Abstract
Hfq is an RNA binding protein involved in posttranscriptional regulation of gene expression in bacteria. It acts by binding to regulatory small RNAs (sRNAs), which confer specificity for the regulation. Recently, orthologues of the Hfq protein were annotated in cyanobacterial genomes, although its capacity to regulate gene expression by interacting with sRNAs has not been yet demonstrated. Anabaena sp. strain PCC 7120 is a filamentous cyanobacterium that, in the absence of combined nitrogen, is able to fix atmospheric nitrogen by differentiating specialized cells called heterocysts. We have generated an hfq knockout mutant of Anabaena sp. PCC 7120. Deletion of this gene results in differentiation of heterocysts in the presence of nitrate, suggesting a defect in nitrate assimilation. We show that hfq mutant cells are affected in transport and use of nitrate and nitrite. An analysis of the expression of several genes in the nir operon, encoding different elements of the nitrate assimilation pathway, demonstrates a downregulation of their transcription in mutant cells. We also observed that genes ntcB and cnaT, involved in the regulation of the nir operon, show a lower expression in cells lacking Hfq. Finally, when hfq was reintroduced in the mutant, heterocyst differentiation was no longer observed in the presence of nitrate. Therefore, our results indicate that the RNA chaperone Hfq is involved in the regulation of the nir operon, although the mechanism for this regulation is still unknown.
INTRODUCTION
The Hfq protein, initially identified as a host factor required for Qβ bacteriophage replication (17), acts as a global posttranscriptional regulator in enterobacteria (34, 47). In Escherichia coli, Hfq has been shown to regulate gene expression by binding to small RNAs (sRNAs). sRNAs regulate gene expression by altering either stability or translation of target mRNAs (22). Hfq binds strongly to the sRNAs at single-stranded AU-rich regions, and the interaction stabilizes many of the sRNAs. Hfq also binds to target mRNAs, promoting sRNA-mRNA duplex formation and, in some cases, enhancing association rates of sRNAs and mRNAs (30, 38, 51). These properties facilitate regulation by sRNAs that otherwise show short, and often incomplete, target complementarity (1, 26, 36, 43). Two possible roles for Hfq have been suggested. In one model, interactions between the RNAs and Hfq increase local concentrations of RNAs, aiding RNA-RNA interaction (22). The second model suggests more of a chaperone role, in which enhanced sRNA-mRNA hybridization may result from the ability of Hfq to alter the secondary structure of either sRNA or mRNA (22).
In 2002, a standard BLAST search identified Hfq in about half of the completed or nearly completed bacterial genomes (44). However, this standard BLAST search did not identify Hfq in cyanobacteria. In 2004, in a new search based on sequence length, amino acid conservation, and sequence similarity, a gene encoding an Hfq orthologue was identified in the cyanobacterium Anabaena sp. strain PCC 7120. Subsequently, a BLAST search using the Anabaena Hfq sequence as a query identified Hfq homologues in a wide variety of unicellular and filamentous cyanobacteria, including Synechocystis sp. strain PCC 6803 and Prochlorococcus (47). The cyanobacterial Hfqs constitute a new group of Hfq proteins, with differences in some of the highly conserved residues present in the Sm-1 motif of the enterobacterial Hfq as well as the signature sequence in the Sm-2 region (4, 47) (Fig. 1A). However, the crystal structures of the Anabaena and Synechocystis Hfq proteins were recently solved (4), and the structures reveal that the cyanobacterial Hfqs are quite similar in structure to other known bacterial Hfq proteins, despite lacking several key sequence elements. They possess variant RNA-binding sites, have a significantly lower affinity for known E. coli sRNAs in vitro, and cannot mediate Hfq-dependent regulation in E. coli in vivo (4).
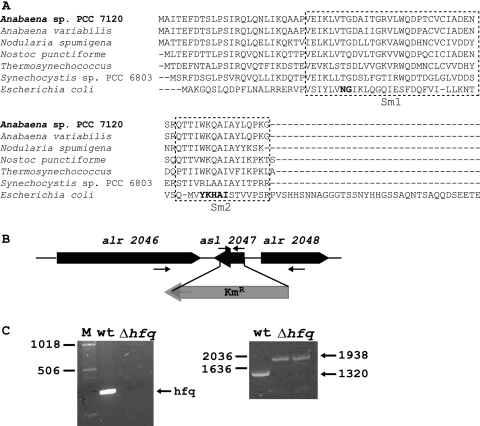
Fig. 1.
Hfq protein in cyanobacteria and preparation of an Δhfq mutant in Anabaena sp. PCC 7120. (A) Cyanobacterial Hfq proteins were aligned with the Anabaena sp. PCC 7120 Hfq protein sequence. The Escherichia coli sequence was also aligned for comparison. Residues belonging to the Sm-1 and Sm-2 motifs are boxed, and residues in bold indicate the signature sequences for the SM motifs. (B) Gene asl2047, encoding Hfq protein in Anabaena sp. PCC 7120, was replaced with a kanamycin resistance cassette (Kmr) by homologous recombination to generate a knockout mutant. (C) A PCR amplifying the hfq gene shows the total absence of this gene in mutant cells (left panel). The correct genomic organization of mutant cells was corroborated using primers located in genes alr2046 and alr2047 (primers shown as arrows in panel B), which generated a 1,938-bp product when genomic DNA of mutant cells was used as the template and a 1,320-bp product when wild-type DNA was used as the template (right panel). Two mutants were analyzed, and the rest of the work was continued with one of them. M, size markers, labeled on the left; wt, wild-type Anabaena sp. PCC 7120.
Inactivation of the hfq gene in diverse eubacteria caused pleiotropic physiological effects (32, 33, 46) and loss of virulence in pathogenic bacteria (9). However, no apparent phenotype emerged from an hfq knockout in Staphylococcus aureus (6). In cyanobacteria, the hfq gene has been inactivated in Synechocystis sp. strain PCC 6803, and the most striking change observed was the loss of motility. Moreover, microarray analyses showed a number of genes whose expression depends on Hfq (10). On the other hand, the implication of the Hfq protein in gene regulation has not been yet demonstrated for Anabaena species.
Anabaena sp. PCC 7120 is a filamentous cyanobacterium able to use different sources of combined nitrogen, including nitrate, nitrite, or ammonium. Additionally, under conditions of depletion of combined nitrogen in the medium, Anabaena is also able to fix atmospheric nitrogen. Nitrogen fixation occurs by differentiation of a specific cell type, called a heterocyst, in which the machinery for nitrogen fixation is confined (15, 16). The assimilation of nitrate by cyanobacteria takes place in three successive steps: (i) nitrate transport, (ii) nitrate reduction to ammonium, through two sequential reactions catalyzed by nitrate reductase and nitrite reductase, and (iii) ammonium incorporation into carbon skeletons, which takes place mainly through the glutamine synthetase-glutamate synthase cycle (15). The genes encoding proteins involved in nitrate assimilation in Anabaena sp. PCC 7120 are expressed as an operon in the order 5′-nirA (encoding nitrite reductase)-nrtABCD (encoding an ABC-type permease for nitrate and nitrite)-narB (encoding nitrate reductase)-3′ (7, 20). In all cyanobacteria tested, the expression of the nir operon has been shown to be subject to negative control by ammonium, and in Anabaena, its activation requires both NtcA and NtcB transcription factors (19). Also, genes all0601 and all0605, encoding CnaT and NirB, respectively, are involved in this regulation (18, 21). NtcA, NtcB, and CnaT are positive regulators of the operon, whereas NirB and NirA are negative regulators of the operon and are involved in the induction of the operon by nitrate observed in N2-fixing cyanobacteria (14, 18).
We have generated an hfq knockout mutant of Anabaena sp. PCC 7120. Deletion of hfq results in a phenotype of heterocyst differentiation in the presence of nitrate, suggesting a defect in nitrate assimilation when Hfq is absent. We have determined nitrate and nitrite reductase activities in the hfq mutant, and we have analyzed the expression of the nir operon in mutant cells. Our results show a downregulation of the nir operon, as well as a defect in nitrate assimilation when the Hfq protein is absent. The hfq mutant was partially complemented by reintroduction of the hfq gene. Our results indicate that the Hfq protein is involved in the regulation of the nir operon in Anabaena.
