Abstract
We recently reported that diadenosine tetraphosphate hydrolase (Ap4A hydrolase) plays a critical role in gene expression via regulation of intracellular Ap4A levels. This enzyme serves as a component of our newly described lysyl tRNA synthetase (LysRS)-Ap4A biochemical pathway that is triggered upon immunological challenge. Here we explored the mechanism of this enzyme's translocation into the nucleus and found its immunologically dependent association with importin beta. Silencing of importin beta prevented Ap4A hydrolase nuclear translocation and affected the local concentration of Ap4A, which led to an increase in microphthalmia transcription factor (MITF) transcriptional activity. Furthermore, immunological activation of mast cells resulted in dephosphorylation of Ap4A hydrolase, which changed the hydrolytic activity of the enzyme.
INTRODUCTION
Diadenosine tetraphosphatase (Ap4A hydrolase) is a Nudix type 2 (nudt2) gene product. This enzyme hydrolyzes Ap4A into AMP and ATP (Ap4A → ATP + AMP). Ap4A hydrolase belongs to the Nudix hydrolase (or MutT) family, which is a group of enzymes containing the Nudix consensus motif GX5EX5[UA]XREX2EEXGU, where X is any amino acid and U is a bulky aliphatic residue (25). Other than the Nudix signature, the enzymes share another common feature—they all hydrolyze nucleoside diphosphate linked to another moiety, X.
In order for Ap4A to be established unambiguously as a second messenger, several criteria had to be fulfilled, including the presence of a metabolizing enzyme in the context of a stimulus. In a previous work, we provided evidence that Ap4A hydrolase is responsible for Ap4A degradation following immunological activation of mast cells (6). We further described the critical role in transcriptional regulation played by Ap4A hydrolase as a component of our newly described lysyl tRNA synthetase (LysRS) biochemical pathway, which is activated upon immunological challenge. Knockdown of Ap4A hydrolase modulated Ap4A accumulation, resulting in changes in the expression of the microphthalmia transcription factor (MITF) and upstream stimulating factor 2 (USF2) target genes. Thus, we provided concrete evidence establishing Ap4A as a second messenger in the regulation of gene expression (6).
In addition to the increased interest in Ap4A hydrolase as the main hydrolyzer of this novel second messenger, a recent paper emphasized the value of this protein not only as an enzyme but also as a potent prognostic factor in human breast carcinoma associated with cell proliferation (29).
In Ehrlich ascites tumor cells, it was found that more than 75% of the whole cellular content of Ap4A hydrolase's substrate (Ap4A) is localized in the nucleus (37). In G1-phase and early S-phase cells of synchronized BHK fibroblast cultures, approximately 90% of the intracellular Ap4A pool is confined to the nuclear compartment. In contrast, Ap4A is distributed nearly equally between the cytoplasm and the nucleus during mid-S phase (37). In light of these findings, the intracellular location of Ap4A hydrolase is of major importance.
Despite extensive studies of eukaryotic Ap4A hydrolase, to date there are only two previous studies—using tomato (14) and Drosophila (39) cells—indicating that Ap4A hydrolase is located in the nucleus. The Caenorhabditis elegans NUDT2 orthologue, Ndx-4, on the other hand, appears to be ribosome associated, similar to Escherichia coli YgdP (23). There are no reports in the literature on the subcellular localization of mammalian Ap4A hydrolase.
Nuclear transport of proteins is mediated by a superfamily of transport receptors known collectively as karyopherins (importins and exportins) (reviewed in reference 30). In the first and best-described mechanism for nuclear import, proteins destined for transport into the nucleus contain amino acid targeting sequences called nuclear localization signals (NLSs) (reviewed in reference 21). The best-characterized transport signal is the classical NLS for nuclear protein import, which consists of either one (monopartite) or two (bipartite) stretches of basic amino acids (19, 32).
Importin beta (karyopherin beta) is a helicoidal molecule constructed of 19 HEAT repeats. Many nuclear pore proteins contain Phe-Gly (FG) sequence repeats that can bind to HEAT repeats within importins, which is important for importin beta-mediated transport (2, 16).
Members of the importin beta family either can bind or transport cargo themselves or can form heterodimers with importin alpha. As part of a heterodimer, importin beta mediates interactions with the nuclear pore complex, while importin alpha acts as an adaptor protein to bind the NLS on the cargo. Most importin beta proteins, however, bind directly to cargoes and therefore do not rely on an adapter (10, 38). NLSs recognized by importin beta are more difficult to identify (30).
As mentioned above, LysRS, which produces Ap4A, can bind directly to the transcription factor where Ap4A is expected to act as a second messenger in a local environment. Therefore, it seems that local regulation of Ap4A levels is extremely important for its efficient activity. Indeed, it was previously shown that local concentration of another second messenger, such as cyclic AMP (cAMP), is essential for its activity. This is achieved mainly through compartment-specific activity of the phosphodiesterase degrading cAMP (15).
In this study, Ap4A hydrolase was found to translocate into the nuclei of mast cells following immunological activation, as do LysRS and MITF. Using a database of putative interacting protein domains, the Nudix motif of Ap4A hydrolase was predicted to interact with importin beta of the karyopherin family. We then obtained evidence that Ap4A hydrolase and importin beta interact in the cell and, moreover, do so in an IgE-antigen (Ag) activation-dependent manner. The Ap4A hydrolase sequence does not contain any predicted NLS. Here we describe a noncanonical mechanism by which this translocation occurs.
In addition, we found that immunological challenge led to dephosphorylation of Ap4A hydrolase, which affected the hydrolytic activity of the enzyme. Thus, by means of nonconventional, Ag-induced regulation, Ap4A hydrolase modulates the transcriptional activity of MITF.
MATERIALS AND METHODS
Cell culture.
RBL-2H3 cells were maintained in RPMI 1640 medium as previously described (31). Bone marrow was isolated from 5- to 6-week-old mice, and bone marrow-derived mast cells (BMMC) were cultured in RPMI medium supplemented with 20 ng/ml interleukin-3 (IL-3) and 20 ng/ml SCF as previously described (33). Cells were generally grown for a minimum of 4 weeks and used when >90% of the population expressed FcεRI. BMMC and rat basophilic leukemia (RBL) cells were sensitized first with anti-DNP IgE monoclonal antibody (SPE-7; Sigma-Aldrich Corp., St. Louis, MO) and then challenged with DNP (Sigma-Aldrich Corp.). IgE antibody was centrifuged (18,000 × g, 5 min) before use to remove aggregates. Cell activation was verified by demonstrating an increased phosphorylation of extracellular signal-regulated kinase (ERK) following the Ag challenge.
