Abstract
Polycomb group (PcG) proteins control the epigenetic inheritance of transcription regulatory states during development. Progression from pluripotency to differentiation requires the concurrent activation and repression of different PcG target genes. We found that REST and nine REST-associated proteins copurified with Cbx family PcG proteins from mouse embryonic stem (ES) cells. REST interacted with Cbx proteins in live cells and coprecipitated with endogenous Ring1b. Endogenous PRC1 subunits occupied all sites tested that were bound by REST in ES cells. Antibodies directed against different PRC1 subunits precipitated proximal versus distal RE1 elements with opposite relative efficiencies, suggesting that PRC1 bound different sites in distinct configurations. Deletion of the amino-terminal region of REST (RestΔN knockout) as well as short hairpin RNA depletion of REST (REST knockdown) in ES cells reduced PRC1 binding at distal RE1 elements and increased PRC1 binding at proximal RE1 elements. RestΔN and PRC1 subunit knockout as well as REST and PRC1 subunit knockdown had similar relative effects on transcription of neuronal genes in ES cells, derepressing genes with distal, but not genes with proximal, RE1 elements. In differentiating neurons, RestΔN knockout reduced PRC1 occupancy and derepressed transcription at distal RE1 elements but increased PRC1 occupancy and repressed transcription at proximal RE1 elements. The opposite effects of REST on PRC1 occupancy at different RE1 elements contributed to the gene-specific control of PRC1 functions during ES cell differentiation.
INTRODUCTION
Epigenetic regulatory factors such as Polycomb group (PcG) proteins are thought to perpetuate genome-wide patterns of transcription when cells divide to produce identical daughter cells. The production of new cell types during development requires the displacement of epigenetic regulatory complexes at some genes and the formation of new epigenetic complexes at other genes in the same cell. The genes where existing epigenetic complexes are displaced and the genes where new epigenetic complexes are formed must be individually specified. Whereas epigenetic mechanisms can maintain transcriptional states, changes in the transcription of specific genes are likely to require sequence-specific DNA recognition. The association of PcG proteins with individual genes must therefore be controlled by gene- and cell-type-specific mechanisms.
Hundreds of genes, many of which encode developmental regulators, are repressed by PcG proteins in mammalian embryonic stem (ES) cells. Different subsets of these genes are derepressed in different cell lineages and at different stages of development. Conversely, new genes are repressed by PcG proteins during differentiation. The mechanisms that determine the opposite changes in the PcG protein associations with different genes during differentiation are largely unknown.
PcG proteins form two classes of Polycomb repressive complexes (PRC1 and PRC2) (28). Previous studies of the regulation of PcG protein occupancy focused on the establishment and maintenance of PRC2 binding (18, 33, 48, 52). The results from those studies do not explain how PcG protein binding at different genes is regulated in opposite ways in the same cells. Moreover, PRC1 can associate with chromatin in cells lacking PRC2 (31, 42, 46, 51). Mechanisms for the regulation of PRC1 binding in the absence of PRC2 are poorly understood.
Each subunit of vertebrate PRC1 is encoded by multiple genes (Cbx, Ring1, Mel18/Bmi1, and Phc families). Previous studies of the PRC1 association with chromatin have focused on PRC1 binding to DNA/RNA and to histones. Mel18 and reconstituted PRC1 can bind DNA directly in vitro (16, 26). Drosophila melanogaster PRC1 can wrap about 400 bp of DNA in a structure predicted to exclude nucleosomes in vitro and cross-links most efficiently to regions depleted of nucleosomes in cells (34). Cbx7 binding to the ANRIL noncoding RNA was previously proposed to recruit PRC1 to the Ink4a/ARF locus (53).
Cbx family proteins can bind H3 trimethylated on K27 in vitro (3, 8). Genome-wide H3 K27 trimethylation correlates with PRC1 occupancy in embryonic stem cells and fibroblasts (5, 6, 29). H3 K27 trimethylation is not essential for the PRC1 association with chromatin (31, 42, 46, 51). It is unclear if changes in H3 K27 trimethylation regulate the association of PRC1 proteins with individual genes, since mechanisms for the gene-specific modulation of H3 K27 trimethylation have not been described. PRC1 subunits copurify with several sequence-specific DNA-binding proteins (13, 38, 45). The respective roles of interactions with DNA, noncoding RNA, histone H3, and DNA-binding proteins in the association of PRC1 proteins with target genes remain incompletely understood.
Here, we identify Cbx family protein interactions with REST (NRSF) and REST-associated proteins in ES cells and in differentiating neurons. REST was originally characterized as a repressor of neuronal genes in nonneuronal cells (10, 47). Subsequent studies have shown that REST regulates neuronal genes during neurogenesis and that it can both activate and repress genes containing RE1 elements (2, 4, 25, 30). REST and Cbx family proteins copurified from ES cell extracts and formed complexes in live cells. REST facilitated PRC1 binding at proximal RE1 elements and inhibited PRC1 binding at distal RE1 elements. REST and PRC1 subunits coregulated neuronal gene transcription in ES cells. REST had opposite effects on PRC1 occupancy as well as on transcription at genes that contained distal versus proximal RE1 elements in differentiating neurons.
MATERIALS AND METHODS
Cell lines and culture conditions.
The ES cell lines expressing Cbx2, Cbx6, Cbx7, and Cbx8 fused to Venus and the FLAG epitope were described previously (43). RestΔN and Rest+/+ ES cells were provided by Helle Jørgensen and Amanda Fisher (MRC) (9, 24). Cbx2−/−, Ring1bfl/fl Ring1a−/−, Ring1bfl/fl, Bmi1−/− Mel18−/−, and control ES cells were provided by Haruhiko Koseki (RIKEN) (12, 13, 27). Embryoid bodies were prepared and induced to differentiate into neuronal stem cells (NSCs) and neurons as described previously (39). The cells were cultured under the conditions described in the supplemental material.
Immunoaffinity purification and mass spectrometry.
Mouse ES cells stably expressing Cbx fusion proteins and the parental PGK12.1 cells were lysed, and nuclei were isolated. Nuclear proteins were extracted, and the extracts were incubated with anti-FLAG M2 beads (catalog number A2220; Sigma). The beads were washed, and bound proteins were eluted by using the FLAG peptide. The eluted fractions were incubated with anti-green fluorescent protein (GFP) agarose beads (catalog number D153-8; MBL International), the beads were washed, and bound proteins were eluted. The purification protocol is described in detail in the supplemental material.
The purified proteins were precipitated and separated by SDS-PAGE. The lanes containing proteins from cells that expressed Cbx2 and Cbx7 fusions were cut into 20 slices. The proteins in each slice were digested with trypsin, and the peptides were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) at the Taplin Mass Spectrometry Facility (Harvard Medical School). The fragmentation patterns of peptide ions were matched to protein sequences by using Sequest.
BiFC analysis of REST interactions with Cbx proteins.
Expression vectors for bimolecular fluorescence complementation (BiFC) analysis were constructed by inserting the sequence encoding the N-terminal 172 amino acids of the Venus fluorescent protein (VN) upstream of the coding sequences of the human Cbx proteins (51) in the pLVX vector. Sequences encoding intact GFP as well as C-terminal amino acids 173 to 238 of yellow fluorescent protein (YFP) (YC) were inserted downstream of the coding sequence of human REST, provided by Gail Mandel (Oregon Health & Science University) (1), in pLVX. Plasmids encoding truncated REST fusion proteins lacking the N-terminal 275 (ΔN1) or 332 (ΔN2) amino acid residues or encompassing amino acid residues 73 to 546 (DBD) (49) were constructed by using the same strategy.
