Abstract
Despite extensive surveillance, food-borne Salmonella enterica infections continue to be a significant burden on public health systems worldwide. As the S. enterica species comprises sublineages that differ greatly in antigenic representation, virulence, and antimicrobial resistance phenotypes, a better understanding of the species' evolution is critical for the prediction and prevention of future outbreaks. The roles that virulence and resistance phenotype acquisition, exchange, and loss play in the evolution of S. enterica sublineages, which to a certain extent are represented by serotypes, remains mostly uncharacterized. Here, we compare 17 newly sequenced and phenotypically characterized nontyphoidal S. enterica strains to 11 previously sequenced S. enterica genomes to carry out the most comprehensive comparative analysis of this species so far. These phenotypic and genotypic data comparisons in the phylogenetic species context suggest that the evolution of known S. enterica sublineages is mediated mostly by two mechanisms, (i) the loss of coding sequences with known metabolic functions, which leads to functional reduction, and (ii) the acquisition of horizontally transferred phage and plasmid DNA, which provides virulence and resistance functions and leads to increasing specialization. Matches between S. enterica clustered regularly interspaced short palindromic repeats (CRISPR), part of a defense mechanism against invading plasmid and phage DNA, and plasmid and prophage regions suggest that CRISPR-mediated immunity could control short-term phenotype changes and mediate long-term sublineage evolution. CRISPR analysis could therefore be critical in assessing the evolutionary potential of S. enterica sublineages and aid in the prediction and prevention of future S. enterica outbreaks.
INTRODUCTION
The Gram-negative bacterial species Salmonella enterica comprises more than 2,500 described serovars, many of which cause disease in humans and a wide range of animals (26). In the United States, S. enterica is responsible for an estimated 1.4 million cases of infection, 15,000 hospitalizations, and more than 400 deaths annually (78) and is considered a category B pathogen and a potential food safety threat by the Centers for Disease Control and Prevention (CDC; http://www.bt.cdc.gov/agent/agentlist-category.asp#b). In the western world, S. enterica populations from livestock animals are the most important source of infection in humans and usually cause a self-limiting gastroenteritis (46). In immunocompromised patient populations, however, the same serovars can cause life-threatening systemic infection that requires immediate and effective antimicrobial therapy (46). In order to monitor populations from clinical, veterinary, and food sources, the Food and Drug Administration maintains an intensive active and passive S. enterica surveillance system that characterizes hundreds of isolates per year by serotyping, pulsed-field gel electrophoresis (PFGE) fingerprinting, and antimicrobial resistance determination. Despite these efforts, S. enterica continues to be an important public health threat in the United States, with several outbreaks occurring every year (http://cdc.gov/salmonella/outbreaks.html). S. enterica outbreaks, as well as individual cases of S. enterica infections, linked to various food sources, including animal products, fruits, and vegetables, can be caused by different S. enterica serovars and often involve multidrug resistance (MDR) phenotypes (4, 20). The evolutionary dynamics responsible for the emergence of new S. enterica serotypes or sublineages, as well as for the acquisition and exchange of virulence and resistance phenotypes between these sublineages, are still largely uncharacterized. A better understanding of the mechanisms that mediate this evolutionary process would aid in the prediction and potentially prevention of future S. enterica outbreaks.
Today, S. enterica isolates, based on immunogenic properties, are commonly classified as serovars, which often share similar phenotypic presentations. For example, S. enterica serovars Typhi and Paratyphi cause typhoid in humans, a serious systemic infection with a high mortality rate (57), whereas S. enterica serovar Typhimurium causes systemic infection in mice but only gastroenteritis in humans and cattle (54). The distribution of reported S. enterica serovars differs among animals, between animals and humans, and among humans from different continents (9, 25, 76). Depending on their host range, serovars have been described as host restricted (e.g., S. Typhi and S. Paratyphi in humans and S. Gallinarum in poultry), host adapted (S. Dublin in cattle and S. Choleraesuis in swine), or ubiquitous (S. Typhimurium and S. Enteritidis) (42, 77). Variations in the severity of disease outcome depending on the infecting S. enterica serovar have also been reported (40). The genetic basis underlying the phenotypic diversity of S. enterica has not been sufficiently investigated, although comparative genome analyses suggested that restriction of S. Typhi and S. Paratyphi A to the human host was accompanied by genome degradation and the accumulation of pseudogenes (17, 33, 53, 56).
In order to better understand the processes that drive the evolution of this species, we sequenced the genomes of 17 nontyphoidal S. enterica strains derived from 12 serovars and performed a comparative analysis of genotype variations within the S. enterica species. The strains sequenced were isolated from human, animal, and food sources. The species S. enterica is taxonomically divided into six subspecies of which only subspecies I (subsp. enterica) is associated with disease in warm-blooded animals. Correspondingly, all isolates selected for this study, as well as all previously sequenced S. enterica isolates, with the exception of S. enterica subsp. arizonae RKS2980, belong to subsp. enterica. All serovars from which isolates were selected for this study were not represented in the public genome sequence databases. The sequenced strains include seven isolates from the most frequently reported Salmonella serovars from human sources as reported by the CDC, including Newport, Heidelberg, Javiana, Typhimurium-like I 4,[5],12:i:−, Saintpaul, Agona, and Hadar (9). Additional strains sequenced were from the serovars Kentucky and Schwarzengrund, which are among the 10 most commonly identified Salmonella serovars from retail meat (20), as well as from serovars Dublin (the second-most-often-isolated serotype from cattle [76]), Virchow, and Weltevreden. A special emphasis was put on the comparison of isolate pairs from the same serovar, representing either phenotypic variation with respect to antimicrobial resistance (Heidelberg, Kentucky, Newport, Schwarzengrund) or genotypic variation within the same serotype as established by PFGE (Newport, Saintpaul).
MATERIALS AND METHODS
S. enterica strains, genome sequencing, and annotation.
Seventeen S. enterica subsp. enterica isolates of 12 serovars were selected for sequencing (Agona, Dublin, Hadar, Heidelberg, I 4,[5],12:i:−, Javiana, Kentucky, Newport, Schwarzengrund, Saintpaul, Virchow, and Weltevreden). Several of these isolates are part of strain collections in more than one laboratory and have received different names, which are shown in Table S1 in the supplemental material. Cultures of all S. enterica single-colony isolates that were sequenced have been deposited at the BEI Resources strain collections (http://www.beiresources.org/). Prior to being used for sequencing library generation, the two Saintpaul isolates, SARA23 and SARA29, were retyped at the Center for Veterinary Medicine of the U.S. Food and Drug Administration according to the WHO Collaborating Centre for Reference and Research on Salmonella guidelines (27). All Newport isolates used for sequencing and comparative analyses were serotyped at the Minnesota Department of Health. All S. enterica isolates were assayed for susceptibility to a panel of 15 antimicrobials used by the National Antimicrobial Resistance Monitoring System program (see Table S2). Antimicrobial MICs were interpreted according to CLSI standards where available (16). Whole-genome, random shotgun plasmid insert libraries of 3 to 5 kbp and 10 to 12 kbp and fosmid insert libraries of 30 to 40 kbp were constructed as previously reported (62) and sequenced using a 3730xl DNA analyzer (Applied Biosystems, Foster City, CA). Between 50,000 and 55,000 sequence reads per genome (coverage, 10× to 12×) were assembled, and for a subset (Table 1), closure was performed as previously described (62).
Table 1.
