Abstract
DNA damage activates signaling pathways that lead to modification of local chromatin and recruitment of DNA repair proteins. Multiple DNA repair proteins having ubiquitin ligase activity are recruited to sites of DNA damage, where they ubiquitinate histones and other substrates. This DNA damage-induced histone ubiquitination is thought to play a critical role in mediating the DNA damage response. We now report that the polycomb protein BMI1 is rapidly recruited to sites of DNA damage, where it persists for more than 8 h. The sustained localization of BMI1 to damage sites is dependent on intact ATM and ATR and requires H2AX phosphorylation and recruitment of RNF8. BMI1 is required for DNA damage-induced ubiquitination of histone H2A at lysine 119. Loss of BMI1 leads to impaired repair of DNA double-strand breaks by homologous recombination and the accumulation of cells in G2/M. These data support a crucial role for BMI1 in the cellular response to DNA damage.
INTRODUCTION
The induction of a DNA break leads to activation of multiple signaling pathways that lead to local modification of chromatin structure and recruitment of DNA repair complexes (18, 22, 55). Histone H2AX is rapidly phosphorylated near sites of DNA breaks by ATM, ATR, and DNA-PK (39, 54) and can spread to encompass a region of chromatin covering several megabases (40, 41).
H2AX phosphorylation facilitates the recruitment of other proteins, including MDC1 (52) and the E3 ubiquitin ligases RNF8 and RNF168, which in turn participate locally in the K63-linked polyubiquitination of histones H2A and H2AX (23, 32, 50, 51). Polyubiquitinated K63-linked histones provide a recognition element that recruits RAP80 through its ubiquitin interaction motifs (28, 49, 56). RAP80 can then promote the recruitment of other DNA repair factors such as BRCA1 and Abraxas, which are essential for efficient repair. RNF8 and RNF168 function are also required for proper localization of 53BP1, although the exact mechanism is unclear (12, 23, 32, 51). 53BP1 recruitment to regions of DNA damage is dependent upon its Tudor domains, which have been found to specifically interact with methylated histone residues (6, 24, 42). A model has been proposed in which RNF8- and RNF168-mediated ubiquitination of histones confers local changes in chromatin structure, leading to exposure of methylated lysine residues in core histones, allowing the subsequent recruitment of 53BP1 (50). Enzymes involved in deubiquitination, such as BRCC36, USP3, and USP28, are also critical for efficient DNA repair, demonstrating that a dynamic regulation of ubiquitin conjugation and hydrolysis is necessary for optimal DNA repair (37, 46, 47, 61).
Polycomb group proteins BMI1 and RING1B/RNF2 form an active heterodimer E3 ligase that catalyzes the monoubiquitination of histone H2A at Lysine 119. (7, 8, 44, 53, 57). This activity is important for BMI1-mediated transcriptional silencing during organism development and cellular differentiation (27, 48, 58). Ubiquitination of H2A at lysine 119 is also induced locally at sites of DNA damage, both at sites of UV lesions and double-strand breaks (DSBs) (4, 33, 59, 62). Since H2A lysine 119 ubiquitination is central to epigenetic regulation during both development and DNA repair, this raises the hypothesis that polycomb group proteins may play a role in DNA repair response. Consistent with this hypothesis, Ring1B/RNF2 has been shown to be required for UV damage-induced H2A-K119 ubiquitination (4), and loss of BMI1 is associated with activation of the DNA repair response and checkpoint function (29).
We report here that BMI1 is directly recruited to the sites of DNA DSBs, where it persists for more than 8 h. The sustained localization of BMI1 to sites of damage is dependent upon ATR/ATM, H2AX phosphorylation, and RNF8 recruitment. BMI1 is required for postdamage ubiquitination of histone H2A at lysine 119 and contributes to efficient homology-mediated repair of DNA breaks. These data implicate polycomb group proteins as part of the ubiquitin ligase cascade involved in DSB-associated histone ubiquitination and support a role for BMI1 in DNA damage response.
MATERIALS AND METHODS
Cell lines and chemicals.
HeLa cells were obtained from the American Type Culture Collection (ATCC; Rockville, MD). Bmi1+/+; Ink4a−/−; Arf−/− and Bmi1−/−; Ink4a−/−; Arf−/− murine embryonic fibroblasts (MEFs) were a gift from Maarten van Lohuizen. Seckel cells were obtained from Coriell Cell Repository (Camden, NJ). H2AX−/− MEFs were a gift from Andre Nussenzweig, 53BP1−/− MEFs were a gift from Junjie Chen, and RNF8−/− MEFs were a gift from Xiaochun Yu. Brca1−/−; p53−/− breast cancer cells and Brca1+/+; p53−/− breast cancer cells were a gift from Jos Jonkers. ATM kinase inhibitor (ATMi; KU-55933) used was obtained from Astra-Zeneca/Kudos Pharmaceuticals (Cambridge, United Kingdom). Aphidicolin was obtained from Sigma-Aldrich (catalog no. A0781).
UVA laser scissors.
For laser scissors analyses, cells were grown at 37°C and 5% CO2 in Lab-Tek chamber slides (Nalge-Nunc International, Naperville, IL) and treated with 10 μM 5-iodo-2-deoxyuridine (Sigma, St. Louis, MO) for 24 h prior to laser irradiation. For micro-irradiation, a LabTek chamber with cells was mounted on the stage of a Zeiss Axiovert 200 microscope integrated with the a PALM Microlaser workstation (PALM Laser Technologies, Bernried, Germany). The cells were visualized under visible light, laser-targeted nuclei were selected using the supplied software (PALM Robo v3.2), and the nuclei were subsequently irradiated with a pulsed solid-state UVA laser (30 Hz, 337 nm) coupled to the bright-field path of the microscope focused through an LD 40× objective lens to yield a spot size of ∼1 μm. The operation was assisted by using the PALM Robo v3.2 software supplied by the manufacturer. The laser output was set to 50%, which was the lowest power that reproducibly gave a focused pH2AX/53BP1-positive nuclear stripe. Typically, an average of 100 cells were micro-irradiated within 2 to 5 min, and each cell/nucleus was exposed to the laser beam for less than 500 ms. Striped cells were immediately returned to a 37°C and 5% CO2 incubator and fixed with CSK buffer-4% paraformaldehyde 15 to 30 min later.
Immunofluorescence.
Cells were washed twice in phosphate-buffered saline (PBS) at room temperature, fixed in 4% paraformaldehyde for 10 min, and permeabilized with 0.05% Triton X-100 in PBS for 10 min. Samples were preincubated with 5% goat serum in PBS for 15 min and then incubated with primary antibodies (see Table S1 in the supplemental material) diluted in PBS and 5% goat serum. The cells were then washed twice with PBS and incubated for 15 min at room temperature in PBS and 5% goat serum before a further incubation for 60 min at room temperature in the dark with the appropriate secondary antibody fluorescein isothiocyanate- and TRITC (tetramethyl rhodamine isothiocyanate)-labeled anti-rabbit or anti-mouse antibody (Jackson Immunoresearch Laboratories) at 1:200. Digital images were obtained with a Nikon eclipse 80i fluorescence microscope. For the uH2A immunostaining, cells were permeabilized prior to fixation by in cytoskeletal (CSK) buffer (100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 10 mM PIPES [pH 6.8]) for 30 s, followed by 30 s in CSK buffer containing 0.5% Triton X-100 and 30 s in CSK buffer without Triton X-100, all on ice. Quantitation of the number of DNA-damage induced foci was performed by calculating the average number of BMI1 foci that colocalized with H2AX foci per cell in each condition analyzed. For quantitation of the laser scissors experimental results, the percentages of cells treated with laser scissors showing a linear region of focal enhancement of signal (“stripes or tracks”) that colocalize with pH2AX or 53BP1 signal were calculated. The data represent the averages and standard errors of three independent experiments, scoring at least 50 cells each.
