (see above).
Construction of B. subtilis β-galactosidase reporter strains to monitor nagA and nagP promoter activity.
Plasmids for the construction of reporter strains were constructed as follows. Fragments of nagA or nagP (including respective dre sites within the upstream region and 33 or 20 codons of the genes, respectively) were amplified with primers A_lacZ_fw and A_lacZ_rev or P_lacZ_fw and P_lacZ_rev, respectively. Fragments were cloned into plasmid pAC7 (66) via EcoRI/BamHI to obtain pAC7-nagA and pAC7-nagP, which encode N-terminal translational fusions of NagA or NagP with β-galactosidase (β-Gal). Strains FT1 (parental strain), FT11 (Pxyl/tet-yvoA), and FT21 (ΔyvoA) were transformed with plasmid pAC7-nagA or pAC7-nagP, and chromosomal integration into the amyE loci was verified using starch agar plates. The obtained strains were designated FT2, FT12, and FT22 in cases of encoded NagA′–β-gal fusions and FT3, FT13, and FT23 in cases of encoded NagP′–β-gal fusions (Table 2).
Enzyme assays.
nagB-encoded GlcN-6-P isomerase activity was measured in a coupled assay. B. subtilis cells were cultivated in CSK minimal medium (36), supplemented with a final concentration of 0.2% (wt/vol) GlcNAc, where appropriate, to an optical density at 600 nm (OD600) of about 0.8. Cells were broken by sonication, and 250 μl of soluble protein extract, adjusted to 1 μg/μl in ZAP buffer (10 mM Tris-HCl, pH 8, 200 mM NaCl, 5 mM DTT) was used. The reaction mixture additionally contained 1 mM GlcN-6-P, 0.2 mM NADP+, 230 mM Na2HPO4, 230 mM NaH2PO4, 3 U of phosphoglucoisomerase, and 2 U Glc-6-P dehydrogenase (both from Sigma, Munich, Germany). Reduction of NADP+ as a means for NagB activity was determined by measuring the rate of change at E340. The specific activity was calculated using the following formula: specific activity = dA/min·1,000/6.3·Pr, whereby dA denotes the change in E340 and Pr denotes the protein concentration (mg/ml). Quantification of β-gal activity of mid-log-growth-phase cells cultured in LB medium supplemented with different antibiotics and/or GlcNAc, where appropriate, was performed according to the method of Miller (38) and adapted as described previously (26).
Microarray experiments.
Transcriptome analyses were conducted with four biological replicates (each) of strains WH558 (wild-type [wt] yvoA) and FT20 (ΔyvoA), which harbor identical resistance markers. Overnight cultures of cells grown in baffled Erlenmeyer flasks containing 20 ml tryptone-yeast extract (TY) broth with kanamycin and chloramphenicol but without GlcNAc were freshly diluted into identically prepared flasks to an OD600 of ∼0.05. Cultures were harvested in mid-log growth phase at OD600 values between 0.71 and 0.79. The further steps, including breaking of cells, RNA preparation, quantity and quality control, synthesis, labeling and purification of cDNA, hybridization, microarray handling, and data evaluation, were taken as described previously (33). Data with Cyber-T (Bayes) P values of <0.01 were judged significant, and genes regulated by a factor of at least 2 are listed in Table 3.
Table 3.
Transcriptome analysis for definition of YvoA (NagR) regulon in B. subtilisa
| Category and KEGG no. | Name(s) | Function | Expression, ΔyvoA/wt | cis-acting element | Pathway | Regulon(s) | Reference(s) | |
|---|---|---|---|---|---|---|---|---|
| Genes upregulated in the ΔyvoA mutant compared to parental strain B. subtilis WH558 | ||||||||
| BSU07700 | nagP | PTS N-acetylglucosamine-specific enzyme IICB component | +4.55 | YvoA | GlcNAc metabolism | YvoA | This study | |
| BSU35010 | nagA | N-Acetylglucosamine-6-phosphate deacetylase | +2.53 | YvoA | GlcNAc metabolism | YvoA | This study | |
| BSU04300 | ydaM | Putative glycosyltransferase associated to biofilm formation | +3.95 | — | Biofilm formation | — | — | |
| BSU33950 | cggR | Central glycolytic genes repressor | +3.49 | CggR | Glycolysis | CggR | 31 | |
| BSU15510 | pyrAA | Carbamoyl phosphate synthase small subunit | +2.85 | PyrR | Pyrimidine biosynthesis | PyrR | 25 | |
| BSU15520 | pyrAB | Carbamoyl phosphate synthase large subunit | +3.44 | PyrR | Pyrimidine biosynthesis | PyrR | 25 | |
| BSU15500 | pyrC | Dihydroorotase | +2.35 | PyrR | Pyrimidine biosynthesis | PyrR | 25 | |
| BSU15540 | pyrD | Dihydroorotate dehydrogenase 1B | +2.30 | PyrR | Pyrimidine biosynthesis | PyrR | 25 | |
| BSU15560 | pyrE | Orotate phosphoribosyltransferase | +2.21 | PyrR | Pyrimidine biosynthesis | PyrR | 25 | |
| BSU15550 | pyrF | Orotidine 5′-phosphate decarboxylase | +2.23 | PyrR | Pyrimidine biosynthesis | PyrR | 25 | |
| BSU15530 | pyrK | Dihydroorotate dehydrogenase electron transfer subunit | +2.24 | PyrR | Pyrimidine biosynthesis | PyrR | 25 | |
| BSU02700 | lip, estA | Secreted triacylglycerol alkaliphilic lipase | +2.01 | — | Triacylglycerol metabolism | — | 15 | |
| BSU37210 | ywjC | Hypothetical protein | +2.60 | — | Unknown function | — | — | |
| Genes downregulated in the ΔyvoA mutant compared to parental strain B. subtilis WH558 | ||||||||
| BSU37310 | fnr | Fnr, fumarate nitrate reductase regulator | −2.63 | Fnr | Anaerobic redox regulator | Fnr | 14 | |
