Abstract
Klebsiella pneumoniae is an opportunistic pathogen which frequently causes hospital-acquired urinary and respiratory tract infections. K. pneumoniae may establish these infections in vivo following adherence, using the type 3 fimbriae, to indwelling devices coated with extracellular matrix components. Using a colony immunoblot screen, we identified transposon insertion mutants which were deficient for type 3 fimbrial surface production. One of these mutants possessed a transposon insertion within a gene, designated mrkI, encoding a putative transcriptional regulator. A site-directed mutant of this gene was constructed and shown to be deficient for fimbrial surface expression under aerobic conditions. MrkI mutants have a significantly decreased ability to form biofilms on both abiotic and extracellular matrix-coated surfaces. This gene was found to be cotranscribed with a gene predicted to encode a PilZ domain-containing protein, designated MrkH. This protein was found to bind cyclic-di-GMP (c-di-GMP) and regulate type 3 fimbrial expression.
INTRODUCTION
Klebsiella pneumoniae is an opportunistic pathogen which is a significant cause of nosocomially acquired infections, including catheter-associated urinary tract infections (CAUTIs) and ventilator-associated pneumonias (5, 15, 28). K. pneumoniae type 3 fimbriae mediate attachment to, and biofilm formation on, extracellular matrix-coated surfaces in vitro (2, 13, 16). In vivo, indwelling devices rapidly become coated with host material, creating an environment that facilitates infection by type 3 fimbria-producing enterobacteria. In addition, most isolates of K. pneumoniae causing nosocomially acquired infections are resistant to multiple antibiotics (22, 23). The ability of K. pneumoniae to form biofilms and the antimicrobial resistance of the organism are factors that make infections by these bacteria very difficult to eradicate.
K. pneumoniae possesses several virulence factors which aid in the ability of the organism to persist and thrive within an animal host. One class of virulence factors, those involved in bacterial adherence, includes the type 1 and type 3 fimbrial adhesins. Previously, the type 1 fimbriae in K. pneumoniae have been shown to play a role in infectivity by using a murine bladder cystitis model in which a type 1 hyperfimbriate strain more readily forms intracellular bacterial communities (IBCs) within bladder umbrella cells (29). The type 3 fimbriae have previously been characterized by our group and have been shown to mediate bacterial adherence in vitro to human extracellular matrix (HECM) components (13). In many strains of K. pneumoniae, this fimbrial type is encoded by a chromosomally borne gene cluster previously shown to be comprised of five genes (Fig. 1A). These genes include determinants encoding the major fimbrial subunit (MrkA), a chaperone-usher system (MrkBC, respectively), the fimbrial tip adhesin (MrkD), and an as-yet-uncharacterized structural component (MrkF) (6, 8).
Fig. 1.
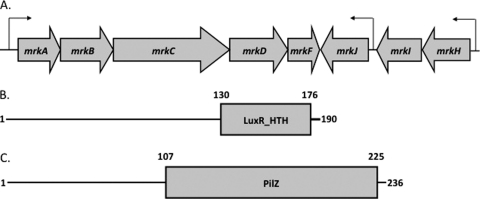
(A) Genetic organization of the mrk gene cluster. Putative promoter regions have been identified by sequence analysis and are indicated by arrows. (B) Predicted domain architecture of MrkI. The site of the mini-Tn5 insertion is within the predicted LuxR-like DNA binding domain (amino acids 130 to 176) in the C-terminal region of the 190-amino-acid MrkI polypeptide. (C) The location of the predicted PilZ c-di-GMP binding site lies within the C-terminal region (amino acids 107 to 225).
Like many other fimbrial types in enterobacteria, the type 3 fimbriae are assembled by the chaperone/usher pathway (8). However, the regulation of type 3 fimbrial expression in K. pneumoniae is poorly understood in comparison to mechanisms of regulation of type 1 and pap fimbrial expression in Escherichia coli and type 1 fimbrial production in Salmonella. Our group recently reported that type 3 fimbrial production is affected by the intracellular concentrations of the second messenger cyclic-di-GMP (c-di-GMP) (14). Mutants of the K. pneumoniae phosphodiesterase MrkJ accumulated intracellular c-di-GMP, which resulted in a type 3 hyperfimbriate phenotype that more readily formed biofilms (14). Fimbrial systems often employ complex regulatory circuits, and it is expected that several as-yet-unidentified regulators govern the expression of type 3 fimbriae. Here we describe a screen which identified a mutant within a putative transcriptional regulator, MrkI, that resulted in decreased type 3 fimbrial expression and biofilm formation. Additionally, we have identified a determinant which is cotranscribed with mrkI and encodes a PilZ-domain containing protein, MrkH, which was shown to bind c-di-GMP and affect type 3 fimbrial expression.
MATERIALS AND METHODS
Strains, plasmids, and DNA manipulations.
The strains and plasmids used in this study are shown in Table 1. To detect the presence of type 3 fimbriae, all strains were grown on either glycerol-Casamino Acids (GCAA) or Luria-Bertani (LB) medium at 37°C unless otherwise stated (6, 7, 11, 16). When necessary, strains were cultured in medium supplemented with antibiotics at the following concentrations: ampicillin (Amp; 100 μg/ml), kanamycin (Kan; 25 μg/ml), spectinomycin (Spec; 100 μg/ml), and tetracycline (Tet; 25 μg/ml).
Table 1.