Chromatin immunoprecipitation (ChIP) assays.
For each experimental condition, HeLa cells growing in 10-cm dishes were treated with 1% formaldehyde for 10 min to cross-link proteins to DNA. Glycine (0.125 M) was added to quench the reaction. The cells were scraped, washed twice in cold PBS, washed for 10 min in solution I (10 mM HEPES [pH 7.5], 10 mM EDTA, 0.5 mM EGTA, 0.75% Triton X-100), and 10 min in solution II (10 mM HEPES [pH 7.5], 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA). The cells were then resuspended in lysis buffer containing protease inhibitors (25 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS], 0.5% deoxycholate) and sonicated by using a Branson Sonifier 450 five times for 10 s each time (30% output) to shear the chromatin to an average size of 0.5 kb. The lysate was precleared overnight with protein A-agarose beads. Immunoprecipitations were performed for 2 h in lysis buffer with antibodies against BMI1, CBX2, and phospho-H2AX proteins (see Table S1 in the supplemental material). Preimmune serum or rabbit anti-human IgG (H+L) antibody (Jackson Immunoresearch Laboratories) was used as a negative control. Complexes were washed twice with radioimmunoprecipitation assay buffer, once in high-salt buffer (50 mM Tris-Cl [pH 8.0], 500 mM NaCl, 0.1% SDS, 0.5% deoxycholate, 1% NP-40, 1 mM EDTA), once in LiCl buffer (50 mM Tris-Cl [pH 8.0], 250 mM LiCl, 1% NP-40, 0.5% deoxycholate, 1 mM EDTA), and twice in TE buffer (10 mM Tris-Cl [pH 8.0], 1 mM EDTA [pH 8.0]). Beads were resuspended in TE containing 50 mg of RNase/ml, followed by incubation for 30 min. Beads were washed with elution buffer (1% SDS, 0.1 M NaHCO3), which was added for 15 min. Cross-links were reversed by adding 200 mM NaCl, followed by incubation for 6 h at 65°C. Samples were deproteinized overnight with proteinase K, and DNA was extracted with phenol-chloroform, followed by ethanol precipitation. Real-time PCR was performed using SYBR Green (ABI) on a Stratagene MX4000 instrument, and quantitative reverse transcription-PCR (RT-PCR) data were analyzed by using the 2ΔΔ−CT method as described previously (30).
Several different primers were used. For the FRA3B distal region, we used the forward primer 5′-CAATGGCTTAAGCAGACATGGT-3′ and the reverse primer 5′-AGTGAATGGCATGGCTGGAATG-3′. For the FRA3B central region, we used the forward primer 5′-TGTTGGAATGTTAACTCTATCCCAT-3′ and the reverse primer 5′-ATATCTCATCAAGACCGCTGCA-3′. The following primers were also utilized: AAVS1 ChIP primers AAVS1F-2127 (5′-CGGTTAATGTGGCTCTGGTT-3′) and AAVS1R-2380 (5′-AGGATCCTCTCTGGCTCCAT-3′); HOXA3 ChIP primers (forward, 5′-TCAGGCAGAAACTGCTGACA-3′; reverse, 5′-ACCTGTCTCTCCCAGGACATT-3′); and MBS85 ChIP primers (forward primer, 5′-GGAGATGAGGATGAGGGAGGTGAAG-3′; reverse primer, 5′-GCTCTTAGGGTTGGAGGACACAGG-3′). The full set of 20 primers used for analysis of the FRA3B-FHIT region is available upon request.
ZFN-driven DNA DSBs.
The zinc finger nuclease (ZFN) pair targeting at the AAVS1 site on human chromosome 19 has been described (20). mRNA encoding the AAVS2 targeting ZFN was provided by Sigma (20). The mRNA was transfected (FuGene6; Roche) in HeLa cells and incubated for 4 to 6 h, followed by the ChIP protocol.
RNA interference.
Small interfering RNAs (siRNAs) targeting BMI1, ATR, RING1B, and a nontargeting control siRNA were purchased from Sigma-Aldrich. siRNA transfection (Fugene6) was performed according to the manufacturer's protocol. The sequences of siRNA 1 and siRNA 2 against BMI1 were 5′-GAC AUU GCA UCU GAU CUG U(dT)(dT)-3′ and 5′-ACA GAU CAG AUG CAA UGU C(dT)(dT)-3′, respectively. Additional siRNA sequences included the following: for ATR, 5′-CAG UGU ACC UGG CGG CAG U(dT)(dT)-3′ and 5′-ACU GCC GCC AGG UAC ACU G(dT)(dT)-3′; and for RING1B, 5′-CCUCUAUACCACCCAUUAA(dT)(dT)-3′ and 5′-UUAAUGGGUGGUAUAGAGG(dT)(dT)-3′.
Western blotting.
Denatured protein lysates (40 μg) in Laemmli loading buffer were analyzed by gradient SDS-PAGE (4 to 15% gels) and transferred to polyvinylidene difluoride membranes. For Ub-H2A analysis, chromatin extracts were prepared, and Western blotting was performed as described previously (57, 60). Details regarding the antibodies used are given in Table S1 in the supplemental material.
Clonogenic survival assay.
Cell survival was assessed by using a standard colony-forming assay. A total of 103 cells were seeded in triplicate dishes 1 day prior to transfection (Fugene6) with either control siRNA or BMI1 siRNA. Cell were treated with mean irradiation doses from 0 to 10 Gy. After 10 days, the plates were fixed and stained with 0.01% crystal violet in 1.5% acetic acid, and colonies larger than 50 cells were counted. The number of surviving colonies was normalized to nonirradiated samples, with the latter value being set to 100%. The data analysis was performed as described in detail elsewhere (14).
Retroviral vectors and infection.
For viral particle packaging, PA Ampho cells were transiently transfected with retroviral constructs of Bmi-1 wild type, and the deletion mutants ΔRF and ΔHT were as described previously (11). Viral supernatant was collected at 48 h posttransfection and was used for infection. Stable pools of infected Bmi−/−; Ink4a−/−; Arf−/− MEFs were selected in the presence of 2 μg of puromycin/ml.
Homology-directed recombination assay.
Homologous recombination (HR) assays were performed as described previously using the DR-U2OS cell line (DR-GFP) (36). To assay the rate of HR, DR-U2OS cells were transfected with control or BMI1-specific siRNA and 48 h later were transfected with an I-SceI-encoding plasmid (or control plasmid) plus additional siRNA. After I-SceI transfections, the cells were then grown for 3 days and harvested. The percentage of green fluorescent protein (GFP)-positive cells was measured by using the Cytomics FC 500 Series flow cytometer (Beckman Coulter). The data were analyzed by using CXP software (Beckman Coulter).
Cell cycle analysis.
Cells were treated with or without siRNA and collected at the indicated time points. For fluorescence-activated cell sorting (FACS) analysis, the cells were ethanol fixed and labeled with propidium iodide (catalog no. R-4875; Sigma), and fluorescence was measured by flow cytometry (Cytomics FC 500 Series; Beckman Coulter) and normalized to the transfection efficiency. The data were analyzed by using CXP software.
RESULTS
BMI1 is recruited to sites of DNA damage.