MATERIALS AND METHODS
Strains and growth conditions.
Anabaena/Nostoc sp. (here referred to as Anabaena sp.) strain PCC 7120 (40) was routinely grown photoautotrophically at 30°C under white light (65 to 105 μE m−2 s−1) with shaking for liquid cultures. The media used for growth were BG11 (NaNO3 as the nitrogen source [39]), BG110 (BG11 lacking nitrate), and BG110NH4+ [BG11 medium lacking nitrate and supplemented with 2.5 mM NH4Cl and 5 mM N-tris(hydroxymethyl)methyl-2-aminoethane sulfonic acid (TES)-NaOH buffer, pH 7.5). For growth on plates, medium solidified with separately autoclaved 1% agar (Difco) was used.
The hfq knockout mutant (the Δhfq strain) was generated by homologous recombination, with the Anabaena sp. PCC 7120 gene asl2047 replaced with the kanamycin resistance cassette C.K1 (13). To do this, a PCR product containing the C.K1 cassette flanked by 500 bp of genes asl2046 and asl2048 was generated and subsequently cloned into the conjugal suicide plasmid pRL277 (3), yielding plasmid pRL277Hfq. This plasmid, after confirmation of its sequence (MWG Biotech, Germany), was transferred to Anabaena sp. PCC 7120 by conjugation (12). Briefly, E. coli HB101 containing plasmid pRL277Hfq and helper plasmid pRL623 (11) was mixed with E. coli ED8654 carrying the conjugative plasmid pRL443 and, thereafter, with Anabaena sp. PCC 7120. The resulting cell suspension was spread onto nitrocellulose filters (filter type, 0.45-μm HATF; Millipore catalog no. HATF08550) set successively on top of solid medium (BG11) supplemented with 5% LB medium (incubated for 24 h), medium not supplemented (incubated for 24 h), and medium supplemented with neomycin (20 μg/ml) and incubated until colonies appeared. Double recombinants were selected by their ability to grow in a sucrose-containing medium (8). The genomic structures of the exconjugants were confirmed by PCR and Southern blot analyses. The Δhfq strain was routinely grown in BG110NH4+.
To generate the complemented mutant (Δhfq all1697::hfq), gene asl2047 was introduced into a neutral site in the genome of the hfq knockout mutant. Gene all1697 was chosen for this purpose, since disruption of this gene has no effect on Anabaena cells (our unpublished results). A PCR product containing gene asl2047 was introduced upstream of a streptomycin-spectinomycin resistance cassette (C.S3), in a vector that contained the C.S3 cassette within the all1697 gene. The whole construct, all1697-hfq-C.S3-all1697, was cloned as an XhoI fragment in vector pRL278 (3), rendering plasmid pRL278all1697-C.S3-hfq. The identity of this plasmid was confirmed by sequencing (MWG Biotech, Germany) and transferred to the Δhfq strain by conjugation, as described above. Exconjugants were selected in BG110NH4+ plates supplemented with neomycin (20 μg/ml) and spectinomycin (5 μg/ml). The genomic structure of the exconjugants was confirmed by PCR.
Growth rates and frequency of heterocysts.
The growth rate constant (μ = ln2/td, where td is the doubling time) was calculated from the increase of protein content determined in 0.2-ml samples of bubbled (1% [vol/vol]) CO2 and filtered air) liquid cultures, taken over time. Protein concentration was determined using a modified Lowry procedure (28).
To analyze heterocyst differentiation in the presence of different nitrogen sources, cells were grown in 150 ml of BG110NH4+ (6 mM NH4+, 12 mM TES, pH 7.5) at 30°C under white light (105 μE m−2 s−1) and bubbled with 1% (vol/vol) CO2 and filtered air. Exponentially growing cells were washed in BG110, resuspended in 50 ml of BG11, BG110, or BG110NH4+ at a concentration of ∼3 μg chlorophyll/ml, incubated at 30°C under white light (105 μE m−2 s−1), and bubbled with 1% (vol/vol) CO2 and filtered air. After 24 h, filaments were carefully collected and visualized under standard light microscopy. To avoid the breakage of the filaments, a drop of 1% low-melting-temperature agarose was mixed with the cell suspension on the slide before visualization. Alcian blue staining was used to facilitate visualization of the heterocysts (35). To calculate the frequency of heterocysts in the wild-type and mutant strains, under different nitrogen sources, over 6,000 cells from each strain from at least two independent experiments were counted in each case.
Determination of nitrogenase activity.
Different strains were grown in 150 ml of BG110NH4+ (6 mM NH4+, 12 mM TES, pH 7.5) at 30°C under white light (105 μE m−2 s−1) and bubbled with 1% (vol/vol) CO2 and filtered air. Exponentially growing cells were washed in BG110, divided in 75 ml of BG11 or BG110 at a concentration of ∼3 μg chlorophyll/ml, and incubated under the same conditions for 24 h. After 24 h, a cell suspension containing 20 μg of chlorophyll was used to determine nitrogenase activity, using the acetylene reduction technique previously described (31). The rest of the culture was used for RNA isolation and Northern blot analysis of the nifHDK gene.
Determination of nitrate and nitrite reductase activities.
Different strains were grown in 150 ml of BG110NH4+ (6 mM NH4+, 12 mM TES, pH 7.5) at 30°C under white light (105 μE m−2 s−1) and bubbled with 1% (vol/vol) CO2 and filtered air. Exponentially growing cells were washed in BG110, divided in 50 ml of BG11, BG110, or BG110NH4+ at a concentration of 4 μg chlorophyll/ml, incubated at 30°C under white light (105 μE m−2 s−1), and bubbled with 1% (vol/vol) CO2 and filtered air. After 4 h of incubation in the different nitrogen sources, 30 ml of cells was collected for RNA isolation. The other 20 ml was washed in BG110 and resuspended in BG110 to a final concentration of 50 μg chlorophyll/ml for determination of nitrate and nitrite reductase activities.
Nitrate reductase (23) and nitrite reductase (24) activities were measured with dithionite-reduced methyl viologen as the reductant, in cells made permeable with mixed alkyltrimethylammonium bromide. The cells added to the enzymatic assay mixtures for nitrate reductase and nitrite reductase contained 5 and 25 μg of chlorophyll, respectively. One unit of activity corresponded to 1 μmol of nitrite produced per min (nitrate reductase) or 1 μmol of nitrite removed per min (nitrite reductase).
Determination of nitrate and nitrite assimilation.
To determine nitrate and nitrite assimilation, different strains were grown in 150 ml of BG110NH4+ (6 mM NH4+, 12 mM TES, pH 7.5) at 30°C under white light (105 μE m−2 s−1) and bubbled with 1% (vol/vol) CO2 and filtered air. Exponentially growing cells were washed in BG110 and resuspended in 50 ml of BG11 at a concentration of 10 μg chlorophyll/ml. Cells were incubated for 4 h and then were washed twice in BG110. For nitrate assimilation, cells were resuspended in 10 ml of 25 mM TES, pH 7.5, at a concentration of 8 to 10 μg chlorophyll/ml, and 100 μM sodium nitrate was added. Aliquots of 1 ml were taken at different times, cells were removed by filtration, and the nitrate remaining in the medium was measured by high-performance liquid chromatography (HPLC). To determine nitrite assimilation, cells were resuspended in either 10 ml of 25 mM TES, pH 7.5, or 10 ml of 25 mM glycine, pH 9.6, at a concentration of 8 to 10 μg chlorophyll/ml, and 100 μM sodium nitrite was added to the cell suspension. Aliquots of 500 μl were taken at different times. Reactions were stopped by the addition of 1 ml of sulfanilamide, and the nitrite concentration in the medium was determined spectrophotometrically by measuring the optical density (OD) at 540 nm, after the addition of 1 ml of N-(10-naphthyl)-ethylenediaminedihydrochloride (N-NED).
