Abstract
Studies of the Escherichia, Neisseria, Thermotoga, and Mycobacteria clustered regularly interspaced short palindromic repeat (CRISPR) subtypes have resulted in a model whereby CRISPRs function as a defense system against bacteriophage infection and conjugative plasmid transfer. In contrast, we previously showed that the Yersinia-subtype CRISPR region of Pseudomonas aeruginosa strain UCBPP-PA14 plays no detectable role in viral immunity but instead is required for bacteriophage DMS3-dependent inhibition of biofilm formation by P. aeruginosa. The goal of this study is to define the components of the Yersinia-subtype CRISPR region required to mediate this bacteriophage-host interaction. We show that the Yersinia-subtype-specific CRISPR-associated (Cas) proteins Csy4 and Csy2 are essential for small CRISPR RNA (crRNA) production in vivo, while the Csy1 and Csy3 proteins are not absolutely required for production of these small RNAs. Further, we present evidence that the core Cas protein Cas3 functions downstream of small crRNA production and that this protein requires functional HD (predicted phosphohydrolase) and DEXD/H (predicted helicase) domains to suppress biofilm formation in DMS3 lysogens. We also determined that only spacer 1, which is not identical to any region of the DMS3 genome, mediates the CRISPR-dependent loss of biofilm formation. Our evidence suggests that gene 42 of phage DMS3 (DMS3-42) is targeted by CRISPR2 spacer 1 and that this targeting tolerates multiple point mutations between the spacer and DMS3-42 target sequence. This work demonstrates how the interaction between P. aeruginosa strain UCBPP-PA14 and bacteriophage DMS3 can be used to further our understanding of the diverse roles of CRISPR system function in bacteria.
INTRODUCTION
Clustered regularly interspaced short palindromic repeats (CRISPRs) are important for the interaction of bacteria with foreign genetic material such as bacteriophage and conjugative plasmids (1, 16). CRISPRs are transcribed as long pre-CRISPR RNAs that are processed into small CRISPR RNAs (crRNAs) by protein products of CRISPR-associated (cas) genes (3, 11). These small crRNAs in association with select Cas proteins have been shown to mediate resistance to bacteriophage infection and plasmid inheritance in bacteria, an observation that has led to the CRISPR systems being labeled as bacterial adaptive immune systems (1, 16).
A CRISPR region is comprised of a locus encoding the precursor CRISPR RNA as well as a suite of cas genes. There have been 8 CRISPR region subtypes identified to date, namely, Escherichia, Yersinia, Neisseria, Desulfovibrio, Thermotoga, Haloarcula, Aeropyrum, and Mycobacteria, each of which has a distinctive CRISPR repeat sequence and at least 2 cas genes found only in that subtype (abbreviated csx, where x denotes the first letter of the subtype) (23). The ubiquitous cas gene, cas1, is the only CRISPR-associated gene found in all subtypes, and it is hypothesized to function in the acquisition of new sequences into CRISPRs (25). There are additional cas genes that are shared between some subtypes, termed “core” cas genes (23). Together, the diversity of subtype-specific and core Cas proteins encoded by CRISPR regions raises the question of whether CRISPRs may play roles in microbial biology other than resistance to invasion by foreign genetic elements. Indeed, there is a growing body of evidence that not all CRISPR regions are capable of conferring resistance to foreign DNA elements (5, 8, 19, 24, 27).
We previously reported that the Yersinia-subtype CRISPR of Pseudomonas aeruginosa strain UCBPP-PA14 (PA14) does not confer any detectable resistance to bacteriophage infection (5) but is instead required for altering group behaviors when this microbe is lysogenized by bacteriophage DMS3 (27). In this previous work, we provided evidence that the cause of biofilm inhibition was not the alteration of growth or the initiation of the lytic cycle by bacteriophage DMS3 (27). Here, we characterize the requirement for components of the CRISPR region in bacteriophage DMS3-dependent inhibition of biofilm formation by P. aeruginosa PA14 and assess the role of the Cas proteins in this bacteriophage-host interaction. Furthermore, we present the first example of a non-identity-mediated interaction between a spacer and bacteriophage that results in a biologically meaningful output. Finally, this is the first report to characterize the target sequence required for a spacer to interact with a chromosomally integrated bacteriophage. In summary, this work dissects the in vivo contributions of each component of the prevalent Yersinia-subtype CRISPR region by using a bacterium-bacteriophage interaction that impacts biofilm formation as the functional readout.
MATERIALS AND METHODS
Strains and media.
Strains, plasmids, and primers used in this study are listed in Table S1 in the supplemental material. The P. aeruginosa strain UCBPP-PA14 (abbreviated P. aeruginosa PA14) was used in this study. P. aeruginosa and Escherichia coli strains were routinely cultured in lysogeny broth (LB) at 37°C. The minimal medium used was M63 supplemented with MgSO4 (1 mM) and arginine (0.4%). Growth media were supplemented with antibiotics at the following concentrations: ampicillin (Ap), 150 μg ml−1 (E. coli); gentamicin (Gm), 10 μg ml−1 (E. coli) and 50 μg ml−1 (P. aeruginosa); and carbenicillin, 50 μg ml−1 (E. coli) and 500 μg ml−1 (P. aeruginosa).
Generating lysogens of P. aeruginosa.
Bacteriophage DMS3 was purified from a lysogenized P. aeruginosa PA14 strain (SMC3884) by passage of an LB-grown culture through a 0.22-μm filter (Millipore, Billerica, MA). Lysogenic strains were created by incubation of 10 μl of purified DMS3 with 10 μl of wild-type (WT) P. aeruginosa PA14 (or mutant) in 500 μl of LB for 2 h at 37°C with shaking. Following incubation, cultures were struck to single colonies on LB agar plates and incubated at 37°C overnight. Single colonies were picked and tested for phage production using the plaque assay described below.
Plaque assay.
DMS3 bacteriophage production was determined using a plaque assay, as described by Budzik et al. (4). Briefly, 100 μl of P. aeruginosa PA14 was added to 3 ml of molten top agar (0.8%) and poured over a prewarmed LB agar plate. Strains to be tested for DMS3 production were grown overnight at 37°C in LB and filter sterilized using a 0.22-μm filter. After solidification of top agar lawns, 5-μl portions of serially diluted filter-sterilized control and test lysates were spotted onto the top agar lawn and incubated at 37°C overnight. Plaques were counted and expressed as numbers of PFU/ml.
Static biofilm assay and quantification.
Biofilm formation at 24 h on polyvinyl chloride (PVC) plastic was assayed essentially as previously described by O'Toole and Kolter (17), except that M63 was supplemented with 0.4% arginine and 1 mM MgSO4. Quantification of biofilm formation was performed as follows: crystal violet (CV) stain was solubilized from PVC-attached cells by using 150 μl of acetic acid (30% in water) per microtiter dish well. After incubation at room temperature for ∼10 min, 100 μl of the acetic acid-solubilized CV was transferred into a fresh, optically clear, flat-bottom microtiter plate and the absorbance measured at 550 nm on a Spectra Max M2 microplate reader (Molecular Devices, Sunnyvale, CA).
Strain construction.
The construction of the strains used in this study is detailed below.
(i) In-frame deletion mutants.
In-frame deletions of the csy3, csy2, csy1, cas3, and cas1 genes and DMS3 gene 42 (designated DMS3-42) were created using constructs pMQ30-Δcsy3, pMQ30-Δcsy2, pMQ30-Δcsy1, pMQ30-Δcas1, pMQ30-Δcas3, and pMQ30-ΔDMS3-42. These constructs were created utilizing the Saccharomyces cerevisiae recombineering technique described by Shanks et al. (21). Constructs were electroporated into E. coli and analyzed by colony PCR or sequencing. Plasmids were propagated in E. coli S17 and conjugated into P. aeruginosa PA14 as previously reported (13). Exconjugants containing an inserted plasmid were selected on gentamicin before counterselection on 5% sucrose. All mutations were confirmed via PCR amplification and sequencing of the mutated region.
(ii) Allelic replacement complementation of Δcas3 mutation.
Allelic replacement complementation of the Δcas3 mutation was performed using constructs pMQ30-cas3 KON (“KON” stands for “knock-on” and distinguishes plasmids used to introduce mutations onto the chromosome from complementation plasmids with similar names), pMQ30-cas3(D124A) KON, and pMQ30-cas3(D576A) KON. Constructs and strains were created in a manner similar to in-frame deletions, except that WT or mutant cas3 was located between the regions of homology.
(iii) Allelic replacement complementation of ΔDMS3-42 mutation.
