Abstract
The conserved RECQ5 DNA helicase is a tumor suppressor in mammalian cells. Defects in RECQ5 lead to the accumulation of spontaneous DNA double-stranded breaks (DSBs) during replication, despite the fact that these cells are proficient in DSB repair by homologous recombination (HR). The reason for this is unknown. Here, we demonstrate that these DSBs are linked to RNA polymerase II (RNAPII)-dependent transcription. In human RECQ5-depleted cells, active RNAPII accumulates on chromatin, and DNA breaks are associated with an RNAPII-dependent transcribed locus. Hence, transcription inhibition eliminates both active RNAPII and spontaneous DSB formation. In addition, the regulatory effect of RECQ5 on transcription and its interaction with RNAPII are enhanced in S-phase cells, supporting a role for RECQ5 in preventing transcription-associated DSBs during replication. Finally, we show that the SET2-RPB1 interaction (SRI) domain of human RECQ5 is important for suppressing spontaneous DSBs and the p53-dependent transcription stress response caused by the stalling of active RNAPII on DNA. Thus, our studies provide novel insights into a mechanism by which RECQ5 regulates the transcription machinery via its dynamic interaction with RNAPII, thereby preventing genome instability.
INTRODUCTION
Human RECQ proteins belong to the highly conserved family of DNA helicases and are named after the Escherichia coli ortholog, RecQ. Perhaps surprisingly, although E. coli and yeast, such as Saccharomyces cerevisiae, contain only one RecQ gene per organism, there are at least five RECQ helicases in humans. They are known as RECQ1, BLM, WRN, RECQ4, and RECQ5. Mutations in the human RECQ proteins lead to distinct clinical syndromes with various physical abnormalities (for a review, see reference 10). Therefore, these proteins have nonredundant functions to ensure proper development. So far, deficiencies in BLM, WRN, and RECQ4 have been associated with Bloom syndrome (BS), Werner syndrome, and Rothmund-Thomson syndrome, respectively. All these patients, in particular those with BS, have a high risk of developing cancers. To date, even though no clinical diseases have been linked to mutations in RECQ5, previously reported genetic studies demonstrated that recq5-deficient mice are highly cancer prone (12). Therefore, it is clear that RECQ5 also functions as a tumor suppressor in the mammalian system.
In mice, a recq5 deficiency leads to genome instability (11, 12). Specifically, embryonic stem cells and primary embryonic fibroblasts derived from recq5−/− mice show a high level of sister chromatid exchanges (SCEs), which are the result of RAD51-dependent homologous recombination (HR) to repair double-stranded breaks (DSBs). The elevated SCE frequency in recq5−/− cells is reminiscent of that observed for cells deficient in BLM, and this observation led to the proposal that RECQ5 and BLM share an antirecombination function (6, 12). Indeed, both BLM and RECQ5 dissociate RAD51 from DNA in vitro (6, 12), and this activity may explain the elevated SCE frequency observed in vivo. However, mouse recq5−/− blm−/− double mutants exhibit an even greater SCE frequency than does either the recq5−/− or the blm−/− single mutant (11), suggesting that RECQ5 has an additional nonredundant role that contributes to SCE formation.
In addition to the elevated-SCE phenotype, mouse recq5−/− cells also show an increase in spontaneous γH2AX foci, which have been widely used as a molecular marker for the presence of DNA breaks (12). Consistent with this observation, spontaneous DNA breaks also accumulate in Drosophila melanogaster recq5 mutant larvae (22). Because mouse recq5−/− cells positive for γH2AX foci also contain PCNA foci, these DNA lesions are thought to be associated with DNA replication (12). The elevated level of DSBs in recq5-deficient cells cannot be explained by a defect in DSB repair, as these cells are proficient in HR, a major pathway for repairing DSBs during replication (11, 12). Most likely, RECQ5 plays a direct role in preventing spontaneous DSB formation. To date, the nature of these DNA breakages remains unknown.
Perhaps most unexpectedly, even though genetic analyses suggest an intimate link between RAD51-dependent recombinational repair and RECQ5, recent biochemical studies revealed that RNA polymerase II (RNAPII) is the primary interacting protein for RECQ5 in human cells (2, 16). Importantly, the interaction with RNAPII is specific to RECQ5 but not other RECQ helicases (2), suggesting a unique role of RECQ5 in RNA metabolism. In agreement with a potential function of RECQ5 in transcription, sequence analyses have identified two regions within RECQ5 that share sequence homology with the protein domains known as the SET2-RPB1 interaction (SRI) and KIX domains, found in the transcription factors SET2 and CBP (4, 15). Nevertheless, the physiological relevance of the interaction with RNAPII for the function of RECQ5 in genome maintenance is not clear.
In the present work, we show that in human RECQ5-depleted cells, spontaneous DSBs accumulate during replication, and their formation depends on RNAPII-associated transcription. Furthermore, we demonstrate that RECQ5 regulates RNAPII on human chromatin and prevents spontaneous DNA breaks. The overexpression of a RECQ5 fragment lacking the SRI domain leads to an accumulation of the transcriptionally active form of RNAPII on human chromatin and further induces the p53-dependent transcriptional stress response. Thus, our data provide novel insights into the function of human RECQ5 in preventing transcription-associated genome instability.
MATERIALS AND METHODS
Cell culture, cell fractionation, FLAG immunoprecipitation, and siRNA.
Human 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (vol/vol) fetal bovine serum (FBS) and streptomycin-penicillin (100 U/ml). 293T cells stably expressing FLAG-tagged RECQ5 constructs were generated as previously described (2). Cell fractionation, FLAG immunoprecipitation (IP), and cell cycle synchronization were performed as described previously (2, 30). For UV treatment, cells were irradiated with 30 J/m2 UV light. For the synchronization of small interfering RNA (siRNA) knockdown cells, fluorescence-activated cell sorter (FACS) analysis was carried out by using a standard propidium iodide method. Twenty hours after siRNA transfection, nocodazole (Noc) (50 ng/ml) was added to the medium. After 20 h of incubation, cells were released by washing twice with complete DMEM. Cells were further incubated in complete DMEM for 3, 10, and 14 h before harvesting. For aphidicolin blocking, aphidicolin (5 μg/ml) was added 20 h after siRNA transfection. FACS analysis was carried out by using a standard propidium iodide method. siRNA knockdown cells were generated by using SMARTpool siRNAs (Dharmacon) for RECQ5 and BLM. siRNAs were transfected into 293T cells by using RNAiMax transfection reagent (Invitrogen) according to the manufacturer's protocol. For the 5,6-dichloro-beta-d-ribofuranosyl benzimidazole (DRB) treatment, cells were treated with 10 μM DRB for 12 h before harvesting.
Antibodies.
