Abstract
Xenorhabdus nematophila, the mutualistic bacterium of the nematode Steinernema carpocapsae, produces the R-type bacteriocin called xenorhabdicin, which is thought to confer a competitive advantage for growth in the insect host. We have identified a P2-like tail synthesis gene cluster (xnp1) that is required for xenorhabdicin production. The xnp1 genes were expressed constitutively during growth and were induced by mitomycin C. Deletion of either the sheath (xnpS1) or fiber (xnpH1) genes eliminated xenorhabdicin production. Production of R-type bacteriocins in a host organism had not been shown previously. We show that xenorhabdicin is produced in the hemocoel of insects infected with the wild type but not with the ΔxnpS1 deletion strain. Xenorhabdicin prepared from the wild-type strain killed the potential competitor Photorhabdus luminescens TT01. P. luminescens was eliminated during coculture with wild-type X. nematophila but not with the ΔxnpS1 strain. Furthermore, P. luminescens inhibited reproduction of S. carpocapsae in insect larvae, while coinjection with wild-type X. nematophila, but not the ΔxnpS1, strain restored normal reproduction, demonstrating that xenorhabdicin was required for killing P. luminescens and protecting the nematode partner. Xenorhabdicin killed X. nematophila from Steinernema anatoliense, demonstrating for the first time that it possesses intraspecies activity. In addition, activity was variable against diverse strains of Xenorhabdus and Photorhabdus and was not correlated with phylogenetic distance. These findings are discussed in the context of the role of xenorhabdicin in the life cycle of the mutualistic bacterium X. nematophila.
INTRODUCTION
Bacteria employ diverse strategies to compete for favorable ecological niches. Production of small-molecule antibiotic compounds and proteinaceous bacteriocins are widespread strategies by which bacteria gain a competitive advantage. Antibiotics, generally derived from secondary metabolic pathways, are active against a broad spectrum of competitors and are usually less active against more closely related species. In contrast, R-type bacteriocins are more active against related species and strains. Besides providing a competitive advantage to producer strains, bacteriocins can also affect microbial community dynamics and enhance the reproductive success of symbiotic partners involved in mutualistic associations (12, 17, 29, 31).
The bacterium Xenorhabdus nematophila engages in a mutualistic association with the entomopathogenic nematode (EPN) Steinernema carpocapsae. The bacteria reside in a specialized region of the anterior gut in the infective juvenile (IJ) form of the nematode. The nematode invades soil-dwelling insects, migrates through the intestine, and penetrates the midgut to enter the hemocoel. There, the nematode releases the bacterium into the hemolymph and together they kill the insect host (9, 11, 15). X. nematophila provides a nutrient base for nematode reproduction and also produces antimicrobial compounds to suppress the growth of potential competitors. After 2 to 3 generations, the nematode develops into the IJ stage, which is colonized by X. nematophila (29, 30).
To date, 20 Xenorhabdus species and more than 50 Steinernema species have been described (20, 33, 35, 38). Bacterial competitors can gain access to the hemocoel by coinvasion of the insect host by different EPN species that occupy the same ecological community (4, 7, 34). Several laboratory studies have shown that interspecies competition occurs between different EPNs invading a common host (2, 3, 19). Reproduction of S. carpocapsae in the insect was shown to be compromised by the presence of nonsymbiont species of Xenorhabdus (29). In addition, the closely related bacterium Photorhabdus luminescens prevented reproduction of S. carpocapsae, while the presence of X. nematophila alleviated the antagonistic effect and restored normal reproduction. Thus, the ability of X. nematophila to eliminate closely related competitors enhances the fitness of its nematode partner. X. nematophila produces a phage tail-like (R-type) bacteriocin called xenorhabdicin, which can kill Xenorhabdus and Photorhabdus species (6, 31). R-type bacteriocins, referred to as R-pyocins, have been extensively studied in free-living pathogens, such as Pseudomonas aeruginosa (23). They are composed of an outer sheath, an inner tube, and associated tail fibers. R-type pyocins bind via tail fibers to lipopolysaccharide (LPS) receptors on the surface of sensitive bacteria (18, 24, 27, 40). It has been shown that recombination of tail fiber proteins broadens R-type bacteriocin specificity (24). Upon tail fiber binding, the tail sheath contracts, causing the tail tube to penetrate into the cytoplasmic membrane, resulting in depolarization and cell death (37).
Cultures of X. nematophila produce xenorhabdicin at high levels upon induction with mitomycin C (6, 26, 36). These proteinaceous structures resemble headless phage tail particles and are composed of a 43-kDa tail sheath and 20-kDa tail tube proteins as well as several other structural proteins (5, 6, 36). So far, xenorhabdicin has been shown to be active against two Xenorhabdus species and two strains of P. luminescens (6, 31). While R-type bacteriocins have been extensively studied in vitro, their function under biologically relevant situations has not been established. In addition, the role they play in the life cycle of a mutualistic bacterium such as X. nematophila has not been studied in detail.
It is becoming increasingly more apparent that the function of R-type bacteriocins in a mutualistic association is more complex than previously recognized. In certain cases, competitive success could be primarily dependent on R-type bacteriocin. In other scenarios the outcome of the competition could be dictated by varied abilities of the competing strains to produce multiple antimicrobial agents, such as antibiotics and colicin-type bacteriocins (1, 8, 17, 22, 28, 32, 39). The range of xenorhabdicin activity against other Xenorhabdus and Photorhabdus strains has not been well studied, and the extent to which other antimicrobial compounds contribute to competitive success in the absence of xenorhabdicin is not known. Several aspects of xenorhabdicin production and function remain unanswered. What genetic loci are required for xenorhabdicin production? Is xenorhabdicin produced in vivo? Is xenorhabdicin necessary and sufficient for elimination of other Xenorhabdus and Photorhabdus strains? What is the spectrum of activity of xenorhabdicin? What are the possible roles of R-type bacteriocins in mutualistic interactions? These questions are addressed in the present study.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains and plasmids used in this study are listed in Table 1. Xenorhabdus sp. and Escherichia coli strains were grown in lysogeny broth (LB; 1% Bacto tryptone, 0.5% yeast extract, 0.5% NaCl, and 0.01 mM MgSO4) at 30°C or 37°C, respectively. Solid medium was obtained by adding 1.5% agar. LB cultures (10 to 100 ml) were inoculated with an overnight culture (50-fold diluted) and monitored for growth based on the optical density at 600 nm (OD600). Graces insect culture medium (Gibco) was used instead of LB for experiments involving injection of cells into insects and for dilutional plating. Growth medium was supplemented with ampicillin (50 μg/ml), chloramphenicol (12.5 μg/ml), and gentamicin (10 μg/ml) as indicated.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source |