| BSU37290 | arfM | Anaerobic respiration and fermentation modulator | −4.10 | Fnr | Anaerobic genes regulator | Fnr/ResDE | 35 | |
| BSU37300 | ywiC | Unknown, putative integral inner membrane protein | −4.13 | Fnr | Unknown | — | — | |
| BSU37280 | narG | Nitrate reductase (alpha subunit) | −2.70 | Fnr | Nitrate reduction | Fnr | 14 | |
| BSU37270 | narH | Nitrate reductase (beta subunit) | −3.13 | Fnr | Nitrate reduction | Fnr | 14 | |
| BSU37250 | narI | Nitrate reductase assembling factor | −3.17 | Fnr | Nitrate reduction | Fnr | 14 | |
| BSU37260 | narJ | Nitrate reductase (gamma subunit) | −2.62 | Fnr | Nitrate reduction | Fnr | 14 | |
| BSU37320 | narK | Nitrite extrusion permease | −3.84 | Fnr | Nitrate extrusion | Fnr | 14 | |
| BSU14160 | ykuO | Hypothetical protein | −5.13 | Fur | Flavodoxins for NO production | Fnr/Fur | 3, 47 | |
| BSU14170 | ykuP | Short-chain flavodoxin | −3.99 | Fur | Flavodoxins for NO production | Fnr/Fur | 3, 47 | |
| BSU32000 | dhbA | 2.3-Dihydroxybenzoate-2.3-dehydrogenase | −4.26 | Fur | Siderophore (Bacillibactin) synthesis | Fnr/Fur | 3, 47 | |
| BSU31970 | dhbB | Isochorismatase | −3.58 | Fur | Siderophore (Bacillibactin) synthesis | Fnr/Fur | 3, 47 | |
| BSU31990 | dhbC | Isochorismate synthase | −2.99 | Fur | Siderophore (Bacillibactin) synthesis | Fnr/Fur | 3, 47 | |
| BSU31980 | dhbE | 2.3-Dihydroxybenzoate-AMP ligase | −3.64 | Fur | Siderophore (Bacillibactin) synthesis | Fnr/Fur | 3, 47 | |
| BSU31960 | dhbF | Siderophore 2.3-dihydroxybenzoate-glycine-threonine trimeric ester | −4.91 | Fur | Siderophore (Bacillibactin) synthesis | Fnr/Fur | 3, 47 | |
| BSU38740 | cydC | ABC transporter (ATP-binding) for cytochrome bd function | −3.32 | YdiH | Low oxygen tension | Fnr/Fur/YdiH | 59 | |
| BSU22070 | xpt | Xanthine phosphoribosyltransferase | −2.59 | — | Purine biosynthesis | — | — | |
| BSU02420 | ybgH, glnT | Glutamine transporter | −2.39 | GlnL | Gln/Glu metabolism | GlnL | 58 | |
| BSU01640 | ybbB, Btr | Bacillibactin transport regulator | −2.38 | Fur | Bacillibactin uptake | Fur | 3 | |
| BSU37410 | albE | Putative hydrolase involved in subtilosin production | −2.02 | AbrB/ResD | Antilisterial bacteriocin (subtilosin) production | ResD/Fnr/AbrB/Spo0A | 69 | |
| BSU37420 | albF | Putative peptidase involved in subtilosin production | −2.37 | AbrB/ResD | Antilisterial bacteriocin (subtilosin) production | ResD/Fnr/AbrB/Spo0A | 69 | |
| BSU01630 | feuA | Iron hydroxamate-binding lipoprotein | −2.27 | Fur | Siderophore uptake | Fur | 3 | |
| BSU01620 | feuB | Iron-uptake protein | −2.00 | Fur | Siderophore uptake | Fur | 3 | |
| BSU01610 | feuC | Iron-uptake protein | −2.02 | Fur | Siderophore uptake | Fur | 3 | |
| BSU32940 | yusV | Iron(III)-siderophore transporter | −2.10 | Fur | Siderophore uptake | Fur | 3 | |
| BSU33320 | fhuD | Ferrichrome ABC transporter | −2.04 | Fur | Siderophore uptake | Fur | 3 | |
| BSU39610 | yxeB | ABC transporter (ferrioxamine binding lipoprotein) | −2.21 | Fur | Siderophore uptake | Fur | 3 | |
| BSU04530 | ydbN | Hypothetical protein | −2.02 | Fur | Unknown function | Fnr/Fur | 3, 47 | |
| BSU32030 | yuiG, bioYB | Putative biotin transporter | −3.77 | — | — | — | — | |
| BSU36550 | spoIIQ | Forespore protein required for alternative engulfment | −2.21 | — | Spore engulfment | — | 30 | |
| BSU34140 | yvfM, ganQ | Arabinogalactan oligomer permease | −2.20 | — | Arabinogalactan metabolism | — | — | |
| BSU30280 | amyD | Carbohydrate ABC transporter | −2.16 | — | Carbohydrate metabolism | — | — | |
| BSU19310 | dhaS | Putative aldehyde dehydrogenase | −2.12 | — | Unknown | — | — | |
| BSU11990 | yjdB | Hypothetical protein | −2.01 | — | Unknown function | — | — |
—, cis-acting element, trans-acting regulator, pathway, or regulon unknown. Genes listed exhibited differential expression by a factor of at least 2, with a Cyber-T (Bayes) P value of <0.01 (see Materials and Methods), between WH558 and FT20 grown without GlcNAc.
RESULTS AND DISCUSSION
In silico identification of YvoA as presumed N-acetylglucosamine utilization regulator in Bacillus species.
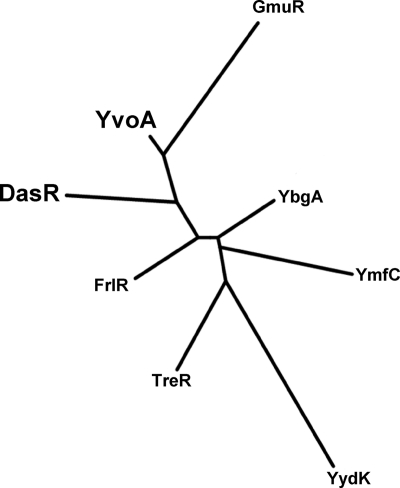
A role of YvoA in regulating GlcNAc metabolism in Bacillus species was predicted from a BLASTP analysis of the S. coelicolor GlcNAc utilization regulator DasR (KEGG identifier [ID] SCO5231). The highest similarity to DasR was indeed shown by the GntR/HutC subfamily (50) transcriptional regulator YvoA, with a sequence identity of 40% and a similarity value of 60%. Phylogeny analyses, either performed on the entire protein sequence or limited to the DNA-binding domain (not shown) of all seven GntR/HutC regulators from B. subtilis and DasR from S. coelicolor, suggested that DasR and YvoA, together with GmuR, the glucomannan utilization repressor (56), had emerged from a common ancestor (Fig. 1). The predisposition of YvoA and DasR to share similar DNA-binding properties was recently confirmed by Resch and collaborators, who showed that YvoA was able to bind with high affinity to the DasR responsive element (dre)-like sequence (ATTGGTATAGATCACT) located at 66 nucleotides [nt] upstream of nagP, coding for the GlcNAc-specific PTS EIIBC component in B. subtilis (49).
Fig. 1.