RNA isolation.
To isolate RNA, cells grown under different nitrogen sources were collected by filtration (filter type, 0.45-μm HA; Millipore catalog no. HAWP05000) and washed in RNase-free TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.5). Cells were then resuspended in 100 μl of a lysozyme solution (3 mg/ml in water). Cell lysis was facilitated by three freeze-thaw cycles, and RNA was isolated with 1 ml of Trizol reagent (Invitrogen), according to the manufacturer's instructions. After isolation, RNA was sequentially extracted with phenol and chloroform-isoamyl alcohol (24:1), precipitated with ethanol, and washed with 70% ethanol. Finally, RNA was resuspended in 30 μl RNase-free water for subsequent uses.
Northern blot and quantitative reverse transcription-PCR (qRT-PCR).
For Northern blot analysis, 5 to 10 μg of total RNA was loaded per lane in a 1% agarose-TBE gel. Transfer and fixation to Hybond-N+ membranes (GE Healthcare) were performed using 0.05 M NaOH. Hybridization was carried out at 65°C. The nifH probe was generated from a PCR product as previously described (48). As a control of RNA loading and transfer efficiency, the filters were hybridized with a probe of the RNase P RNA gene (rnpB) from strain PCC 7120 amplified by PCR with primers Universal and Reverse and plasmid pT7-7120 as the template (49). All probes were labeled with [α-32P]dCTP using the Ready-to-Go labeling kit (GE Healthcare). Images of radioactive filters were obtained with a Cyclone storage phosphor system (Packard).
For qRT-PCR, 10 μg of total RNA was treated with 2 U of RQ1 DNase (Promega), in 20 μl, for 1 h at 37°C. The reaction was then stopped with 2 μl of the provided stop buffer and heated for 10 min at 70°C. Five micrograms of this treated RNA was used for reverse transcription, using 3 μg of random hexamer primers (Invitrogen) and 100 U of Superscript-II reverse transcriptase (Invitrogen), by following the manufacturer's instructions. An RT control reaction, in which the RT enzyme was absent, was always included to rule out amplification of contaminant DNA.
Quantitative PCR (qPCR) was performed using the Bio-Rad IQ5 apparatus and the Quantimix Easy SYG kit (Biotools, Madrid, Spain), which uses SYBR green I for detection of amplification. Five microliters of a 1:10 dilution of the generated cDNA was used as a template in a 20-μl reaction mixture. Primer pairs to amplify different portions of the transcripts under study were designed using the free ProbeFinder software (Roche Applied Science) and were used at a 0.4 μM final concentration. The sequences of the oligonucleotides used are available upon request.
Cycle conditions were as follows: 95°C, 10 min; 40 times at 95°C, 15 s; 60°C, 1 min. CT (cycle threshold) values were determined using the IQ5 optical system software (Bio-Rad) with automatic baseline and threshold determinations. Triplicate experiments were run for each reaction, and the CT value is averaged from them. The absence of primer dimers was corroborated by running a dissociation curve analysis at the end of each experiment to determine the melting temperature of the amplicon. The correct size of the amplicon was confirmed by 1% agarose electrophoresis.
For quantification, the efficiency of each primer pair was determined to be between 90% and 110%, by following the instructions for efficiency determination described by Applied Biosystems (Guide to Performing Relative Quantification of Gene Expression Using Real-Time Quantitative PCR [company manual]). These efficiencies indicate that the amount of DNA is doubled in each PCR cycle and allows for direct comparison between different genes. Relative RNA levels were determined using the ΔΔCT method as described in the above-mentioned guide. Briefly, each gene CT value is normalized to the CT value for the internal control (rnpB) (49), which gives the ΔCT value. This value is then related to a reference gene, giving us the ΔΔCT value. Since the amount of DNA doubles in each PCR cycle, the relative amount of input cDNA can be determined by using the formula 2−ΔΔCT. Each ΔΔCT determination was performed at least in three different RNA samples, and the values shown are the average of these replicates.
Determination of RNA stability.
To determine RNA stability, different strains were grown in 150 ml of BG110NH4+ (6 mM NH4+, 12 mM TES, pH 7.5) at 30°C under white light (105 μE m−2 s−1) and bubbled with 1% (vol/vol) CO2 and filtered air. Exponentially growing cells were washed in BG110 and resuspended in 50 ml of BG11, at a concentration of 10 μg chlorophyll/ml. Cells were incubated in this medium under white light and bubbled with 1% (vol/vol) CO2 and filtered air, for 4 h, and transcription was stopped by adding rifampin (200 μg/ml) to the cell suspension. After addition of rifampin, 3 ml of cells was collected by filtration at different time points to isolate RNA, as previously described. The abundance of different RNAs was determined by quantitative RT-PCR, as described above. Results were normalized using rnpB as the internal control (ΔCT) and related to time zero for reference (ΔΔCT).
RESULTS
Generation of an hfq knockout mutant.
The cyanobacterial Hfq protein has been suggested to form a specialized subfamily of Hfq proteins that binds relatively weakly to A/U-rich tracks of regulatory RNAs (4) and is rather different from the enterobacterial Hfq protein (Fig. 1A). The cyanobacterial Hfq protein is considerably shorter than the E. coli Hfq protein and lacks some of the signature residues for the Sm-1 and Sm-2 motifs (Fig. 1A) (47). To investigate the function of the predicted Hfq orthologue from Anabaena sp. PCC 7120 (47), gene asl2047, encoding Hfq, was replaced with a kanamycin resistance cassette (Fig. 1B), as described in Materials and Methods. The identity of the mutants, from here on named the Δhfq strains, as well as complete segregation, was corroborated by Southern blot (not shown) and PCR (Fig. 1C) analyses.
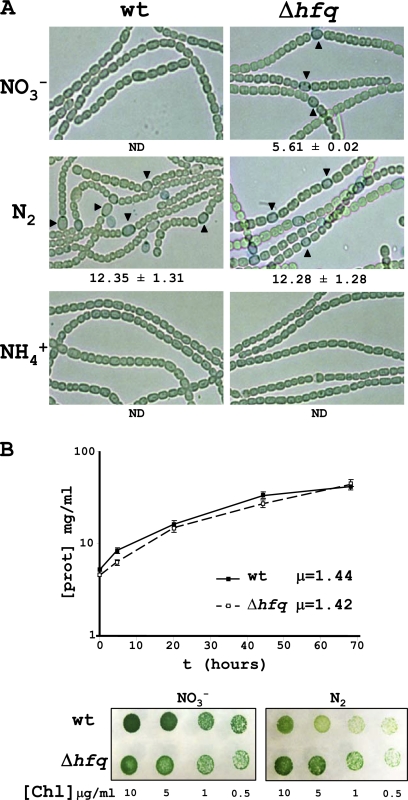
In order to assess possible altered growth characteristics, wild-type and mutant cells were analyzed under different growth conditions. A remarkable phenotype was observed when cells were grown in the presence of nitrate as the nitrogen source. Under these conditions, the Δhfq strain, unlike the wild-type strain, differentiates heterocysts (Fig. 2A), with a morphology similar to that observed in wild-type cells grown in a medium without combined nitrogen (Fig. 2A). The frequency of heterocysts was calculated to be 5.61% for Δhfq cells in the presence of nitrate (Fig. 2A), while the wild-type strain does not differentiate heterocysts in the presence of nitrate, and we could observe only a single heterocyst in approximately 6,000 cells counted.
Fig. 2.
Phenotype and growth rates for the Anabaena Δhfq mutant strain. (A) Anabaena wild type (wt) and Δhfq cells grown in the presence of different nitrogen sources. Numbers below each panel indicate the percentages of of heterocysts. Heterocysts were not observed when cells were grown in an ammonium-containing medium or in wild-type cells grown in the presence of nitrate after analysis of many cells, so the frequency of heterocysts could not be determined in those cells (ND). Visualization of heterocysts, indicated by triangles, was improved by alcian blue staining. (B) A representative example of a growth curve for wild-type and mutant cells grown in the presence of nitrate is shown, with specific growth rates (μ, day−1) calculated for both. Growth in plates was also tested. Seven microliters of a suspension with the indicated concentration of cells was spotted in BG11 or BG110 plates and incubated for 8 days at 30°C under white light. No significant differences in growth were observed. prot, protein; Chl, chlorophyll.