Allelic replacement complementation of the ΔDMS3-42 mutation was performed similarly to that of the Δcas3 mutation, except constructs pMQ30-DMS3-42 KON, pMQ30-DMS3-42(T2G) KON, pMQ30-DMS3-42(C234T) KON, pMQ30-DMS3-42(G235T) KON, pMQ30-DMS3-42(C238T) KON, pMQ30-DMS3-42(C238G) KON, pMQ30-DMS3-42(C243T) KON, pMQ30-DMS3-42(C246T) KON, pMQ30-DMS3-42(C249T) KON, pMQ30-DMS3-42(C252T) KON, pMQ30-DMS3-42(T254C) KON, pMQ30-DMS3-42(C256A C258A) KON, pMQ30-DMS3-42(G261T) KON, pMQ30-DMS3-42(A262T) KON, pMQ30-DMS3-42(C663T) KON, and pMQ30-DMS3-42(G681T) KON were utilized. Constructs and strains were created in a manner similar to in-frame deletions, except that WT or mutant DMS3-42 was located between the regions of homology. The loss of DMS3-42 gene function resulted in a complete loss of phage DMS3 production, and the successful complementation of ΔDMS3-42 mutants was assayed by the renewed production of DMS3 bacteriophage, as measured by plaque assay.
(iv) Allelic replacement complementation of the ΔCRISPR2 mutation.
Allelic replacement complementation of the ΔCRISPR2 mutation was performed using constructs pMQ30-CRISPR2 KON, pMQ30-Δspacer20 KON, pMQ30-Δspacer17 KON, pMQ30-Δspacers7-21 KON, pMQ30-Δspacers2-21 KON, pMQ30-Δspacers1-21 KON, pMQ30-Δspacers1-2 KON, pMQ30-spacers1 SalI KON, and pMQ30-spacer 1 BamHI KON. S. cerevisiae recombineering techniques were also used to make these constructs. Primers CRISPR2 KON for and CRISPR2 KON rev were used in conjunction with mutation-specific forward and reverse primers, which annealed within the CRISPR region (see Fig. S1 in the supplemental material). The only exception to this was pMQ30-CRISPR2 KON, which utilized only CRISPR2 KON for and CRISPR2 KON rev primer set (see Fig. S1). Internal primers contained sequence identity to the spacer and repeat to the left or right of the desired mutation, allowing for specificity of amplification. Internal spacers for all deletion constructs contained the full sequence of P. aeruginosa strain PA2192 CRISPR1 spacer 8 (CGC ATT GCC AGG ATC AGC ACG TCG ACC ATC TT) at one end of the primer, while point mutation primers harbored the desired point mutations within the spacer 1 sequence. The PA2192 spacer contained a SalI restriction site, simplifying the identification of properly recombineered plasmids. When these primers were utilized in PCR, we obtained 2 products, each with sequence identity either upstream or downstream of CRISPR2 (see Fig. S1). These products recombined with each other at the PA2192 spacer sequence and at the vector pMQ30, creating the desired CRISPR2 allelic replacement construct. A diagram of the PCR products generated for representative CRISPR2 mutations is shown in Fig. S1.
(v) Extrachromosomal complementation constructs.
Extrachromosomal complementation constructs pMQ70-csy4His, pMQ70-csy4His(H29A), pMQ70-csy4His(H29K), pMQ70-E. coli csy4His, pMQ70-E. coli csy4His(H29A), pMQ70-csy3, pMQ70-csy2, pMQ70-csy1, pMQ70-cas3, pMQ70-cas3(D124A), and pMQ70-cas3(D576A) were created using recombineering as previously described (21). The pMQ70 derivative plasmid pDPM73 (14) was utilized as the vector and digested with XhoI and EcoRI (New England BioLabs, Beverly, MA). Forward and reverse primers (see Table S1 in the supplemental material) were used to PCR amplify wild-type and mutant genes, which were then utilized in yeast recombination reactions to form complementation constructs. Plasmids were purified from yeast cells, electroporated into E. coli Top10 cells (Invitrogen, Carlsbad, CA), prepared using the Qiagen spin miniprep kit (Valencia, CA), and electroporated into P. aeruginosa PA14 strains as previously described (6). Plasmid selection was maintained with 50 μg ml−1 carbenicillin both on LB plates and in LB liquid cultures.
Northern blot analysis.
Northern blot assays were performed to determine the ability of various deletion and point mutations to process the native full-length CRISPR transcript into small crRNAs in vivo. Cultures grown overnight in LB medium either supplemented with carbenicillin and arabinose (harboring complementation constructs) or not supplemented (deletion or chromosomal allelic replacement strains) were pelleted and resuspended in 100 μl of TE (Tris-EDTA) buffer containing 400 μg/ml lysozyme for 3 to 5 min at room temperature. Following lysozyme-mediated cell lysis, total RNA was purified using a mirVana microRNA (miRNA) isolation kit (Ambion, Austin, TX) by essentially following the manufacturer's instructions for total RNA extraction. Two 10% TBE (Tris-borate-EDTA)-urea polyacrylamide gels (Bio-Rad, Hercules, CA) were loaded with 10 μg of total RNA per sample lane and resolved at 50 V. Following electrophoresis, one gel was transferred to a nylon membrane (Thermo Scientific, Rockford, IL) for Northern analysis while the other was stained with ethidium bromide and photographed under UV light to serve as an additional loading control. Primers Northern Marker/probe For (TGG ATT CAT GTA TCA GGC ATC GTA CTC GGC AAT TTC TTA GCT GCC TAC ACG GC) and Northern Marker/probe Rev2 (ATC CTC GAA CAG CCC GGG TTC GGT CGC CTC CAG TTC ACT GC) were utilized in PCRs to synthesize a probe comprising the last 4 spacers and 3 repeats of P. aeruginosa PA14 CRISPR2. The random primed DNA labeling kit (Roche, Indianapolis, IN) and [α-32P]dCTP (3,000 Ci mmol−1; PerkinElmer, Waltham, MA) were utilized according to the manufacturer's specifications to radioactively label the probe. mirVana-suggested small RNA (sRNA) prehybridization, hybridization, and wash solutions were utilized at 65°C, 37°C, and 37°C respectively. Under these hybridization conditions, this probe hybridized with the conserved repeat region of all CRISPR-derived small crRNAs regardless of spacer sequence. Following the third 15-min wash, membranes were covered with plastic wrap and exposed to a storage phosphor screen (GE Healthcare, Piscataway, NJ) overnight. After exposure, phosphor screens were read on a Storm 860 (Molecular Devices, Sunnyvale, CA) and processed using Image Quant software v5.2 (Molecular Devices, Sunnyvale, CA).
Detection of Cas3 and penta-His-tagged Cas proteins.
The following section describes the methods used in the Western blot studies.
(i) Penta-His-tagged-protein detection.
C-terminal penta-His epitope tag-fused Csy4 and E. coli Csy4, Csy2, and Csy1 proteins were expressed under an arabinose-inducible pBad promoter. To detect the level of His-tagged proteins, cultures were grown with shaking overnight in 5 ml of LB medium containing 50 μg ml−1 carbenicillin and 0.2% arabinose. Overnight cultures (100 μl) were boiled in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer and resolved using SDS-PAGE. His-tagged Csy4 was detected via Western blot analysis using penta-His antibody (Qiagen, Valencia, CA), per the manufacturer's instructions.
(ii) Cas3 level determination.
Cas3 levels were determined after overnight growth in M63 minimal medium supplemented with 0.4% arginine and 1 mM MgSO4. M63 medium was utilized because less background was observed after growth in minimal medium than after that in LB medium. Similar to Western blot analyses detecting the His tag epitope, 100-μl portions of overnight cultures were boiled in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer and resolved using SDS-PAGE. Cas3 proteins were detected via Western blot analysis using polyclonal rabbit antibodies against Cas3 (Josman Inc., Napa, CA).
RESULTS
P. aeruginosa cas genes involved in bacteriophage DMS3-mediated biofilm inhibition.
The laboratory strain P. aeruginosa PA14 harbors two repeat-rich CRISPR loci flanking a series of 6 cas genes. Four of these cas genes are unique to Yersinia-subtype CRISPR regions, while two are core cas genes found in multiple subtypes (Fig. 1A). We previously reported that infection of P. aeruginosa PA14 with lysogenic bacteriophage DMS3 results in disruption of biofilm formation and that the biofilm phenotype is restored in lysogenic strains with gene disruptions in either CRISPR2, csy1, csy2, csy3, csy4, or cas3 (27). To further dissect this bacteriophage-host interaction and to determine the contribution of each component of the Yersinia-subtype CRISPR region to DMS3-mediated biofilm inhibition, we systematically deleted each of the components of the Csy-type CRISPR system.