|---|---|---|
| Xenorhabdus and Photorhabdus strains | ||
| X. nematophila 19061 | S. carpocapsae, USA | Laboratory stock |
| X. nematophila 19061::ΔxnpS1 | S. carpocapsae, USA | This study |
| X. nematophila 19061::ΔxnpS2 | S. carpocapsae, USA | This study |
| X. nematophila 19061::ΔxnpS1S2 | S. carpocapsae, USA | This study |
| X. nematophila 19061::ΔxnpH1 | S. carpocapsae, USA | This study |
| X. nematophila | S. anatoliense, Jordan | P. Stock |
| X. nematophila | S. websteri, Peru | P. Stock |
| X. beddingii Q58 | Steinernema sp. M, Australia | ATCC 49542 |
| X. bovienii | S. feltiae, USA | P. Stock |
| X. bovienii | S. intermedium, USA | P. Stock |
| X. bovienii | S. jollieti, USA | H. Goodrich-Blair |
| X. bovienii | S. kraussei, Canada | P. Stock |
| X. bovienii | S. oregonense, USA | P. Stock |
| X. bovienii | S. puntauvense, Costa Rica | P. Stock |
| X. bovienii | S. weiseri, Turkey | P. Stock |
| X. cabanillasii | S. riobrave, USA | P. Stock |
| X. innexi | S. scapterisci, Uruguay | DMS 16336 |
| X. japonica | S. kushidai, Japan | DMS 16522 |
| X. poinarii | S. glaseri, USA | DMS 4768 |
| X. szentirmaii | S. rarum, Argentina | A. Fodor |
| P. luminescens subsp. laumondii | H. bacteriophora, Trinidad and Tobago, TT01 | T. Ciche |
| P. luminescens subsp. luminescens | H. bacteriophora, Australia, Hb | Laboratory stock |
| P. luminescens subsp. luminescens | NA, NA, Hm | Laboratory stock |
| P. luminescens subsp. akhurstii | NA, Egypt, S1 | H. El-Sadawy |
| P. luminescens subsp. akhurstii | NA, Egypt, RM1 | H. El-Sadawy |
| E. coli strains | ||
| E. coli S17(λpir) | recA thi pro hsd(r− m+); RP4-2Tc::Mu Km::Tn7 in the chromosome | Laboratory stock |
| E. coli S17 (λpir/pER2) | Host strain containing suicide vector pER2 | D. Saffarini |
| E. coli S17 (λpir/pER2-ΔxnpS1::Cmr) | Host strain containing suicide vector pER2-ΔxnpS1::Cmr | This study |
| E. coli S17 (λpir/pER2-ΔxnpS2) | Host strain containing suicide vector pER2-ΔxnpS2 | This study |
| E. coli S17 (λpir/pER2-ΔxnpH1) | Host strain containing suicide vector pER2-ΔxnpH1 | This study |
| Plasmids | ||
| pSTBlue-1 | Cloning vector; Ampr Kmr | Novagen |
| pUC18 | Cloning vector; Ampr | Invitrogen |
| pER2 | Suicide vector; p15A, oriR6k, sacB, Mob+ Gmr | D. Saffarini |
| pUC18-ΔxnpS1::Cmr | Cloning vector containing xnpS1::Cmr fragment | This study |
| pSTBlue-1-ΔxnpS2 | Cloning vector containing xnpS2 fragment | This study |
| pSTBlue-1-ΔxnpH1 | Cloning vector containing xnpH1 fragment | This study |
| pER2-ΔxnpS1::Cmr | pER2 containing xnpS1::Cmr fragment for construction of ΔxnpS1 strain | This study |
| pER2-ΔxnpS2 | pER2 containing xnpS2 fragment for construction of ΔxnpS2 strain | This study |
| pER2-ΔxnpH1 | pER2 containing xnpH1 fragment for construction of ΔxnpH1 strain | This study |
For Xenorhabdus and Photorhabdus strains, the description in the middle column provides information on the vector host, geographic region, and strain name (when available).
NA, information not available.
xnp loci analysis.
The genomic location of xnpS1 and xnpS2 in X. nematophila 19061 was determined using the BLASTP algorithm in the MaGe platform (http://www.genoscope.cns.fr/agc/microscope/mage/viewer.php). Genes upstream and downstream of xnpS1 and xnpS2 were annotated based on analysis against public databases and the enterobacteria phage P2 genome sequence (NCBI accession number NC_001895), using the BLASTP and BLAST2 algorithms.
RNA purification from X. nematophila 19061 uninduced and mitomycin C-induced cultures.
Wild-type X. nematophila 19061 was grown in 20 ml of LB, and cell pellets were collected at the indicated growth phase. Mitomycin C was added (5 μg/ml, final concentration) during mid-log phase to determine whether it induced gene expression. Cell pellets were collected before induction (0 h) and 1 h and 2.5 h after induction. Total RNA was extracted with TRIzol reagent (Invitrogen) according to standard procedures. Final RNA pellets were resuspended in 30 μl of nuclease-free H2O. The purity and concentration of the RNA were determined by measuring the optical density at 260 nm and 280 nm. Total RNA was digested with RNase-free DNase reagent (Promega) to a final concentration of 100 ng/μl.
RT-PCR and quantitative RT-PCR (qRT-PCR) analyses.
Reverse transcription-PCR (RT-PCR; 25-μl final volume) was carried out using the AccessQuick RT-PCR system (Promega) and included 10 pmol of each forward and reverse gene-specific primer (see Table S1 in the supplemental material) and 200 ng of RNA. cDNA synthesis was carried out for 45 min at 50°C followed by the PCR (20 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min). Primers for recA were used as internal controls for uninduced RNA samples. For negative-control experiments, reactions were conducted without reverse transcriptase.
qRT-PCR was performed with the SuperScript III Platinum two-step qRT-PCR kit with SYBR green (Invitrogen). First-strand cDNA synthesis was carried out according to the manufacturer's protocol. This reaction mixture included 800 ng of DNase-treated RNA, reverse transcriptase, and random hexamers in a final volume of 20 μl. The cDNA synthesis product was treated with 2 U of RNase H and then diluted 1:16 with nuclease-free H2O. The qPCR samples contained 2.5 μl of diluted and RNase H-treated cDNA and a 10 μM concentration (each) of forward and reverse qPCR primers (see Table S1), in a final volume of 25 μl. Reactions were carried out in the DNA Engine Opticon 2 system under the following conditions: 50°C for 2 min and 95°C for 2 min, followed by qPCR (25 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 40 s). Each reaction was carried out in triplicate. To detect any possible DNA contamination, cDNA was replaced with DNase-treated RNA. Experimental samples included cDNA collected before mitomycin C induction (0 h) and 1 h and 2.5 h after mitomycin C addition. 16S rRNA expression was used as the internal reference for all reactions. Cycle threshold (CT), reaction efficiencies, and melting curve analysis were performed by using the Opticon Monitor analysis software, version 3.1. The expression levels of xnpS1, xnpT1, and xnpH1 transcripts were normalized to that of the 16S rRNA internal reference (ΔCT). The level of mitomycin C induction at 1 h and 2.5 h was calculated relative to the uninduced (0 h) level of expression (ΔΔCT). The following equations were employed to determine the change in transcript expression: at each time point, ; ; .
Construction of ΔxnpS1, ΔxnpS2, ΔxnpS1S2, and ΔxnpH1 deletion strains.