Plasmids, strains, and oligonucleotides used in this study
| Strain/plasmid/oligonucleotide | Description/sequence (5′–3′)a | Source/reference |
|---|---|---|
| Strains | ||
| IApc35 | Plasmid-cured variant of IA565, type 3 fimbria positive | 12 |
| IApc35 mrkI::Knr | Kanr; IApc35 mrkI insertion mutant, type 3 fimbria negative | This study |
| IApc35 ΔmrkHI | IApc35 mrkHI deletion mutant, type 3 fimbria negative | This study |
| BL21 AI | Protein expression strain | Invitrogen (Carlsbad, CA) |
| NEB 5-α | General E. coli cloning strain | NEB (Ipswich, MA) |
| S17-1 λpir | E. coli donor strain | 3 |
| SM10 λpir | E. coli donor strain | 19 |
| Plasmids | ||
| pACYC184ΔCmr | Tetr Cams; empty vector control for complementation constructs | This study |
| pACYCmrkH | Tetr; mrkH complementation vector | This study |
| pACYCmrkI | Tetr; mrkI complementation vector | This study |
| pACYCmrkHI | Tetr; mrkHI complementation vector | This study |
| pACYCmrkHR113AmrkI | Tetr; mutated mrkHI complementation vector | This study |
| pDEST17 | Ampr; 6×His-tagged Gateway expression vector | Invitrogen |
| pDESTmrkH | Ampr; 6×His-tagged MrkH expression construct | This study |
| pDESTmrkHR113A | Ampr; 6×His-tagged MrkH(R113A) expression construct | This study |
| pDS132 | Camr; sacB suicide vector | 26 |
| pDS132mrkI::knR | Camr Kanr; construct used to make IApc35 mrkI::Knr | This study |
| pDS132ΔmrkHI | Camr Specr; construct used to make IApc35 ΔmrkHI | This study |
| pGEM-T Easy | Ampr; subcloning vector | Promega (Madison, WI) |
| pTrc99A | Ampr; general subcloning vector | Amersham-Pharmacia (Piscataway, NJ) |
| pTrc99APmrkA-lacZ | Ampr; pTrc99A-based reporter construct | This study |
| pUTminiTn5-Kn | Ampr Kanr; mini-Tn5 delivery plasmid | 3 |
| Oligonucleotides | ||
| CNM003 | GTGCAGCGTCGCCGCGATCCCGCCTTTCGT TTAGCCCATGAGCATGACTTTTATTGC | |
| CNM004 | GCAATAAAAGTCATGCTCATGGGCTAAACGAAAGGCGGGATCGCGGCGACGCTGCAC |
Cam, chloramphenicol. Underlining indicates the site of substitution.
Plasmid and genomic DNA preparations, restriction enzyme digestions, and PCR procedures were performed by conventional techniques using commercially available material. All manipulations of DNA were performed according to the manufacturers' instructions.
Construction and screening of mini-Tn5 transposon library.
Conjugation of K. pneumoniae IApc35 with E. coli S17-1 λpir carrying the plasmid pUTminiTn5-Kn was performed as previously described by our group (2). Conjugants were selected on M9 minimal medium supplemented with kanamycin to prevent growth of both the donor and recipient strains. Subsequently, appropriate dilutions of bacterial suspensions in phosphate-buffered saline (PBS) were plated on M9 minimal medium and incubated overnight at 37°C. Bacterial colonies were screened for the production of surface-associated type 3 fimbriae using conventional immunoblotting techniques and monospecific fimbrial antiserum at a dilution of 1:40,000 and for subsequent development with goat anti-rabbit serum conjugated to alkaline phosphatase (4, 20). All colonies that did not react with the fimbrial serum were isolated, retested for lack of reactivity with fimbria-specific antiserum, and stored at −80°C. Insertions of the mini-Tn5 into genes encoding the structural and assembly components of the mrkABCDF cluster were identified by standard PCR procedures and not examined further.
Mapping of the mini-Tn5 insertion site.
Genomic DNA was isolated from nonfimbrial mutants, restricted with SphI, and ligated into SphI-digested pACYC184. The nucleotide sequence of the inserted DNA fragment was determined, and K. pneumoniae-derived sequences flanking the transposon were identified. Subsequently, the location of these sequences on the K. pneumoniae genome was identified using the genome sequence of K. pneumoniae MGH 78578, available online (http://genome.wustl.edu/genomes). The nucleotide sequences flanking the mini-Tn5 were determined in mutants and were found not to have resulted in large rearrangements of the DNA during transposition.
Construction of defined site-directed mutants.
Approximately 1-kb regions of DNA flanking the mrkI and the mrkHI genes were cloned into the vector pGEM-T Easy. Fragments were ligated together, incorporating an internal XbaI restriction site, into which a kanamycin resistance determinant was introduced for the construction of the MrkI mutant only. K. pneumoniae-derived DNA was excised from the pGEM-T Easy recombinant plasmids using SacI and SphI. These fragments were ligated into either the suicide vector pDS132 (for mrkI) or pDS132-specR (for mrkHI). The resulting plasmids, pDS132mrkI::knR and pDS132ΔmrkHI, were transformed into the permissive host E. coli SM10 λpir and subsequently introduced into K. pneumoniae IApc35 via conjugation. Transconjugants were selected on either LB-Kan/Amp or LB-Spec/Amp plates, followed by counterselection on 5% sucrose plates (17, 26). Characterization of mrkI insertion or mrkHI deletion mutants was performed using standard PCR techniques.
Detection of type 3 fimbriae.
Surface production of fimbrial appendages was detected using monospecific fimbrial antiserum as described elsewhere by our group (11, 14). Aerobic cultures were grown at 37°C overnight on either LB agar or as 25-ml LB cultures grown in a 125-ml flask shaken at 220 rpm. Anaerobic and microaerophilic cultures were grown either on LB agar in anaerobic Bio-Bag type A environmental chambers (Becton-Dickinson, Sparks, MD) or as static LB broth cultures, respectively. When necessary, fimbriae were observed by transmission electron microscopy using formaldehyde-fixed bacteria stained with uranyl acetate as previously described (29).
Transcription of mrk.
Expression of mrk genes in K. pneumoniae strains grown under aerobic or anaerobic conditions was determined by quantitative reverse transcription-PCR (qRT-PCR) as previously described (14). Comparison of gene expression between strains grown aerobically and anaerobically was done following cDNA synthesis from equal concentrations of total cellular RNA. Also, the cloned mrk genes in the K. pneumoniae IApc35 ΔmrkHI mutant were assayed for mrkA expression under aerobic conditions using qRT-PCR.
In addition, the ability of the cloned mrkH, mrkI, and mrkHI genes and their derivatives to affect expression of mrkA was determined using a plasmid-borne reporter fusion, pTrc99APmrkA-lacZ, in an E. coli host. This fusion was constructed by cloning a XbaI/HindIII-tailed 444-bp fragment of DNA immediately upstream of mrkA, and possessing the promoter region, into those respective sites in pTrc99A containing a promoterless lacZ gene.