The antibodies (Abs) used were rabbit anti-FLAG polyclonal antibody (pAb) (Sigma), mouse anti-pSer2 monoclonal antibody (MAb) H5 (Abcam), mouse anti-RPB1 MAb A10 (Santa Cruz), goat antiactin (Santa Cruz), mouse anti-γH2AX MAb (Biolegend), mouse anti-phospho-p53 (Ser15P) 16G8 (Cell Signaling), goat anti-BLM (C-18; Santa Cruz), mouse anti-p53 MAb sc-126 (Santa Cruz), rat antibromodeoxyuridine (anti-BrdU) MCA2060 (AbD; Serotec), rhodamine-conjugated goat anti-mouse Ab, and DyLight 488-conjugated goat anti-rat Ab (Jackson ImmunoResearch). Mouse anti-pSer5 MAb 4H8 was generously provided by Jesper Svejstrup (Cancer Research UK). Mouse anti-PCNA MAb PC10 and rabbit anti-RAD51 pAb were gifts from Stephen West (Cancer Research UK). Rabbit anti-RECQ5 pAb was generated by using a synthetic peptide containing RECQ5651-660.
Plasmids.
The pCMV-FLAG-RECQ5 and pCMV-FLAG-RECQ51-542 constructs were generated as described previously (2). To generate pCMV-FLAG-RECQ51-668, pCMV-FLAG-RECQ5FL was cut with SmaI and XcmI and recircularized. To generate pCMV-FLAG-RECQ51-899, a PCR fragment produced using 5′-AACATGAGACATTCCGG-3′ and 5′-TTTTCTCGAGTTAAGCCGTGGGATTCAAGG-3′ was digested with XcmI and XhoI and cloned into XcmI-XhoI-digested plasmid pCMV-FLAG-RECQ5FL. To generate pCMV-FLAG-RECQ5451-991, a PCR fragment made using 5′-TTTTCTCGAGAGCAGCTGGAGCAAGACC-3′ and 5′-CCCGGCTCCATCTTCCTC-3′ was digested with EcoRV and XhoI and cloned into EcoRV-XhoI-digested plasmid pCMV-FLAG-RECQ5541-991. The four oligonucleotides used for site-directed mutagenesis to generate SMARTpool siRNA-resistant RECQ5 cDNA were 5′-GCTTTGATTCAGGACCAAGTGGACCACCTCCTGACGTTGAAAGTACGAGTAAGTTCCCTGAACTCGAAG-3′, 5′-CTGCGGCATTGTGTACTGCAGAACCCGCGAAGCCTGCGAACAGCTGGCCATAGAGCTC-3′, 5′-CAGGGCTGAAGGCCTCTGAAAGGACCTTAGTCCAAAATGACTGGATGGAGGAGAAGGTCC-3′, and 5′-TGGCAGGGATGGGAAGCCTTCGTGGTGTAGGTTGTACTACTCCAGGAATGACCGGGAC-3′.
Analysis of genomic DNA integrity.
High-molecular-weight fragments were analyzed as described previously (20, 24). Briefly, 48 h after siRNA transfection, 1 × 106 to 2 × 106 cells were collected, washed three times with cold phosphate-buffered saline (PBS), embedded in a low-melting-point agarose block, and lysed at 37°C for 48 h in lysis buffer (100 mM Tris-HCl [pH 8.5], 5 mM EDTA, 200 mM NaCl, 0.2% SDS, 0.2 mg/ml proteinase K). Blocks were inserted into a 0.5% agarose gel for standard agarose gel electrophoresis at 35 V for 48 h at 4°C. DNAs were visualized by ethidium bromide staining. For Southern blot analysis, EcoRI-digested genomic DNA (25 μg) was separated in 0.7% agarose gels in Tris-acetate-EDTA (TAE) buffer overnight at 35 V. The separated DNA was transferred onto Hybond N+ membranes overnight under alkaline conditions with buffer (0.4 N NaOH, 1 M NaCl). The membrane was incubated with prehybridization buffer (6× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA {pH 7.7}], 0.5% SDS, 5× Denhardt's solution, 100 μg/ml salmon sperm DNA) for 2 h at 42°C, followed by hybridization buffer (50% formamide, 6× SSPE, 0.5% SDS, 100 μg/ml salmon sperm DNA) containing 32P-labeled DNA oligonucleotides at 42°C overnight. The membranes were washed with 0.2% SDS and 0.1× SSPE and analyzed by radiography. The oligonucleotides used for Southern analysis were 5′-CGG CCC CTC CAT CGT CCA CCG CAA ATG CTT CTA GGC GGA CTA TGA CTT AGT TGC GTT ACA CCC TTT CTT GAC-3′ for ACTB, 5′-TTC CTC CCT TCC TCC AGA TCC CTT TGA AGT ACA GGT GAA AGC GGG CTG TGA GCT GCA TTC TGG AAA GAG CCC-3′ for CD1C, and 5′-ATC CGC TAA GGA GTG TGT AAC AAC TCA CCT GCC GAA TCA ACT AGC CCT GAA AAT GGA TGG CGC TGG AGC CGT C-3′ for ribosomal DNA (rDNA). For detecting centromeric DNA of chromosomes 13 and 21, PCR products generated from primers 5′-CTT CTG TCT AGA TTT TAG A-3′ and 5′-CAT AGA GAT GAA CAT GG-3′ were used as probes.
Neutral comet assays.
DNA breaks were analyzed by using neutral comet assays according to the manufacturer's instructions (Trevigen, United Kingdom). Briefly, after siRNA transfection and synchronization, cells were harvested and resuspended in PBS at a density of 1 × 105 cells/ml. Samples were fixed in low-melting-point agarose and spread evenly onto a precoated slide. Samples were allowed to dry at 4°C for 30 min, followed by cell lysis in prechilled lysis solution (Trevigen, United Kingdom) for 30 min at 4°C in the dark. Slides were then washed in 1× Tris-borate-EDTA (TBE) buffer and electrophoresed at a constant voltage of 1 V/cm for 30 min. Slides were immersed in 70% ethanol for 5 min, air dried, stained using SYBR green, and air dried for at least 24 h before viewing with a fluorescent microscope. The comet tail moment was recorded by randomly counting about 200 cells per slide using Comet Assay IV software. Two-tailed t tests were used to evaluate the significance of the data. P values of <0.05 were considered statistically significant (marked with asterisks in the figures).
Immunofluorescent microscopy.