Primers used for the construction of the deletions strains are listed in Table S1 of the supplemental material. Primers with restriction enzyme sites (SphI and XbaI) were designed to PCR amplify the upstream and downstream chromosomal flanking regions of xnpS1 and xnpH1. For the construction of the ΔxnpS1 strain, primers with compatible restriction enzyme sites were designed to PCR amplify the chloramphenicol resistance cassette from the pKnock-Cm vector. Primers for xnpS2 and xnpH1 included internal overlapping sequences. The xnpS1 fragments ligated to the chloramphenicol resistance cassette were cloned into pUC18, and the xnpH1 fragment was cloned into pSTBlue-1 by using SphI and XbaI restriction sites. The blunt-ended xnpS2 fragment was cloned into the EcoRV site of the pSTBlue-1 cloning vector. The resulting ΔxnpS1::Cmr, ΔxnpS2, and ΔxnpH1 inserts were screened by PCR. All inserts were digested with SphI and XbaI restriction enzymes and cloned into the suicide vector pER2, which is gentamicin resistant. The pER2 plasmid constructs were transformed into E. coli S17(λpir) and transferred by conjugation to X. nematophila ATCC 19061 wild-type strain. X. nematophila exconjugants containing the first allelic exchange were screened on LB agar supplemented with ampicillin-gentamicin. Colonies were picked into 2 ml of LB and grown for 2.5 h, and 100 μl of undiluted or 10-fold-diluted culture was plated on sucrose LB agar (5% sucrose, no NaCl, no MgSO4) and incubated at 30°C. Sucrose-resistant colonies were plated on LB agar plates supplemented with ampicillin-chloramphenicol (for the ΔxnpS1::Cmr strain) or ampicillin (for the ΔxnpS2 and ΔxnpH1 strains). The second allelic exchange and gene replacement were confirmed by sensitivity to gentamicin as well as by PCR screening.
Xenorhabdicin preparations from uninduced X. nematophila cultures.
Overnight cultures were diluted to an OD600 of 0.5, and 200 μl was used to inoculate 10 ml of LB. Cultures were incubated at 30°C for 24 h. Cells were removed by centrifugation, and the supernatants containing xenorhabdicin were passed through a 0.45-μm-pore-size filter and concentrated to 1 ml by passage through an Amicon Ultra-15 centrifugal filter (3,000 nominal molecular weight limit; Millipore) at an Rmax of 2,790 × g. Proteins were collected by ultracentrifugation of 500 μl (transmission electron microscopy [TEM] analysis) or 400 μl (SDS-PAGE analysis) of cell-free supernatants at an Rmax of 287,582 × g at 4°C for 15 min. Protein pellets for TEM analysis were resuspended in 50 μl of 50 mM Tris-HCl (pH 8.7), and a 5-fold dilution was negatively stained with 0.8% phosphotungstate. Protein pellets resuspended in 25 μl of Laemmli buffer were resolved by 15% SDS-PAGE and visualized by Coomassie blue staining.
PEG preparations of xenorhabdicin from induced X. nematophila cultures.
Overnight cultures were normalized, and a 50-fold dilution was used to inoculate 100 ml of LB. Cells were grown at 30°C to an OD600 of 0.5 to 0.6 when mitomycin C was added to a final concentration of 5 μg/ml, and cultures were incubated for 18 h. RNase A and DNase I (1 μg/ml, final concentration of each) were added, and cultures were incubated for 30 min at 37°C and then centrifuged to remove cellular debris. The resulting supernatants were sterilized by filtration (0.45-μm-pore-size filter) and incubated for 1 h at 4°C in the presence of 1 M NaCl (final concentration). After centrifugation, polyethylene glycol 8000 (PEG 8000) was added to culture supernatants to a final concentration of 10% (wt/vol) and incubated at 4°C for 10 to 12 h. Precipitated structures, including xenorhabdicin, flagella, and outer membrane vesicles, were collected by centrifugation at Rmax of 13,776 × g, 4°C, for 15 min, resuspended in 3 ml of LB, and centrifuged again to remove insoluble material. The final supernatant containing the xenorhabdicin preparation was sterilely filtered and stored at 4°C. For SDS-PAGE analysis, 50 μl of the xenorhabdicin preparation was ultracentrifuged at an Rmax of 287,582 × g, 4°C, for 15 min, and pellets resuspended in 30 μl of Laemmli buffer were applied to 15% SDS-PAGE gels and visualized by Coomassie blue staining.
TEM analysis of xenorhabdicin from Manduca sexta insect hemolymph.
X. nematophila wild-type and ΔxnpS1 strains were grown in 5 ml of Graces insect medium to an OD600 of 0.2. Cells were diluted 105-fold in Graces and injected into fourth-instar M. sexta insect larvae. After 28 h the insects were surface sterilized and hemolymph was collected on ice into a 1.5-ml tube. After a brief centrifugation to remove hemocytes, 500 μl of hemolymph supernatant was ultracentrifuged, and the resulting pellet was resuspended in 50 μl of 50 mM Tris-HCl (pH 8.7), and an 8-fold dilution was negatively stained with 0.8% phosphotungstate and analyzed by TEM.
Microplate assay of xenorhabdicin activity.
Xenorhabdus and Photorhabdus strains were separately subcultured in 5 ml of LB and grown at 30°C to an OD600 of 0.5 to 0.6. Cultures were diluted 1,200-fold, and 100 μl was mixed with 50 μl of each PEG-precipitated xenorhabdicin preparation in a 96-well microplate. For cells grown without addition of xenorhabdicin, 50 μl of LB was added instead of the xenorhabdicin preparation. Three replicates were included for experiments performed with P. luminescens subsp. laumondii TT01, and all other experiments were performed in duplicate. The xenorhabdicin bactericidal assay was carried out using 100 μl of undiluted P. luminescens TT01 culture. Microplate cultures were incubated at 30°C with shaking. The OD600 was measured at 0 h and after 18 to 24 h of incubation.
Ex vivo competition assay.
Overnight cultures of X. nematophila 19061 wild type, ΔxnpS1, and P. luminescens TT01 were normalized to an OD600 of 1.0. A 50-fold dilution of each culture was used to inoculate 5 ml of LB. Cultures were incubated at 30°C to an OD600 of 0.4. The competitions were established by coinoculating at equal volumes wild-type and ΔxnpS1 strains of X. nematophila with P. luminescens TT01 in a microplate. The competitions were plated in triplicate on LB agar at 0 h, 3 h, 6 h, and 20 h of incubation at 30°C with shaking. The ratio (percent) of P. luminescens TT01 to X. nematophila was determined at each time point by colony morphology and pigmentation. This experiment was performed two times.
In vivo competition in Galleria mellonella.
Aliquots (150 to 200 μl) of overnight LB cultures of either X. nematophila 19061 wild type, the ΔxnpS1 strain, or P. luminescens TT01 were inoculated into 5 ml of LB, and cultures were grown to mid-log phase at 30°C. An aliquot of each culture was normalized to an OD600 of 0.34. Individual cultures and 1:1 competition mixtures (19061:TT01 and ΔxnpS1:TT01) were diluted with Graces insect medium (Gibco) to a final concentration of ∼200 cells/μl. Aposymbiotic S. carpocapsae IJs were surface sterilized (13) and combined with diluted bacterial cultures for a final mixture of 20 IJs and 200 bacterial cells per 20 μl. Individual G. mellonella were injected with 20 μl containing the IJ-bacteria mixture. After insect death, cadavers were placed in a water trap and IJ emergence was monitored for 20 days, at which time the water was poured into a graduated cylinder and water was added to a final volume of 20 ml. Six 1-μl drops were counted under a binocular microscope to calculate the number of IJs/ml. Two water traps were used for the individual injection controls (19061, ΔxnpS1, and TT01), and four water traps were used for each competition condition. For the injection control treatments, the average number of IJs/ml was determined. For each competition condition, the water traps were analyzed individually.