Phylogeny analysis of GntR/HutC subfamily regulators of B. subtilis and DasR from S. coelicolor. Proteins used as a training set to generate the tree are as follows: (i) GmuR, the regulator of glucommanan catabolism (YdhQ; NP_388466) (56), (ii) FrlR, possibly involved in regulation of utilization of sugar amines according to the SubtiWiki database (YurK; NP_391136) (28), (iii) YbgA, which presumably represents a repressor of the gamAP operon encoding glucosamine catabolism enzymes (NP_388119) (48), (iv) TreR, the trehalose utilization repressor (NP_388663) (60), (v) YmfC (NP_389563), (vi) YydK (NP_391893), (vii) YvoA (NP_391383), and (viii) DasR, the GlcNAc utilization regulator from S. coelicolor (NP_629378) (51). The phylogenic tree was generated via the Phylogeny.fr platform (16). Note that YvoA and DasR emerge together from a common ancestor.
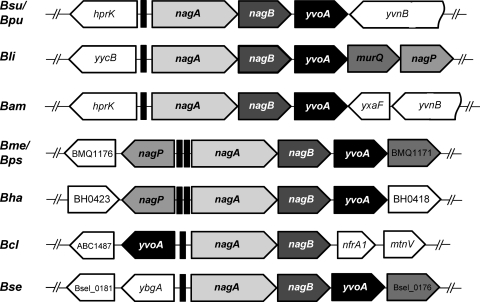
Examination of gene organization around the yvoA locus also provided unambiguous indications for its presumed implication in GlcNAc utilization (62). Indeed, in yvoA-containing Bacillus species (except Bacillus clausii), this gene is the third element of a putative tricistronic operon that includes in the first two positions nagA (the GlcNAc-6-P deacetylase gene) and nagB, which encodes the enzyme that catalyzes the deamination and isomerization of GlcNAc-6-P to Fru-6-P (Fig. 2). Interestingly, other GlcNAc metabolism-related genes are located in the vicinity of the nagAB-yvoA locus in other Bacillus species and therefore are presumably part of the YvoA regulon. This is the case for nagP, which is the fifth element of the putative nagAB-yvoA operon in Bacillus licheniformis and is positioned divergently from nagA in Bacillus halodurans, Bacillus pseudofirmus, and Bacillus megaterium (Fig. 2). In addition, murQ, which encodes an N-acetylmuramic acid-6-phosphate (MurNAc-6-P) etherase converting MurNAc-6-P to GlcNAc-6-P and d-lactate, is located between yvoA and nagP in B. licheniformis. The observed synteny of yvoA loci in Bacillus species thus strengthens the presumed role of YvoA in controlling key genes of the GlcNAc metabolism.
Fig. 2.
Synteny of the yvoA locus in Bacillus species. Gene organization around the various yvoA loci was deduced from the complete genome sequences retrieved from NCBI. Note the integration of murQ and nagP into the putative nagAB-yvoA operon in several species. Abbreviations: nagA, GlcNAc-6-P deacetylase gene; nagB, GlcN-6-P isomerase gene; nagP, encoding sugar phosphotransferase EIIBC component for GlcNAc transport and phosphorylation; murQ, MurNAc-6-P etherase gene. Black rectangles upstream of nagA orthologues represent identified dre-like sequences putatively bound by YvoA. Bsu, B. subtilis; Bpu, B. pumilis; Bli, B. licheniformis; Bam, B. amyloliquefaciens; Bme, B. megaterium; Bps, B. pseudofirmus; Bha, B. halodurans; Bcl, B. clausii; Bse, B. selenitireducens.
Identification of new YvoA-binding sequences and glucosamine-6-phosphate as YvoA DNA-binding activity modulator.
The YvoA binding site verified by Resch and colleagues—ATTGGTATAGATCACT, termed drenagP (49)—fits 13 out of the 16 nucleotides of the consensus of the DasR responsive element dreStreptomyces (AGTGGTCTAGACCACT) deduced from previous S. coelicolor studies (11). Exploiting the PREDetector software program (24) to identify further putative YvoA binding sites revealed another DNA sequence (GCTGGTCTAGATCACT) that also fits 13 out of the 16 nucleotides of dreStreptomyces. We designated this site, located at position −70 in the upstream region of the nagAB-yvoA region, drenagA. We could identify similar dre sequences upstream of the nagAB or nagP gene in all Bacillus species which possess an YvoA ortholog and for which genome data are available (see Table S1 in the supplemental material).
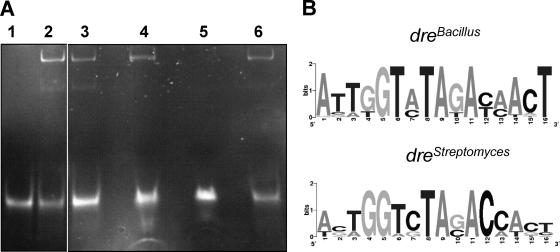
In order to confirm the interaction of YvoA with the newly found dre-like motif, we used DNA fragments containing drenagA and dreStreptomyces for qualitative binding assays with purified YvoA. As depicted in Fig. 3 A, migration of drenagA fragment during electrophoresis was clearly retarded in the presence of YvoA (which also held true for EMSA with dreStreptomyces; not shown). In order to determine which low-molecular-weight effector(s) could modulate the DNA-binding capability of YvoA, the regulator was incubated with drenagA in the presence of each of the four different aminosugar compounds GlcNAc, GlcNAc-6-P, GlcN-6-P, and GlcN. Subsequent EMSA analyses resulted in the observation that among the tested compounds only GlcN-6-P was able to abolish YvoA binding to drenagA (Fig. 3A). GlcN-6-P was also reported to inhibit the DNA-binding capability of DasR in S. coelicolor (51), whereas isothermal calorimetry titration measurements for binding of GlcN-6-P to YvoA conducted by Resch and colleagues had been inconclusive (49). Interestingly, the crystal structure in this study had been solved for YvoA in complex with GlcNAc-6-P, which had not appeared to interfere with dre binding in EMSA. These data fit a model that suggests that upon GlcNAc-6-P-binding, only one half-side of the YvoA dimer is detached from the dre sequence, which, however, does not abolish the YvoA-DNA complex. It is plausible that GlcN-6-P could cause a different allosteric mechanism through which effector binding modulates DNA affinity. Depending on the intracellular concentrations of the various aminosugar compounds (GlcNAc, GlcNAc-6-P, or GlcN-6-P), the effect on YvoA DNA binding would be different with appropriate expression adjustment of YvoA-dependent genes. Compilation of known and predicted YvoA-binding sites (see the table in the supplemental material) led us to identify ATTGGTATAGACAACT as an YvoA consensus sequence (dreBacillus) which is not a perfect palindrome and differs from the Streptomyces dre consensus sequence at positions 2 (G → T), 7 (C → A), and 13 (C → A). Graphical representations of Streptomyces and Bacillus dre sites were generated using the Weblogo software application and are displayed in Fig. 3B.
Fig. 3.