Biofilm formation assays.
The ability of K. pneumoniae strains to form biofilms on solid surfaces was determined as previously described (14, 24, 25).
Mutation of the MrkH c-di-GMP binding site.
Arginine-113 of MrkH was mutated to alanine using overlapping oligonucleotides. Using primers CNM003 and CNM004 which contained the desired mutation and pACYCmrkHI as template, the FailSafe PCR enzyme mix (Epicentre, Madison, WI) was used with 18 cycles of the following reaction: 95°C for 30 s, 55°C for 1 min, and 68°C for 5 min. The resulting plasmid PCR product was digested with DpnI for 1 h at 37°C and then transformed into chemically competent E. coli DH5α (Invitrogen, Carlsbad, CA). The appropriate mutation in the resulting plasmid and the absence of any additional mutations in pACYCmrkHR113AmrkI were verified by DNA sequencing.
Purification of MrkH and MrkH(R113A).
The K. pneumoniae IApc35 mrkH and mrkH(R113A) genes were amplified from pACYCmrkHI and pACYCmrkHR113AmrkI, respectively, by standard PCR procedures and cloned into the Gateway vector pENTR/D-Topo (Invitrogen). These genes were subsequently integrated into the Gateway-compatible destination vector pDEST17 to construct genes encoding His-tagged fusion proteins and introduced into the expression strain BL21-AI. E. coli BL21-AI transformants carrying either pDEST17mrkH or pDEST17mrkHR113A were used to prepare native MrkH or MrkH(R113A), respectively, by Ni-nitrilotriacetic acid (NTA) affinity chromatography by following the manufacturer's instructions (Qiagen, Valencia, CA). Successful purification of both MrkH and MrkH(R113A) was assessed by 12% SDS-PAGE and Western blotting using anti-6×His antibody (Qiagen). Additionally, MrkI was purified as a maltose binding protein (MBP)-fusion gene product (MBP-MrkI) using a commercially available system (NEB, Ipswich, MA).
Binding of c-di-GMP to MrkH.
Generation of 32P-labeled c-di-GMP was performed as previously described (9, 10). The c-di-GMP binding assay was based on that described by Hickman and Harwood (9). A 20-μl mixture of 0.2 mM protein and 2.0 μM [32P]c-di-GMP in binding buffer (40 mM Tris [pH 7.8], 10 mM magnesium acetate, 50 mM KCl) was incubated on ice for 25 min. The reaction mixtures were then brought to a 100-μl volume with binding buffer and immediately loaded onto a slot blot apparatus (PR 600 slot blot; Hoefer Scientific) containing a 0.2-μM nitrocellulose membrane (0.45 mM Protran BA85; Whatman), followed by a wash using 1.0 ml cold binding buffer. The membrane was removed and scanned on a phosphorimager (Packard Instant Imager; Packard Instrument Company) to measure radioactive counts of membrane-bound [32P]c-di-GMP. For the competition assay, a 10-fold excess (20 μM) of cold c-di-GMP was added to the reaction mixture (Biolog, Bremen, Germany). In additional experiments, an equal amount of [α-32P]GTP was substituted for [32P]c-di-GMP to further assess MrkH binding specificity. Additionally, MBP-MrkI was examined for the ability to bind [32P]c-di-GMP. Reaction mixtures containing purified LacZα or protein buffer alone were used as controls.
RESULTS
Immunoblotting of a mini-Tn5 transposon bank of insertion mutants in K. pneumoniae IApc35.
More than 21,000 insertion mutants were screened for their ability to produce surface-associated type 3 fimbriae. Of these, 11 (0.05%) mutants consistently failed to react with fimbria-specific antiserum, even after growth on GCAA agar, which favors the phenotypic expression of these fimbriae (7). Following mapping of the insertion site of the mini-Tn5, three of the mutants were shown to have the transposon inserted into genes that are part of the previously described mrk gene cluster. Therefore, eight mutations in genes that do not encode either structural or assembly components of the type 3 fimbrial system were isolated. The insertion sites of these mutants are currently being identified, and one of these is described below.
K. pneumoniae IApc35 MrkI and MrkHI mutants do not produce surface-associated fimbriae.
Of the eight mutants isolated that possess the transposon in a gene distinct from the mrkABCDF cluster, one of these was further characterized. The site of insertion in this mutant was found to be in a gene encoding a putative transcriptional regulator and annotated KPN_03273 on the K. pneumoniae MGH 78578 genome (GenBank accession number CP000647). The predicted size of this gene is 573 bp, encoding a gene product of 190 amino acids. BLAST analyses of this gene product suggested that it belongs to a family of regulators characterized by a LuxR-like DNA binding domain spanning amino acids 130 to 176 in its C-terminal region (Fig. 1B). The precise site of insertion of the mini-Tn5 was within the predicted DNA binding region encoding amino acid 151. We previously named this gene mrkI, which is located between mrkH and mrkJ, though the K. pneumoniae MGH 78578 genome lacks the correct annotation for mrkH (14). The mrkHIJ genes are located adjacent to the previously characterized mrk gene cluster and exhibit opposite transcriptional polarity to these genes (Fig. 1A). Using intergenic RT-PCR, it was found that mrkH, mrkI, and mrkJ are cotranscribed (data not shown). mrkH is predicted to encode a protein containing a PilZ c-di-GMP binding domain at its C terminus and an N terminus that exhibits little homology to currently characterized domains (Fig. 1C).