RESULTS
P2-like phage clusters in X. nematophila.
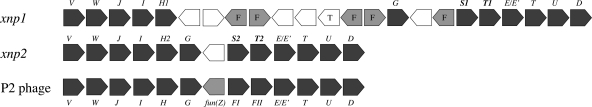
Xenorhabdicin was previously characterized as an R-type phage tail structure (5, 6, 26, 36). To identify the gene cluster encoding xenorhabdicin proteins, the X. nematophila genome was searched for P2-like phage genes. Two distinct P2-like phage clusters were identified (Fig. 1). One cluster, named xnp1 (Xenorhabdus nematophila P2-phage 1), was located at 1.05 Mb and contained tail synthesis genes but lacked genes for capsid synthesis and replication (25). The genes encoding the phage sheath, tube, and fiber proteins were named xnpS1, xnpT1, and xnpH1, respectively. Unlike the contiguous arrangement of the P2 phage genes, a DNA segment of lambdoid origin was identified downstream of xnpH1 (Fig. 1). This DNA segment contained multiple smaller genes with sequence homology to portions of the C terminus of the main tail fiber gene xnpH1. A P2 phage gpG-like gene and a transposase gene were also located in this region. This organization resembles that of the pts remnant P2 phage cluster of P. luminescens TT01, which encodes the R-type photorhabdicin produced by this strain (10). The second P2-like cluster, xnp2, was located at 3.03 Mb. It more closely resembled an intact prophage, containing contiguously arranged genes for tail and capsid synthesis, replication, lysogeny, and lysis (Fig. 1). The sheath, tube, and fiber genes in this cluster were named xnpS2, xnpT2, and xnpH2, respectively.
Fig. 1.
P2-like phage loci in X. nematophila 19061. Comparison of the tail synthesis region of P2-like phage loci in X. nematophila to the tail synthesis region of bacteriophage P2. Shown are tail-related genes (dark gray boxes), additional phage-related genes (light gray boxes), a predicted transposase (T) gene, and other annotated genes (white boxes). Genes encoding the tail sheath protein (xnpS), tail tube protein (xnpT), and tail fiber protein (xnpH) are identified according to their loci, S1 or S2, T1 or T2, and H1 or H2, respectively. Also shown are smaller genes with sequence similarities to the main tail fiber gene xnpH1 (F; light gray). The diagram is not to scale.
Xenorhabdicin is encoded by the xnp1 cluster.
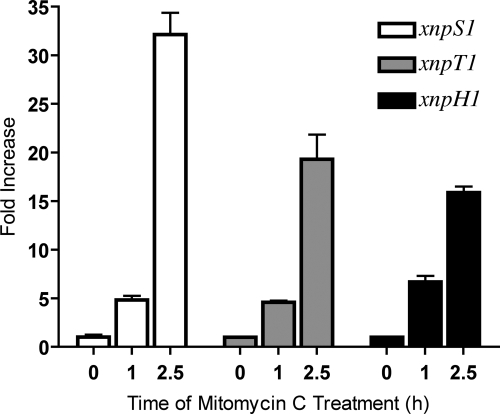
To determine whether genes from the xnp1 and/or xnp2 clusters were involved in xenorhabdicin production, unique sets of primers were designed to assess their expression by RT-PCR. xnpS1, xnpT1, and xnpH1 were found to be expressed constitutively throughout early to late exponential growth (see Fig. S1 in the supplemental material). In contrast, the xnpS2, xnpT2, and xnpH2 genes were not expressed during any phase of growth (data not shown). Since xenorhabdicin had been shown to be inducible, the expression of xnp1 and xnp2 genes was analyzed in the presence of mitomycin C (6). RT-PCR (data not shown) and qRT-PCR (Fig. 2) analyses revealed that xnpS1, xnpT1, and xnpH1 were expressed at higher levels in cells exposed to mitomycin C for 1 h and 2.5 h than in uninduced cells. In contrast, xnpS2 and xnpH2 were not induced by mitomycin C, while xnpT2 was expressed at barely detectable levels (data not shown). These findings suggested that the xnp1 cluster was primarily responsible for the production of xenorhabdicin in X. nematophila.
Fig. 2.
xnp1 gene expression is induced by mitomycin C treatment. Results shown are for qRT-PCR analysis of xnp1 gene expression before treatment with mitomycin C (0 h) and 1 h and 2.5 h after induction. For each gene, time zero was arbitrarily set to 1. The fold changes at stimulated time points (1 h and 2.5 h) are relative to the untreated sample (0 h).
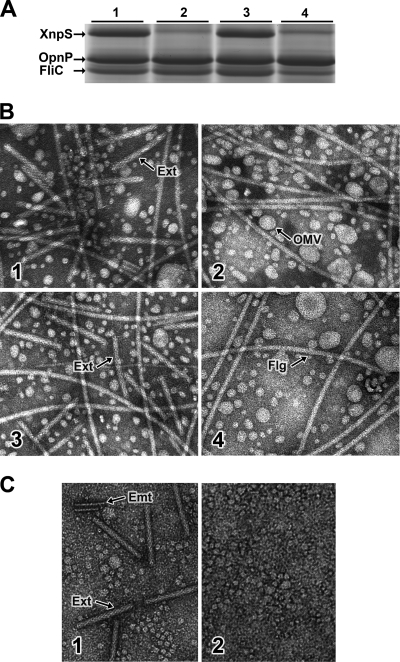
To confirm that the xnp1 cluster was required for xenorhabdicin production, we created strains in which either the xnpS1 or xnpS2 sheath gene was deleted. In addition, a ΔxnpS1S2 deletion strain was constructed. The strains were grown overnight in LB broth under uninducing conditions, and xenorhabdicin was obtained from cell-free supernatants by high-speed ultracentrifugation. SDS-PAGE analysis (Fig. 3A) showed that the sheath protein was absent in the ΔxnpS1 (lane 2) and ΔxnpS1S2 (lane 4) strains but was produced in the ΔxnpS2 strain (lane 3). The 40.6-kDa and 33.3-kDa proteins that migrated below XnpS1 were previously identified as the outer membrane porin, OpnP, and the flagellin protein, FliC, respectively (16, 21). The presence of OpnP in the preparations suggested that outer membrane vesicles had pelleted with the phage tail structures during ultracentrifugation. Xenorhabdicin production was also assessed by transmission electron microscopy (Fig. 3B). Phage tail structures were present in the wild-type strain (panel 1). Flagellar structures were also observed in these preparations, consistent with the finding that X. nematophila grown in LB becomes motile during late exponential growth (16). Round structures that were presumably outer membrane vesicles were also present in the sample. In contrast, phage tail structures were not present in preparations of the ΔxnpS1 (Fig. 3B, panel 2) and ΔxnpS1S2 (panel 4) strains but were present in preparations of the ΔxnpS2 strain (panel 3). Taken together, these results confirmed that the xnp1 locus is required for the production of xenorhabdicin, while the xnp2 locus is not involved in xenorhabdicin production.
Fig. 3.