YvoA DNA-binding abilities. (A) EMSA displaying binding of YvoA to drenagA and identification of GlcN-6-P as a DNA-binding inhibitor. Twenty picomoles of double-stranded DNA fragment was electrophoresed alone (lane 1) or after incubation with 10 pmol of YvoA monomers (lane 2) and a final concentration of 100 mM GlcNAc (lane 3), GlcNAc-6-P (lane 4), GlcN-6-P (lane 5), or GlcN (lane 6), respectively. (B) Weblogo representation of Streptomyces and Bacillus consensus dre sites. Note main differences at positions 2 (G → T), 7 (C → A), and 13 (C → A) of the dre sites. Weblogo representation of dreBacillus was generated with all YvoA-binding sequences upstream on Bacillus species nagA and nagP genes (see Table S1 in the supplemental material), and the Weblogo dreStreptomyces representation was generated with dre sites identified upstream of the S. coelicolor nagB, nagA, dasA (12), and PTS genes involved in GlcNAc uptake.
YvoA represses expression of the N-acetylglucosamine-induced genes nagP, nagA, and nagB.
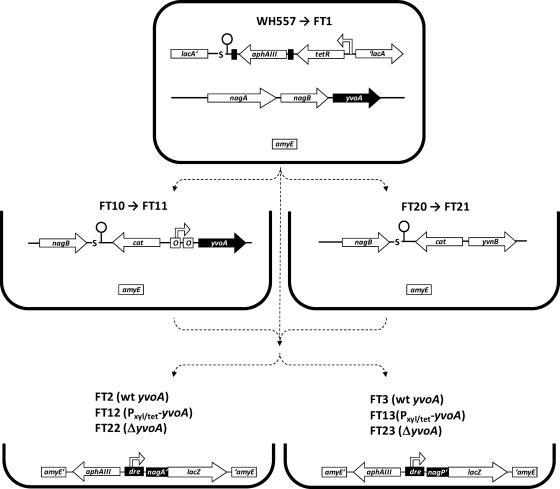
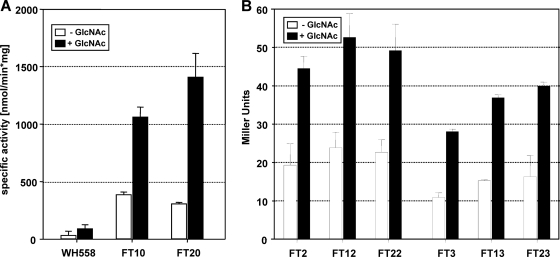
To gain insights into the regulatory role of YvoA in B. subtilis in vivo, we first constructed two new strains on the basis of the yvoA-proficient strain WH557 (Fig. 4). To enable artificial repression of yvoA, strain FT10 was constructed, in which we placed yvoA expression under the control of the TetR-controlled promoter Pxyl/tet (6, 21). In addition, FT20, which bears a complete yvoA deletion, was generated. In order to test the assumption of YvoA-dependent regulation of nagA, nagB, and nagP in vivo, the activity of NagB, catalyzing the deamination and isomerization of GlcN-6-P to Fru-6-P (63), was quantified using B. subtilis FT10 (Pxyl/tet-yvoA), FT20 (ΔyvoA), and WH558 (wt yvoA, comparable to the parental strain, WH557, but with a kanamycin resistance marker identical to that of FT10 and FT20). Observed NagB activities, displayed in Fig. 5 A, were all elevated among the strains with deleted or repressed yvoA in comparison to findings for WH558, suggesting a transcriptional repressor role for YvoA. According to expectations, WH558 cells displayed higher values upon growth with GlcNAc, apparently due to release of YvoA from drenagA. Interestingly, however, higher NagB activities in the presence of GlcNAc were also observed for strains FT10 and FT20, which can be expected to produce only minute amounts of YvoA or no YvoA at all, respectively (see below).
Fig. 4.
Schematic representation of selected strains constructed in this study. Parent strain WH557 bears an unaffected yvoA gene and a chromosomally integrated tetR gene. FT1 is isogenic to WH557 but lacks the kanamycin resistance gene aphAIII after excision by Cre recombinase. Strains FT10 and FT20 are derived from WH557 and were also treated with Cre to yield FT11 and FT21, respectively. FT2, FT3, FT12, FT13, FT22, and FT23 are constructs allowing reporter gene assays of the nagA or the nagP promoter(s).
Fig. 5.
(A) NagB assay. The activity of the glucosamine-6-phosphate isomerase NagB was assayed in strains with either WH558 (yvoA proficient), FT20 (ΔvyoA), or FT10 (repressed yvoA). White or black bars denote specific activities of cells grown without or with GlcNAc, respectively. The error bars represent the standard deviations of biological triplicate determinations. (B) β-Galactosidase assays of nagA′-lacZ and nagP′-lacZ fusions. Transcription of nagA (left part) and nagP (right part) was indirectly assayed in strains with either pristine yvoA (FT2 and FT3) or ΔvyoA (FT22 and FT23) or transcriptionally repressed yvoA (FT12 and FT13) by means of translational fusions with lacZ. White or black bars denote specific activities of cells grown without or with GlcNAc, respectively. The error bars represent the standard deviations of biological triplicate determinations.
To further validate regulation of catabolic nag genes, vectors carrying translational lacZ fusions to nagA or nagP were constructed. After elimination of the kanamycin resistance markers from WH557, FT10, and FT20 by Cre recombinase, the plasmids were chromosomally integrated into the obtained strains FT1 (wt yvoA), FT11 (Pxyl/tet-yvoA), and FT21 (ΔyvoA). The obtained descendants bearing the nagA′-lacZ fusion were designated FT2, FT12, and FT22, and those with nagP′-lacZ were termed FT3, FT13, and FT23 (Fig. 4). Levels of β-Gal in these strains were found to be very low, but the same tendency as in NagB assays was still observed, i.e., higher activities when yvoA was inactivated by deletion or repression (Fig. 5B). Moreover, nagA and nagP promoters' activities were still higher when strains were grown in the presence of GlcNAc, as previously observed in NagB assays (Fig. 5A). Since GlcNAc-dependent regulation was not abolished in strains with low amounts of or no YvoA, we anticipate the coexistence of (an)other system(s) of nag genes' control. Early articles described that the activity of enzymes involved in GlcNAc transport, deamination, deacetylation and isomerization was elevated in the presence of the aminosugar but decreased under glucose conditions (4, 10, 39). These observations give reason to assume a glucose carbon catabolite repression system (reviewed in reference 22) of higher hierarchy than a GlcNAc-specific mode of control. In fact, nagAB has been described to be repressed by Glc, with possible involvement of the pleiotropic regulator CcpA (8).
Expression profiling using DNA microarrays for definition of the YvoA regulon in B. subtilis.
