Abstract
Glycosylation of proteins is known to impart novel physical properties and biological roles to proteins from both eukaryotes and prokaryotes. In this study, gel-based glycoproteomics were used to identify glycoproteins of the potential biothreat agent Burkholderia pseudomallei and the closely related but nonpathogenic B. thailandensis. Top-down and bottom-up mass spectrometry (MS) analyses identified that the flagellin proteins of both species were posttranslationally modified by novel glycans. Analysis of proteins from two strains of each species demonstrated that B. pseudomallei flagellin proteins were modified with a glycan with a mass of 291 Da, while B. thailandensis flagellin protein was modified with related glycans with a mass of 300 or 342 Da. Structural characterization of the B. thailandensis carbohydrate moiety suggests that it is an acetylated hexuronic acid. In addition, we have identified through mutagenesis a gene from the lipopolysaccharide (LPS) O-antigen biosynthetic cluster which is involved in flagellar glycosylation, and inactivation of this gene eliminates flagellar glycosylation and motility in B. pseudomallei. This is the first report to conclusively demonstrate the presence of a carbohydrate covalently linked to a protein in B. pseudomallei and B. thailandensis, and it suggests new avenues to explore in order to examine the marked differences in virulence between these two species.
INTRODUCTION
Burkholderia pseudomallei is a Gram-negative saprophytic rod which is particularly prevalent in the soil and standing water in southeast Asia and northern Australia (11, 13). It is the causative agent of melioidosis, a severe infectious disease in which infection is probably due to soil or water contamination of skin abrasions or inhalation from an environmental source (32). B. pseudomallei is also highly infectious to a large number of animal species, particularly BALB/c mice, for which the median lethal doses are approximately 10 bacteria following aerosol infection and 1,000 bacteria following intraperitoneal infection (51). B. pseudomallei can disseminate from sites of localized infection, such as the lungs or the skin, to virtually any other organ of the body (14). Acute septicemic melioidosis, the most severe form of disease, is responsible for much morbidity and mortality, especially in northeastern Thailand. Here, the incidence of melioidosis is approximately 3.6 to 5.5 cases per 100,000 annually, with mortality rates of up to 50% in adults and 35% in children (8, 56). The disease may also manifest as a chronic or subclinical infection and remain undetected for a number of years until activated by a traumatic event or a decrease in immunocompetence (12). Because of these factors, melioidosis is today regarded as an emerging infectious disease and as a potential bioterrorism threat (42).
Burkholderia thailandensis is closely related to B. pseudomallei, and for a time the two were considered to be the same species because of their similarity. However, genetic and biochemical differences between them, not the least of which is their relative virulences in humans and animals, have now been identified. B. thailandensis is generally not considered to be a human or animal pathogen, although sporadic cases of infection, usually following particularly traumatic events involving significant exposure, have been reported (19). The reasons for the differing virulences of B. pseudomallei and B. thailandensis are not well understood, although the absence of most of the capsule gene cluster from B. thailandensis is thought to be a significant factor (39).
Glycosylation of proteins is known to impart novel physical properties and biological roles to proteins from both eukaryotes and prokaryotes. Glycoproteins of bacteria have received considerable attention recently; most notable are those identified in pathogenic species and localized on the bacterial cell surface, where they may be involved in interactions with the host. Examples of surface-associated glycoproteins in Gram-negative bacteria are the pilins of Pseudomonas aeruginosa (7, 47, 55) and Neisseria spp. (49), the adhesins TibA and AIDA-1 of Escherichia coli (2, 33) and HMW1 of Haemophilus influenzae (21), and the flagellins of P. aeruginosa (6), Helicobacter pylori (26, 46), Clostridium botulinum (52), and Campylobacter jejuni/Campylobacter coli (17). Although the full significance of glycosylation of these proteins has yet to be defined, there are a number of reports describing the contribution of these modifications to virulence and colonization (1, 21, 22).
There are some reports of glycoproteins in B. pseudomallei and B. thailandensis, although work to conclusively prove the existence and characters of these glycoproteins has not been forthcoming. For example, there are reports of an acid phosphatase glycoprotein in B. pseudomallei (28–30, 38) and, based on comparison with the E. coli TibA/TibC and AIDA-I/AAH systems, indications that the BimA protein of B. mallei is a probable glycoprotein modified by BimC (44). Also, monoclonal antibodies which appear to react to proteins susceptible to both proteinase K and periodic acid treatment have been generated, suggesting that the antigen is a glycoprotein (59). Since glycoproteins of other bacteria have been identified as being important as virulence factors and as being highly immunogenic, we aimed to identify glycoproteins of B. pseudomallei and B. thailandensis to determine their potential role in virulence in these species and determine their utility as vaccine candidates. Thus, we used gel-based glycoproteomics to identify putative glycoproteins of B. pseudomallei and B. thailandensis followed by top-down and bottom-up mass spectrometry to analyze the glycan modification. We show that the flagellins of both strains are modified with O-linked glycan moieties and that the glycans decorating the flagella of each species are dissimilar. In addition, we have identified through mutagenesis a gene from the lipopolysaccharide (LPS) O-antigen biosynthetic cluster which is involved in flagellar glycosylation.
MATERIALS AND METHODS
Bacterial strains.
The strains used in this study are detailed in Table 1. E. coli TOP10 and S17-1 (λ-Pir), B. pseudomallei K96243 (25) and NCTC10276, and B. thailandensis E264 and E27 (48). All strains were grown in Luria-Bertani (LB) broth at 37°C with shaking for 18 h or on LB agar at 37°C for 18 h unless otherwise stated. When appropriate, chloramphenicol, gentamicin, and kanamycin at 50 μg/ml were added to the media. B. pseudomallei was handled under containment level 3 (CL3) conditions. Prior to removal of any CL3-derived material (e.g., protein extracts) to CL2 laboratories for analysis, a sterility check was performed.
Table 1.
Bacterial strains, primers, and plasmids used in this study
| Strain or plasmid | Descriptiona | Source/reference |
|---|---|---|
| E. coli strains | ||
| TOP10 | Used for cloning and blue-white screening | Invitrogen |
| S17-1 (λ-Pir) | Mobilizing strain; RP4-2 Tc::Mu Km::Tn7 (λ-Pir) phoA20 thi-1 rpsE rpoB; Tpr Strr Spr Kms | 37 |
| B. pseudomallei strains | ||
| K96243 | Clinical isolate; Genr Strr Chls | Siriraj Hospitalb |
| K96243::pDM4-wcbH | K96243 derivative; merodiploid strain; wcbH::pDM4-wcbH (sacBR oriT oriR6K Chlr); Genr Strr Chlr | This study |
| K96243::pDM4-rmlB | K96243 derivative; merodiploid strain; rmlB::pDM4-rmlB (sacBR oriT oriR6K Chlr); Genr Strr Chlr | This study |
| K96243::pDM4-rfbH | K96243 derivative; merodiploid strain; rfbH::pDM4-rfbH (sacBR oriT oriR6K Chlr); Genr Strr Chlr | This study |
| K96243::pDM4-udg2 | K96243 derivative; merodiploid strain; udg2::pDM4-udg2 (sacBR oriT oriR6K Chlr); Genr Strr Chlr | This study |
| K96243 ΔwcbH | K96243 derivative; unmarked deletion strain; ΔwcbH (nucleotides 491-1262); Genr Strr Chls | This study |
| K96243 ΔrmlB | K96243 derivative; unmarked deletion strain; ΔrmlB (nucleotides 96-969); Genr Strr Chls | This study |
| K96243 ΔrfbH | K96243 derivative; unmarked deletion strain; ΔrfbH (nucleotides 296-1119); Genr Strr Chls | This study |
| K96243 Δudg2 | K96243 derivative; unmarked deletion strain; Δudg2 (nucleotides 365-1209); Genr Strr Chls | This study |
| 10276 | Clinical isolate | NCTC |
| B. thailandensis strains | ||
| E27 | Environmental isolate | 48 |
| E264 | Environmental isolate; Thailand | ATCC |
| Plasmids | ||
| pDM4 | Mobilisable suicide vector; sacBR oriT oriR6K Chlr | 36 |
| pDM4-wcbH | pDM4 vector containing the BPSL2800 (wcbH) deletion construct | This study |
| pDM4-rmlB | pDM4 vector containing the BPSL2686 (rmlB) deletion construct | This study |
| pDM4-rfbH | pDM4 vector containing the BPSS0421 (rfbH) deletion construct | This study |
| pDM4-udg | pDM4 vector containing the BPSS1833 (udg2) deletion construct | This study |
Genr, gentamicin resistance; Strr, streptomycin resistance; Chlr, chloramphenicol resistance; Chls, chloramphenicol sensitivity; Tpr, trimethoprim resistance; Spr, spectinomycin resistance; Kms, kanamycin sensitivity.