For γH2AX immunostaining, cells were grown on coverslips for 1 to 2 days, washed twice with PBS, fixed in 3.7% paraformaldehyde for 10 min at room temperature, and permeabilized with 0.2% Triton X-100 for 10 min on ice. The coverslips were washed three times with PBS, followed by sequential incubations with 1 μg/ml of mouse anti-γH2AX antibody for 4 h at room temperature and 30 μg/ml of rhodamine-conjugated goat anti-mouse antibody for 1 h at room temperature in the dark. After the final wash with PBS, the coverslips were mounted in antifade solution (Invitrogen). For γH2AX and chlorodeoxyuridine (CIdU) double staining, cells grown on coverslips were incubated with 10 μM CIdU (Sigma) for 15 min, washed with PBS, fixed with 3% paraformaldehyde for 30 min, and permeabilized with 0.1% Triton X-100 for 30 min at room temperature. The cells were immunostained with mouse anti-γH2AX antibody and rhodamine-conjugated goat anti-mouse antibody as described above. The cells were further treated with ice-cold methanol for 15 min at −20°C and with 2 N HCl for 1 h at 37°C and washed three times with 0.1 M sodium borate (pH 8.5), followed by immunostaining with rat anti-BrdU antibodies (1:250) and DyLight 488-conjugated anti-rat antibodies (1:250) for 1 h each at room temperature.
Chromatin immunoprecipitation (ChIP).
Cells were fixed with 1% formaldehyde, incubated for 10 min, and neutralized with 1.25 M glycine for 5 min at room temperature. Cells were collected, resuspended in SDS lysis buffer (1% SDS, 10 mM EDTA [pH 8.0], 50 mM Tris [pH 8.1]), and incubated on ice for 10 min, followed by three 3-s sonications. After centrifugation at 13,000 rpm for 10 min at 4°C, the supernatant was collected and precleared with protein A-agarose. The supernatant was diluted with 10 volumes of IP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris [pH 8.0], 167 mM NaCl plus phenylmethylsulfonyl fluoride [PMSF]) and incubated overnight at 4°C with mouse anti-phospho-Ser5 (pSer5) (4H8), followed by incubation with protein A-agarose for 2 h at 4°C. After extensive washing, the bound protein-DNA complexes were eluted with elution buffer (0.1 M NaHCO3, 1% SDS). NaCl was added to the eluent to a final concentration of 0.2 M to reverse-cross-link at 65°C for 4 h. The sample was then treated with proteinase K (50 μg/ml) for 1 h at 45°C. DNA was concentrated by ethyl alcohol (EtOH) precipitation and purified (Qiagen PCR purification kit). PCRs were performed by using the following primers: 5′-CGT GGA CAT CCG CAA AGA CCT G-3′ and 5′-GCC GGC CTT GCA CAT GCC GGA-3′ for the 5′ untranslated region (5′UTR) and 5′-ACG AAA CTA CCT TCA ACT CCA TCA TG-3′ and 5′-TCC CTC CTC AGA TCA TTG CTC CTC C-3′ for the 3′UTR.
RESULTS
Replication-associated spontaneous DSBs in human recq5 cells.
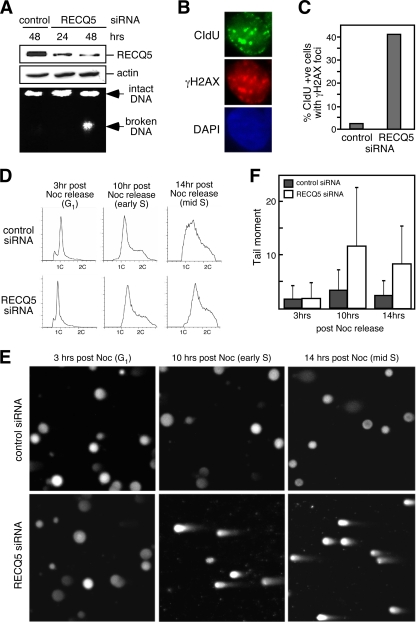
To study RECQ5 in human cells, we utilized siRNA to generate RECQ5-deficient 293T cells. We confirmed that 48 h after siRNA transfection, more than 80% of the RECQ5 proteins were depleted (Fig. 1A, top). To determine if human RECQ5 is also required for preventing spontaneous DSBs, chromosomes were isolated from the RECQ5-depleted cells and analyzed by standard agarose gel electrophoresis. Unlike the control knockdown cells, in which the majority of chromosomes remained intact and stayed in the well during electrophoresis, RECQ5 knockdown cells showed an accumulation of high-molecular-weight fragments (broken DNA) (Fig. 1A, bottom). Since these cells were not treated with DNA-damaging agents, these DNA lesions were spontaneous DNA breaks which likely arose from normal cellular processes such as DNA replication. Indeed, consistent with previously reported observations made for mouse recq5-deficient cells (12), we found that all the γH2AX focus-positive cells also contained CIdU-labeled foci, indicating that these cells were undergoing DNA synthesis (Fig. 1B). We further demonstrated that these γH2AX foci colocalized with the DNA synthesis foci, suggesting that the DNA breaks were formed at replication forks. In addition, γH2AX foci were found in approximately 40% of CIdU-positive cells after RECQ5 knockdown, a significant increase from the 5% found for the control knockdown cells (Fig. 1C). To further confirm that DSBs accumulate during S phase in the absence of RECQ5, cells were synchronized to the G2/M-phase transition by serum starvation, followed by nocodazole (Noc) treatment. FACS analysis indicated that at 3, 10, and 14 h after release from Noc blocking, cells were in G1, early S, and mid-S phases, respectively (Fig. 1D). Neutral comet assays revealed that broken chromosomes were present in RECQ5 knockdown cells during S phase but not G1 phase (Fig. 1E and F). Under the same conditions, control knockdown cells showed little accumulation of broken DNA during S phase.
Fig. 1.
Human RECQ5 prevents DNA breaks during S phase. (A) Human 293T cells were transfected with control or RECQ5 siRNA for 24 or 48 h. Expression levels of RECQ5 (top) or actin (center) were determined by Western blot analysis of whole-cell extracts. Chromosomal DNA integrity was analyzed by standard agarose gel electrophoresis (bottom). (B) Representation of RECQ5 knockdown cells with γH2AX foci (red) and CIdU replication foci (green). (C) Quantification of the percentage of CIdU-positive cells containing γH2AX foci transfected with either control or RECQ5 siRNA. (D) Flow cytometry of the control (top panels) and RECQ5 knockdown cells (bottom panels) 3, 10, or 14 h after release from nocodazole (Noc) treatment. (E) Representative images of the neutral comet assays using control and RECQ5 knockdown cells harvested at the 3rd, 10th, or 14th hour after Noc treatment. (F) Average tail moment measured from >200 cells per sample from panel E.
RECQ5 regulates RNAPII on human chromatin during S phase.
