Abstract
In response to stresses, Mycobacterium cells become dormant. This process is regulated by the DosR transcription factor. In Mycobacterium tuberculosis, the dormancy regulon is well characterized and contains the dosR gene itself and dosS and dosT genes encoding DosR kinases, nitroreductases (acg; Rv3131), diacylglycerol acyltransferase (DGAT) (Rv3130c), and many universal stress proteins (USPs). In this study, we apply comparative genomic analysis to characterize the DosR regulons in nine Mycobacterium genomes, Rhodococcus sp. RHA1, Nocardia farcinica, and Saccharopolyspora erythraea. The regulons are highly labile, containing eight core gene groups (regulators, kinases, USPs, DGATs, nitroreductases, ferredoxins, heat shock proteins, and the orthologs of the predicted kinase [Rv2004c] from M. tuberculosis) and 10 additional genes with more restricted taxonomic distribution that are mostly involved in anaerobic respiration. The largest regulon is observed in M. marinum and the smallest in M. abscessus. Analysis of large gene families encoding USPs, nitroreductases, and DGATs demonstrates a mosaic distribution of regulated and nonregulated members, suggesting frequent acquisition and loss of DosR-binding sites.
INTRODUCTION
Mycobacterium tuberculosis is a dangerous human pathogen infecting nearly one-third of the human population (1). An important feature of tuberculosis is the prevalence of latent infection without disease manifestation (14). During latency, M. tuberculosis is believed to exist in a state of nonreplicating persistence with low metabolic activity (40, 46, 49). Understanding the mechanisms used by M. tuberculosis to exist in this state and to switch to a metabolically active, infectious form is an important problem in tuberculosis research.
M. tuberculosis is an obligate aerobe; under hypoxia, it ceases growth, decreases protein and RNA synthesis, and enters a dormant state (48, 49). Low oxygen tension, nitric oxide, and carbon monoxide activate a three-component regulatory system comprised of two sensor kinases, DosS (Rv3132c) and DosT (Rv2027c), and a response regulator, DosR (Rv3133c) (21, 34, 38, 46–48). DosR-DosS is the dormancy regulon, which has been studied in M. tuberculosis, M. bovis (19), and M. smegmatis (5, 25). For M. tuberculosis and M. bovis, not only DosS, but also DosT, was shown to be a sensor kinase for DosR (17, 18, 42).
DosR activates transcription of genes that allow bacteria to survive long periods of anaerobiosis and hence may be important for long-term survival within the host during latent infection (3, 7, 13, 18, 34, 35).
Both kinases, DosT and DosS, respond to nitric oxide, but DosT is more important at the early stage of hypoxia. When oxygen becomes limited, DosT becomes less prominent in the regulatory cascade and DosS alone maintains induction of the dormancy regulon (18, 22, 23, 50). As with many other two-component systems, the dosRS operon is autoregulated (2). The DosR-binding motif, an 18-bp palindrome, has been found by experiment (45) and by computational analysis of eight DosR-regulated promoters from M. tuberculosis (13).
In this study, we characterized the dormancy regulons in nine Mycobacterium spp., Nocardia farcinica, Rhodoccocus sp. strain RHA1, and Saccharopolyspora erythraea. Using positional weight matrices (PWMs) to search for candidate sites in these genomes, and applying consistency filtering, we identified new regulon members that are absent in M. tuberculosis. Analyzing a large group of genomes at appropriate phylogenetic distances allowed us to describe duplications of the regulatory system, coevolution of the kinases (DosT and DosS) and the regulators (DosR), their autoregulation, and the chromosomal neighborhood of their genes, yielding new regulon members. We also were able to describe the evolution of DosR regulation in three large gene families, universal stress proteins (USPs), nitroreductases, and diacilglycerol acyltransferases (DGATs).
MATERIALS AND METHODS
The complete genomes of M. tuberculosis H37Rv, M. bovis AF2122, M. ulcerans Agy99, M. marinum M, M. avium 104, M. vanbaalenii PYR1, M. smegmatis strain MC2 155, Mycobacterium sp. strain MCS, M. abscessus, Rhodococcus sp. RHA1, N. farcinica, and S. erythraea NRRL 2338 were downloaded from GenBank (4). The phylogenetic tree for all studied species (see Fig. S1 in the supplemental material) was obtained from Microbes Online (MO) (http://microbesonline.org/cgi-bin/speciesTree.cgi?taxId = 405948).
We used locus tags from the GenBank genome entries as gene identifiers. They have the following abbreviations for the beginning of gene names in M. tuberculosis H37Rv, M. ulcerans Agy99, M. marinum M, M. avium 104, M. vanbaalenii PYR1, M. smegmatis strain MC2 155, Mycobacterium sp. MCS, M. abscessus, Rhodococcus sp. RHA1, N. farcinica, S. erythraea NRRL 2338, M. leprae TN, and Corynebacterium diphtheriae NCTC 13129, respectively: Rv, MUL, MMAR, MAV, Mvan, MSMEG, Mmcs, MAB, RHA1, nfa, SACE, ML, and DIP.
Protein similarity searches were performed using the Smith-Waterman algorithm implemented in the GenomeExplorer program (30). Orthologous proteins were defined by bidirectional best hits (43) and validated, if necessary, by construction of phylogenetic trees.