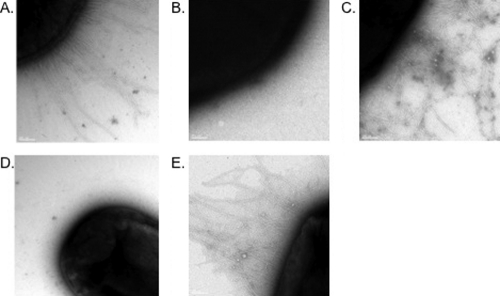
MrkI and MrkHI mutants of K. pneumoniae IApc35 were constructed by conventional techniques. Neither of the K. pneumoniae IApc35 mrkI::Knr and IApc35 ΔmrkHI mutants produce surface-associated type 3 fimbriae following growth under aerobic conditions (Table 2). Interestingly, the MrkI mutant does express type 3 fimbriae when grown anaerobically or microaerophilically as either agar or static broth cultures, respectively, while the ΔmrkHI mutant remains nonfimbriate under either condition (Table 2). The mutants can be complemented to restore fimbrial production by transformation with the cloned genes (Table 2). Electron microscopy confirmed the absence of fimbriae on the MrkI and MrkHI mutants and many fimbriae on the surfaces of complemented strains (Fig. 2). Interestingly, in a ΔmrkHI background, introduction of a plasmid solely expressing mrkI was unable to complement fimbrial expression, while a plasmid expressing only mrkH was able to restore type 3 fimbriation (Table 3). Also, overexpression of mrkH in an mrkI mutant background was able to restore fimbrial expression despite the absence of mrkI (Table 3).
Table 2.
Type 3 fimbrial production of Klebsiella strains
| Culture condition | Serum titera |
||||
| IApc35 | IApc35 mrkI::Knr | IApc35 mrkI::Knr plus mrkI | IApc35 ΔmrkHI | IApc35 ΔmrkHI plus mrkHI | |
|---|---|---|---|---|---|
| Aerobic (shaken flask) | 5,120 | <40 | 40,960 | <40 | 40,960 |
| Aerobic (agar grown) | 5,120 | <40 | 40,960 | <40 | 40,960 |
| Microaerophilic (static tube) | 40,960 | 20,480 | ND | <40 | 40,960 |
| Anaerobic (agar grown) | 5,120 | 10,240 | ND | <40 | 40,960 |
The serum titer represents the reciprocal of the anti-MrkA serum dilution needed to produce visible agglutination. The lowest dilution of serum used was 1:40. ND, not determined.
Fig. 2.
Fimbrial production by K. pneumoniae strains. (A) K. pneumoniae IApc35; (B) MrkI mutant; (C) MrkI mutant transformed with the cloned mrkI gene; (D) MrkHI mutant; (E) complemented MrkHI mutant carrying cloned mrkHI.
Table 3.
Phenotypic complementation analyses
| Strain (plasmid) | Serum titer |
|---|---|
| IApc35 (pACYCΔCmr) | 5,120 |
| IApc35 mrkI::Knr (pACYCΔCmr) | <40 |
| IApc35 mrkI::Knr (pACYCmrkH) | 40,960 |
| IApc35 mrkI::Knr (pACYCmrkI) | 40,960 |
| IApc35 mrkI::Knr (pACYCmrkHI) | 40,960 |
| IApc35 ΔmrkHI (pACYCΔCmr) | <40 |
| IApc35 ΔmrkHI (pACYCmrkH) | 40,960 |
| IApc35 ΔmrkHI (pACYCmrkI) | <40 |
| IApc35 ΔmrkHI (pACYCmrkHI) | 40,960 |
MrkI and MrkHI mutants are affected in mrkA gene transcription.
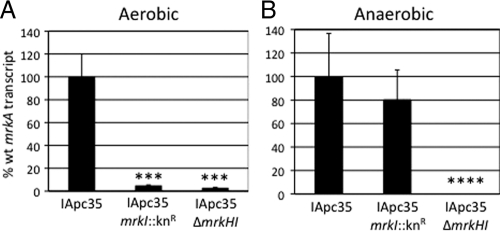
Using qRT-PCR analysis of RNA extracted from aerobically grown agar cultures, it was found that both IApc35 mrkI::Knr and IApc35 ΔmrkHI have significantly reduced levels of mrkA transcription. Levels of the mrkA transcript in the IApc35 mrkI::Knr strain were approximately 20-fold lower than those in IApc35. Similarly, using RNA extracted from IApc35 ΔmrkHI, a significant decrease in mrkA expression was observed, with a 33-fold reduction in transcription compared to that of the parental strain (Fig. 3A). Since MrkI mutants assemble surface-associated fimbriae when grown anaerobically, mrkA expression under these conditions was determined. K. pneumoniae IApc35 mrkI::Knr grown anaerobically exhibited levels of mrkA expression that were indistinguishable from those of the parental IApc35 strain. mrkA transcription levels in the IApc35 ΔmrkHI mutant were significantly lower than those of the parental strain and were reduced by approximately 2,000-fold (Fig. 3B). Also, we examined a possible autoregulatory role of MrkI on mrkHI transcription and observed no decrease in gene expression in the MrkI mutant (data not shown).
Fig. 3.
qRT-PCR of mrkA encoding the major fimbrial subunit in K. pneumoniae strains. Aerobic (A) and anaerobic (B) mrkA transcription in both IApc35 mrkI::Knr and IApc35 ΔmrkHI are shown as the relative decreases in transcription compared to that shown by the parental strain. Statistical significance was determined using Student's t test (***, P value < 0.001; ****, P value < 0.0001).
Expression of mrk genes is increased following anaerobic growth.
Quantitative RT-PCR analysis using RNA extracted from K. pneumoniae IApc35 cultures grown anaerobically indicated increased mrkA, mrkH, and mrkI expression compared to that of cultures incubated aerobically. Increased expression levels of approximately 285-, 77-, and 91-fold were observed for mrkA, mrkH, and mrkI, respectively (Table 4).
Table 4.
Aerobic and anaerobic transcriptions of mrk genes in parental IApc35
| Transcript (condition) | Fold change (± SD) |
|---|---|
| mrkA (aerobic) | 1.0 (± 0.1) |
| mrkA (anaerobic) | 284.78 (± 47.75)a |
| mrkH (aerobic) | 1.0 (± 0.15) |
| mrkH (anaerobic) | 76.93 (± 24.45)a |
| mrkI (aerobic) | 1.0 (±0.15) |
| mrkI (anaerobic) | 90.65 (± 12.34)a |
P value < 0.0001.
MrkI and MrkHI mutants have a decreased ability to form biofilms on an abiotic surface.