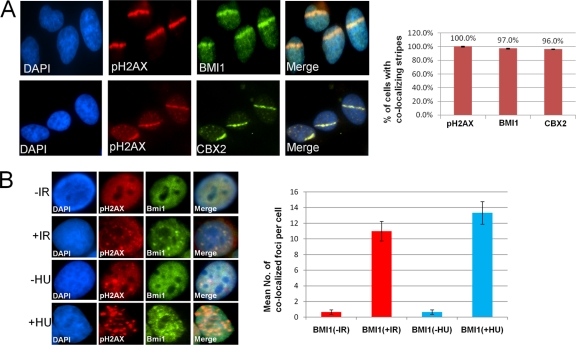
Local micro-irradiation using a UV laser scissors approach was used to induce focal regions of DSBs in HeLa cell nuclei (41). The linear regions of induced DNA breaks were visualized by immunofluorescence imaging using antibodies to phosphorylated histone H2AX (pH2AX). A subset of endogenous BMI1 localizes to the sites of DNA damage in these cells, as seen by colocalization of BMI1 immunofluorescence (IF) signal with pH2AX (Fig. 1 A). Knockdown of BMI1 leads to loss of BMI1 IF signal, demonstrating the specificity of the IF signal (see Fig. S1C in the supplemental material). A BMI1-eGFP fusion protein is recruited to laser scissors-induced DNA damage sites when introduced into HeLa cells (see Fig. S1B in the supplemental material). Chromobox protein 2 (CBX2), a polycomb group protein known to interact with BMI1 (2), also localizes to sites of laser scissors-induced DNA damage (Fig. 1A). Localization of BMI1 at laser scissors-induced DNA damage can be seen as early as 2 min after damage, peaks at 10 min to 1 h after damage, and can still be detected up to 24 h after initiation of DNA damage (see Fig. S1A in the supplemental material).
Fig. 1.
BMI1 and CBX2 are recruited to sites of DNA damage. (A) HeLa cells were subjected to laser scissors-induced DNA damage and, after 30 min, localization of pH2AX and either BMI1 (top panel) or CBX2 (bottom panel) was assayed by IF. The percentage of cells with colocalization of pH2AX with BMI1 or CBX2 is indicated (right panel). (B) The top panels show HeLa cells treated with either 0 Gy of IR (−IR) or 10 Gy of IR (+IR). After 8 h, the localization of BMI1 and pH2AX was assayed by IF. In the bottom panels, HeLa cells were treated with either no or 3 mM HU for 2 h, and localization of BMI1 and 53BP1 was assayed by IF. In the right panel, the mean numbers of BMI1 foci colocalizing with pH2AX foci per cell are plotted for each condition. The data shown are means ± the standard error of the mean (SEM) from at least three separate experiments.
To determine whether BMI1 was recruited to DSBs occurring in response to alternative forms of genotoxic stress, we investigated BMI1 localization to replication-stress-induced DSBs and to ionizing radiation (IR)-induced foci. BMI1 is recruited to pH2AX foci induced by hydroxyurea (HU) treatment in HeLa cells (Fig. 1B). BMI1 was also recruited to IR-induced nuclear foci with partial colocalization seen with pH2AX foci in HeLa cells treated with 10 Gy (Fig. 1B). These data demonstrate that BMI1 can be recruited to DSBs induced by different conditions, including UV laser scissors, HU-induced collapsed replication forks, and IR.
BMI is recruited to both stressed replication forks and enzymatically induced DNA DSBs.
To introduce DNA lesions into specific genomic regions, two complementary methods were used: (i) aphidicolin treatment to induce stalled replication forks and DNA breaks at known endogenous fragile sites (17) and (ii) introduction of ZFN to create an enzymatic DNA DSB at a defined genomic location (35). The localization of BMI1 and other repair factors to these specific sites can then be investigated by ChIP assay.
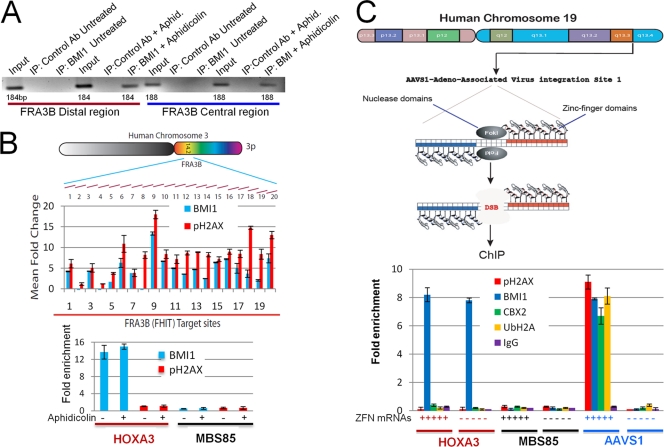
To determine whether BMI1 is recruited to stalled replication forks induced at the FHIT region in the common fragile site FRA3B, HeLa cells were incubated with 0.5 μM aphidicolin or buffer alone for 24 h. The cells were then processed for ChIP by using either a control antibody or a BMI1-specific antibody. Primers specific to two separate regions in FRA3B fragile locus were used to determine whether BMI1 occupies the FRA3B locus. As shown in Fig. 2A (see also Fig. S2A in the supplemental material), BMI1 does not occupy the FRA3B locus in untreated cells but is recruited to this site after aphidicolin treatment. To better define the localization of BMI1 to this site, a set of primers that cover ∼18 kb of the FRA3B locus covering FHIT region was used to determine the presence of BMI1 and pH2AX before and after aphidicolin treatment. Primers for a region of HOXA3 gene were used as a positive control for BMI1 in HeLa cells (5; Gaetano Gargiulo and Maarten van Lohuizen, unpublished data), and primers for a region of the MBS85 gene on chromosome 19 were used as a negative control. All ChIP signals were quantitated by real-time PCR, and the change in signal induced by aphidicolin treatment at each site was determined. As seen in Fig. 2B, aphidicolin treatment induces an increase in pH2AX across the FHIT region of FRA3B; BMI1 occupancy is also increased in a manner that parallels pH2AX. BMI1 was found to occupy the HOXA3 region regardless of aphidicolin treatment. Neither pH2AX nor BMI1 is recruited to the nonfragile MBS85 locus after aphidicolin treatment. These results demonstrate that BMI1 is specifically recruited to the sites of DNA breaks associated with aphidicolin-induced replication stress at FRA3B.
Fig. 2.
(A) HeLa cells were incubated with 0.5 μM aphidicolin or carrier alone for 24 h and then processed for ChIP using either irrelevant IgG or BMI1-specific antibodies. PCR was performed with two different sets of primers specific for the FRA-3B locus. (B) HeLa cells were mock treated or treated with 0.5 μM aphidicolin for 24 h and then subjected to ChIP using antibodies to BMI1 (blue bars) or pH2AX (red bars). Quantitative real-time PCR was performed with sets of primers that covered ∼18 kb of the FRA-3B locus, as shown. The relative fold increase in signal between untreated cells and aphidicolin-treated cells is shown for each primer pair for the ChIP using BMI1 antibody (blue bars) or pH2AX antibody (red bars) shown in the upper panel. The lower panel shows the relative occupancy of BMI1 and pH2AX at the positive control locus (HOXA3) and at a nonfragile site (MBS85) in both untreated and aphidicolin-treated cells. The data shown are means of at least three experiments ± the SEM. (C) A scheme of the ZFN strategy to create specific DNA DSBs is shown in the upper panel. HeLa cells were transfected with mRNA encoding a ZFN targeting the AAVS1 site or mock transfected and processed for ChIP using antibodies to pH2AX, BMI1, CBX2, or UbH2A or an irrelevant IgG and primers to either the flanking region of AAVS1, to MBS85 site on chromosome 19, or to the HOXA3 locus. The ChIP signal was quantitated by RT-PCR, and the lower panel shows the relative fold enrichment for each condition. Each point is the mean of at least three experiments ± the SEM.