Ap4A hydrolase activity assay.
The assay mixture contained 35 mM HEPES-KOH (pH 7.8), 5 mM magnesium acetate, 10 μM Ap4A (Sigma), 15 μl reconstituted ATP monitoring reagent (Perkin-Elmer), and 15 μl of extract and was incubated at room temperature. The initial rate of increase in luminescence was measured using a luminometer as previously described (1).
Gel electrophoresis and Western blotting.
Protein concentration was determined using a Tecan microplate reader (Tecan, Research Triangle Park, NC). Equal amounts of protein from each sample were resolved by either 10% or 15% SDS-PAGE under reducing conditions and transferred to polyvinylidene difluoride (PVDF) membranes. Visualization of reactive proteins was performed by enhanced chemiluminescence.
The antibody to Ap4A hydrolase was custom-made using a specially designed determinant (FKEMKATLQEGHQFLC) (Hy Laboratories Ltd., Israel). Antibody against importin beta was purchased from Abcam (Cambridge, MA). Antibody against the Myc tag was purchased from Santa Cruz Biotechnology.
Immunoprecipitation.
Immunoprecipitation of specific proteins from RBL cells was carried out as previously described (22).
siRNA.
Cells were transfected with a small interfering RNA (siRNA) duplex consisting of two complementary 21-nucleotide RNA strands with 3′-dTdT overhangs (Qiagen Inc., CA) in order to downregulate Ap4A hydrolase, importin beta, importin 5, and importin 7. siRNAs were designed to be complementary to nucleotide sequences found in rat mRNA for Ap4A hydrolase, and a nontargeting nucleotide sequence was used as the control (NT siRNA).
Transfection.
Amaxa Nucleofector technology (Amaxa, Cologne, Germany) was used to transfect cells. A total of 2 × 106 cells were transfected with 3 μg of the selected siRNA oligonucleotide according to the manufacturer's protocol. Briefly, the cells were resuspended in 100 μl Ingenio solution (Mirus, WI), either DNA (plasmid) or RNA was added, and the mixture was transferred to an electroporation cuvette. Electroporation was performed using the T-20 program.
Real-time quantitative PCR.
MITF-responsive gene transcription was measured using real-time quantitative PCR. mRNAs of the target genes were quantified by SYBR green incorporation (ABgene SYBR green ROX mix; ABgene). Real-time PCR was performed on a Rotor-Gene sequence detection system (Corbett, Australia). The genes whose mRNA levels were quantified by real-time PCR were the rat Kit, granzyme B (GrB), and β-actin genes.
TC-FlAsh-based protein detection.
TC-FlAsH in-cell tetracysteine tag (TC tag) detection uses biarsenical labeling reagents to bind and detect proteins containing a tetracysteine motif (11). The biarsenical labeling reagents are nonfluorescent until they bind the tetracysteine motif, at which time they become highly fluorescent. The FlAsH-EDT2 labeling reagent binds the TC tag through four covalent bond formations—the two arsenic groups of the FlAsH-EDT2 reagent each bind two thiols in the TC tag sequence. Upon binding, the FlAsH-EDT2 reagent is converted to a highly fluorescent state that can be detected at the appropriate emission peak.
The TC tag (Cys-Cys-Pro-Gly-Cys-Cys) is encoded in the pcDNA6.2/TC-Tag-DEST vector. When fused to a gene of interest, the TC tag allows the expressed fusion protein to be recognized specifically by a biarsenical labeling reagent.
Cloning of the gene of interest into the vector is performed using Gateway technology, which is a universal cloning method that takes advantage of the site-specific recombination properties of bacteriophage lambda.
Briefly, BMMC were transiently transfected with the TC tag expression construct and plated on the day prior to the experiment at ∼90% confluence in a chambered coverslip. On the day of the experiment, growth medium was aspirated, replaced with Opti-MEM I containing 2 μM FlAsH, and incubated for 1 h. After incubation, cells were washed 3 times with BAL buffer (supplied by Invitrogen in the labeling kit).
ImageStream flow cytometry.
An ImageStream instrument automatically acquires up to six different spatially registered images (bright-field, dark-field, and four fluorescent images) per cell at rates on the order of thousands of objects per minute, using a digital charge-coupled device (CCD) camera. The digital imagery obtained is analyzed using the IDEAS statistical image analysis program, which provides tools for the objective numerical scoring and discrimination of cells based on the characteristics of their imagery. The ability to numerically score large numbers of automatically acquired images is ideally suited to the analysis of nuclear translocation within primary immune system cells.
2D electrophoresis.
Two-dimensional (2D) electrophoresis was performed as previously described (13).
Cells were solubilized in 2D lysis buffer {7 M urea, 2 M thiourea, 4% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 100 mM dithiothreitol (DTT)}. Cell lysates were loaded onto immobilized pH gradient (IPG) strip gels (linear pH gradient of pH 3 to 10; 7 cm). Isoelectric focusing (IEF) was performed at 4,000 V until the total volt-hours reached 10 kV-h, using a Protean IEF cell (Bio-Rad). Following two-step equilibration with 375 mM Tris-HCl (pH 8.8), 6 M urea, 2% SDS, 20% glycerol, 2% DTT, and 2.5% iodoacetamide, the IPG strips were embedded on top of 8% SDS-PAGE gels and sealed with 2% agarose. Proteins were separated based on their molecular weight.
Nuclear protein extraction.
Subcellular protein fractionation was performed using a ProteoJET cytoplasmic and nuclear protein extraction kit (Fermentas) following the procedures suggested by the manufacturer. Briefly, 10 volumes of cell lysis buffer (containing protease inhibitors, phosphatase inhibitors, and DTT) was added to 1 volume of packed cells. Cells were vortexed for 10 s and then set on ice for 10 min. The cytoplasmic fraction was separated from nuclei by centrifugation at 500 × g for 7 min at 4°C. Isolated nuclear pellets were washed twice with 500 μl of nuclear washing buffer. Nuclear pellets were resuspended with ice-cold nuclear storage buffer (containing protease inhibitors, phosphatase inhibitors, and DTT), and a 1/10 volume of nuclear lysis reagent was added to the suspension. Separation of the nucleus and the cytosol was verified using anti-tubulin as a cytoplasmic marker and antilamin as a nuclear marker.