General genome features of the newly and previously sequenced S. enterica strains used in this study
| Serovar and strain | Source | Location | Yr | No. of contigs | Sequence location and length (bp)a | GenBankb accession no. |
|---|---|---|---|---|---|---|
| Agona SL483 | Human | WI | 2003 | 0 | Chromosome, 4,798,660 | CP001138 |
| Unknown plasmid, 37,978 | CP001137 | |||||
| Dublin SL477 | Human | MO | 2004 | 0 | Chromosome, 4,842,908 | CP001144 |
| Virulence plasmid, 74,551 | CP001143 | |||||
| Hadar SL485 | Human | MA | 2005 | 49 | Chromosome, 4,789,824 | ABFG00000000 |
| Unknown plasmid, ∼5 | ||||||
| Heidelberg | ||||||
| SL476 | Ground turkey | MN | 2003 | 0 | Chromosome, 4,888,768 | CP001120 |
| Unknown plasmid, 91,374 | CP001118 | |||||
| Unknown plasmid, 3,373 | CP001119 | |||||
| SL486 | Human | GA | 2005 | 45 | Chromosome, 4,690,574 | ABEL00000000 |
| Unknown plasmid, ∼37 | ||||||
| I 4,[5],12:i:− SL474 | Human | IA | 2002 | 107 | Chromosome, 4,801,982 | ABAO00000000 |
| Virulence plasmid, ∼98 | ||||||
| Javiana SL478 | Human | GA | 2004 | 19 | Chromosome, 4,546,826 | ABEH00000000 |
| Kentucky | ||||||
| SL475 | Chicken breast | GA | 2003 | 1 | Chromosome, 4,934,032 | ABAK00000000 |
| Res./Vir. plasmid, 146,811 | CP001122 | |||||
| Resistance plasmid, 101,461 | CP001121 | |||||
| Unknown plasmid, 46,121 | CP001123 | |||||
| SL479 | Human | WI | 2002 | 8 | Chromosome, 4,698,474 | ABEI00000000 |
| Newport | ||||||
| SL254 | Human | MN | 2000 | 0 | Chromosome, 4,827,641 | CP001113 |
| Resistance plasmid, 176,473 | CP000604 | |||||
| Unknown plasmid, 3,605 | CP001112 | |||||
| SL317 | Human | MN | 2002 | 63 | Chromosome, 4,950,279 | ABEW00000000 |
| Saintpaul | ||||||
| SARA23 | Human | PA | 2 | Chromosome, 4,726,546 | ABAM00000000 | |
| SARA29 | Human | FL | 173 | Chromosome, 4,849,571 | ABAN00000000 | |
| Unknown plasmid, ∼58 | ||||||
| Unknown plasmid, ∼18 | ||||||
| Schwarzengrund | ||||||
| SL473 | Dehydrated chili | Thailand | 2002 | 0 | Chromosome, 4,709,075 | CP001127 |
| Resistance plasmid, 110,227 | CP001125 | |||||
| Unknown plasmid, 4,585 | CP001126 | |||||
| SL480 | Human | OR | 2000 | 61 | Chromosome, 4,688,585 | ABEJ00000000 |
| Resistance plasmid, ∼71 | ||||||
| Unknown plasmid, ∼2 | ||||||
| Unknown plasmid, ∼2 | ||||||
| Virchow SL491 | Human | CTc | 2005 | 3 | Chromosome, 4,867,937 | ABFH00000000 |
| Unknown plasmid, 5,880 | CP001148 | |||||
| Unknown plasmid, 3,176 | CP001149 | |||||
| Weltevreden SL484 | Scallops | Indonesia | 2002 | 77 | Chromosome, 4,950,708 | ABFF00000000 |
| Unknown plasmid, ∼51 | ||||||
| Unknown plasmid, ∼47 | ||||||
| 62:z4,z23:− RKS2980 | NA d | NA | NA | 0 | Chromosome, 4,600,800 | CP000880 |
| Choleraesuis SC-B67 | Human | Taiwan | 2002 | 0 | Chromosome, 4,755,700 | AE017220 |
| Resistance plasmid, 138,742 | AY509004 | |||||
| Virulence plasmid, 49,558 | AY509003 | |||||
| Enteritidis P125109 | Human | United Kingdom | NA | 0 | Chromosome, 4,685,848 | AM933172 |
| Virulence plasmid, unknown | ||||||
| Gallinarum 287/91 | Chicken | Brasil | NA | 0 | Chromosome, 4,658,697 | AM933173 |
| Virulence plasmid, unknown | ||||||
| Paratyphi A | ||||||
| AKU_12601 | Human | NA | NA | 0 | Chromosome, 4,581,797 | FM200053 |
| Resistance plasmid, 212,711 | AM412236 | |||||
| ATCC 9150 | Laboratory strain | NA | NA | 0 | Chromosome, 4,585,229 | CP000026 |
| Paratyphi B SPB7 | Human | Malaysia | NA | 0 | Chromosome, 4,858,887 | CP000886 |
| Paratyphi C RKS4594 | Human | France | 1988 | 0 | Chromosome, 4,833,080 | CP000857 |
| Virulence plasmid, 55,414 | CP000858 | |||||
| Typhi | ||||||
| CT18 | Human | Vietnam | 1993 | 0 | Chromosome, 4,809,037 | AL513382 |
| Resistance plasmid, 218,160 | AL513383 | |||||
| Unknown plasmid, 106,516 | AL513384 | |||||
| Ty2 | NA | NA | 1970s | 0 | Chromosome, 4,791,961 | AE014613 |
| Typhimurium LT2 | Human | NA | 1940s | 0 | Chromosome, 4,857,432 | AE006468 |
| Virulence plasmid, 93,939 | AE006471 |
Chromosome and plasmid sizes were calculated including linker sequences. Plasmid contigs were identified by BLAST from the draft assemblies. Plasmid phenotypes are shown as involved in virulence, antimicrobial resistance, both, or unknown.
Closed plasmids have separate GenBank accession numbers, whereas predicted plasmid contigs are referenced with the draft genome accession number.
Prior to the onset of illness, the patient had visited a farm in India and had exposure to farm animals.
NA, not available.
Genome reconstructions and annotations.
The genomes of six S. enterica isolates were sequenced to completion; draft genome sequences were generated for 11 more isolates (Table 1). Eleven complete S. enterica genome sequences from the NCBI genome database (http://www.ncbi.nlm.nih.gov/sites/genome) were used for comparative analysis. To facilitate genome comparisons, contigs from the draft assemblies were aligned to scaffolds and concatenated using linker sequences in order to generate “pseudochromosomes.” Linker sequences (NNNNNCACACACTTAATTAATTAAGTGTGTGNNNNN) encode start and stop codons in each frame and orientation, allowing Gene Finder tools to call truncated genes at all contig ends. Contig scaffolds were built based on NUCmer (45) nucleotide sequence comparisons of each contig against the chromosome sequence of the most closely related phylogenetic neighbor. Phylogenetic relationships were determined based on the multilocus sequence typing (MLST) system from University College Cork (http://mlst.ucc.ie/). Contigs from the draft assemblies that did not match related chromosomal sequences were BLASTed against the NCBI nucleotide sequence database. Contigs that showed similarity to plasmids from Enterobacteriaceae were kept separately and treated as plasmid sequences; all others (mostly matching sequences encoding phage-related functions) were attached with linker sequences to the ends of their corresponding pseudochromosome. All pseudochromosomes were checked for misassemblies or artificial genome rearrangements by comparison to optical maps (Opgen) that were generated for each genome as previously described (43). Chromosomes and pseudochromosomes were processed with the automated CloVR-Microbe pipeline (http://clovr.org/methods/clovr-microbe/), which includes steps for gene finding, homology searches, and automatic annotation as described for the IGS Annotation Engine (http://ae.igs.umaryland.edu/).
Comparative genome analysis.
Whole-genome nucleotide sequence comparisons were performed with the Mugsy tool, which aligns bacterial chromosomes as chains of syntenic blocks, without using a reference (3). All chromosomes and pseudochromosomes from the sequenced S. enterica genomes were aligned with Mugsy by using the “−distance 1000” and “−minlength 100” options, which specify the maximum genomic distance between adjacent anchors and the minimum block length, respectively. From the resulting output, syntenic blocks of sequence fragments larger than 10 kbp that aligned across all 28 S. enterica genomes were selected and concatenated by using customized Perl scripts that are available upon request. The resulting core alignment was filtered using the Gblocks tool, which eliminates poorly aligned positions and divergent alignment regions and thereby significantly improves the phylogenetic analysis (72). Gblocks was run with the options “b1 = 15” (minimum number of sequences for a conserved position) and “b2 = 24” (minimum number of sequences for a gap-flanking position). Phylogenetic trees were predicted based on the resulting filtered S. enterica core alignment (2,087,548-bp total length) using FastTree (60). If block sizes smaller than 10 kbp from the Mugsy output were used, FastTree failed to calculate phylogenetic trees, possibly due to the large size of the resulting alignment. MLST phylogenetic trees were generated based on Muscle (18) multiple-sequence alignments, PAUP* (71), and MrBayes (63) tree predictions by using maximum-likelihood calculations and Bayesian inference, respectively. Dendrograms for the two-gene MLST comparison of S. Newport strains were generated with PAUP* using maximum-parsimony methods. One mdh and three overlapping mutS gene fragments were sequenced and concatenated (total fragment length, 1,460 bp). repA genes for the plasmid comparison were identified by BLAST, corresponding protein sequences were aligned with Muscle, and a phylogenetic tree, including bootstrap values, was predicted with FastTree.
BLAST score ratio comparisons (61) between all newly and previously sequenced S. enterica chromosomes were carried out. Briefly, each translated coding sequence (CDS) from one chromosome is compared using BLASTp against the translated CDS from all other chromosomes. The resulting BLAST score is normalized against the BLAST score of the comparison of each protein against itself. CDS with a BLAST score ratio of <0.8 were considered to be absent from a genome. Customized Perl scripts were used to determine the distribution of each CDS across all chromosomes. Pseudogene fractions among the CDS considered absent based on BLAST score ratio analyses were determined using tBLASTn searches (options: -F F, -e 1.0e-20) of the protein sequences of the missing CDS against host (pseudo)chromosomes. Single or concatenated matches with a coverage of 80% or more over the protein sequence query were considered pseudogenes.