Using crystal violet plate assays, it was shown that K. pneumoniae IApc35 mrkI::Knr has a decreased ability to form a biofilm on plastic surfaces compared to that of the parental strain. When the cloned mrkI gene was reintroduced into the IApc35 mrkI::Knr strain, full restoration of biofilm formation was observed, as shown in Fig. 4A. Also, using these assays, the IApc35 mrkI::Knr mutant transformed with an empty vector had a significantly decreased ability to form biofilms, whereas the parental IApc35 strain carrying the same plasmid is a biofilm producer. Likewise, as shown in Fig. 4B, the mrkHI deletion mutant exhibited a significantly reduced (approximately 7-fold) ability to form a biofilm compared to that exhibited by the parental strain. Restoration of biofilm formation was achieved by complementation with the cloned mrkHI determinants, and such transformants also exhibited an increased ability to form biofilms compared to that exhibited by the parental strain (approximately 2-fold). Transformants of the MrkHI mutant possessing an empty cloning vector did not demonstrate biofilm formation.
Fig. 4.
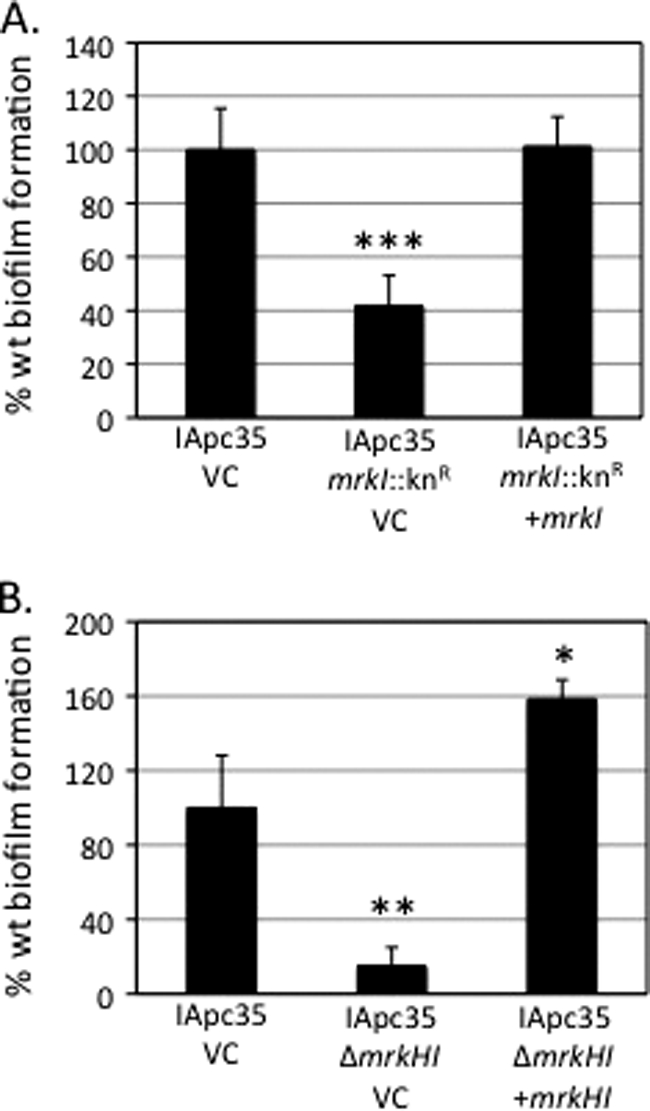
Biofilm phenotypes of K. pneumoniae strains. (A) Biofilm formation of parental IApc35 and the MrkI mutant carrying the empty vector (VC) compared to that of the complemented MrkI mutant on an abiotic surface. (B) Decreased biofilm formation of the IApc35 MrkHI mutant compared to those of parental IApc35 and the MrkHI mutant complemented with plasmid-borne mrkHI. Statistical significance was determined using Student's t test (*, P value < 0.05; **, P value < 0.01; ***, P value < 0.001).
Mutation of a conserved PilZ residue results in the inability to induce type 3 fimbria production.
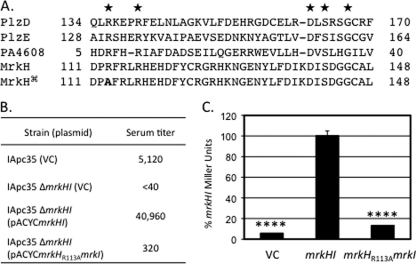
Alignment of the PilZ domain in MrkH with other PilZ domain-containing proteins revealed complete conservation of five residues which have previously been shown to be important in the ability of the PilZ domain to bind c-di-GMP (27) (Fig. 5A). Replacement of the conserved arginine-113 with alanine on a plasmid-borne copy of mrkH was performed and confirmed by nucleotide sequencing. Introduction of this plasmid, pACYCmrkHR113AmrkI, into IApc35 ΔmrkHI did not restore type 3 fimbrial expression compared to that observed using the parental pACYCmrkHI plasmid (Fig. 5B). This significant reduction in type 3 fimbrial production was further investigated using the reporter plasmid pTrc99APmrkA-lacZ in an E. coli background. When the parental plasmid pACYCmrkHI was introduced into this E. coli strain, an increase in β-galactosidase production was observed compared to that observed in the strain carrying the empty vector (pACYC184ΔCmr), as shown in Fig. 5C. In contrast, when pACYCmrkHR113AmrkI transformants were assayed, a significant decrease in β-galactosidase activity was observed compared to that of the parental plasmid (Fig. 5C).
Fig. 5.
Analysis of the R113A mutation in MrkH. (A) Alignment of previously characterized PilZ domain-containing proteins with MrkH. Conserved residues shown to affect c-di-GMP binding are indicated with stars. The alignment of the R113A mutant is also indicated (). (B) Effect of the R113A mutant on type 3 fimbria production. Values are reciprocals of serum titers needed to cause visible agglutination. (C) Use of a PmrkA-lacZ fusion to examine the ability of MrkHI to induce expression in E. coli transformants. Statistical significance was determined using Student's t test (****, P value < 0.0001).
