Abstract
Microcin C (McC), a natural antibacterial compound consisting of a heptapeptide attached to a modified adenosine, is actively taken up by the YejABEF transporter, after which it is processed by cellular aminopeptidases, releasing the nonhydrolyzable aminoacyl adenylate, an inhibitor of aspartyl-tRNA synthetase. McC analogues with variable length of the peptide moiety were synthesized and evaluated in order to characterize the substrate preferences of the YejABEF transporter. It was shown that a minimal peptide chain length of 6 amino acids and the presence of an N-terminal formyl-methionyl-arginyl sequence are required for transport.
INTRODUCTION
In the current ongoing quest for new antibiotics, aminoacyl-tRNA synthetases (aaRSs) have been regarded as promising targets (5, 11, 14). The natural antibiotic microcin C (McC) (Fig. 1, compound 1a) targets an aaRS and has been envisaged as a lead compound for further development as an antibacterial agent (17). McC consists of a heptapeptide that is covalently linked through a phosphoramidate bond to adenosine, with, in addition, an aminopropyl moiety esterified to the phosphoramidate linker (4). Once inside a sensitive cell, McC is processed by peptide deformylase and several peptidases that remove the N-terminal formyl group and the peptide part, respectively (7). As a result of intracellular processing, the active compound (compound 2), a modified nonhydrolysable aspartyl-adenylate, is released. Processed McC is a potent inhibitor of aspartyl-tRNA synthetase (AspRS) (8).
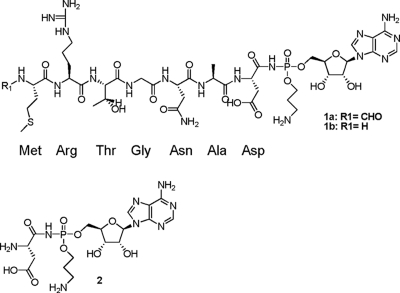
Fig. 1.
Microcin C (compound 1a), the deformylated variant (compound 1b), and processed McC (compound 2).
McC penetrates the outer membrane of the Escherichia coli cell mostly through the OmpF porin, but also through other, yet-unidentified transport systems (M. Novikova, A. Metlitskaya, and K. Severinov, unpublished data), and is subsequently transported through the inner membrane by the YejABEF transporter (10). YejABEF is the only complex responsible for McC transport, since yej mutants are highly resistant to McC and its maturation intermediates and chemical analogues. While intact McC inhibits the growth of sensitive E. coli cells at low micromolar concentrations, processed McC does not affect cell growth, even at millimolar concentrations. Thus, the peptide chain enables McC to function through a Trojan-horse mechanism by promoting active uptake via the YejABEF transporter. The recently improved synthetic approach for the production of McC analogues has led us to investigate the uptake properties of the Yej transporter in more detail. The results obtained could be important for further drug development, where peptides function as carrier moieties for drugs that otherwise would not be able to penetrate the bacterial membranes. Here, we used a number of McC analogues truncated either from their C- or N-terminal sides, or otherwise modified, to determine the minimal peptide chain length sufficient for facilitated transport by Yej.
MATERIALS AND METHODS
Chemistry.
Reagents and solvents were purchased from commercial suppliers (Acros, Sigma-Aldrich, Bachem, and Novabiochem) and used as provided unless otherwise indicated. N,N-Dimethylformamide (DMF) and tetrahydrofuran (THF) were analytical grade and were stored over 4-Å molecular sieves for drying. For reactions involving 9-fluorenylmethoxy carbonyl (Fmoc)-protected amino acids and peptides, DMF was used for peptide synthesis (low amine content). All other solvents used for reactions were analytical grade and were used as provided. Reactions were carried out in oven-dried glassware under a nitrogen atmosphere, and the reaction mixtures were stirred at room temperature unless otherwise indicated.