Conserved positional clusters of genes were analyzed using the “gene context” tool of the Microbes Online platform (11).
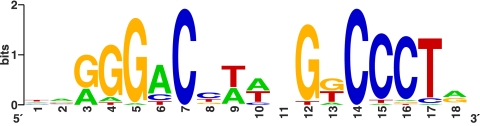
From the literature, we compiled a set of DosR-regulated genes from M. tuberculosis (Rv0079, Rv0571c, Rv1733c, Rv1734, Rv1737c, Rv1813c, Rv1997, Rv2007c, Rv2031, Rv2626c, Rv2627, and Rv3134) (13, 31, 34, 47) and supplemented them with most of their orthologs from M. ulcerans, M. avium, M. smegmatis, and M. vanbaalenii (see Table S1 in the supplemental material). We then applied SignalX (29) to identify the candidate DosR-regulatory motif (Fig. 1) and to construct a PWM. Sequence logos were constructed using the WebLogo tool (9).
Fig. 1.
Logo of the DosR-binding motif, based on 36 sites (see Table S1 in the supplemental material), as described in Materials and Methods.
The constructed PWM was used to scan all studied genomes with the GenomeExplorer software (30) and to identify candidate sites in the regions from −400 to 10 bp relative to translation start sites, excluding the coding regions of upstream genes.
We predicted genes or operons to be DosR regulated if the orthologous operons had candidate sites in at least three genomes. Given that, sites found in closely related genomes, those of M. tuberculosis and M. bovis, were counted as one.
Multiple sequence alignments and phylogenetic trees were obtained from the MO server (11). For multiple-sequence alignments, MO uses the MUSCLE algorithm (12) with the Trim MSA option and for tree construction the PhyML (15) option. Phylogenetic trees were visualized using the Dendroscope package (20).
To identify members of large protein families, we collected annotated DGATs, nitroreductases, and USPs in the genomes of interest and found their homologs using the FastBLAST homology search tool from MO with the default search parameters and an E value threshold of 10−30 for DGATs and nitroreductases and 10−10 for USPs. After that, we collected all identified proteins and used them as the input in the next iteration. Iterations were repeated until convergence.
The protein sequences of all two-component histidine kinases and two-component transcriptional regulators of M. tuberculosis H37Rv, M. ulcerans Agy99, M. marinum M, M. avium 104, M. vanbaalenii PYR1, M. smegmatis strain MC2 155, Mycobacterium sp. MCS, M. abscessus, Rhodococcus sp. RHA1, N. farcinica, S. erythraea NRRL 2338, M. leprae TN, and C. diphtheriae NCTC 13129 were downloaded from the Microbial Signal Transduction database (MiSTDB2) (44).
ClustalX (24) was used to produce multiple-sequence alignments and phylogenetic trees.
All predicted DosR-binding sites, with GeneID numbers, scores, and distances from the gene start site are available in the RegPrecise database (32).
RESULTS
Identification of the DosR regulators and DosT and DosS kinases.
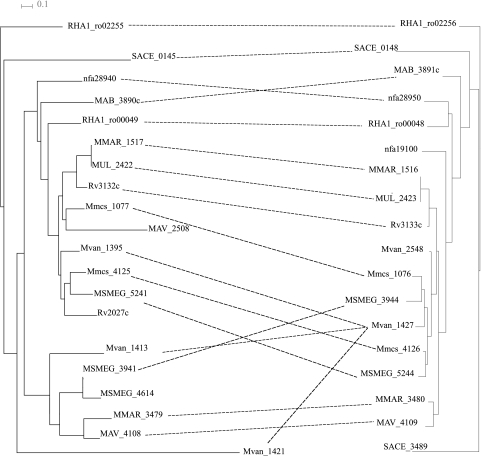
It is known that M. tuberculosis has two histidine kinases, DosS (encoded by Rv3132c) and DosT (encoded by Rv2027c) (18, 34, 47), and that M. smegmatis has two DosR-DosS pairs (5). Therefore, we started with the identification of DosR and DosS homologs in all genomes and the construction of phylogenetic trees (Fig. 2).
Fig. 2.
Phylogenetic tree of the DosR and DosS homologs. Regulators (right-hand tree) and kinases (left-hand tree) whose genes are adjacent and likely form operons are linked by dashed lines. Pairs of regulators and kinases in the same locus but not immediately adjacent are linked by dashed lines.
The species tree (see Fig. S1 in the supplemental material) shows three major groups of genomes: Mycobacteriaceae and Nocardiaceae, Corynebacteriaceae, and Saccharopolyspora. We downloaded the protein sequences of all two-component histidine kinases and two-component transcriptional regulators of M. tuberculosis H37Rv, M. ulcerans Agy99, M. marinum M, M. avium 104, M. vanbaalenii PYR1, M. smegmatis strain MC2 155, Mycobacterium sp. MCS, M. abscessus, Rhodococcus sp. RHA1, N. farcinica, S. erythraea NRRL 2338, M. leprae TN, and C. diphtheriae NCTC 13129 from the Microbial Signal Transduction database (MiSTDB2) (44). For these histidine kinases and transcriptional regulators, phylogenetic trees were constructed (see Fig. S2 and S3, respectively, in the supplemental material). These trees clearly demonstrate the absence of candidate DosR and DosS proteins in M. leprae and C. diphtheriae, and this implies that the Corynebacteriaceae and M. leprae lack the DosRS system. Moreover, all DosS and DosR homologs form separate clades in the respective trees (see Fig. S2 and S3 in the supplemental material). However, all other Mycobacteriaceae and Nocardiaceae, as well as S. erythraea, have the DosRS regulatory systems, but the numbers and compositions of the regulators vary.