To determine whether BMI1 localizes to sites regions of enzymatically generated DNA DSBs, a ZFN strategy (35) was used to generate a DNA DSB at a specific endogenous site in the human genome (Fig. 2C). HeLa cells were either mock transfected or transfected with mRNAs encoding a pair of ZFNs targeting the adeno-associated virus insertion site 1 (AAVS1) on human chromosome 19. After 6 to 8 h, cells were then processed for ChIP using antibodies to pH2AX, BMI1, or CBX2 and primers for a region flanking the ZFN targeting site. Primers for the HOXA3 gene and the MBS85 gene were used as positive and negative controls for BMI1. All ChIP signals were quantitated by real-time PCR. As shown in Fig. 2C, introduction of the AAVS1-targeted ZFN led to increased pH2AX ChIP signal at the target site. Increased occupancy of BMI1 and CBX2 was also present at the AAVS1 site after introduction of ZFN (Fig. 2C), a finding consistent with BMI1 and CBX2 occupying the flanking region of chromatin upon induction of DNA DSBs. BMI1 was present at the HOXA3 site both in the absence and in the presence of AAVS1-targeted ZFN. Neither BMI1 nor H2AX demonstrated increased occupancy at the MBS85 site upon introduction of the AAVS1-targeted ZFN. Together, these data show that induction of a specific DNA break at the AAVS1 site using targeted ZFNs led to specific localization of pH2AX, BMI1, and CBX2 to the flanking chromatin region.
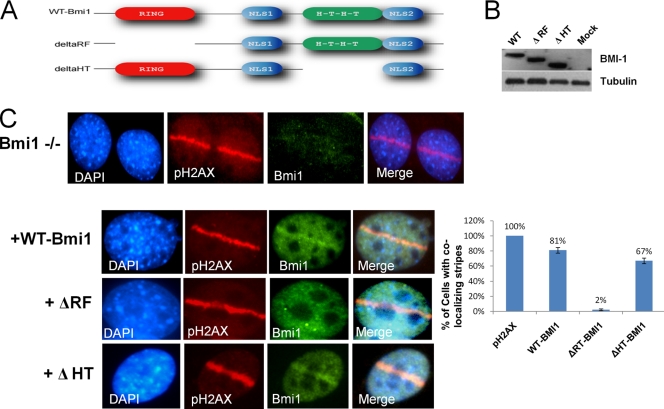
The RING domain of BMI1 is required for localization to DNA breaks.
In culture, MEFs derived from Bmi1−/− mice rapidly enter cellular senescence, which can be rescued by codeletion of the Ink4a/Arf locus (26). Bmi1−/−; Ink4a−/−; Arf−/− MEFs were reconstituted with empty vector, vector encoding wild-type BMI1 (wt-BMI1), or mutants with a deletion of the RING domain (Δ-RF) or a deletion of the helix-turn-helix domain (Δ-HT); Western blots showed equal expression of the mutant constructs (Fig. 3 B). As shown in Fig. 3C, both wt-BMI1 and Δ-HT mutant localized to sites of laser scissor-induced DNA breaks, but the Δ-RT mutant fails to localize. These data indicate that the RING finger of BMI1 is required for recruitment to the sites of DNA breaks.
Fig. 3.
(A) Diagrams of wt-Bmi1 and mutant Bmi1 constructs are shown. (B) Empty vector or vectors encoding wt-Bmi1, Δ-RF Bmi1, or Δ-HT Bmi1 were introduced into Bmi1−/−; Ink4a−/− Arf−/− MEFs. Western blots with an antibody that can recognize both wild-type and mutant constructs are shown. (C) The cells were then treated with laser scissors and processed for IF analysis using antibodies to Bmi1 and pH2AX. A bar graph shows the percentages of cells with a laser scissors-induced region of pH2AX that had colocalizing regions of Bmi1 staining in each condition. Only cells with both vector expression and evidence of laser scissors damage were assessed. The data are plotted as means ± the SEM of three separate experiments.
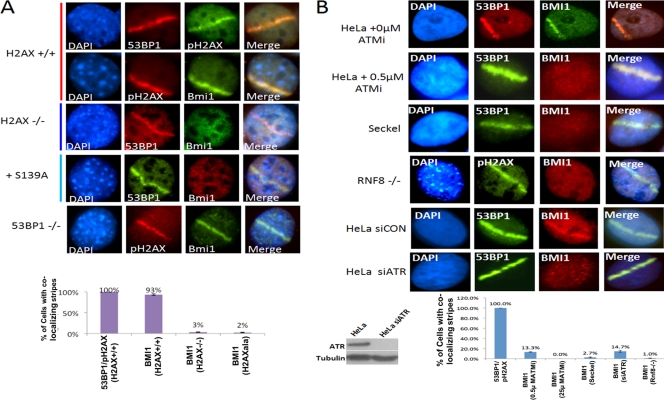
Sustained localization of BMI1 to sites of DNA damage is dependent on H2AX phosphorylation and ATM/ATR.
To determine the signaling events required for localization of BMI1 to sites of DNA damage, laser scissors were used to induce DNA DSBs in H2AX−/− MEFs, and wild-type controls. Induction of DNA DSBs by laser scissors leads to no detectable induction of pH2AX in H2AX−/− MEFs; however, 53BP1 localization is still detectable at 30 min (see Fig. S2B in the supplemental material). Localization of Bmi1 to DNA breaks at 30 min after DNA damage is greatly reduced in H2AX−/− MEFs compared to wild-type MEFs (Fig. 4 A). BMI1 localization to laser scissors-induced DNA breaks is also impaired in H2AX−/− MEFs reconstituted with S139A H2AX mutant (9) (Fig. 4A). This finding indicates that phosphorylation of H2AX at serine 139 is required for proper localization of BMI1 to sites of DNA damage.
Fig. 4.
(A) Bmi1+/+; Ink4a−/−; Arf−/− MEFs, Bmi1−/−; Ink4a−/− Arf−/− MEFs, H2AX−/− MEFs, or H2AX−/− MEFs expressing S139A mutant H2AX were treated with laser scissors and, after 30 min, processed for IF using antibody to BMI1 and either 53BP1 or pH2AX as indicated. A bar graph shows the percentages of cells with laser scissors-induced damage showing colocalization of BMI1 with 53BP1 in each MEF strain. The data plotted are the mean ± the SEM of three separate experiments. (B) In the top two panels, HeLa cells were treated with either 0 or 0.5 μM ATMi and subjected to laser scissors and, after 30 min, processed for IF using antibodies to BMI1 or 53BP1. In the middle two panels, Seckel cells or Rnf8−/− MEFs were treated with laser scissors and processed for IF using the antibodies indicated. In the bottom two panels, HeLa cells were treated with either control siRNA or ATR-specific siRNA and then treated with lasers scissors and processed for IF using antibodies to BMI1 and 53BP1. Western blots showing the effect of siATR on ATR protein levels are shown below the panels. Bar graph shows quantitation of percentage of cells that had clear colocalization of BMI1 with 53BP1 or pH2AX. The data are plotted as means ± the SEM of at least three separate experiments.
To better determine the effect of loss of H2AX on the dynamics of BMI1 recruitment, the localization of BMI1 to laser scissors-induced DNA damage was analyzed in H2AX+/+ and H2AX−/− MEFs at multiple time points. As shown in Fig. S3 in the supplemental material, BMI1 localization is significantly reduced in H2AX−/− cells starting as early as 10 min after DNA damage and abolished by 30 min. Of note, some BMI1 signal is still present at laser scissor tracks in H2AX−/− MEFs at very early time points (5 to 10 min), suggesting that initial localization of BMI1 to damage sites may be partly independent of H2AX phosphorylation (see Fig. S3 in the supplemental material). However, sustained localization (>20 min) is clearly dependent upon H2AX phosphorylation.