Analysis of xenorhabdicin production in X. nematophila 19061. (A) SDS-PAGE analysis of uninduced xenorhabdicin preparations from wild-type (lane 1), ΔxnpS1 (lane 2), ΔxnpS2 (lane 3), and ΔxnpS1S2 (lane 4) strains. The tail sheath (XnpS), outer membrane porin (OpnP), and flagellum (FliC) proteins are indicated with arrows. (B) TEM analysis of xenorhabdicin preparations from wild-type (panel 1), ΔxnpS1 (panel 2), ΔxnpS2 (panel 3), and ΔxnpS1S2 (panel 4) strains, showing extended tails (Ext), outer membrane vesicles (OMV), and flagellar (F) structures. (C) Analysis of hemolymph collected from M. sexta after injection with wild-type (panel 1) and ΔxnpS1 (panel 2) strains, showing extended tails (Ext) and empty sheath (Emt) structures.
In vivo production of xenorhabdicin.
Xenorhabdicin production had only been examined under laboratory culture conditions and was assumed to occur in vivo. To determine whether xenorhabdicin was indeed produced in vivo, hemolymph of Manduca sexta infected with either wild type or the ΔxnpS1 strain was analyzed by TEM for the production of phage tail structures (Fig. 3C). Phage tail structures were produced in insects infected with the wild-type strain (panel 1) but not the with the xenorhabdicin mutant strain (panel 2). These findings showed that X. nematophila xenorhabdicin can be produced in the insect hemocoel, where it is presumably involved in interspecies competition.
Analysis of mitomycin C-induced xenorhabdicin preparations.
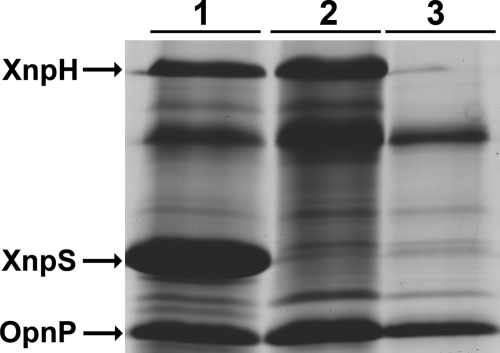
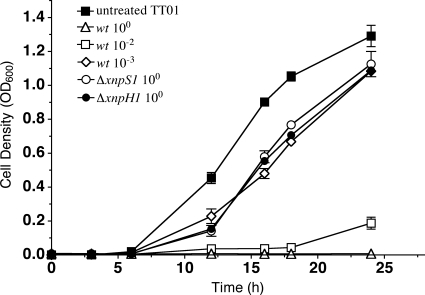
Xenorhabdicin preparations were previously shown to be active against P. luminescens, Xenorhabdus beddingii, and Xenorhabdus bovienii (S. feltiae) and were inactive against Xenorhabdus innexi and Xenorhabdus cabanillasii (6, 31). Whether components in the preparations other than phage tail structures contributed to antimicrobial activities could not be addressed, because xenorhabdicin-minus strains were unavailable. The ΔxnpS1 strain allowed us to address this question. Preparations of the wild-type strain induced with mitomycin C during early exponential phase contained predominantly the XnpS1 tail sheath protein and OpnP derived from outer membrane vesicles (Fig. 4, lane 1). N-terminal sequence analysis of the 78.5-kDa protein identified it as the tail fiber encoded by xnpH1 (data not shown). This was confirmed by deletion of xnpH1 (lane 3). As expected, preparations from the ΔxnpS1 strain lacked XnpS1 (lane 2), consistent with the finding that phage tail structures were not produced by this mutant strain. XnpH was present in these preparations, suggesting that xnpH1 was expressed and fiber structures were assembled in the ΔxnpS1 strain. Phage tail structures were not present in xenorhabdicin preparations from the ΔxnpH1 strain as determined SDS-PAGE analysis (Fig. 4, lane 3) and TEM (data not shown), indicating that deletion of xnpH1 affected the production of xenorhabdicin. Antimicrobial activities of preparations derived from wild-type and ΔxnpS1 strains were assessed using P. luminescens TT01 as the test organism. Growth inhibition was monitored over a 24-h time course (Fig. 5). Treatment with undiluted and 100-fold-diluted xenorhabdicin preparations from X. nematophila wild type completely inhibited growth of P. luminescens, while activity was not detectable with preparations diluted 1,000-fold. As expected, undiluted xenorhabdicin prepared from the ΔxnpS1 and ΔxnpH1 strains was inactive, since phage tail structures were not present in preparations derived from these strains.
Fig. 4.
Deletion of xnpS1 and xnpH1 eliminated tail sheath (XnpS) and tail fiber (XnpH) protein production. Xenorhabdicin preparations from wild-type (lane 1), ΔxnpS1 (lane 2), and ΔxnpH1 (lane 3) strains are shown. The proteins identified are as follows: tail fiber (XnpH), tail sheath (XnpS), porin (OpnP). Other proteins found in purified xenorhabdicin preparations have been described previously (36).
Fig. 5.
Xenorhabdicin preparations inhibit the growth of P. luminescens. Assays performed with xenorhabdicin preparations from the wild-type strain included undiluted and 10−2 and 10−3 dilutions. Assays were performed with undiluted preparations from the ΔxnpS1 and ΔxnpH1 strains. Results with the untreated TT01 culture grown without added xenorhabdicin are also shown.
Role of xenorhabdicin in interspecies competition.
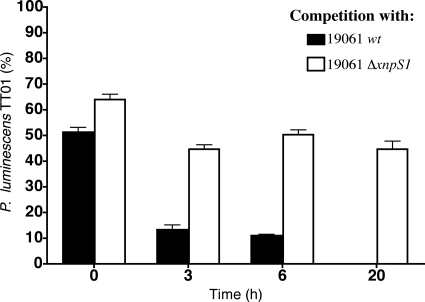
It had been speculated that xenorhabdicin production allowed X. nematophila to outcompete other Xenorhabdus and Photorhabdus strains (6, 29, 31). The availability of the ΔxpnS1 strain allowed us to directly address this question. We used P. luminescens TT01 as the competitor, since it forms red-orange raised colonies that can be clearly distinguished from X. nematophila and it is sensitive to xenorhabdicin. Exponentially growing cultures of P. luminescens, wild-type X. nematophila, and the ΔxnpS1 strain were normalized to OD600 of 0.4, and equal volumes of either wild-type X. nematophila and P. luminescens or the ΔxnpS1 strain and P. luminescens were combined. The percentage of P. luminescens in the coculture was determined by dilutional plating and counting pigmented raised colonies. P. luminescens initially represented 52% of the coculture with wild-type X. nematophila (Fig. 6). After 3 h the level of P. luminescens was reduced to 13%, and after 6 h the level was 11%. By 20 h X. nematophila had completely outcompeted P. luminescens. In contrast, P. luminescens initially represented 62% of the coculture with the ΔxnpS1 strain, and the levels did not significantly decrease over the 20-h incubation period. Thus, in the absence of xenorhabdicin production, X. nematophila was unable to eliminate the competitor, indicating that production of xenorhabdicin was required to confer a competitive advantage over P. luminescens TT01. Interestingly, after 20 h the total cell concentration in the coculture with the ΔxnpS1 strain was 33% of the concentration of wild-type X. nematophila. The reduction in total cell concentration suggested that both P. luminescens and the ΔxnpS1 strain produced additional antimicrobial compounds that adversely affected the growth of the other species (interference competition).
Fig. 6.
Xenorhabdicin is required to eliminate P. luminescens during ex vivo competition. Cocultures of TT01 with either the wild-type strain of X. nematophila or ΔxnpS1 strain were inoculated into LB broth, and the percentage of TT01 was determined at each time point. The data represent the means and standard errors of triplicate experimental samples.