The 1H and 13C nuclear magnetic resonance (NMR) spectra of the compounds dissolved in dimethyl sulfoxide-d6 (DMSO-d6) or D2O were recorded on a Bruker UltraShield Avance 300-MHz or 500-MHz spectrometer. The chemical shifts are expressed as δ values in parts per million, using the residual solvent peaks (for DMSO, 1H, 2.50 ppm; 13C, 39.60 ppm; for D2O, 1H, 4.79 ppm) as a reference. Coupling constants are given in hertz. The peak patterns are indicated by the following abbreviations: bs, broad singlet; d, doublet; m, multiplet; q, quadruplet; s, singlet; and t, triplet. High-resolution mass spectra were recorded on a quadrupole time-of-flight mass spectrometer (Q-Tof-2; Micromass, Manchester, United Kingdom) equipped with a standard ESI (electrospray ionization) interface; samples were infused in 2-propanol–H2O (1:1) at 3 μl · min−1.
For thin-layer chromatography (TLC), precoated aluminum sheets were used (Silica gel 60 F254; Merck). The spots were visualized by UV light. Column chromatography was performed on ICN silica gel, 60 Å, 60 to 200 μm. Preparative high-performance liquid chromatography (HPLC) of peptides was done using a Waters Xbridge C18 (19- by 150-mm) column connected to a Waters 1525 binary HPLC pump and a Waters 2487 dual-absorbance detector. The final products were purified using a PLRP-S 100-Å column connected to a Merck-Hitachi L6200A intelligent pump. Eluent compositions are expressed as volume/volume. Purity was checked by analytical HPLC on an Inertsil ODS-3 (C18; 4.6- by 100-mm) column connected to a Shimadzu LC-20AT pump using a Shimadzu SPD-20A UV detector. Recordings were performed at 254 nm and 214 nm. Synthesis of all compounds was accomplished using the conditions described in the legend to Fig. 2. More detailed methods can be found in the supplemental material.
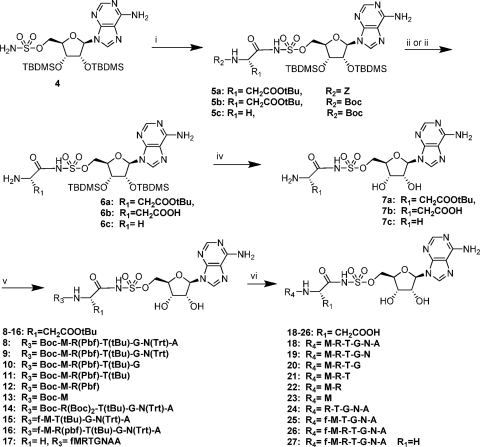
Fig. 2.
General reaction scheme for the various McC analogues. (i) N-α-CBZ-l-aminoacyl-(tBu or Boc)-succinimide, DBU in DMF, 6 h, room temperature (RT). (ii) For R2 = Z group, H2, Pd/C in MeOH, 3 h, RT. (iii) For R2 = Boc group of TFA/H2O (5:2), 4 h, 0°C to RT. (iv) Et3N.3HF in THF, 16 h, RT. (v) Protected peptide (1 eq.), HOBt (4 eq.), DIC (4 eq.) and DIEA (2 eq.) in DMF, 16 h, RT. R3 may be either a Boc or formyl group. (vi) TFA/thioanisole/H2O (90/2.5/7.5), 2 h, RT.
Biology: whole-cell activity determinations.
Bacteria were grown overnight in LB medium and cultured again the following day in fresh LB medium or LB medium containing 5 mM (l)-arabinose. Compounds were titrated in a 96-well plate using LB medium either with or without 5 mM l-arabinose to dilute the compounds. To each well, 85 μl LB medium with or without 5 mM l-arabinose was added to a total volume of 90 μl. Next, 10 μl of bacterial cell culture grown to an optical density at 600 nm (OD600) of 0.1 was added. The cultures were next placed into a Tecan Infinite M200 incubator and shaken at 37°C; then, the OD600 was determined after 8 h. All experiments were performed in triplicate.
The bacterial strains used for the evaluations were as follows: E. coli Ara-Yej (BW39758), expressing the yejABEF transporter upon l-arabinose induction; E. coli K-12 (BW28357), used as the wild-type control; E. coli ΔyejA, lacking subunit A of the YejABEF transporter; and E. coli ΔABN, lacking all three peptidase genes, pepA, pepB, and pepN.
Aminoacylation experiments.