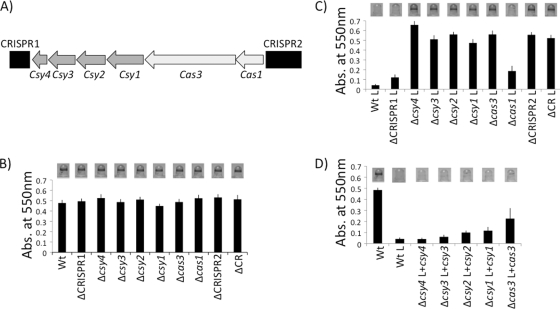
Fig. 1.
Deletion and complementation analysis of P. aeruginosa PA14 CRISPR region. (A) Chromosomal organization of the CRISPR genomic region, including the two CRISPRs and CRISPR-associated (cas) genes. CRISPRs are shown as black boxes, Yersinia-subtype cas genes are depicted as dark gray arrows, and core cas genes found in multiple CRISPR subtypes are diagrammed as light gray arrows. (B) Biofilm phenotypes of wild-type and deletion mutants in the absence of bacteriophage DMS3 (nonlysogens). A representative image of the biofilm formed by each strain is shown above the quantification of results for 8 independent wells. The ΔCRISPR1 strain has only CRISPR1 deleted, the ΔCRISPR2 strain has only CRISPR2 deleted, and the ΔCR strain has both CRISPRs and the cas genes they flank deleted. (C) Biofilm phenotypes of bacteriophage DMS3-infected (DMS3 lysogens) wild-type and mutant strains. A representative image of the biofilm formed by each strain is shown above the quantification of results for 8 independent wells. An “L” following a strain designation, in this panel and panel D, indicates a strain lysogenized with DMS3. (D) Complementation analysis of the csy4, csy3, csy2, csy1, and cas3 mutants in DMS3 lysogens. A representative image of the biofilm formed by each strain is shown above the quantification of results for 8 independent wells.
To clearly establish the role of each cas gene in this biofilm phenotype, we made in-frame deletions of each cas gene, complemented these deletions, and determined their biofilm phenotypes in nonlysogenized and lysogenized strain backgrounds (Fig. 1B to D). We also deleted each CRISPR or the entire CRISPR region, including both CRISPRs and all cas genes, and performed a similar analysis. Deletion of either CRISPR, any cas gene, or the entire CRISPR region has no effect on biofilm formation in the absence of bacteriophage DMS3 (i.e., nonlysogens) (Fig. 1B). This observation indicates that neither the cas genes nor either CRISPR is required for biofilm formation under these conditions.
When these mutants are lysogenized by bacteriophage DMS3, however, a biofilm-defective phenotype can be observed in some strains (Fig. 1C). As expected, the wild-type P. aeruginosa PA14 is unable to form a biofilm when lysogenized by DMS3 (27). Similarly, when lysogenized, the ΔCRISPR1 and Δcas1 mutant strains are also unable to form a biofilm, indicating that these loci do not contribute to the DMS3-dependent loss of biofilm formation. These findings are consistent with our previous study indicating that CRISPR1 and cas1 are dispensable for the DMS3-dependent biofilm phenotype (27). In contrast, when lysogenized by DMS3, the ΔCRISPR2, Δcsy1, Δcsy2, Δcsy3, Δcsy4, and Δcas3 mutant strains, as well as the strain lacking the entire region (ΔCR strain), regain the ability to form a biofilm (Fig. 1C, compare to the WT lysogen).
Each biofilm-positive cas mutant phenotype could be completely or partially complemented with plasmids expressing penta-His-tagged proteins (Fig. 1D). We noticed poor complementation of the Δcas3 mutant by the plasmid-borne cas3 gene (Fig. 1D, far right); however, as shown below, we could complement the cas3 mutation by replacing the mutant allele on the chromosome with the WT gene. A mutation in CRISPR2 could also be complemented by the reintroduction of the CRISPR at the native locus (see below).
Together, these data confirm and extend our previous findings (27) and allow us to address the contribution of each component of the CRISPR region to the inhibition of biofilm formation in the DMS3-lysogenized strains.
Requirement for cas gene products in small crRNA production in vivo.
Functional CRISPR regions are transcribed as a large RNA that is subsequently processed into mature small crRNAs by selected Cas proteins (Fig. 2A) (3, 10). To determine which cas gene products are involved in processing the CRISPR transcripts into small crRNAs in vivo, we utilized Northern blot analysis with a probe constituting the 4 spacers and 3 repeats closest to the cas1 gene (Fig. 2A, dashed black bar).
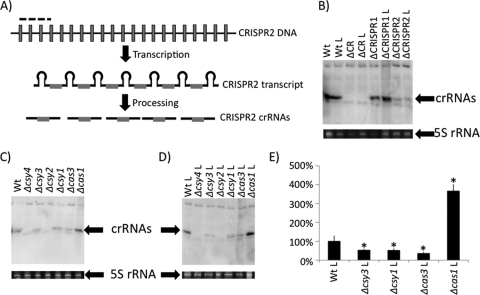
Fig. 2.
Characterization of small crRNA production by CRISPR region mutants. (A) Diagram showing CRISPR DNA transcribed into a long CRISPR transcript, which is processed into mature crRNAs by Cas proteins. The portion of CRISPR2 utilized as the probe for Northern blot analysis is denoted by a dashed line. Unique spacer sequences are shown as gray boxes, while repeat sequences are represented by black lines. (B) Detection of in vivo small crRNA production by WT, ΔCRISPR1, ΔCRISPR2, and ΔCRISPR region nonlysogens and lysogens by using Northern blot analysis. Also shown is the 5S rRNA band for each of the corresponding RNA preparations used in the Northern blot analysis. (C) Shown is a representative Northern blot (top) of crRNA production by each cas mutant in the absence of DMS3. Also shown is the 5S rRNA band for each of the corresponding RNA preparations used in the Northern blot analysis. (D) Shown is a representative Northern blot (top) of crRNA production by cas mutants in the presence of DMS3 (lysogens). Also shown is the 5S rRNA band for each of the corresponding RNA preparations used in the Northern blot analysis. (E) Quantification of representative data shown in panel D. Shown is an average calculated from three replicate experiments. The bars indicate standard deviation. *, Significantly different from the WT, P < 0.05. An “L” following the genotype designation indicates a strain lysogenized with DMS3.
The Northern assay probe, which contains both repeat and spacer sequences, cross-hybridizes specifically with the conserved repeat sequence retained in small crRNAs derived from the Yersinia-subtype CRISPR region (regardless of spacer content) as previously observed (5) (Fig. 2B, compare WT and ΔCR lanes). Furthermore, we can detect small crRNAs derived from both CRISPR1 and CRISPR2, although CRISPR2-derived small RNAs are either more prevalent or preferentially hybridize to this probe (Fig. 2B, compare WT, ΔCRISPR1, and ΔCRISPR2 lanes). Finally, comparison of crRNA production from the lysogenized and nonlysogenized strains indicates that levels of the crRNA do not change upon lysogeny by DMS3 (Fig. 2B).
Analysis of crRNA production in the individual cas mutant backgrounds revealed that only Δcsy4 and Δcsy2 were absolutely required for small crRNA production, although the Δcsy3, Δcsy1, and Δcas3 mutants showed significantly decreased levels of crRNAs compared to the WT (Fig. 2C to E).
While it was expected that csy4 would be required for crRNA production, because its product has been shown to have nuclease activity that can cleave CRISPR transcripts in vitro (11), our data also indicate the involvement of the csy1, csy2, and csy3 (csy1-3) and cas3 genes and their products in crRNA production. The prediction by others (23) that Csy1 to Csy4 (Csy1-4) may be forming a complex led us to assess the stability of Csy4 in each cas mutant background. To this end, we expressed penta-His-tagged Csy4 in each mutant background but observed no alteration in the stability of this nuclease in any cas or csy mutant (data not shown). These data indicate that the products of csy1-3 and cas3 genes may be involved in CRISPR transcript processing or crRNA stabilization independent of Csy4 stability.
Catalytic residues of Csy4 are required for CRISPR transcript processing in vivo.
A recent report by Doudna and colleagues showed the Csy4 protein could process full-length CRISPR RNA in vitro, and furthermore, they identified residues critical for this in vitro activity (11). The data presented in Fig. 2C and D and in Fig. 3, lanes 1 and 2, show that the Δcsy4 mutant lacks any detectable small crRNA production in vivo, consistent with the published biochemical studies (11).
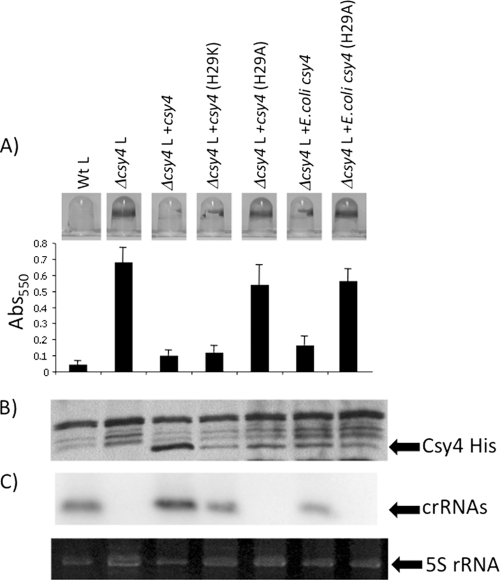
Fig. 3.