The above findings suggested that xenorhabdicin possesses bactericidal activity toward P. luminescens. This activity was confirmed by adding xenorhabdicin to a culture of exponentially growing P. luminescens (see Fig. S2 in the supplemental material). Untreated cells grew to a final optical density of 1.4. In contrast, the optical density of the cell culture treated with xenorhabdicin decreased by 90% after 6 h and was below the level of detection by 24 h.
Role of xenorhabdicin in interspecies competition and nematode reproduction in vivo.
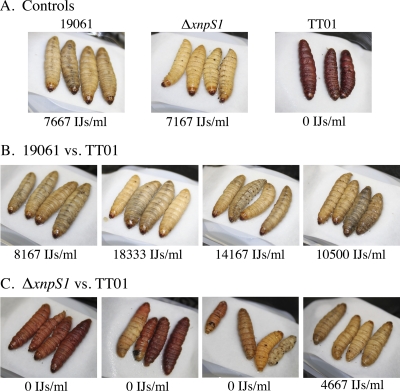
To determine if xenorhabdicin was required to counter the antagonistic effects of P. luminescens TT01 on the reproduction of S. carpocapsae, in vivo competition studies were conducted. G. mellonella larvae injected with aposymbiotic S. carpocapsae and either wild-type X. nematophila or the ΔxnpS1 strain were cream colored and resulted in normal nematode reproduction (Fig. 7A). In contrast, insects injected with P. luminescens and aposymbiotic S. carpocapsae were red and the presence of P. luminescens inhibited nematode reproduction in these insects. Coinjection of wild-type X. nematophila together with P. luminescens and aposymbiotic S. carpocapsae produced cream-colored cadavers and allowed for normal reproduction, indicating that X. nematophila suppressed the growth of P. luminescens and alleviated the antagonistic effect on nematode reproduction (Fig. 7B). In contrast, coinjection of the ΔxnpS1 strain with P. luminescens and aposymbiotic S. carpocapsae produced a mixture of pigmented cadavers (Fig. 7C). In 2 infections almost all of the cadavers were red, while in the third infection most of the cadavers were pigmented, with one cream-colored cadaver. Nematodes were unable to reproduce in these insects. These results indicated that xenorhabdicin is a primary factor in protecting the nematode partner from the detrimental effects of P. luminescens on reproduction. Interestingly, the fourth infection resulted in cream-colored cadavers, in which nematodes were able to reproduce, albeit at lower levels, suggesting that X. nematophila produces antimicrobial factors besides xenorhabdicin that may be active against P. luminescens.
Fig. 7.
Xenorhabdicin is required for protection of the nematode host, as shown by coinjection of X. nematophila wild-type and ΔxnpS1 strains with P. luminescens TT01 and aposymbiotic S. carpocapsae. (A) Representative nematode trap for the individual strain injection controls. (B) Nematode traps for the X. nematophila wild-type strain and TT01 competition. (C) Nematode traps for the ΔxnpS1 strain and TT01 competition.
Sensitivity of diverse Xenorhabdus and Photorhabdus strains to xenorhabdicin.
While xenorhabdicin has been proposed to confer a competitive advantage over related bacteria, to date only two Xenorhabdus species and two P. luminescens strains were shown to be sensitive (6, 31). To determine the range of activity of xenorhabdicin, we assessed the sensitivity of 15 different Xenorhabdus species/strains and 5 strains of P. luminescens in an in vitro xenorhabdicin assay (Table 2). These strains allowed us to study three different levels of competition: intraspecies (X. nematophila strains), interspecies (other Xenorhabdus species), and intergenus (Photorhabdus strains). We found that X. nematophila derived from S. anatoliense was highly sensitive, providing the first demonstration of intraspecies activity of xenorhabdicin. In contrast, activity against X. nematophila isolated from S. websteri was very low. X. japonica and five different strains of X. bovienii were highly sensitive to xenorhabdicin, while X. bovienii (S. oregonense) displayed low sensitivity. Interestingly, X. bovienii (S. feltiae), which is closely related to the highly sensitive strain X. bovienii (S. weiseri), displayed a low level of sensitivity. Several other Xenorhabdus species (X. poinarii, X. cabanillasii, and X. szentirmaii) also displayed low xenorhabdicin sensitivity. Finally, several strains of the sister taxon, Photorhabdus, were highly sensitive, while one strain (P. luminescens RM1) displayed low sensitivity. Together, these findings indicated that the sensitivity to xenorhabdicin was variable and not positively correlated with either phylogenetic distance or geographic location.
Table 2.
Sensitivities of Xenorhabdus and Photorhabdus strains to xenorhabdicin
| Straina | % inhibition |
|---|---|
| High-sensitivity strains | |
| X. nematophila (S. anatoliense) | 97 |
| X. japonica | 100 |
| X. bovienii (S. puntauvense) | 100 |
| X. bovienii (S. intermedium) | 100 |
| X. bovienii (S. jollieti) | 80 |
| X. bovienii (S. kraussei) | 100 |
| X. bovienii (S. weiseri)+ | 100 |
| P. luminescens TT01+ | 100 |
| P. luminescens Hb | 98 |
| P. luminescens Hm | 82 |
| P. luminescens S1 | 84 |
| Low-sensitivity strains | |
| X. nematophila (S. websteri)+ | 23 |
| X. nematophila 19061 | 0 |
| X. bovienii (S. oregonense) | 14 |
| X. bovienii (S. feltiae) | 0 |
| X. beddingii+ | 22 |
| X. poinarii | 0 |
| X. cabanillasii | 0 |
| X. szentirmaii | 10 |
| X. innexi | 24 |
| P. luminescens RM1 | 15 |
+, xnpS1-independent activity was detectable.
DISCUSSION
While R-type bacteriocins have been characterized in free-living pathogenic bacteria, their role in mutualistic bacteria has not been well studied (14, 24). In X. nematophila, xenorhabdicin may function to eliminate related bacteria that antagonize growth of its nematode partner and that compete for nutrient resources of the hemocoel and are able to colonize the nematode. Here we identified a P2-like tail synthesis cluster (xnp1) that contained tail sheath (xnpS1), tube (xnpT1), and fiber (xnpH1) genes required for xenorhabdicin production. The xenorhabdicin genes were expressed constitutively during growth in broth culture. Furthermore, phage tail structures were produced in the hemolymph of insects infected with wild-type X. nematophila but not in insects infected with the ΔxnpS1 strain. To our knowledge, this is the first time an R-type bacteriocin has been shown to be produced in a host organism.
In nature, insect hosts invaded by S. carpocapsae can be coinfected by other species of nematodes (19). During coinfection, X. nematophila encounters competitors such as P. luminescens, which is highly sensitive to xenorhabdicin (6, 31) (present study), whereas X. nematophila is resistant to photorhabdicin, the R-type bacteriocin produced by P. luminescens (10). P. luminescens inhibited reproduction of S. carpocapsae in vivo, while the presence of X. nematophila allows nematodes to reproduce normally (31). Furthermore, S. carpocapsae has been shown to outcompete H. bacteriophora during coinfection of G. mellonella (2, 3). Together these findings predict that xenorhabdicin is required for X. nematophila to gain a competitive advantage over P. luminescens. The availability of a xenorhabdicin-minus strain allowed us to directly address this prediction. We showed here that P. luminescens TT01 was eliminated when coincubated with wild-type X. nematophila but not with the ΔxnpS1 strain, demonstrating that xenorhabdicin is required for successful competition.