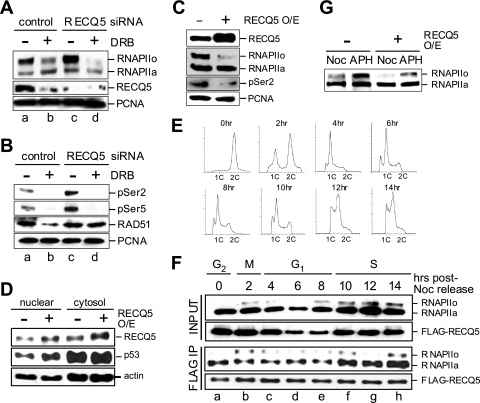
The timing of DSB formation coincides with DNA replication, suggesting that RECQ5 likely interacts with replication proteins to maintain the integrity of replication forks. However, recently reported biochemical analyses showed that RECQ5 predominately forms a stable protein complex with RNAPII in human cells (2, 16), and this interaction inhibits the in vitro RNAPII-dependent transcription reaction (3). To further elucidate the role of the RECQ5-RNAPII interaction in vivo, we analyzed the role of the RECQ5 deficiency in the RNAPII-dependent transcription cycle. During transcription activation, the promoter-bound RNAPII, known as RNAPIIa, is hyperphosphorylated by CDK7/9 at Ser2 and Ser5 of the C-terminal domain (CTD) repeats to generate active RNAPII (RNAPIIo). We found that in the absence of RECQ5, there was a significant increase in the level of RNAPII on human chromatin (Fig. 2A, compare lanes a and c). The increase of the RNAPIIo population was further confirmed by using anti-phospho-Ser2 (H5) and anti-phospho-Ser5 (4H8) antibodies (Fig. 2B, compare lanes a and c). Furthermore, the overexpression of RECQ5 by 2- to 4-fold in human 293T cells reduced the amount of RNAPII on DNA, and the inhibitory effect was most obvious for RNAPIIo (Fig. 2C). Since the inhibition of RNAPII-dependent transcription is known to cause p53 nuclear retention (for a review, see reference 21), we examined the distribution of p53 in cells overexpressing RECQ5 and observed an increase in the nuclear p53 level (Fig. 2D). These data together support a role for RECQ5 as a negative regulator for RNAPII in vivo.
Fig. 2.
Human RECQ5 regulates transcription in vivo. (A) 293T cells were first transfected with either control or RECQ5 siRNA for 36 h before the addition of the transcription inhibitor DRB for a further 12 h of incubation. Cells were fractionated to obtain the soluble chromatin-bound fraction. The amounts of chromatin-bound RNAPIIo, RNAPIIa, and RECQ5 were analyzed by Western blotting using rabbit anti-RPB1 (A10) and anti-RECQ5 pAbs. PCNA is shown as a loading control. (B) The levels of the chromatin-bound RNAPIIo in control and RECQ5 siRNA knockdown cells were further confirmed by Western blotting using mouse anti-phospho-Ser2 (H5) and anti-phospho-Ser5 (4H8). The amount of chromatin-bound RAD51 was detected by using rabbit anti-RAD51 pAb. PCNA is shown as a loading control. (C) The levels of the chromatin-bound RNAPIIo and RNAPIIa in 293T cells (left) and 293T cells with RECQ5 overexpression (O/E) (right) were analyzed by Western blotting using anti-RPB1 (A10) (second panel) and anti-phospho-Ser2 (H5) (third panel) antibodies. RECQ5 overexpression was confirmed by Western blotting using rabbit anti-RECQ5 pAb (top). (D) Western blot analysis of the nuclear and cytosolic p53 (second panel) from cells with or without RECQ5 overexpression (top). Actin is a control for protein loading. (E) Flow cytometry of 293T-FLAG-RECQ5 cells at different cell cycle stages after release from serum starvation and nocodazole double synchronization. (F) Western blot analysis of the total chromatin-bound RNAPII and FLAG-RECQ5 input (top panels) and the amount of RNAPII being coimmunopurified with FLAG-RECQ5 (bottom panels) at different cell cycle stages from panel E. (G) Western blot analysis of the chromatin-bound RNAPIIo and RNAPIIa from S-phase (aphidicolin [APH]) or M-phase (Noc) cells with or without RECQ5 overexpression.
While RECQ5 may have separate roles in transcription and replication, it is also possible that the involvement of RECQ5 in regulating RNAPII is linked to its function in suppressing DSBs during replication. To test the latter possibility, we analyzed the complex formation by RECQ5 and RNAPII during different cell cycle stages. We found that the interaction of RECQ5 with RNAPII can be detected throughout the cell cycle (Fig. 2E and F), consistent with the reduction in the amount of chromatin-bound RNAPIIo in cells overexpressing RECQ5 at both M and S phases (Fig. 2G). Interestingly, the interaction of RECQ5 with RNAPII was enhanced during S phase (Fig. 2F, third panel). This increase may be required for regulating the higher level of RNAPIIo molecules on DNA during S phase (Fig. 2F, top, and G). Together, our data demonstrate that during DNA replication, RECQ5 functions both in preventing DSB formation and in regulating RNAPII. Surprisingly, even though both RAD51 and PCNA were shown previously to interact with RECQ5 (12, 17), neither the depletion nor the overexpression of RECQ5 exhibited any significant effect on the chromatin binding of these proteins (Fig. 2A, B [compare lanes a and c], and C).
Spontaneous DSBs in human recq5 cells are linked to transcription.
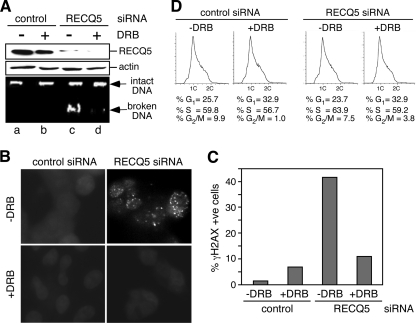
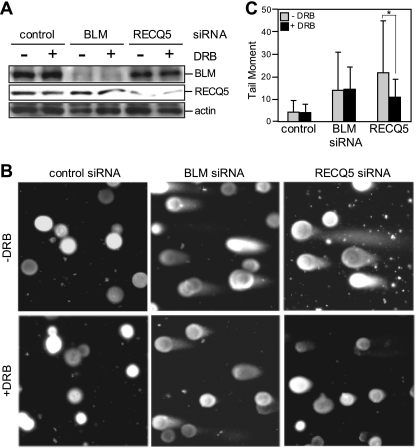
Given that the interaction of RECQ5 with RNAPII is enhanced during DNA replication, we next tested if the replication-associated DSBs found in the RECQ5-depleted cells were linked to transcription. For this, we examined the effect of the transcription inhibitor 5,6-dichloro-beta-d-ribofuranosyl benzimidazole (DRB) on RECQ5-dependent DSB accumulation. We discovered that transient incubation with DRB not only eliminated DNA-bound RNAPIIo (Fig. 2A and B, compare lanes c and d) but also led to a significant reduction in the amount of broken DNA (Fig. 3A, compare lanes c and d) and γH2AX foci (Fig. 3B and C). Since FACS analysis showed little change in the cell cycle distribution upon DRB treatment (Fig. 3D), this decrease in DSB accumulation was not due to the lower number of S-phase cells. To further confirm that the DRB effect is specific to RECQ5 knockdown cells, we analyzed the effect of DRB treatment on BLM-depleted cells by neutral comet assays (Fig. 4A). Indeed, even though a BLM deficiency also led to the accumulation of spontaneous DSBs, DRB treatment significantly reduced the tail moment only in RECQ5-depleted cells (P = 0.000596) but not control or BLM knockdown cells (Fig. 4B and C).