Supplied by S. Songsivilai, Department of Immunology, Faculty of Medicine, Siriraj Hospital, Mahidol University, Bangkok, Thailand.
Protein isolation and 2D gel electrophoresis.
Proteins for two-dimensional (2D) gel electrophoresis were recovered using the ReadyPrep sequential extraction kit (Bio-Rad) according to the manufacturer's instructions. For 2D gel electrophoresis, a 150-μl aliquot of ReadyPrep extract was mixed with 200 μl of rehydration buffer {8 M urea, 2% [wt/vol] 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS], 0.5% [vol/vol] immobilized pH gradient buffer [pH 4 to 7], 200 μg ml−1 trace of bromophenol blue} and applied to pH 4 to 7 Immobiline DryStrips. The strips were rehydrated at 20°C for 12 h by using an IPGphor isoelectric focusing system (GE Healthcare) before protein separation at 500 V for 1 h, 1,000 V for 1 h, and 8,000 V for 10 h at 20°C by using a 50-μA current per strip. Following separation, the strips were equilibrated in buffer 1 (6 M urea, 75 mM Tris-HCl, pH 8.8, 30% glycerol, 2% SDS, 10 mg ml−1 dithiothreitol) and then buffer 2 (6 M urea, 75 mM Tris-HCl, pH 8.8, 30% glycerol, 2% SDS, 0.002% bromophenol blue, 25 mg ml−1 iodoacetamide) for 15 min each at room temperature. Second-dimension separation was performed on a Multiphor II electrophoresis system at 15°C using ExcelGel Gradient XL 12-14 gels with a two-step program of 200 V, 200 mA, and 20 W for 40 min followed by 800 V, 40 mA, and 40 W for 160 min.
Glycoprotein staining.
Following electrophoresis, 2D gels containing Burkholderia proteins were stained for glycoproteins by using the ProQ Emerald 300 kit (Invitrogen) according to the manufacturer's instructions. Stained gels were visualized using a 300-nm-UV transilluminator. Gels were subsequently stained for total protein by using the ProteoSilver kit (Sigma-Aldrich) according to the manufacturer's instructions.
Flagellar isolation and purification.
Agar plates were inoculated with B. pseudomallei or B. thailandensis and incubated at 37°C for 18 h. The growth from four plates was scraped into 1 ml of 50 mM sodium phosphate buffer (pH 7.0), and the flagella were sheared from the bacterial surface by passage 20 times through a 25-gauge needle (B. thailandensis) or vortexing of the suspension for 5 min (B. pseudomallei). The homogenate was centrifuged at 12,000 × g at 4°C for 20 min to remove the cells from the supernatant and the flagellar proteins were precipitated using 10% ammonium sulfate overnight at room temperature followed by centrifugation at 12,000 × g at 4°C for 20 min. The protein pellet was resuspended in 50 mM sodium phosphate buffer (pH 7.0) and dialyzed overnight at 4°C against milliQ water. Protein purity was assessed by SDS-PAGE using 4% stacking and 20% separating Novex Tris-glycine gels (Invitrogen).
Mass spectrometry.
Mass spectrometry studies of intact flagellin proteins were carried out as described previously, with some modifications (45, 52). A 50-μl aliquot of protein-containing solution was injected onto a protein microtrap (Michrom Bioresources Inc., Auburn, CA) connected to a gradient high-pressure liquid chromatography (HPLC) pump (Agilent 1100 HPLC). An HPLC gradient of 5 to 60% acetonitrile with 0.2% formic acid (1 ml/min) over 60 min was used to resolve the protein mixture. A precolumn splitter was used to direct approximately 60 μl/min of the HPLC mobile phase through the trap and into the electrospray interface of the QTOF2 hybrid quadrupole time of flight mass spectrometer (Micromass, Manchester, United Kingdom) to allow real-time monitoring of ion elution profiles. Intact masses of proteins were calculated by spectral deconvolution, using MaxEnt (Waters, Beverly, MA) or similar software. In tandem mass spectrometry (MS/MS) mode, an abundant multiply charged protein ion was selected. The collision energy was increased incrementally, and labile, protein-associated fragment ions were observed.
Solution enzymatic digests.
To identify the types and locations of glycosylation sites, flagellin (50 to 200 μg) was digested with trypsin or Asp-N (Promega, Madison, WI) at a ratio of 30:1 (protein to enzyme, vol/vol) in 50 mM ammonium bicarbonate at 37°C overnight, as described previously (52). Protein digests were analyzed by nano-liquid chromatography-tandem mass spectrometry (nLC-MS/MS) using either a Q-TOF Ultima hybrid quadrupole time-of-flight mass spectrometer (Waters, Milford, MA) or LTQ XL linear ion trap mass spectrometer (Thermo Fisher Scientific, Ottawa, ON, Canada) coupled to a nanoAcuity ultrahigh-pressure liquid chromatography (UPLC) system (Waters, Milford, MA). Tandem mass spectra (MS/MS spectra) were acquired automatically on doubly, triply, and quadruply charged ions.
Construction and verification of unmarked deletion mutants.
We generated a range of allelic exchange mutants which contained unmarked in-frame deletions of genes involved in polysaccharide (PS) biosynthesis from the four polysaccharide clusters identified in B. pseudomallei (43). Briefly, the deletion construct was made by PCR amplifying the upstream (F For and F Rev primers) and downstream (B For and B Rev primers) regions flanking the gene by using the primers listed in Table 2. These PCR products were digested with XbaI and BglII and ligated into BglII-digested pDM4 by using three-way ligation. These plasmids were conjugated into B. pseudomallei K96243 by using a membrane filter technique with E. coli S17-1 (λ-Pir) as the donor strain. Merodiploid integrants were selected using ampicillin to screen out E. coli and chloramphenicol to select for pDM4-containing strains. Chloramphenicol-resistant colonies were subcultured onto LB agar lacking sodium chloride but containing 10% sucrose to enrich for excision of the integrated vector DNA, resulting in either a wild-type or deleted allele (3, 35). Colonies were screened to identify deletion mutants by PCR and Southern hybridization. Reverse transcription-PCR (RT-PCR) using the method of Rodrigues et al. (41) was used to confirm that deletion did not affect transcription of adjacent genes.
Table 2.