Xenorhabdicin was also required in vivo for X. nematophila to gain a competitive advantage over P. luminescens. Interestingly, in some cases, even in the absence of xenorhabdicin, cadavers remained cream colored, and nematodes were able to reproduce. In these cases it appears that nonxenorhabdicin antimicrobial activity is produced at sufficient levels to allow X. nematophila to outcompete P. luminescens. X. nematophila produces nonxenorhabdicin antimicrobial compounds and colicin-type bacteriocins (xenocin) that are active against other species and strains of Xenorhabdus and Photorhabdus (8, 32) (unpublished data). Thus, while xenorhabdicin provides a competitive advantage against P. luminescens nonxenorhabdicin, molecules produced by X. nematophila may also participate in interspecies competition and have a more primary role against other competitors. It was shown previously that coinjection of insects with X. nematophila and Photorhabdus asymbiotica resulted in reduced virulence relative to single-strain injections (interference competition), consistent with the idea that both species were inhibiting each other (22). Whether P. luminescens TT01 produces diffusible antimicrobial activity against X. nematophila remains to be determined. In addition, some strains of Photorhabdus produce colicin-type bacteriocins (lumicin) that exhibit antimicrobial activity against other strains of Photorhabdus and E. coli (28). It is not presently known whether P. luminescens TT01 produces lumicins that are active against X. nematophila.
Examination of 15 strains of Xenorhabdus revealed that X. nematophila (S. anatoliense) was highly sensitive to xenorhabdicin. This is the first demonstration of xenorhabdicin exhibiting intraspecies activity. We have found that X. nematophila (S. anatoliense) can colonize S. carpocapsae nematodes (unpublished data). In the event of coinfection of the insect by both nematodes (S. carpocapsae and S. anatoliense), xenorhabdicin, besides conferring a competitive advantage for nutrient resources, may eliminate X. nematophila (S. anatoliense), preventing it from colonizing S. carpocapsae. While most of the X. bovienii and Photorhabdus strains tested were sensitive to xenorhabdicin, variability was noted even among closely related strains. For example, X. bovienii (S. weiseri) and X. bovienii (S. feltiae) are sister taxa; however, the former was highly sensitive to xenorhabdicin while the latter displayed low sensitivity. Four of the Photorhabdus luminescens strains tested were highly sensitive, while P. luminescens subsp. akhurstii RM1 displayed low sensitivity. These findings show that xenorhabdicin activity is not dictated by bacterial or nematode phylogeny and suggest that sensitivity to xenorhabdicin is determined by structural components of the LPS receptor molecule (18). If this were the case, then the structure of LPS of sensitive strains would be predicted to be similar, while the LPS structure of less sensitive strains would be more divergent. Furthermore, mutations or modifications to the LPS structure could alter sensitivity to xenorhabdicin.
The C-terminal region of tail fiber proteins determines target cell specificity of R-type bacteriocins (24). In the xnp1 locus, 5 smaller genes with sequence homology to portions of the C-terminal domain of the main tail fiber gene, xnpH1, are located between xnpH1 and xnpS1. Similarly, the pts cluster required for the production of photorhabdicin in P. luminescens contains multiple fiber genes in the same location (10). Variation at the C terminus of fiber proteins created by DNA inversion or recombination, or replacement of the native fiber gene with one from a related strain, alters the spectrum of activity of the respective R-type bacteriocins (24, 27, 40). Recombination between the C-terminal domain of xnpH1 and tail fiber genes adjacent to xnpH1 in a subpopulation of cells could expand the range of xenorhabdicin activity and enhance the ability of X. nematophila to compete against other Xenorhabdus and Photorhabdus strains and possibility less related bacteria.
X. nematophila is equipped with an arsenal of antimicrobial products it can use for successful competition in the hemocoel environment of the insect. Here we show that xenorhabdicin provides a competitive advantage against P. luminescens TT01 and likely other microbial competitors and is essential for optimal reproduction of its nematode host. Several questions concerning in vivo production and regulation of xenorhabdicin remain unanswered. How is the cost of bacteriocin production that involves cell lysis balanced with the need to reproduce and express virulence factors? Is xenorhabdicin production regulated at the level of xnp1 gene expression and/or the level of cell lysis? Addressing these and other questions will further expand our understanding of the function of R-type bacteriocins in mutualistic bacteria.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to H. Owen for providing technical expertise with the electron microscopy, to P. Stock, H. Goodrich-Blair, and H. El-Sadawy for providing bacterial strains, and to K. Reddy and S. Dukhvir for help with the in vivo competition experiments. We also thank C. Wimpee and D. Park and members of the Forst laboratory for contributing ideas and suggestions to this project.
This work was supported by NSF grant NSF0919912.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 20 May 2011.
REFERENCES
- 1. Akhurst R. J. 1982. Antibiotic activity of Xenorhabdus spp., bacteria symbiotically associated with insect pathogenic nematodes of the families Heterorhabditidae and Steinernematidae. J. Gen. Microbiol. 1982:3061–3065 [DOI] [PubMed] [Google Scholar]
- 2. Alatorre-Rosas R., Kaya H. K. 1990. Interspecific competition between entomopathogenic nematodes in the genera Heterorhabditis and Steinernema for an insect host in sand. J. Invertebr. Pathol. 55:179–188 [Google Scholar]
- 3. Alatorre-Rosas R., Kaya H. K. 1991. Interactions between two entomopathogenic nematode species in the same host. J. Invertebr. Pathol. 57:1–6 [Google Scholar]
- 4. Amarasinghe L. D., Hominick W. M., Briscoe B. R., Reid A. P. 1994. Occurrence and distribution of entomopathogenic nematodes in Sri Lanka. J. Helminthol. 68:277–286 [Google Scholar]
- 5. Baghdiguian S., Boyer-Giglio M. H., Thaler J. O., Bonnot G., Boemare N. 1993. Bacteriocinogenesis in cells of Xenorhabdus nematophilus and Photorhabdus luminescens: Enterobacteriaceae associated with entomopathogenic nematodes. Biol. Cell 79:177–185 [Google Scholar]
- 6. Boemare N. E., Boyer-Giglio M. H., Thaler J. O., Akhurst R. J., Brehelin M. 1992. Lysogeny and bacteriocinogeny in Xenorhabdus nematophilus and other Xenorhabdus spp. Appl. Environ. Microbiol. 58:3032–3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Campbell J. F., Lewis E., Yoder F., Gaugler R. 1995. Entomopathogenic nematode TPI 68592732 (Heterorhabditidae and Steinernematidae) seasonal population dynamics and impact on insect populations in turfgrass. Biol. Control 5:598–606 [Google Scholar]