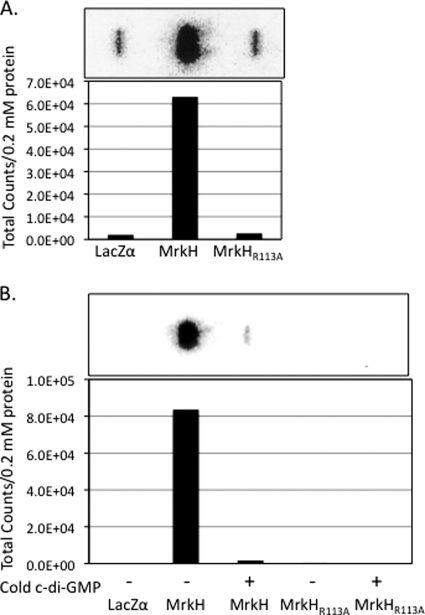
MrkH binds c-di-GMP.
To determine whether MrkH is capable of binding, c-di-GMP filter binding assays were used as previously described (9). Purification of 6×His-tagged MrkH and MrkH(R113A) was performed, and protein purity was determined by Western blot analysis. Equimolar amounts of MrkH proteins immobilized on nitrocellulose were used in the c-di-GMP binding assays. Bound [32P]c-di-GMP was determined by phosphorimaging, and those results are shown in Fig. 6. MrkH bound radiolabeled c-di-GMP, whereas the MrkH(R113A) protein did not (Fig. 6A). Inhibition of MrkH-mediated binding of labeled c-di-GMP was achieved by competition with an unlabeled nucleotide (Fig. 6B). Additionally, binding assays were also performed using [α-32P]GTP, but neither MrkH nor MrkH(R113A) bound this nucleotide (data not shown).
Fig. 6.
Ability of MrkH to bind [32P]c-di-GMP. (A) Filter binding assay using purified LacZα, MrkH, and MrkH(R113A) proteins as targets for binding. Graph represents total specific counts detected from assays represented above the graph. (B) Filter binding assays of binding reactions with (+) or without (−) the addition of unlabeled c-di-GMP.
MrkH and MrkHI activate the mrkA promoter.
To examine whether MrkH, MrkI, or MrkHI were sufficient to induce transcription from the mrkA promoter, β-galactosidase assays were used. Plasmids comprised of the same vector backbone, carrying either mrkH or mrkI alone or mrkHI together, were introduced into E. coli NEB 5-α transformed with a PmrkA-lacZ reporter fusion. The strain carrying both the reporter fusion and mrkH alone was found to exhibit a significant increase (approximately 114-fold) in transcriptional activity from the mrkA promoter compared to that exhibited by a transformant possessing the cloning vector alone. When mrkI alone was introduced into the strain carrying the reporter plasmid, no increase in β-galactosidase production was seen compared to that exhibited by transformants without mrkI. When a plasmid carrying both mrkH and mrkI was transformed into the reporter strain, a significant increase in mrkA transcription, compared to that exhibited by the strain carrying mrkH alone, was observed (approximately 8-fold) (Fig. 7).
Fig. 7.
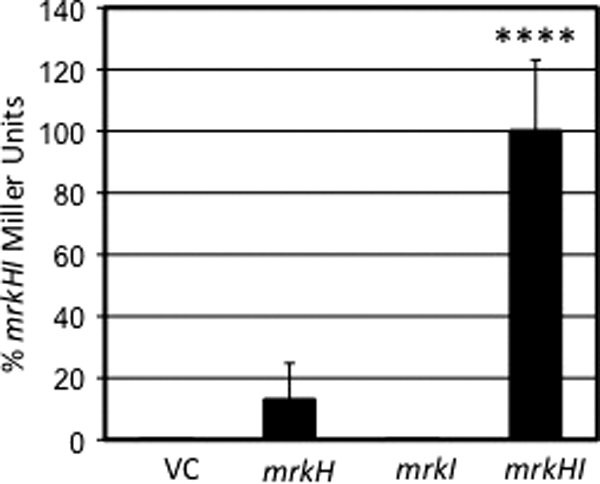
Ability of cloned mrkH, mrkI, and mrkHI to induce transcription of a PmrkA-lacZ reporter in E. coli compared to that of a vector control (VC). Statistical significance was determined using Student's t test (****, P value < 0.0001).
Similarly qRT-PCR analysis of K. pneumoniae IApc35 ΔmrkHI transformed with the mrkI, mrkH, and mrkHI genes also indicated that MrkH but not MrkI could affect mrkA expression. The MrkHI mutant transformed with mrkI alone did not exhibit any increase in mrkA expression compared to that exhibited by mutants transformed with the empty vector. However, transformation with a plasmid bearing the mrkHI genes resulted in a 36-fold increase in mrkA transcription compared to that seen with MrkH alone (P < 0.001).
DISCUSSION
K. pneumoniae type 3 fimbriae play an important role in the ability of the bacteria to bind to, and subsequently form biofilms on, HECM-coated surfaces. Both the fimbrial adhesin (MrkD) and the polymerized fimbrial shaft protein (MrkA) play important roles in this function (12, 13, 16, 30). We have previously proposed that MrkD facilitates the adherence of the organism to specific collagen molecules that form part of the HECM. However, fimbriate bacteria that possess no functional adhesin also form biofilms on abiotic surfaces (12, 14, 16). Consequently, the production of type 3 fimbriae could lead to the initiation of biofilm formation on inserted devices such as catheters shortly after insertion and also after these devices become coated in situ with host factors. The genetic regulation of mrk gene expression is poorly understood but, like other enterobacterial fimbrial systems, is likely to involve complex regulatory circuits.