Phenotype characterizations.
The Biolog phenotype microarray system allows the testing of nearly 1,200 metabolic and chemical sensitivity phenotypes in a single run within 48 h (32). Here, the Biolog system was used to assay the utilization of different carbon sources by all newly sequenced S. enterica isolates in comparison to that by S. Typhimurium LT2 as described previously (7). Briefly, cells in a defined medium containing one of two reducible dyes were inoculated into 96-well plates that contained various carbon substrates. The plates were incubated at 37°C in the Biolog chamber, where cell growth and respiration led to reduction of the tetrazolium dye. Color intensities, which are proportional to bacterial growth, were recorded periodically (every 15 min) by a charge-coupled device camera and analyzed by the Biolog software, yielding a quantitative analysis of all collected data. Two independent assays were carried out with two different dyes for each isolate.
Plasmid transfer experiments.
Conjugative plasmid transfers between the two plasmid-carrying S. Schwarzengrund strains and plasmid-free strains S. Newport SL317, S. Kentucky SL479, and Escherichia coli DH10B were performed as described previously (22). Briefly, donor strains with antimicrobial resistance plasmids and rifampin-resistant recipient strains were grown to mid-log phase and harvested by centrifugation, and 108 CFU of the donor and recipient were mixed and spotted onto LB agar plates. Conjugation mixtures were incubated for 3 h at 37°C, after which the cells were resuspended in phosphate-buffered saline, diluted, and plated onto LB agar containing appropriate media to counterselect for the donor (100 μg/ml rifampin) and the plasmid resistance marker (10 μg/ml tetracycline).
RESULTS AND DISCUSSION
Of the 17 genomes that were submitted to the NCBI genome database, 6 were sequenced and fully closed while 11 were sequenced to draft level with between 2 and 173 contigs (Table 1). Unless specified otherwise, the use of the serovar designations will refer to all newly or previously sequenced S. enterica isolates from this serovar.
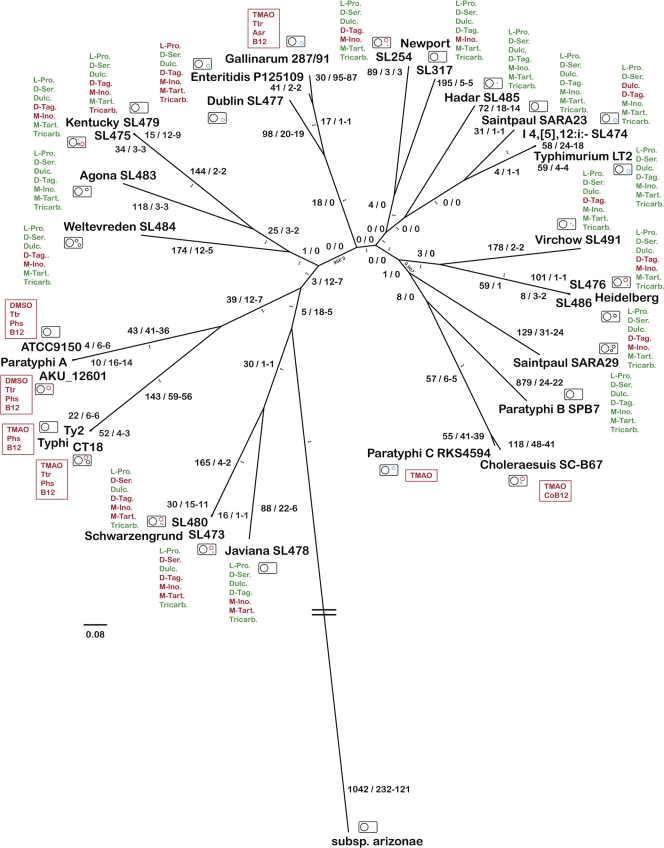
The chromosome sizes range from ∼4,546 kbp (Javiana draft chromosome, 19 contigs) to ∼4,951 kbp (Weltevreden draft chromosome, 77 contigs). In comparison, the two host-restricted Paratyphi A strains are among the isolates with the smallest chromosomes (4,582 kbp and 4,585 kbp, respectively). The other two human host-restricted typhoid strains S. Typhi Ty2 and CT18, however, have intermediate chromosome sizes (4,792 kbp and 4,809 kbp), suggesting that host adaptation and increased pathogenicity in S. enterica serovars are not necessarily reflected in smaller genome sizes. Yet it could be argued that the two Typhi and Paratyphi A strains are at different stages of their evolutionary paths toward specialization to a host-adapted pathogenic lifestyle, and thus, differences in chromosomal size are reflective of the degree of genome decay. In this case, the different evolutionary stages would, however, not be reflected in the pseudogene contents, which are similar in the two serovars (Fig. 1) (33).
Fig. 1.
Phylogenetic tree of all sequenced S. enterica strains based on whole-chromosome alignments. Values at branch tips are the identified plasmid content and type, color coded as associated with predicted antimicrobial resistance (red), virulence (blue), both (pink), or unknown (black) phenotypes. Plasmids smaller than 10 kbp are shown as black dots. Chromosomal CDS whose presence or absence is unique to the members of each phylogenetic branch, relative to all other chromosomes, and pseudogenes are shown as large numbers (e.g., for S. enterica subsp. arizonae, 1042/232-121 indicates 1,042 unique present genes and 232 unique absent genes of which 121 are predicted unique pseudogenes), and bootstrap values are shown as small numbers. Negative and positive Biolog phenotypes are shown in red and green, respectively, and abbreviated as L-Pro. (l-proline), D-Ser. (d-serine), Dulc. (dulcitol), D-Tag. (d-tagatose), M-Ino. (myo-inositol), M-Tart. (meso-tartaric acid), and Tricarb. (tricarballylic acid). Additional negative phenotypes, as predicted from the comparison of CDS by BLAST score ratio, are boxed and abbreviated as DMSO (anaerobic DMSO reduction), TMAO (anaerobic TMAO reduction), Ttr (anaerobic tetrathionate reduction), Phs (anaerobic thiosulfate reduction), Asr (anaerobic sulfite reduction), and CoB12 (cobalamin biosynthesis).
Phylogenetic relationship of all S. enterica genomes.
In silico MLST using seven gene fragments with a combined length of 3,340 bp (http://mlst.ucc.ie/), was applied to construct a phylogenetic tree of all 28 newly and previously sequenced S. enterica isolates (see Fig. S1 in the supplemental material). This tree, however, contains several branches with low bootstrap support (<50%) and places the two sequenced Typhi strains, which have a unique ancestor in common with the two Paratyphi A strains, on a long separate phylogenetic branch, indicating that the Typhi strains evolved faster from the last common ancestor than the Paratyphi A strains did. This difference in branch length is largely due to 17 single-nucleotide polymorphisms (SNPs) and six gaps in the 400-bp alignment of the purE gene fragment compared to 61 SNPs and the same number of gaps over the entire 3,340-bp MLST fragment between Typhi CT18 and Typhimurium. As MLST schemes that use a limited number of housekeeping genes can be sensitive to sequence variations caused by homologous recombination, a phylogenetic tree was also constructed based on multiple sequence alignments of a 2.088-Mbp core chromosome shared by all 28 sequenced S. enterica genomes (Fig. 1). We expect the genome-based phylogenetic reconstruction to be less influenced by horizontal gene transfer and recombinatorial events than that based on the 3.3-kbp MLST fragment. In contrast to the MLST tree, unique branches of similar lengths are observed in the core chromosome-based tree that indicate equivalent evolutionary rates for the Typhi and Paratyphi A serovars.
The S. enterica serotyping-based strain classification is, in some instances, supported by the core chromosome tree, as most pairs of isolates of the same serovar (Heidelberg, Kentucky, Paratyphi A, Schwarzengrund, or Typhi) are more closely related to each other than to isolates from other serovars (Fig. 1). Interestingly, isolates of serovars Choleraesuis and Paratyphi C, Gallinarum and Enteritidis, and Typhimurium and I 4,[5],12:i:−, respectively, are predicted to share recent common ancestries comparable to those of the serovar isolate pairs mentioned above. This observation is in agreement with previous findings that were based on shared gene (Paratyphi C and Choleraesuis [50]) and pseudogene (Gallinarum and Enteritidis [73]) contents. Except for Typhimurium and I 4,[5],12:i:−, related strains of different serotypes are characterized by different host-adapted pathogenic lifestyles. That is, whereas S. Paratyphi C is a human-adapted typhoid agent (38), S. Choleraesuis is primarily a swine pathogen (11). Likewise, S. Gallinarum is a poultry-restricted pathogen (66) and S. Enteritidis is considered a ubiquitous serovar (73). Within a closely related chromosomal background, these strains evolved into different serovars, suggesting that host adaptation exerts strong selection pressure on the genetic loci defining the serotype, namely, the O lipopolysaccharide and the H flagellar antigens (9). An overview of the differences between the gene clusters encoding O- and H-antigen biosynthesis between the S. enterica chromosomes is given in Table S3 in the supplemental material.