Statistical analysis.
Analysis of variance (ANOVA) was performed when appropriate, with differences being considered significant at P values of <0.05.
RESULTS
Ap4A hydrolase translocates into the nuclei of BMMC.
We recently showed that LysRS, the main enzyme that produces Ap4A, translocates into the nucleus in activated mast cells (40). We hypothesized that Ap4A production occurs in the nuclei of activated mast cells, with concurrent Ap4A accumulation and LysRS nuclear localization. Evidence has previously been presented for a predominantly nuclear location of Ap4A itself (37). Since we have previously shown that Ap4A hydrolase is part of the transcriptional network regulating MITF and USF2, we examined whether aggregation of FcεRI by IgE-Ag causes translocation of Ap4A hydrolase from the cytosol to the nuclear compartment.
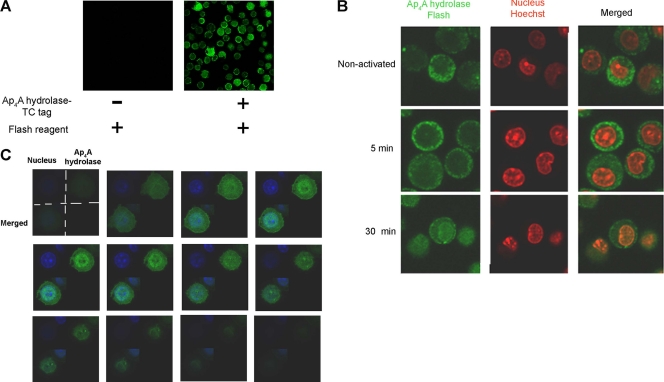
In order to determine whether Ap4A hydrolase nuclear translocation occurs in activated mast cells, murine Ap4A hydrolase was cloned into pcDNA6.2/TC-Tag-DEST, which encodes the six-amino-acid TC tag, by using the unique Gateway recombination system. By means of the FlAsh labeling system, as described in detail in Materials and Methods, we determined the localization of Ap4A hydrolase-TC tag in BMMC. The FlAsh reagent remained nonfluorescent in control cells but gained significant fluorescence in cells which were transfected with the Ap4A hydrolase-TC tag vector (Fig. 1A).
Fig. 1.
Ap4A hydrolase translocates into the nuclei of immunologically activated BMMC. (A) BMMC were transfected with an Ap4A hydrolase-TC tag-expressing vector. Transfected cells exhibited fluorescence at the predicted wavelength, while no fluorescence was detected in nontransfected cells. (B) BMMC were transfected with Ap4A hydrolase-TC tag. The cells were immunologically activated for specific times, plated on chambered coverslips, and labeled with 2 μM FlAsH. The cell nuclei were stained with Hoechst stain. Confocal microscopy was used to evaluate Ap4A hydrolase subcellular localization. One representative field of three is presented. (C) BMMC were transfected with Ap4A hydrolase-TC tag and activated for 30 min. z-Stack analysis of Ap4A hydrolase was performed using confocal microscopy to obtain sequential z-axis images.
BMMC were transiently transfected with Ap4A hydrolase-TC tag, plated on chambered coverslips, and stained with the biarsenical dye FlAsH. Distinct localization patterns were observed for Ap4A hydrolase-TC-FlAsh (green), as can be seen in Fig. 1B. Ap4A hydrolase was found solely in the cytoplasm in quiescent cells and in cells activated for 5 min, whereas 30 min following activation, Ap4A hydrolase was also found in the nucleus (Fig. 1B). While the subcellular localization of Ap4A hydrolase was cytoplasmic both in quiescent cells and in cells activated for 5 min, different distribution patterns were observed. In cells activated for 5 min, Ap4A hydrolase was closer to the cell membrane and nonuniformly distributed, with accumulations at specific foci.
In order to verify the entrance of the Ap4A hydrolase into the nuclei of activated mast cells, 12 sequential z-stack images of the same field are presented. It is clear that 30 min after activation, Ap4A hydrolase was present within the nuclei of BMMC rather than surrounding the nuclei from the outside (Fig. 1C).
Similarly, Ap4A hydrolase was found to translocate into the nucleus in immunologically activated RBL cells (Fig. 2A and B). In order to quantitatively assess this nuclear internalization of Ap4A hydrolase, ImageStream imaging flow cytometry was used. The ImageStream instrument has been utilized previously by others to demonstrate quantitative measurement of nuclear translocation events with regard to NF-κB (12), IRF-7 (12), RAF1 (35), and others.
Fig. 2.
Identification of Ap4A hydrolase nuclear translocation in RBL cells following IgE-Ag activation. (A) RBL cells were either quiescent or sensitized with IgE and then were challenged with antigen for 20 or 60 min. Immunostaining was performed with anti-Ap4A hydrolase. The cells were analyzed by confocal laser scanning microscopy. Images for one representative experiment of three are presented. (B) One representative cell from the field is shown at high magnification. Using Photoshop picture-editing software, the percent brightness parameter was determined at specific intervals along the x axis. (C) RBL cells were activated (or not) with IgE, challenged with Ag for specific times, and then stained for Ap4A hydrolase by anti-Ap4A hydrolase followed by Cy3-conjugated anti-rabbit, together with PI staining to visualize nuclei. Nuclear translocation was evaluated by ImageStream flow cytometry. For each treatment, 5,000 cells were collected. Positive single cells were gated, and nuclear translocation of Ap4A hydrolase was plotted. The relative translocated population size is indicated in the upper right corner. (D) Representative images of untranslocated (left) and translocated (right) Ap4A hydrolase (Cy3 column), as well as PI and merged images, for the 15-min treatment.
In this study, cell nuclei were stained with propidium iodide (PI) while Ap4A hydrolase was stained with anti-Ap4A hydrolase, using a fluorescent Cy3-conjugated antibody as a secondary antibody. To assess Ap4A hydrolase nuclear translocation, the localization of Ap4A hydrolase and PI was measured and compared (giving a similarity score) for each RBL cell, with areas positively stained for PI considered nuclear. The “Hyd translocation” gate represents positive correlation for the PI-Cy3 similarity score, and the relative population size of cells that fall within this gate is indicated in the upper right corner of the plots (Fig. 2C). Representative images of cells that fall within and to the left of the gate are shown (Fig. 2D). As shown in Fig. 2C, the localization of Ap4A hydrolase in the nuclei was increased 2-fold 15 min following activation.