In the absence of combined nitrogen (BG110), mutant cells differentiate heterocysts with a frequency similar to that of wild-type cells (Fig. 2A), and no heterocysts were observed in ammonium for wild-type or mutant cells. Calculations based on growth curves of the wild-type and Δhfq cells determined specific growth rates of 1.44 day−1 for wild-type cells and 1.42 day−1 for mutant cells in BG11 (Fig. 2B), indicating that Δhfq cells are not impaired in growth in the presence of nitrate, although they differentiate heterocysts. In the absence of combined nitrogen, wild-type and mutant cells also showed similar growth rates, indicating that heterocysts developed in BG110 are functional to allow normal growth in a medium without nitrogen source (Fig. 2B). This was further confirmed by nitrogenase activity measurements (Fig. 3B).
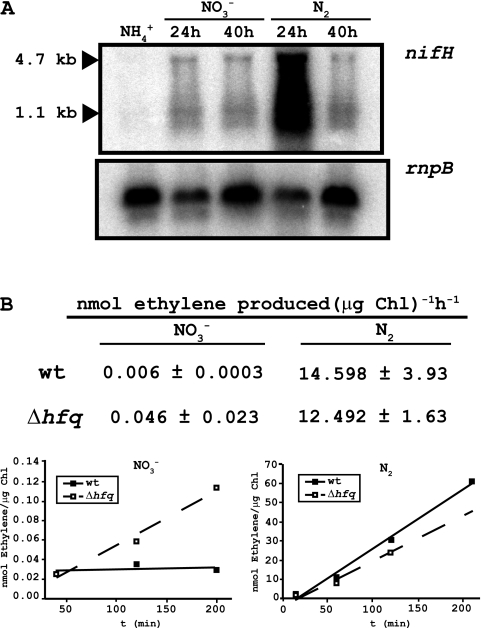
Fig. 3.
Analysis of the expression of the nifHDK operon and determination of nitrogenase activity. (A) Northern blot analysis of the expression of the nifHDK operon in Δhfq cells, using a nifH probe. Expression is not observed when cells are grown in ammonium. However, expression is observed after 24 or 40 h of incubation in either nitrate or N2. The sizes of previously identified nifH and nifHDK transcripts are indicated on the left. Hybridization with an rnpB gene probe was used as a loading and transfer control. (B) Nitrogenase activity was determined for wild-type and mutant cells incubated in the presence of nitrate or N2 for 24 h. Data are the means of results of three independent experiments, with standard deviations shown as errors. A representative kinetic of ethylene production, for cells incubated in the presence of nitrate or N2, is shown in the lower part of the figure.
Heterocysts are specialized cells able to fix atmospheric nitrogen in the absence of combined nitrogen, and in order to further assess the functionality of the heterocysts developed by Δhfq cells, we performed an analysis of the expression of the nifHDK operon in mutant cells. The nifHDK operon, encoding the nitrogenase enzyme complex, is expressed in Anabaena sp. strain PCC 7120 under oxic conditions exclusively in the heterocysts (50). As shown in Fig. 3A, expression of the operon is observed in mutant cells incubated in nitrate for 24 or 40 h. When cells are incubated in the absence of combined nitrogen, a much higher peak of expression is shown after 24 h, with some expression remaining after 40 h. Expression is considerably higher in the absence of combined nitrogen after 24 h than in the presence of nitrate. As expected, no expression of the nifHDK operon is observed when cells are incubated in the presence of ammonium, consistent with the absence of heterocysts in filaments incubated in ammonium (Fig. 3A). It has been reported that Anabaena wild-type cells do not show any expression of the nifHDK genes either in nitrate or ammonium, showing expression only when cells are incubated in the absence of combined nitrogen (50). These results indicate that heterocysts in the Δhfq strain incubated in the presence of nitrate are able to express, to some extent, the genes responsible for formation of the nitrogenase complex. We also analyzed the nitrogenase activity of wild-type and Δhfq cells, either in BG11 or BG110. Results showed that when incubated in the absence of combined nitrogen, wild-type and mutant cells have similar nitrogenase activities (Fig. 3B). However, when incubated in the presence of nitrate, Anabaena wild-type cells did not show any nitrogenase activity whereas mutant cells showed very low, but detectable, nitrogenase activity (Fig. 3B). These results indicate that in the Δhfq strain, the heterocysts observed are somewhat functional, since expression of nifHDK is observed and nitrogenase activity is detectable.
The observation that the Δhfq mutant strain developed heterocysts in the presence of nitrate suggested either that heterocyst differentiation was deregulated in this strain or that Δhfq cells are unable to use nitrate. In fact, differentiation of heterocysts in the presence of nitrate has been reported for mutants altered in the genes responsible for the assimilation of nitrate, such as ntcB, cnaT, and moeA (19, 21, 37). Thus, to investigate a possible defect in nitrate assimilation, we performed enzymatic assays to determine the capacity of utilization of nitrate by Δhfq cells.
Nitrate and nitrite assimilation in Δhfq cells.
The assimilation of nitrate by Anabaena sp. PCC 7120 requires the effective transport of nitrate through the membrane and the reduction of nitrate to ammonium, in two sequential reactions. To check the capacity of the Δhfq mutant cells to utilize nitrate, both nitrate and nitrite reductase activities were measured as described in Materials and Methods.
As previously described for wild-type cells (20), maximal activities were observed in the presence of nitrate (Table 1). Δhfq mutant cells showed about half the nitrate reductase activity of the wild-type cells in all three media. Nitrite reductase activity is null when cells are incubated in dinitrogen or ammonium for wild-type and mutant cells. When incubated in nitrate, mutant cells show a 7-fold reduction, compared to the wild type, in nitrite reductase activity. These results indicated that both nitrate and nitrite reductase activities are impaired in Anabaena cells lacking Hfq.
Table 1.
Nitrate and nitrite reductase activities of wild-type Anabaena sp. PCC 7120 and Δhfq mutant strain in the presence of different nitrogen sourcesa
| Strain | Nitrate reductase (mU/mg of protein) from indicated nitrogen source |
Nitrite reductase (mU/mg of protein) from indicated nitrogen source |
||||
|---|---|---|---|---|---|---|
| Nitrate | Dinitrogen | Ammonium | Nitrate | Dinitrogen | Ammonium | |
| Wild type | 104 ± 15.8 | 20.5 ± 4.5 | 9.6 ± 4.4 | 14.34 ± 2.8 | ND | ND |
| Δhfq mutant | 52.32 ± 8.74 | 11 ± 4.5 | 3.81 ± 0.7 | 2.05 ± 0.75 | ND | ND |
Cells were grown in the presence of ammonium, washed twice with nitrogen-free medium, and then transferred to medium with the indicated nitrogen source. Enzymatic activities were measured after 4 h of incubation in the indicated nitrogen source. The values shown are the mean ± standard deviation of results from three independent experiments for nitrite reductase determination, four experiments for nitrate reductase when cells were grown in ammonium or N2, and five experiments for nitrate reductase when cells were grown in the presence of nitrate. ND, not detectable.