To assess the degree of inhibition of the aminoacylation reaction, in vitro tests were performed using the relevant S30 cell extracts.
Preparation of S30 cell extracts.
Cells were grown in 50 ml LB medium. After centrifugation at 3,000 × g for 10 min, the supernatant was discarded and the pellet was resuspended in 40 ml buffer containing Tris · HCl or HEPES · KOH (pH = 8.0; 20 mM), MgCl2 (10 mM), and KCl (100 mM). The cell suspension was centrifuged again at 4,800 rpm. This procedure was repeated 2 times. The pellet was resuspended in 1 ml of a buffer comprised of Tris · HCl or HEPES · KOH (pH = 8.0; 20 mM), MgCl2 (10 mM), KCl (100 mM), and dithiothreitol (DTT) (1 mM) and kept at 0°C. Then, the cells were sonicated for 10 s and left at 0°C for 10 min. This procedure was repeated 5 to 8 times. The lysate was centrifuged at 15,000 × g for 30 min at +4°C.
tRNA aminoacylation reaction.
To 1 μl of solution containing inhibitor, 3 μl of E. coli S30 extracts was added. Next, 16 μl of the following aminoacylation mixture was added: Tris · HCl (30 mM; pH 8.0), DTT (1 mM), bulk E. coli tRNA (5 g/liter), ATP (3 mM), KCl (30 mM), MgCl2 (8 mM), and the specified radiolabeled amino acid (40 μM). The reaction products were precipitated in cold 10% trichloroacetic acid (TCA) on Whatman 3MM papers 5 min after the aminoacylation mixture was added. The aminoacylation reaction was carried out at room temperature. Depending on whether processing was needed, variable time intervals were included between the addition of the cell extract and the addition of the aminoacylation mixture. After thorough washing with cold 10% TCA, the papers were washed twice with acetone and dried on a heating plate. Following the addition of scintillation liquid, the amount of radioactivity was determined in a scintillation counter.
RESULTS
Design, synthesis, and inhibitory activity of McC analogues in cell extracts.
Acyladenylates are the natural reaction intermediates of aaRSs. Therefore, compounds mimicking acyladenylates could inhibit charging of tRNAs with their cognate amino acids, thus leading to cessation of mRNA translation. A crucial portion of such an inhibitor scaffold is the linker between the amino acid and the adenosine moiety, which needs to be metabolically stable (13). Many different linkers have been developed as surrogates for the natural labile acylphosphate linkage (2, 15, 18). Aminoacyl-sulfamoyl adenosines were found to be the most potent analogues and proved to be nanomolar inhibitors of their corresponding aaRSs (3, 12, 16). When trying to mimic natural McC, we therefore opted for aminoacyl-sulfamoyl adenosines, as they are more stable and readily synthesized than aminoacyl-phosphoramidate-adenosine analogues, as found in natural McC (17). Following a recently developed method for the synthesis of McC analogues (Fig. 2), a series of compounds were created that were truncated from their C termini (compounds 18 to 23 in Fig. 2; each compound contains an aspartate as the ultimate C-terminal residue and lacks the N-formyl group) or from their N termini (compound 24). To evaluate the role of arginine at the second position of the peptide, compound 25 was created. The expected positive effect of the presence of an N-formyl moiety could be studied via the “full analogue” (compound 26).
In cell extracts prepared from McC-sensitive E. coli, the addition of these compounds inhibited the tRNAAsp aminoacylation reaction (Fig. 3). No inhibition was observed in extracts prepared from cells lacking aminopeptidases A, B, and N with compounds 1a and 1b or 18 to 26. In contrast, the addition of compound 7b to mutant cell extracts led to tRNAAsp aminoacylation inhibition. This is an expected result, since D-SA (where D refers to aspartic acid and SA to sulfamoyl adenosine), is a nonhydrolyzable aspartyl-adenylate that can inhibit AspRS directly, i.e., without processing.