HEK293T cells were cotransfected with 0.25 μg pVN-CbxN (n = 2, 4, 6, 7, or 8), 0.25 μg pREST-YC, and 0.5 μg pCDNA3(+) or 0.1 μg Jun-cyan fluorescent protein (CFP), used as a transfection control. The cells were imaged by using fluorescence microscopy, and the fluorescence intensities were measured by using flow cytometry as described in the supplemental material.
Chromatin immunoprecipitation and immunoblotting.
Chromatin immunoprecipitation (ChIP), immunoprecipitation, and immunoblotting were performed as described previously (43), with the antibodies and modifications described in the supplemental material. The efficiencies of chromatin immunoprecipitation were quantified relative to a standard curve prepared using input chromatin. The sequences of the primers used for ChIP analysis are listed in Table S2 in the supplemental material.
REST, Rcor1, and PRC1 subunit knockdown and transcript analysis.
PGK12.1 mouse ES cells were infected by pGIPZ lentiviral vectors containing short hairpin RNA (shRNA) sequences (see Table S3 in the supplemental material). Infected cells were selected by culture in the presence of puromycin.
Total RNA was isolated from the cells indicated. The RNA was reverse transcribed, and the cDNA levels were measured by quantitative PCR using the primers listed in Table S4 in the supplemental material.
RESULTS
Identification of Cbx protein interaction partners.
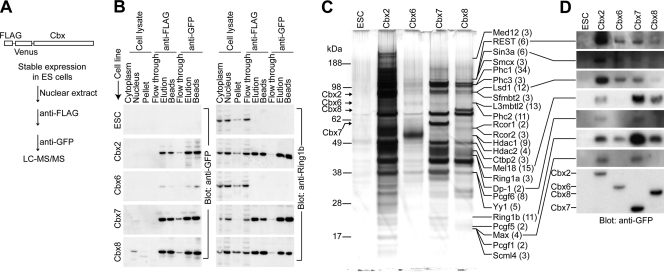
To identify Cbx protein interaction partners that could regulate the PRC1 association with individual genes, we purified proteins associated with different Cbx family fusion proteins from mouse ES cells through sequential immunoaffinity chromatography (Fig. 1A). The Cbx fusion proteins were enriched up to 310,000-fold, and Ring1b was coenriched up to 1,000-fold (Fig. 1B). No histone H3 was detected in the purified fractions by immunoblotting or by mass spectrometry, indicating that the copurification was not due to contamination by total chromatin (data not shown).
Fig. 1.
Purification of proteins associated with Cbx family members from mouse ES cells. (A) Flow diagram for immunoaffinity purification and identification of proteins associated with Cbx family members from mouse ES cells that expressed Cbx fusion proteins (43). (B) The enrichments of Cbx fusion proteins and of Ring1b during purification from the cell lines indicated to the left of each blot were evaluated. An equal proportion of each fraction (indicated above the lanes) was analyzed by immunoblotting using anti-GFP (left) and anti-Ring1b (right) antibodies. Multiple exposures of the blots were scanned, and the total protein concentrations were measured to calculate the fold enrichment of each Cbx protein and of Ring1b at each stage of purification. The final fold enrichments of each Cbx protein and of Ring1b were 310,000 and 240 (Cbx2), 1,000 and 1.5 (Cbx6), 12,000 and 1,000 (Cbx7), and 6,600 and 390 (Cbx8), respectively, relative to the cell lysates. ESC, parental ES cell line used to generate the ES cell lines expressing Cbx fusions. (C) SDS-PAGE analysis of the fractions purified from cells that expressed the Cbx fusion proteins indicated above the lanes or the parental cells (ESC). The mobilities of PRC1- and REST-associated proteins identified by mass spectrometry are indicated by the labels at the right of the images. The number of unique peptides corresponding to each protein is indicated in parentheses after the name of each protein. (D) Fractions purified from cells that expressed the Cbx fusions indicated above the lanes were analyzed by immunoblotting using antibodies directed against the proteins connected to the lines on the left of the images. The Cbx fusions were detected by using anti-GFP antibodies (bottom). Equal proportions of the fractions purified from cells that expressed Cbx2, Cbx7, and Cbx8 fusions and from the parental cells (ESC) were analyzed. A 10-fold-larger proportion of the fraction purified from cells that expressed the Cbx6 fusion was analyzed to compensate for the loss of the Cbx6 fusion during the isolation of nuclei.
LC-MS/MS analysis of proteins that copurified with Cbx2 and Cbx7 identified a total of 66 unique peptides derived from REST and 9 proteins that were previously reported to interact with REST (1, 2, 17, 21, 36, 40) (Fig. 1C). Immunoblotting confirmed that REST, Sin3a, and Lsd1, as well as known PRC1 subunits and previously identified Ring1b interaction partners, copurified with Cbx family proteins (Fig. 1D).
REST interactions with Cbx proteins in living cells and cell extracts.
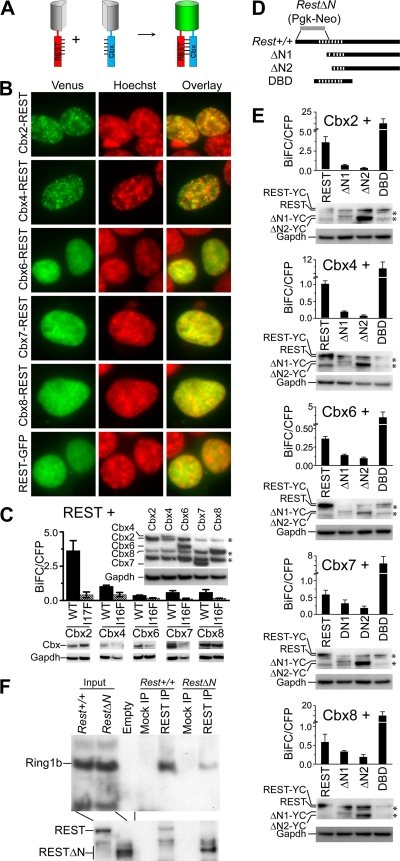
To determine if REST formed complexes with Cbx proteins in living cells, we used bimolecular fluorescence complementation (BiFC) analysis to investigate their interactions (20) (Fig. 2A). REST formed BiFC complexes with all Cbx proteins albeit with different efficiencies (Fig. 2B and C). The higher efficiency of Cbx2-REST BiFC complex formation correlated with the larger amount of REST that copurified with Cbx2 (compare Fig. 2C and 1D). The differences in BiFC complex formation by different Cbx proteins were not caused by differences in the levels of fusion protein expression (Fig. 2C, inset). BiFC complexes formed by REST with different Cbx proteins were enriched in subnuclear foci, whereas the bulk of REST was uniformly distributed in the nucleoplasm (Fig. 2B).
Fig. 2.