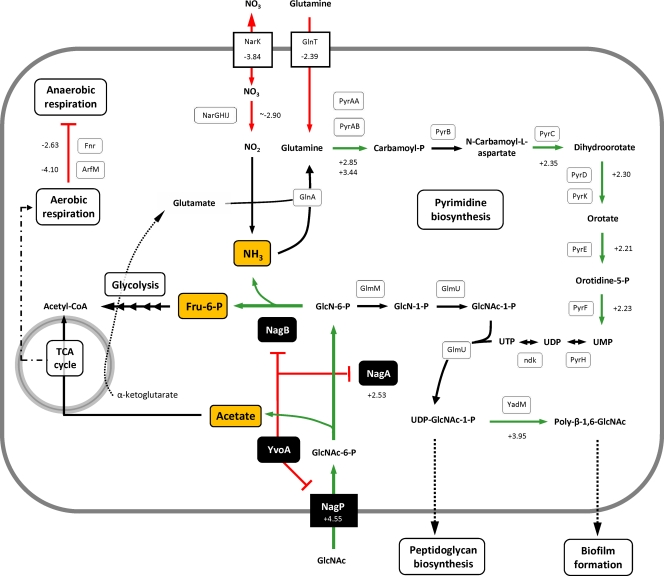
Changes in the transcriptome of yvoA mutant strain FT20 compared to that of the B. subtilis parental strain WH558 were analyzed using whole-genome DNA microarrays and total RNA of cells harvested in mid-log growth phase. Table 3 lists genes the mRNA levels of which were significantly altered by a factor of at least 2 (further transcriptome data are listed in Table S2 in the supplemental material), and Fig. 6 summarizes main YvoA-related pathways. Three different groups of yvoA-dependent genes were distinguished by their expression patterns. The first group consists of genes that are repressed by YvoA and that have a significant dre site within their upstream region. The second and third groups consist of genes repressed or induced by YvoA, respectively, but with no obvious YvoA-binding site detected.
Fig. 6.
Summary of pathways influenced by YvoA (NagR). Genes (or their deduced proteins) that possess an YvoA-binding sequence within their upstream region are represented in black. Final end products of complete GlcNAc hydrolysis are highlighted in orange, i.e., acetate, NH3, and Fru-6-P. Arrows in green or red indicate genes, the expression of which is up- or downregulated, respectively, in strain FT20 (ΔyvoA mutant) compared to expression in the parental strain B. subtilis WH558. Metabolic fluxes are represented by arrows; lines with T ends indicate transcriptional repression. “+” and “−” values indicate the expression ratio between strains FT20 and WH558 deduced from microarray analyses (for a complete set of values and protein functions, see Table 3).
Group 1 includes only nagP and nagA, with a 4.6- and 2.5-fold upregulation in the yvoA mutant background compared to the parental strain, respectively. The distribution of predicted YvoA-binding sites was limited to nagP and the nagAB-yvoA locus within the B. subtilis chromosome, which contrasts with the situation in streptomycetes, where hundreds of dre sites for the YvoA ortholog DasR have been identified in the S. coelicolor genome (S. Rigali, personal communication). The limited number of predicted dre sites in B. subtilis also suggests that YvoA might be a less-prominent regulator than DasR and may fulfill more confined tasks, i.e., the uptake and subsequent utilization of GlcNAc. However, yvoA deletion results in the modulation of the expression of many other important genes (Table 3; also see below). The absence of dre sites within their upstream region suggests that the yvoA deletion may thus indirectly alter their expression, possibly by affecting intracellular levels of Fru-6-P, acetate, and NH3, which are the final products of complete GlcNAc catabolism and are key molecules involved in numerous biological processes. At this stage, based upon the compilation of in silico, in vitro, and in vivo data obtained in this study, it seems timely to rename YvoA “NagR,” for N-acetylglucosamine utilization regulator.
Group 2 contains genes upregulated in strain FT20 but without exhibiting significant NagR-binding sites. ydaM (BSU04300) is 4.0-fold-upregulated in the nagR mutant background and could be directly related to GlcNAc metabolism, since it encodes a protein 52% similar to N-acetylglucosamine transferase PgaC of Actinobacillus pleuropneumoniae, responsible for the synthesis of poly-β-1,6-N-acetyl-d-glucosamine (9). Such polymeric substances, called polysaccharide intercellular adhesin (PIA), which can be partly deacetylated, are major components of bacterial biofilms in diverse eubacteria (20). One can speculate that inactivation of nagR would mimic signaling abundance of GlcNAc, which in turn would activate expression of genes for PIA synthesis. Interestingly, this coincides with the activation of the pyrimidine biosynthesis pathway in mutant FT20 (average, 2.5-fold upregulated), which leads to UTP, which is used to activate GlcNAc-1-P via GlmU, the UDP-N-acetylglucosamine diphosphorylase. The generated UDP-GlcNAc-1-P is used for anabolic purposes, such as cell wall biosynthesis or PIA-dependent biofilm formation.
Another important gene of group 2 is cggR, the central glycolytic gene repressor, controlling expression of the five downstream genes of its operon, namely, gapA, pgk, tpi, pgm, and eno, gathering the steps of interconversion of the triose phosphates from dihydroxyacetone phosphate to phosphoenolpyruvate (17). The repressor activity of CggR is inhibited by fructose-1,6-biphosphate (FBP) resulting from phosphorylation of Fru-6-P by phosphofructokinase (pfk). Pfk enzymatic activity is a paradigm of allosteric regulation, where the intracellular pool of its substrate Fru-6-P plays a crucial regulatory role. Fru-6-P is generated by NagB from GlcN-6-P. We demonstrated in this study that the deletion of nagR results in NagB overproduction, which would increase the intracellular pool of FBP by allosteric activation of Pfk due to an excess of Fru-6-P. The overload of FBP would abolish the DNA-binding activity of CggR, which would prevent repression of its own expression, explaining the 3.5-fold upregulation in the ΔyvoA mutant. Surprisingly, downstream genes of the cggR operon do not exhibit significant changes in their expression pattern, probably due to a complex and multiple level of control of this operon, as reported earlier (31, 32, 37).
Genes of group 3, downregulated in the ΔnagR strain without apparent cognate binding sites, are estimated to be indirectly controlled, because HutC/GntR regulators like NagR are almost exclusively repressor proteins.