Fig. 3.
Transcription-dependent DSB formation in human recq5 cells. (A) The chromosomal DNA integrity of control and RECQ5 knockdown cells treated or not treated with DRB was analyzed by standard agarose gel electrophoresis. (B) Immunofluorescent detection of γH2AX foci in control (left panels) and RECQ5 knockdown (right panels) cells with or without DRB treatment. The images shown in all panels except RECQ5 siRNA without DRB treatment are overexposed to show the background signal. (C) Quantification of the percentage of γH2AX-positive cells from panel B. More than 150 cells were analyzed per sample. (D) Flow cytometry of control (left panels) and RECQ5 knockdown (right panels) cells treated or not treated with DRB.
Fig. 4.
RNAPII-dependent genome instability is associated with RECQ5 but not BLM. (A) Western blot analysis of the BLM and RECQ5 protein levels in cells with and without DRB treatment. (B) Representative images for the neutral comet assays using control and BLM and RECQ5 knockdown cells with and without DRB treatments, as shown in panel A. (C) Average tail moment measured from >200 cells per sample from B. *, P = 0.000596.
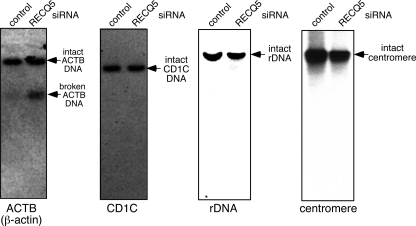
Based on our data, we expected that an actively transcribed gene locus would be more prone to DNA breaks than a nontranscribed chromosomal region. To test this hypothesis, we carried out a Southern blot analysis to assess the integrity of the ACTB (β-actin) gene locus, as it was previously reported that the ACTB mRNA level is elevated in human RECQ5-depleted cells (16). A restriction-digested genomic DNA containing the intact ACTB gene from both control and RECQ5 knockdown cells was recognized by a 32P-labeled oligonucleotide containing the ACTB sequence (Fig. 5, left). However, in addition to the intact ACTB DNA, fragmented ACTB DNA was also detected in the RECQ5 knockdown cells. In contrast, DNA fragmentation was not seen in either the transcriptionally silent CD1C gene locus or the rDNA locus, which depends on RNA polymerase I for transcription (Fig. 5, second and third panels) (for a review of tissue-specific CD1C gene expression, see reference 26). In addition, noncoding regions, such as centromeres, remain intact in RECQ5 knockdown cells (Fig. 5, right). Therefore, these data together allow us to conclude that in the absence of RECQ5, RNAPII-dependent transcription contributes to spontaneous DSB formation.
Fig. 5.
DSB analysis of the transcribed and nontranscribed chromosomal regions in human RECQ5 knockdown cells. Shown are data from Southern blot analyses of the EcoRI-digested genomic DNA from control and RECQ5 knockdown 293T cells. After Southern transfer, one membrane was first probed with a 32P-labeled ACTB oligonucleotide (left), stripped thoroughly, and then reprobed with CD1C oligonucleotide (second panel). Separate membranes containing EcoRI-digested DNA were probed with rDNA or centromere oligonucleotides (right two panels). The EcoRI site is absent in ACTB gene, CD1C gene, and rDNA repeats.
It is interesting that there appears to be a specific DNA break site in the ACTB gene locus, resulting in a defined ACTB DNA fragment in the Southern blot analysis. Based on the size of the fragmented ACTB DNA, we estimated that the break site was located close to the 3′UTR of the ACTB gene. This observation is reminiscent of the transcription-dependent DNA break found near the 3′ end of the ACTB gene due to R-loop formation in chicken DT40 cells deficient in the ASF splicing factor (20). To test if this DNA break is due to a specific transcriptional block near the 3′UTR, we carried out a ChIP analysis to determine the distribution of RNAPIIo at the 5′UTR and 3′UTR in RECQ5-depleted cells. We found that while there was an increase in the RNAPIIo level at both the 5′UTR and 3′UTR of the ACTB gene in RECQ5 knockdown cells, the increase was more significant at the 5′UTR (4-fold increase, compared to 2-fold at the 3′UTR) (see Fig. S1 in the supplemental material), suggesting that there may be a sequence-dependent transcriptional block near the 3′UTR of the ACTB gene, making this region susceptible to genome instability. If this were the case, one would expect that not all transcribed genes are prone to DNA breaks. Indeed, no significant DNA breaks were detected in the highly transcribed LDHA and RPL19 genes (data not shown). This is consistent with the fact that not all RNAPII molecules are associated with RECQ5, and there is likely a specific subset of gene loci that are regulated by RECQ5. Further detailed genome-wide analyses will be necessary to identify this set of genes.
The SRI domain of RECQ5 prevents the accumulation of RNAPIIo on chromatin.
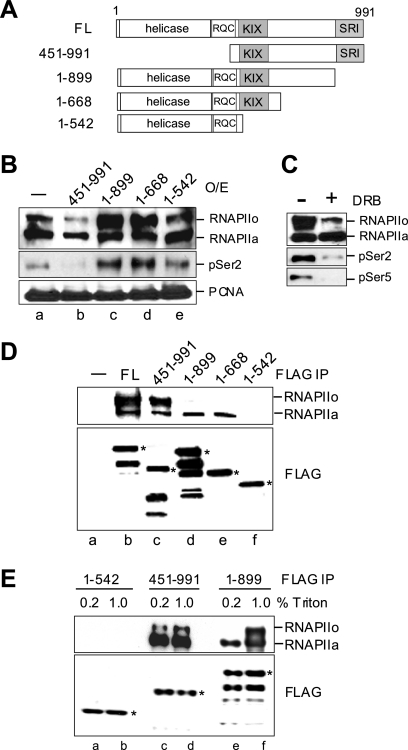
We next wished to determine which domains of RECQ5 are required for transcription suppression. For this, we overexpressed different RECQ5 fragments in 293T cells and analyzed the effect of each overexpression on the level of chromatin-bound RNAPIIo (Fig. 6A). We have shown that the overexpression of the full-length (FL) RECQ5 protein led to a decrease in the level of the chromatin-bound RNAPIIo (Fig. 2C). Here, we found that the overexpression of the RECQ5 fragment at positions 451 to 991 (RECQ5451-991), which interacts with both RNAPIIo and RNAPIIa (Fig. 6D, lane c), had the same effect (Fig. 6B, compare lanes a and b). On the other hand, the overexpression of RECQ51-542, which contains the helicase domain but does not interact with RNAPII (Fig. 6D, lane f) (2), did not affect the level of chromatin-bound RNAPIIo (Fig. 6B, lane e). This result indicates that the interaction with RNAPII, but not the conserved helicase domain of RECQ5, is required for transcription regulation in vivo. Perhaps most unexpectedly, the overexpression of either RECQ51-668 or RECQ51-899 led to a significant accumulation of RNAPIIo on DNA, suggesting that the last 100 amino acids of RECQ5 are crucial for transcription regulation (Fig. 6B, lanes c and d). These RNAPII molecules that accumulated on DNA were recognized by both phospho-Ser2 (pSer2) and phospho-Ser5 (pSer5) antibodies and could be suppressed by the addition of DRB, confirming that they were indeed RNAPIIo phosphorylated by CDK7/9 (Fig. 6C).