All genomes studied further had at least one pair of the dosR and dosS homologs that belong to the same locus (Fig. 2). Furthermore, some genomes had an additional regulator, kinase, or both.
In S. erythraea, we identified two homologs of dosR and one homolog of dosS. The gene SACE_3489 encodes a homolog of dosR adjacent to, and likely forming an operon with, a sensor hystidine kinase gene, SACE_3490, that is homologous to dosS only in the kinase domain, and even these domains belong to different PFAM families. Hence, we do not expect the DosRS functionality for this pair. At a different locus, we found a sensor kinase gene, SACE_0145, homologous to dosS and a transcription factor, encoded by SACE_0148, homologous to DosR. Thus, these genes likely encode the DosS-DosR pair in S. erythraea.
In addition to M. smegmatis, the DosRS systems (both kinases and transcription factors) are duplicated in M. vanbaalenii, Mycobacterium sp. MCS, and M. marinum. Interestingly, two histidine kinase genes (MSMEG_5241 and Mmcs_4125) form a clade with dosT, and their regulators (MSMEG_5244 and Mmcs_4126) also form one clade. Two other kinase genes from this clade, dosT (Rv2027c) and Mvan_1395, do not have DosR-like regulators in the respective loci. The Mvan_1395 and Mvan_1427 genes are positionally close but do not form a compact locus; hence, it is not clear whether they are functionally related. If this is indeed the third regulator-kinase pair from the DosT clade, we can suggest a loss of the second regulator, but not the kinase, in the M. tuberculosis and M. bovis genomes.
In M. marinum, the second regulator-kinase pair (MMAR_3479 and MMAR_3480) does not belong to the dosT clade but forms a new clade with MAV_4108 and MAV_4109. This is one more example of DosRS system duplication.
The species tree (see Fig. S1 in the supplemental material) indicates that M. marinum and M. ulcerans are closely related. However, we did not detect orthologs for the MMAR_3479-MMAR_3480 pair in M. ulcerans. M. marinum has two DosRS systems, which distinguishes it from the closely related M. ulcerans and M. tuberculosis genomes with only one such system. One of the DosRS systems in M. marinum is orthologous to the DosRS systems of M. tuberculosis and M. ulcerans and the second to the M. avium regulator-kinase pair (Fig. 2). These facts suggest a recent horizontal transfer or an ancestral duplication with subsequent lineage-specific losses of paralogs. Analysis of additional genomes is required to resolve these possibilities.
Even mycobacteria closely related to M. tuberculosis do not have exactly the same DosR-DosS-DosT system. Sequence analysis alone is not sufficient to predict the exact roles of homologous sensors, but there are only two kinase genes, MAV_2508 and MSMEG_4614, that, like dosT from M. tuberculosis, do not have a regulator gene in their immediate neighborhoods. MSMEG_4614 is a recently duplicated paralog of MSMEG_3941 and does not have any potential regulon members in its genome neighborhood, whereas MAV_2508 is adjacent to two USP genes, MAV_2506 and MAV_2507, and the nitrite reductase gene MAV_2505 that are DosR regulated (see below).
Comparative analysis of DosR regulons.
After completing identification of the DosRS systems in all studied genomes, we performed characterization of the regulons. The DosR-binding motif, represented by a logo (Fig. 1) and a PWM (see Materials and Methods), roughly coincides with published predicted (13) and experimentally discovered (45) motifs. Using the constructed PWM (see Materials and Methods), we screened all genomes and identified operons with upstream candidate DosR-binding sites.
We used the threshold of 3.7 to define binding sites. This threshold allowed us to pick up all known DosR-binding sites in M. tuberculosis, and the number of false-positive predictions was not large. Indeed, this cutoff produced about 50 sites per Mycobacteriaceae genome and 90 sites per Nocardiaceae genome. More than one-third of the sites found were likely real, based on the consistency filtering procedure and the functions of the regulated genes. In the cases of unresolved coparalogs, we assumed that the consistency condition was met if at least one of the paralogs had a candidate site.
To improve the quality of predictions, the consistency filtering procedure (see Materials and Methods) was applied by pairwise comparison of all genomes to identify genes and operons having candidate binding sites in at least three genomes. (The list of genes can be found at http://regprecise.lbl.gov/RegPrecise/project.jsp?project_id=1189 and in Tables S2 and S3 in the supplemental material. These tables contain the same data as the RegPrecise link.) Most of the genes in the list belong to the dormancy regulon based on their functions, but we also found several genes with conserved sites encoding proteins whose functions had no obvious relation to dormancy.