In order to identify regulatory elements of the type 3 fimbrial operon, we constructed a mini-Tn5 transposon library in K. pneumoniae IApc35. This strain is a plasmid-cured derivative of the clinical isolate K. pneumoniae IA565, produces high levels of type 3 fimbriae, and forms robust biofilms on abiotic surfaces (12). It possesses only one chromosomally borne copy of the mrk gene cluster. One nonfimbriate mutant from this library possessed a transposon insertion within a gene encoding a putative transcriptional regulator, which we have previously termed mrkI (14). This gene is predicted to encode, by comparison to families of functional proteins, a protein which contains only one identifiable domain, a LuxR-like DNA binding domain in its C-terminal region. The N-terminal region of MrkI exhibits little relatedness to any characterized protein domains and therefore has no readily identifiable receiver domain. Interestingly, mrkI is located between two genes, as follows: the first, which we have named mrkH, is predicted to encode a protein which contains a C-terminal c-di-GMP binding domain (PilZ), and the second, mrkJ, is a gene which we have previously shown to produce a functional phosphodiesterase which modulates the intracellular levels of c-di-GMP within K. pneumoniae (14). A defined MrkI mutant of strain IApc35 was constructed and, like the original transposon mutant, was found to be unable to assemble type 3 fimbriae. Also, we found that mrkI is cotranscribed with mrkH. This is consistent with the observation that the only promoter identified by sequence analysis, which is likely to drive mrkI transcription, lies upstream of mrkH. In addition to mrkHI cotranscription, we also found that mrkJ transcription can also occur from the mrkH promoter, though the mrkI and mrkHI mutations were not found to significantly alter the levels of mrkJ transcription (data not shown). Therefore, it is possible that transcription of mrkJ can also occur from a promoter immediately upstream of it. Deletion of mrkHI, like the single mrkI mutation, resulted in the decreased ability of K. pneumoniae to produce surface-associated type 3 fimbriae. Repeated attempts were made to construct an mrkH deletion mutant but proved unsuccessful. The precise reason for this is unclear but suggests that such a mutation may be lethal, even though deletion of mrkHI together and reintroduction of mrkI alone is not.
Interestingly, we also demonstrated that the MrkI mutant was nonfimbriate only when cultured under aerobic conditions. When these strains were grown anaerobically on agar or microaerophilically as deep static broth cultures, the mutants exhibited fimbrial titers equivalent to or higher than those observed for the parental strains grown aerobically. The MrkHI mutant, in contrast, was consistently nonfimbriate under both aerobic and anaerobic conditions. It is possible that the MrkI mutant is fimbriate when grown anaerobically due to increased expression of mrkH under these conditions, which facilitates fimbria production independently of MrkI. Consequently, during anaerobic growth, MrkH and MrkI are likely to facilitate increased mrkA expression, resulting in a strongly fimbriate phenotype. However, in the absence of MrkI, the increased MrkH production, compared to that exhibited by bacteria grown aerobically, may enable MrkH to interact with an orphan activator to facilitate mrkA transcription. Also, it is possible that K. pneumoniae, in response to different environmental conditions, produces different regulators that can interact with MrkH to modulate surface expression of type 3 fimbriae. Currently, we are investigating the interaction between MrkH and MrkI in the presence and absence of c-di-GMP. However, MrkH may act as a protein with a more general function of sensing the intracellular concentrations of c-di-GMP. This is supported by the observation that unlike many organisms in which c-di-GMP serves a regulatory role, all sequenced K. pneumoniae genomes (K. pneumoniae MGH78578, K. pneumoniae NTUH-K2044, and K. pneumoniae 342) possess only one PilZ domain-containing protein (MrkH) that is not predicted to act as a cellulose synthase. This is not unique within the Enterobacteriaceae, as it appears that many members of this family contain only the PilZ domain carrying protein YcgR. However, the previously described N-terminal YcgR domain of these proteins exhibits no relatedness to that of MrkH. Currently, no other c-di-GMP binding proteins have been characterized in K. pneumoniae, so it is possible that c-di-GMP sensing is a major function of MrkH.
The ability of MrkH and MrkI to facilitate transcription of mrkA in an E. coli transformant that possesses no mrk genes of its own and the DNA binding domain present in MrkI led us to speculate that MrkI binds the promoter region of mrkA. However, we were not able to demonstrate binding in vitro using gel mobility shift assays (data not shown). The fact that MrkA can be made in the absence of MrkI under specific growth conditions makes it less surprising that it was not possible to demonstrate this interaction. However, it is possible that MrkI binds to this region, and we were unable to replicate in vitro the conditions for binding in vivo. Additional bacterial factors may be required for this activity. The results shown by E. coli transformants are consistent with the observation that MrkH and MrkI affect transcription, as detected by qRT-PCR in K. pneumoniae. The presence of MrkH alone facilitates detectable levels of mrkA transcription in both E. coli and K. pneumoniae transformants, but this is significantly lower than that observed when both MrkH and MrkI are present. The MrkI mutant of K. pneumoniae is able to produce MrkH, but this mutant is phenotypically nonfimbriate and exhibits no mrkA transcription. The level of mrkA transcription in E. coli transformants possessing only MrkH could simply be due to the relatively high concentrations of MrkH produced by the cloned gene. Since analysis of the K. pneumoniae genome indicates that the only PilZ-possessing protein in these bacteria that is not involved in cellulose metabolism is MrkH, it is possible that this protein acts as a c-di-GMP signaling adaptor for many systems that are regulated by the intracellular concentrations of the molecule. Consequently, its effect on gene transcription may depend on the intracellular concentrations of MrkH. In the absence of MrkI, high levels of MrkH may more weakly interact, directly or indirectly, with additional regulators. Further studies will be required to investigate this hypothesis.
It is becoming increasingly clear that the genetic regulation of fimbrial genes encoding appendages assembled by the chaperone-usher pathway is subject to a complex regulatory circuit involving different families of DNA binding proteins (1, 18, 21). The type 3 fimbrial system, a fimbrial type commonly observed to be produced by enterobacteria associated with nosocomially acquired infections, is also likely to be regulated by multiple gene products. The identification of these regulatory factors will facilitate an understanding of type 3 fimbria production and its role in host cell interaction. Due to the multitiered regulatory networks that govern other fimbrial systems, it is possible that MrkH and MrkI regulate type 3 fimbrial expression by acting upstream of a primary regulator. However, the location of mrkHIJ immediately adjacent to mrkABCDF may indicate an evolutionary selection for these two gene clusters. Currently, efforts are under way to further investigate the role of both MrkH and MrkI as fimbrial regulators and also to determine whether MrkI regulates expression of nonfimbrial genes.
ACKNOWLEDGMENTS
We thank Carrie Harwood (University of Washington, Seattle, WA) for kindly providing the WspR protein to synthesize labeled c-di-GMP.