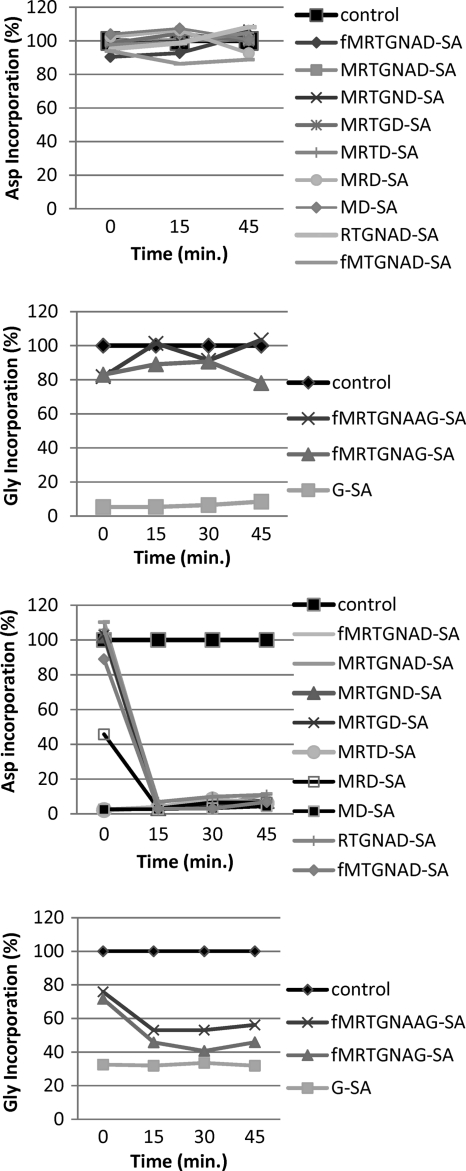
Fig. 3.
(Top two panels) Asp-RS and Gly-RS inhibition in S30 extracts of McC-resistant E. coli cells lacking peptidases A, B, and N. (Bottom two panels) Asp-RS and Gly-RS inhibition in S30 extracts of McC-sensitive E. coli cells. In all tests, the different extracts were incubated with the respective AspRS or GlyRS inhibitors, taking samples at different time points for evaluation of the aminoacylation reaction. The amounts of aminoacylated tRNAAsp or tRNAGly were measured via scintillation counting.
McC analogues targeting AspRS and carrying peptide chains longer than 7 amino acids were found to be significantly less stable under acidic deprotection conditions during their synthetic assembly and thus proved hard to synthesize, which could be attributed to intramolecular cyclization, resulting in N3-5′-cycloadenosine (12). Hence, to evaluate McC analogues carrying a peptide longer than the usual heptapeptide, fMRTGNAAG-SA (compound 17) was synthesized, with incorporation of an additional internal alanine and substitution of the C-terminal aspartate for glycine. This compound could be synthesized using a nonprotected peptide, albeit with a low yield. In cell extracts, the addition of this compound inhibited the tRNAGly aminoacylation reaction (Fig. 3, lower panel). Except for compound 7c, no inhibition was observed in extracts prepared from cells lacking aminopeptidases A, B, and N. Like D-SA, described above, compound 7c does not require processing and therefore can inhibit tRNAGly formation directly. The results thus indicate that compound 17 targets GlyRS through formation of glycyl sulfamoyl adenosine generated by processing in McC-sensitive E. coli cell extracts. The control heptapeptidic analogue (fMRTGNAG-SA; compound 27) likewise targets GlyRS.
Antibacterial activity of McC analogues with truncated or elongated peptide chains.
The ability of new compounds to inhibit the growth of McC-sensitive E. coli was evaluated next. The assay used to determine the sensitivities to various compounds consisted of measuring the optical densities reached by identical cell cultures in wells of microtiter plates in the presence of various concentrations of inhibitors. The assay has proven to be highly sensitive and reproducible and is superior to standard tests based on the determination of growth inhibition zones on plates.