REST interactions with Cbx proteins in living cells and with Ring1b in native cell extracts. (A) Principle for BiFC analysis of REST interactions with Cbx proteins. REST and Cbx proteins fused to fluorescent protein fragments were coexpressed. The interaction of REST with Cbx proteins was detected based on the facilitated association of the fluorescent protein fragments. (B) The subnuclear distributions of complexes formed by the Cbx family proteins and REST indicated at the left of the images were visualized by using BiFC analysis (green) in live HEK293T cells stained by Hoechst dye (red). Different exposure times were used to compensate for differences in fluorescence intensities of BiFC complexes formed by different Cbx proteins with REST. The images are representative of the majority of cells in each population. (C) Efficiencies of BiFC complex formation by REST with wild-type (WT) and mutant (I17F or I16F) Cbx proteins measured by flow cytometry. The bars show the mean fluorescence intensities of 20,000 cells in each of two independent experiments. The levels of expression of different Cbx fusion proteins were compared by analyzing equal amounts of the cell extracts by immunoblotting using anti-GFP antibodies (inset above bars) (*, cross-reactive proteins). The levels of wild-type and mutant Cbx fusion protein expression were similarly compared (below the graph). The levels of Gapdh were measured to determine the amount loaded into each lane. (D) The diagrams indicate the truncated REST variants used to map interactions with Cbx proteins. The stripes indicate the zinc finger DNA-binding domain. The region replaced by Pgk-Neo in RestΔN knockout cells is indicated by a gray bar (9). (E) Efficiencies of BiFC complex formation by the Cbx proteins indicated above the bars with the truncated REST proteins indicated below the bars measured by flow cytometry. The bars show the mean fluorescence intensities of 20,000 cells in each of two independent experiments. The levels of expression of full-length and truncated REST fusions were measured by immunoblotting using anti-REST antibody (below each graph) (*, cross-reactive proteins). Gapdh was measured to determine the amount loaded into each lane. (F) Analysis of endogenous REST and Ring1b interactions in ES cell extracts. Extracts from Rest+/+ and RestΔN ES cells were precipitated by using anti-REST antibodies or beads alone (mock). The precipitates were analyzed by immunoblotting using antibodies directed against the proteins indicated at the left of the image. The input lanes contained 5% of the extract.
The specificity of BiFC complex formation by REST with Cbx family proteins was evaluated by mapping the region in each Cbx protein and in REST required for fluorescence complementation. Single-amino-acid substitutions in the chromodomains of Cbx proteins (51) as well as N-terminal truncations of REST mimicking the deletion in RestΔN knockout mice (9) reduced BiFC complex formation up to 10-fold (Fig. 2C and E). These mutations did not alter the levels of fusion protein expression (Fig. 2C and E, blots below the graphs). A REST fragment encompassing the zinc finger DNA-binding domain (DBD) formed BiFC complexes with all Cbx proteins (Fig. 2D and E).
Complex formation by endogenous REST and PRC1 was tested by an analysis of Ring1b coprecipitation with REST from ES cell extracts. Ring1b was selected because Cbx2 and Cbx7 comigrated with the heavy and light chains, respectively, and antibodies against Ring1b were the most sensitive among those available against PRC1 subunits. Endogenous Ring1b was coprecipitated with REST from extracts of wild-type ES cells (Fig. 2F). The efficiency of Ring1b coimmunoprecipitation was markedly reduced in extracts from RestΔN knockout ES cells. Ring1b was expressed at the same level in these cells, and RESTΔN was precipitated at least as efficiently as wild-type REST by anti-REST antibodies. BiFC complex formation by REST and Cbx proteins and the coprecipitation of endogenous REST and Ring1b indicate that these proteins reside in the same complexes, but these observations do not demonstrate that the proteins contact each other directly.
To investigate potential functions of the interactions between REST and PRC1, we examined (i) if REST and PRC1 bound to sequences containing RE1 elements in ES cells, (ii) if RESTΔN knockout or REST knockdown affected PRC1 binding to RE1 elements, (iii) if REST or PRC1 knockdown or knockout affected the transcriptional activities of genes containing RE1 elements, and (iv) if RESTΔN knockout affected PRC1 binding or gene transcription in differentiating neurons. Since REST has been implicated in the regulation of neuron-specific gene expression, we focused on neuronal genes that were predicted to be occupied by REST and PRC1 in ES cells.
REST and PRC1 binding at distal versus proximal RE1 elements.
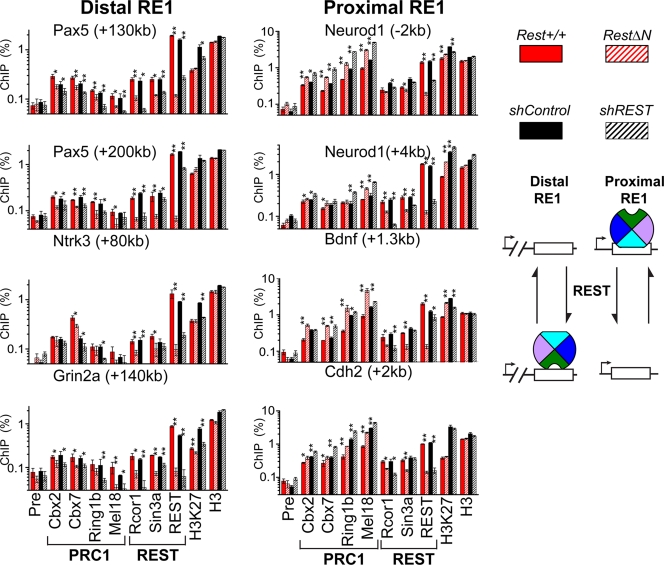
We tested REST and PRC1 subunit binding at both distal RE1 elements located far from the transcription start site (>5 kb away) as well as proximal RE1 elements located near the transcription start site (<5 kb away) using chromatin immunoprecipitation from ES cell extracts. PRC1 occupied every RE1 element tested that was occupied by REST in ES cells (Fig. 3 and 4). Antibodies directed against different PRC1 subunits precipitated proximal versus distal RE1 elements with opposite relative efficiencies. Distal RE1 elements were precipitated with lower efficiencies by anti-Mel18 or anti-Ring1b antibodies than by anti-Cbx2 and anti-Cbx7 antibodies (Fig. 3, left column). In contrast, proximal RE1 elements were precipitated with higher efficiencies by anti-Mel18 and anti-Ring1b than by anti-Cbx2 or anti-Cbx7 antibodies (Fig. 3, right column). Most RE1 elements were precipitated more efficiently by antibodies directed against Cbx2 and Cbx7 than by antibodies directed against Rcor1 or Sin3a, which were previously identified REST interaction partners (1, 17, 21, 36). The Oct3/4 and Gapdh (glyceraldehyde-3-phosphate dehydrogenase) promoter regions were not precipitated by the antibodies used (data not shown).
Fig. 3.
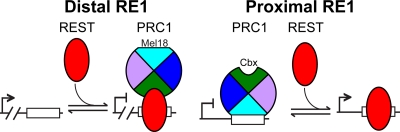
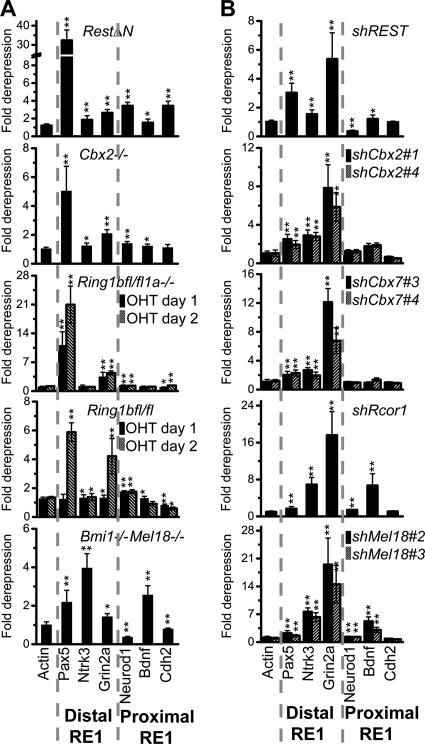
Effects of RestΔN knockout and REST knockdown on PRC1 subunit, Rcor1, Sin3a, and REST binding at distal versus proximal RE1 elements. The precipitation of the RE1 elements indicated above each graph by antibodies directed against the proteins and histone modifications indicated below the bars was measured by ChIP analysis. The efficiencies of precipitation were compared between Rest+/+ control and RestΔN knockout ES cells as well as between shControl and shREST knockdown ES cells. The data were plotted using a logarithmic scale to accommodate the data using different antibodies in the same graph (Pre, preimmune serum). Cbx2 and Cbx7 were precipitated using rabbit sera, whereas Ring1b and Mel18 were precipitated using purified IgG, which produced lower background signal. The left column corresponds to distal RE1 sites (>5 kb from transcription start), and the right column corresponds to proximal RE1 sites (<5 kb from transcription start). Proteins that are associated with PRC1 or with REST are indicated by the brackets below the graphs. The results shown are representative of data from two experiments using independently cultured ES cells (*, P < 0.05; **, P < 0.01). The difference between the effects of the RestΔN knockout as well as shREST knockdown on PRC1 occupancy at each distal versus each proximal RE1 element was statistically significant (P < 0.05 by a pairwise-difference test). The diagrams to the right of the graphs show the effects of REST on PRC1 (multicolored wheel) binding at distal versus proximal RE1 elements. The opposite rotational orientations of PRC1 represent distinct configurations of PRC1 binding.