Ap4A hydrolase associates with importin beta upon immunological activation.
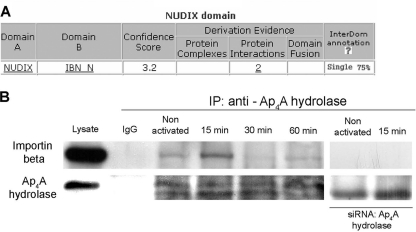
Though molecules smaller than ∼20 to 40 kDa can passively diffuse through the nuclear pore complex (NPC) (reviewed in references 27 and 30), the movement of Ap4A hydrolase into the nuclei of activated cells seems to be regulated tightly, as it is stimulus dependent and demonstrates specific kinetics. Since Ap4A hydrolase does not contain any known NLS and all attempts to identify this type of motif in the Ap4A hydrolase sequence by use of bioinformatic tools of NLS prediction were in vain, we hypothesized that following mast cell activation, Ap4A hydrolase may not directly associate with one of the importins but may use a third party as a carrier protein to enter the nucleus in a “piggyback” process. There is almost no literature regarding Ap4A hydrolase-associated proteins in cells, so we used another bioinformatic tool, InterDom, to predict which proteins might interact with Ap4A hydrolase (28). As shown in Fig. 3A, we found that the Nudix motif of Ap4A hydrolase was predicted to interact with the N-terminal domain of importin beta (IMP_B) of the karyopherin family. The Nudix motif (Nudix box) is comprised of the 23-amino-acid sequence GX5EX5[UA]XREX2EEXGU, where U is an aliphatic, hydrophobic residue (PROSITE accession no. PS00893). This sequence is located in a loop-helix-loop structural motif, and the Glu residues in the core of the motif play an important role in binding essential divalent cations.
Fig. 3.
Substantial association between Ap4A hydrolase and importin beta in RBL cells. (A) The protein sequence of Ap4A hydrolase was subject to domain-domain association prediction analysis using bioinformatic tools. The Nudix domain was predicted to associate with the importin beta N-terminal domain (IBN_N) by the putative interaction database InterDom (http://interdom.i2r.a-star.edu.sg/). (B) Coimmunoprecipitation analysis demonstrating the time course of Ap4A hydrolase-importin beta interaction following IgE-Ag stimulation, using anti-Ap4A hydrolase antibody for immunoprecipitation (IP) and either anti-importin beta or anti-Ap4A hydrolase for immunoblotting. The specific Ap4A hydrolase band was verified by a control experiment using siRNA to knock down Ap4A hydrolase (right lanes). Data for one representative experiment of six are shown.
In order to experimentally confirm the existence of an in vivo Ap4A hydrolase-importin beta complex, coimmunoprecipitation of Ap4A hydrolase with importin beta was performed in quiescent and activated cells (Fig. 3B). The recovered and resolved immune complexes showed that Ap4A hydrolase was associated with importin beta in an IgE-Ag activation-dependent manner. The observed association between the two proteins peaked 15 to 60 min after stimulation. Knockdown of Ap4A hydrolase by an RNA interference (RNAi) silencing approach (siRNA) abolished coimmunoprecipitation of importin beta.
Nuclear translocation of Ap4A hydrolase in activated mast cells is mediated by importin beta.
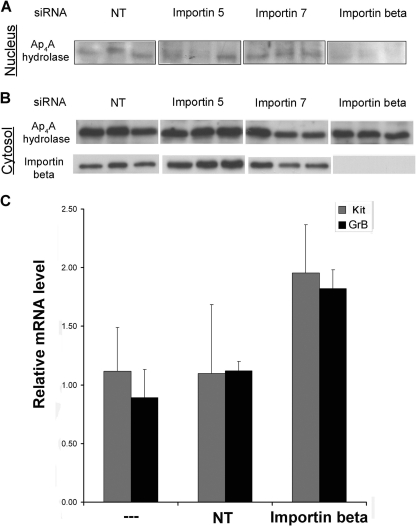
In order to verify the essential role played by importin beta in nuclear translocation of Ap4A hydrolase, we used an siRNA approach to target expression of importin beta. Cells were transfected with various siRNA oligonucleotides, each targeting a different member of the importin family, and the nuclear extracts from the respective cells were analyzed for the presence of Ap4A hydrolase by using anti-Ap4A hydrolase. While silencing of either importin 5 or 7 as well as transfection with a nontargeting (NT) siRNA did not interfere with translocation to the nucleus, Ap4A hydrolase was not detected in nuclei of cells that were treated with siRNA against importin beta (Fig. 4A).
Fig. 4.
Importin beta-mediated Ap4A hydrolase nuclear translocation is involved in MITF transcriptional activity regulation. (A and B) RBL cells were transfected with siRNA oligonucleotides against importin beta, 5, and 7. NT siRNA was used as a control. Nuclear fractions were isolated from cells activated with IgE and antigen for 30 min. Nuclear extracts (A) and cytosolic extracts (B) were analyzed by Western blotting with anti-Ap4A hydrolase antibody. Silencing of importin beta was verified by Western blotting with anti-importin beta. Each lane represents an independent experiment. (C) RBL cells were transfected with either importin beta siRNA or NT siRNA. Control and transfected cells were activated with IgE-Ag. Twenty-four hours later, cells were lysed and mRNA quantitation for GrB and Kit was performed by SYBR green incorporation into real-time PCR with RBL cells. Expression levels were normalized to those of the β-actin housekeeping gene. Results are presented as relative quantities, with the untreated cell level arbitrarily set to 1. The means and standard errors of the means for three experiments are shown.
Nuclear translocation of Ap4A hydrolase via association with importin beta plays a role in the regulation of MITF transcriptional activity.
We previously provided evidence that Ap4A hydrolase is responsible for Ap4A degradation in immunologically activated mast cells and, moreover, that Ap4A hydrolase plays a critical role in the transcriptional regulation of MITF (6). Here we further explored the effect of Ap4A hydrolase nuclear translocation on MITF transcriptional activity.