From the 31 genes in this category, the presence of the fumarate nitrate reductase regulator Fnr (14), required for the adaptation of B. subtilis to anaerobic conditions, required particular consideration. When grown under oxygen limitation, B. subtilis can use nitrate as a final electron acceptor (40), and this physiological adaptation requires Fnr. Fifteen other genes of group 3 belong to the Fnr regulon (Table 3), and these are as follows: (i) narG, narH, narI, narJ, and narK, involved in nitrate reduction and nitrate extrusion (25a), respectively; (ii) dhbA, dhbB, dhbC, dhbE, and dhbF, for siderophore bacillibactin synthesis (55); (iii) ykuO and ykuP, for flavodoxins involved in nitric oxide production (65); (iv) cydC, for the ABC transporter (ATP-binding component) for cytochrome bd production (67); (v) arfM, the anaerobic respiration and fermentation modulator; and (vi) ydbN (hypothetical protein). A plausible explanation for such a drastic repression of the Fnr regulon could be the nature of the end products of GlcNAc catabolism produced by NagA and NagB, which are overexpressed in the ΔnagR mutant. Indeed, acetate, NH3, and Fru-6-P are very strong signals of efficient catabolism requiring oxidative phosphorylation. We expect that the oxygen tension would be high and as a consequence the path for using nitrate as an alternative electron acceptor in anaerobic respiration would be severely repressed. This hypothesis is also supported by the downregulation of the albE and albF genes, involved in antibacterial subtilosin production (69), which in a wild-type background are induced in response to nutrient starvation and by oxygen limitation (41).
Some NagR-dependent genes of the Fnr regulon, namely, the dhb and yku operons as well as cydC, were previously reported to also belong to the ferric uptake repressor (Fur) regulon (47), which controls the transcriptional response to iron starvation (3). Although expression of the fur gene itself is not strongly affected (1.4-fold upregulated in the nagR mutant), a total of 16 Fur-repressed genes were downregulated in strain FT20 (Table 3). The relationship between the Fnr and Fur regulons could be attributed to the inactivation of Fur repression by nitric oxide (NO), as reported for Salmonella enterica serovar Typhimurium (13). NO is thought to mimic or cause iron deficiency by complexing with free iron, leading to depletion of the cellular iron pool and subsequent activation of the Fur regulon. In the nagR mutant, the downregulation of Fnr would reduce intracellular amounts of NO. In turn, less ferric iron could be complexed by NO, so elevated intracellular levels of ferric iron would lead to formation of the Fe-Fur complex, which represses the iron starvation stimulon.
Finally, microarray data highlighted nagR mutant downregulated genes involved in carbohydrate transport (ganQ and amyD), spore engulfment (spoIIQ), biotin uptake (bioYB), or aldehyde dehydrogenation (dhaS) or with unknown function (yjdB), but their association to any GlcNAc-related process is too speculative to generate discussion. Of note, the methionine aminopeptidase-encoding gene yflG, previously reported as moderately controlled by NagR (68), did not match our criteria of an expression profile in strain FT20 significantly different from that in the parental strain based upon our microarray data.
Conclusions.
So far, the mode of control of GlcNAc uptake and metabolism in B. subtilis has been considerably less well analyzed than is the case for E. coli (2) and the Gram-positive high-G+C model bacterium S. coelicolor (43, 51). According to earlier work and to data presented here, the current knowledge on GlcNAc-dependent pathways in B. subtilis is summarized in Fig. 6. NagR represses expression of the three GlcNAc-inducible genes nagA, nagB, and nagP by directly binding a 16-bp sequence identified within their upstream regions, and this interaction is abolished upon GlcN-6-P binding to the regulator. The verification of connections highlighted from our microarray data between NagR and major regulatory proteins of B. subtilis, such as Fnr, Fur, and CggR—and their associated biological processes—requires further experimental support.
The result of the computational prediction of full-length NagR-binding sites indicated that these are found only scarcely in the B. subtilis genome, which strongly suggests indirect regulation by end products of GlcNAc hydrolysis rather than by NagR/dre-dependent direct cis/trans control. However, Resch and collaborators demonstrated that once bound to GlcNAc-6-P, NagR is still able to bind half-side dre in vitro. This is compatible with a model in which the regulator acts as both an activator and a repressor beyond the canonical mutually exclusive effector-bound or DNA-bound regulator logic (49). If GlcNAc-6-P should prove capable of inducing a jumping-jack-like motion of NagR (as proposed by Resch et al.) in vivo or if the NagR/GlcNAc-6-P complex is still able to recognize and bind half-side dre, this automatically raises the question of how widespread the distribution of half-NagR-binding sites in the B. subtilis chromosome is. A preliminary prediction of this 8-bp motif obviously gave reason to assume a much larger putative dre half-side-dependent NagR regulon, if this mode of control held true. Demonstration of how (and if) NagR could behave as a repressor and activator depending on the nutritional context will require significant additional experiments, which are necessary for understanding the complex response to GlcNAc in Bacillus species.
Supplementary Material
ACKNOWLEDGMENTS
We thank Annette Kamionka for help with YvoA purification, Hildegard Stork for support with enzyme assays, Martina Kolb for cloning of lacZ fusions and measurements, and Anne Winter and Klaus Pfleiderer for further technical support. Jörg Stülke is acknowledged for fruitful discussions and the gift of pAC7. We are grateful to Anne de Jong, Aldert Zomer, and Siger Holsappel for in silico work and array data evaluation and to Markus Resch for providing a template for Fig. 6.
This study was supported by the Deutsche Forschungsgemeinschaft through the Sonderforschungsbereich SFB 473 (Schaltvorgänge der Transkription) and through grant BE4038/1. S.R. is an F.R.S.-FNRS research associate.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 20 May 2011.