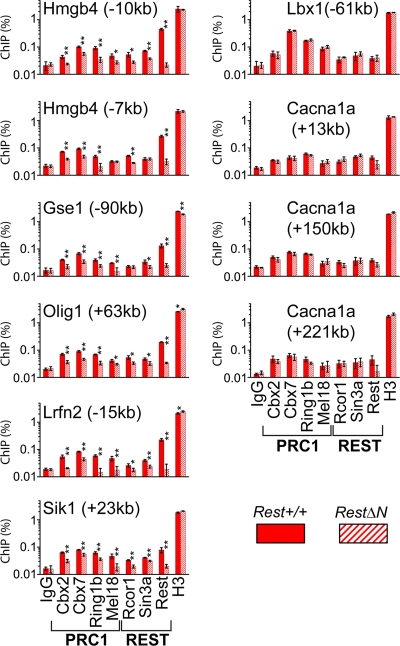
Fig. 4.
Effects of RestΔN knockout on PRC1 binding at sites occupied by REST versus sites not occupied by REST in ES cells. The effects of the RestΔN knockout on PRC1 binding at distal RE1 elements (left column) and at sites not occupied by REST (right column) were compared in ES cells, as described in the legend of Fig. 3. The efficiencies of precipitation of the regions indicated above each graph by antibodies directed against the proteins indicated below the bars are shown for Rest+/+ and RestΔN knockout cells. Monoclonal anti-Ring1b antibody was used. The bars show the amounts of input chromatin precipitated on a logarithmic scale (*, P < 0.05; **, P < 0.01).
We hypothesize that the differences in the relative efficiencies of precipitation of different RE1 elements by antibodies directed against different PRC1 subunits reflect differences in the efficiencies of cross-linking of different PRC1 subunits to these RE1 elements. Thus, the higher efficiencies of proximal than of distal RE1 element precipitation by anti-Mel18 antibodies compared to anti-Cbx2 and anti-Cbx7 antibodies indicate that Mel18 cross-linked more efficiently to proximal than to distal RE1 elements. The opposite relative efficiencies of precipitation of distal versus proximal RE1 elements by antibodies directed against different PRC1 subunits suggest that PRC1 occupied these elements in distinct configurations (Fig. 3 and see Discussion).
Effects of REST on PRC1 binding at distal versus proximal RE1 elements.
We examined the effects of RestΔN knockout on Rcor1, Sin3a, and PRC1 subunit binding at distal and proximal RE1 elements. RestΔN knockout altered PRC1 occupancy at most RE1 elements that were occupied by REST in Rest+/+ ES cells (14 of 17 RE1 elements tested). Unexpectedly, RestΔN knockout had opposite effects on PRC1 occupancy at distal versus proximal RE1 elements. RestΔN knockout reduced PRC1 binding at distal RE1 elements but increased PRC1 binding at proximal RE1 elements in the same cells (Fig. 3, compare solid and striped red bars in the left [distal] and the right [proximal] columns). The magnitudes of the changes in PRC1 binding varied at different RE1 elements and among different PRC1 subunits. Nevertheless, the RestΔN knockout had consistently opposite effects on PRC1 binding at distal versus proximal RE1 elements in the same cells. The parallel changes in binding by different PRC1 subunits at each RE1 site corroborate the specificities of the antibodies for PRC1 subunits. Consistent results were obtained using both monoclonal mouse and polyclonal rabbit antibodies directed against Ring1b (Fig. 3 and 4). The opposite effects of RestΔN knockout on PRC1 binding at proximal versus distal RE1 elements were specific to PRC1 subunits, since REST depletion reduced Rcor1 as well as Sin3a binding at both proximal and distal RE1 elements (Fig. 3).
To establish if the opposite changes in PRC1 binding at distal versus proximal RE1 elements in RestΔN knockout ES cells were a consequence of the REST mutation, we examined the effects of REST knockdown on PRC1 binding. The expression of shRNA targeting REST (shREST) reduced the levels of REST transcript and protein as well as REST binding at all of the RE1 elements examined (Fig. 3 and see Fig. 6A). REST knockdown had quantitatively smaller, but qualitatively equivalent, effects on PRC1 binding at RE1 elements compared to RestΔN knockout (Fig. 3, black bars). The consistent results obtained using unrelated methods to deplete REST support the interpretation that both the decrease in PRC1 binding at distal RE1 elements and the increase in PRC1 binding at proximal RE1 elements were consequences of REST depletion. The RestΔN knockout ES cells and the ES cells used to generate the REST knockdown cell line were derived independently in different laboratories (9, 37) and were grown in the presence and absence of feeder cells, respectively. The effects of REST depletion on PRC1 binding were consistent in cells of different genetic and epigenetic backgrounds grown under different culture conditions.
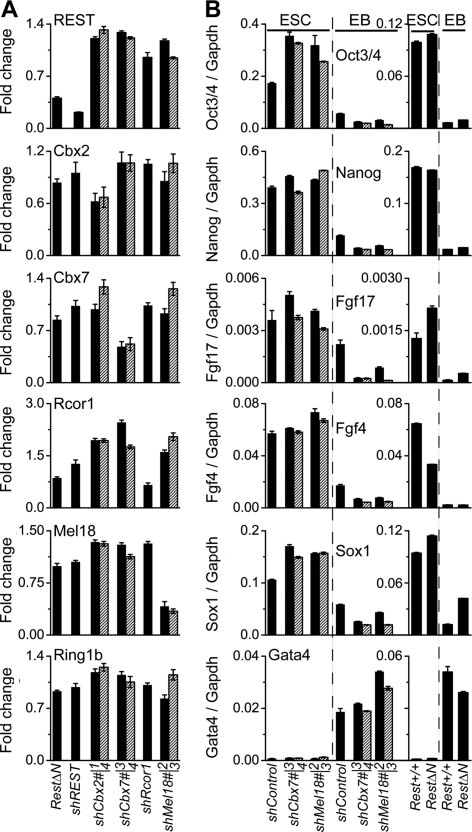
Fig. 6.
Selectivity of REST, Rcor1, and PRC1 subunit knockdown and RestΔN knockout. (A) Effects of RestΔN knockout and of the shRNA constructs indicated below the bars on the REST and PRC1 subunit transcripts indicated above the bars. The bars show the ratios between the transcript levels in RestΔN knockout cells and those in Rest+/+control cells or between the transcript levels in cells expressing the knockdown shRNA and those in cells expressing the control shRNA. Two different shRNAs directed against different sequences in Cbx2, Cbx7, and Mel18 were used (solid bars and striped bars). The primers used to detect REST transcripts also detect transcripts from the RestΔN knockout locus. The results shown represent the means of data from two or more independent experiments. (B) Effects of the RestΔN knockout and of the shRNA constructs indicated below the bars on the levels of the pluripotency- and differentiation-associated transcripts indicated above the bars. The transcript levels were measured in ES cells (ESC) and in embryoid bodies (EB) and were normalized relative to Gapdh levels.