Fig. 6.
The SRI domain of RECQ5 is required for regulation and interaction with RNAPIIo. (A) Schematic diagram of the full-length (FL) RECQ5 and RECQ5 fragments. The conserved helicase and RecQ C-terminal (RQC) domains are shown. Also, the RNAPII interaction domains KIX and SRI are shown in gray. (B) Chromatin-bound RNAPIIo, RNAPIIa, and PCNA from cells overexpressing different RECQ5 fragments were analyzed by Western blotting. (C) Chromatin-bound RNAPIIo and RNAPIIa from 293T cells overexpressing RECQ51-899 with or without DRB treatment were analyzed by Western blotting. (D) FL FLAG-RECQ5 and fragments were immunopurified from 293T cells. The presence of RNAPII (top) and FLAG proteins (bottom) in the immunopurified RECQ5 complexes was analyzed by Western blotting using anti-RPB1 antibody A10 and anti-FLAG MAb. The asterisks indicate the expected FLAG-RECQ5 proteins on SDS-PAGE gels. (E) FLAG-RECQ5 fragments were immunopurified from 293T cells in the presence of 0.2 or 1.0% Triton X. The copurified RNAPII was analyzed by Western blotting using anti-RPB1 antibody A10. The asterisks indicate the expected FLAG-RECQ5 proteins on SDS-PAGE gels.
The SRI domain is required for the stable interaction of RECQ5 with RNAPIIo.
Interestingly, recently reported sequence analyses revealed that the C-terminal end of RECQ5 shares sequence homology with the SRI domain of the Set2 histone methyltransferase, which is known to play a role in transcription (4, 15). Since Set2 interacts specifically with RNAPIIo via its SET2 domain (19), the lack of the SRI domain in RECQ51-668 or RECQ51-899 likely alters their interactions with RNAPII. Consistent with this hypothesis, we found that only RNAPIIa, but not RNAPIIo, was copurified with RECQ51-668 and RECQ51-899 from 293T cell extracts (Fig. 6D, lanes d and e). Interestingly, in contrast to our observation, recent reports demonstrated the copurification of RNAPIIo with a similar RECQ5 fragment lacking the SRI domain (15, 16, 18). Upon closer examination, we found that a significantly higher concentration of detergent was present during the FLAG immunoprecipitation procedure in those previous reports. Indeed, we found that a high concentration of detergent (1% Triton), which is conducive for charge-charge interactions, promoted the interaction of RNAPIIo with RECQ51-899 (Fig. 6E, compare lanes e and f). Since CTD hyperphosphorylation significantly alters the charge environment of RNAPII, it is possible that the SRI domain has a special role in stabilizing the interaction of RECQ5 with the highly negatively charged RNAPIIo. Previously, we identified that the central region of RECQ5 is required for the interaction with both RNAPIIo and RNAPIIa (2, 3). This region was later shown to exhibit sequence homology with the KIX domain, often found in transcription factors such as CBP (15). Here, we found that a high concentration of Triton has no effect on RECQ51-542 (Fig. 6E, compare lanes a and b), which lacks both KIX and SRI domains (Fig. 6A). This result indicates that the hydrophobic environment created by detergent replaces the function of the SRI domain in stabilizing the interaction between the KIX domain of RECQ5 and the highly negatively charged RNAPIIo. Hence, we conclude that RECQ5 interacts differentially with RNAPIIo and RNAPIIa via the SRI domain. Importantly, the weakened interaction with RNAPIIo due to the lack of the SRI domain most likely contributes to the accumulation of chromatin-bound RNAPIIo in cells overexpressing RECQ51-668 or RECQ51-899 (Fig. 6B, lanes c and d).
The SRI domain is important for preventing the transcription-associated stress response and genome instability.
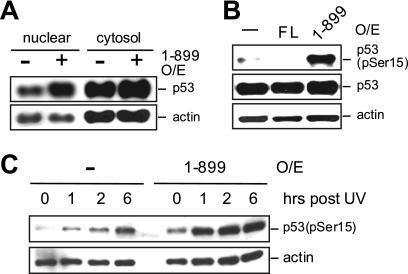
The elevated level of chromatin-bound RNAPIIo suggests an upregulation of transcription in cells overexpressing RECQ51-899. However, similarly to FL RECQ5 (Fig. 2D), the overexpression of RECQ51-899 also induced the nuclear retention of p53 (Fig. 7A). While the increase in the level of nuclear p53 could be a consequence of transcription inhibition prior to the initiation step, it was shown that a transcription blockage that stalls RNAPIIo at the elongation step also causes p53 to accumulate in the nucleus (for a review, see reference 21). Nevertheless, only inhibition at the elongation step, but not prior to transcription initiation, further leads to the ATR-dependent phosphorylation of p53 at Ser15 (7). Interestingly, we found that the overexpression of RECQ51-899, but not FL RECQ5, induced p53 phosphorylation at Ser15 (Fig. 7B). In addition, UV radiation, which generates DNA damage known to stall RNAPIIo during transcription, further exacerbated the p53-dependent transcription stress response in cells overexpressing RECQ51-899 (Fig. 7C). Therefore, these data together indicate that the increased level of chromatin-bound RNAPIIo in cells overexpressing RECQ51-899 is a result of transcription elongation blockage leading to the accumulation of stalled RNAPIIo molecules on DNA.
Fig. 7.