Among the 17 newly sequenced S. enterica genomes, the Saintpaul and Newport serovars represent two cases where serotyping only poorly reflects evolutionary relationships, as determined by whole-chromosome alignments. The two S. Saintpaul strains do not have a unique serovar-specific ancestor in common, suggesting that this serotype evolved at least twice in independent events or that the genetic loci responsible for the Saintpaul serotype have been transferred horizontally. Instead, Saintpaul SARA23 is most closely related to Typhimurium and I 4,[5],12:i:−, whereas Saintpaul SARA29 evolved on a separate phylogenetic branch, showing no close relationship to any of the other sequenced isolates but having an ancestor in common with serovars Choleraesuis and Paratyphi B and C.
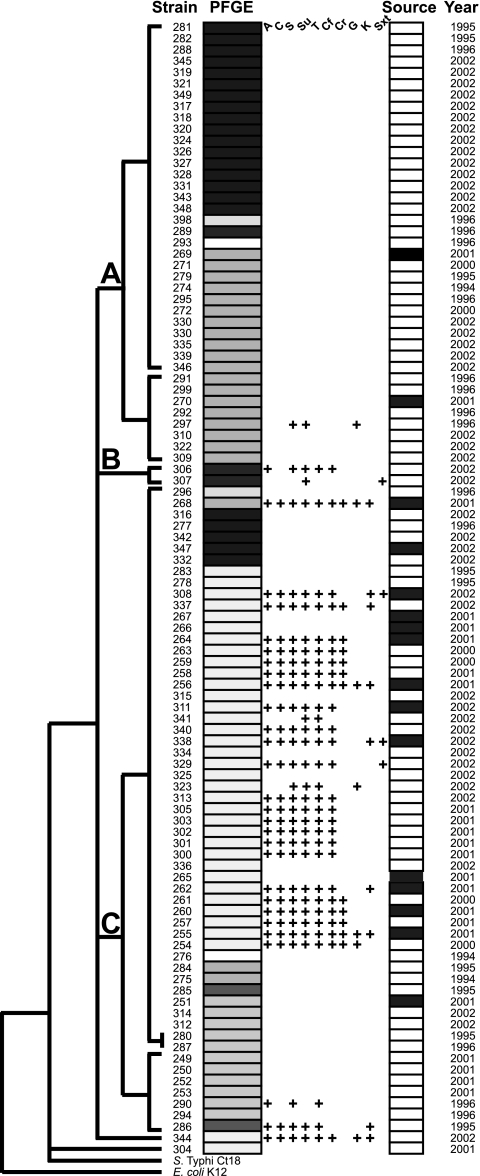
The two Newport strains, while having a Newport-specific ancestor in common, have evolved on separate branches that are longer than those separating many other isolates from different serotypes, e.g., Gallinarum, Enteritidis, and Dublin. Different sublineages within the Newport serotype have been identified and studied previously, based on PFGE pattern analysis (30) and MLST (65). We verified the existence of at least two sublineages within the Newport serovar using sequence analysis of the mdh and mutS genes (Fig. 2). The two Newport sublineages represented by strains SL254 and SL317 differ substantially with respect to the prevalence of antimicrobial resistance phenotypes, with Newport SL317 belonging to a cluster with a low prevalence of resistance phenotypes and Newport SL254 belonging to a cluster of isolates with a high prevalence of MDR (Fig. 2). The high prevalence of MDR-AmpC-SN resistance phenotypes (resistance to ACSSuT [ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, and tetracycline], additional resistance to amoxicillin-clavulanate and ceftiofur, and decreased susceptibility to ceftriaxone [MIC, ≥2 μg/ml]) in S. Newport have been described before (80). Resistance phenotypes in S. Newport are often plasmid encoded, and MDR plasmids of the IncA/C type have been identified in many Newport isolates from agricultural sources (48, 79). We have previously shown that Newport SL317 is less efficient as a recipient of plasmids by conjugation than Kentucky SL479 or E. coli DH10B (22). It is therefore plausible that as-yet-unidentified factors controlling plasmid acquisition and/or other modes of horizontal gene transfer play a role in determining the ability of a genome to acquire antimicrobial resistance and/or other phenotypes. The clustered regularly interspaced short palindromic repeats (CRISPR) system (41), which is discussed below, potentially provides such a mechanism, and differences in the CRISPR locus might be responsible for the observed evolutionary split of serovar Newport into different sublineages.
Fig. 2.
Characterization of S. Newport sublineages. One hundred S. Newport isolates obtained from the Minnesota Department of Health, including sequenced strains SL254 and SL317, were typed based on a two-gene (mdh, mutS, 1,460 bp) MLST scheme (dendrogram) and PFGE (70). Antimicrobial resistance patterns are shown in the middle columns. Abbreviations: A, ampicillin; C, chloramphenicol; S, streptomycin; Su, sulfisoxazole; T, tetracycline; Cf, cephalothin; Cr, ceftriaxone; G, gentamicin; K, kanamycin; Sxt, trimethoprim-sulfamethoxazole. Isolate sources are indicated by white (human isolate) and black (animal reservoir) bars.
Plasmid content.
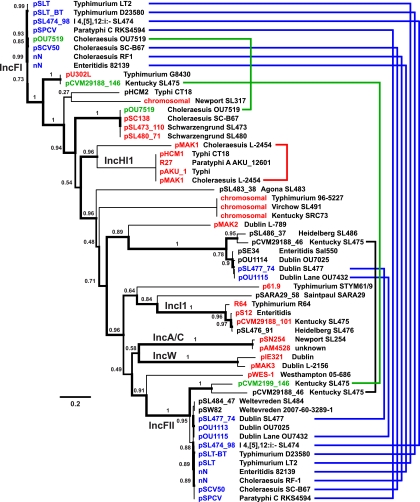
Of the 28 sequenced S. enterica genomes, 20 harbor between one and three plasmids or plasmid contigs (Table 1), including seven antimicrobial resistance plasmids and eight virulence plasmids. Those were of two types, (i) most notably, the Salmonella plasmid virulence (spv) type (28), responsible for predicted systemic infection phenotypes in serovars Typhimurium, I 4,[5],12:i:−, Choleraesuis, Paratyphi C, Gallinarum, Enteritidis, and Dublin, and (ii) the avian pathogenic E. coli (APEC) virulence type, responsible for chicken colonization and extraintestinal disease phenotypes in serovar Kentucky SL475 (22, 39). RepA-based phylogeny was used to compare all 38 sequenced repA+ S. enterica plasmids or plasmid contigs (Fig. 3). The majority of the known S. enterica virulence plasmids belong to the IncFI and IncFII incompatibility groups, including spv and APEC virulence plasmids, whereas antimicrobial resistance plasmids were identified in all incompatibility groups, including IncF, IncA/C, IncHI1, and IncI1. In several cases (Fig. 3), multiple repA genes were found on the same plasmid or on chromosomes, suggesting a dynamic S. enterica plasmid evolution that includes frequent plasmid cointegration events and integrations into the host chromosomes.
Fig. 3.
Phylogenetic relationships of S. enterica plasmids based on RepA protein sequence alignments. Confidence values are reflected in the line widths of the corresponding branches. Names of plasmids with predicted virulence, antimicrobial resistance, or both phenotypes in combination are shown in blue, red, and green, respectively. Lines on the right side connect repA genes that were identified on the same plasmid or plasmid contig.
The Weltevreden draft genome contains two contigs (47 kbp and 51 kbp) with high similarity (99% nucleotide identity over 25 kbp and 51 kbp, respectively) to the recently sequenced plasmid pSW82 from Weltevreden strain 2007-60-3289-1 (accession no. FR775255). Both pSW82 and the concatenated sequence of the two contigs from the Weltevreden SL484 draft genome contain two CDS (13.3 kbp and 8.4 kbp) for a nonribosomal peptide synthetase/polyketide synthase (NRPS/PKS) of unknown function. In addition, both plasmids harbor a conjugative plasmid transfer operon, which is truncated in pSW82 (traDISTX) compared to the SL484 plasmid (traABCDEFGHKLNRSTUVWX, trbBCI). To our knowledge, this is the first time that an NRPS/PKS system has been identified on a potentially conjugative plasmid in S. enterica and in the family Enterobacteriaceae. Both Weltevreden plasmids carry repA genes of the IncFII type (Fig. 3) similar to one of two plasmid origins found on all spv virulence plasmids, except pOU7519 from Choleraesuis OU7519 (accession no. EU219534).