As mentioned above, there are no DosR/DosS regulators in the Corynebacteriaceae and M. leprae species. Nevertheless, we scanned these genomes with the DosR PWM. M. leprae has 14 sites scoring above the threshold (we used the same cutoff, 3.7, as for other genomes), but none of these sites were conserved in other Mycobacterium spp. No conserved sites were found in the Corynebacteriaceae. These observations prove the absence of the same dormancy regulation in the Corynebacteriaceae and M. leprae.
The largest number of candidate DosR-regulated operons (34 operons) was identified in M. marinum, with only 5 DosR-regulated operons observed in M. abscessus and with M. tuberculosis having an intermediate number of 24 DosR-regulated operons. Interestingly, all genes that are predicted to be DosR regulated in M. abscessus have regulated orthologs in most studied genomes, specifically, encoding DosR, DosS, nitroreductase (MAB_3903), USPs (MAB_2489 and MAB_3904c), and the ortholog of Rv2004c (MAB_3902c). These genes seem to form the minimal dormancy regulon.
Cores of DosR regulons.
In most studied genomes, the regulon core contains orthologous groups whose members are regulated by DosR. In addition to autoregulated dosRS operons, the core contains genes encoding proteins involved in anaerobic metabolism, such as nitroreductases, ferredoxin (fdxA), and diacylglycerol acyltransferases, as well as USPs and heat shock protein (hspX) (Table 1).
Table 1.
Core of the DosR regulon
| Gene function | No. of paralogous proteinsa |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MT | MB | MU | MMAR | MAV | MSMEG | M.MCS | MVan | MABS | NF | RH | SE | |
| DosR | 1/1 | 1/1 | 1/1 | 2/2 | 2/2 | 2/3 | 2/2 | 2/2 | 1/1 | 1/2 | 2/2 | 1/2 |
| DosS | 2/2 | 2/2 | 1/1 | 2/2 | 2/2 | 1/2 | 2/2 | 2/3 | 1/1 | 1/1 | 1/2 | 1/1 |
| USP | 5/9 | 5/9 | 3/9 | 8/14 | 3/6 | 6/14 | 6/18 | 8/16 | 2/6 | 1/6 | 7/21 | 10/28 |
| DGAT | 1/13 | 1/13 | 2/12 | 6/22 | 0/10 | 3/7 | 4/15 | 3/13 | 0/6 | 0/5 | 3/15 | 0/1 |
| Nitroreductase | 2/7 | 2/7 | 2/10 | 5/16 | 3/12 | 3/18 | 5/14 | 2/14 | 1/10 | 2/9 | 3/11 | 3/14 |
| Rv2004c | 1/1 | 1/1 | 0/1 | 1/1 | 0/0 | 1/1 | 1/1 | 1/1 | 1/1 | 0/0 | 1/1 | 1/2 |
| FdxA | 1/1 | 1/1 | 1/1 | 1/2 | 0/1 | 0/1 | 1/2 | 1/1 | 0/1 | 0/0 | 0/3 | 0/1 |
| HcpX | 1/2 | 1/2 | 0/1 | 1/2 | 0/1 | 1/2 | 0/1 | 1/3 | 1/1 | 0/0 | 0/0 | 1/2 |
Each cell shows the number of DosR-regulated paralogous proteins (numerator) and the overall number of all paralogous proteins from the given family (denominator).
The gene for the hypothetical protein Rv2004c from M. tuberculosis is also part of the DosR regulon (34). We confirmed a strong DosR-binding site in M. tuberculosis upstream of the USP-Rv2004c operon. The orthologs of this protein in M. abscessus, M. smegmatis, and Mycobacterium sp. MCS form regulated divergons with nitroreductase genes. The Rv2004c orthologs in M. vanbaalenii and S. erythraea also have upstream candidate DosR-binding sites. We scanned this protein against the PFAM database and determined that it belongs to the Uma3 family (26) and combines an unknown domain (COG2187) with a typical P-loop kinase domain (COG0645). It is widespread in bacteria, especially in the cyanobacteria and proteobacteria. Therefore, we expect that the product of Rv2004c is a histidine kinase.
The sizes of gene families, e.g., universal stress protein genes, vary between genomes, and the numbers of regulated family members may also be quite different, e.g., ranging from 1 USP in N. farcinica to 14 in S. erythraea.
The gene fdxA, encoding ferredoxin, is present in all studied mycobacteria except M. abscessus and in other studied Actinobacteria, but only in M. tuberculosis, M. marinum, Mycobacterium sp. MCS, and M. vanbaalenii does this gene have candidate DosR-binding sites (see Tables S2 and S3 in the supplemental material).
DosR regulation of regulators, kinases, and genes in adjacent loci.
Previous analyses of the DosR regulons have demonstrated that many DosR-regulated genes are clustered on the chromosome (47). We analyzed a total of 24 of the 22-kbp DNA loci centered at all dosR and dosS homologs. Most of these loci contain additional DosR-regulated genes (see Fig. S4 in the supplemental material). For example, the DosR segment in M. tuberculosis (locus 11 in Fig. S4 in the supplemental material) contains three regulated operons. Two of these operons form a divergon (Rv3130c and Rv3131), and the third operon (Rv3134-Rv3134-Rv3132) contains usp (Rv3134) and the dosRS genes (Rv3133c-Rv3132c). The highest numbers of DosR-binding sites (10) and DosR-regulated operons (12) were observed in M. smegmatis (locus 18). It is clear that nitroreductase (MSMEG_3943) and the ortholog of Rv2004c (MSMEG_3942) are both under DosR regulation (see Fig. S4 and Tables S2 and S3 in the supplemental material). MSMEG_3942 and a dosS homolog, MSMEG_3941, form an operon (Fig. 2; see Fig. S4 in the supplemental material).