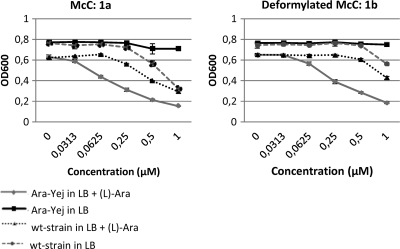
Since intracellular transport by YejABEF may limit cell sensitivity to McC and its derivatives, a new E. coli Ara-Yej tester strain (BW39758) was developed (K. A. Datsenko and B. L. Wanner, personal communication). In this strain, the genomic yejABEF operon is under the control of the arabinose-inducible araBAD promoter. We expected that the strain would be relatively resistant to McC in the absence of inducer but might be hypersensitive in its presence. To test this expectation, the sensitivities of Ara-Yej cells and control wild-type E. coli K-12 BW28357 cells to McC (compound 1a) were determined (Fig. 4). Uninduced Ara-Yej cells were practically resistant to up to 1 μM McC, the highest concentration used in the experiment. In contrast, the growth of wild-type cells was visibly inhibited under these conditions. The addition of 5 mM l-arabinose to the medium had little effect on the McC sensitivity of wild-type cells but dramatically increased the sensitivity of the Ara-Yej strain to a level above that of wild-type cells at matching McC concentrations (pronounced growth inhibition in the presence of 0.25 μM McC). The results thus indicate that (i) induced E. coli Ara-Yej is a superior strain for measuring sensitivity to McC and related compounds (by comparing induced and uninduced cell growth in the presence of a given concentration of inhibitor tested) and (ii) the amount of the YejABEF transporter produced by wild-type E. coli during growth in rich medium indeed limits cell sensitivity to McC and, presumably, the natural YejABEF substrates are taken up less efficiently, as well. The latter finding is of potential interest, since it has been shown that the levels of the yejABEF transcript are subject to negative regulation by a small RNA, rydC (1), which is involved in stress response.
Fig. 4.
Antibacterial activity of McC (compound 1a) and its nonformylated variant (compound 1b) assayed with wild-type E. coli in the presence and absence of added l-arabinose and with induced and uninduced Ara-Yej cells.
In agreement with previous findings, we also observed that an McC derivative lacking the N-terminal formyl group (compound 1b) was less effective against wild-type cells than the formylated variant, 1a (Fig. 4). This was likewise demonstrated with the formylated synthetic McC analogue (compound 26) versus the nonformylated variant (compound 18). The effect was also observed (though it was less pronounced) in the case of the induced Ara-Yej strain.
The antibacterial activities of various McC analogues were next determined. The activities of new compounds were compared to that of MRTGNAD-SA (compound 18 in Fig. 5), a previously characterized synthetic McC analogue containing a sulfamoyl bond instead of the natural phosphoamide and lacking the aminopropyl moiety. Both formylated (compound 26) and nonformylated (compound 3) MRTGNAD-SA variants were approximately 10 times less effective than McC. This difference is likely due to the lack of the aminopropyl group, which was shown to increase the efficiency of target inhibition by processed McC (9). Interestingly, uninduced Ara-Yej cells were partially inhibited by compound 18 at concentrations above 2.5 μM. This could be explained by low expression levels of the araBAD::yej fusion caused by traces of arabinose in the LB medium or by the presence of a yet-unknown additional, low-affinity transporter.
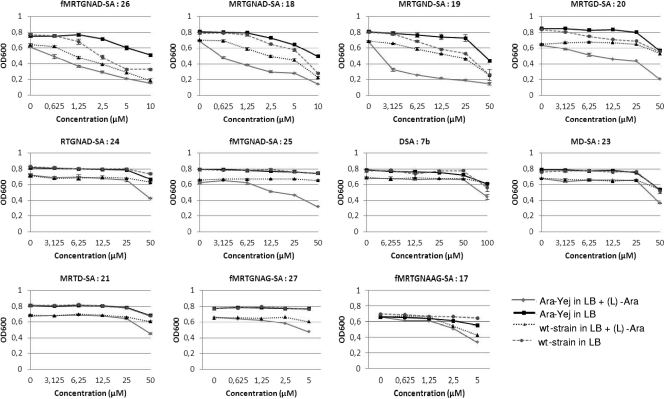
Fig. 5.
Antibacterial activities of various shortened McC analogues (compounds 23 to 26) targeting AspRS and of heptamer (compound 27) and octamer (compound 17) variants targeting GlyRS and assayed with wild-type E. coli in the presence and absence of added l-arabinose and with induced and uninduced Ara-Yej cells.