Oligonucleotide primers used in this study
| Primer | Sequence (5′-3′)a |
|---|---|
| WcbH F For | TCTAGACGAACTGCCTGGCGAATATGG |
| WcbH F Rev | AGATCTCTCGAACATCGGAAACGAGGC |
| WcbH B For | AGATCTGTGTCCGGCACTACCATCGAT |
| WcbH B Rev | TCTAGACGATATCGATGCGTAGAGACG |
| RmlB F For | TCTAGACGTGGCCATAGAAGATGT |
| RmlB F Rev | AGATCTCGCTGGTATCTCGACCAT |
| RmlB B For | AGATCTGCTTATCGACGTTCAGCA |
| RmlB B Rev | TCTAGAGTCATTCGAGCGACCAAC |
| RfbH F For | TCTAGACCGACAAGTGCTACGAGAACC |
| RfbH F Rev | AGATCTACCGGAGTTCACCGTGATCAC |
| RfbH B For | AGATCTGATTTGCTCGATTACCTGAGC |
| RfbH B Rev | TCTAGACGTCGATAAGCCATGCGAACG |
| Udg2 F For2 | TCTAGACGCAGTTCATCAATGTGC |
| Udg2 F Rev | AGATCTCACCGTCGACTTGTCGACGA |
| Udg2 B Rev | AGATCTGTCACCGAATGGAAGGCGTTC |
| Udg2 B Rev2 | TCTAGAGCTGCACGTTGTAATGCT |
Underlined sequences indicate either XbaI or BglII restriction sites.
Motility assay.
B. pseudomallei strains were screened for their motility phenotypes by transferring individual colonies to plates containing LB broth with 0.3% agar. Motility plates were incubated for 24 h at 37°C, and the motility phenotype of each mutant was assessed by measuring the diameter of the zone of growth.
RESULTS
Isolation and identification of glycoproteins.
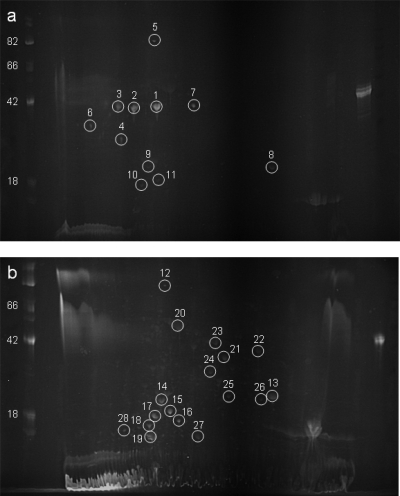
Previous work has demonstrated the utility of separating protein samples by acrylamide gel electrophoresis with subsequent detection using periodate to oxidize the carbohydrate groups for the identification of novel glycoproteins (31, 58). This methodology was used to resolve putative glycoproteins from B. pseudomallei and B. thailandensis prior to their analysis using mass spectrometry. Considerable differences were observed between the glycoprotein profiles of B. pseudomallei K96243 and B. thailandensis E264 following separation by 2D electrophoresis (Fig. 1). The B. pseudomallei K96243 profile was dominated by three intensely reactive protein spots of approximately 40 kDa, with pI values between 4 and 5 (spots 1 to 3). In contrast, these three spots were absent from the B. thailandensis E264 profile, which was instead dominated by a series of intensely reactive spots of between 15 and 25 kDa, with pI values between 4 and 5 (spots 14 to 19).
Fig. 1.
Identification of glycoproteins in B. pseudomallei and B. thailandensis. Protein extracts from B. pseudomallei K96243 (a) and B. thailandensis E264 (b) were separated by 2D electrophoresis and stained with ProQ Emerald 300. Indicated spots were identified by peptide mass fingerprinting following tryptic digestion. Marker sizes are indicated in kDa.
Twenty-eight proteins were excised from the gel and identified by nLC-MS/MS of their tryptic digests. The identities of the isolated proteins from B. pseudomallei and B. thailandensis can be seen in Table 3. Several of the putative glycoprotein spots provided more than one protein identification, making it difficult to determine which of the proteins were modified with glycan. During protein identification, peptides with anomalously high molecular weights which could not be identified by database searching of MS/MS spectra were observed in samples identified as containing flagellin protein FliC. The major flagellin filament protein has previously been identified as being glycosylated in a range of other Gram-negative and -positive organisms (6, 17, 26, 46) and having importance in virulence and serospecificity. Additionally, methods for the purification of native flagella to facilitate glycopeptide analysis exist. Based on this knowledge, efforts focused on the major flagellin filament protein FliC. The other putative glycoproteins on this list were not examined further to verify the presence of a covalently attached carbohydrate.
Table 3.
Putative glycoproteins of B. pseudomallei and B. thailandensis
| Species and spot no. | Open reading frame | Predicted function | Accession no. | % cover | Size (kDa) | pI |
|---|---|---|---|---|---|---|
| B. pseudomallei K96243 | ||||||
| 1 | BPSL3319 | Major flagellar filament protein (FliC) | YP_109915 | 61 | 39.2 | 5.05 |
| BPSL3021 | Cell division protein (FtsA) | YP_109617 | 57 | 43.8 | 4.96 | |
| BPSL2697 | Chaperonin (GroEL) | YP_109293 | 46 | 57.1 | 5.13 | |
| 2 | BPSL3319 | Major flagellar filament protein (FliC) | YP_109915 | 61 | 39.2 | 5.05 |
| BPSL2697 | Chaperonin (GroEL) | YP_109293 | 59 | 57.1 | 5.13 | |
| BPSL2270 | Phosphopyruvate hydratase (Eno) | YP_108866 | 37 | 45.7 | 4.81 | |
| 3 | BPSS2288 | HSP20 family protein | YP_112291 | 28 | 16.0 | 5.14 |
| BPSL1744 | Ornithine carbamoyltransferase (ArcB) | YP_108344 | 16 | 37.9 | 6.17 | |
| 4 | BPSL3319 | Major flagellar filament protein (FliC) | YP_109915 | 20 | 39.2 | 5.05 |
| 5 | BPSL2697 | Chaperonin (GroEL) | YP_109293 | 57 | 57.1 | 5.13 |
| 6 | BPSL3319 | Major flagellar filament protein (FliC) | YP_109915 | 19 | 39.2 | 5.05 |
| 7 | BPSL0779 | Succinyl-CoA synthetase subunit β (SucC) | YP_107404 | 25 | 41.2 | 5.25 |
| 8 | BPSL2298 | Phasin-like protein (PhaP) | YP_108894 | 80 | 19.8 | 5.96 |
| 9 | BPSL2298 | Phasin-like protein (PhaP) | YP_108894 | 56 | 19.8 | 5.96 |
| 10 | BPSL2298 | Phasin-like protein (PhaP) | YP_108894 | 69 | 19.8 | 5.96 |
| 11 | BPSS2288 | HSP20 family protein | YP_112291 | 60 | 16.0 | 5.14 |
| B. thailandensis E264 | ||||||
| 12 | BTH_I1458 | Chaperonin (GroEL) | YP_442004 | 34 | 57.1 | 5.13 |
| 13 | BTH_I1867 | Phasin-like protein (PhaP) | YP_442397 | 72 | 20.0 | 5.96 |
| 14 | BTH_I1867 | Phasin-like protein (PhaP) | YP_442397 | 72 | 20.0 | 5.96 |
| BTH_I1458 | Chaperonin (GroEL) | YP_442004 | 24 | 57.1 | 5.13 | |
| 15 | BTH_I1867 | Phasin-like protein (PhaP) | YP_442397 | 73 | 20.0 | 5.96 |
| 16 | BTH_I1867 | Phasin-like protein (PhaP) | YP_442397 | 73 | 20.0 | 5.96 |
| 17 | BTH_I1867 | Phasin-like protein (PhaP) | YP_442397 | 72 | 20.0 | 5.96 |
| 18 | BTH_I1867 | Phasin-like protein (PhaP) | YP_442397 | 73 | 20.0 | 5.96 |
| 19 | BTH_II2321 | Stress response protein | YP_440508 | 17 | 57.1 | 11.4 |
| 20 | BTH_I3070 | Translation elongation factor (Tuf-1) | YP_443574 | 59 | 42.9 | 5.36 |
| BTH_I1458 | Chaperonin (GroEL) | YP_442004 | 38 | 57.1 | 5.13 | |
| 21 | BTH_I1458 | Chaperonin (GroEL) | YP_442004 | 46 | 57.1 | 5.13 |
| 22 | BTH_I1159 | Thiol peroxidase (Tpx) | YP_441706 | 56 | 17.3 | 5.80 |
| 23 | BTH_I0410 | Glutathione synthase (GshB) | YP_440968 | 48 | 34.6 | 5.49 |
| 24 | BTH_I1867 | Phasin-like protein (PhaP) | YP_442397 | 73 | 20.0 | 5.96 |
| 25 | BTH_I3042 | RNA polymerase, α subunit (RpoA) | YP_443546 | 52 | 35.6 | 5.76 |
| 26 | BTH_I3077 | Ribosomal protein L7/L12 (RplL) | YP_443581 | 57 | 12.5 | 4.90 |
| BTH_I1867 | Phasin-like protein (PhaP) | YP_442397 | 53 | 20.0 | 5.96 | |
| 27 | BTH_I1458 | Chaperonin (GroEL) | YP_442004 | 73 | 57.1 | 5.13 |
| 28 | BTH_I1458 | Chaperonin (GroEL) | YP_442004 | 23 | 57.1 | 5.13 |
Mass spectrometry and top-down analyses of flagellins.