This work was supported by a grant from the NIH to S.C. (R01 AI050011). J.G.J. was supported by the NIH Mechanisms of Parasitism Training Grant (T32 AI007511).
Footnotes
Published ahead of print on 13 May 2011.
REFERENCES
- 1. Blomfield I. C. 2001. The regulation of pap and type 1 fimbriation in Escherichia coli. Adv. Microb. Physiol. 45:1–49 [DOI] [PubMed] [Google Scholar]
- 2. Boddicker J. D., Anderson R. A., Jagnow J., Clegg S. 2006. Signature-tagged mutagenesis of Klebsiella pneumoniae to identify genes that influence biofilm formation on extracellular matrix material. Infect. Immun. 74:4590–4597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de Lorenzo V., Timmis K. N. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386–405 [DOI] [PubMed] [Google Scholar]
- 4. Dickson C. 2008. Protein techniques: immunoprecipitation, in vitro kinase assays, and Western blotting. Methods Mol. Biol. 461:735–744 [DOI] [PubMed] [Google Scholar]
- 5. Frank D. N., Wilson S. S., St. Amand A. L., Pace N. R. 2009. Culture-independent microbiological analysis of foley urinary catheter biofilms. PLoS One 4:e7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gerlach G. F., Allen B. L., Clegg S. 1988. Molecular characterization of the type 3 (MR/K) fimbriae of Klebsiella pneumoniae. J. Bacteriol. 170:3547–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gerlach G. F., Allen B. L., Clegg S. 1989. Type 3 fimbriae among enterobacteria and the ability of spermidine to inhibit MR/K hemagglutination. Infect. Immun. 57:219–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gerlach G. F., Clegg S., Allen B. L. 1989. Identification and characterization of the genes encoding the type 3 and type 1 fimbrial adhesins of Klebsiella pneumoniae. J. Bacteriol. 171:1262–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hickman J. W., Harwood C. S. 2008. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 69:376–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hickman J. W., Tifrea D. F., Harwood C. S. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. U. S. A. 102:14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hornick D. B., Allen B. L., Horn M. A., Clegg S. 1992. Adherence to respiratory epithelia by recombinant Escherichia coli expressing Klebsiella pneumoniae type 3 fimbrial gene products. Infect. Immun. 60:1577–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hornick D. B., Thommandru J., Smits W., Clegg S. 1995. Adherence properties of an mrkD-negative mutant of Klebsiella pneumoniae. Infect. Immun. 63:2026–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jagnow J., Clegg S. 2003. Klebsiella pneumoniae MrkD-mediated biofilm formation on extracellular matrix- and collagen-coated surfaces. Microbiology 149:2397–2405 [DOI] [PubMed] [Google Scholar]
- 14. Johnson J. G., Clegg S. 2010. Role of MrkJ, a phosphodiesterase, in type 3 fimbrial expression and biofilm formation in Klebsiella pneumoniae. J. Bacteriol. 192:3944–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jones R. N. 2010. Microbial etiologies of hospital-acquired bacterial pneumonia and ventilator-associated bacterial pneumonia. Clin. Infect. Dis. 51(Suppl. 1):S81–S87 [DOI] [PubMed] [Google Scholar]
- 16. Langstraat J., Bohse M., Clegg S. 2001. Type 3 fimbrial shaft (MrkA) of Klebsiella pneumoniae, but not the fimbrial adhesin (MrkD), facilitates biofilm formation. Infect. Immun. 69:5805–5812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Link A. J., Phillips D., Church G. M. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228–6237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mikkelsen H., Ball G., Giraud C., Filloux A. 2009. Expression of Pseudomonas aeruginosa CupD fimbrial genes is antagonistically controlled by RcsB and the EAL-containing PvrR response regulators. PLoS One 4:e6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Miller V. L., Mekalanos J. J. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moravek M., et al. 2004. Colony immunoblot assay for the detection of hemolysin BL enterotoxin producing Bacillus cereus. FEMS Microbiol. Lett. 238:107–113 [DOI] [PubMed] [Google Scholar]
- 21. Nicastro G. G., Boechat A. L., Abe C. M., Kaihami G. H., Baldini R. L. 2009. Pseudomonas aeruginosa PA14 cupD transcription is activated by the RcsB response regulator, but repressed by its putative cognate sensor RcsC. FEMS Microbiol. Lett. 301:115–123 [DOI] [PubMed] [Google Scholar]
- 22. Nordmann P., Cuzon G., Naas T. 2009. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 9:228–236 [DOI] [PubMed] [Google Scholar]
- 23. Ohana S., et al. 2005. Spread of a Klebsiella pneumoniae strain producing a plasmid-mediated ACC-1 AmpC beta-lactamase in a teaching hospital admitting disabled patients. Antimicrob. Agents Chemother. 49:2095–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. O'Toole G. A., Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295–304 [DOI] [PubMed] [Google Scholar]
- 25. O'Toole G. A., et al. 1999. Genetic approaches to study of biofilms. Methods Enzymol. 310:91–109 [DOI] [PubMed] [Google Scholar]
- 26. Philippe N., Alcaraz J. P., Coursange E., Geiselmann J., Schneider D. 2004. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51:246–255 [DOI] [PubMed] [Google Scholar]
- 27. Pratt J. T., Tamayo R., Tischler A. D., Camilli A. 2007. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J. Biol. Chem. 282:12860–12870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ronald A. 2002. The etiology of urinary tract infection: traditional and emerging pathogens. Am. J. Med. 113(Suppl. 1A):14S–19S [DOI] [PubMed] [Google Scholar]
- 29. Rosen D. A., et al. 2008. Utilization of an intracellular bacterial community pathway in Klebsiella pneumoniae urinary tract infection and the effects of FimK on type 1 pilus expression. Infect. Immun. 76:3337–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schurtz T. A., Hornick D. B., Korhonen T. K., Clegg S. 1994. The type 3 fimbrial adhesin gene (mrkD) of Klebsiella species is not conserved among all fimbriate strains. Infect. Immun. 62:4186–4191 [DOI] [PMC free article] [PubMed] [Google Scholar]