The presence or absence of specific plasmid types or plasmid-encoded antimicrobial resistance phenotypes in the sequenced S. enterica strains does not correlate with the evolutionary relationship of their host chromosomes, as similar resistance plasmids are found in isolates from different serovars (Fig. 3). Resistance plasmids can be exchanged among phylogenetically unrelated S. enterica strains (22, 79), and identical resistance gene cassettes can be shared between different plasmid backbones (24). Virulence plasmids of the spv type, however, are present in only seven members of three phylogenetic clusters, (i) Typhimurium and I 4,[5],12:i:−, (ii) Choleraesuis and Paratyphi C, and (iii) Gallinarum, Enteritidis, and Dublin. Although the results presented here are biased by the selection of sequenced S. enterica isolates, this could indicate that spv virulence plasmids are more stably maintained than known resistance plasmids and are therefore likely to play important roles in long-term species evolution.
Gene acquisition and loss.
BLAST score ratio comparisons (61) between all sequenced S. enterica chromosomes were carried out in order to investigate the distribution of CDS across all genomes. With this method, using one chromosome as a reference, the presence or absence of each gene across all other chromosomes can be determined (see Fig. S2 in the supplemental material). This information was used to map protein-coding characteristics onto the phylogenetic tree (Fig. 1), i.e., shared gene presence or absence, including pseudogenes, among all members of a phylogenetic branch, thereby assigning specific functions to different evolutionary sublineages within the S. enterica species. The same information was also used to investigate the presence or absence of specific metabolic or other functions across all S. enterica chromosomes independently of their phylogenetic relationships (see Table S3).
All 28 S. enterica genomes have a core chromosome of 2,276 CDS and contain genomic islands that are unique or shared with only a subset of the other chromosomes and are located predominantly at a few conserved locations (see Fig. S2 in the supplemental material). Most S. enterica chromosomes for which the phylogenetic tree indicates recent common ancestry; i.e., isolates which are connected by relatively short unique branches are characterized by CDS whose presence or absence is unique to these isolates, suggesting that S. enterica evolution into phylogenetic sublineages is accompanied by gene acquisition and/or loss (Fig. 1).
A large fraction of the CDS unique to members of a phylogenetic branch are predicted to originate from chromosomally integrated prophage elements, whereas more of the absent CDS are annotated with metabolic functions. Of the CDS that are unique to members of a single serotype (with the exception of the Newport and Saintpaul serovars), 3,020 CDS (76%) are annotated as “hypothetical proteins,” compared to only 78 (10%) of the absent CDS. None of the predicted antimicrobial resistance determinants are shared among phylogenetically related S. enterica chromosomes (see Table S4 in the supplemental material), which is in accordance with the assumption that antimicrobial resistance determinants in S. enterica are acquired mostly via horizontal gene transfer (22). It further supports the conclusion made above, based on plasmid type comparisons, that the emergence of specific resistance phenotypes is not correlated with the evolutionary history of S. enterica.
Loss or acquisition of metabolic functions. (i) Biolog phenotype data.
All 17 newly sequenced S. enterica isolates and Typhimurium LT2 were phenotypically characterized for their metabolic abilities to grow on different carbon sources by using the Biolog Phenotype MicroArrays system (see Table S5 in the supplemental material). Phenotypic variations even between closely related S. enterica isolates were observed for seven carbon substrates (Fig. 1). For three of these compounds (l-proline, galactitol, and myo-inositol), incomplete gene sets for substrate uptake or degradation could be identified based on BLAST score ratios (see Table S3). These phenotypic analyses suggest that during the evolution from the last common ancestor, the ability to utilize various carbon sources was either lost or acquired on multiple independent occasions. On the genotypic level, a previous microarray analysis of the presence of Typhimurium LT2 genes in representative Salmonella genomes from all seven subspecies came to similar conclusions based on the observation that genes were not distributed according to species phylogeny (59).
(ii) Anaerobic metabolism.
Porwollik et al. (2002) identified a set of genes specific for S. enterica compared to two E. coli strains (K12 and O157:H7), Klebsiella pneumoniae MGH 78578, and Yersinia pestis CO92 (59). This set included CDS from the ttr, phs, and asr operons involved in the dissimilatory reduction of oxidized sulfur compounds under anaerobic conditions (2, 15, 31, 37). Since CDS of these operons were found to be absent from several S. enterica genomes analyzed by BLAST score ratio in this study, the presence of a larger group of CDS involved in anaerobic respiration was examined across all sequenced genomes (see Table S3 in the supplemental material). The results of this analysis indicate that seven sequenced S. enterica isolates from five serovars (Typhi, Paratyphi A and C, Choleraesuis, and Gallinarum) have lost the ability to reduce at least one of the following oxidized substrates in order to maintain a respiratory metabolism under anaerobic conditions: tetrathionate (Typhi, Paratyphi A, and Gallinarum), thiosulfate (Typhi and Paratyphi A), sulfite (Gallinarum), dimethyl sulfoxide (DMSO; Paratyphi A), and trimethylamine N-oxide (TMAO; Typhi, Choleraesuis, Gallinarum, and Paratyphi C). In addition, the gene cluster encoding malate dehydratase FumB, which can provide fumarate as an electron acceptor under anaerobic conditions (75), is incomplete in serovars Typhi, Paratyphi A, Javiana, Schwarzengrund, and Saintpaul SARA29. Similarly, the nitrate-inducible formate dehydrogenase, which serves as a major electron donor during anaerobic respiration (5), is predicted to be inactive in serovars Choleraesuis, Typhi, and Paratyphi A, B, and C. As the enzymes for tetrathionate, thiosulfate, and DMSO reduction use a molybdenum cofactor, the cbi operon of coenzyme B12 biosynthesis was examined and found to be incomplete in serovars Typhi and Paratyphi A, Choleraesuis, and Gallinarum. Other gene clusters for coenzyme B12-dependent reactions, which are incomplete in several S. enterica genomes include the 1,3-propanediol utilization operon pdu (Typhi, Paratyphi A, Choleraesuis, Gallinarum, and Newport SL317) and the ethanolamine utilization operon eut (Choleraesuis and Paratyphi C).
It is interesting that S. enterica isolates associated with the highest number of predicted deficiencies in anaerobic respiration, vitamin B12 uptake or biosynthesis, and cobalamin-dependent enzymatic reactions belong to the S. enterica group of host-restricted (Typhi; Paratyphi A, B, and C; and Gallinarum) or host-adapted (Choleraesuis) serovars. It was previously shown that Typhimurium strains defective in vitamin B12 biosynthesis show no decrease in virulence (6, 64). A recent study also suggested that ethanolamine and propanediol utilization allows S. Typhimurium to occupy specific niches within food environments and harvest substrate sources such as egg yolk or whole milk (68). The ability to maintain a broad substrate spectrum and respiratory metabolism under anaerobic conditions, which requires an extensive repertoire of genes coding for enzymes, transporters, and cofactor biosynthesis, seems to be less important for sublineages specialized on host-adapted pathogenic lifestyles than for ubiquitous S. enterica sublineages with human, animal, and environmental reservoirs such as, for example, Newport, Heidelberg, and Schwarzengrund.
Antimicrobial resistance.
Antimicrobial resistance determinants corresponding to observed phenotypes (see Table S2 in the supplemental material) were identified in all newly sequenced S. enterica isolates (see Table S4). The genetic background of the resistance loci suggests that all resistance phenotypes were acquired by horizontal gene transfer, with the exception of resistance to quinolones in Schwarzengrund and Virchow.
Nonsynonymous mutations in the gyrA (Ser83Phe, Asp87Gly) and parC (Ser80Arg) genes encoding DNA gyrase and DNA topoisomerase IV subunits, respectively, were identified in both Schwarzengrund isolates, which are resistant to nalidixic acid (a narrow-spectrum quinolone) and ciprofloxacin (an expanded-spectrum fluoroquinolone). These mutations are known to mediate quinolone resistance in S. enterica and E. coli (44, 47). S. enterica strains of serotype Schwarzengrund were responsible for the first recognized outbreak of fluoroquinolone-resistant salmonellosis in the United States (55). The parC Ser80Arg mutation, found in Schwarzengrund but not in any other S. enterica genome examined in the present study, is thought to occur rarely among S. enterica serovars (34). Previously, this parC mutation was found in concert with the same two gyrA mutations in three other Schwarzengrund isolates (34), suggesting that this serovar may be more likely to manifest increased quinolone resistance. The same two gyrA mutations (Ser83Phe, Asp87Gly) were also identified in Choleraesuis SC-B67, a fluoroquinolone-resistant strain isolated in Taiwan in 2002 (10), where emerging fluoroquinolone resistance phenotypes caused significant concerns about the treatment of systemic nontyphoidal salmonellosis (12). Virchow SL491, which shows resistance to nalidixic acid but not ciprofloxacin, carries a distinct single mutation in gyrA (Asp87Asn).