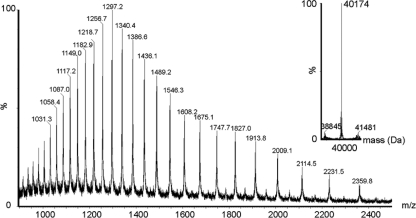
Brett and colleagues have previously described the recovery and characterization of flagellin proteins from B. pseudomallei 319a (4), and a modified method was used here to recover FliC protein. Following protein recovery, tandem mass spectrometry of tryptic digests of purified B. pseudomallei and B. thailandensis flagellins confirmed the identities of the FliC proteins. The observed masses of flagellin proteins from B. thailandensis strains E264 and E27 were obtained by rapid LC-MS into a QTOF2 electrospray ionization (ESI) mass spectrometer. The protein mass spectrum from each strain showed an envelope of multiply charged protein ions. From this, the reconstructed molecular mass profile was calculated. Representative data are shown in Fig. 2 for the reconstructed molecular mass profile of B. thailandensis E264 flagellin. This showed a single major peak at 40,174 Da (±50 Da). Two peaks of much lower intensity were observed at 38,845 Da and 41,481 Da. B. thailandensis E264 FliC had a predicted mass of 38,809 Da calculated from the translated DNA sequence and the minor protein peak observed at 38,845 Da likely represents this unmodified protein. The major observed protein peak has a mass excess of 1,365 Da, approximately 3.4% of the total protein. The reconstructed molecular mass profile of B. thailandensis E27 similarly showed prominent peaks at 38,524 Da and 41,780 Da. B. pseudomallei flagellin was not isolated in sufficient quantities for intact mass analysis; therefore, a bottom-up approach was used to further characterize proteins from this species.
Fig. 2.
Electrospray mass spectrometry of intact flagellin proteins from B. thailandensis E264. The inset shows the reconstructed molecular mass profile showing a major peak at 40,174 Da and two minor peaks at 38,845 Da and 41,481 Da.
Examination of intact proteins by top-down MS has been shown to readily identify labile glycan oxonium ions found on flagellin glycoproteins (45). Flagellin protein from B. thailandensis E264 was thus examined for the presence of glycan marker ions. MS/MS spectra of multiply charged B. thailandensis E264 flagellin ions revealed intense ions of m/z 301.1 and 343.1. Analysis of B. thailandensis E27 flagellin showed an identical pattern of protein-related fragment ions.
Mass spectrometry analyses of flagellin trypsin digests.
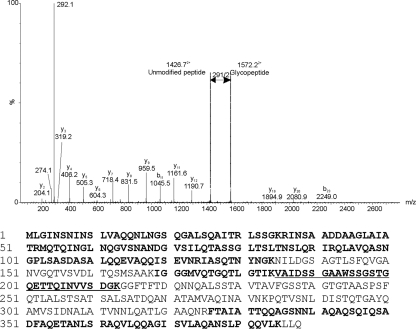
Many of the MS/MS spectra of tryptic digests of the purified flagellin proteins could be readily assigned to the predicted amino acid sequence, but a number of peptide spectra that did not match any expected tryptic peptides were observed. Manual inspection of these MS/MS spectra identified peptide y and b ions, in addition to further peaks that were not related to peptide fragment ions. For example, within the B. pseudomallei K96243 flagellin tryptic digest, the MS/MS spectrum of the triply charged ion at m/z 1145.6 (giving an observed peptide mass of 3,434.8 Da) gave a typical peptide MS/MS spectrum, with a clear series of type y and b ions, the sequence of which (185VAIDSSGAAWSSGSTGQETTQINVVSDGK213) corresponds to the central region of B. pseudomallei K96243 FliC. The peptide has a theoretical mass of 2,851.4 Da, observed in the MS/MS spectrum in Fig. 3. The difference between the observed and predicted peptide masses was 582.4 Da, likely to be a glycan. A neutral loss of 145.5 from the doubly charged ion series corresponded to a putative glycan oxonium ion at m/z 292.1 (indicated by an arrow in Fig. 3). The low-m/z region of the spectrum is dominated by the carbohydrate oxonium ion (m/z 292.1) and what could be a fragmentation component following neutral loss of water (m/z 274.1). These data suggest that this peptide is modified with two glycans of residue mass 291 Da.
Fig. 3.
Mass spectrometry analyses of tryptic peptides from flagellin proteins of B. pseudomallei K96243. The nLC-MS/MS spectrum of the triply protonated T185–213 glycopeptide ion of m/z 1145.6 from B. pseudomallei is shown. The spectrum is dominated in the low-molecular-m/z region by the carbohydrate oxonium ion (m/z 292.1) and what could be a fragmentation component (m/z 274.1). Loss of m/z 1452+ in the high-molecular-m/z region of the spectrum corresponds to loss of glycan of residue mass 291.1. From the observed mass of the glycopeptide, a mass excess of 582 Da was observed, corresponding to two glycans of 291 Da. Below the graph is the amino acid sequence of the B. pseudomallei K96243 FliC protein. The sequence of the T185–213 glycopeptide is underlined. Sequences in bold indicate unmodified peptides observed in tryptic digests.
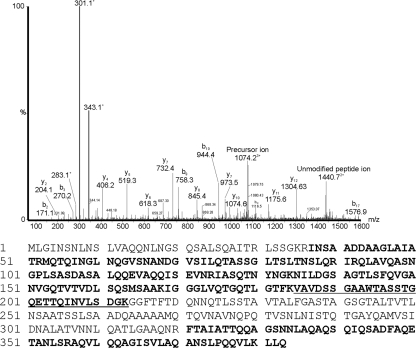
Unmatched MS/MS spectra were also observed with the B. thailandensis flagellin tryptic digests. For example, tandem mass spectra of triply charged ions at m/z 1074.2 and 1061.2 gave typical peptide MS/MS spectra representing the same peptide but with distinct glycan modifications. There was a clear series of peptide y and b ions, the sequence of which (185VAVDSSGAAWTASSTGQETTQINVLSDGK213) corresponded to the central region of B. thailandensis E264 FliC and is equivalent to the glycopeptide sequence identified from B. pseudomallei K96243. The unmodified peptide ion (theoretical mass of 2,879.4 Da) was observed in the doubly charged form at m/z 1440.7 in both spectra. The total mass of the glycopeptide represented by m/z 1061.2 corresponded to the mass of the peptide with an additional 300 Da. Observed within this spectrum was an ion at m/z 301.1, likely corresponding to the glycan oxonium ion (data not shown). In contrast, the total mass of the glycopeptide represented by m/z 1074.2 corresponded to the mass of the peptide with an additional 342 Da. Observed within the spectrum was an ion at m/z 343.1, likely corresponding to the glycan oxonium ion (Fig. 4). Also observed in this glycopeptide MS/MS spectrum were putative glycan-related fragment ions at m/z 283.1 and 301.1. These similarities suggest that the 342-Da glycan is likely the same as the 300-Da glycan with the addition of an acetyl group (42 Da).