In vivo analysis of P. aeruginosa and E. coli Csy4. (A) Shown are the abilities of P. aeruginosa and E. coli plasmid-expressed WT and Csy4 mutants to complement the intact biofilm phenotype resulting from a Δcsy4 mutation in the presence of bacteriophage DMS3 in comparison to a WT lysogen. An “L” following the genotype designation indicates a strain lysogenized with DMS3. A representative image of the biofilm formed by each strain is shown above the quantification of results of 8 independent wells. (B) Western blot analysis of strain indicated in panel A. The His-tagged Cys4 protein was detected using anti-His antibody. (C) Northern blot analysis of small crRNA production by each mutant indicated in panel A. In this experiment, the three spacers and two repeats closest to the cas1 gene of CRISPR2 of P. aeruginosa PA14 was used as the probe (see Fig. 2A). Also shown at the bottom of this panel is the 5S rRNA band for each of the corresponding RNA preparations used in the Northern blot analysis.
To confirm and extend these studies, we assessed the ability of mutant Csy4 proteins, with point mutations in known catalytic sites, to mediate CRISPR processing and to impact biofilm formation in DMS3 lysogens. We constructed penta-His-tagged constructs expressing the WT and mutant Csy4 proteins and assessed the ability of the mutants to complement the Δcsy4 mutation (Fig. 3).
Based on published biochemical data for Csy4 (11), we mutated the catalytic H29 residue of the P. aeruginosa Csy4 protein to lysine or alanine (H29K or H29A) to assess the role of the in vitro-documented nuclease activity on crRNA production and biofilm formation in vivo. A construct expressing penta-His-tagged Csy4 was able to complement the Δcsy4 mutation, resulting in Δcsy4 lysogens regaining the ability to disrupt biofilm formation (Fig. 3A, compare Δcsy4 L to Δcsy4+csy4 L). Mutation of P. aeruginosa Csy4 catalytic residue H29 to lysine has been shown to be active in vitro (11), and the mutant protein retained the ability to complement biofilm inhibition, while the in vitro catalytically dead H29A mutant (11) was unable to complement the biofilm inhibition phenotype (Fig. 3A). Western blot analysis showed that the noncomplementing Csy4(H29A) mutant protein, while less stable than WT Csy4, was similar in stability to the complementing H29K mutant protein, indicating that the decreased level of the H29A mutant protein could not explain the lack of complementation (Fig. 3B). Finally, Northern blot analysis demonstrated that the Csy4(H29K) mutant protein retained the ability to process the CRISPR transcript into small crRNAs, while the H29A mutants are not functional for production of small crRNAs (Fig. 3C).
While it has been shown that a Csy4 protein from E. coli can process a P. aeruginosa CRISPR transcript in vitro (11), to our knowledge no study has looked at the ability of a cas protein from one bacterial species to cross-complement another species in vivo. To this end, E. coli UTI89 csy4 was amplified and cloned into the same vector backbone that was used for P. aeruginosa Csy4 expression. The E. coli Csy4 protein is 49% identical and 64% similar to Csy4 of P. aeruginosa. We found that the E. coli Csy4 protein was able to restore biofilm inhibition to P. aeruginosa Δcsy4 mutant lysogen (Fig. 3A). Further, the E. coli Csy4(H29A) mutant protein, like that of P. aeruginosa, was unable to complement the biofilm inhibition phenotype. Western and Northern blot analyses demonstrated that the E. coli Csy4 protein is produced and functional in P. aeruginosa and that the WT but not the H29A mutant is able to process the P. aeruginosa CRISPR transcript into correctly sized crRNAs in vivo (Fig. 3B).
The Yersinia-subtype CRISPR region codes for a predicted Cas3 protein but lacks an apparent Cas2 homolog.
The core CRISPR-associated gene cas3 is found in 6 of 8 CRISPR region subtypes and is not predicted to be complexed with Csy proteins based on a published study (23). A recent study demonstrated that Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in vitro (22).
We compared the sequences and predicted domains of Cas3 homologs from 3 different CRISPR region subtypes to lend insight into Cas3 function. Alignment of Cas3 proteins from pseudomonads and E. coli strains harboring different CRISPR region subtypes, including P. aeruginosa PA14 (Yersinia subtype), E. coli UTI89 (Yersinia subtype), P. aeruginosa PA2192 (Escherichia subtype), Pseudomonas mendocina (Escherichia subtype), and Pseudomonas stutzeri (Desulfovibrio subtype), are shown in Fig. 4.
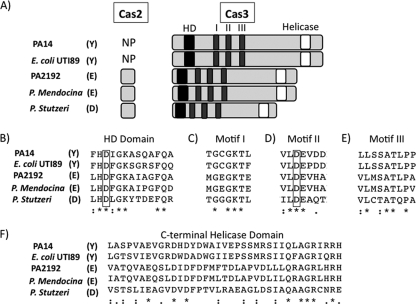
Fig. 4.
Conservation and domain architecture of Cas3 from multiple CRISPR region subtypes. (A) Columns (from left to right): bacterial strain harboring CRISPR region, subtype of CRISPR regions (Y, Yersinia; E, Escherichia; D, Desulfovibrio), presence or absence of cas2 sequence (NP, not present), and the cas3-encoded product. Black boxes denote HD domains, dark gray boxes represent conserved motifs of DEXD/H domains, and white boxes show location of C-terminal helicase domains. (B to F) Conservation of Cas3 amino acid sequence around each domain/motif. Stars represent residues that are 100% conserved, and black dots represent highly conserved sequences. Boxes are drawn around key residues chosen for mutational analysis.
The Cas2 protein is coded for in some organisms by a gene adjacent to the cas3 gene (Fig. 4A), and in a previous study, Makarova and colleagues reported that Yersinia-subtype CRISPR regions harbored in bacteria such as P. aeruginosa PA14 and E. coli UTI89 have N-terminal Cas2-like domains fused to the Cas3 protein (15, 23). However, while both P. aeruginosa PA14 and E. coli UTI89 Yersinia-subtype Cas3 proteins were larger than the Cas3 proteins of other subtypes (Fig. 4A), protein sequence analysis provided no evidence of a fusion between Cas2 and Cas3 proteins (Fig. 4A; also see Fig. S2A and B in the supplemental material). For example, none of the residues previously published to be well conserved in Cas2 (2) (see Fig. S2A, highlighted residues) are conserved in the N termini of Yersinia-subtype Cas3 proteins. Further, only one residue is 100% conserved between the previously studied Cas2 homologs and the P. aeruginosa and E. coli Yersinia-subtype Cas3 proteins (see Fig. S2A), and while this residue was previously implicated in Cas2 nuclease activity (2), its conservation is likely a result of the CLUSTAL alignment algorithm attempting to obtain at least some level of identity/similarity between these clearly distinct protein sequences. While it is formally possible that the N termini of Yersinia-subtype Cas3 proteins share a tertiary structure and/or biochemical activity with Cas2, we could not identify any DNA or protein sequences indicative of the presence of a predicted cas2 homolog in Yersinia-subtype CRISPR regions.
Predicted domain structure and conservation of Cas3.
To investigate the function of Cas3 in bacteriophage DMS3-mediated biofilm inhibition, we first inspected the protein sequence for conserved domains. The Cas3 proteins share three conserved domains regardless of subtype in which they are found: an N-terminal HD domain, a central DEXD domain with multiple conserved motifs, and a C-terminal helicase domain (Fig. 4A).
HD domains are common yet poorly understood globular domains that have been shown to participate in metal-dependent phosphohydrolysis of nucleotides such as cyclic di-GMP, ppGpp, dGTP, and deoxyribonucleotides and have a highly conserved HD motif (26).
In bacteria, DEXD/H proteins have been shown to play roles in ribosome biogenesis, mRNA processing and decay, and translation initiation (12). Nine motifs are found in canonical DEXD/H proteins, labeled Q, I, Ia, Ib, II, III, IV, V, and VI (12). Our alignments show conserved motifs I, II, and to a lesser degree III (Fig. 4C to E). Motif I (Walker A motif) mediates interactions with ATP through hydrogen bonding of the β and γ phosphates of ATP with the conserved lysine (K426 of Cas3) and an Mg2+ ion coordinated by the neighboring threonine (T427) (12). The initial aspartate (D576) and glutamate (E577) of the DEXD/H sequence of motif II (Walker B motif) are responsible for hydrolysis of ATP and also coordinate the same Mg2+ as T427 (12). The terminal aspartate (D579) interacts with the conserved serine (S607) and alanine (A608) of motif III, which do not directly interact with ATP but are believed to mediate ATP binding and helicase activities (12). Finally, the C-terminal helicase domain is known to interact with the DEXD/H domain, aiding in both substrate binding and ATP hydrolysis (20).