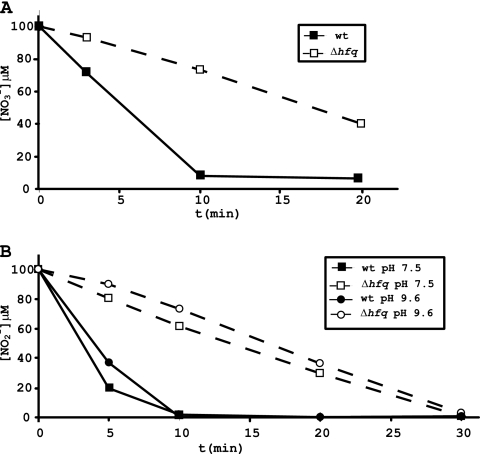
The uptake of nitrate and nitrite in wild-type and mutant cells was also measured (Fig. 4). Results show that whereas wild-type cells are able to take up 100 μM sodium nitrate added to the medium in 10 min, Δhfq cells take up about 60 μM sodium nitrate after 20 min (Fig. 4A). Uptake of nitrate requires passage of the molecule through the membrane, either by passive diffusion or active transportation, and subsequent utilization of nitrate by nitrate reductase. Since nitrate reductase activity is reduced 2-fold in mutant cells (Table 1), one possible explanation is that the defect in uptake observed for mutant cells is due to a defect in nitrate reductase activity. However, we observed that, at 10 min, 80% of the added nitrate still remains in the medium for mutant cells, whereas almost none is detected in the medium for wild-type cells. Since nitrate reductase activity could explain only a defect of 50% in uptake, these data suggest that both transport and reduction are affected.
Fig. 4.
Nitrate and nitrite assimilation in wild-type Anabaena sp. PCC 7120 and the Δhfq mutant strain. To determine nitrate (A) and nitrite (B) assimilation in Anabaena wild-type or Δhfq cells, 100 μM sodium nitrate or sodium nitrite was added to the medium and incorporation was measured over a time course. Nitrite assimilation was measured at pH 7.5 and pH 9.6. Assimilation was determined in three independent experiments, with similar results, and a representative example is shown.
Nitrite uptake was determined at pH 7.5 and 9.6. At pH 9.6, nitrite is ionized and cannot go through the membrane by passive diffusion; therefore, its uptake requires an active transport through the membrane. However, at pH 7.5, both mechanisms, passive diffusion and active transport, are used to introduce nitrite into the cell (5). There is a significant defect in nitrite uptake for Δhfq cells at both pHs (Fig. 4B). Uptake for wild-type cells at pH 7.5 is slightly better that at pH 9.6, as expected, and uptake of 100 μM sodium nitrite is completed after 10 min at either pH. Δhfq cells also show a slightly better incorporation at pH 7.5 than at pH 9.6. A defect in nitrite assimilation is clearly observed in these cells, with less than 40 μM sodium nitrite taken up after 10 min at pH 7.5 (Fig. 4B, white squares) and less than 25 μM taken up at pH 9.6, when active transport is necessary (Fig. 4B, white circles). These data indicate that Δhfq cells show a defect in both transport and reduction of nitrite.
Expression of genes involved in nitrate assimilation.
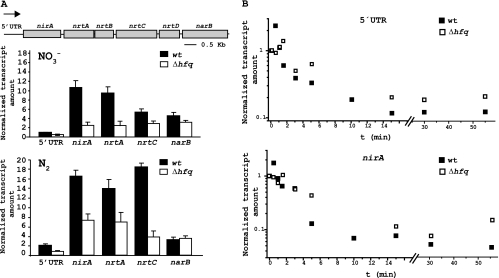
Genes involved in nitrate assimilation in Anabaena sp. PCC 7120 are grouped in the so-called nir operon. The operon contains the genes for nitrite reductase (nirA), nitrate reductase (narB), and four components of a multicomponent transporter (nrtA, -B, -C, and -D) (Fig. 5A). It also contains an unusually long 5′ untranslated region (5′ UTR), of 460 nucleotides (20). Transcription of the nir operon takes place from an NtcA activated promoter and requires both NtcA and NtcB transcription factors for activation (19, 20). CnaT and NirB, encoded by genes all0601 and all0605, respectively, are also involved in this regulation (18, 21).
Fig. 5.
Expression and stability of transcripts corresponding to the nir operon in wild-type Anabaena sp. PCC 7120 and the Δhfq mutant strain. (A) The genomic organization of the Anabaena sp. PCC 7120 nir operon is shown in the upper part of the figure. nirA codes for nitrite reductase, narB for nitrate reductase, and the nrt genes encode different subunits of a multicomponent transporter. An arrow indicates the direction of transcription. Quantitative RT-PCR was used to determine the transcript amount of some regions of the nir operon in wild-type and Δhfq strains, grown in the presence of nitrate or N2. Data are normalized using an internal control (rnpB), and results are presented relative to the transcript amount of the 5′ UTR in wild-type cells in nitrate. Results are the average of at least three independent experiments, with standard deviations shown as error bars. (B) Determination of RNA stability of some regions of the nir operon. Transcription of the nir operon was induced by transferring cells to a nitrate-containing medium. After 4 h of induction, transcription was stopped by adding rifampin to the medium (t0), and cells were collected at different time points. The abundance of different regions of the nir operon transcript was determined by qRT-PCR for wild-type and Δhfq cells. rnpB was used as an internal control, and data were normalized for each strain to t0. Experiments were run in triplicate, and representative examples of the stability of the 5′ UTR region and nirA mRNA are shown.
Since Anabaena Δhfq mutant cells showed defects in nitrate assimilation, the expression of the nir operon in these cells was analyzed. Expression of the operon under different nitrogen conditions was assessed by qRT-PCR, using primer pairs corresponding to different parts of the operon (nirA, nrtA, nrtC, and narB genes, as well as the 5′ UTR region; see Materials and Methods). As previously described (20), amplification was not observed when cells were incubated in ammonium (data not shown). Our results show that expression of the operon, for most of the primer pairs used in the study, is significantly higher in N2 than in the presence of nitrate (Fig. 5A), in either wild-type or mutant cells. Similar results were observed by other authors, who described that nir operon transcription takes place at the highest levels in N2-fixing cultures (7). Comparing wild-type and mutant cells, we detected a lower level of amplification for mutant cells, with a decrease of more than 2-fold for most of the regions of the operon examined (Fig. 5A). These results are consistent with the reduction in nitrate and nitrite assimilation observed in the Δhfq strain. The only gene of the operon for which the decrease in transcript amount observed in mutant cells is not that prominent, either in nitrate or N2, is narB. Our data also show that the amount of the transcript corresponding to the 5′ UTR region is lower than that corresponding to the coding genes in both wild-type and mutant cells. In the presence of nitrate, transcript abundance of different parts of the operon decreases as the operon reaches its 3′ end. In the presence of N2, transcripts corresponding to different genes in the operon show similar levels of abundance, except for the already mentioned 5′ UTR and the narB gene.
In order to test whether the differences we observed in transcript amounts between wild-type and mutant cells were due to a difference in transcription or stability, we determined the stabilities of different regions of the nir operon transcript in wild-type and Δhfq cells. To do so, transcription of the nir operon was induced by transferring cells from an ammonium-containing medium to a nitrate-containing medium. After 4 h of incubation in the nitrate-containing medium, transcription was stopped by adding rifampin to the medium (see Materials and Methods). Samples were taken at different time points, and transcript levels were determined by qRT-PCR. Figure 5B shows representative examples of the decay curves for regions of the operon transcript corresponding to the 5′ UTR and nirA gene. Stability was also determined for nrtA and nrtC with similar results (not shown). A similar mRNA decay curve was observed in both the wild-type and mutant strains, indicating similar stabilities for RNAs in both strains (Fig. 5B). These data indicate that the transcript of the nir operon in the wild-type strain is not more stable than that in the mutant strain, and thus the decrease in transcript levels observed for Δhfq cells (Fig. 5A) is likely due to a decrease in the transcription of the nir operon in the Δhfq strain.