- 8. Fodor A., et al. 2010. Comparative analysis of antibacterial activities of Xenorhabdus species on related and non-related bacteria in vivo. J. Microbiol. Antimicrob. 2:36–46 [Google Scholar]
- 9. Forst S., Dowds B., Boemare N., Stackebrandt E. 1997. Xenorhabdus and Photorhabdus spp.: bugs that kill bugs. Annu. Rev. Microbiol. 51:47–72 [DOI] [PubMed] [Google Scholar]
- 10. Gaudriault S., et al. 2004. Identification of a P2-related prophage remnant locus of Photorhabdus luminescens encoding an R-type phage tail-like particle. FEMS Microbiol. Lett. 233:223–231 [DOI] [PubMed] [Google Scholar]
- 11. Goodrich-Blair H., Clarke D. J. 2007. Mutualism and pathogenesis in Xenorhabdus and Photorhabdus: two roads to the same destination. Mol. Microbiol. 64:260–268 [DOI] [PubMed] [Google Scholar]
- 12. Hawlena H., Bashey F., Mendes-Soares H., Lively C. M. 2010. Spiteful Interactions in a natural population of the bacterium Xenorhabdus bovienii. Am. Nat. 175:374–381 [DOI] [PubMed] [Google Scholar]
- 13. Heungens K., Cowles C. E., Goodrich-Blair H. 2002. Identification of Xenorhabdus nematophila genes required for mutualistic colonization of Steinernema carpocapsae nematodes. Mol. Microbiol. 45:1337–1353 [DOI] [PubMed] [Google Scholar]
- 14. Ishii S. I., Nishi Y., Egami F. 1965. The fine structure of a pyocin. J. Mol. Biol. 13:428–431 [DOI] [PubMed] [Google Scholar]
- 15. Kaya H. K., Gaugler R. 1993. Entomopathogenic nematodes. Annu. Rev. Entomol. 38:181–206 [Google Scholar]
- 16. Kim D. J., Boylan B., George N., Forst S. 2003. Inactivation of ompR promotes precocious swarming and flhDC expression in Xenorhabdus nematophila. J. Bacteriol. 185:5290–5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kirkup B. C., Riley M. A. 2004. Antibiotic-mediated antagonism leads to a bacterial game of rock-paper-scissors in vivo. Nature 428:412–414 [DOI] [PubMed] [Google Scholar]
- 18. Kohler T., Donner V., van Delden C. 2010. Lipopolysaccharide as shield and receptor for R-pyocin-mediated killing in Pseudomonas aeruginosa. J. Bacteriol. 192:1921–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koppenhofer A. M., Kaya H. K. 1996. Coexistence of two steinernematid nematode species (Rhabditida:Steinernematidae) in the presence of two host species. Appl. Soil Ecol. 4:221–230 [Google Scholar]
- 20. Lee M. M., Sicard M., Skeie M., Stock S. P. 2009. Steinernema boemarei n. sp. (Nematoda: Steinernematidae), a new entomopathogenic nematode from southern France. Syst. Parasitol. 72:127–141 [DOI] [PubMed] [Google Scholar]
- 21. Leisman G. B., Waukau J., Forst S. A. 1995. Characterization and environmental regulation of outer membrane proteins in Xenorhabdus nematophilus. Appl. Environ. Microbiol. 61:200–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Massey R. C., Buckling A., French-Constant R. 2004. Interference competition and parasite virulence. Proc. R. Soc. Lond. B 271:785–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Michel-Briand Y., Baysse C. 2002. The pyocins of Pseudomonas aeruginosa. Biochimie 84:499–510 [DOI] [PubMed] [Google Scholar]
- 24. Nguyen H. A., et al. 2001. DNA inversion in the tail fiber gene alters the host range specificity of carotovoricin Er, a phage-tail-like bacteriocin of phytopathogenic Erwinia carotovora subsp. carotovora Er. J. Bacteriol. 183:6274–6281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nilsson A. S., Haggard-Ljungquist E. 2007. Evolution of P2-like phages and their impact on bacterial evolution. Res. Microbiol. 158:311–317 [DOI] [PubMed] [Google Scholar]
- 26. Poinar G. O., Hess R. T., Thomas G. 1980. Isolation of defective bacteriophages from Xenorhabdus spp. (Enterobacteriaceae). IRCS Med. Sci. 8:141 [Google Scholar]
- 27. Scholl D., et al. 2009. An engineered R-type pyocin is a highly specific and sensitive bactericidal agent for the food-borne pathogen Escherichia coli O157:H7. Antimicrob. Agents Chemother. 53:3074–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sharma S., et al. 2002. The lumicins: novel bacteriocins from Photorhabdus luminescens with similarity to the uropathogenic-specific protein (USP) from uropathogenic Escherichia coli. FEMS Microbiol. Lett. 214:241–249 [DOI] [PubMed] [Google Scholar]
- 29. Sicard M., et al. 2004. When mutualists are pathogens: an experimental study of the symbioses between Steinernema (entomopathogenic nematodes) and Xenorhabdus (bacteria). J. Evol. Biol. 17:985–993 [DOI] [PubMed] [Google Scholar]
- 30. Sicard M., et al. 2003. Effect of native Xenorhabdus on the fitness of their Steinernema hosts: contrasting types of interaction. Parasitol. Res. 91:520–524 [DOI] [PubMed] [Google Scholar]
- 31. Sicard M., Tabart J., Boemare N. E., Thaler O., Moulia C. 2005. Effect of phenotypic variation in Xenorhabdus nematophila on its mutualistic relationship with the entomopathogenic nematode Steinernema carpocapsae. Parasitology 131:687–694 [DOI] [PubMed] [Google Scholar]
- 32. Singh J., Banerjee N. 2008. Transcriptional analysis and functional characterization of a gene pair encoding iron-regulated xenocin and immunity proteins of Xenorhabdus nematophila. J. Bacteriol. 190:3877–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stock S. P., Goodrich-Blair H. 2008. Entomopathogenic nematodes and their bacterial symbionts: the inside out of a mutualistic association. Symbiosis 46:61–64 [Google Scholar]
- 34. Stuart R. J., Gaugler R. 1994. Patchiness of populations of entomopathogenic nematodes. J. Invertebr. Pathol. 64:39–45 [Google Scholar]
- 35. Tailliez P., Pages S., Ginibre N., Boemare N. 2006. New insight into diversity in the genus Xenorhabdus, including the description of ten novel species. Int. J. Syst. Evol. Microbiol. 56:2805–2818 [DOI] [PubMed] [Google Scholar]
- 36. Thaler J. O., Baghdiguian S., Boemare N. 1995. Purification and characterization of xenorhabdicin, a phage tail-like bacteriocin, from the lysogenic strain F1 of Xenorhabdus nematophilus. Appl. Environ. Microbiol. 61:2049–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Uratani Y., Hoshino T. 1984. Pyocin R1 inhibits active transport in Pseudomonas aeruginosa and depolarizes membrane potential. J. Bacteriol. 157:632–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Uribe-Lorio L., Mora M., Stock S. P. 2007. Steinernema costaricense n. sp. and S. puntauvense n. sp. (Rhabditida: Steinernematidae), two new entomopathogenic nematodes from Costa Rica. Syst. Parasitol. 68:167–182 [DOI] [PubMed] [Google Scholar]
- 39. Webster J. M., Chen G., Hu K., Li J. 2002. Bacterial metabolites, p. 99–114. In Gauler R. (ed.), Entomopathogenic nematology. CABI Publishing, Wallingford, United Kingdom [Google Scholar]
- 40. Williams S. R., Gebhart D., Martin D. W., Scholl D. 2008. Retargeting R-type pyocins to generate novel bactericidal protein complexes. Appl. Environ. Microbiol. 74:3868–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.