Fig. 4.
Mass spectrometry analyses of tryptic peptides from flagellin proteins of B. thailandensis E264. nLC-MS/MS spectrum of the triply protonated T185–213 glycopeptide ion of m/z 1074.2 from B. thailandensis. The spectrum is dominated in the low-molecular-m/z region by the carbohydrate oxonium ion (m/z 343.1) and what could be fragmentation components (m/z 301.1 and 283.1). From the observed mass of the glycopeptide, a mass excess of 342 Da was observed, corresponding to a single glycan modification. Below the graph is the amino acid sequence of the B. thailandensis E264 FliC protein. The sequence of the T185–211 glycopeptide is underlined. Sequences in bold indicate unmodified peptides observed in tryptic digests.
To determine whether the differences observed between B. pseudomallei K9624 and B. thailandensis E264 represent strain-specific differences, flagellin proteins from an additional strain of each species were analyzed. B. pseudomallei 10276 flagellin protein was observed to contain glycan oxonium ions of m/z 292.1, indicating the presence of glycan similar to that observed in B. pseudomallei K96243 flagellin proteins. Similarly, B. thailandensis E27 flagellin proteins were observed to contain glycan oxonium ions of m/z 301.1 and 343.1, indicating the presence of a glycan similar to those observed in B. thailandensis E264 flagellin proteins and supporting data from top-down analysis. It was not possible to identify the m/z 301.1 and 343.1 glycans in the B. pseudomallei samples or the m/z 292.1 glycan in the B. thailandensis samples, indicating that, in these four strains at least, these glycans are species specific and mutually exclusive.
Determination of glycan attachment sites.
In our recent work, we have had success using electron transfer dissociation to map the linkage sites of bacterial O-linked carbohydrates (52). Therefore, in the current study, we used this approach to determine the site of modification of the peptide 185VAVDSSGAAWTASSTGQETTQINVLSDGK213 from B. thailandensis E264 and E27. These attempts were unsuccessful due to the size of the peptide and lack of charged amino acid residues. Experiments using the equivalent peptides cleaved with Asp-N were similarly unsuccessful. However, beta elimination of the Asp-N-digested peptide 188DSSGAAWTASSTGQETTQINVLS210 did allow the site of modification to be mapped to either serine 197 or 198, establishing these as O-linked glycans. Closely overlapping and weak fragment ions within the spectrum prohibited definitive assignment of the beta-eliminated amino acid residue.
Genetic strategies using defined point mutations to unequivocally determine the glycan attachment site were pursued but were not successful. However, expression of the B. pseudomallei K96243 fliC gene in B. thailandensis E264 yielded protein which was glycosylated (data not shown). The B. pseudomallei protein lacks S197, and thus, it must be concluded that, in these recombinant proteins at least, S198 was the residue used for glycan attachment. It should be noted, however, that there are other sequence differences between B. pseudomallei and B. thailandensis at this location which may influence the attachment site and that in the absence of defined mutations, these data can only be considered suggestive.
Structural characterization of unusual O-linked glycan moieties by MS/MS analysis.
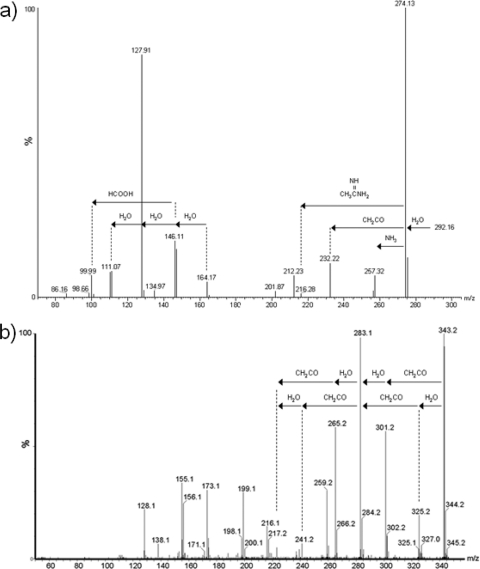
Thibault and colleagues (50) have previously shown the utility of tandem mass spectrometry to obtain information about the glycan modifications present on glycopeptides of C. jejuni. Similar methods were used to obtain second-generation ion spectra of the 291-Da and 342-Da glycans present on the B. pseudomallei K96243 and B. thailandensis E264 FliC proteins, respectively (Fig. 5). A series of ions were formed from m/z 292.1, representing neutral losses of molecules such as water and acetyl. The m/z 343.1 ion from B. thailandensis yielded a spectrum which was dissimilar from that of the m/z 292.1 spectrum but which included neutral losses of molecules such as water and acetyl. Clear evidence that the m/z 343.1 ion is at least doubly acetylated, in contrast to m/z 292.1, which appears to contain a single acetyl group, can be seen.
Fig. 5.
Electrospray mass spectrometry of B. pseudomallei K96243 and B. thailandensis E27 glycans. Second-generation product ion spectra of ions obtained from in-source dissociation of multiply charged flagellin ions. MS/MS spectra were acquired by increasing the radio frequency (RF) lens 1 voltage from 40 to 90 V, forming fragment ions in the orifice/skimmer region of the mass spectrometer and promoting the formation of labile ions from the intact flagellin MS/MS spectrum of B. pseudomallei K96243 m/z 292.1 (a) and B. thailandensis E264 m/z 343.1 (b). This yielded a series of predominant fragment ions which did not correspond to peptide type y and b ions. Plausible neutral losses of molecules such as acetyl groups and water are indicated.
The accurate mass of the m/z 343.1 glycan oxonium ion was determined from MS/MS spectra of the tryptic flagellin peptide T185–213 (i.e., residues 185 to 213). The peptide fragment ion y5 (predicted m/z, 519.2778; observed m/z, 519.2777) was used as an internal reference. The glycan oxonium ion was observed at m/z 343.1410. From this, the top-ranked plausible elemental formula was calculated to be C15H23N2O7 (predicted m/z, 343.1499). Comparison of the glycan fragment ions with previous studies of bacterial glycans showed some similarities with N-acetyl hexuronic acids (52, 54). In particular glycan fragment ions at m/z 259.2, 241.2, 223.2, and 200.1 were common to both the di-N-acetyl hexuronic acid and this unknown glycan.
Involvement of rmlB in glycosylation.
B. pseudomallei contains four gene clusters with predicted roles in polysaccharide biosynthesis (43). Previous work has identified two of these clusters as being responsible for capsule and LPS O-antigen synthesis, but the function of the remaining two clusters remains unclear. To determine whether these clusters had a role in flagellar glycosylation, unmarked in-frame deletion mutants were constructed to inactivate a gene in each of these four clusters. The genes selected for inactivation were those previously identified by Sarkar-Tyson and colleagues (43), with the exception that inactivation of the LPS O-antigen cluster was accomplished by deletion of the rmlB gene (BPSL2686) rather than wbiA. The rmlB mutant was determined to be lacking O antigen by silver staining and Western blotting using specific monoclonal antibodies. Flagellin protein recovered from the ΔwcbH, ΔBPSS0421, and ΔBPSS1833 mutants displayed no differences relative to the parental wild-type protein. In contrast, flagellin protein recovered from the ΔrmlB mutant displayed no evidence of the 291-Da glycan, and the glycopeptide seen in the parental strain was observed only as an unmodified peptide (data not shown).