The transcript levels of two MITF target genes, encoding GrB (17) and Kit (36), were measured in immunologically activated RBL cells in which importin beta was knocked down by siRNA. A significant accumulation of GrB mRNA and a moderate increase in Kit mRNA were observed in RBL cells, concurrent with reduced expression of importin beta and reduced nuclear translocation of Ap4A hydrolase (Fig. 4B). Thus, nuclear translocation of Ap4A hydrolase via its association with importin beta is clearly involved in the regulation of MITF transcriptional activity.
Ap4A hydrolase is subject to dephosphorylation upon immunological activation.
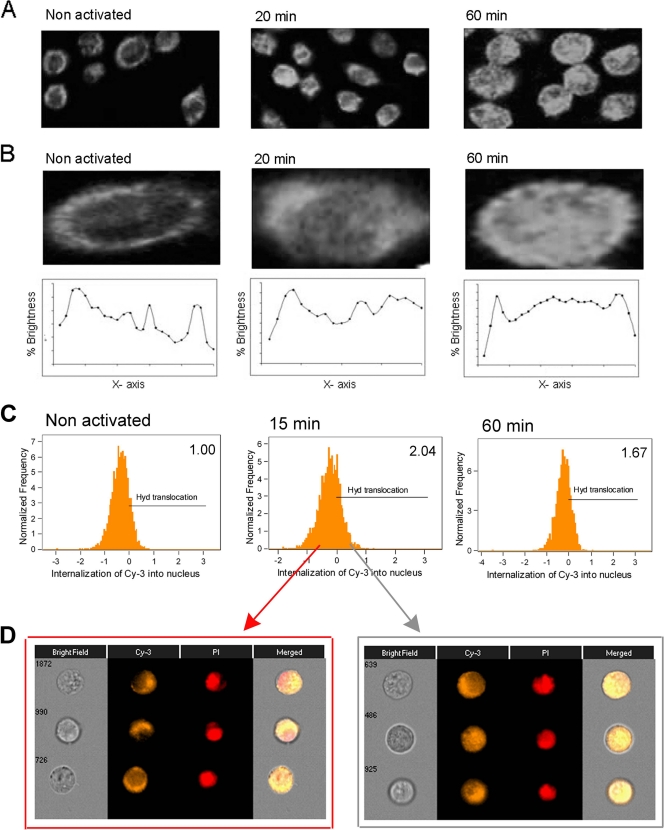
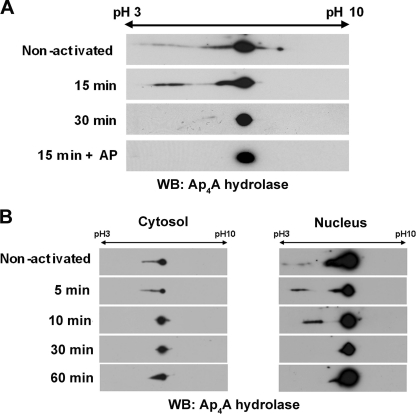
Recently, we found that LysRS translocates into the nuclei of activated mast cells in a serine phosphorylation-dependent manner (40) via the mitogen-activated protein kinase (MAPK)/ERK pathway. In order to determine whether the phosphorylation status of Ap4A hydrolase changes upon IgE-Ag activation, we used 2D gel electrophoresis. Ap4A hydrolase was immunoprecipitated from nonstimulated or IgE-Ag-activated mast cells, and its phosphorylation was identified by a more acidic pI compatible with phosphorylation (as seen by a shift to the left of the gel). This species was detected in unstimulated cells and cells that were activated for short periods (up to 15 min), whereas in cells activated with IgE-Ag for 30 min the enzyme appeared to be dephosphorylated (Fig. 5A). This phosphorylation was totally blocked by the presence of alkaline phosphatase (AP) (Fig. 5A). Thus, Ap4A hydrolase is dephosphorylated upon immunological activation.
Fig. 5.
Ap4A hydrolase is subject to dephosphorylation upon IgE-Ag activation. (A) Lysates from IgE-Ag-activated and nonactivated RBL cells were subjected to 2D electrophoresis on a pH 3 to 10 gradient in an 8% polyacrylamide gel. The gel was blotted with anti-Ap4A hydrolase antibody. WB, Western blot. (B) Nuclear and cytoplasmic fractions were isolated from RBL cells activated with IgE and antigen. The subcellular extracts were analyzed by 2D electrophoresis with anti-Ap4A hydrolase antibody.
We then determined whether the dephosphorylation of Ap4A hydrolase (Fig. 5A) correlated with the enzyme's nuclear translocation. Nuclear and cytoplasmic fractions were isolated from IgE-Ag-activated RBL cells. The subcellular extracts were analyzed by 2D gel electrophoresis with anti-Ap4A hydrolase antibody. No significant difference in phosphorylation status was detected in the cytosolic and nuclear fractions. Similar to the case with whole-cell lysates, phosphorylation was detected in both the cytosol and nuclei of nonactivated cells as well as in cells that were activated for less than 30 min, while the dephosphorylated form of the hydrolase was detected in both the cytosol and nuclei of cells 30 and 60 min following activation (Fig. 5B).
We subsequently determined the amino acid residue(s) subject to the dephosphorylation process. Immunoprecipitation of potential phosphorylated proteins with phospho-specific antibodies and immunoblot analysis with anti-Ap4A hydrolase demonstrated dephosphorylation of Ap4A hydrolase on serine residues, but not on threonine residues, 60 min after cell activation.
Ser108 phosphorylation of Ap4A hydrolase increases its enzymatic activity.