We examined the specificity of the effects of RestΔN knockout on PRC1 binding at RE1 sites occupied by REST in wild-type ES cells versus sites not occupied by REST. RestΔN knockout reduced binding by all PRC1 subunits at distal RE1 sites occupied by REST in wild-type ES cells (Fig. 4, left column). In contrast, the RestΔN knockout had little effect on PRC1 binding at sites that were not occupied by REST in wild-type ES cells (Fig. 4, right column). The lower efficiencies of distal RE1 element precipitation by anti-Ring1b and anti-Mel18 antibodies were also selectively reduced at RE1 sites occupied by REST but not at RE1 sites that were not occupied by REST in wild-type ES cells, indicating that they reflected specific binding. The effects of RestΔN knockout on PRC1 binding were therefore specific to sites occupied by REST in wild-type ES cells.
The concordant effects of RestΔN knockout and REST knockdown on PRC1 binding at proximal versus distal RE1 elements and the absence of these effects at sites not occupied by REST in ES cells are consistent with direct effects of REST on PRC1 binding at RE1 elements. RestΔN knockout and REST knockdown had little effect on the levels of expression of PRC1 subunit transcripts or proteins (see Fig. 6A and data not shown). RestΔN knockout and REST knockdown also had little effect on ES cell pluripotency (see Fig. 6B and data not shown).
There was a perfect correlation between the opposite effects of REST depletion on PRC1 occupancy at proximal versus distal RE1 elements and the opposite relative efficiencies of precipitation of these RE1 elements by antibodies directed against different PRC1 subunits (Fig. 3 and 4). We hypothesize that the opposite effects of REST on PRC1 occupancy at distal versus proximal RE1 elements were due to distinct configurations of PRC1 binding at these elements (see Fig. 8 and Discussion). At each RE1 element, both RestΔN knockout and REST knockdown had parallel effects on binding by all PRC1 subunits tested (Fig. 3 and 4). REST depletion therefore did not alter the configuration of PRC1 binding at individual RE1 elements. Instead, REST depletion had opposite effects on the levels of different configurations of PRC1 binding at distal versus proximal RE1 elements.
Fig. 8.
Model for the effects of REST on PRC1 binding at distal versus proximal RE1 elements. The opposite rotational orientations of the multicolored wheel represent different configurations of PRC1 binding at distal versus proximal RE1 elements. We hypothesize that the opposite effects of REST on PRC1 binding at distal versus proximal RE1 elements are due to the distinct configurations of PRC1 binding. The data do not allow a determination of whether PRC1 interacts with REST at proximal RE1 elements.
PRC1 occupancy at many genes correlates with H3 K27 trimethylation in ES cells (29). There was no change in H3 K27 trimethylation at the RE1 elements examined in RestΔN knockout or REST knockdown cells that correlated with the changes in PRC1 occupancy (Fig. 3). There was also no consistent difference in H3 K27 trimethylation at distal versus proximal RE1 elements. We also detected no changes in H3 K4 or H3 K9 methylation at RE1 elements in REST knockdown cells that correlated with the changes in PRC1 occupancy (data not shown). There was also no significant difference in RE1 element precipitation by anti-H3 antibodies from Rest+/+ versus RestΔN knockout cells or from control shRNA (shControl) versus REST knockdown (shREST) cells (Fig. 3). It is therefore unlikely that the effects of either RestΔN knockout or REST knockdown on PRC1 binding at RE1 elements were mediated by changes in histone modifications.
Effects of REST versus PRC1 subunit depletion on neuronal gene transcription in ES cells.
To investigate potential effects of REST interactions with PRC1 on transcriptional activity, we examined the transcription of genes containing RE1 elements in ES cells depleted of REST or PRC1 subunits. The knockout or knockdown of REST or PRC1 subunits derepressed closely overlapping subsets of neuronal genes. RestΔN as well as Cbx2−/−, Ring1bfl/fl Ring1a−/−, and Ring1bfl/fl knockouts derepressed mainly genes containing distal RE1 elements (Fig. 5A). Bmi1−/− Mel18−/− knockout also derepressed transcription of Bdnf, Mash1, and Pcdhb3. Likewise, knockdown of REST, Cbx2, or Cbx7 derepressed mainly genes containing distal RE1 elements (Fig. 5B). The knockdown of Rcor1 or Mel18 also derepressed transcription of Bdnf, Mash1, and Pcdhb3 (data not shown). The differences between the effects of RestΔN knockout and REST knockdown on transcription may be due to differences in the genetic backgrounds or the conditions used to culture these cells.
Fig. 5.
Coregulation of neuronal genes by REST and PRC1 subunits in ES cells. (A) Derepression of the transcripts indicated below the bars in knockout ES cells containing the mutations indicated above the bars. Transcripts in Ring1bfl/fl Ring1a−/− and Ring1bfl/fl ES cells were analyzed 24 and 48 h after the addition of 4-hydroxytamoxifen (OHT). The bars show the ratio between the transcript levels in the mutant cells relative to untreated cells or cells isolated from wild-type control mice. The transcript levels were normalized relative to the levels of Gapdh transcripts in the same cells. The results shown are representative of data from two experiments using independently cultured ES cells (*, P < 0.05; **, P < 0.01). (B) Derepression of the transcripts indicated below the bars in ES cells expressing the shRNAs indicated above the bars. Two different shRNAs directed against different sequences in Cbx2, Cbx7, and Mel18 were used. The bars show the ratios between the transcript levels in cells expressing the knockdown shRNA and cells expressing the control shRNA. The transcript levels were normalized relative to the levels of Gapdh transcripts in the same cells. The results shown are representative of data from two experiments using independently cultured ES cells (*, P < 0.05; **, P < 0.01).
The derepression of genes containing distal RE1 elements by REST, Rcor1, and PRC1 subunit depletion suggests that these proteins act in concert to repress these genes. In contrast, the lack of a derepression of most genes containing proximal RE1 sites is consistent with the independent repression of these genes by REST or PRC1 complexes. REST, Cbx2, Cbx7, or Ring1a/b depletion had effects on neuronal gene transcription that were more similar to each other than to the effects of Rcor1, Mel18, or Bmi1 depletion and vice versa. This observation suggests that the functions of individual PRC1 subunits and REST-associated proteins are more closely related to each other than are the functions of different PRC1 subunits or those of REST and Rcor1. The similar relative effects of REST and PRC1 subunit depletion on the levels of transcription of the neuronal genes examined corroborate the functional significance of REST interactions with PRC1 subunits.
We examined whether indirect effects of REST or PRC1 depletion contributed to the coregulation of neuronal gene transcription by REST and PRC1 subunits. RestΔN knockout and REST knockdown had little effect on the levels of PRC1 subunit transcripts or proteins (Fig. 6A and data not shown). Conversely, knockdown of PRC1 subunits selectively reduced the levels of the targeted transcripts and had little effect on the level of REST transcripts. RestΔN knockout and PRC1 subunit knockdown did not reduce the levels of pluripotency-associated transcripts in ES cells (Fig. 6B). There was also no detectable change in the levels of the Oct3/4 or Nanog protein expressed in individual cells detected by immunofluorescence analysis or in the alkaline phosphatase activities of these cells (data not shown). RestΔN knockout and PRC1 subunit knockdown did not prevent pluripotency-associated gene repression or differentiation-associated transcript induction in embryoid bodies (Fig. 6B). RestΔN knockout did not inhibit ES cell self-renewal or differentiation, consistent with previous findings (7, 23, 24). It is therefore unlikely that indirect effects of the depletion of REST or PRC1 contributed to the coregulation of neuronal gene transcription by REST and PRC1 subunits.