Only two loci, one in M. smegmatis (locus 21) and one in S. erythraea (locus 1), contain neither candidate DosR-binding sites nor homologs of dormancy-related genes. In M. smegmatis, the histidine kinase gene MSMEG_4614 is a recently duplicated paralog of MSMEG_3941 (Fig. 2) and does not have a corresponding transcription factor. As mentioned above, in S. erythraea, the SACE_0145-SACE_0148 pair (locus 3), but not the SACE_3489-SACE_3490 pair, seems to be the DosR-DosS pair. The former pair has a predicted DosR-binding site.
In most cases, dosR and dosS homologs are cotranscribed and coregulated. Sometimes, e.g., in M. smegmatis (locus 18), they form divergent operons but likely share common DosR-binding sites in the intergenic region. In M. vanbaalenii (locus 16), the dosR and dosS homologs belong to different DosR-regulated operons in the same locus. Interestingly, all DosR orthologs but those of S. erythraea (loci 1 and 3) and N. farcinica (locus 7) are under DosR regulation. Finally, most kinases that are not colocalized with dosR homologs (dosT, MAV_2508, and Mvan_1395) are also regulated by DosR. We conclude that conserved DosR regulation of these kinases in different genomes is a strong indicator of their functional relevance.
Evolution of large protein families.
The identified DosR regulons contain many paralogs. For example, some 22-kbp loci centered at dosR contain up to four DosR-regulated USPs (see Fig. S4 in the supplemental material). We identified additional DosR-regulated USPs, nitroreductases, and DGATs (Table 1; see Tables S2 and S3 in the supplemental material). Conversely, many other USPs, nitroreductases, and DGATs do not seem to be regulated by DosR. To study the evolution of DosR regulation in these large gene families, we constructed phylogenetic trees and analyzed the distribution of regulated genes. We began the analysis with known USPs, nitroreductases, and DGATs and subsequently ran an iterative FastBlast search as described in Materials and Methods.
In M. tuberculosis, we detected 13 candidate DGATs, but only one of these DGAT genes (Rv3130c) had a strong DosR-binding site. Close homologs of this gene have candidate DosR-binding sites in most mycobacterial genomes. For example, M. smegmatis and Mycobacterium sp. MCS have two DosR-regulated DGATs (MSMEG_5242 and MSMEG_3948) (see Fig. S5 in the supplemental material). M. marinum has six DGAT genes that seem to be DosR regulated. Four of them, MMAR_1519, MMAR_3406, MMAR_2649, and MMAR_3403, have strong candidate DosR-binding motifs, while another two, MMAR_ 5271 and MMAR_2073, belong to an operon with DosR sites (see Fig. S4 and Tables S2 and S3 in the supplemental material).
We also identified candidate DosR-binding sites upstream of DGAT precursors in M. marinum (MMAR_2649), M. ulcerans (MUL_3125), M. vanbaalenii (Mvan_4103), M. smegmatis (MSMEG_3933), Mycobacterium sp. MCS (Mmcs_3408, Mmcs_3409, and Mmcs_3541), and Rhodococcus sp. RHA1 (RHA1_ro00023, RHA1_ro00024, RHA1_ro08645, and RHA1_ro05356) (see Tables S2 and S3 and Fig. S5 in the supplemental material). In total, 12 genomes of interest contain 132 DGATs, and 23 of them are DosR regulated (Table 1; see Fig. S5 in the supplemental material).
The DosR-regulated DGATs are localized at three distinct clades on the DGAT phylogenetic tree (see Fig. S5 in the supplemental material). The fact that DosR regulation of DGATs survives operon rearrangements demonstrates the importance of DGATs for the dormancy process.
In contrast, about half of the genes (64 out of 156) encoding universal stress proteins are DosR regulated (Table 1; see Fig. S6 in the supplemental material). All studied genomes have USP genes in the DosR regulons; moreover, all functional dosRS loci (see Fig. S4 in the supplemental material) also contain USP genes.
The phylogenetic tree for an additional DosR-regulated gene family (nitroreductase genes) contains members of the PF00881 PFAM family (see Fig. S7 in the supplemental material). One representative, MSMEG_6505, was recently confirmed as a NfnB family nitroreductase (27). Therefore, we expect nitroreductase functionality from all proteins of the tree. In addition, it is possible to distinguish the clade where most genes are DosR regulated. The studied genomes contain 142 homologous nitroreductases, with 33 of them being members of the DosR regulons.
Therefore, we were able to observe the coexistence of regulated and nonregulated paralogs in the same genome; rearrangement of operons retaining regulation, especially of single-copy genes; and, on the other hand, relatively weak phylogenetic clustering of regulated and nonregulated genes in large families, indicating easy loss and gain of regulatory sites following duplication.
Peripheries of DosR regulons.