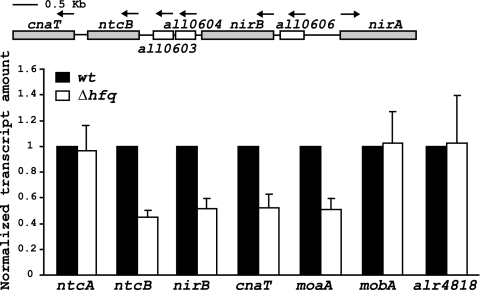
Because regulation of the nir operon is mediated by NtcA, NtcB, CnaT, and NirB (18, 19, 21), we decided to analyze the expression of the corresponding genes in Δhfq cells (Fig. 6). RNA from cells incubated in nitrate was isolated and used as a template for qRT-PCR (see Materials and Methods). Results show that ntcB, cnaT, and nirB transcript levels are about 50% in the Δhfq strain relative to the level in the wild type (Fig. 6). Because NtcB and CnaT are positive regulators of the nir operon, these results are consistent with the observed downregulation of the nir operon in Δhfq cells. NirB is required to achieve optimal nitrite reductase activity, and it has been proposed to act as a chaperone or scaffold for expression of maximal nitrite reductase activity (45). The decrease in nirB expression observed in the Δhfq mutant strain (Fig. 6), together with the reduced levels of nirA RNA (Fig. 5), could explain the 90% decrease observed in nitrite reductase activity in the mutant, compared to that in wild-type cells (Table 1). On the other hand, transcript levels for the ntcA gene were similar for mutant and wild-type cells (Fig. 6). NtcA is a major transcriptional regulator in Anabaena, and it is essential for the transcription of genes required for heterocyst development. Since the hfq mutant is capable of developing functional heterocysts in an N2-containing medium, this result was not surprising and suggests that regulation of the ntcA gene is not affected by Hfq.
Fig. 6.
Expression of other genes related to nitrate assimilation in wild-type Anabaena sp. PCC 7120 and the Δhfq mutant strain. Genomic organization of Anabaena sp. PCC 7120 genes ntcB, cnaT, and nirB, with respect to the nir operon, is shown in the upper part of the figure. Genes analyzed in this study are represented by gray rectangles. Arrows indicate the direction of transcription. Quantitative RT-PCR was used to determine the abundance of ntcA, nirB, ntcB, and cnaT in Anabaena wild-type cells and the Δhfq strain, incubated for 4 h in a nitrate-containing medium. Genes involved in cofactor biosynthesis, moaA, mobA, and alr4818, were also analyzed. Data were normalized using an internal control (rnpB), and results are presented relative to the expression of each gene in wild-type cells. Results are the average of at least three independent experiments, with standard deviations shown as error bars.
Nitrate and nitrite reductase enzymes require complex metal- containing prosthetic groups for their correct function (14). Nitrate reductase contains a molybdenum cofactor (Mo cofactor), in the form of Mo-bis-molybdopterin guanine dinucleotide (25, 41). The biosynthesis of the Mo cofactor requires the product of at least seven genes (moaA, moaC, moaD, moaE, moeA, moeB, and mobA) (29). Nitrite reductase contains a siroheme prosthetic group (14), and the cysG gene encoding a general siroheme-synthesizing enzyme is clustered with the nir gene in some cyanobacteria (e.g., Synechococcus sp. strains WH 8102 and WH 8103 [2, 14]).
Therefore, we decided to analyze the expression of genes moaA, mobA (encoded by all3865 and all0961, respectively) and alr4818. ORF alr4818 has 34% identity to a siroheme synthase gene from Photorhabdus luminescens. The results shown in Fig. 6 demonstrate that expression of moaA is reduced 50% in the Δhfq mutant. However, genes mobA and alr4818 show similar transcript levels in wild-type and Δhfq mutant cells.
Complementation of Δhfq phenotype by ectopic expression of hfq.
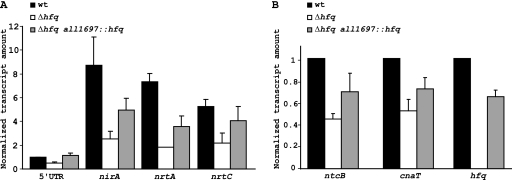
To confirm that the phenotype shown by the Δhfq strain was exclusively a result of the mutation of the hfq gene, we complemented the Δhfq mutation by inserting the hfq gene (asl2047) in a neutral position in the Anabaena genome. The neutral location used for insertion was gene all1697, whose disruption has no effect on Anabaena sp. PCC 7120 (our unpublished results). The fragment inserted contained, in addition to asl2047, the flanking genes alr2046 and alr2048 (Fig. 1B). Therefore, it is expected to include the asl2047 promoter and transcriptional terminator. A PCR product containing this region was cloned into a suitable vector containing a streptomycin-spectinomycin resistance cassette interrupting gene all1697, as described in Materials and Methods. The resulting vector was conjugated into strain Δhfq, and conjugants were selected by their capacity to grow in a neomycin-spectinomycin-containing medium. Identity of the conjugants, as well as the presence of an hfq gene, was corroborated by PCR and Southern blot analyses (not shown). This strain, named the Δhfq all1697::hfq strain, did not show the characteristic heterocysts, in the presence of nitrate, of the Δhfq mutant (data not shown). Nitrate reductase activity (81.02 ± 5.2 mU/mg of protein) and nitrite reductase activity (14.22 ± 2.05 mU/mg of protein) were also determined. Nitrite reductase activities are similar in the complemented strain and wild-type cells (Table 1), and although nitrate reductase activity is lower than in wild-type cells, it is significantly higher than the activity determined for Δhfq cells (Table 1). The expression of some of the nir operon genes in the complemented mutant was also analyzed by quantitative RT-PCR, using RNAs isolated from cells incubated in the presence of nitrate. Results show a higher level of the operon transcript in the Δhfq all1697::hfq strain than in the Δhfq strain, although both transcripts levels are lower than wild-type levels (Fig. 7A). We also analyzed expression of the regulatory genes ntcB and cnaT (Fig. 7B) and observed an increase in transcript levels in the complemented mutant, compared to that of the Δhfq mutant. Finally, we analyzed expression of the hfq gene in the Δhfq all1697::hfq mutant strain, showing that the hfq transcript level is around 60% of the wild-type transcript (Fig. 7B). These results suggest that the phenotype observed for the Δhfq mutant is, indeed, due to the absence of the Hfq protein in these cells.
Fig. 7.
Analysis of an hfq complemented mutant. (A) Quantitative RT-PCR was used to determine the abundance of some regions of the nir operon transcript in the Hfq complemented mutant incubated in the presence of nitrate. Data were normalized to an internal control (rnpB), and results are presented relative to the expression of 5′UTR in wild-type cells. (B) The expression of the regulatory genes ntcB and cnaT, as well as the hfq gene, was also determined. Data are normalized to rnpB and are presented relative to the expression in wild-type cells. Results are the average of at least three independent experiments, with standard deviations shown as error bars.
DISCUSSION
Here we show that deletion of the hfq gene in Anabaena sp. PCC 7120 results in a phenotype of heterocyst differentiation in nitrate-containing media. The same phenotype was previously reported for strains unable to use nitrate, such as the ntcB, cnaT, and moeA mutants (19, 21, 37), so the observed phenotype, which could be restored by introducing the hfq gene back in the mutant (strain Δhfq all1697::hfq), suggested a defect in nitrate assimilation. Although our results clearly show that Δhfq cells are defective in the use of nitrate, with impaired nitrate and nitrite reductase activities (Table 1) and a defect in nitrate and nitrite transport (Fig. 4), the possibility that the heterocyst-forming phenotype observed in Δhfq cells might also be due to a deregulation of some of the genes involved in heterocyst differentiation cannot be excluded.
Heterocysts differentiated in the mutant strain, either in BG11 or BG110, appear to be morphologically identical to heterocysts developed in the wild type in the absence of combined nitrogen (Fig. 2A)., The frequency of heterocysts in the mutant strain is similar to that of wild-type cells in the absence of combined nitrogen. However, the frequency of heterocysts in the mutant strain incubated in the presence of nitrate is approximately half of that observed in BG110 (5.61% versus 12.28%) (Fig. 2A). Expression of the nifHDK operon in mutant cells occurs both in nitrate and in N2 (Fig. 3A). A measure of nitrogenase activity demonstrates that mutant cells show wild-type levels of nitrogenase activity in N2, indicating that the heterocysts developed in N2 are functional (Fig. 3B). In the presence of nitrate, mutant cells show very low, although detectable, nitrogenase activity (Fig. 3B). This low level of nitrogenase activity suggests that mutant cells are growing at the expense of nitrate, metabolized through the nitrate reductase pathway. This pathway is impaired in Δhfq cells, as demonstrated by a reduction in nitrate and nitrite reductase activities, and because of such a reduction, suboptimal utilization of nitrate might originate the heterocyst-forming phenotype shown by mutant cells in nitrate. This hypothesis is consistent with the observation that the number of heterocysts developed by the Δhfq strain in the presence of nitrate is about half that observed in N2. In contrast, the ntcB and cnaT mutants, which have been described to be unable to use nitrate, showed similar frequencies of heterocysts in nitrate and N2 (reported previously to be 8 to 9%) (19, 21).