Inactivation of rmlB in B. thailandensis was attempted using a number of mutational strategies, but no mutants were obtained, and thus it could not be ascertained whether the B. thailandensis glycan uses the same synthesis pathway as the B. pseudomallei glycan. It is possible that, in the absence of the capsular polysaccharide present in B. pseudomallei, inactivation of rmlB is a lethal mutation in B. thailandensis.
Flagellar glycosylation is needed for full motility but not for filament assembly.
Correct glycosylation of the major flagellar filament protein of both C. jejuni (20) and Clostridium difficile (53) has been shown to be necessary for correct assembly of the flagella and therefore motility. To determine whether deletion of rmlB and the attendant lack of glycosylation affect the motility of B. pseudomallei, motility assays were performed. These indicated that, while the parental strain had good motility, with an average halo diameter of 6.92 cm (±0.68 cm), the ΔrmlB mutant displayed a significant reduction in motility, with an average halo diameter of 2.52 cm (±0.53 cm). Transmission electron microscopy following uranyl acetate staining revealed that the ΔrmlB mutant had flagellar filaments on its surface, and these were similar to those seen on wild-type organisms (data not shown), indicating that the motility defects observed were not due to loss of the flagellar filament itself.
DISCUSSION
The results presented here show that, in common with an increasing number of bacterial pathogens, B. pseudomallei and B. thailandensis are capable of increasing their repertoire of protein diversity through posttranslational glycosylation. Such modifications have been postulated to occur in B. pseudomallei, notably in reports of an acid phosphatase glycoprotein (29, 30) and reports of a glycoprotein known as antigen 8 (28, 38). However, this report is the first to conclusively demonstrate the presence of a carbohydrate covalently linked to a B. pseudomallei protein. Two-dimensional gel electrophoresis and subsequent staining for carbohydrates allowed the identification of a range of putative glycoproteins from both B. pseudomallei and B. thailandensis. Interestingly, there was only limited overlap between the two gene lists, despite the close genetic relationship of B. pseudomallei and B. thailandensis. Indeed, the 2D gels from these two species display almost mutually exclusive patterns of glycosylation. However, it should be noted that the method used here to identify candidate glycoproteins is unlikely to recognize all glycoproteins, since the chemistry of glycol staining relies upon the presence of an oxidizable aldehyde on the glycan moiety. Given the diversity of bacterial carbohydrates, it is impossible to predict the reactivity of protein-modifying glycans.
The identification of the major flagellar filament protein FliC as a putative glycoprotein is not without precedent. This protein has been shown to be glycosylated in a range of other bacteria, such as Helicobacter pylori (26, 46), Clostridium botulinum (52), C. difficile (53), Campylobacter jejuni/Campylobacter coli (17), and Pseudomonas aeruginosa (6). Previous work (4) has indicated that the FliC proteins of four B. pseudomallei strains have experimentally determined masses some 3.3 kDa larger than expected, and it was suggested that posttranslational modifications might explain this discrepancy. The lack of sequence data for the fliC genes of these strains makes it difficult to be certain that these strains do not have genes encoding proteins of this size. However, given that the sequences of 16 B. pseudomallei fliC genes in the NCBI database (accessed 16 November 2010) are all identical and encode proteins of 39.2 kDa, it is likely that these experimentally derived data reveal a glycosylated protein and indicate that flagellar glycosylation is a common phenomenon in B. pseudomallei.
It was possible to use tandem mass spectrometry to provide information on the general structures of the glycans isolated from these FliC proteins, and both samples provided clear fragmentation profiles of their respective glycans. The 291.1-Da glycan from B. pseudomallei K96243 and 10276 contained peaks which indicated neutral losses of multiple water molecules, at least one acetyl group (CH2CO), formic acid (HCOOH), and possibly evidence of an acetamidino group [CH3C( NH)NH2] and ammonia. The 342.1-Da glycan from B. thailandensis E264 and E27 contained peaks which indicated neutral losses of multiple water molecules and acetyl groups. Comparison of the 342.1-Da glycan profile with that of an N-linked glycan recovered from the flagellin protein of the marine archaeon Methanococcus voltae (54) and an O-linked glycan recovered from the flagellin protein of C. botulinum (52) reveals a degree of similarity between the profiles. This might suggest that the 342.1-Da glycan present on the B. thailandensis flagellin protein is an acetylated uronic acid. Uronic acids are components of many surface-associated glycoconjugates, including a number of well-characterized lipopolysaccharides and capsular polysaccharides. It is of interest that serotype O5 strain Pseudomonas aeruginosa contains di-N-acetyl uronic acid derivatives (40), and there have been reports of uronic acids present in capsule extracts of B. pseudomallei (27).
NH)NH2] and ammonia. The 342.1-Da glycan from B. thailandensis E264 and E27 contained peaks which indicated neutral losses of multiple water molecules and acetyl groups. Comparison of the 342.1-Da glycan profile with that of an N-linked glycan recovered from the flagellin protein of the marine archaeon Methanococcus voltae (54) and an O-linked glycan recovered from the flagellin protein of C. botulinum (52) reveals a degree of similarity between the profiles. This might suggest that the 342.1-Da glycan present on the B. thailandensis flagellin protein is an acetylated uronic acid. Uronic acids are components of many surface-associated glycoconjugates, including a number of well-characterized lipopolysaccharides and capsular polysaccharides. It is of interest that serotype O5 strain Pseudomonas aeruginosa contains di-N-acetyl uronic acid derivatives (40), and there have been reports of uronic acids present in capsule extracts of B. pseudomallei (27).
The flagella of a range of different bacterial species have been shown to have a role in virulence, through either motility directly or adherence (1, 22). Glycosylation is known to have a role here, since inactivation of glycan synthesis machinery has been shown to eliminate motility and attenuate strains of C. jejuni (22). In line with this, inactivation of rmlB is seen here to result in nonglycosylated FliC in B. pseudomallei and also to abrogate motility. Interestingly, however, ΔrmlB mutants still have flagellar filaments on their surfaces visible by transmission electron microscopy, indicating that the loss of motility is not due to defects in export and assembly but rather in the function of the flagellar filament itself. It should be noted, however, that the rmlB mutation also results in loss of O-antigen synthesis and it thus cannot be discounted that the defects in motility are due to defects in membrane structure. There is some debate as to whether the flagella of B. pseudomallei are virulence determinants (9, 10, 15, 57), especially given that the closely related but aflagellate B. mallei is highly pathogenic to humans, but they are likely to play some role in disease, as evidenced by the ability of antibodies raised against flagellar protein to provide passive protection in a diabetic rat model (4). Additionally, the glycopeptide present in B. pseudomallei and B. thailandensis is predicted to be surface exposed and thus exposed to the immune response. The presence of glycans on such surface-exposed flagellar peptides has been shown to affect serospecificity in other bacteria (17, 34). This raises the possibility that glycosylation of flagellar filaments in Burkholderia species is a mechanism used to evade detection by the immune system or to modulate virulence and could potentially go some way to explaining the differing virulence of these two species. Recent work has indicated that monoclonal antibodies which recognize proteins susceptible to both proteinase K and periodic acid treatment can be identified in B. pseudomallei but not in B. thailandensis (59). This suggests that the glycans recognized by these monoclonal antibodies are missing from B. thailandensis proteins and indicates that the differences in glycosylation observed here in the flagella and on a wider scale by 2D electrophoresis are real and not an experimental artifact. Both B. pseudomallei and B. thailandensis are readily recovered from environmental samples, especially paddy fields. It is expected that in this environment, the flagella may play a role in allowing swimming to acquire access to new resources. Hence, it is entirely possible that the differential flagellar glycosylation observed with these strains might have a role in determining environmental fitness.