We utilized published structure/function and mutagenesis studies of these domains (12, 26, 28), and our alignments, to guide the analysis of Cas3's role in phage-mediated biofilm suppression and small crRNA production presented in the next section.
Cas3 requires both its HD and DEXD/H domains for bacteriophage-mediated inhibition of biofilm formation but does not impact small crRNA production.
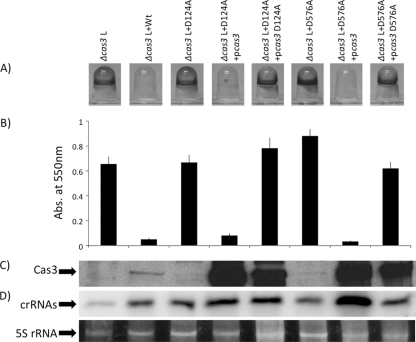
Deletion analysis of the CRISPR region in P. aeruginosa PA14 showed that a Δcas3 mutant produces less mature crRNA than the wild type (Fig. 2C to E). However, we could not complement small crRNA production in a Δcas3 mutant by expression of Cas3-His from a plasmid (data not shown), and we could only partially rescue the biofilm phenotype of a Δcas3 mutant by expression of Cas3-His from a plasmid (Fig. 1D). Expression of the native Cas3 protein from the same vector used to express Cas3-His also did not fully complement the Δcas3 mutant. Together, these observations suggested either that plasmid-encoded Cas3 was not functioning properly or that the Δcas3 mutation was having polar impacts on downstream genes in the cas gene cluster. As an alternative approach, we repaired the Δcas3 deletion mutation by placing the WT, untagged cas3 gene back into its native locus. This chromosomal copy completely restored small crRNA production and biofilm inhibition to WT levels in the presence of bacteriophage DMS3 (Fig. 5A, compare Δcas3 L versus Δcas3 L+WT).
Fig. 5.
Requirement of the D124 and D576 residues of Cas3 for in vivo function. (A) Shown are the biofilm phenotypes of strains carrying the WT Cas3, as well as the Cas3(D124A) and Cas3(D576A) mutants, in the presence of bacteriophage DMS3. A representative picture of the biofilm formed by each strain is shown. An “L” following a genotype designation indicates a strain lysogenized with DMS3. (B) Quantification of biofilm formed by each strain. The data shown are average values from eight independent wells. (C) Western analysis of WT and mutant Cas3 proteins for each strain indicated, as detected by an anti-Cas3 antibody. (D) Northern blot analysis of small crRNA production of each strain. As in Fig. 2 and 3, the three spacers and two repeats closest to the cas1 gene of CRISPR2 of P. aeruginosa PA14 were used as the probe. Also shown at the bottom of this panel is the 5S rRNA band for each of the corresponding RNA preparations used in the Northern blot analysis.
We next made mutations in the HD (D124A) and DEXD/H (D576A) domains of Cas3 to assess the roles of these domains in CRISPR transcript processing and bacteriophage DMS3-mediated biofilm inhibition. In contrast to the WT Cas3 protein, expression of the Cas3(D124A) and Cas3(D576A) mutant proteins from their native chromosomal loci were both unable to complement the biofilm phenotype (Fig. 5A and B, lanes 3 and 6) but, surprisingly, fully restored WT crRNA production (Fig. 5D, lanes 3 and 6).
Complicating our interpretation of these studies was the fact that Western blot analysis showed that WT Cas3 was more abundant than both the D124A and D576A mutants (Fig. 5C), indicating that the mutations impacted protein stability. To address this issue, we overexpressed WT Cas3 and the mutant proteins from a plasmid and did so in both cas3(D124A) and cas3(D576A) chromosomal mutant backgrounds. Overexpression of the WT protein in the strains carrying the cas3(D124A) and cas3(D576A) chromosomal mutations completely restored biofilm inhibition and CRISPR processing to the WT phenotype (Fig. 5, compare lane 1 to lanes 4 and 7). In contrast, high-level overexpression of the Cas3(D124A) and Cas3(D576A) mutant proteins could not restore the WT biofilm-inhibited phenotype, despite the fact that these strains were producing small crRNAs at levels equal to or greater than those of the WT (Fig. 5, compare lane 2 to lanes 5 and 8).
Taken together, these data suggest that Cas3 is indeed required for bacteriophage-mediated loss of biofilm formation and that HD and DEXD/H are required for this phenotype, but that neither domain plays a discernible role in the production and/or stability of small crRNAs.
CRISPR2 content required for bacteriophage DMS3-dependent biofilm inhibition.
We next turned our attention to understanding the role of the Yersinia-subtype CRISPR2 in mediating bacteriophage-dependent loss of biofilm formation. Current models suggest that CRISPR-mediated resistance to bacteriophage and plasmids requires 100% identity between a spacer sequence within the bacterial CRISPR and the corresponding portion of the plasmid or bacteriophage genome (known as a protospacer) (23). This model, along with our previous observation that P. aeruginosa PA14 CRISPR2 was required for loss of biofilm formation (27), suggested that CRISPR2 spacers (17 and 20) harboring identity to DMS3 were responsible for CRISPR-mediated biofilm inhibition.
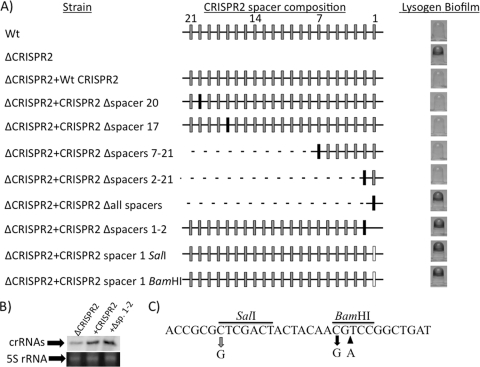
We first established that we could complement the strain lacking CRISPR2 by reintroducing the WT CRISPR2 region back onto the chromosome in the ΔCRISPR2 mutant (Fig. 6A, compare rows 1 to 3). These data indicated that we had the tools to also engineer variants of CRISPR2.
Fig. 6.
CRISPR2 spacer content required for bacteriophage-mediated biofilm inhibition. (A) The left column shows the name of the WT or CRISPR2 mutant strain diagrammed in the center column. The CRISPR2 schematic representation depicts repeat sequences as black lines, deleted sequence as dotted lines, WT spacers of PA14 CRISPR2 as gray boxes, P. aeruginosa strain 2192 CRISPR1 spacer 8 as black boxes, and point mutations within spacer 1 as white boxes. Wild-type PA14 CRISPR2 spacer numbers are shown in intervals of 7 at the top of the center column, allowing orientation of various spacer mutations. The right column provides a representative image of the biofilm phenotype of each DMS3-infected WT and mutant strain. (B) Northern blot analysis demonstrating that the Δspacers1-2 (Δsp. 1–2) mutant produces a level of small crRNAs similar to that of the WT CRISPR2-complemented strain. The three spacers and two repeats closest to the cas1 gene of CRISPR2 of P. aeruginosa PA14 were used as the probe. Also shown at the bottom of this panel is the 5S rRNA band for each of the corresponding RNA preparations used in the Northern blot analysis. (C) Shown is the sequence of CRISPR2 spacer 1. The gray arrow shows the site of the mutation caused by introduction of the SalI site, and the black arrows show the site of the mutation (left arrow) and base insertion (right arrow) caused by introduction of the BamHI site. The black bar indicates the nucleotides comprising the restriction sites.
To determine the requirement for spacers 17 and 20 (i.e., those spacers with high sequence identity to regions of bacteriophage DMS3) in CRISPR-mediated biofilm inhibition, we replaced each spacer with a sequence lacking high identity to the DMS3 genome, namely, spacer 8 from Yersinia-subtype CRISPR1 of P. aeruginosa strain PA2192. The PA2192 spacer shared only 8 nucleotide residues at identical locations with that of CRISPR2 spacer 17 and shared 14 residues with spacer 20. These CRISPR mutants with altered spacers 17 or 20 retained the ability to disrupt biofilm formation in the presence of DMS3, indicating that neither of these sequences is required for this phenotype (Fig. 6A).