Antimicrobial resistance-conferring plasmids, identified in Newport SL254 and Kentucky SL475, were described in detail in previous publications (22, 23, 79). Both Schwarzengrund isolates carry MDR plasmids that share a backbone with pSC138 from Choleraesuis SC-B67 but show no significant sequence homology to any other previously sequenced plasmid, except for the resistance-encoding region (see Fig. S3 in the supplemental material). These two plasmids carry a type IV secretion system-like conjugal transfer operon, which is absent from pSC138 and most closely related to a chromosomally integrated transfer gene cluster from the endophyte K. pneumoniae 342 (CP000964). Both Schwarzengrund plasmids could be transferred to E. coli DH10B but not at detectable rates to Kentucky SL479 or Newport SL317 (see Table S6). The high prevalence of fluoroquinolone-resistant Choleraesuis strains in Asia (12, 13) and previously raised concerns about the spread of MDR Schwarzengrund strains from chickens to persons in Thailand and from imported Thai food products to persons in Denmark and the United States (1) raises the somber prospect that resistance determinants from the horizontal gene pool are circulating around the globe and are potentially able to transfer to pathogenic strains in the United States.
Chromosomally integrated resistance islands are found in Hadar, Heidelberg SL476, Kentucky SL475, and Virchow. Virchow SL491 was the first strain identified in the United States with the rmtC gene, which confers resistance to aminoglycosides (21). This gene is located within a resistance fragment for which the presence of a repA (Fig. 3) and other plasmid-related CDS (traN, traGH for conjugative plasmid mating pair stabilization and sex pilus assembly functions) suggests at least a partially conjugative (plasmid) origin. This fragment also contains a mercury(II) resistance-encoding partial transposon, Tn512, which was first identified on an IncP plasmid from a clinical Pseudomonas aeruginosa isolate and characterized based on its ability to perform resolvase-independent random transposition (58).
Kentucky SL475 carries a total of four copies—two complete (tnpA–blaCMY-2-–blc–sugE) and two truncated (tnpA–blaCMY-2) copies—of a previously described transposon-like resistance element (69). The blaCMY-2 gene encodes a cephalomycinase that exhibits extended resistance to many β-lactams, including narrow-, expanded-, and broad-spectrum cephalosporins, whereas SugE has been shown to confer decreased susceptibility to quaternary ammonium compounds, including common disinfectants such as cetylpyridinium chloride (14, 81). The close physical proximity of sugE to the blaCMY-2 β-lactamase gene is of particular public health significance, as it points to the potential coselection of linked CDS carried by extrachromosomal elements and their role in the spread of MDR phenotypes among veterinary and human pathogens.
CRISPR-mediated control of horizontal gene transfer.
CRISPR elements, identified in most sequenced archaea (∼90%) and many bacteria (∼40%) (67), including E. coli and S. enterica (74), provide acquired immunity against viruses and plasmids by targeting nucleic acid in a sequence-specific manner (35). CRISPR loci typically contain a CRISPR array, consisting of several noncontiguous direct repeats separated by stretches of variable sequences (CRISPR spacers), which is often located adjacent to CRISPR-associated (cas) gene clusters. CRISPR systems perform two processes. (i) During immunization, exogenous viral or plasmid DNA is recognized by the Cas complex, which integrates a novel CRISPR spacer unit into the CRISPR array (36, 52). (ii) During immunity, the CRISPR array is transcribed and used by the Cas complex to target invading DNA or RNA, based on sequence homology between the CRISPR spacer and the invading nucleic acids (29, 51).
To determine whether differences in the CRISPR setup, i.e., the number and/or specificity of CRISPR spacers, could account for variations in observed S. enterica plasmid and phage contents, CRISPR loci from all sequenced S. enterica genomes were identified and comparatively analyzed (see Table S7 in the supplemental material). S. enterica subsp. arizonae does not contain a CRISPR locus, and CRISPR loci from S. Javiana and S. Paratyphi B are truncated and most likely nonfunctional, as they lack most of the cas/cse (Cas subtype E. coli) locus found in the other chromosomes. The numbers of both CRISPR arrays and CRISPR spacers within those arrays vary considerably among the sequenced S. enterica strains (see Table S7). Previous studies have suggested frequent gain and loss of the CRISPR/cas locus by homologous recombination (74). The third CRISPR locus in Newport SL317, which is a 100% identical truncated copy of one of the other two loci, is a likely result of such a recombination event.
Phylogenetic distances between the sequenced S. enterica isolates are partially reflected in the composition of CRISPR arrays (see Fig. S4 in the supplemental material). Isolates from the same serovar have identical CRISPR loci (Heidelberg and Schwarzengrund) or share at least most of their CRISPR spacers (Kentucky, Paratyphi A, and Typhi), with the exception of the two Newport and Saintpaul strains. The phylogenetically related Typhimurium and Typhimurium-like Salmonella I 4,[5],12:i:− isolates, as well as Gallinarum and Enteritidis, also show overlapping CRISPR spacer sets. In addition, several phylogenetically more distantly related Heidelberg, Typhimurium, I 4,[5],12:i:−, Newport SL317, and Hadar isolates share a significant subset of their CRISPR spacers (see Fig. S4).
To determine whether CRISPR could be involved in the control of horizontal gene transfer of the sequenced S. enterica plasmids or prophages, BLAST comparisons of all CRISPR spacers were carried out. BLAST searches (>30-nucleotide length, <4 nucleotide mismatches) identified 14 matches between CRISPR spacers and S. enterica non-CRISPR chromosomal regions (see Table S8 in the supplemental material). With one exception, all CRISPR spacers match CDS predicted to be part of prophage elements. All spv plasmid-containing S. enterica isolates carry a CRISPR spacer that matches a noncoding chromosomal region in the two Typhi strains for which there is no indication of a phage, plasmid, or other horizontal gene transfer origin. A single match between a CRISPR spacer and any plasmid sequence from the NCBI database was found between Saintpaul SARA23 and the pefC gene on the spv type virulence plasmids, which encodes a fimbrial usher protein (e.g., P37868). On the spv plasmids, pefC is genetically linked to the IncFI origin of replication, because pefC is found only on those virulence plasmids that carry both IncFI ori and IncFII ori (Typhimurium, I 4,[5],12:i:−, Choleraesuis, Paratyphi C, and Enteritidis) and not on those that carry just the IncFII ori (Dublin). Saintpaul SARA23 has a recent ancestor in common with the spv virulence plasmid-carrying Typhimurium and I 4,[5],12:i:− strains (Fig. 1). It is therefore conceivable that CRISPR-mediated incompatibility with the spv virulence plasmid prevented the Saintpaul SARA23 ancestor from adapting to the same pathogenic lifestyle as the ancestor of Typhimurium and I 4,[5],12:i:−. This CRISPR-mediated incompatibility could have led to the emergence of a new phylogenetic sublineage within the S. enterica species, which is now represented by Saintpaul SARA23.
The identified matches between CRISPR spacers and actual S. enterica plasmids and prophages indicate that the CRISPR system is potentially involved in controlling plasmid- and phage-mediated horizontal gene transfer, which has been shown to be associated with virulence functions in S. enterica, suggesting an important impact of the CRISPR system on S. enterica species evolution and the emergence of new S. enterica sublineages.
Conclusion.
The 28 completely sequenced S. enterica genomes from 21 serotypes analyzed here represent a broad coverage of the phenotypic and genotypic diversity of the species, ranging from susceptible to MDR, from mildly to highly pathogenic, and from human to animal and food isolates. Whole-chromosome alignments were used to draw a comprehensive phylogenetic map of the S. enterica species, indicating a continuing evolution into several separate sublineages. The comparison of phylogenetic, genotypic, and phenotypic data suggests that phenotypes are frequently lost and/or acquired in different S. enterica sublineages and thus correlate poorly with the evolutionary relationships within this species. Classification schemes that rely solely on phenotype data (e.g., serotyping) will therefore perform poorly in predicting evolutionary relationships.
Plasmid- and phage-mediated horizontal gene transfer events play an important role in the short-time acquisition of new phenotypes, including antimicrobial resistance and virulence, as documented by the identification of unrelated resistance plasmids and chromosomally integrated resistance islands, the combined resistance-virulence plasmid in Kentucky SL475 (22), and prophages containing virulence genes in S. Typhimurium (19). Plasmid-mediated phenotypes seem no less stable than chromosomally encoded phenotypes, as correlations between phylogeny and spv plasmid presence indicate that these plasmids play an important role in S. enterica sublineage evolution.