Interestingly, deletion of the rmlB gene in B. pseudomallei resulted in nonglycosylated FliC protein and nonmotile bacteria. The rmlB gene is located within the gene cluster responsible for LPS O-antigen synthesis, and disruption of this gene has previously been shown to eliminate O-antigen synthesis (16). The gene itself is predicted to be a dTDP-d-glucose 4,6-dehydratase involved in the conversion of d-glucose-1-phosphate into dTDP-l-rhamnose, which is then presumably epimerized into talose for incorporation into the LPS O antigen (16). It is unlikely that the glycan present on FliC protein is actually the LPS O antigen, since previous work has demonstrated that purified flagella from B. pseudomallei 319a do not react to anti-LPS monoclonal antibodies (5), and the flagella purified here did not show a reaction to anti-LPS monoclonal or polyclonal sera capable of recognizing B. pseudomallei LPS (data not shown), but without full structural information about the glycan, it is impossible to say. One possible explanation for this phenomenon is that deletion of rmlB depletes the sugar nucleotide pool required for glycosylation. At this moment, it is unclear at which point downstream of rmlB the glycosylation machinery operates, and the genetic basis of the machinery which attaches the glycan to FliC is also unclear. The two unassigned polysaccharide gene clusters (type III O-PS and type IV O-PS, according to the scheme of Sarkar-Tyson et al. [43]) and the capsule cluster would seem to be uninvolved, since deletion of genes in these clusters has no effect on FliC glycosylation (data not shown). However, it is still possible that other genes in these clusters play a role in glycosylation. Work is under way to provide answers to these questions. It was not possible to ascertain whether rmlB plays a role in the synthesis of the glycan in B. thailandensis, since, despite repeated attempts using a number of strategies, mutants could not be successfully constructed. This possibly suggests that in B. thailandensis, which lacks the protective polysaccharide capsule normally present in B. pseudomallei, the rmlB mutation is lethal, although further work would need to be done to confirm this fact.
These data have particular relevance for the generation of recombinant subunit vaccines. Considerable work has been done to identify protein targets to act as a subunit vaccine for B. pseudomallei and B. mallei (4, 5, 18, 23, 24). However, relatively few of these proteins provide suitable protection, despite being highly immunogenic. It is possible that some of these candidates are glycoproteins and need the glycan, something which is missing from proteins recombinantly produced in E. coli, for full immune recognition. Moreover, these data indicate that there is a range of other glycoproteins present in both B. pseudomallei and B. thailandensis, examination of which may elucidate information about the lifestyle and virulence mechanisms of these important organisms.
ACKNOWLEDGMENTS
This work was supported by funding from the United Kingdom Ministry of Defense and the National Research Council Canada.
We thank Luc Tessier for technical assistance with mass spectrometers.
This paper is published with the permission of the Defense Science and Technology Laboratory on behalf of the controller of HMSO.
Footnotes
Published ahead of print on 20 May 2011.
REFERENCES
- 1. Arora S. K., Neely A. N., Blair B., Lory S., Ramphal R. 2005. Role of motility and flagellin glycosylation in the pathogenesis of Pseudomonas aeruginosa burn wound infections. Infect. Immun. 73:4395–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Benz I., Schmidt M. A. 2001. Glycosylation with heptose residues mediated by the aah gene product is essential for adherence of the AIDA-I adhesin. Mol. Microbiol. 40:1403–1413 [DOI] [PubMed] [Google Scholar]
- 3. Blomfield I. C., Vaughn V., Rest R. F., Eisenstein B. I. 1991. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol. Microbiol. 5:1447–1457 [DOI] [PubMed] [Google Scholar]
- 4. Brett P. J., Mah D. C., Woods D. E. 1994. Isolation and characterization of Pseudomonas pseudomallei flagellin proteins. Infect. Immun. 62:1914–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brett P. J., Woods D. E. 1996. Structural and immunological characterization of Burkholderia pseudomallei O-polysaccharide-flagellin protein conjugates. Infect. Immun. 64:2824–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brimer C. D., Montie T. C. 1998. Cloning and comparison of fliC genes and identification of glycosylation in the flagellin of Pseudomonas aeruginosa a-type strains. J. Bacteriol. 180:3209–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Castric P., Cassels F. J., Carlson R. W. 2001. Structural characterization of the Pseudomonas aeruginosa 1244 pilin glycan. J. Biol. Chem. 276:26479–26485 [DOI] [PubMed] [Google Scholar]
- 8. Cheng A. C., Currie B. J. 2005. Melioidosis: epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 18:383–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chua K. L., Chan Y. Y., Gan Y. H. 2003. Flagella are virulence determinants of Burkholderia pseudomallei. Infect. Immun. 71:1622–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chuaygud T., Tungpradabkul S., Sirisinha S., Chua K. L., Utaisincharoen P. 2008. A role of Burkholderia pseudomallei flagella as a virulent factor. Trans. R. Soc. Trop. Med. Hyg. 102:S140–S144 [DOI] [PubMed] [Google Scholar]
- 11. Currie B. J. 2003. Melioidosis: an important cause of pneumonia in residents of and travellers returned from endemic regions. Eur. Respir. J. 22:542–550 [DOI] [PubMed] [Google Scholar]
- 12. Currie B. J., Fisher D. A., Anstey N. M., Jacups S. P. 2000. Melioidosis: acute and chronic disease, relapse and re-activation. Trans. R. Soc. Trop. Med. Hyg. 94:301–304 [DOI] [PubMed] [Google Scholar]
- 13. Dance D. A. 1990. Melioidosis. Rev. Med. Microbiol. 1:143–150 [Google Scholar]
- 14. Dance D. A. 1996. Melioidosis, p. 925–930In Cook G. C. (ed.), Manson's tropical diseases, 20th ed. W. B. Saunders Company Ltd., London, England [Google Scholar]
- 15. DeShazer D., Brett P. J., Carlyon R., Woods D. E. 1997. Mutagenesis of Burkholderia pseudomallei with Tn5-OT182: isolation of motility mutants and molecular characterization of the flagellin structural gene. J. Bacteriol. 179:2116–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. DeShazer D., Brett P. J., Woods D. E. 1998. The type II O-antigenic polysaccharide moiety of Burkholderia pseudomallei lipopolysaccharide is required for serum resistance and virulence. Mol. Microbiol. 30:1081–1100 [DOI] [PubMed] [Google Scholar]
- 17. Doig P., Kinsella N., Guerry P., Trust T. J. 1996. Characterization of a post-translational modification of Campylobacter flagellin: identification of a sero-specific glycosyl moiety. Mol. Microbiol. 19:379–387 [DOI] [PubMed] [Google Scholar]
- 18. Fernandes P. J., Guo Q., Waag D. M., Donnenberg M. S. 2007. The type IV pilin of Burkholderia mallei is highly immunogenic but fails to protect against lethal aerosol challenge in a murine model. Infect. Immun. 75:3027–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Glass M. B., et al. 2006. Pneumonia and septicemia caused by Burkholderia thailandensis in the United States. J. Clin. Microbiol. 44:4601–4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goon S., Kelly J. F., Logan S. M., Ewing C. P., Guerry P. 2003. Pseudaminic acid, the major modification on Campylobacter flagellin, is synthesized via the Cj1293 gene. Mol. Microbiol. 50:659–671 [DOI] [PubMed] [Google Scholar]