We next deleted CRISPR2 spacers 7 to 21 (Δspacers7-21), thereby removing spacers 17 and 20 simultaneously, and replaced this region with the single P. aeruginosa PA2192 spacer 8. We found that none of these spacers were required for DMS3-dependent biofilm inhibition (Fig. 6A). It is important to note that spacer 8 of P. aeruginosa PA2192 is introduced here and in all the deletion constructs below to streamline the screening of candidate CRISPR mutants, as outlined in Materials and Methods.
We subsequently removed spacers 2 to 21 (Δspacers2-21), and the strain still retained DMS3-mediated biofilm inhibition, indicating that only CRISPR2 spacer 1 was necessary for this phenotype (Fig. 6A). Deletion of all the spacers and introduction of just spacer 8 of P. aeruginosa PA2192 (Δall spacers) did restore biofilm formation, indicating that spacer 1 was critical for DMS3-mediated biofilm inhibition. Unfortunately, multiple PCR primers failed to amplify the correct fragments required to create a CRISPR2 spacer 1 in-frame replacement construct and the corresponding mutant. However, we were able to replace both spacers 1 and 2 with spacer 8 of P. aeruginosa PA2192, and this mutant lacking only spacers 1 and 2 (Δspacers1-2) behaved like a CRISPR2 deletion; that is, this mutation restored the ability of P. aeruginosa PA14 to form a biofilm when lysogenized by bacteriophage DMS3 (Fig. 6A). It is also important to note that in nonlysogens, none of these CRISPR2 mutants displayed a biofilm phenotype (data not shown). Together, these data indicate that spacer 1 is critical for the CRISPR-dependent loss of biofilm formation in DMS3 lysogens.
To make certain that the biofilm-positive Δspacers1-2 mutant was still able to transcribe and process the remaining CRISPR2 spacers, we analyzed small crRNA production. Northern blot analysis of ΔCRISPR2, ΔCRISPR2+CRISPR2, and ΔCRISPR2+Δspacers1-2 mutants (Fig. 6B) shows that while the ΔCRISPR2+Δspacers1-2 mutant produces crRNA at a level similar to that of the WT complement, it cannot inhibit biofilm formation.
Although we were unable to delete spacer 1 (see above), we were able to create point mutations within the spacer sequence by introducing unique restriction sites. We introduced a BamHI restriction enzyme site into spacer 1, resulting in a 2-bp mutation near the center of the spacer (adding one nucleotide to the spacer length), while the addition of a SalI site altered only a single nucleotide near the end of the spacer (the location of point mutations are denoted by arrows in Fig. 6C). Neither the SalI nor BamHI point mutations were able to inhibit biofilm formation in the presence of bacteriophage DMS3, indicating that spacer 1 requires these nucleotides for function (Fig. 6, compare rows 1, 2, 10, and 11).
Together, these analyses demonstrate that the CRISPR-mediated interaction between P. aeruginosa PA14 and bacteriophage DMS3 is dependent only on CRISPR2 spacer 1, and furthermore, that there are sequence determinants of spacer 1 important for its function.
A role of DMS3 gene 42 in DMS3-mediated biofilm inhibition.
Comparison of PA14 CRISPR2 spacer 1 against the NCBI database by using BLASTN identified no sequences with 100% identity, thus providing no obvious putative targets for this spacer (i.e., protospacers). The bacteriophage DMS3 sequence with the highest overall identity to spacer 1 was a portion of DMS3-42 (see Table S2 in the supplemental material), which was identical at 27 of 32 nucleotides (Fig. 7A). To verify that bacteriophage DMS3 had not been mutated in this region, we resequenced DMS3-42 in the lysogen and found no alteration from the previously reported DMS3 genome sequence (27). These data indicated that a non-identity-mediated interaction between spacer 1 and DMS3-42 might be responsible for the impact on biofilm formation.
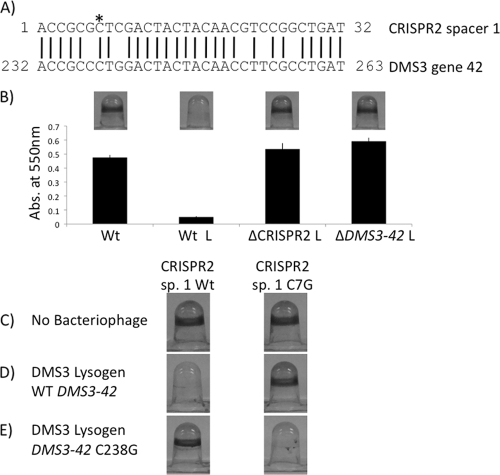
Fig. 7.
DMS3-42 is targeted by CRISPR2 spacer 1. (A) Alignment of the CRISPR2 spacer 1 sequence with the complementary portion of the DMS3-42 sequence. The DMS3-42 numbering is measured from the putative translational start site. The asterisk denotes the location of spacer 1 C7G point mutation. (B) The biofilm phenotypes of WT nonlysogen, as well as the DMS3 infected-WT, ΔCRISPR2 and ΔDMS3-42 mutants. A representative image of the biofilm formed by each strain is shown above the quantification of results for 8 independent wells. An “L” following a strain designation indicates a strain lysogenized with DMS3. (C) Representative images of the biofilm phenotypes of the WT and the spacer 1 (sp. 1) C7G point mutant (harboring a SalI restriction site) in the absence of bacteriophage. (D) Biofilm phenotypes of the WT and the spacer 1(C7G) mutant lysogenized by bacteriophage DMS3. (E) Biofilm phenotypes of the WT and spacer 1(C7G) mutant bacteria lysogenized by DMS3-42(C238G) mutant bacteriophage DMS3.
We employed several approaches to address whether non-identity-mediated interaction between spacer 1 and DMS3-42 might be responsible for the inhibition of biofilm formation. First, we deleted the DMS3-42 gene from the lysogen and observed restoration of biofilm formation; that is, the lysogen lacking the DMS3-42 gene showed a biofilm phenotype identical to that of a strain carrying a deletion of the CRISPR2 region (Fig. 7B), confirming a role for the DMS3-42 gene in DMS3-mediated biofilm inhibition. Of note is that the deletion of the DMS3-42 gene resulted in a lysogen that produced no detectable phage particles, as determined by PFU assay, indicating that the DMS3-42 gene product is critical for the DMS3 phage life cycle.
Second, we exploited the mutation in spacer 1 of CRISPR2 caused by introduction of the SalI restriction site (Fig. 6A and C), which resulted in a transversion of the C7 position of this spacer to a guanine nucleotide (Fig. 7A, asterisk). To determine whether a compensatory mutation within the DMS3-42 gene could restore the biofilm inhibitory phenotype to a strain carrying the spacer 1(C7G) mutation, we constructed the corresponding C238G point mutation in the DMS3-42 gene and introduced this mutation onto the chromosome of the ΔDMS3-42 strain. The DMS3-42(C238G) mutation was able to restore biofilm inhibition in the presence of the spacer 1(C7G) mutation but not in the presence of the WT spacer 1 sequence (Fig. 7C, D, and E).
It should be noted that the DMS3-42(C238G) mutation is predicted to introduce an L80V mutation in the DMS3-42 protein sequence. However, this mutation still retained the ability to produce approximately WT levels of bacteriophage particles as measured by plaque assay (data not shown), indicating that this mutation did not eliminate the function of the DMS3-42 protein.
Previously, we reported that both Pseudomonas aeruginosa PA14 and other clinical isolates with functional CRISPR regions did not mediate resistance to bacteriophage harboring sequences 100% identical to spacers within their CRISPRs (5). An efficiency-of-plating (EOP) assay was utilized to determine if WT and spacer 1(C7G) mutant CRISPR regions were equally susceptible to WT and DMS3-42(C238G) mutant bacteriophages, respectively. Interestingly, WT and spacer 1(C7G) mutant strains were equally susceptible to both WT and DMS3-42(C238G) mutant bacteriophages, with measured EOPs of 1.1 ± 0.02 and 1.7 ± 0.89, respectively, confirming that the Yersinia-subtype CRISPR region plays no detectable role in bacteriophage resistance.
Taken together, these data indicate that the interaction between spacer 1 and a nonidentical region of the DMS3-42 sequence is required for phage DMS3-mediated biofilm inhibition but not for bacteriophage resistance.
The DMS3-42 protein is not required for phage-mediated biofilm inhibition.
The requirement for DMS3-42 in DMS3-mediated biofilm inhibition raised the question of whether the DMS3-42 protein has a role in this CRISPR-dependent phenotype. To assess the contribution of the DMS3-42 protein to biofilm inhibition, we mutated the predicted DMS3-42 translational start site by introducing a T2G point mutation into the chromosomal copy of the DMS3-42 gene (Fig. 8A and B). This DMS3-42(T2G) mutant, like the ΔDMS3-42 deletion mutant, does not produce detectable levels of bacteriophage as measured by the plaque assay (not shown), corroborating that the DMS3-42 protein is required for the DMS3 life cycle and that the T2G mutation likely abrogated production of this protein. However, the DMS3-42(T2G) lysogen was still unable to form a biofilm, in contrast to the ΔDMS3-42 lysogen, which shows a WT level of biofilm formation (Fig. 8C, compare column 5 to columns 1, 2, and 3). Together, these data indicate that while DMS3-42 protein is required for the production of viable DMS3 phage particles, this protein is not required for phage-mediated biofilm inhibition.