REFERENCES
- 1. Alice A. F., Perez-Martinez G., Sanchez-Rivas C. 2003. Phosphoenolpyruvate phosphotransferase system and N-acetylglucosamine metabolism in Bacillus sphaericus. Microbiology 149:1687–1698 [DOI] [PubMed] [Google Scholar]
- 2. Alvarez-Anorve L. I., Calcagno M. L., Plumbridge J. 2005. Why does Escherichia coli grow more slowly on glucosamine than on N-acetylglucosamine? Effects of enzyme levels and allosteric activation of GlcN6P deaminase (NagB) on growth rates. J. Bacteriol. 187:2974–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baichoo N., Wang T., Ye R., Helmann J. D. 2002. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 45:1613–1629 [DOI] [PubMed] [Google Scholar]
- 4. Bates C. J., Pasternak C. A. 1965. Further studies on the regulation of amino sugar metabolism in Bacillus subtilis. Biochem. J. 96:147–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bertram R., Kolb M., Hillen W. 2009. In vivo activation of tetracycline repressor by Cre/lox-mediated gene assembly. J. Mol. Microbiol. Biotechnol. 17:136–145 [DOI] [PubMed] [Google Scholar]
- 6. Bertram R., Köstner M., Müller J., Vazquez Ramos J., Hillen W. 2005. Integrative elements for Bacillus subtilis yielding tetracycline-dependent growth phenotypes. Nucleic Acids Res. 33:e153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bhavsar A. P., Brown E. D. 2006. Cell wall assembly in Bacillus subtilis: how spirals and spaces challenge paradigms. Mol. Microbiol. 60:1077–1090 [DOI] [PubMed] [Google Scholar]
- 8. Blencke H. M., et al. 2003. Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways. Metab. Eng. 5:133–149 [DOI] [PubMed] [Google Scholar]
- 9. Bossé J. T., et al. 2010. Regulation of pga operon expression and biofilm formation in Actinobacillus pleuropneumoniae by sigmaE and H-NS. J. Bacteriol. 192:2414–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clarke J. S., Pasternak C. A. 1962. The regulation of amino sugar metabolism in Bacillus subtilis. Biochem. J. 84:185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Colson S., et al. 2007. Conserved cis-acting elements upstream of genes composing the chitinolytic system of streptomycetes are DasR-responsive elements. J. Mol. Microbiol. Biotechnol. 12:60–66 [DOI] [PubMed] [Google Scholar]
- 12. Colson S., et al. 2008. The chitobiose-binding protein, DasA, acts as a link between chitin utilization and morphogenesis in Streptomyces coelicolor. Microbiology 154:373–382 [DOI] [PubMed] [Google Scholar]
- 13. Crawford M. J., Goldberg D. E. 1998. Regulation of the Salmonella typhimurium flavohemoglobin gene. A new pathway for bacterial gene expression in response to nitric oxide. J. Biol. Chem. 273:34028–34032 [DOI] [PubMed] [Google Scholar]
- 14. Cruz Ramos H., et al. 1995. Anaerobic transcription activation in Bacillus subtilis: identification of distinct FNR-dependent and -independent regulatory mechanisms. EMBO J. 14:5984–5994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dartois V., Baulard A., Schanck K., Colson C. 1992. Cloning, nucleotide sequence and expression in Escherichia coli of a lipase gene from Bacillus subtilis 168. Biochim. Biophys. Acta 1131:253–260 [DOI] [PubMed] [Google Scholar]
- 16. Dereeper A., et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Doan T., et al. 2003. The Bacillus subtilis ywkA gene encodes a malic enzyme and its transcription is activated by the YufL/YufM two-component system in response to malate. Microbiology 149:2331–2343 [DOI] [PubMed] [Google Scholar]
- 18. Dobrogosz W. J. 1968. Effect of amino sugars on catabolite repression in Escherichia coli. J. Bacteriol. 95:578–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ettner N., et al. 1996. Fast large-scale purification of tetracycline repressor variants from overproducing Escherichia coli strains. J. Chromatogr. A 742:95–105 [DOI] [PubMed] [Google Scholar]
- 20. Flemming H. C., Wingender J. 2010. The biofilm matrix. Nat. Rev. Microbiol. 8:623–633 [DOI] [PubMed] [Google Scholar]
- 21. Geissendörfer M., Hillen W. 1990. Regulated expression of heterologous genes in Bacillus subtilis using the Tn10 encoded tet regulatory elements. Appl. Microbiol. Biotechnol. 33:657–663 [DOI] [PubMed] [Google Scholar]
- 22. Görke B., Stülke J. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6:613–624 [DOI] [PubMed] [Google Scholar]
- 23. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 24. Hiard S., et al. 2007. PREDetector: a new tool to identify regulatory elements in bacterial genomes. Biochem. Biophys. Res. Commun. 357:861–864 [DOI] [PubMed] [Google Scholar]
- 25. Hobl B., Mack M. 2007. The regulator protein PyrR of Bacillus subtilis specifically interacts in vivo with three untranslated regions within pyr mRNA of pyrimidine biosynthesis. Microbiology 153:693–700 [DOI] [PubMed] [Google Scholar]
- 25a. Hoffmann T., Troup B., Szabo A., Hungerer C., Jahn D. 1995. The anaerobic life of Bacillus subtilis: cloning of the genes encoding the respiratory nitrate reductase system. FEMS Microbiol. Lett. 131:219–225 [DOI] [PubMed] [Google Scholar]
- 26. Kamionka A., Bertram R., Hillen W. 2005. Tetracycline-dependent conditional gene knockout in Bacillus subtilis. Appl. Environ. Microbiol. 71:728–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kraus A., Hueck C., Gärtner D., Hillen W. 1994. Catabolite repression of the Bacillus subtilis xyl operon involves a cis element functional in the context of an unrelated sequence, and glucose exerts additional xylR-dependent repression. J. Bacteriol. 176:1738–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lammers C. R., et al. 2010. Connecting parts with processes: SubtiWiki and SubtiPathways integrate gene and pathway annotation for Bacillus subtilis. Microbiology 156:849–859 [DOI] [PubMed] [Google Scholar]
- 29. Leibig M., et al. 2008. Marker removal in staphylococci via Cre recombinase and different lox sites. Appl. Environ. Microbiol. 74:1316–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Londono-Vallejo J. A., Frehel C., Stragier P. 1997. SpoIIQ, a forespore-expressed gene required for engulfment in Bacillus subtilis. Mol. Microbiol. 24:29–39 [DOI] [PubMed] [Google Scholar]
- 31. Ludwig H., et al. 2001. Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon. Mol. Microbiol. 41:409–422 [DOI] [PubMed] [Google Scholar]
- 32. Ludwig H., Rebhan N., Blencke H. M., Merzbacher M., Stülke J. 2002. Control of the glycolytic gapA operon by the catabolite control protein A in Bacillus subtilis: a novel mechanism of CcpA-mediated regulation. Mol. Microbiol. 45:543–553 [DOI] [PubMed] [Google Scholar]
- 33. Lulko A. T., Buist G., Kok J., Kuipers O. P. 2007. Transcriptome analysis of temporal regulation of carbon metabolism by CcpA in Bacillus subtilis reveals additional target genes. J. Mol. Microbiol. Biotechnol. 12:82–95 [DOI] [PubMed] [Google Scholar]
- 34. Mack D., et al. 1996. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178:175–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marino M., Ramos H. C., Hoffmann T., Glaser P., Jahn D. 2001. Modulation of anaerobic energy metabolism of Bacillus subtilis by arfM (ywiD). J. Bacteriol. 183:6815–6821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martin-Verstraete I., Stülke J., Klier A., Rapoport G. 1995. Two different mechanisms mediate catabolite repression of the Bacillus subtilis levanase operon. J. Bacteriol. 177:6919–6927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meinken C., Blencke H. M., Ludwig H., Stülke J. 2003. Expression of the glycolytic gapA operon in Bacillus subtilis: differential syntheses of proteins encoded by the operon. Microbiology 149:751–761 [DOI] [PubMed] [Google Scholar]