The SRI domain of human RECQ5 prevents the p53-induced transcription stress response. (A) Western blot analysis of nuclear and cytosolic p53 from cells with or without overexpression of RECQ51-899. Actin is a control for protein loading. (B) Western blot analysis of the phosphorylated p53 in whole-cell extracts obtained from the parental 293T cells and cells overexpressing either FL RECQ5 or RECQ51-899. (C) Western blot analysis of phosphorylated p53 at different time points after UV radiation from cells with or without an overexpression of RECQ51-899.
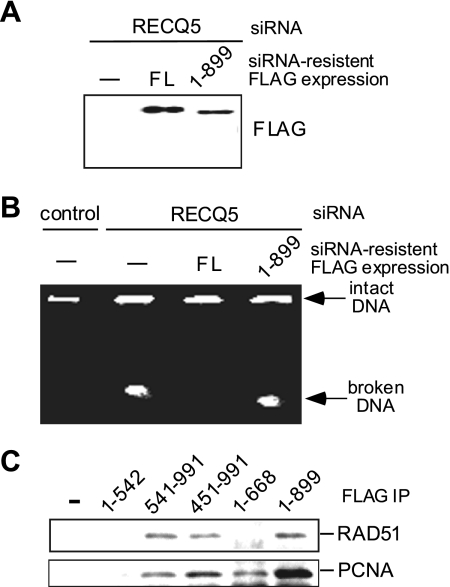
Given that the expression of RECQ51-899 leads to RNAPIIo accumulation on DNA and the p53-dependent stress response, we next examined the ability of RECQ51-899 to suppress DSB formation in RECQ5 knockdown cells. As expected, the expression of an siRNA-resistant FL FLAG-RECQ5 construct efficiently inhibited DSB formation in RECQ5 knockdown cells (Fig. 8A and B). On the other hand, broken chromosomes were still present in RECQ5 knockdown cells complemented with an siRNA-resistant FLAG-RECQ51-899 construct. We further confirmed that the inability of RECQ51-899 to complement the DSB phenotype was not due to a defect in previously known interactions of RECQ5, such as those with PCNA and RAD51 (Fig. 8C) (17, 25). Our analyses together demonstrate the importance of the SRI domain of human RECQ5 in maintaining genome stability during transcription.
Fig. 8.
The SRI domain of human RECQ5 prevents transcription-associated DNA breaks. (A) Western blot analysis of the expressions of the siRNA-resistant FL FLAG-RECQ5 and FLAG-RECQ51-899 in 293T RECQ5 knockdown cells. (B) The chromosomal integrities of the control (left lane) and RECQ5 knockdown cells (second lane from the left) were compared to those of RECQ5 knockdown cells complemented with either siRNA-resistant FL FLAG-RECQ5 FL (third lane from the left) or FLAG-RECQ51-899 (right lane). (C) Western blot analysis of the presence of RAD51 (top) and PCNA (bottom) in the FLAG-immunoprecipitated complexes from cells expressing FLAG-RECQ5 fragments from 293T cells.
DISCUSSION
Even though transcription is required for cell viability, this essential cellular process is known to trigger DNA lesions in various organisms (1, 5, 8, 9, 14, 20, 27, 28, 31). These transcription-initiated DNA lesions play a role in causing genome instability during cell growth and should be avoided. In this work, we demonstrate the critical connection between a RECQ5 deficiency and transcription-associated DSBs. We show that the DSBs formed during DNA replication depend on transcription in human recq5-deficient cells, and this observation provides the first functional link between the roles of RECQ5 in genome maintenance and transcription. The increasing amount of transcription-associated DSBs during replication is expected to be repaired by HR. Therefore, it is possible that the elevated SCE frequency in recq5 cells is a consequence of the increase in numbers of HR events to repair these spontaneous DSBs.
Current models for transcription-associated genome instability suggest that this phenomenon is intimately linked to DNA replication (5, 8, 9, 27, 28). For example, a head-on collision between transcription machinery and a replication fork during S phase may cause the fork to stall and collapse, leading to DSB formation. Consistent with the collision model, in bacteria, transcription in the opposite direction of the replication fork slows down the replication elongation rate compared to co-oriented transcription and replication (28). Alternatively, an extensive RNA-DNA hybrid, or R loop, formed during transcription can initiate recombination possibly by stalling replication forks or triggering single-stranded DNA breaks (SSBs) at the nontranscribed strand. These SSBs can then be converted to DSBs at replication forks. Indeed, genetic evidence from previously reported studies of the THO transcription elongation complex and ASF/SF2 splicing factor indicates that cells have evolved a defense mechanism to protect DNA integrity during transcription by regulating R-loop formation (1, 14, 20). It was further proposed that the function of ASF/SF2 in R-loop suppression depends on topoisomerase I (27). Interestingly, mammalian recq5 cells show hypersensitivity to the topoisomerase I inhibitor camptothecin but not to other DNA-damaging agents (13). It is yet to be determined if the camptothecin hypersensitivity in the absence of RECQ5 relates to its function in preventing transcription-associated genome instability.
While spontaneous DNA breaks associated with a RECQ5 deficiency depend on transcription, we found that not all transcribed genes are equally prone to DNA breaks in these cells. One possible explanation for this finding is that if RECQ5 functions to inhibit transcription near active replication sites to avoid transcription-replication collision, perhaps only transcription in the opposite orientation relative to the replication fork is subject to RECQ5 regulation. Alternatively, if extensive R-loop formation is responsible for the spontaneous DSBs in recq5 cells, only genes containing a high G density may be prone to DNA breaks (23). A comprehensive genome-wide analysis of the RNAPII distribution, replication pulse sites, and DNA break sites in recq5 cells will be necessary to address this question.
During the course of evolution, not only has the RecQ gene amplified to multiple homologs in humans, but different RECQ helicases have also acquired distinct protein domains that reflect their nonredundant cellular functions. Indeed, the N terminus of human RECQ4, which shares a limited homology with the yeast Sld2 replication factor, physically links RECQ4 to the MCM replicative helicase complex to function in DNA replication (30). RECQ5, on the other hand, acquires two protein domains, KIX and SRI, found in enzymes involved in RNA metabolism (4, 15). We show that the SRI domain of human RECQ5 is critical for maintaining genome integrity by preventing the accumulation of DNA-bound RNAPIIo and DSBs. Interestingly, the exogenous expression of RECQ51-899 lacking the SRI domain exhibits a dominant negative effect over wild-type RECQ5 in regulating RNAPII. It is possible that in normal cells, the interaction of RNAPII with wild-type RECQ5 prevents the transition of RNAPII from the promoter-bound RNAPIIa to active RNAPIIo. However, when RECQ51-899 is also present in cells, RECQ51-899 may compete with wild-type RECQ5 to interact with RNAPIIa but may fail to prevent transcription initiation, leading to aberrant transcription elongation and RNAPIIo stalling on DNA. Further structural analyses will likely provide important molecular insights into the regulation of RNAPII by the SRI domain of RECQ5 to prevent transcription-associated genome instability.