Sustained localization of BMI1 to sites of laser scissors-induced DNA damage (evaluated at 30 min) is also reduced in Rnf8−/− MEFs (Fig. 4B) but not impaired in 53bp1−/− MEFs (Fig. 4A) or Brca1 mutant cells (see Fig. S4 in the supplemental material). BMI1 protein levels in Rnf8−/− MEFs are comparable to wild-type MEFs, demonstrating that the loss of BMI1 signal at sites of DNA damage is not due to a change in total protein levels (see Fig. S5A in the supplemental material). These data together suggest that sustained BMI1 recruitment at DNA damage sites requires both H2AX phosphorylation and RNF8 recruitment but is independent of 53BP1 and BRCA1.
To determine the role of ATM activation in BMI1 recruitment, experiments were performed with a specific ATM inhibitor (ATMi; KU55933) (19). Treatment with this ATMi leads to loss of phosphorylated ATM-serine 1981 signal and significant reduction in H2AX phosphorylation at sites of induced DNA damage (see Fig. S5B in the supplemental material). ATMi treatment also led to the loss of BMI1 localization to sites of laser scissors-induced DNA damage (Fig. 4B), without affecting BMI1 protein levels (see Fig. S5A in the supplemental material). This observation suggests that ATM activity is required for sustained BMI1 recruitment to DNA damage. BMI1 localization to laser scissors induced DNA breaks was also reduced in the ATR mutant Seckel cells (Fig. 4B). To confirm the role of ATR in BMI1 localization, HeLa cells were treated with either control siRNA or ATR-specific siRNA. Treatment with ATR-specific siRNA led to both reduction of ATR protein and reduced localization of BMI1 to laser scissors-induced DNA breaks (Fig. 4B). As reported previously, 53BP1 localization is partly independent of ATM, ATR, and H2AX (9, 43), as shown by its presence at laser scissors-induced damage in cells treated with ATMi and in ATR mutant Seckel cells (Fig. 4B). Of note, BMI1 protein is present in Seckel cells and not altered in HeLa cells upon treatment with ATR knockdown (see Fig. S5A in the supplemental material). Together, these data suggest that activation of ATM and ATR, and subsequent phosphorylation of H2AX, contributes to sustained localization of BMI1 to the sites of laser scissors-induced DNA damage.
To determine whether the localization of BMI1 was dependent upon poly(ADP-ribose) polymerase (PARP), the dynamics of BMI1 localization to laser scissors-induced damage was determined in wild-type and Parp1−/− MEFs (see Fig. S6 in the supplemental material). BMI1 localization was still present in Parp1−/− MEFs at both early (5 min) and late (30 min) time points, suggesting that BMI1 localization to sites of DNA damage is independent of PARP1.
BMI1 is required for laser scissors-induced ubiquitination of H2A at lysine 119.
Ubiquitination of histone H2A at lysine 119 (Ub-H2A-K119) is induced by laser scissors-associated DNA breaks, as shown by IF with a specific antibody that recognizes Ub-H2A-K119 (Fig. 5A). Similarly, induction of a DNA DSB at the AAVS1 site using a targeted ZFN in HeLa cells induced increased Ub-H2A-K119 occupancy in flanking chromatin, as assayed by ChIP (Fig. 2C). Thus, DNA DSBs induced either by laser scissors or by targeted nuclease activity can induce local H2A-K119 ubiquitination.
Fig. 5.
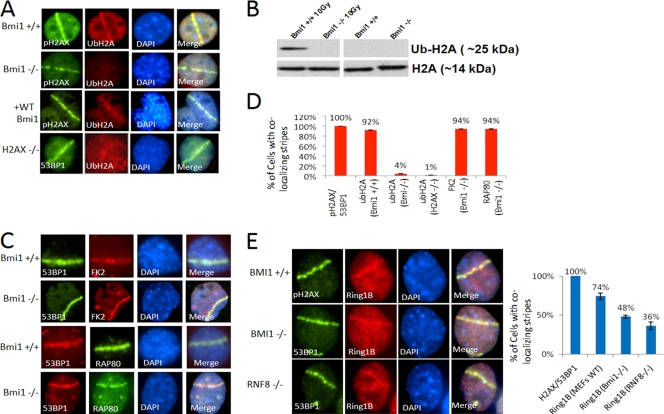
(A) Bmi1+/+; Ink4a−/−; Arf−/− MEFs, Bmi1−/−; Ink4a−/− Arf−/− MEFs, Bmi1−/−; Ink4a−/− Arf−/− MEFs reconstituted with wt-BMI1, or H2AX−/− MEFs were treated with laser scissors and, after 30 min, processed for IF using antibody to Ub-H2A-K119 and either 53BP1 or pH2AX as indicated. (B) Bmi1+/+; Ink4a−/−; Arf−/− MEFs or Bmi1−/−; Ink4a−/− Arf−/− MEFs were treated with 10 Gy of IR or mock treated and, after 30 min, chromatin extracts were prepared and processed for Western blotting with antibody to Ub-H2AK119 or antibody to histone H2A. (C) Bmi1−/−; Ink4A−/− Arf−/− MEFs were treated with laser scissors and, after 30 min, processed for IF with antibodies to polyubiquitin (FK2) or RAP80 and 53BP1 as indicated. (D) Bar graphs show the percentages of the indicated cell types with laser scissors-induced regions of pH2AX or 53BP1 that also show colocalization with Ub-H2A-K119, FK1, or RAP80. (E) Bmi1+/+; Ink4a−/− Arf−/− MEFs, Bmi1−/−; Ink4a−/− Arf−/− MEFs, or Rnf8−/− MEFs were treated with laser scissors and, after 30 min, processed for IF using antibodies to RING1B and either pH2AX or 53BP1 as indicated. The percentages of cells with laser scissors-induced damage showing colocalization of RING1B with pH2AX or 53BP1 for each MEF strain is shown in the bar graph. All data are plotted as means ± the SEM of at least three independent experiments.
To evaluate the role of BMI1 in DNA damage-induced ubiquitination, experiments were performed in Bmi1−/− cells. In Bmi1−/−; Ink4a−/−; Arf−/− MEFs, there is dramatic reduction of Ub-H2A-K119 induced by laser scissors at 30 min compared to Bmi1+/+; Ink4a−/−; Arf−/− MEFs (Fig. 5A). A similar loss of laser scissors-induced H2A-K119 ubiquitination is seen in H2AX−/− MEFs (Fig. 5A). We also confirmed prior reports demonstrating that Rnf8−/− cells are impaired in the ubiquitination of H2A-K119 at sites of DNA breaks (59) (see Fig. S7 in the supplemental material). Reintroduction of wt-BMI1 into Bmi1−/−; Ink4a−/−; Arf−/− MEFs restores Ub-H2A-K119 signal at laser scissors-induced DNA damage (Fig. 5A).
IR treatment with 10 Gy induces an increase in total Ub-H2A-K119 in Bmi1+/+; Ink4a−/−; Arf−/− cells (Fig. 5B and see Fig. S9 in the supplemental material) as demonstrated by Western blotting. The IR-induced increase in Ub-H2A-K119 is abolished in Bmi1−/−; Ink4a−/−; Arf−/− MEFs (Fig. 5B). General polyubiquitination at the sites of breaks, as labeled by the FK2 antibody (15, 16), remains intact in Bmi1−/−; Ink4a−/−; Arf−/− cells, as does localization of RAP80 and 53BP1 (Fig. 5C). These data demonstrate that, at sites of DNA damage, intact BMI1 is required for ubiquitination of H2A-K119 but is not required for polyubiquitination of other substrates or for RAP80 or 53BP1 recruitment.