- 38. Miller J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 39. Mobley H. L., Doyle R. J., Streips U. N., Langemeier S. O. 1982. Transport and incorporation of N-acetyl-d-glucosamine in Bacillus subtilis. J. Bacteriol. 150:8–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nakano M. M., Dailly Y. P., Zuber P., Clark D. P. 1997. Characterization of anaerobic fermentative growth of Bacillus subtilis: identification of fermentation end products and genes required for growth. J. Bacteriol. 179:6749–6755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nakano M. M., Zheng G., Zuber P. 2000. Dual control of sbo-alb operon expression by the Spo0 and ResDE systems of signal transduction under anaerobic conditions in Bacillus subtilis. J. Bacteriol. 182:3274–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nothaft H., et al. 2003. The phosphotransferase system of Streptomyces coelicolor is biased for N-acetylglucosamine metabolism. J. Bacteriol. 185:7019–7023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nothaft H., et al. 2010. The permease gene nagE2 is the key to N-acetylglucosamine sensing and utilization in Streptomyces coelicolor and is subject to multi-level control. Mol. Microbiol. 75:1133–1144 [DOI] [PubMed] [Google Scholar]
- 44. Parche S., Schmid R., Titgemeyer F. 1999. The phosphotransferase system (PTS) of Streptomyces coelicolor identification and biochemical analysis of a histidine phosphocarrier protein HPr encoded by the gene ptsH. Eur. J. Biochem. 265:308–317 [DOI] [PubMed] [Google Scholar]
- 45. Plumbridge J. 2001. Regulation of PTS gene expression by the homologous transcriptional regulators, Mlc and NagC, in Escherichia coli (or how two similar repressors can behave differently). J. Mol. Microbiol. Biotechnol. 3:371–380 [PubMed] [Google Scholar]
- 46. Pomerantsev A. P., Sitaraman R., Galloway C. R., Kivovich V., Leppla S. H. 2006. Genome engineering in Bacillus anthracis using Cre recombinase. Infect. Immun. 74:682–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reents H., Münch R., Dammeyer T., Jahn D., Hartig E. 2006. The Fnr regulon of Bacillus subtilis. J. Bacteriol. 188:1103–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Reizer J., et al. 1999. Novel phosphotransferase system genes revealed by genome analysis—the complete complement of PTS proteins encoded within the genome of Bacillus subtilis. Microbiology 145(Pt. 12):3419–3429 [DOI] [PubMed] [Google Scholar]
- 49. Resch M., Schiltz E., Titgemeyer F., Muller Y. A. 2010. Insight into the induction mechanism of the GntR/HutC bacterial transcription regulator YvoA. Nucleic Acids Res. 38:2485–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rigali S., Derouaux A., Giannotta F., Dusart J. 2002. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J. Biol. Chem. 277:12507–12515 [DOI] [PubMed] [Google Scholar]
- 51. Rigali S., et al. 2006. The sugar phosphotransferase system of Streptomyces coelicolor is regulated by the GntR-family regulator DasR and links N-acetylglucosamine metabolism to the control of development. Mol. Microbiol. 61:1237–1251 [DOI] [PubMed] [Google Scholar]
- 52. Rigali S., et al. 2004. Extending the classification of bacterial transcription factors beyond the helix-turn-helix motif as an alternative approach to discover new cis/trans relationships. Nucleic Acids Res. 32:3418–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rigali S., et al. 2008. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 9:670–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Robinson C., Rivolta C., Karamata D., Moir A. 1998. The product of the yvoC (gerF) gene of Bacillus subtilis is required for spore germination. Microbiology 144(Pt. 11):3105–3109 [DOI] [PubMed] [Google Scholar]
- 55. Rowland B. M., Grossman T. H., Osburne M. S., Taber H. W. 1996. Sequence and genetic organization of a Bacillus subtilis operon encoding 2,3-dihydroxybenzoate biosynthetic enzymes. Gene 178:119–123 [DOI] [PubMed] [Google Scholar]
- 56. Sadaie Y., Nakadate H., Fukui R., Yee L. M., Asai K. 2008. Glucomannan utilization operon of Bacillus subtilis. FEMS Microbiol. Lett. 279:103–109 [DOI] [PubMed] [Google Scholar]
- 57. Saier M. H., Jr., et al. 2002. Transport capabilities encoded within the Bacillus subtilis genome. J. Mol. Microbiol. Biotechnol. 4:37–67 [PubMed] [Google Scholar]
- 58. Satomura T., et al. 2005. Enhancement of glutamine utilization in Bacillus subtilis through the GlnK-GlnL two-component regulatory system. J. Bacteriol. 187:4813–4821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schau M., Eldakak A., Hulett F. M. 2004. Terminal oxidases are essential to bypass the requirement for ResD for full Pho induction in Bacillus subtilis. J. Bacteriol. 186:8424–8432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schöck F., Dahl M. K. 1996. Expression of the tre operon of Bacillus subtilis 168 is regulated by the repressor TreR. J. Bacteriol. 178:4576–4581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Simoni R. D., Roseman S., Saier M. H., Jr 1976. Sugar transport. Properties of mutant bacteria defective in proteins of the phosphoenolpyruvate:sugar phosphotransferase system. J. Biol. Chem. 251:6584–6597 [PubMed] [Google Scholar]
- 62. Stülke J., Hillen W. 2000. Regulation of carbon catabolism in Bacillus species. Annu. Rev. Microbiol. 54:849–880 [DOI] [PubMed] [Google Scholar]
- 63. Vincent F., Davies G. J., Brannigan J. A. 2005. Structure and kinetics of a monomeric glucosamine 6-phosphate deaminase: missing link of the NagB superfamily? J. Biol. Chem. 280:19649–19655 [DOI] [PubMed] [Google Scholar]
- 64. Vincent F., Yates D., Garman E., Davies G. J., Brannigan J. A. 2004. The three-dimensional structure of the N-acetylglucosamine-6-phosphate deacetylase, NagA, from Bacillus subtilis: a member of the urease superfamily. J. Biol. Chem. 279:2809–2816 [DOI] [PubMed] [Google Scholar]
- 65. Wang Z. Q., et al. 2007. Bacterial flavodoxins support nitric oxide production by Bacillus subtilis nitric-oxide synthase. J. Biol. Chem. 282:2196–2202 [DOI] [PubMed] [Google Scholar]
- 66. Weinrauch Y., Msadek T., Kunst F., Dubnau D. 1991. Sequence and properties of comQ, a new competence regulatory gene of Bacillus subtilis. J. Bacteriol. 173:5685–5693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Winstedt L., Yoshida K., Fujita Y., von Wachenfeldt C. 1998. Cytochrome bd biosynthesis in Bacillus subtilis: characterization of the cydABCD operon. J. Bacteriol. 180:6571–6580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. You C., et al. 2005. The two authentic methionine aminopeptidase genes are differentially expressed in Bacillus subtilis. BMC Microbiol. 5:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zheng G., Yan L. Z., Vederas J. C., Zuber P. 1999. Genes of the sbo-alb locus of Bacillus subtilis are required for production of the antilisterial bacteriocin subtilosin. J. Bacteriol. 181:7346–7355 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.