Phylogenetic distances are, to a certain extent, reflected in conserved CRISPR locus compositions, including CRISPR-associated genes and target-specific CRISPR spacers. Correspondingly, the existence of sublineages within a serotype, such as those seen in Newport and Saintpaul, is reflected in different CRISPR locus compositions. Matches between CRISPR spacers and plasmid and prophage regions suggest the involvement of CRISPR in the control of horizontal gene transfer, with likely consequences for both short-term phenotype changes and long-term sublineage evolution in S. enterica. It is tempting to speculate about short-term phenotype changes, which could be determined by the receptiveness toward plasmid acquisition, as in Newport SL317, which belongs to a sublineage within the Newport serovar with a relatively low prevalence of antimicrobial resistance phenotypes and, in previous experiments, has been shown to be less efficient as a plasmid recipient than Kentucky SL479 (22). Long-term sublineage evolution, on the other hand, could be determined by the CRISPR-mediated incompatibility with plasmids that are key for the adaption to pathogenic lifestyles, such as seen in the separation of spv plasmid-containing Typhimurium and I 4,[5],12:i:− serovars and the plasmid-free Saintpaul SARA23 sublineage. Although the data presented here are very preliminary, CRISPR configurations could therefore be critical in assessing the evolutionary potential of S. enterica strains and thus aid in efforts to monitor, predict, and prevent the future emergence of S. enterica outbreak strains. While frequent variations in CRISPR array compositions between closely related S. enterica isolates contraindicate the use of these elements alone for isolate or sublineage typing, a recent (2011) study by Liu et al. showed that an MLST scheme that combines CRISPR loci and virulence genes (sseL, fimH) performed well in differentiating outbreak isolates from the same Salmonella serovars (49).
Based on the identification of S. enterica CDS whose presence or absence characterizes members of different phylogenetic branches within the species, the following two mechanisms seem to play a major role in S. enterica species evolution, (i) the loss of relatively well-characterized CDS with known metabolic functions and (ii) the horizontal acquisition via phage or plasmid transfer of CDS or pseudogenes with mostly unknown functions of which a small fraction are involved in antimicrobial resistance and virulence. This would be in accordance with the assumption that the sequenced S. enterica genomes represent sublineages within the S. enterica species that are in the process of evolving from a metabolically more versatile ancestor to a more specialized lifestyle. The genomes of the more specialized and host-adapted serotypes (Typhi, Paratyphi, Gallinarum, and Choleraesuis) show the most significant loss of metabolic functions in gene-by-gene comparisons and might be reflective of a more advanced stage of the evolutionary process.
Supplementary Material
ACKNOWLEDGMENTS
We thank Samuel Angiuoli, David Rasko, Jason Sahl, and James White at the Institute for Genome Sciences, Baltimore, MD, for thoughtful feedback and for suggestions, as well as Scott Durkin at the J. Craig Venter Institute, Rockville, MD. We thank the Minnesota Department of Health for graciously providing the S. Newport strains used in the analysis.
Sequencing of the 17 S. enterica genomes was supported with federal funds from the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health, Department of Health and Human Services, under NIAID contract N01-AI-30071.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 20 May 2011.
REFERENCES
- 1. Aarestrup F. M., et al. 2007. International spread of multidrug-resistant Salmonella Schwarzengrund in food products. Emerg. Infect. Dis. 13:726–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alami N., Hallenbeck P. C. 1995. Cloning and characterization of a gene cluster, phsBCDEF, necessary for the production of hydrogen sulfide from thiosulfate by Salmonella typhimurium. Gene 156:53–57 [DOI] [PubMed] [Google Scholar]
- 3. Angiuoli S. V., Salzberg S. L. 2011. Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics 27:334–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anonymous. 2010. Surveillance for foodborne disease outbreaks—United States, 2007. MMWR Morb. Mortal. Wkly. Rep. 59:973–979 [PubMed] [Google Scholar]
- 5. Barrett E. L., Riggs D. L. 1982. Salmonella typhimurium mutants defective in the formate dehydrogenase linked to nitrate reductase. J. Bacteriol. 149:554–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Björkman J., Rhen M., Anderson D. I. 1996. Salmonella typhimurium cob mutants are not hyper-virulent. FEMS Microbiol. Lett. 139:121–126 [DOI] [PubMed] [Google Scholar]
- 7. Bochner B. R. 2003. New technologies to assess genotype-phenotype relationships. Nat. Rev. Genet. 4:309–314 [DOI] [PubMed] [Google Scholar]
- 8. Boyd E. F., et al. 1993. Salmonella reference collection B (SARB): strains of 37 serovars of subspecies I. J. Gen. Microbiol. 139(Pt. 6):1125–1132 [DOI] [PubMed] [Google Scholar]
- 9. CDC 2008. Public Health Laboratory Information System (PHLIS): Salmonella surveillance, annual summary, 2006. CDC, Atlanta, GA: http://www.cdc.gov/ncidod/dbmd/phlisdata/salmonella.htm [Google Scholar]
- 10. Chiu C. H., et al. 2004. Isolation of Salmonella enterica serotype Choleraesuis resistant to ceftriaxone and ciprofloxacin. Lancet 363:1285–1286 [DOI] [PubMed] [Google Scholar]
- 11. Chiu C. H., et al. 2005. The genome sequence of Salmonella enterica serovar Choleraesuis, a highly invasive and resistant zoonotic pathogen. Nucleic Acids Res. 33:1690–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chiu C. H., et al. 2002. The emergence in Taiwan of fluoroquinolone resistance in Salmonella enterica serotype Choleraesuis. N. Engl. J. Med. 346:413–419 [DOI] [PubMed] [Google Scholar]
- 13. Chiu C. H., Wu T. L., Su L. H., Liu J. W., Chu C. 2004. Fluoroquinolone resistance in Salmonella enterica serotype Choleraesuis, Taiwan, 2000-2003. Emerg. Infect. Dis. 10:1674–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chung Y. J., Saier M. H., Jr 2002. Overexpression of the Escherichia coli sugE gene confers resistance to a narrow range of quaternary ammonium compounds. J. Bacteriol. 184:2543–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clark M. A., Barrett E. L. 1987. The phs gene and hydrogen sulfide production by Salmonella typhimurium. J. Bacteriol. 169:2391–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. CLSI 2010. Twentieth informational supplement. CLSI document M100-S20. CLSI, Wayne, PA [Google Scholar]
- 17. Deng W., et al. 2003. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J. Bacteriol. 185:2330–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Edgar R. C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ehrbar K., Hardt W. D. 2005. Bacteriophage-encoded type III effectors in Salmonella enterica subspecies 1 serovar Typhimurium. Infect. Genet. Evol. 5:1–9 [DOI] [PubMed] [Google Scholar]
- 20. FDA 2010. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): Retail Meat Annu. Report, 2007. FDA, Bethesda MD: http://www.fda.gov/downloads/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/UCM166132.pdf [Google Scholar]
- 21. Folster J. P., Rickert R., Barzilay E. J., Whichard J. M. 2009. Identification of the aminoglycoside resistance determinants armA and rmtC among non-Typhi Salmonella isolates from humans in the United States. Antimicrob. Agents Chemother. 53:4563–4564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fricke W. F., et al. 2009. Antimicrobial resistance-conferring plasmids with similarity to virulence plasmids from avian pathogenic Escherichia coli strains in Salmonella enterica serovar Kentucky isolates from poultry. Appl. Environ. Microbiol. 75:5963–5971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fricke W. F., et al. 2009. Comparative genomics of the IncA/C multidrug resistance plasmid family. J. Bacteriol. 191:4750–4757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fricke W. F., et al. 2008. Insights into the environmental resistance gene pool from the genome sequence of the multidrug-resistant environmental isolate Escherichia coli SMS-3-5. J. Bacteriol. 190:6779–6794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Galanis E., et al. 2006. Web-based surveillance and global Salmonella distribution, 2000-2002. Emerg. Infect. Dis. 12:381–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Grassl G. A., Finlay B. B. 2008. Pathogenesis of enteric Salmonella infections. Curr. Opin. Gastroenterol. 24:22–26 [DOI] [PubMed] [Google Scholar]
- 27. Grimont P. A. D., Weill F. X. 2007. Antigenic formulae of the Salmonella serovars, 9th ed. WHO Collaborating Centre for Reference and Research on Salmonella, Pasteur Institute, Paris, France [Google Scholar]
- 28. Gulig P. A. 1990. Virulence plasmids of Salmonella typhimurium and other salmonellae. Microb. Pathog. 8:3–11 [DOI] [PubMed] [Google Scholar]
- 29. Hale C. R., et al. 2009. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139:945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Harbottle H., White D. G., McDermott P. F., Walker R. D., Zhao S. 2006. Comparison of multilocus sequence typing, pulsed-field gel electrophoresis, and antimicrobial susceptibility typing for characterization of Salmonella enterica serotype Newport isolates. J. Clin. Microbiol. 44:2449–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hensel M., Hinsley A. P., Nikolaus T., Sawers G., Berks B. C. 1999. The genetic basis of tetrathionate respiration in Salmonella typhimurium. Mol. Microbiol. 32:275–287 [DOI] [PubMed] [Google Scholar]