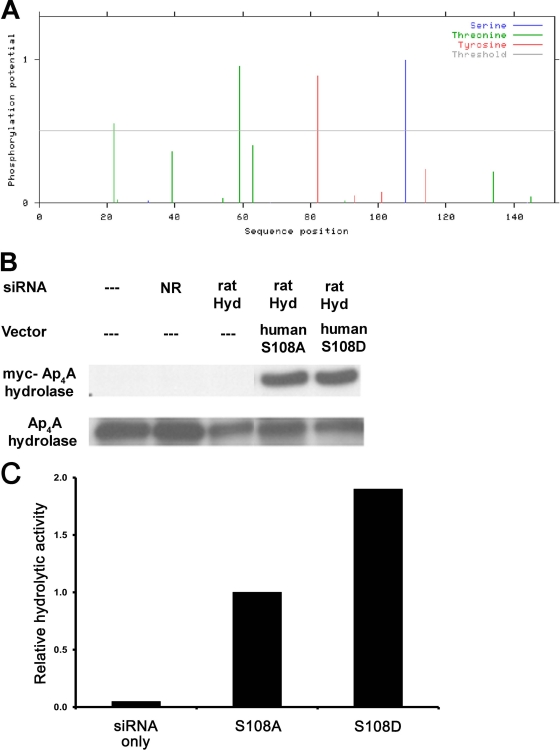
In order to specifically predict which serine residues may be subject to phosphorylation, the web-based tool NetPhos was used (3). Only one serine residue, S108, in the human Ap4A (hAp4A) hydrolase sequence was significantly predicted to be phosphorylated (Fig. 6A). Based on this observation, fused Myc-tagged constructs of mutated hAp4A hydrolase were constructed (S108A and S108D mutants). Each of the constructs was administered to the cells after the endogenous Ap4A hydrolase was silenced by siRNA. Under these conditions, no loss of the exogenous hAp4A hydrolase occurred (Fig. 6B). Assessment of Ap4A hydrolytic activity was carried out on extracts derived from immunologically activated RBL cells that were transfected with each of the constructs. As shown in Fig. 6C, the S108A mutant hAp4A hydrolase, in which the residue at position 108 was not phosphorylated, exhibited reduced Ap4A degradation in these transfected RBL cells, suggesting that phosphorylation of this residue is critical for Ap4A hydrolase enzymatic activity.
Fig. 6.
Phosphorylation status of serine 108 in Ap4A hydrolase is involved in the enzymatic activity of Ap4A metabolism. (A) Putative serine, threonine, and tyrosine phosphorylation sites in the Ap4A hydrolase protein sequence, as predicted by the NetPhos 2.0 program (www.cbs.dtu.dk/services/NetPhos/). (B) RBL cells were transfected with rat Ap4A hydrolase siRNA. Twenty-four hours later, cells were transfected with human Ap4A hydrolase S108A and S108D variants. Next, the cells were incubated with IgE and challenged with DNP for 30 min. The cell extracts were analyzed by Western blotting with anti-Myc or anti-Ap4A hydrolase antibodies. (C) RBL cell lysates from panel B were subjected to an enzymatic activity assay. Ap4A hydrolase hydrolytic activity was determined as the rate of synthetic Ap4A degradation, which was evaluated by chemiluminescence detection of ATP as described in Materials and Methods. Data for one representative experiment of three are shown.
DISCUSSION
We previously demonstrated that LysRS forms a complex with MITF and its repressor Hint-1, which is released from the complex by its binding to the LysRS product Ap4A, enabling MITF to transcribe its target genes (40).
Recently, we also provided evidence that Ap4A hydrolase is responsible for Ap4A degradation following immunological activation of mast cells and thus established Ap4A as a novel second messenger (6). In that study, we described the key role played by Ap4A hydrolase in regulation of both MITF and USF2 transcriptional activity.
The intracellular compartmentalization of Ap4A in mammalian cells during various growth and cell cycle stages has been studied, and the majority of cellular Ap4A is localized in the nuclei, especially during G1 and early S phase (37). As mentioned above, we previously reported the nuclear function of Ap4A in the transcription regulation network in activated mast cells. Moreover, we showed that the nuclear translocation kinetics of the main Ap4A producer, LysRS, corresponded with the synthesis of this nucleotide (40). Therefore, we hypothesized that at least some of the Ap4A hydrolase, which in this context is the major degrading enzyme of Ap4A, also changes its subcellular localization upon immunological challenge.
Though bioinformatic analysis did not predict an NLS for Ap4A hydrolase, in the present work we characterized the mechanism of its nuclear import. We found that the 17-kDa Ap4A hydrolase translocates to the nucleus following immunological activation of mast cells (Fig. 1 and 2). Proteins smaller than 40 kDa have been proposed to diffuse freely through the nuclear pores. However, we showed in mast cells that Ap4A hydrolase actively translocates to the nucleus in a stimulus-dependent manner rather than passively diffusing into this compartment.
Stimulus-dependent nuclear translocation of signaling proteins is critical for time-regulated processes such as cell cycle and transcription. In the first and best-described mechanism for nuclear import, proteins destined for transport into the nucleus contain amino acid targeting sequences called nuclear localization signals (NLSs) (reviewed in reference 21), which consist of either one (monopartite) or two (bipartite) stretches of basic amino acids (19, 32).
Unexpectedly, however, many of the proteins involved in the aforementioned functions, including cyclins (26), ERKs (41), SMAD transcription factors (24), and Ap4A hydrolase, do not contain the canonical NLS. ERKs, MEKs, and SMADs were recently found to contain a specific 3-amino-acid motif, SPS, which was termed the nuclear translocation sequence (NTS). Upon stimulation, this motif is phosphorylated, resulting in nuclear translocation of these proteins in an importin 7-dependent manner (8). Though the sequence of Ap4A hydrolase does not contain either SPS or the similarly functioning TPT motif, the description of the above-mentioned mechanism led us to hypothesize that it was possible that Ap4A hydrolase translocated in a nonclassical (non-NLS) manner via an importin family member.
As shown in Fig. 3, the Nudix motif of Ap4A hydrolase was indeed predicted to interact directly with the N-terminal domain of importin beta. This association was verified using coimmunoprecipitation of the two proteins, and moreover, silencing of Ap4A hydrolase prevented coimmunoprecipitation of importin beta (Fig. 3B) and silencing of importin beta prevented Ap4A hydrolase nuclear translocation (Fig. 4A).
Importin beta has previously been described to mediate the nuclear translocation of numerous components of cyclin-dependent kinase (Cdk)/cyclin complexes, such as cyclin B1 (26) and Cdc7 protein (20). Cyclin B1 performs cardinal roles in the eukaryotic cell division cycle by phosphorylating key cellular substrates, and Cdc7 is essential for the initiation of DNA replication (4). It has been reported that importin beta binds directly to Cdc7, which does not have a classical NLS, via the kinase insert II domain, promoting its nuclear import. Immunodepletion of importin beta, but not importin alpha, abrogated Cdc7 nuclear import (20).