REST regulation of transcription and PRC1 occupancy at RE1 elements during neuronal differentiation.
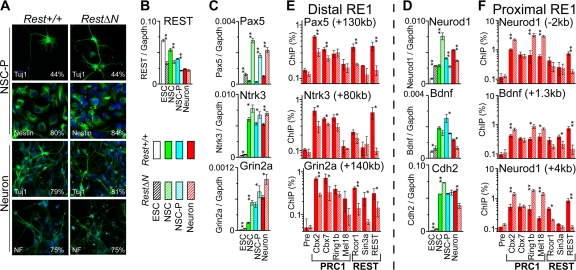
We investigated neuronal gene transcription and PRC1 occupancy during Rest+/+ and RestΔN knockout ES cell differentiation into neurons. Embryoid bodies were differentiated into neuronal stem cells (NSCs), stimulated to proliferate (NSC-P), and induced to form differentiating neurons (39). To establish the homogeneity of the cells at each stage of differentiation, we stained cells at each stage of differentiation using antibodies that detect markers of neuronal stem cells (nestin) and differentiating neurons (Tuj1 and neurofilament). At each stage of differentiation, Rest+/+ control and RestΔN knockout cells expressed markers of neuronal differentiation in nearly identical proportions of the cells and had indistinguishable morphologies (Fig. 7A). These results suggest that RestΔN knockout did not affect the in vitro differentiation of ES cells into neurons.
Fig. 7.
Effects of RestΔN knockout on PRC1 binding and neuron-specific gene transcription during neuronal differentiation. (A) Embryoid bodies were differentiated into neuronal stem cells (NSCs), stimulated to proliferate (NSC-P), and induced to form differentiating neurons (Neuron) (39). NSC-P (top) and differentiating neurons (bottom) derived from Rest+/+ control (left) and RestΔN knockout (right) ES cells were stained using Tuj1, antinestin, and antineurofilament (NF) antibodies (green) and Hoechst dye (blue). The images are representative of all cells in each population. The numbers indicate the percentages of cells stained by the antibodies. (B to D) Neuronal gene transcription in Rest+/+ control (solid) and RestΔN knockout (striped) cells at each stage of differentiation. (E and F) Precipitation of the RE1 elements indicated above each graph from differentiating Rest+/+ control and RestΔN knockout neurons by antibodies directed against the proteins indicated below the bars. The results shown are representative of data from two independent experiments with differentiating neurons and one experiment with neuronal stem cells (*, P < 0.05; **, P < 0.01). ChIP analysis of the Cdh2 RE1 site could not be performed with differentiating neurons due to the limited number of cells and the low sensitivity of detection at this site.
The transcription of genes containing distal versus proximal RE1 elements was differentially regulated during Rest+/+ versus RestΔN knockout ES cell differentiation into neurons. Genes with distal RE1 elements (Pax5, Ntrk3, and Grin2a) were transcribed at higher levels in differentiating RestΔN knockout neurons than in differentiating Rest+/+ control neurons (Fig. 7C, compare solid red and striped bars). In the same cells, genes with proximal RE1 elements (Neurod1, Bdnf, and Cdh2) were transcribed at lower levels in differentiating RestΔN knockout neurons than in differentiating Rest+/+ control neurons (Fig. 7D). The levels of REST as well as truncated RESTΔN transcripts were reduced 2- to 3-fold during neuronal differentiation (Fig. 7B). Thus, RestΔN knockout had opposite effects on the transcription of genes containing distal versus proximal RE1 elements in differentiating neurons.
We examined whether the difference in the effects of RestΔN knockout on the transcription of genes containing distal versus proximal RE1 elements in differentiating neurons was related to differences in PRC1, Rcor1, or Sin3a binding at these elements. PRC1 binding at distal RE1 elements was reduced by RestΔN knockout in differentiating neurons (Fig. 7E). In contrast, proximal RE1 element binding by most PRC1 subunits was increased by RestΔN knockout in differentiating neurons (Fig. 7F). Cbx7 binding at proximal RE1 elements was not affected by RestΔN knockout in differentiating neurons, suggesting that RestΔN knockout also affected the composition of PRC1 at proximal RE1 elements. In contrast to the opposite effects of RestΔN knockout on PRC1 subunit occupancy at distal versus proximal RE1 elements, RestΔN knockout reduced Rcor1 and Sin3a occupancy at all RE1 elements with detectable binding in differentiating neurons (Fig. 7E and F).
The opposite effects of RestΔN knockout on the transcription of genes containing distal versus proximal RE1 elements in differentiating neurons correlated with the opposite effects of RestΔN knockout on PRC1 occupancy at these elements. The increased derepression of genes containing distal RE1 elements in RestΔN knockout cells compared to Rest+/+ cells was consistent with the lower PRC1 occupancy at these elements in neurons lacking REST (Fig. 7C and E). Conversely, the reduced derepression of genes containing proximal RE1 elements in RestΔN knockout cells was consistent with the higher PRC1 occupancy at these elements in neurons lacking REST (Fig. 7D and F). These results also suggest that PRC1 was a more potent repressor than REST at genes containing proximal RE1 elements in differentiating neurons compared to ES cells. The opposite effects of RestΔN knockout both on PRC1 occupancy at proximal versus distal RE1 elements and on the transcriptional activities of genes containing these elements implicate REST interactions with PRC1 in the gene-specific regulation of transcription during neuronal differentiation.
DISCUSSION
Integration of DNA sequence-dependent and epigenetic transcription regulation.
The interactions between REST and Cbx family proteins provide a potential link between DNA sequence-dependent and epigenetic transcription regulatory networks. REST regulates neuronal gene expression in ES cells and during neuronal differentiation. Whereas REST undergoes rapid proteolytic turnover upon the induction of ES cell differentiation, neuronal genes are not derepressed until several days after the initiation of differentiation (2). The maintenance of neuronal gene repression over many cell generations after the degradation of the bulk of REST is reminiscent of the classical epigenetic regulation of Hox gene expression by Drosophila PcG (32).
Many interaction partners for both REST and PRC1 subunits have been identified (1, 2, 13, 17, 21, 36, 38, 40, 45). Interactions between REST and PRC1 have not been reported previously. One possible reason for this is that proteins associated with Cbx proteins have not been purified from ES cells. It is also possible that the interaction of REST with PRC1 depends on experimental conditions.
In RestΔN knockout mice, several brain regions are disorganized at embryonic day 9.25 (E9.25) (9). At this stage some neuron-specific genes are misregulated, but no defect in neurogenesis was reported previously (9). These observations are consistent with our findings that RestΔN knockout does not affect ES cell pluripotency or neuronal differentiation but alters neuron-specific gene expression in ES cells and in differentiating neurons. The bulk of REST is degraded during ES cell differentiation, but the remaining REST binds to neuronal genes in the cerebral cortex of E12.5 mice (2). This is consistent with our finding that REST binds to RE1 elements in differentiating neurons derived from ES cells. The effects of RestΔN knockout on PRC1 occupancy in differentiating neurons could be either a direct consequence of REST binding to RE1 elements in these neurons or a result of the epigenetic inheritance of differences in PRC1 occupancy established in ES cells. Previous studies documented the roles of Ring1b and Bmi1 in neuronal stem cell proliferation and differentiation (11, 14, 15, 19, 35, 44). We hypothesize that cross talk between the REST and PRC1 networks coordinates neuronal gene regulation and other developmental programs.