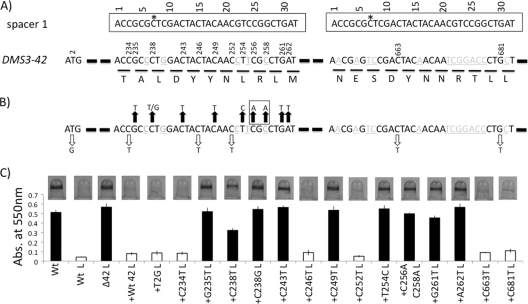
Fig. 8.
Non-identity-mediated targeting of CRISPR2 spacer 1 to DMS3-42. (A) Shown is a diagram of CRISPR2 spacer 1 (top) and selected regions of the DMS3-42 gene (bottom). The putative ATG translational start site of DMS3-42 is shown to the left, while the two regions of complementarity between CRISPR2 spacer 1 and DMS3-42 are shown to the right (with the flanking gene sequence denoted by a thick dashed line). CRISPR2 spacer 1 sequence is boxed and aligned with the two complementary regions of DMS3-42. Light gray, underlined DMS3-42 nucleotides are not identical between CRISPR2 spacer 1 and DMS3-42. The two CRISPR2 spacer 1 sequences have been numbered for ease of analysis, and an asterisk placed at the site of the C7G point mutation. The locations of point mutations made in the DMS3-42 gene are shown above the DMS3-42 nucleotide sequence. The putative amino acid sequence of the regions of complementarity is shown below DMS3-42 nucleotide sequence. (B) DMS3-42 point mutations that retain or abolish CRISPR2 spacer 1-mediated biofilm inhibition. Black arrows signify point mutations that restored biofilm formation in the presence of DMS3, while white arrows denote those that had no detectable effect on biofilm phenotype. The C256/C258 double mutant is boxed. (C) Representative image of the biofilm phenotype of each indicated strain from the quantification of results for at least 7 biofilm wells. White bars signify strains that retain the phenotype of the WT lysogen, that is, no biofilm formation, while black bars denote strains that show restored biofilm formation.
Nucleotide requirements of the DMS3-42 target sequence.
The finding that CRISPR2 spacer 1 interacts with gene DMS3-42 indicates that 100% identity between spacer and target is not required; however, the observation that the CRISPR2 spacer 1(C7G) mutation is sufficient to completely abolish biofilm inhibition is consistent with a requirement for complementarity at some nucleotides. To better understand the sequence required for spacer 1 targeting of DMS3-42, multiple point mutations were introduced in the two regions of complementarity between spacer 1 and DMS3-42. The first region of complementarity is identical to CRISPR2 spacer 1 at 27 of 32 nucleotides, while the second region shares 19 of 32 bases (Fig. 8A). Care was taken to introduce point mutations that would not affect the DMS3-42 protein sequence, when possible, and so maintain DMS3-42 protein function.
Mutations were created in at least one nucleotide of every codon within the first complementarity region, while two selected mutations were made within the second region of lower complementarity (Fig. 8A). Although we attempted to make single-base-pair mutations that maintained the DMS3-42 protein sequence, four point mutations resulted in amino acid substitutions (G235T→A79S, C238G→L80V, T254C→L85P, and A262T→M88L). Only the DMS3-42(T254C→L85P) mutant was unable to produce bacteriophage particles and therefore likely had a marked negative effect on DMS3-42 protein function (data not shown). The C256A C258A double mutation was created to test the requirement of C256 (C258 already differs from CRISPR2 spacer 1) while retaining the WT amino acid sequence of the DMS3-42 protein.
The ability of each DMS3-42 mutant to form a biofilm was assayed and quantified (Fig. 8C). The C234T, C246T, C252T, C663T, and G681T mutations all retained the ability to inhibit biofilm formation, indicating that they are not required for CRISPR2 spacer 1 interaction with DMS3-42 (Fig. 8B and C, white arrows and bars). However, the G235T, C238T, C238G, C243T, C249T, T254C, G261T, and A262T mutants and the C256A C258A double mutant were all unable to suppress biofilm formation, implicating them in CRISPR2 spacer 1 targeting (Fig. 8B and C, black arrows and bars). Finally, the observation that C238T and C238G mutants have different abilities to suppress biofilm formation (Fig. 8C) is consistent with some nucleotide substitutions being tolerated more than others at certain positions.
DISCUSSION
Our work has shown that the Yersinia-subtype CRISPR region of P. aeruginosa PA14 plays no detectable role in viral immunity but instead is required for bacteriophage DMS3-dependent inhibition of biofilm formation by P. aeruginosa. This finding is in contrast to the work reported for other CRISPR subtypes (1, 3). Because of the distinct function proposed for the Yersinia-subtype CRISPR region, we sought to identify the components of this specific CRISPR system that are required to mediate DMS3-dependent biofilm inhibition. While our findings in some cases confirmed previous reports from other groups, this work also provides new insight into which components of the Yersinia-subtype CRISPR region are required for bacteriophage DMS3-mediated biofilm inhibition.
We are the first to demonstrate the in vivo requirement for both csy4 and csy2 in the production and/or stability of small crRNA. Decreased crRNA levels were also observed in the csy1 and csy3 mutant backgrounds, indicating that their protein products aid in crRNA production or stability. Others have speculated that the Csy proteins form a complex involved in crRNA processing (23); however, the similar stabilities of tagged Csy4 in the csy1-3 mutants indicate roles for these Csy proteins other than stabilizing Csy4 in vivo. An important caveat of this conclusion is that, to assess its stability, Csy4 was expressed from a plasmid rather than its native locus; thus, it is still formally possible that Csy1-3 may impact the stability of the Csy4 protein in the native context.
We would like to stress that this study does not contradict previous work demonstrating that Csy4 is necessary and sufficient to process CRISPR transcript in vitro (11). In their study, Haurwitz and colleagues showed that coexpression of Csy4 and its cognate CRISPR transcript in E. coli results in robust crRNA production (11). Taken together, these data suggest that no other Cas proteins are required for crRNA production. The work here indicates that a mutation in the csy2 gene results in no detectable crRNA in vivo, while mutating the csy1 or csy3 gene results in a marked decrease in crRNA levels. It is possible that in its native context in P. aeruginosa, Csy4 does require accessory factors to produce crRNA, or alternatively, that the Csy1-3 proteins are required to stabilize the crRNA produced by Csy4. Going forward, the tools developed here can help elucidate the in vivo function of each of the Cas proteins in small crRNA production.
Determining the function of the few conserved cas gene products found in multiple CRISPR region subtypes is key to understanding shared CRISPR region mechanisms. The core cas gene cas3 is found in 6 of 8 CRISPR region subtypes (23), and has been shown recently to be a single-stranded DNA nuclease and ATP-dependent helicase (22). Our studies indicate that Cas3 requires both its HD and DEXD/H domains to function in biofilm inhibition, and mutations similar to those analyzed here (Fig. 4 and 5) were reported to disrupt the nuclease and helicase activities of Cas3 in vitro (22). Thus, our in vivo studies corroborate the conclusions drawn from the published Cas3 in vitro studies (22). Furthermore, the mutants with point mutations in the Cas3 HD and DEXD/H domains are the only biofilm-positive cas mutants we found to have crRNA production equivalent to that of the WT strain, indicating that in the context of the bacteriophage-CRISPR interaction in vivo, the Cas3 HD and DEXD/H domains function in a process other that of small crRNA production, and we favor the model that Cas3 maybe an effector protein.
Considerable effort was also taken to identify the previously proposed Cas2-Cas3 fusion protein; however, no evidence was found supporting a Cas2 protein encoded within the Yersinia-subtype CRISPR region. This analysis indicates that Cas2 is not found in all CRISPR region subtypes, as has been previously reported (15, 23).