As can be seen, removal of the penultimate McC peptide amino acid (Ala6) (compound 19) had little effect on antibacterial activity. The result thus indicates that the Yej transporter can recognize McC derivatives with a 6-amino-acid peptide rather than the 7-amino-acid peptide found in natural McC. Removal of 2 amino acids (Asn5 and Ala6; compound 20) strongly decreased antibacterial activity, which, however, remained yejABEF dependent. Removal of additional internal amino acids of the McC peptide abolished the activity (compounds 21 to 23 [data not shown]).
In contrast to the single amino acid deletion at the C terminus, a variant lacking the N-terminal methionine (compound 24) was biologically inactive, indicating that YejABEF recognizes not just the length, but also the sequence of the peptide attached to the AMP analogue. Compound 25, lacking Arg2 and containing formylated N-terminal methionine, likewise was very poorly active.
For each compound, control experiments with McC-resistant cells lacking one of the yej genes or the pepA, -B, and -N genes were also performed (the sensitivities of these cells were determined in a standard plate assay) on M63 agar plates. Both control cells were fully resistant to all compounds, with the exception of compound 23 (MD-SA), which showed minor activity against Δyej cells but no activity against ΔABN cells, and compound 7b (D-SA), which is active at concentrations of >100 μM against both the wild-type and mutant cells, as expected (17).
Elongation of the peptide chain by a single amino acid residue, as in compound 17, did not affect its potency. In fact, this compound was slightly more active than the corresponding compound 27 containing the heptapeptide (Fig. 4). As expected, compound 17 proved inactive against E. coli cells with a disrupted yej gene or cells without functional aminopeptidases A, B, and N and is therefore acting through a Trojan-horse mechanism.
To confirm the mechanism of action, a series of in vitro aminoacylation experiments were performed. As can be observed from Fig. 3, all shortened compounds inhibit tRNA aminoacylation reactions after a 15-min incubation in cell extract, sufficient time for processing of natural McC. On the other hand, no inhibition was observed in cell extracts prepared from ΔABN cells. This demonstrates again that all of the shortened compounds, with the exception of D-SA and G-SA, required the action of PepA, PepB, or PepN for release of the inhibitor.
The physiological substrate of the YejABEF transporter (other than McC) is not yet known. Since our data indicate that McC uptake is peptide length dependent, we attempted to estimate the optimal length of peptide substrates transported by YejABEF by setting up in vivo competition between McC and several MccA-based peptides. To this end, fixed amounts of McC were deposited on lawns of sensitive cells, along with increasing concentrations of peptides. The expectation was that at some point the peptide would outcompete McC, rendering the cells resistant and leading to the disappearance of growth inhibition zones. In all, five peptides were tried (MRTGNAN, GMRTGNAN, GGMRTGNAN, G3MRTGNAN, and G6MRTGNAN). The results indicated that the minimal peptide concentrations needed for complete protection from 13 μM McC (with no inhibition zone observed) were ∼7 mM for the peptides MRTGNAN and G6MRTGNAN but three times lower, or ∼2 mM, for the other three peptides. The results are consistent with increased potency of an octapeptide-based GSA inhibitor and indicate that the preferred peptide length for YejABEF-mediated uptake is more than 7 but less than 13 amino acids.
DISCUSSION
As shown previously, the McC peptide part plays an important role in endowing the intracellularly active compound 2 with its antibacterial activity. Without the peptide being recognized by the YejABEF transporter, no activity at micromolar concentrations can be demonstrated, as the phosphoamidated nucleoside analogue is not efficiently taken up by the target cells. This has been demonstrated clearly using cells carrying mutations in the yej genes coding for the transporter, which proved resistant to McC, highlighting the necessity of the transporter for the internalization and antibacterial activity of compounds 1a and 1b (10). Likewise, this has been demonstrated for different sulfamoylated nucleoside analogues, like compound 18, which display strong inhibitory activity in vitro against cognate aaRSs but proved to be rather weak inhibitors in cellular antibacterial screens. Site-specific mutagenesis of the mccA gene coding for the peptide moiety of McC and providing substitution of the internal amino acid positions generated multiple active McC variants. Only the C-terminal asparagine, which is converted to aspartic acid upon maturation of McC, proved indispensable (6). The N-terminal formyl methionine was not included in the analysis, since it is essential for initiation of translation of mccA mRNA. Chemical manipulation of sulfamoylated congeners of McC allows the replacement of the last aspartic acid, enabling inhibition of different aaRSs (17) and probing of the importance of the N-terminal methionine residue. Hence, to further delineate the recognition elements for uptake by the YejABEF transporter, different truncated and modified peptides were introduced here to determine the minimal chain length and best composition for efficient uptake.