In addition to interfering with Ap4A hydrolase nuclear translocation, silencing of importin beta led to an increase in MITF transcriptional activity (Fig. 4). This finding adds strength to the notion that the LysRS-Ap4A pathway is central in the regulation of MITF transcriptional activity.
Posttranslational modifications, e.g., the phosphorylation of nuclear import substrates, is required for optimal interaction with importin alpha (7, 9). ERK phosphorylation at the SPS motif was found to be critical for its association with importin 7 (8). Additionally, for Trx, it was demonstrated that specific S-nitros(yl)ation of the Cys residue is critical for its nuclear import (34).
We recently showed that LysRS nuclear translocation occurred as a result of serine 207 phosphorylation via the MAPK/ERK pathway. Furthermore, phosphorylation of the serine 207 residue was found to be a prerequisite for Ap4A synthesis, as a LysRS S207A variant that was linked to a strong NLS did not cause induction of Ap4A synthesis, despite its nuclear localization (40).
Here we hypothesized that Ap4A hydrolase may also be subject to a posttranslational modification, such as phosphorylation, which modulates its intracellular location and/or activity. We found that Ap4A hydrolase was phosphorylated in resting mast cells, while dephosphorylation was observed in activated cells (Fig. 5). Moreover, the phosphorylated form of Ap4A hydrolase was found to have a higher Ap4A hydrolysis rate than the nonphosphorylated form, suggesting that its dephosphorylation is involved in the maintenance of the Ap4A level in cells upon activation. It is important that although the S108A mutant of Ap4A hydrolase also exhibited hydrolytic activity, it was observed to be 2-fold lower than that of the S108D mutant, suggesting that phosphorylation of Ser108 only partially affects the enzyme's activity and that it may be regulated by additional modifications which have yet to be investigated.
Ap4A hydrolase phosphorylation seems to peak 15 min after activation. The positive correlation between phosphorylation status and Ap4A hydrolysis rate is in accordance with the Ap4A level, which drops rapidly 15 min after exposure to Ag (6).
In summary, this study demonstrates nonconventional, Ag-induced regulation of Ap4A hydrolase (Fig. 7). As the importance of Ap4A as a second messenger and as a signaling molecule involved in gene transcription is uncovered, a better understanding of the mechanisms regulating its cellular levels is important to shed light on its function(s). The findings reported herein may also contribute to future emphasis on the manipulation of Ap4A levels in diseases.
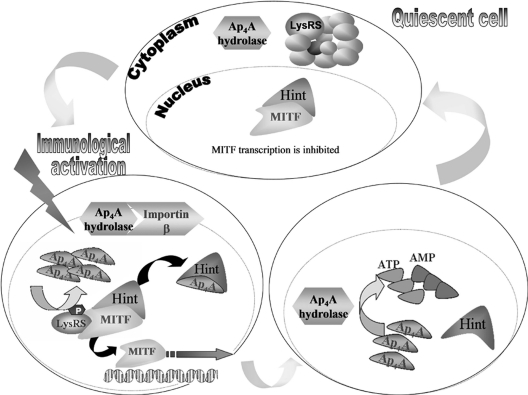
Fig. 7.
Proposed model for involvement of Ap4A hydrolase in transcription regulation. Upon immunological activation, Ap4A hydrolase associates with importin beta and is translocated into the nucleus when Ap4A levels are elevated. Ap4A binds to Hint-1, dissociating it from MITF. With the repression removed, transcription factors are free to transcribe their target genes. The presence of Ap4A hydrolase in the nucleus leads to hydrolysis of the accumulated Ap4A into AMP and ATP, decreasing its levels to that found in resting cells. When Ap4A levels decrease, Hint-1 reassociates with MITF.
ACKNOWLEDGMENTS
This work was supported by the United States Binational Science Foundation (E.R.), the Israeli Academy of Science (E.R.), the German-Israel Foundation for Scientific Research and Development (E.R.), DKFZ-MOST cooperation (E.R.), the National Medical Research Council of Singapore (D.M.K. and E.R.), the Morasha Foundation Fund (H.N.), the Acceleration Research of KOSEF (S.K.), the 21st Frontier Functional Proteomics Research (S.K.), and the CREATE project of the National Research Foundation of Singapore (E.R. and M.K.). I.C.-L. and Z.Y. were supported by the Canadian Friends of Hebrew University.
We have no conflicting financial interests.
Footnotes
Published ahead of print on 14 March 2011.
REFERENCES
- 1. Abdelghany H. M., et al. 2001. Cloning, characterisation and crystallisation of a diadenosine 5′, 5‴-P(1),P(4)-tetraphosphate pyrophosphohydrolase from Caenorhabditis elegans. Biochim. Biophys. Acta 1550:27–36 [DOI] [PubMed] [Google Scholar]
- 2. Bayliss R., Littlewood T., Strawn L. A., Wente S. R., Stewart M. 2002. GLFG and FxFG nucleoporins bind to overlapping sites on importin-beta. J. Biol. Chem. 277:50597–50606 [DOI] [PubMed] [Google Scholar]
- 3. Blom N., Gammeltoft S., Brunak S. 1999. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294:1351–1362 [DOI] [PubMed] [Google Scholar]
- 4. Bousset K., Diffley J. F. 1998. The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev. 12:480–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reference deleted.