RING1B and MEL18 are recruited to sites of DNA damage.
BMI1 interacts with RING1B (RNF2) to form a heterodimer that is required for PRC1-mediated histone ubiquitination (3, 7). RING1B has been shown to be required for UV damage-induced H2A-K119 ubiquitination (4). RING1B also localizes to sites of DNA DSBs induced by laser scissors (Fig. 5E). Similar to BMI1, the localization of RING1B to laser scissors tracks is sustained, being present at 30 min after laser treatment. This localization of RING1B is still present, although at decreased frequency, in Bmi1−/−; Ink4a−/−; Arf−/− cells and in Rnf8−/− cells, suggesting that RING1B can be recruited to DNA breaks independently of BMI1 (Fig. 5E). Similarly, knockdown of RING1B does not abolish localization of BMI1 to sites of laser scissors-induced DNA damage (see Fig. S8 in the supplemental material), suggesting that BMI1 can be recruited independently of RING1B.
MEL18, a polycomb factor with high homology to BMI1, is also recruited to sites of laser scissors induced DNA damage (see Fig. S8 in the supplemental material). Unlike BMI1, this localization is transient, being present only at early time points (5 to 10 min) after laser treatment. MEL18 is not detectable at DNA damage tracks at later time points (>20 min) where BMI1 is clearly present. RING1A, a homolog of RING1B, was not detectable at UV laser scissors-induced DNA damage at any time point (see Fig. S8 in the supplemental material).
Loss of BMI1 is associated with DNA damage-induced checkpoint activation and the accumulation of cells in G2/M.
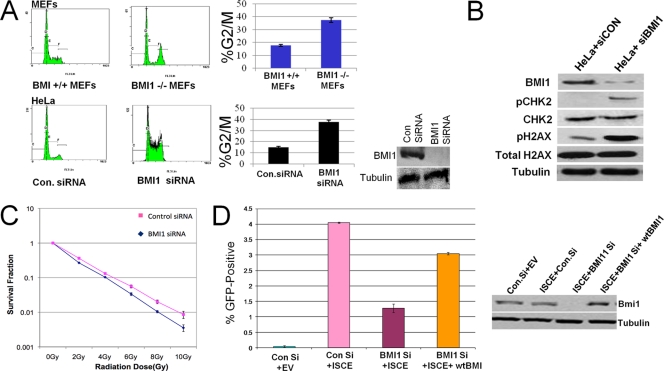
Cell cycle analysis demonstrated that Bmi1−/−; Ink4a−/−; Arf−/− MEFs showed increased accumulation of cells in G2/M compared to Bmi1+/+; Ink4a−/−; Arf−/− MEFs (Fig. 6A). Treatment of HeLa cells with a BMI1-specific siRNA led to a similar accumulation of cells in G2/M phase of the cell cycle compared to cells treated with control siRNA (Fig. 6A). siRNA-mediated knockdown of BMI1 in HeLa cells also results in increased levels of H2AX phosphorylation that is accompanied by increased phosphorylation of the checkpoint kinase CHK2 (Fig. 6B). These data are consistent with the loss of BMI1, leading to the induction of spontaneous DNA damage and subsequent DNA-damage-dependent G2/M checkpoint activation.
Fig. 6.
(A) Loss of BMI1 leads to accumulation of cells in G2/M associated with DNA damage-associated checkpoint activation. In the top panels, wild-type MEFs or Bmi1−/−; Ink4a−/− Arf−/− MEFs were harvested, stained with propidium iodide, and analyzed by FACS for DNA content. Plots of the FACS analysis are shown in the left panels, and quantitation of the percentages in G2/M is shown in the bar graph. In the bottom panels, HeLa cells were treated either with control siRNA or BMI1-specific siRNA and then processed for DNA content-based cell cycle analysis as described. The percentages in G2/M are plotted. The data shown are means ± the SEM of three separate experiments. In the inset, a Western blots shows the expression of BMI1 and tubulin in cells treated with control or specific siRNA. (B) HeLa cells were treated either with control siRNA or BMI1-specific siRNA. Protein extracts were then subjected to Western blotting with antibodies for BMI1, phoshpo-CHK2-S19 (pCHK2), total CHK2 (CHK2), pH2AX, or tubulin. (C) Control siRNA- or BMI1 siRNA-treated HeLa cells were exposed to various doses of IR, and colony formation was assayed, plotted, and normalized to that of untreated cells. (D) U20S cells contain an integrated tandem GFP reporter of HR was treated with control siRNA, BMI1-specific siRNA, or BMI1-specific siRNA plus wt-BMI1. The cells were then transfected with an I-SceI-expressing vector. Cells transfected with an empty vector are also shown as a negative control. After 24 h, the percentages of cells expressing GFP were measured by flow cytometry. Each data point represents the mean ± the SEM of three separate experiments. Western blots showing the expression of BMI1 in control cells, BMI1 siRNA-treated cells, and BMI1 siRNA-treated cells reconstituted with wt-BMI1 are shown to the right.
BMI1 is required for efficient HR-mediated repair of DNA DSBs.
Knockdown of BMI1 by siRNA in HeLa cells leads to decreased survival after treatment with ionizing radiation, as measured by clonogenic assay (Fig. 6C), demonstrating that the loss of BMI1 leads to increased sensitivity to IR-induced DNA breaks. To investigate the role of BMI1 in HR-mediated repair of DNA DSBs, experiments were performed with U20S-DR cells containing a single-copy integrated tandem GFP-based reporter of HR (36). Knockdown of BMI1 in these cells leads to a clear decrease in the efficacy of HR-mediated repair, as measured by this assay (Fig. 6C). This reduction in HR-mediated repair induced by BMI1 siRNA could be rescued by the introduction of a vector encoding wild-type murine Bmi1, which is not targeted by the siRNA used (Fig. 6C).
DISCUSSION
We have shown that BMI1 is recruited to sites of DNA damage. Several types of DNA lesions are able to recruit BMI, including DNA DSBs induced by laser scissors, ZFNs, and IR, and collapsed replication forks induced by HU and aphidicolin. BMI1 is rapidly localized to sites of laser scissors induced breaks, and remains present for up to 24 h (see Fig. S1 in the supplemental material). The sustained localization of BMI1, assayed at 30 min after DNA damage, is dependent on H2AX phosphorylation and intact ATM/ATR (Fig. 3). Sustained localization of BMI1 is also reduced in Rnf8−/− MEFs but is intact in 53bp1−/− cells and in Brca1 mutant cells. This suggests that sustained BMI1 localization to DNA damage sites requires signaling through ATM/ATR pathway but is independent of 53BP1 and proximal to BRCA1.
Of note, BMI1 localization is still present at very early time points (5 to 10 min) after DNA damage in H2AX−/− MEFs (see Fig. S3 in the supplemental material). This observation suggests that initial transient recruitment of BMI1 may occur independently of H2AX phosphorylation. Recent reports suggest that some polycomb factors, including MEL18, may be recruited transiently to sites of DNA damage in a PARP-dependent fashion (10). BMI1 localization to laser scissors-induced DNA damage is still present in PARP1 deficient cells, both at early and late time points (see Fig. S6 in the supplemental material). Thus, initial, transient recruitment of BMI1 to DNA breaks may be independent of both PARP1 and H2AX; however, sustained localization of BMI1, and subsequent ubiquitination of H2A-K119, requires intact ATM/ATR, H2AX phosphorylation, and RNF8 recruitment. The differing dynamics of MEL18 and BMI1 to sites of DNA damage suggest that MEL18 and BMI1 may be differentially regulated in the DNA repair process. Thus, different polycomb factors may be present in several independent complexes and function in different signaling pathways during the DNA damage response.