Interestingly, even though the interaction between RECQ5 and RNAPII is conserved in chicken DT40, the RECQ5 deletion in these cells exhibits little of the genomic instability phenotype (15, 29). Unlike mammalian RECQ5, the SRI domain of chicken RECQ5 plays no role in genome maintenance, and an elevated SCE frequency is seen only for the recq5−/− blm−/− double mutant but not the recq5−/− single mutant. Since only RECQ5 but not BLM associates with RNAPII (2, 15), it is not clear if the increased SCE frequency found for the DT40 double mutant is related to the function of RECQ5 in transcription. Nevertheless, HR-dependent DSB repair can lead to a non-SCE outcome; as such, DSB formation may not necessarily result in an increase in SCE frequency. Alternatively, it is also possible that redundant pathways to RECQ5 in transcription regulation may exist in chicken DT40 cells. As a result, the RECQ5 deficiency has little consequence for genome stability in these cells.
In summary, our data elucidating the critical connection between RECQ5 deficiency and transcription-associated DSBs in human cells suggest that this is a highly complex defense mechanism involving not only RNA-processing factors but also enzymes that are better known for their link to DNA repair. Importantly, we unveil the dynamic interactions of the two distinct domains of RECQ5 with RNAPII that ultimately govern the ability of RECQ5 to regulate RNAPII and suppress transcription-associated DNA breaks.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Leslie Warner postdoctoral fellowship to M.L. Y.L. is supported by grants from the NIH, Yale Cancer Center, Breast Cancer Alliance, and Elsa U. Pardee Foundation.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 14 March 2011.
REFERENCES
- 1. Aguilera A. 2002. The connection between transcription and genomic instability. EMBO J. 21:195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aygün O., Svejstrup J., Liu Y. 2008. A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc. Natl. Acad. Sci. U. S. A. 105:8580–8584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aygün O., et al. 2009. Direct inhibition of RNA polymerase II transcription by RECQL5. J. Biol. Chem. 284:23197–23203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aygün O., Svejstrup J. Q. 2010. RECQL5 helicase: connections to DNA recombination and RNA polymerase II transcription. DNA Repair (Amst.) 9:345–353 [DOI] [PubMed] [Google Scholar]
- 5. Azvolinsky A., Giresi P. G., Lieb J. D., Zakian V. A. 2009. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol. Cell 34:722–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bugreev D. V., Yu X., Egelman E. H., Mazin A. V. 2007. Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev. 21:3085–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Derheimer F. A., et al. 2007. RPA and ATR link transcriptional stress to p53. Proc. Natl. Acad. Sci. U. S. A. 104:12778–12783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gottipati P., Cassel T. N., Savolainen L., Helleday T. 2008. Transcription-associated recombination is dependent on replication in mammalian cells. Mol. Cell. Biol. 28:154–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gottipati P., Helleday T. 2009. Transcription-associated recombination in eukaryotes: link between transcription, replication and recombination. Mutagenesis 24:203–210 [DOI] [PubMed] [Google Scholar]
- 10. Hickson I. D. 2003. RecQ helicases: caretakers of the genome. Nat. Rev. Cancer 3:169–178 [DOI] [PubMed] [Google Scholar]
- 11. Hu Y., et al. 2005. RecQL5 and Blm RecQ helicases have non-redundant roles in suppressing crossovers. Mol. Cell. Biol. 25:3431–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu Y., et al. 2007. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 21:3073–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hu Y., Lu X., Zhou G., Barnes E. L., Luo G. 2009. Recql5 plays an important role in DNA replication and cell survival after camptothecin treatment. Mol. Biol. Cell 20:114–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huertas P., Aguilera A. 2003. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell 12:711–721 [DOI] [PubMed] [Google Scholar]
- 15. Islam M. N., Fox III D., Guo R., Enomoto T., Wang W. 2010. RecQL5 promotes genome stabilization through two parallel mechanisms—interacting with RNA polymerase II and acting as a helicase. Mol. Cell. Biol. 30:2460–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Izumikawa K., et al. 2008. Association of human DNA helicase RecQ5beta with RNA polymerase II and its possible role in transcription. Biochem. J. 413:505–516 [DOI] [PubMed] [Google Scholar]
- 17. Kanagaraj R., Saydam N., Garcia P. L., Zheng L., Janscak P. 2006. Human RECQ5beta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 34:5217–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kanagaraj R., et al. 2010. RECQ5 helicase associates with the C-terminal repeat domain of RNA polymerase II during productive elongation phase of transcription. Nucleic Acids Res. 38:8131–8140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kizer K. O., et al. 2005. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell. Biol. 25:3305–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li X., Manley J. L. 2005. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122:365–378 [DOI] [PubMed] [Google Scholar]
- 21. Ljungman M. 2007. The transcription stress response. Cell Cycle 6:2252–2257 [DOI] [PubMed] [Google Scholar]
- 22. Nakayama M., et al. 2009. Loss of RecQ5 leads to spontaneous mitotic defects and chromosomal aberrations in Drosophila melanogaster. DNA Repair (Amst.) 8:232–241 [DOI] [PubMed] [Google Scholar]
- 23. Roy D., Lieber M. R. 2009. G clustering is important for the initiation of transcription-induced R-loops in vitro, whereas high G density without clustering is sufficient thereafter. Mol. Cell. Biol. 29:3124–3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sakahira H., Enari M., Ohsawa Y., Uchiyama Y., Nagata S. 1999. Apoptotic nuclear morphological change without DNA fragmentation. Curr. Biol. 9:543–546 [DOI] [PubMed] [Google Scholar]
- 25. Schwendener S., et al. 2010. Physical interaction of RECQ5 helicase with RAD51 facilitates its anti-recombinase activity. J. Biol. Chem. 21:15739–15745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sugita M., Brenner M. B. 2000. T lymphocyte recognition of human group 1 CD1 molecules: implications for innate and acquired immunity. Semin. Immunol. 12:511–516 [DOI] [PubMed] [Google Scholar]
- 27. Tuduri S., et al. 2009. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell Biol. 11:1315–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang J. D., Berkmen M. B., Grossman A. D. 2007. Genome-wide coorientation of replication and transcription reduces adverse effects on replication in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 104:5608–5613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang W., et al. 2003. Functional relation among RecQ family helicases RecQL1, RecQL5, and BLM in cell growth and sister chromatid exchange formation. Mol. Cell. Biol. 23:3527–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu X., Rochette P. J., Feyissa E. A., Su T. V., Liu Y. 2009. MCM10 mediates RECQ4 association with MCM2-7 helicase complex during DNA replication. EMBO J. 28:3005–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yu K., Chedin F., Hsieh C. L., Wilson T. E., Lieber M. R. 2003. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 4:442–451 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.