Four important results were obtained in this work. First, it was shown that the YejABEF transporter is able to transport larger McC analogues across the bacterial inner membrane, but there seems to be no recognition of compounds with peptide chains of less than 6 amino acids. We were not able to investigate to what extent the maximal chain length can be extended, due to difficulties in synthesizing larger compounds. However, from a pharmaceutical perspective, this is obviously of minor importance. Second, if the N-terminal formyl group is excluded from the McC analogue, a slightly lower activity is observed. This observation is also consistent with earlier data that showed that McC (compound 1a) is slightly more active than nonformylated McC (compound 1b). Third, our data show that if the N-terminal amino acid of the hexapeptidyl McC analogue is different from methionine, little or no activity is observed. In contrast, a hexapeptidyl analogue carrying methionine at the N terminus (compound 19) proved active, although a 5-fold-higher concentration was required to achieve the activity levels of parent compound 26. This suggests that the N-terminal methionine or, more exactly, formyl-methionine (see above) is important to recognition by the YejABEF transporter. Finally, upon exclusion of arginine from the hexapeptide moiety, as in compound 25, the activity is dramatically decreased compared to compound 26. Therefore, the presence of an N-terminal formyl-methionine in combination with arginine seems to be important for uptake by the YejABEF transporter. One therefore could speculate that a positive charge is beneficial at this position. However, the latter result is in contrast with the McC variants obtained by mutagenesis, where Ala2, Ser2, and Trp2 variants also proved active (6). In addition, and more curiously, compounds 23 and 7b displayed antibacterial activity, albeit at high concentrations, which may be explained by their small size and hence their ability to diffuse through the bacterial membrane. The lowest concentrations for all new products at which inhibition can be detected for the wild-type E. coli strain can be found in Table 1, highlighting the different points made above. The results obtained here delineate the recognition properties of the YejABEF transporter, which proved indispensable for the antibacterial potentials of different McC analogues. The Trojan-horse mechanism of McC action, mediated by the transporter, also paves the way for improving the uptake, and hence the biological activities, of different toxic entities with otherwise low in vitro and/or in vivo activity.
Table 1.
Lowest concentrations of all analogues at which clear inhibition could be observed against wild-type E. coli K-12 (BW28357) cells
| Compound (no.) | Lowest inhibitory concn (μM) |
|---|---|
| McC (1a) | <0,25 |
| Deformylated McC (1b) | 0,25 |
| fMRTGNAD-SA (26) | <0,63 |
| MRTGNAD-SA (18) | 0,63 |
| MRTGND-SA (19) | 3,13 |
| MRTGD-SA (20) | 6,25 |
| MRTD-SA (21) | 50 |
| MD-SA (23) | 50 |
| D-SA (7b) | 100 |
| RTGNAD-SA (24) | 50 |
| fMTGNAD-SA (25) | 12,5 |
| fMRTGNAG-SA (27) | 5 |
| fMRTGNAAG-SA (17) | 5 |
Supplementary Material
ACKNOWLEDGMENTS
G.H.M.V. is the recipient of a Belgian Agency for Innovation by Science and Technology (IWT) fellowship (SB 81116). Work in the Severinov laboratories was supported by a TCF grant from Rutgers University and by a Molecular and Cellular Biology Program grant from the Russian Academy of Sciences Presidium. This work was partially supported by NIH grant RC1GM09047 to B.L.W.
We are indebted to Roger Busson for assistance in interpreting some of the NMR spectra, and we thank Chantal Biernaux for assistance in editing the manuscript.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org.
Published ahead of print on 20 May 2011.