- 6. Carmi-Levy I., Yannay-Cohen N., Kay G., Razin E., Nechushtan H. 2008. Diadenosine tetraphosphate hydrolase is part of the transcriptional regulation network in immunologically activated mast cells. Mol. Cell. Biol. 28:5777–5784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chook Y. M., Blobel G. 2001. Karyopherins and nuclear import. Curr. Opin. Struct. Biol. 11:703–715 [DOI] [PubMed] [Google Scholar]
- 8. Chuderland D., Konson A., Seger R. 2008. Identification and characterization of a general nuclear translocation signal in signaling proteins. Mol. Cell 31:850–861 [DOI] [PubMed] [Google Scholar]
- 9. Conti E., Uy M., Leighton L., Blobel G., Kuriyan J. 1998. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell 94:193–204 [DOI] [PubMed] [Google Scholar]
- 10. Fried H., Kutay U. 2003. Nucleocytoplasmic transport: taking an inventory. Cell. Mol. Life Sci. 60:1659–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gaietta G., et al. 2002. Multicolor and electron microscopic imaging of connexin trafficking. Science 296:503–507 [DOI] [PubMed] [Google Scholar]
- 12. George T. C., Morrissey P. J., Cui C., Singh S., Fitzgerald Bocarsly P. 2009. Measurement of cytoplasmic to nuclear translocation. Curr. Protoc. Cytom. 2009:Unit 9.28 [DOI] [PubMed] [Google Scholar]
- 13. Han J. M., et al. 2008. AIMP2/p38, the scaffold for the multi-tRNA synthetase complex, responds to genotoxic stresses via p53. Proc. Natl. Acad. Sci. U. S. A. 105:11206–11211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hause B., Feussner K., Wasternack C. 1997. Nuclear location of a diadenosine 5′,5‴-P1,P4-tetraphosphate (Ap4A) hydrolase in tomato cells grown in suspension cultures. Bot. Acta 110:452–457 [Google Scholar]
- 15. Houslay M. D. 2010. Underpinning compartmentalised cAMP signalling through targeted cAMP breakdown. Trends Biochem. Sci. 35:91–100 [DOI] [PubMed] [Google Scholar]
- 16. Isgro T. A., Schulten K. 2007. Association of nuclear pore FG-repeat domains to NTF2 import and export complexes. J. Mol. Biol. 366:330–345 [DOI] [PubMed] [Google Scholar]
- 17. Ito A., et al. 1998. Systematic method to obtain novel genes that are regulated by mi transcription factor: impaired expression of granzyme B and tryptophan hydroxylase in mi/mi cultured mast cells. Blood 91:3210–3221 [PubMed] [Google Scholar]
- 18. Reference deleted.
- 19. Kalderon D., Roberts B. L., Richardson W. D., Smith A. E. 1984. A short amino acid sequence able to specify nuclear location. Cell 39:499–509 [DOI] [PubMed] [Google Scholar]
- 20. Kim B. J., Lee H. 2006. Importin-beta mediates Cdc7 nuclear import by binding to the kinase insert II domain, which can be antagonized by importin-alpha. J. Biol. Chem. 281:12041–12049 [DOI] [PubMed] [Google Scholar]
- 21. Lange A., et al. 2007. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J. Biol. Chem. 282:5101–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Levy C., Nechushtan H., Razin E. 2002. A new role for the STAT3 inhibitor, PIAS3: a repressor of microphthalmia transcription factor. J. Biol. Chem. 277:1962–1966 [DOI] [PubMed] [Google Scholar]
- 23. Li S., et al. 2004. A map of the interactome network of the metazoan C. elegans. Science 303:540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Massague J., Seoane J., Wotton D. 2005. Smad transcription factors. Genes Dev. 19:2783–2810 [DOI] [PubMed] [Google Scholar]
- 25. McLennan A. G. 2006. The Nudix hydrolase superfamily. Cell. Mol. Life Sci. 63:123–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moore J. D., Yang J., Truant R., Kornbluth S. 1999. Nuclear import of Cdk/cyclin complexes: identification of distinct mechanisms for import of Cdk2/cyclin E and Cdc2/cyclin B1. J. Cell Biol. 144:213–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mosammaparast N., Pemberton L. F. 2004. Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 14:547–556 [DOI] [PubMed] [Google Scholar]
- 28. Ng S. K., Zhang Z., Tan S. H., Lin K. 2003. InterDom: a database of putative interacting protein domains for validating predicted protein interactions and complexes. Nucleic Acids Res. 31:251–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oka K., et al. 2011. Nudix-type motif 2 (NUDT2) in human breast carcinoma: a potent prognostic factor associated with cell proliferation. Int. J. Cancer 128:1770–1782 [DOI] [PubMed] [Google Scholar]
- 30. Pemberton L. F., Paschal B. M. 2005. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 6:187–198 [DOI] [PubMed] [Google Scholar]
- 31. Razin E., et al. 1999. Suppression of microphthalmia transcriptional activity by its association with protein kinase C-interacting protein 1 in mast cells. J. Biol. Chem. 274:34272–34276 [DOI] [PubMed] [Google Scholar]
- 32. Robbins J., Dilworth S. M., Laskey R. A., Dingwall C. 1991. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64:615–623 [DOI] [PubMed] [Google Scholar]
- 33. Saitoh S., et al. 2000. LAT is essential for FceRI-mediated mast cell activation. Immunity 12:525–535 [DOI] [PubMed] [Google Scholar]
- 34. Schroeder P., Popp R., Wiegand B., Altschmied J., Haendeler J. 2007. Nuclear redox-signaling is essential for apoptosis inhibition in endothelial cells—important role for nuclear thioredoxin-1. Arterioscler. Thromb. Vasc. Biol. 27:2325–2331 [DOI] [PubMed] [Google Scholar]
- 35. Smith J., et al. 2009. Retinoic acid induces nuclear accumulation of Raf1 during differentiation of HL-60 cells. Exp. Cell Res. 315:2241–2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tsujimura T., et al. 1996. Involvement of transcription factor encoded by the mi locus in the expression of c-kit receptor tyrosine kinase in cultured mast cells of mice. Blood 88:1225–1233 [PubMed] [Google Scholar]
- 37. Weinmann-Dorsch C., Grummt F. 1986. Diadenosine tetraphosphate (Ap4A) is compartmentalized in nuclei of mammalian cells. Exp. Cell Res. 165:550–554 [DOI] [PubMed] [Google Scholar]
- 38. Weis K. 2003. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell 112:441–451 [DOI] [PubMed] [Google Scholar]
- 39. Winward L., Whitfield W. G., Woodman T. J., McLennan A. G., Safrany S. T. 2007. Characterisation of a bis(5′-nucleosyl)-tetraphosphatase (asymmetrical) from Drosophila melanogaster. Int. J. Biochem. Cell Biol. 39:943–954 [DOI] [PubMed] [Google Scholar]
- 40. Yannay-Cohen N., et al. 2009. LysRS serves as a key signaling molecule in the immune response by regulating gene expression. Mol. Cell 34:603–611 [DOI] [PubMed] [Google Scholar]
- 41. Yoon S., Seger R. 2006. The extracellular signal-regulated kinase: multiple substrates regulate diverse cellular functions. Growth Factors 24:21–44 [DOI] [PubMed] [Google Scholar]