Some genes apparently demonstrated sporadic occurrence of candidate DosR-binding sites. While some of these sites may be false positives, simultaneous occurrence of spurious sites upstream of orthologous genes in relatively distant genomes is unlikely.
DosR regulation of the nitrate/nitrite transporter narK2 in M. tuberculosis has been demonstrated in several experimental studies (13, 41), but in addition to this genome, candidate sites were observed only in M. bovis, despite the fact that the gene is present in five genomes. This is an example of a peripheral member of the DosR regulon. The divergent gene for the nitrate/nitrite transporter (Rv1738), encoding a hypothetical protein, is present in nine of the studied genomes, and DosR regulation is conserved in seven genomes (see Table S4 in the supplemental material).
In E. coli, Pseudomonas aeruginosa, Bacillus subtilis, and many other bacteria, the nitrate/nitrite transporters are involved in anaerobic respiration and are regulated by FNR and NarP (16, 28, 36, 37). Thus, it is understandable that the genes that are involved in anaerobic respiration in mycobacteria belong to the anaerobic dormancy regulon (DosR) and have predicted DosR-binding sites in their upstream regions. In this way, we detected DosR-binding sites upstream of ribonucleotide reductase genes (Rv0570, Mb0585, Mmcs_1085, and Mvan_1398), cytochrome bd ubiquinol oxidases, and ABC-type transport systems (Mmcs_3036-Mmcs_3040, Mvan_2830-Mvan_2826, and SACE_0142-SACE_0144).
Three genes encoding cupredoxin domain proteins, Mvan_0508, Mvan_3637, and Mvan_5414, also have candidate DosR-binding sites. Cupredoxin proteins are a part of the electron transport chain (6), and their genes form operons with various membrane protein genes.
Many phosphofructokinase PfkB genes (Rv2029c, Mb2054c, Mmcs_3412, Mvan_1394, MMAR_3482, and RHA_ro00056) are DosR regulated and belong to the dosS loci (see Fig. S4 in the supplemental material). Pfk is an important enzyme of glycolysis (39). Rv2030c and its orthologs (Mb2056c, MMAR_3483, and Mvan_1393) encoding phosphoribosyltransferase form operons with genes for phosphofructokinase and heat shock protein HspX in M. tuberculosis, M. bovis, and M. marinum. In M. vanbaalenii, the hspX gene is located separately, but the regulation is conserved. We detected other phosphoribosyltransferase genes (Rv0571c, Mb0586c, MUL_3276, and MMAR_2069) that do not form operons with other genes (singleton operons) but are likely DosR regulated. Other genes that are likely regulated by DosR are shown in Tables S2 to S4 in the supplemental material and in the RegPrecise database (http://regprecise.lbl.gov/RegPrecise/project.jsp?project_id=1189).
The next group of genes that are necessary for the cell during dormancy are involved in fatty acid metabolism. DGATs are enzymes required for the formation of triacylglycerols (TAGs), which are a common and efficient form of energy storage in organisms that may be utilized during long-term survival (10).We have predicted DosR regulation of DGATs. We also found that desaturases are likely regulated in M. avium (MAV_1793), M. ulcerans (MUL_3646), M. marinum (MMAR_3704), M. abscessus (MAB_3354), N. farcinica (nfa6080), and Rhodococcus sp. RHA1 (RHA1_ro02258) (Table 1; see Tables S2 to S4 in the supplemental material). Desaturase genes of another class (sterol-c5-desaturases), erg3, are predicted to be DosR regulated in M. tuberculosis (Rv1814), M. bovis (Mb1844), M. ulcerans (MUL_3066), and M. marinum (MMAR_2691).
The periphery of the dormancy regulon also contains several acyltransferases (Rv1734c, Mb1763c, Mvan_1410, and RHA1_ro00054).
The functions of most other genes that have conserved candidate DosR-binding sites are unknown, and the encoded proteins are hypothetical (see Tables S2 to S4 in the supplemental material).
DISCUSSION
We began by characterizing the DosR regulons in nine Mycobacteriaceae species, two Nocardia species, and S. erythraea. Starting from the phylogenetic analysis of all DosR and DosS homologs, we detected duplication of the DosRS system in M. marinum, M. smegmatis, M. vanbaalenii, and Rhodococcus sp. RHA1.
We observed that the proteins upregulated in M. tuberculosis during dormancy, such as USPs, nitroreductases, DGATs, heat shock proteins, and ferredoxins, belong to the DosR regulon in most of the genomes analyzed in our study, forming the regulon core. We also detected regulon members that are specific for smaller groups of genomes, with most of them actively involved in anaerobic respiration. Interestingly, the DosR-regulated genes are often colocalized on the chromosome, forming loci consisting of several regulated genes and operons. We also note that autoregulation was conserved in all studied species but S. erythraea.
We performed functional evolutionary analysis of the regulators and large families of regulated proteins. Using phylogenetic trees in conjunction with the DosR motif search for genes from three different protein families, USPs, nitroreductases, and DGATs, we detected the coexistence of regulated and nonregulated paralogs in the same genome; the absence of clustering of regulated versus nonregulated genes on the trees, and operon rearrangements without loss of regulation. While some of the identified sites may be false positives, it is unlikely that all of them are; there are reasons why candidate sites should occur upstream of members of the same protein family. Also, sporadic, phylogenetically inconsistent regulation is a typical feature of large protein families (33).