DNA damage-induced ubiquitination of H2A-K119 is dependent on BMI1. However, DNA damage-induced polyubiquitination of other substrates is not impaired and RAP80 recruitment is intact in Bmi1−/− cells (Fig. 5). This suggests that, unlike RNF8, BMI1 is specifically required for H2A-K119 ubiquitination but not required for polyubiquitination of other substrates. Western blot data suggest that the monoubiquitination of H2A-K119 is the predominant modification induced by DNA damage at this residue; however, we cannot exclude the possibility that polyubiquitination of H2A-K119 may also be present. These data are consistent with the presence of multiple, topologically distinct ubiquitin species at DSBs (34). Since BMI1 is required for efficient HR-mediated repair (Fig. 6), these data further suggest that BMI1-mediated H2AK119 ubiquitination at sites of DNA breaks plays a role in facilitating HR-mediated repair.
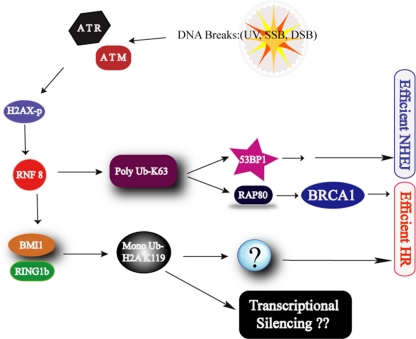
These data support a model in which DNA damage-induced activation of ATM and ATR leads to local H2AX phosphorylation and recruitment of MDC1 and RNF8. These events are required for sustained localization of BMI1 to sites of DNA breaks, where, together with RING1B, BMI1 ubiquitinates H2A-K119 (Fig. 7) and contributes to efficient HR-mediated DNA repair. The exact function of ubiquitinated H2A-K119 in facilitating HR-mediated DNA repair remains to be determined. It is also possible that other substrates of BMI1 ubiquitin-ligase activity, such as TOP2, may also play a role in the DNA repair pathway (1). Since H2A-K119 ubiquitination has a role in transcriptional silencing, BMI1-associated H2A-K119 ubiquitination at DNA breaks may also play a role in mediating local transcriptional silencing during the DNA repair response (45).
Fig. 7.
Model showing the role for Bmi1 in the DNA damage response pathway. See the text for details.
A role for BMI1 in the DNA damage response has been previously reported (10, 13, 25, 29). BMI1 was shown to modulate mitochondrial production of reactive oxygen species (ROS). Loss of BMI1 leads to greater generation of ROS-mediated DNA damage, resulting in checkpoint activation (29). BMI1 has also been recently reported to localize at sites of DNA damage, interact with ATM, and contribute to radioresistance in glioblastoma cells (13). Our work demonstrates a direct and specific role of BMI1 in local H2A-K119 ubiquitination at sites of DNA damage and functional role of BMI1 in HR-mediated DNA repair. We show that cells lacking BMI1 are sensitive to ionizing radiation (Fig. 6) (13, 25). How much of this phenotype is due to direct impairment of HR-mediated repair and how much is due to the potential role of BMI1 in regulating ROS metabolism is not clear at present. It is also possible the increased ROS-mediated DNA damage seen in cells lacking BMI1 may be partly due to impaired DNA repair pathways.
Unlike the findings presented here, Facchino et al. found that localization of BMI1 to sites of IR-induced damage was not dependent upon ATM activation. The differences between our findings and those of Facchino et al. may be due to different cell types analyzed and differing methods of generating and visualizing DNA breaks. Our data are in agreement with Ismail et al., who recently reported that BMI1 was recruited to sites of DNA damage and involved in histone ubiquitination (25). These researchers found that an initial recruitment of BMI1 is still present in H2AX−/− cells 5 min after induction of damage. Our data also show that initial recruitment is partly independent of H2AX. However, we demonstrate that both sustained localization of BMI1 at time points greater than 20 min, and H2A-K119 ubiquitination is abolished in H2AX−/− cells. These data suggest that there may be biphasic recruitment of BMI1, with an immediate transient phase that is independent of H2AX and a later sustained phase that is dependent upon ATM/ATR and H2AX phosphorylation.
We have demonstrated that BMI1, CBX2, and RING1B can all localize to sites of DNA breaks. It is not clear what other polycomb-related proteins are involved in the DNA repair response. Both PHF1 and heterochromatin protein 1 (HP1) family members have been shown to be recruited to sites of UV damage and DNA DSBs in human cells (21, 31, 55). The histone methyltransferase EZH2 has also been shown to be recruited to sites of DNA breaks induced by ISCE1-induced DNA breaks in mammalian cells (38). A recent report has demonstrated that a variety of polycomb proteins, including MEL18, CBX6, CBX7, and CBX8, are recruited transiently to sites of DNA damage in a PARP-dependent manner (10). It is possible that distinct complexes of polycomb group proteins may be involved in transient early PARP-dependent DNA damage response and the sustained ATM/ATR-dependent DNA damage response.
BMI1 and related polycomb group proteins are thought to play an essential role in mediating chromatin compaction and transcriptional silencing during development. The recruitment of BMI1 to sites of DNA breaks suggests that similar effects may be occurring near sites of DNA damage. DNA DSBs have been shown to induce local ATM-dependent transcriptional silencing in nearby chromatin regions that is associated with H2A-K119 ubiquitination and chromatin compaction (45). Transcriptional silencing associated with DNA methylation of CpG islands and associated changes in histone methylation also been shown to occur near sites of DNA breaks (38). The data present here would suggest a role for BMI1 and related polycomb proteins in mediating these processes. Thus, BMI1 and associated polycomb proteins may link chromatin changes associated with DNA repair response to localized transcriptional repression near sites of DNA damage. Whether this can result in long-term epigenetic changes at sites of DNA damage, i.e., a chromatin memory of prior to DNA damage, under certain conditions remains to be determined.
BMI1 has been implicated in playing a crucial role in stem cell maintenance. Loss of BMI1 is associated with cell cycle arrest and premature senescence in many cell types (53). Dysfunction of DNA repair factors also induces similar checkpoint activation and cellular senescence, due to the accumulation of unrepaired DNA lesions. It is possible that at least part of the phenotype associated with the loss of BMI1 is due to the role of BMI1 in the DNA damage response.
In summary, these data demonstrate that BMI1 is recruited to sites of DNA damage, where it is required for ubiquitination of histone H2A. This function of BMI1 is downstream of ATR/ATM and RNF8 and is required for efficient HR-mediated DNA repair. These findings may give new insight into the role of BMI1 in mediating chromatin changes at sites of DNA damage and the impact of its dysfunction in both stem cells and somatic cells.
Supplementary Material
ACKNOWLEDGMENTS
We thank Junjie Chen, Andre Nussenzweig, Jos Jonkers, Xiaochun Yu, Maria Jasin, Goberdhan Dimri, and H. Koseki for generously providing cell lines and antibodies. We thank Gaetano Gargiulo for sharing the primer sequence for interrogating HOXA3 in HeLa cells. We thank Ajay Vithala for technical help and Arumugham Raghunathan and Sigma Life Science for providing a beta unit of the CompoZr targeted integration kit (AAVS1) for our research. We thank Bing Xia, Roger Greenberg, Yuri Schwartz, and Vincenzo Pirrotta for valuable discussions.
S.G. was supported by the NIH, the NJCCR, and the CINJ Foundation. J.O. was supported by a predoctoral grant from the NJCCR and the DOD.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 7 March 2011.