When analyzed by qRT-PCR, expression of the genes in the nir operon (nirA, nrtA, nrtC, and narB) is reduced in cells lacking Hfq (Fig. 5A). Results in Fig. 5B show that the transcripts are not more stable in wild-type than in mutant cells, indicating that the differences we observed are due to differences in transcription rather than stability. Our results show that expression of the nir operon is higher in N2 than in the presence of nitrate, although nitrite and nitrate reductase activities reach a maximum when cells are grown in the presence of nitrate (Fig. 5 and Table 1). Similar expression results were described previously for the wild-type strain, in the context of a mutagenesis strategy (7). However, more recent results indicated that abundance of the nir operon RNAs, as measured by Northern blotting, is higher when cells are grown in the presence of nitrate (20).
In the presence of nitrate, transcript levels for different genes in the nir operon decrease as the operon reaches its 3′ end. This might be due to a reduced stability of genes close to the 3′ end of the operon and is consistent with similar results previously described for Anabaena wild-type cells (20), by using a different method. Moreover, some previous results also suggested uncoupled regulation of the nir operon, proposing that nirA and narB are independently regulated (7). In fact, the narB gene is the only one in which differences between wild-type and hfq mutant cells are not observed in cells grown in N2 and only slightly detected in cells incubated in the presence of nitrate. This observation is consistent with the suggested independent regulation of the narB gene.
In the presence of nitrate, the transcript amount determined for the region of the operon corresponding to the nirA gene, encoding nitrite reductase, is 4-fold lower in hfq mutant cells than in wild-type cells (Fig. 5A). However, nitrite reductase activity is 10-fold lower in mutant cells (Table 1). It has been reported that the nirB gene product is required to attain maximum levels of nitrite reductase, and its inactivation provokes an imbalance between nitrate and nitrite reduction (18, 45). In Δhfq mutant cells, levels of nirB transcript amount are about 50% of that in wild-type cells (Fig. 6), so the combined effect of reduced nirA and nirB mRNAs could explain the decrease in nitrite reductase activity observed. Moreover, we cannot rule out the possibility that a posttranscriptional regulation of the nir genes or the nitrite reductase protein could occur and be dependent on Hfq.
Concerning genes related to synthesis of cofactors, no differences in expression are observed for gene alr4818, which might be involved in synthesizing the prosthetic group required for nitrite reductase function (Fig. 6). A downregulation is observed for gene moaA, required in the first step of the biosynthesis of the Mo cofactor of nitrate reductase (Fig. 6). However, no changes in expression are observed for gene mobA, required in the last step of the biosynthesis pathway of the Mo cofactor (Fig. 6). In some bacteria, such as E. coli, there are five known genes (moa, mob, mod, moe, mog) responsible for Mo cofactor biosynthesis (42). In Anabaena, genes moaA and mobA do not belong to the same operon, and they do not appear to form an operon with any of the other genes involved in Mo cofactor biosynthesis. Therefore, it seems possible that they are independently regulated. Our results suggest that in Anabaena, the Hfq protein is involved in the regulation of moaA but not mobA. How Hfq might affect regulation of nirB or moaA is unknown and might be an indirect effect of the downregulation of the nir operon.
Finally, we observed a downregulation of the expression of genes ntcB and cnaT (Fig. 6). NtcB and CnaT are positive regulators of the nir operon, and strains lacking any of these genes show heterocyst differentiation in nitrate-containing medium (19, 21). Moreover, the cnaT mutant was observed to revert frequently, giving rise to cells able to use nitrate as a nitrogen source (21). Δhfq mutant cells show the same phenotype as the ntcB and cnaT mutants. Moreover, we have also observed reversion in the Δhfq heterocyst-forming phenotype when the cells are extensively grown in nitrate, an observation similar to the reversion observed in the cnaT mutant (21). These results suggest that the differentiation of heterocysts in nitrate shown by Δhfq cells might be due to the downregulation of ntcB and cnaT, suggesting an indirect effect of Hfq on the nir operon expression. On the other hand, it has been recently described that the absence of the Hfq protein in E. coli provokes the decrease of some mRNAs without altering their stability (27). The authors propose a mechanism in which Hfq affects transcription, suggesting a role in the elongation step, with interaction of Hfq with the nascent transcript helping to overcome transcription pauses and to prevent preliminary transcript release. We cannot rule out the possibility that something similar happens in Anabaena and that Hfq has a more general role in regulation by affecting transcription directly. Finally, the observed downregulation of genes involved in nitrate assimilation is not a general feature of Δhfq mutant cells, since the expression of genes ntcA (encoding the global nitrogen regulator NtcA), all0961 (mobA), alr4818 (cysG), or rnpB, used as an internal control for quantitative RT-PCR analyses, is not affected by the absence of Hfq.
This study shows that the protein Hfq has an important regulatory role for the assimilation of nitrate in Anabaena sp. PCC 7120, thus integrating a new regulatory element in an already very complex system. The mechanism for this regulation, as well as the possible implication of sRNAs, remains to be investigated.
ACKNOWLEDGMENTS
We are grateful to Carlos Parejo for technical assistance with nitrate determination by HPLC. We thank J. E. Frías for providing protocols for the determination of nitrate and nitrite reductase activities, A. M. Muro-Pastor for critical reading of the manuscript, and both of them for helpful discussions. We also thank anonymous reviewers for their helpful comments.
This work was supported by the Ministerio de Ciencia e Innovación and the European Regional Fund (BFU2007-60651 and BFU2010-14821) and the Plan Andaluz de Investigación (BIO215). E.P-F. was supported by an I3P postdoctoral contract from the Consejo Superior de Investigaciones Científicas.
Footnotes
Published ahead of print on 20 May 2011.