The only Mycobacteriaceae genome that lacks the DosR-DosS system is that of M. leprae. It is well known that M. leprae has lost a large fraction of the genome (8). In particular, it lacks most USPs and DGATs. Therefore, we do not expect the common mycobacterial dormancy strategy to be conserved in M. leprae.
The Corynebacteriaceae lack the DosRS system. However, our observations about colocalization of DosR-regulated genes allowed us to predict the presence of dormancy regulons in other Rhodococcus species, Nocardiodes sp. JS614, Nakamurella multiparita, Thermobifida fusca, Janibacter sp. HTCC2649, Arthrobacter sp,. and some other actinobacteria (data not shown). Indeed, we observed that in these species the dosR and dosS orthologs are colocalized with USPs and other proteins involved in anaerobic respiration, such as pyruvate phosphate dikinases, flavohemoproteins, and phosphoketolases. Hence, we plan to study the regulation of dormancy in a number of other actinobacteria genomes.
Supplementary Material
ACKNOWLEDGMENTS
This study was partially supported by RFBR (09-04-92745, 08-04-01000, and 10-04-00431), RAS (Program in Molecular and Cellular Biology), and the Ministry of Science and Education (2.740.11.0101).
We are grateful to Gregory Dolganov for useful discussion.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 20 May 2011.
REFERENCES
- 1. Aristoff P. A., Garcia G. A., Kirchhoff P. D., Hollis Showalter H. D. 2010. Rifamycins—obstacles and opportunities. Tuberculosis (Edinburgh) 90:94–118 [DOI] [PubMed] [Google Scholar]
- 2. Bagchi G., Chauhan S., Sharma D., Tyagi J. S. 2005. Transcription and autoregulation of the Rv3134c-devR-devS operon of Mycobacterium tuberculosis. Microbiology 151:4045–4053 [DOI] [PubMed] [Google Scholar]
- 3. Bartek I. L., et al. 2009. The DosR regulon of M. tuberculosis and antibacterial tolerance. Tuberculosis (Edinburgh) 89:310–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benson D. A., Karsch-Mizrachi I., Lipman D. J., Ostell J., Wheeler D. L. 2007. GenBank. Nucleic Acids Res. 35:D21–D25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berney M., Cook G. M. 2010. Unique flexibility in energy metabolism allows mycobacteria to combat starvation and hypoxia. PLoS One 5:e8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bertini I., Sigel A., Sigel H. 2001. Handbook on metalloproteins. Marcel Dekker, New York, NY [Google Scholar]
- 7. Boon C., Dick T. 2002. Mycobacterium bovis BCG response regulator essential for hypoxic dormancy. J. Bacteriol. 184:6760–6767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cole S. T., et al. 2001. Massive gene decay in the leprosy bacillus. Nature 409:1007–1011 [DOI] [PubMed] [Google Scholar]
- 9. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Daniel J., et al. 2004. Induction of a novel class of diacylglycerol acyltransferases and triacylglycerol accumulation in Mycobacterium tuberculosis as it goes into a dormancy-like state in culture. J. Bacteriol. 186:5017–5030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dehal P. S., et al. 2010. MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res. 38:D396–D400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Edgar R. C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Florczyk M. A., et al. 2003. A family of acr-coregulated Mycobacterium tuberculosis genes shares a common DNA motif and requires Rv3133c (dosR or devR) for expression. Infect. Immun. 71:5332–5343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gomez J. E., McKinney J. D. 2004. M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis (Edinburgh) 84:29–44 [DOI] [PubMed] [Google Scholar]
- 15. Guindon S., Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 16. Hartig E., Schiek U., Vollack K. U., Zumft W. G. 1999. Nitrate and nitrite control of respiratory nitrate reduction in denitrifying Pseudomonas stutzeri by a two-component regulatory system homologous to NarXL of Escherichia coli. J. Bacteriol. 181:3658–3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Honaker R. W., Dhiman R. K., Narayanasamy P., Crick D. C., Voskuil M. I. 2010. DosS responds to a reduced electron transport system to induce the Mycobacterium tuberculosis DosR regulon. J. Bacteriol. 192:6447–6455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Honaker R. W., Leistikow R. L., Bartek I. L., Voskuil M. I. 2009. Unique roles of DosT and DosS in DosR regulon induction and Mycobacterium tuberculosis dormancy. Infect. Immun. 77:3258–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Honaker R. W., et al. 2008. Mycobacterium bovis BCG vaccine strains lack narK2 and narX induction and exhibit altered phenotypes during dormancy. Infect. Immun. 76:2587–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huson D. H., et al. 2007. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinform. 8:460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kendall S. L., et al. 2004. The Mycobacterium tuberculosis dosRS two-component system is induced by multiple stresses. Tuberculosis (Edinburgh) 84:247–255 [DOI] [PubMed] [Google Scholar]