REFERENCES
- 1. Antal M., Bordeau V., Douchin V., Felden B. 2005. A small bacterial RNA regulates a putative ABC transporter. J. Biol. Chem. 280:7901–7908 [DOI] [PubMed] [Google Scholar]
- 2. Balg C., et al. 2007. Synthesis of beta-ketophosphonate analogs of glutamyl and glutaminyl adenylate, and selective inhibition of the corresponding bacterial aminoacyl-tRNA synthetases. Bioorg. Med. Chem. 15:295–304 [DOI] [PubMed] [Google Scholar]
- 3. Cisar J. S., Ferreras J. A., Soni R. K., Quadri L. E., Tan D. S. 2007. Exploiting ligand conformation in selective inhibition of non-ribosomal peptide synthetase amino acid adenylation with designed macrocyclic small molecules. J. Am. Chem. Soc. 129:7752–7753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guijarro J. I., et al. 1995. Chemical structure and translation inhibition studies of the antibiotic microcin C7. J. Biol. Chem. 270:23520–23532 [DOI] [PubMed] [Google Scholar]
- 5. Hurdle J. G., O'Neill A. J., Chopra I. 2005. Prospects for aminoacyl-tRNA synthetase inhibitors as new antimicrobial agents. Antimicrob. Agents Chemother. 49:4821–4833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kazakov T., Metlitskaya A., Severinov K. 2007. Amino acid residues required for maturation, cell uptake, and processing of translation inhibitor microcin C. J. Bacteriol. 189:2114–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kazakov T., et al. 2008. Escherichia coli peptidase A, B, or N can process translation inhibitor microcin C. J. Bacteriol. 190:2607–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Metlitskaya A., et al. 2006. Aspartyl-tRNA synthetase is the target of peptide nucleotide antibiotic Microcin C. J. Biol. Chem. 281:18033–18042 [DOI] [PubMed] [Google Scholar]
- 9. Metlitskaya A., et al. 2009. Maturation of the translation inhibitor microcin C. J. Bacteriol. 191:2380–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Novikova M., et al. 2007. The Escherichia coli Yej transporter is required for the uptake of translation inhibitor microcin C. J. Bacteriol. 189:8361–8365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ochsner U. A., Sun X., Jarvis T., Critchley I., Janjic N. 2007. Aminoacyl-tRNA synthetases: essential and still promising targets for new anti-infective agents. Expert Opin. Invest. Drugs 16:573–593 [DOI] [PubMed] [Google Scholar]
- 12. Peterson E. M., Brownell J., Vince R. 1992. Synthesis and biological evaluation of 5′-sulfamoylated purinyl carbocyclic nucleosides. J. Med. Chem. 35:3991–4000 [DOI] [PubMed] [Google Scholar]
- 13. Qiao C., et al. 2007. 5′-O-[(N-acyl)sulfamoyl]adenosines as antitubercular agents that inhibit MbtA: an adenylation enzyme required for siderophore biosynthesis of the mycobactins. J. Med. Chem. 50:6080–6094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schimmel P., Tao J., Hill J. 1998. Aminoacyl tRNA synthetases as targets for new anti-infectives. FASEB J. 12:1599–1609 [PubMed] [Google Scholar]
- 15. Somu R. V., et al. 2006. Rationally designed nucleoside antibiotics that inhibit siderophore biosynthesis of Mycobacterium tuberculosis. J. Med. Chem. 49:31–34 [DOI] [PubMed] [Google Scholar]
- 16. Ueda H., et al. 1991. X-ray crystallographic conformational study of 5′-O-[N-(L-alanyl)-sulfamoyl]adenosine, a substrate analogue for alanyl-tRNA synthetase. Biochim. Biophys. Acta 1080:126–134 [DOI] [PubMed] [Google Scholar]
- 17. Van de Vijver P., et al. 2009. Synthetic microcin C analogs targeting different aminoacyl-tRNA synthetases. J. Bacteriol. 191:6273–6280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vannada J., et al. 2006. Design, synthesis, and biological evaluation of beta-ketosulfonamide adenylation inhibitors as potential antitubercular agents. Organic Lett. 8:4707–4710 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.