Several studies of genome-wide PRC1 subunit as well as REST occupancy in ES cells have been conducted (5, 22, 29, 41). More than 200 genes are predicted to be occupied by both REST and PcG proteins based on genome-wide ChIP analyses (see Table S1 in the supplemental material). The degree of REST and PRC1 co-occupancy is difficult to estimate based on the genome-wide ChIP data, since the target genes identified in different experiments for each of these proteins overlap only in part. The opposite effects of REST on PRC1 binding at different distances from transcription start sites predict that REST and PRC1 subunits bind synergistically at only a subset of RE1 elements. Sequences recognized by REST, as well as other transcription regulatory proteins, are overrepresented in the CpG islands of genes occupied by PcG proteins (29).
Different configurations of PRC1 binding at distal versus proximal RE1 elements.
PRC1 occupied RE1 elements at different distances from the transcription start site in distinct configurations, as reflected by the opposite relative efficiencies of distal versus proximal RE1 element precipitation by antibodies directed against different PRC1 subunits. The parallel changes in RE1 element precipitation by these antibodies caused by RestΔN knockout in ES cells suggest that the core subunit composition of PRC1 was the same at proximal and distal RE1 elements. We hypothesize that PRC1 engaged proximal RE1 elements through direct DNA binding in ES cells and bound distal RE1 elements indirectly through interactions with REST (Fig. 8).
Opposite effects of REST on PRC1 occupancy at proximal versus distal RE1 elements.
The opposite effects of REST on PRC1 occupancy at distal versus proximal RE1 elements correlated with distinct configurations of PRC1 binding. We hypothesize that the opposite effects of REST on PRC1 occupancy at distal versus proximal RE1 elements were due to distinct interactions between PRC1 and DNA at these RE1 elements (Fig. 8). The decrease in PRC1 occupancy at distal RE1 elements upon REST depletion is consistent with the stabilization of PRC1 binding at these elements through interactions with REST. Conversely, the increase in PRC1 occupancy at proximal RE1 elements upon REST depletion is consistent with the displacement of PRC1 binding at these elements by competition with REST for a common interaction partner. Such common interaction partners could include DNA (16, 26), RNA (3, 50, 53), or other proteins. REST was not essential for PRC1 binding at distal RE1 elements, nor did REST eliminate PRC1 binding at proximal RE1 elements. However, REST depletion resulted in a 4-fold change on average in the ratio of PRC1 binding to distal versus proximal RE1 elements. REST was therefore a major modulator of PRC1 binding at RE1 elements. The molecular difference(s) between proximal and distal RE1 elements that cause the distinct configurations of PRC1 binding and the opposite effects of REST on PRC1 occupancy remains to be identified.
Effects of REST interactions with PRC1 on neuronal gene transcription.
The similar relative effects of REST, Cbx2, Cbx7, and Ring1b depletion as well as of Rcor1 and Mel18 depletion on neuron-specific gene transcription indicate that the transcriptional activities of these proteins are interrelated. The functions of REST were most closely related to those of Cbx2, Cbx7, and Ring1b, whereas the functions of Rcor1 were more closely related to those of Mel18. Genes containing distal RE1 elements were derepressed by either REST, Rcor1, or PRC1 depletion, suggesting that these proteins act in concert to repress transcription. In contrast, most genes containing proximal RE1 elements were not derepressed by the depletion of either REST, Rcor1, or PRC1 subunits, consistent with the redundant repression of these genes by complexes containing either REST or PRC1. Previous studies of the transcriptomes of RestΔN knockout and REST knockdown ES cells found similar proportions of upregulated and downregulated genes but only 87 transcripts that changed more than 1.4-fold in both RestΔN knockout and REST knockdown cells (24). These results are consistent with the redundant regulation of a large proportion of the genes bound by REST in ES cells. Approximately one-third of the genes whose expression changed more than 1.4-fold contain proximal RE1 elements. Distal RE1 elements have been identified in many genes (41), but the proportion of the deregulated genes that contain distal RE1 elements cannot be determined since the distance over which REST can affect transcription is not known.
The opposite effects of RestΔN knockout on PRC1 occupancy as well as on transcription at genes containing distal versus proximal RE1 elements in differentiating neurons are consistent with a direct role of PRC1 in the regulation of the transcription of these genes by REST. The same region of REST was required both for interactions with Cbx proteins and for the enhancement of PRC1 binding and repression at distal RE1 elements as well as for the inhibition of PRC1 binding and activation at proximal RE1 elements. The regions of REST and Cbx proteins required for BiFC complex formation overlapped with the regions required for DNA binding and H3 interactions, respectively (51). It is therefore possible that REST interactions with Cbx proteins were stabilized by an association with chromatin.
RestΔN knockout had little effect on the levels of PRC1 subunit expression and no significant effect on PRC1 binding at sites that were not occupied by REST in wild-type ES cells. RestΔN knockout also had no detectable effect on ES cell pluripotency, nor did it affect the in vitro differentiation of ES cells into neurons. The limited effects of RestΔN knockout on ES cell pluripotency and on neuronal differentiation make these model systems ideal for investigations of the roles of REST in PRC1 binding and in neuronal gene transcription.
The derepression of genes containing distal RE1 elements and the repression of genes containing proximal RE1 elements in RestΔN knockout cells indicate that REST can both repress and activate genes in differentiating neurons. This observation is consistent with data from previous reports of REST activation and repression of different genes in differentiating neurons (4, 25, 30). REST can therefore simultaneously enhance PRC1 binding and repression at some genes and inhibit PRC1 binding and repression at other genes in the same cells.
Supplementary Material
ACKNOWLEDGMENTS
We thank Haruhiko Koseki for providing Cbx2−/−, Ring1bfl/fl Ring1a−/−, Ring1bfl/fl, and Bmi1−/− Mel18−/− ES cells; Helle Jørgensen for providing RestΔN ES cells; Gail Mandel for providing the REST cDNA; Steven Gygi and the Taplin mass spectrometry facility for LC-MS/MS analysis; Lingqun Liu for bioinformatic analysis; and members of the Kerppola laboratory, in particular Claudius Vincenz, for sharing reagents and for constructive criticisms.
This work was funded by the NIDA (grant R01 DA030339).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 14 March 2011.