- 32. Ho T. D., Slauch J. M. 2001. OmpC is the receptor for Gifsy-1 and Gifsy-2 bacteriophages of Salmonella. J. Bacteriol. 183:1495–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Holt K. E., et al. 2009. Pseudogene accumulation in the evolutionary histories of Salmonella enterica serovars Paratyphi A and Typhi. BMC Genomics 10:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hopkins K. L., Arnold C., Threlfall E. J. 2007. Rapid detection of gyrA and parC mutations in quinolone-resistant Salmonella enterica using pyrosequencing technology. J. Microbiol. Methods 68:163–171 [DOI] [PubMed] [Google Scholar]
- 35. Horvath P., Barrangou R. 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327:167–170 [DOI] [PubMed] [Google Scholar]
- 36. Horvath P., et al. 2008. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 190:1401–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huang C. J., Barrett E. L. 1991. Sequence analysis and expression of the Salmonella typhimurium asr operon encoding production of hydrogen sulfide from sulfite. J. Bacteriol. 173:1544–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jacobs M. R., Koornhof H. J., Crisp S. I., Palmhert H. L., Fitzstephens A. 1978. Enteric fever caused by Salmonella paratyphi C in South and South West Africa. S. Afr. Med. J. 54:434–438 [PubMed] [Google Scholar]
- 39. Johnson T. J., et al. 2010. Horizontal gene transfer of a ColV plasmid has resulted in a dominant avian clonal type of Salmonella enterica serovar Kentucky. PLoS One 5:e15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jones T. F., et al. 2008. Salmonellosis outcomes differ substantially by serotype. J. Infect. Dis. 198:109–114 [DOI] [PubMed] [Google Scholar]
- 41. Karginov F. V., Hannon G. J. 2010. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol. Cell 37:7–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kingsley R. A., Baumler A. J. 2000. Host adaptation and the emergence of infectious disease: the Salmonella paradigm. Mol. Microbiol. 36:1006–1014 [DOI] [PubMed] [Google Scholar]
- 43. Kotewicz M. L., Mammel M. K., LeClerc J. E., Cebula T. A. 2008. Optical mapping and 454 sequencing of Escherichia coli O157: H7 isolates linked to the US 2006 spinach-associated outbreak. Microbiology 154:3518–3528 [DOI] [PubMed] [Google Scholar]
- 44. Kumagai Y., et al. 1996. Quinolone-resistant mutants of Escherichia coli DNA topoisomerase IV parC gene. Antimicrob. Agents Chemother. 40:710–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kurtz S., et al. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Layton A. N., Galyov E. E. 2007. Salmonella-induced enteritis: molecular pathogenesis and therapeutic implications. Expert Rev. Mol. Med. 9(18):1–17 [DOI] [PubMed] [Google Scholar]
- 47. Levy D. D., Sharma B., Cebula T. A. 2004. Single-nucleotide polymorphism mutation spectra and resistance to quinolones in Salmonella enterica serovar Enteritidis with a mutator phenotype. Antimicrob. Agents Chemother. 48:2355–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lindsey R. L., Fedorka-Cray P. J., Frye J. G., Meinersmann R. J. 2009. Inc A/C plasmids are prevalent in multidrug-resistant Salmonella enterica isolates. Appl. Environ. Microbiol. 75:1908–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu F., et al. 2011. Novel virulence gene and clustered regularly interspaced short palindromic repeat (CRISPR) multilocus sequence typing scheme for subtyping of the major serovars of Salmonella enterica subsp. enterica. Appl. Environ. Microbiol. 77:1946–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu W. Q., et al. 2009. Salmonella paratyphi C: genetic divergence from Salmonella choleraesuis and pathogenic convergence with Salmonella typhi. PLoS One 4:e4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Marraffini L. A., Sontheimer E. J. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Marraffini L. A., Sontheimer E. J. 2010. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 11:181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McClelland M., et al. 2004. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat. Genet. 36:1268–1274 [DOI] [PubMed] [Google Scholar]
- 54. McClelland M., et al. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856 [DOI] [PubMed] [Google Scholar]
- 55. Olsen S. J., et al. 2001. A nosocomial outbreak of fluoroquinolone-resistant salmonella infection. N. Engl. J. Med. 344:1572–1579 [DOI] [PubMed] [Google Scholar]
- 56. Parkhill J., et al. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848–852 [DOI] [PubMed] [Google Scholar]
- 57. Parry C. M., Hien T. T., Dougan G., White N. J., Farrar J. J. 2002. Typhoid fever. N. Engl. J. Med. 347:1770–1782 [DOI] [PubMed] [Google Scholar]
- 58. Petrovski S., Stanisich V. A. 2010. Tn502 and Tn512 are res site hunters that provide evidence of resolvase-independent transposition to random sites. J. Bacteriol. 192:1865–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Porwollik S., Wong R. M., McClelland M. 2002. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc. Natl. Acad. Sci. U. S. A. 99:8956–8961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Price M. N., Dehal P. S., Arkin A. P. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rasko D. A., Myers G. S., Ravel J. 2005. Visualization of comparative genomic analyses by BLAST score ratio. BMC Bioinformatics 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Read T. D., et al. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81–86 [DOI] [PubMed] [Google Scholar]
- 63. Ronquist F., Huelsenbeck J. P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574 [DOI] [PubMed] [Google Scholar]
- 64. Sampson B. A., Gotschlich E. C. 1992. Elimination of the vitamin B12 uptake or synthesis pathway does not diminish the virulence of Escherichia coli K1 or Salmonella typhimurium in three model systems. Infect. Immun. 60:3518–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sangal V., et al. 2010. Evolution and population structure of Salmonella enterica serovar Newport. J. Bacteriol. 192:6465–6476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shivaprasad H. L. 2000. Fowl typhoid and pullorum disease. Rev. Sci. Tech. 19:405–424 [DOI] [PubMed] [Google Scholar]
- 67. Sorek R., Kunin V., Hugenholtz P. 2008. CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 6:181–186 [DOI] [PubMed] [Google Scholar]
- 68. Srikumar S., Fuchs T. M. 2011. Ethanolamine utilization contributes to proliferation of Salmonella enterica serovar Typhimurium in food and in nematodes. Appl. Environ. Microbiol. 77:281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Su L. H., et al. 2006. Distribution of a transposon-like element carrying bla(CMY-2) among Salmonella and other Enterobacteriaceae. J. Antimicrob. Chemother. 57:424–429 [DOI] [PubMed] [Google Scholar]
- 70. Swaminathan B., Barrett T. J., Hunter S. B., Tauxe R. V. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Swofford D. L. 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland, MA [Google Scholar]
- 72. Talavera G., Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56:564–577 [DOI] [PubMed] [Google Scholar]
- 73. Thomson N. R., et al. 2008. Comparative genome analysis of Salmonella Enteritidis PT4 and Salmonella Gallinarum 287/91 provides insights into evolutionary and host adaptation pathways. Genome Res. 18:1624–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Touchon M., Rocha E. P. 2010. The small, slow and specialized CRISPR and anti-CRISPR of Escherichia and Salmonella. PLoS One 5:e11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tseng C. P. 1997. Regulation of fumarase (fumB) gene expression in Escherichia coli in response to oxygen, iron and heme availability: role of the arcA, fur, and hemA gene products. FEMS Microbiol. Lett. 157:67–72 [DOI] [PubMed] [Google Scholar]
- 76. USDA 2008. National Antimicrobial Resistance Monitoring System: enteric bacteria. NARMS 2007 animal arm annual report. USDA, Washington, DC: http://www.ars.usda.gov/SP2UserFiles/Place/66120508/NARMS/NARMS_2007 /NARMSAnimalArm2007.pdf [Google Scholar]
- 77. Uzzau S., et al. 2000. Host adapted serotypes of Salmonella enterica. Epidemiol. Infect. 125:229–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Voetsch A. C., et al. 2004. FoodNet estimate of the burden of illness caused by nontyphoidal Salmonella infections in the United States. Clin. Infect. Dis. 38(Suppl. 3):S127–S134 [DOI] [PubMed] [Google Scholar]
- 79. Welch T. J., et al. 2007. Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS One 2:e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Whichard J. M., et al. 2010. Evaluation of antimicrobial resistance phenotypes for predicting multidrug-resistant Salmonella recovered from retail meats and humans in the United States. J. Food Prot. 73:445–451 [DOI] [PubMed] [Google Scholar]
- 81. Zhao S., et al. 2009. Beta-lactam resistance in Salmonella strains isolated from retail meats in the United States by the National Antimicrobial Resistance Monitoring System between 2002 and 2006. Appl. Environ. Microbiol. 75:7624–7630 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.