REFERENCES
- 1. Arluison V., et al. 2007. Spectroscopic observation of RNA chaperone activities of Hfq in post-transcriptional regulation by a small non-coding RNA. Nucleic. Acids Res. 35:999–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bird C., Wyman M. 2003. Nitrate/nitrite assimilation system of the marine picoplanktonic cyanobacterium Synechococcus sp. strain WH 8103: effect of nitrogen source and availability on gene expression. Appl. Environ. Microbiol. 69:7009–7018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Black T. A., Cai Y., Wolk C. P. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 9:77–84 [DOI] [PubMed] [Google Scholar]
- 4. Bøggild A., Overgaard M., Valentin-Hansen P., Brodersen D. E. 2009. Cyanobacteria contain a structural homologue of the Hfq protein with altered RNA-binding properties. FEBS J. 276:3904–3915 [DOI] [PubMed] [Google Scholar]
- 5. Böhme H. 1986. Inhibition of nitrogen fixation by nitrite in Anabaena variabilis. Arch. Microbiol. 146:99–103 [Google Scholar]
- 6. Bohn C., Rigoulay C., Bouloc P. 2007. No detectable effect of RNA-binding protein Hfq absence in Staphylococcus aureus. BMC Microbiol. 7:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cai Y., Wolk C. P. 1997. Nitrogen deprivation of Anabaena sp. strain PCC 7120 elicits rapid activation of a gene cluster that is essential for uptake and utilization of nitrate. J. Bacteriol. 179:258–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cai Y. P., Wolk C. P. 1990. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 172:3138–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chao Y., Vogel J. 2010. The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 13:24–33 [DOI] [PubMed] [Google Scholar]
- 10. Dienst D., et al. 2008. The cyanobacterial homologue of the RNA chaperone Hfq is essential for motility of Synechocystis sp. PCC 6803. Microbiology 154:3134–3143 [DOI] [PubMed] [Google Scholar]
- 11. Elhai J., Vepritskiy A., Muro-Pastor A. M., Flores E., Wolk C. P. 1997. Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J. Bacteriol. 179:1998–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Elhai J., Wolk C. P. 1988. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 167:747–754 [DOI] [PubMed] [Google Scholar]
- 13. Elhai J., Wolk C. P. 1988. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene 68:119–138 [DOI] [PubMed] [Google Scholar]
- 14. Flores E., Frías J. E., Rubio L. M., Herrero A. 2005. Photosynthetic nitrate assimilation in cyanobacteria. Photosynth. Res. 83:117–133 [DOI] [PubMed] [Google Scholar]
- 15. Flores E., Herrero A. 1994. Assimilatory nitrogen metabolism and its regulation, p. 487–517 In Bryant D. A. (ed.), The molecular biology of the cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 16. Flores E., Herrero A. 2010. Compartmentalized function through cell differentiation in filamentous cyanobacteria. Nat. Rev. Microbiol. 8:39–50 [DOI] [PubMed] [Google Scholar]
- 17. Franze de Fernandez M. T., Eoyang L., August J. T. 1968. Factor fraction required for the synthesis of bacteriophage Qβ-RNA. Nature 219:588–590 [DOI] [PubMed] [Google Scholar]
- 18. Frías J. E., Flores E. 2010. Negative regulation of expression of the nitrate assimilation nirA operon in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 192:2769–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Frías J. E., Flores E., Herrero A. 2000. Activation of the Anabaena nir operon promoter requires both NtcA (CAP family) and NtcB (LysR family) transcription factors. Mol. Microbiol. 38:613–625 [DOI] [PubMed] [Google Scholar]
- 20. Frías J. E., Flores E., Herrero A. 1997. Nitrate assimilation gene cluster from the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 179:477–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Frías J. E., Herrero A., Flores E. 2003. Open reading frame all0601 from Anabaena sp. strain PCC 7120 represents a novel gene, cnaT, required for expression of the nitrate assimilation nir operon. J. Bacteriol. 185:5037–5044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gottesman S. 2004. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 58:303–328 [DOI] [PubMed] [Google Scholar]
- 23. Herrero A., Flores E., Guerrero M. G. 1984. Regulation of nitrate reductase cellular levels in the cyanobacteria Anabaena variabilis and Synechocystis sp. FEMS Microbiol. Lett. 26:21–25 [Google Scholar]
- 24. Herrero A., Guerrero M. G. 1986. Regulation of nitrite reductase in the cyanobacterium Anacystis nidulans. J. Gen. Microbiol. 132:2463–2468 [Google Scholar]
- 25. Jepson B. J., et al. 2004. Tuning a nitrate reductase for function. The first spectropotentiometric characterization of a bacterial assimilatory nitrate reductase reveals novel redox properties. J. Biol. Chem. 279:32212–32218 [DOI] [PubMed] [Google Scholar]
- 26. Kawamoto H., Koide Y., Morita T., Aiba H. 2006. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol. Microbiol. 61:1013–1022 [DOI] [PubMed] [Google Scholar]
- 27. Le Derout J., Boni I. V., Regnier P., Hajnsdorf E. 2010. Hfq affects mRNA levels independently of degradation. BMC Mol. Biol. 11:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Markwell M. A., Haas S. M., Bieber L. L., Tolbert N. E. 1978. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal. Biochem. 87:206–210 [DOI] [PubMed] [Google Scholar]
- 29. Mendel R. R., Hansch R. 2002. Molybdoenzymes and molybdenum cofactor in plants. J. Exp. Bot. 53:1689–1698 [DOI] [PubMed] [Google Scholar]
- 30. Moller T., et al. 2002. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell 9:23–30 [DOI] [PubMed] [Google Scholar]
- 31. Montesinos M. L., Herrero A., Flores E. 1995. Amino acid transport systems required for diazotrophic growth in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 177:3150–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Muffler A., Fischer D., Hengge-Aronis R. 1996. The RNA-binding protein HF-I, known as a host factor for phage Q.β RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev. 10:1143–1151 [DOI] [PubMed] [Google Scholar]
- 33. Muffler A., Traulsen D. D., Fischer D., Lange R., Hengge-Aronis R. 1997. The RNA-binding protein HF-I plays a global regulatory role which is largely, but not exclusively, due to its role in expression of the σS subunit of RNA polymerase in Escherichia coli. J. Bacteriol. 179:297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nogueira T., Springer M. 2000. Post-transcriptional control by global regulators of gene expression in bacteria. Curr. Opin. Microbiol. 3:154–158 [DOI] [PubMed] [Google Scholar]
- 35. Olmedo-Verd E., Flores E., Herrero A., Muro-Pastor A. M. 2005. HetR-dependent and -independent expression of heterocyst-related genes in an Anabaena strain overproducing the NtcA transcription factor. J. Bacteriol. 187:1985–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rajkowitsch L., Schroeder R. 2007. Dissecting RNA chaperone activity. RNA 13:2053–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ramaswamy K. S., Endley S., Golden J. W. 1996. Nitrate reductase activity and heterocyst suppression on nitrate in Anabaena sp. strain PCC 7120 require moeA. J. Bacteriol. 178:3893–3898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rasmussen A. A., et al. 2005. Regulation of ompA mRNA stability: the role of a small regulatory RNA in growth phase-dependent control. Mol. Microbiol. 58:1421–1429 [DOI] [PubMed] [Google Scholar]
- 39. Rippka R., Deruelles J., Waterbury J. B., Herdman M., Stainer R. Y. 1979. Generic assignments, strain stories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1–61 [Google Scholar]
- 40. Rippka R., Herdman M. 1992/93. Pasteur Culture Collection of cyanobacterial strains catalogue and taxonomic handbook, I. Institute Pasteur, Paris, France [Google Scholar]
- 41. Rubio L. M., Flores E., Herrero A. 2002. Purification, cofactor analysis, and site-directed mutagenesis of Synechococcus ferredoxin-nitrate reductase. Photosynth. Res. 72:13–26 [DOI] [PubMed] [Google Scholar]
- 42. Shanmugam K. T., et al. 1992. Proposed nomenclature for the genes involved in molybdenum metabolism in Escherichia coli and Salmonella typhimurium. Mol. Microbiol. 6:3452–3454 [DOI] [PubMed] [Google Scholar]
- 43. Soper T. J., Woodson S. A. 2008. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA 14:1907–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sun X., Zhulin I., Wartell R. M. 2002. Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res. 30:3662–3671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Suzuki I., Horie N., Sugiyama T., Omata T. 1995. Identification and characterization of two nitrogen-regulated genes of the cyanobacterium Synechococcus sp. strain PCC7942 required for maximum efficiency of nitrogen assimilation. J. Bacteriol. 177:290–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tsui H. C., Leung H. C., Winkler M. E. 1994. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 13:35–49 [DOI] [PubMed] [Google Scholar]
- 47. Valentin-Hansen P., Eriksen M., Udesen C. 2004. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 51:1525–1533 [DOI] [PubMed] [Google Scholar]
- 48. Valladares A., Maldener I., Muro-Pastor A. M., Flores E., Herrero A. 2007. Heterocyst development and diazotrophic metabolism in terminal respiratory oxidase mutants of the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 189:4425–4430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vioque A. 1997. The RNase P RNA from cyanobacteria: short tandemly repeated repetitive (STRR) sequences are present within the RNase P RNA gene in heterocyst-forming cyanobacteria. Nucleic Acids Res. 25:3471–3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wolk C. P., Ernst A., Elhai J. 1994. Heterocyst metabolism and development, p. 769–823 In Bryant D. A. (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 51. Zhang A., Wassarman K. M., Ortega J., Steven A. C., Storz G. 2002. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell 9:11–22 [DOI] [PubMed] [Google Scholar]