REFERENCES
- 1. Andres M. E., et al. 1999. CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc. Natl. Acad. Sci. U. S. A. 96:9873–9878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ballas N., Grunseich C., Lu D. D., Speh J. C., Mandel G. 2005. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 121:645–657 [DOI] [PubMed] [Google Scholar]
- 3. Bernstein E., et al. 2006. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol. Cell. Biol. 26:2560–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bessis A., Champtiaux N., Chatelin L., Changeux J. P. 1997. The neuron-restrictive silencer element: a dual enhancer/silencer crucial for patterned expression of a nicotinic receptor gene in the brain. Proc. Natl. Acad. Sci. U. S. A. 94:5906–5911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boyer L. A., et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441:349–353 [DOI] [PubMed] [Google Scholar]
- 6. Bracken A. P., Dietrich N., Pasini D., Hansen K. H., Helin K. 2006. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 20:1123–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buckley N. J., Johnson R., Sun Y. M., Stanton L. W. 2009. Is REST a regulator of pluripotency? Nature 457:E5–E6 [DOI] [PubMed] [Google Scholar]
- 8. Cao R., et al. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298:1039–1043 [DOI] [PubMed] [Google Scholar]
- 9. Chen Z. F., Paquette A. J., Anderson D. J. 1998. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat. Genet. 20:136–142 [DOI] [PubMed] [Google Scholar]
- 10. Chong J. A., et al. 1995. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell 80:949–957 [DOI] [PubMed] [Google Scholar]
- 11. Cui H., et al. 2006. Bmi-1 regulates the differentiation and clonogenic self-renewal of I-type neuroblastoma cells in a concentration-dependent manner. J. Biol. Chem. 281:34696–34704 [DOI] [PubMed] [Google Scholar]
- 12. Elderkin S., et al. 2007. A phosphorylated form of mel-18 targets the Ring1B histone H2A ubiquitin ligase to chromatin. Mol. Cell 28:107–120 [DOI] [PubMed] [Google Scholar]
- 13. Endoh M., et al. 2008. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development 135:1513–1524 [DOI] [PubMed] [Google Scholar]
- 14. Fasano C. A., et al. 2007. shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell Stem Cell 1:87–99 [DOI] [PubMed] [Google Scholar]
- 15. Fasano C. A., et al. 2009. Bmi-1 cooperates with Foxg1 to maintain neural stem cell self-renewal in the forebrain. Genes Dev. 23:561–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Francis N. J., Saurin A. J., Shao Z. H., Kingston R. E. 2001. Reconstitution of a functional core polycomb repressive complex. Mol. Cell 8:545–556 [DOI] [PubMed] [Google Scholar]
- 17. Grimes J. A., et al. 2000. The co-repressor mSin3A is a functional component of the REST-CoREST repressor complex. J. Biol. Chem. 275:9461–9467 [DOI] [PubMed] [Google Scholar]
- 18. Hansen K. H., et al. 2008. A model for transmission of the H3K27me3 epigenetic mark. Nat. Cell Biol. 10:1291. [DOI] [PubMed] [Google Scholar]
- 19. Hirabayashi Y., et al. 2009. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron 63:600–613 [DOI] [PubMed] [Google Scholar]
- 20. Hu C. D., Chinenov Y., Kerppola T. K. 2002. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9:789–798 [DOI] [PubMed] [Google Scholar]
- 21. Huang Y., Myers S. J., Dingledine R. 1999. Transcriptional repression by REST: recruitment of Sin3A and histone deacetylase to neuronal genes. Nat. Neurosci. 2:867–872 [DOI] [PubMed] [Google Scholar]
- 22. Johnson R., et al. 2008. REST regulates distinct transcriptional networks in embryonic and neural stem cells. PLoS Biol. 6:e256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jorgensen H. F., Chen Z. F., Merkenschlager M., Fisher A. G. 2009. Is REST required for ESC pluripotency? Nature 457:E4–E5 [DOI] [PubMed] [Google Scholar]
- 24. Jorgensen H. F., et al. 2009. REST selectively represses a subset of RE1-containing neuronal genes in mouse embryonic stem cells. Development 136:715–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kallunki P., Edelman G. M., Jones F. S. 1998. The neural restrictive silencer element can act as both a repressor and enhancer of L1 cell adhesion molecule gene expression during postnatal development. Proc. Natl. Acad. Sci. U. S. A. 95:3233–3238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kanno M., Hasegawa M., Ishida A., Isono K., Taniguchi M. 1995. mel-18, a Polycomb group-related mammalian gene, encodes a transcriptional negative regulator with tumor suppressive activity. EMBO J. 14:5672–5678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Katoh-Fukui Y., et al. 1998. Male-to-female sex reversal in M33 mutant mice. Nature 393:688–692 [DOI] [PubMed] [Google Scholar]
- 28. Kerppola T. K. 2009. Polycomb group complexes—many combinations, many functions. Trends Cell Biol. 19:692–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ku M., et al. 2008. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 4:e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kuwabara T., Hsieh J., Nakashima K., Taira K., Gage F. H. 2004. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell 116:779–793 [DOI] [PubMed] [Google Scholar]
- 31. Leeb M., et al. 2010. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev. 24:265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lewis E. B. 1978. A gene complex controlling segmentation in Drosophila. Nature 276:565–570 [DOI] [PubMed] [Google Scholar]
- 33. Margueron R., et al. 2009. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461:762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mohd-Sarip A., et al. 2006. Architecture of a polycomb nucleoprotein complex. Mol. Cell 24:91–100 [DOI] [PubMed] [Google Scholar]
- 35. Molofsky A. V., et al. 2003. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature 425:962–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Naruse Y., Aoki T., Kojima T., Mori N. 1999. Neural restrictive silencer factor recruits mSin3 and histone deacetylase complex to repress neuron-specific target genes. Proc. Natl. Acad. Sci. U. S. A. 96:13691–13696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Norris D. P., et al. 1994. Evidence that random and imprinted Xist expression is controlled by preemptive methylation. Cell 77:41–51 [DOI] [PubMed] [Google Scholar]
- 38. Ogawa H., Ishiguro K., Gaubatz S., Livingston D. M., Nakatani Y. 2002. A complex with chromatin modifiers that occupies E2F-and Myc-responsive genes in G(0) cells. Science 296:1132–1136 [DOI] [PubMed] [Google Scholar]
- 39. Okabe S., Forsberg-Nilsson K., Spiro A. C., Segal M., McKay R. D. 1996. Development of neuronal precursor cells and functional postmitotic neurons from embryonic stem cells in vitro. Mech. Dev. 59:89–102 [DOI] [PubMed] [Google Scholar]
- 40. Ooi L., Wood I. C. 2007. Chromatin crosstalk in development and disease: lessons from REST. Nat. Rev. Genet. 8:544–554 [DOI] [PubMed] [Google Scholar]
- 41. Otto S. J., et al. 2007. A new binding motif for the transcriptional repressor REST uncovers large gene networks devoted to neuronal functions. J. Neurosci. 27:6729–6739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Puschendorf M., et al. 2008. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat. Genet. 40:411–420 [DOI] [PubMed] [Google Scholar]
- 43. Ren X., Vincenz C., Kerppola T. K. 2008. Changes in the distributions and dynamics of polycomb repressive complexes during embryonic stem cell differentiation. Mol. Cell. Biol. 28:2884–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roman-Trufero M., et al. 2009. Maintenance of undifferentiated state and self-renewal of embryonic neural stem cells by Polycomb protein Ring1B. Stem Cells 27:1559–1570 [DOI] [PubMed] [Google Scholar]
- 45. Sanchez C., et al. 2007. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol. Cell. Proteomics 6:820–834 [DOI] [PubMed] [Google Scholar]
- 46. Schoeftner S., et al. 2006. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 25:3110–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schoenherr C. J., Anderson D. J. 1995. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science 267:1360–1363 [DOI] [PubMed] [Google Scholar]
- 48. Sing A., et al. 2009. A vertebrate polycomb response element governs segmentation of the posterior hindbrain. Cell 138:885–897 [DOI] [PubMed] [Google Scholar]
- 49. Tapia-Ramirez J., Eggen B. J., Peral-Rubio M. J., Toledo-Aral J. J., Mandel G. 1997. A single zinc finger motif in the silencing factor REST represses the neural-specific type II sodium channel promoter. Proc. Natl. Acad. Sci. U. S. A. 94:1177–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tsai M. C., et al. 2010. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329:689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vincenz C., Kerppola T. K. 2008. Different polycomb group CBX family proteins associate with distinct regions of chromatin using nonhomologous protein sequences. Proc. Natl. Acad. Sci. U. S. A. 105:16572–16577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Woo C. J., Kharchenko P. V., Daheron L., Park P. J., Kingston R. E. 2010. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell 140:99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yap K. L., et al. 2010. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 38:662–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.