- 22. Kim M. J., Park K. J., Ko I. J., Kim Y. M., Oh J. I. 2010. Different roles of DosS and DosT in the hypoxic adaptation of Mycobacteria. J. Bacteriol. 192:4868–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kumar A., Toledo J. C., Patel R. P., Lancaster J. R., Jr., Steyn A. J. 2007. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc. Natl. Acad. Sci. U. S. A. 104:11568–11573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Larkin M. A., et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 25. Lee J. M., et al. 2008. O2- and NO-sensing mechanism through the DevSR two-component system in Mycobacterium smegmatis. J. Bacteriol. 190:6795–6804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leipe D. D., Koonin E. V., Aravind L. 2003. Evolution and classification of P-loop kinases and related proteins. J. Mol. Biol. 333:781–815 [DOI] [PubMed] [Google Scholar]
- 27. Manina G., et al. 2010. Biological and structural characterization of the Mycobacterium smegmatis nitroreductase NfnB, and its role in benzothiazinone resistance. Mol. Microbiol. 77:1172–1185 [DOI] [PubMed] [Google Scholar]
- 28. Melville S. B., Gunsalus R. P. 1996. Isolation of an oxygen-sensitive FNR protein of Escherichia coli: interaction at activator and repressor sites of FNR-controlled genes. Proc. Natl. Acad. Sci. U. S. A. 93:1226–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mironov A. A., Koonin E. V., Roytberg M. A., Gelfand M. S. 1999. Computer analysis of transcription regulatory patterns in completely sequenced bacterial genomes. Nucleic Acids Res. 27:2981–2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mironov A. A., Vinokurova N. P., Gelfand M. S. 2000. Software for analysis of bacterial genomes. Mol. Biol. 34:222–231 [PubMed] [Google Scholar]
- 31. Mishra S. 2009. Function prediction of Rv0079, a hypothetical Mycobacterium tuberculosis DosR regulon protein. J. Biomol. Struct. Dyn. 27:283–292 [DOI] [PubMed] [Google Scholar]
- 32. Novichkov P. S., et al. 2010. RegPrecise: a database of curated genomic inferences of transcriptional regulatory interactions in prokaryotes. Nucleic Acids Res. 38:D111–D118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Panina E. M., Mironov A. A., Gelfand M. S. 2001. Comparative analysis of FUR regulons in gamma-proteobacteria. Nucleic Acids Res. 29:5195–5206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Park H. D., et al. 2003. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48:833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rachman H., et al. 2006. Unique transcriptome signature of Mycobacterium tuberculosis in pulmonary tuberculosis. Infect. Immun. 74:1233–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ravcheev D. A., Gerasimova A. V., Mironov A. A., Gelfand M. S. 2007. Comparative genomic analysis of regulation of anaerobic respiration in ten genomes from three families of gamma-proteobacteria (Enterobacteriaceae, Pasteurellaceae, Vibrionaceae). BMC Genomics 8:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reents H., et al. 2006. Bacillus subtilis Fnr senses oxygen via a [4Fe-4S] cluster coordinated by three cysteine residues without change in the oligomeric state. Mol. Microbiol. 60:1432–1445 [DOI] [PubMed] [Google Scholar]
- 38. Roberts D. M., Liao R. P., Wisedchaisri G., Hol W. G., Sherman D. R. 2004. Two sensor kinases contribute to the hypoxic response of Mycobacterium tuberculosis. J. Biol. Chem. 279:23082–23087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ronimus R. S., Morgan H. W. 2001. The biochemical properties and phylogenies of phosphofructokinases from extremophiles. Extremophiles 5:357–373 [DOI] [PubMed] [Google Scholar]
- 40. Schnappinger D., et al. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sohaskey C. D., Wayne L. G. 2003. Role of narK2X and narGHJI in hypoxic upregulation of nitrate reduction by Mycobacterium tuberculosis. J. Bacteriol. 185:7247–7256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sousa E. H., Tuckerman J. R., Gonzalez G., Gilles-Gonzalez M. A. 2007. DosT and DevS are oxygen-switched kinases in Mycobacterium tuberculosis. Protein Sci. 16:1708–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tatusov R. L., Koonin E. V., Lipman D. J. 1997. A genomic perspective on protein families. Science 278:631–637 [DOI] [PubMed] [Google Scholar]
- 44. Ulrich L. E., Zhulin I. B. 2010. The MiST2 database: a comprehensive genomics resource on microbial signal transduction. Nucleic Acids Res. 38:D401–D407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vasudeva-Rao H. M., McDonough K. A. 2008. Expression of the Mycobacterium tuberculosis acr-coregulated genes from the DevR (DosR) regulon is controlled by multiple levels of regulation. Infect. Immun. 76:2478–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Voskuil M. I. 2004. Mycobacterium tuberculosis gene expression during environmental conditions associated with latency. Tuberculosis (Edinburgh) 84:138–143 [DOI] [PubMed] [Google Scholar]
- 47. Voskuil M. I., et al. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198:705–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Voskuil M. I., Visconti K. C., Schoolnik G. K. 2004. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis (Edinburgh) 84:218–227 [DOI] [PubMed] [Google Scholar]
- 49. Wayne L. G., Sohaskey C. D. 2001. Nonreplicating persistence of Mycobacterium tuberculosis. Annu. Rev. Microbiol. 55:139–163 [DOI] [PubMed] [Google Scholar]
- 50. Yukl E. T., et al. 2011. Nitric oxide dioxygenation reaction in DevS and the initial response to nitric oxide in Mycobacterium tuberculosis. Biochemistry 15:1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.