- 21. Grass S., et al. 2003. The Haemophilus influenzae HMW1 adhesin is glycosylated in a process that requires HMW1C and phosphoglucomutase, an enzyme involved in lipooligosaccharide biosynthesis. Mol. Microbiol. 48:737–751 [DOI] [PubMed] [Google Scholar]
- 22. Guerry P., et al. 2006. Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol. Microbiol. 60:299–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harding S. V., et al. 2007. The identification of surface proteins of Burkholderia pseudomallei. Vaccine 25:2664–2672 [DOI] [PubMed] [Google Scholar]
- 24. Harland D. N., et al. 2007. Identification of a LolC homologue in Burkholderia pseudomallei, a novel protective antigen for melioidosis. Infect. Immun. 75:4173–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Holden M. T., et al. 2004. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc. Natl. Acad. Sci. U. S. A. 101:14240–14245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Josenhans C., Vossebein L., Friedrich S., Suerbaum S. 2002. The neuA/flmD gene cluster of Helicobacter pylori is involved in flagellar biosynthesis and flagellin glycosylation. FEMS Microbiol. Lett. 210:165–172 [DOI] [PubMed] [Google Scholar]
- 27. Kawahara K., Dejsirilert S., Ezaki T. 1998. Characterization of three capsular polysacharides produced by Burkholderia pseudomallei. FEMS Microbiol. Lett. 169:283–287 [DOI] [PubMed] [Google Scholar]
- 28. Khrapova N. P., Tikhonov N. G., Prokhvatilova Y. V. 1998. Detection of glycoprotein of Burkholderia pseudomallei. Emerg. Infect. Dis. 4:336–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kondo E., Petkanchanapong V., Naigowit P., Kurata T., Kanai K. 1991. Demonstration of acid phosphatase activity in antigenic glycoprotein fractions obtained from the culture filtrate of Pseudomonas pseudomallei. Jpn. J. Med. Sci. Biol. 44:213–224 [DOI] [PubMed] [Google Scholar]
- 30. Kondo E., Wangroongsaub P., Naigowit P., Kanai K. 1994. Separation of antigenic glycoprotein fractions from cell-free homogenate of Pseudomonas pseudomallei and characterization as tyrosine phosphatase. Southeast Asian J. Trop. Med. Public Health 25:436–442 [PubMed] [Google Scholar]
- 31. Küster B., Krogh T. N., Mortz E., Harvey D. J. 2001. Glycosylation analysis of gel-separated proteins. Proteomics 1:350–361 [DOI] [PubMed] [Google Scholar]
- 32. Leelarasamee A., Bovornkitti S. 1989. Melioidosis: review and update. Rev. Infect. Dis. 11:413–425 [DOI] [PubMed] [Google Scholar]
- 33. Lindenthal C., Elsinghorst E. A. 1999. Identification of a glycoprotein produced by enterotoxigenic Escherichia coli. Infect. Immun. 67:4084–4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Logan S. M., Kelly J. F., Thibault P., Ewing C. P., Guerry P. 2002. Structural heterogeneity of carbohydrate modifications affects serospecificity of Campylobacter flagellins. Mol. Microbiol. 46:587–597 [DOI] [PubMed] [Google Scholar]
- 35. Logue C. A., Peak I. R. A., Beacham I. R. 2009. Facile construction of unmarked deletion mutants in Burkholderia pseudomallei using sacB counter-selection in sucrose-resistant and sucrose-sensitive isolates. J. Microbiol. Methods 76:320–323 [DOI] [PubMed] [Google Scholar]
- 36. Milton D. L., O'Toole R., Horstedt P., Wolf-Watz H. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178:1310–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Penfold R. J., Pemberton J. M. 1992. An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene 118:145–146 [DOI] [PubMed] [Google Scholar]
- 38. Piven N. N., et al. 1996. The immunogenicity and heterogeneity of Pseudomonas pseudomallei surface antigen 8. Zh. Mikrobiol. Epidemiol. Immunobiol. 1996:75–78 (In Russian.) [PubMed] [Google Scholar]
- 39. Reckseidler S. L., DeShazer D., Sokol P. A., Woods D. E. 2001. Detection of bacterial virulence genes by subtractive hybridization: identification of capsular polysaccharide of Burkholderia pseudomallei as a major virulence determinant. Infect. Immun. 69:34–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rocchetta H. L., Burrows L. L., Lam J. S. 1999. Genetics of O-antigen biosynthesis in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 63:523–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rodrigues F., et al. 2006. Global map of growth-regulated gene expression in Burkholderia pseudomallei, the causative agent of melioidosis. J. Bacteriol. 188:8178–8188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rotz L. D., Khan A. S., Lillibridge S. R., Ostroff S. M., Hughes J. M. 2002. Public health assessment of potential biological terrorism agents. Emerg. Infect. Dis. 8:225–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sarkar-Tyson M., et al. 2007. Polysaccharides and virulence of Burkholderia pseudomallei. J. Med. Microbiol. 56:1005–1010 [DOI] [PubMed] [Google Scholar]
- 44. Schell M. A., et al. 2007. Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol. Microbiol. 64:1466–1485 [DOI] [PubMed] [Google Scholar]
- 45. Schirm M., Schoenhofen I. C., Logan S. M., Waldron K. C., Thibault P. 2005. Identification of unusual bacterial glycosylation by tandem mass spectrometry analyses of intact proteins. Anal. Chem. 77:7774–7782 [DOI] [PubMed] [Google Scholar]
- 46. Schirm M., et al. 2003. Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol. Microbiol. 48:1579–1592 [DOI] [PubMed] [Google Scholar]
- 47. Smedley J. G., et al. 2005. Influence of pilin glycosylation on Pseudomonas aeruginosa 1244 pilus function. Infect. Immun. 73:7922–7931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Smith M. D., Wuthiekanun V., Walsh A. L., White N. J. 1995. Quantitative recovery of Burkholderia pseudomallei from soil in Thailand. Trans. R. Soc. Trop. Med. Hyg. 89:488–490 [DOI] [PubMed] [Google Scholar]
- 49. Stimson E., et al. 1995. Meningococcal pilin: a glycoprotein substituted with digalactosyl 2,4-diacetamido-2,4,6-trideoxyhexose. Mol. Microbiol. 17:1201–1214 [DOI] [PubMed] [Google Scholar]
- 50. Thibault P., et al. 2001. Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J. Biol. Chem. 276:34862–34870 [DOI] [PubMed] [Google Scholar]
- 51. Titball R. W., et al. 2008. Burkholderia pseudomallei: animal models of infection. Trans. R. Soc. Trop. Med. Hyg. 102(Suppl. 1):S111–S116 [DOI] [PubMed] [Google Scholar]
- 52. Twine S. M., et al. 2008. Flagellar glycosylation in Clostridium botulinum. FEBS J. 275:4428–4444 [DOI] [PubMed] [Google Scholar]
- 53. Twine S. M., et al. 2009. Motility and flagellar glycosylation in Clostridium difficile. J. Bacteriol. 191:7050–7062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Voisin S., et al. 2005. Identification and characterization of the unique N-linked glycan common to the flagellins and S-layer glycoprotein of Methanococcus voltae. J. Biol. Chem. 280:16586–16593 [DOI] [PubMed] [Google Scholar]
- 55. Voisin S., et al. 2007. Glycosylation of Pseudomonas aeruginosa strain Pa5196 type IV pilins with mycobacterium-like alpha-1,5-linked d-Araf oligosaccharides. J. Bacteriol. 189:151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. White N. J. 2003. Melioidosis. Lancet 361:1715–1722 [DOI] [PubMed] [Google Scholar]
- 57. Wikraiphat C., et al. 2009. Comparative in vivo and in vitro analyses of putative virulence factors of Burkholderia pseudomallei using lipopolysaccharide, capsule and flagellin mutants. FEMS Immunol. Med. Microbiol. 56:253–259 [DOI] [PubMed] [Google Scholar]
- 58. Zacharius R. M., Zell T. E., Morrison J. H., Woodlock J. J. 1969. Glycoprotein staining following electrophoresis on acrylamide gels. Anal. Biochem. 30:148–152 [DOI] [PubMed] [Google Scholar]
- 59. Zou N., et al. 2008. Relationship between antigenicity and pathogenicity for Burkholderia pseudomallei and Burkholderia mallei revealed by a large panel of mouse MAbs. Hybridoma (Larchmt.) 27:231–240 [DOI] [PubMed] [Google Scholar]