Broadly accepted CRISPR doctrine states that CRISPR/cas interactions require 100% identity between the spacer and target (protospacer). Our comprehensive deletion analysis of CRISPR2 clearly demonstrates that while CRISPR2 spacer 20 has 100% identity to bacteriophage DMS3, this spacer plays no role in the CRISPR-mediated biofilm phenotype. Unexpectedly, our data indicate that spacer 1 of CRISPR2, which is not 100% identical to any sequence in the GenBank sequence database, mediates the bacteriophage DMS3-dependent inhibition of biofilm formation. We also demonstrate that CRISPR2 spacer 1 impacts biofilm formation through interaction with the DMS3-42 gene and that this interaction does not require DMS3-42 protein production. Interestingly, DMS3-42 harbors two regions of complementarity to CRISPR2 spacer 1; however, only one seems to be required for the CRISPR2 spacer 1-mediated loss of biofilm formation. If the preexisting mismatches between spacer 1 and the DMS3-42 sequence and the thorough mutational analysis presented here are combined, we have analyzed half of the nucleotides within the region of greatest complementarity between CRISPR2 spacer 1 and DMS3-42, thereby identifying 8 nucleotides required for the interaction between CRISPR2 spacer 1 and DMS3-42. Together, these data challenge current CRISPR models invoking the need for 100% identity between spacers and targets. Our findings are consistent with a recent study published while this paper was under review outlining an Escherichia-subtype CRISPR region interacting with a plasmid that lacked 100% identity to any spacer sequences (18).
There are several models that could explain how the spacer 1 interaction with DMS3-42 might impact the DMS3 lysogen. The spacers encoded by the Mycobacteria- and Neisseria-subtype CRISPRs have been postulated to direct a DNA nuclease against invading foreign DNA, resulting in cleavage of the target DNA (9, 16). We do not favor such a DNA-targeting model at this time, because CRISPR-mediated cleavage of the lysogenized DMS3 would negatively impact viable counts of the lysogen and phage titers and/or alter resistance to DMS3 infection—we have not observed any of these phenotypes in our studies (5, 27). Alternatively, the Yersinia-subtype CRISPR region may be modulating production and/or stability of transcripts produced by the lysogen. In the case of DMS3-42, which is located within a predicted ∼17-kb operon including 20 other putative genes, any impact on message stability mediated by spacer 1 has the potential to positively or negatively impact the expression of one or more proteins encoded in this region. Our ongoing studies aim to understand the mechanism by which spacer 1-mediated effects impede biofilm formation.
The seminal work of Barrangou et al. (1) and Deveau et al. (7) in Streptococcus thermophilus concluded that spacer-mediated resistance of S. thermophilus to bacteriophages φ858 and φ2972 requires 100% identity between the spacer and its bacteriophage target (7). However, the analysis that produced this conclusion included only bacteriophage mutants selected to become resistant to the spacer sequence and excluded all sequence changes that do not alter resistance. Our comprehensive mutagenesis studies here show that a single-point mutation may or may not disrupt spacer-mediated processes, and we suggest that the model invoking the requirement for a 100% match between spacer and target must be revisited and vetted more thoroughly.
It is widely accepted that CRISPR can be transcribed into a large transcript and processed into small crRNAs by Cas proteins. However, it is becoming increasingly clear that CRISPR regions of different subtypes may utilize these crRNAs differently. This observation is perhaps less striking when it is considered that only the cas1 gene is found in all CRISPR region subtypes. Additionally, experimental evidence has shown that the Staphylococcus epidermidis and S. thermophilus CRISPR regions (Mycobacteria and Neisseria subtypes, respectively) target DNA, while that of Pyrococcus furiosus (Thermotoga subtype) targets RNA in vitro (9, 10, 16), indicating the possibility for different output activities for these CRISPR subtypes. In this study, we demonstrate CRISPR-mediated interaction with a lysogenized bacteriophage, which could have a profound implication for a number of environmental and pathogenic bacteria that have their phenotypes modulated by integrated bacteriophages. Given that the DMS3 phage studied here shares high sequence similarity to Mu phage and replicates via a transposition-based mechanism, we would not be surprised if the Yersinia-subtype CRISPR might also target transposons. Together, these findings lead us to assert that each CRISPR region should not be assumed to have a role in resistance to phage infection until such a conclusion is experimentally validated. It is critical that novel CRISPR/cas interactions not be dismissed out of hand as nonfunctional or degenerate simply because they do not fit previous models for other subtypes. We propose that a CRISPR region should be considered functional if the region is demonstrated experimentally to be transcribed and processed into small crRNAs. Finally, this work indicates that some CRISPR/cas interactions may require considerably less than 100% identity between a CRISPR spacer and its “target,” forcing us to reevaluate the use of bioinformatics as the sole mechanism of determining spacer-targeted interactions.
Supplementary Material
ACKNOWLEDGMENTS
We thank B. Wiedenheft and J. A. Doudna for providing unpublished data and antibodies that greatly assisted in the completion of these studies. We also thank Michael Zegans and John Hammond for scientific discussions.
This work was supported by the T32 AI007519 predoctoral fellowship to K.C.C. and NIH grants R01AI083256 and R21AI068662 to G.A.O.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 11 March 2011.
REFERENCES
- 1. Barrangou R., et al. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712 [DOI] [PubMed] [Google Scholar]
- 2. Beloglazova N., et al. 2008. A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. J. Biol. Chem. 283:20361–20371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brouns S., et al. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321:960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Budzik J., Rosche W., Rietsch A., O'Toole G. 2004. Isolation and characterization of a generalized transducing phage for Pseudomonas aeruginosa strains PAO1 and PA14. J. Bacteriol. 186:3270–3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cady K. C., et al. 2011. Prevalence, conservation and functional analysis of Yersinia and Escherichia CRISPR regions in clinical Pseudomonas aeruginosa isolates. Microbiology 157:430–437 [DOI] [PubMed] [Google Scholar]
- 6. Choi K., Kumar A., Schweizer H. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64:391–397 [DOI] [PubMed] [Google Scholar]
- 7. Deveau H., et al. 2008. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 190:1390–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Díez-Villaseñor C., Almendros C., García-Martínez J., Mojica F. 2010. Diversity of CRISPR loci in Escherichia coli. Microbiology 156:1351–1361 [DOI] [PubMed] [Google Scholar]
- 9. Garneau J., et al. 2010. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468:67–71 [DOI] [PubMed] [Google Scholar]
- 10. Hale C., et al. 2009. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139:945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Haurwitz R., Jinek M., Wiedenheft B., Zhou K., Doudna J. 2010. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329:1355–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Iost I., Dreyfus M. 2006. DEAD-box RNA helicases in Escherichia coli. Nucleic Acids Res. 34:4189–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kuchma S., et al. 2010. Cyclic-di-GMP-mediated repression of swarming motility by Pseudomonas aeruginosa: the pilY1 gene and its impact on surface-associated behaviors. J. Bacteriol. 192:2950–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. MacEachran D., et al. 2007. The Pseudomonas aeruginosa secreted protein PA2934 decreases apical membrane expression of the cystic fibrosis transmembrane conductance regulator. Infect. Immun. 75:3902–3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Makarova K., Grishin N., Shabalina S., Wolf Y., Koonin E. 2006. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct 1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marraffini L., Sontheimer E. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. O'Toole G., Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295–304 [DOI] [PubMed] [Google Scholar]
- 18. Perez-Rodriguez R., et al. 2011. Envelope stress is a trigger of CRISPR RNA-mediated DNA silencing in Escherichia coli. Mol. Microbiol. 79:584–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pougach K., et al. 2010. Transcription, processing and function of CRISPR cassettes in Escherichia coli. Mol. Microbiol. 77:1367–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schütz P., et al. 2010. Comparative structural analysis of human DEAD-box RNA helicases. PLoS One 5:e12791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shanks R., Caiazza N., Hinsa S., Toutain C., O'Toole G. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl. Environ. Microbiol. 72:5027–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sinkunas T., et al. 2011. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 30:1335–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van der Oost J., Jore M., Westra E., Lundgren M., Brouns S. 2009. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem. Sci. 34:401–407 [DOI] [PubMed] [Google Scholar]
- 24. van der Ploeg J. 2009. Analysis of CRISPR in Streptococcus mutans suggests frequent occurrence of acquired immunity against infection by M102-like bacteriophages. Microbiology 155:1966–1976 [DOI] [PubMed] [Google Scholar]
- 25. Wiedenheft B., et al. 2009. Structural basis for DNase activity of a conserved protein implicated in CRISPR-mediated genome defense. Structure 17:904–912 [DOI] [PubMed] [Google Scholar]
- 26. Yakunin A., et al. 2004. The HD domain of the Escherichia coli tRNA nucleotidyltransferase has 2′,3′-cyclic phosphodiesterase, 2′-nucleotidase, and phosphatase activities. J. Biol. Chem. 279:36819–36827 [DOI] [PubMed] [Google Scholar]
- 27. Zegans M., et al. 2009. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J. Bacteriol. 191:210–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zimmerman M., Proudfoot M., Yakunin A., Minor W. 2008. Structural insight into the mechanism of substrate specificity and catalytic activity of an HD-domain phosphohydrolase: the 5′-deoxyribonucleotidase YfbR from Escherichia coli. J. Mol. Biol. 378:215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.