REFERENCES
- 1. Alchanati I., et al. 2009. The E3 ubiquitin-ligase Bmi1/Ring1A controls the proteasomal degradation of Top2α cleavage complex: a potentially new drug target. PLoS One 4:e8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alkema M. J., et al. 1997. Identification of Bmi1-interacting proteins as constituents of a multimeric mammalian polycomb complex. Genes Dev. 11:226–240 [DOI] [PubMed] [Google Scholar]
- 3. Ben-Saadon R., Zaaroor D., Ziv T., Ciechanover A. 2006. The polycomb protein Ring1B generates self atypical mixed ubiquitin chains required for its in vitro histone H2A ligase activity. Mol. Cell 24:701–711 [DOI] [PubMed] [Google Scholar]
- 4. Bergink S., et al. 2006. DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Genes Dev. 20:1343–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Birney E., et al. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447:799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Botuyan M. V., et al. 2006. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127:1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buchwald G., et al. 2006. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 25:2465–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cao R., Tsukada Y., Zhang Y. 2005. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell 20:845–854 [DOI] [PubMed] [Google Scholar]
- 9. Celeste A., et al. 2003. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 5:675–679 [DOI] [PubMed] [Google Scholar]
- 10. Chou D. M., et al. 2010. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. U. S. A. 107:18475–18480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dimri G. P., et al. 2002. The Bmi-1 oncogene induces telomerase activity and immortalizes human mammary epithelial cells. Cancer Res. 62:4736–4745 [PubMed] [Google Scholar]
- 12. Doil C., et al. 2009. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136:435–446 [DOI] [PubMed] [Google Scholar]
- 13. Facchino S., Abdouh M., Chatoo W., Bernier G. 2010. BMI1 confers radioresistance to normal and cancerous neural stem cells through recruitment of the DNA damage response machinery. J. Neurosci. 30:10096–10111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Franken N. A., Rodermond H. M., Stap J., Haveman J., van Bree C. 2006. Clonogenic assay of cells in vitro. Nat. Protoc. 1:2315–2319 [DOI] [PubMed] [Google Scholar]
- 15. Fujimuro M., Sawada H., Yokosawa H. 1994. Production and characterization of monoclonal antibodies specific to multi-ubiquitin chains of polyubiquitinated proteins. FEBS Lett. 349:173–180 [DOI] [PubMed] [Google Scholar]
- 16. Fujimuro M., Yokosawa H. 2005. Production of antipolyubiquitin monoclonal antibodies and their use for characterization and isolation of polyubiquitinated proteins. Methods Enzymol. 399:75–86 [DOI] [PubMed] [Google Scholar]
- 17. Glover T. W., Berger C., Coyle J., Echo B. 1984. DNA polymerase alpha inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Hum. Genet. 67:136–142 [DOI] [PubMed] [Google Scholar]
- 18. Harper J. W., Elledge S. J. 2007. The DNA damage response: ten years after. Mol. Cell 28:739–745 [DOI] [PubMed] [Google Scholar]
- 19. Hickson I., et al. 2004. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 64:9152–9159 [DOI] [PubMed] [Google Scholar]
- 20. Hockemeyer D., et al. 2009. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 27:851–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hong Z., et al. 2008. A polycomb group protein, PHF1, is involved in the response to DNA double-strand breaks in human cell. Nucleic Acids Res. 36:2939–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huen M. S., Chen J. 2008. The DNA damage response pathways: at the crossroad of protein modifications. Cell Res. 18:8–16 [DOI] [PubMed] [Google Scholar]
- 23. Huen M. S., et al. 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131:901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huyen Y., et al. 2004. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432:406–411 [DOI] [PubMed] [Google Scholar]
- 25. Ismail I. H., Andrin C., McDonald D., Hendzel M. J. 2010. BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J. Cell Biol. 191:45–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jacobs J. J., Kieboom K., Marino S., DePinho R. A., van Lohuizen M. 1999. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 397:164–168 [DOI] [PubMed] [Google Scholar]
- 27. Kallin E. M., et al. 2009. Genome-wide uH2A localization analysis highlights Bmi1-dependent deposition of the mark at repressed genes. PLoS Genet. 5:e1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim H., Chen J., Yu X. 2007. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 316:1202–1205 [DOI] [PubMed] [Google Scholar]
- 29. Liu J., et al. 2009. Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature 459:387–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 31. Luijsterburg M. S., et al. 2009. Heterochromatin protein 1 is recruited to various types of DNA damage. J. Cell Biol. 185:577–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mailand N., et al. 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131:887–900 [DOI] [PubMed] [Google Scholar]
- 33. Marteijn J. A., et al. 2009. Nucleotide excision repair-induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response. J. Cell Biol. 186:835–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Messick T. E., Greenberg R. A. 2009. The ubiquitin landscape at DNA double-strand breaks. J. Cell Biol. 187:319–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miller J. C., et al. 2007. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 25:778–785 [DOI] [PubMed] [Google Scholar]
- 36. Nakanishi K., et al. 2005. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc. Natl. Acad. Sci. U. S. A. 102:1110–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nicassio F., et al. 2007. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr. Biol. 17:1972–1977 [DOI] [PubMed] [Google Scholar]
- 38. O'Hagan H. M., Mohammad H. P., Baylin S. B. 2008. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet. 4:e1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Paull T. T., et al. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10:886–895 [DOI] [PubMed] [Google Scholar]
- 40. Rappold I., Iwabuchi K., Date T., Chen J. 2001. Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage-signaling pathways. J. Cell Biol. 153:613–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rogakou E. P., Boon C., Redon C., Bonner W. M. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146:905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sanders S. L., et al. 2004. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119:603–614 [DOI] [PubMed] [Google Scholar]
- 43. Schultz L. B., Chehab N. H., Malikzay A., Halazonetis T. D. 2000. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 151:1381–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schwartz Y. B., Pirrotta V. 2007. Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 8:9–22 [DOI] [PubMed] [Google Scholar]
- 45. Shanbhag N. M., Rafalska-Metcalf I. U., Balane-Bolivar C., Janicki S. M., Greenberg R. A. 2010. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141:970–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shao G., et al. 2009. The Rap80-BRCC36 de-ubiquitinating enzyme complex antagonizes RNF8-Ubc13-dependent ubiquitination events at DNA double strand breaks. Proc. Natl. Acad. Sci. U. S. A. 106:3166–3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shao G., et al. 2009. MERIT40 controls BRCA1-Rap80 complex integrity and recruitment to DNA double-strand breaks. Genes Dev. 23:740–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Simon J. A., Kingston R. E. 2009. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell. Biol. 10:697–708 [DOI] [PubMed] [Google Scholar]
- 49. Sobhian B., et al. 2007. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 316:1198–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stewart G. S. 2009. Solving the RIDDLE of 53BP1 recruitment to sites of damage. Cell Cycle. 8:1532–1538 [DOI] [PubMed] [Google Scholar]
- 51. Stewart G. S., et al. 2009. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136:420–434 [DOI] [PubMed] [Google Scholar]
- 52. Stewart G. S., Wang B., Bignell C. R., Taylor A. M., Elledge S. J. 2003. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421:961–966 [DOI] [PubMed] [Google Scholar]
- 53. Valk-Lingbeek M. E., Bruggeman S. W., van Lohuizen M. 2004. Stem cells and cancer; the polycomb connection. Cell 118:409–418 [DOI] [PubMed] [Google Scholar]
- 54. van Attikum H., Gasser S. M. 2005. The histone code at DNA breaks: a guide to repair? Nat. Rev. Mol. Cell. Biol. 6:757–765 [DOI] [PubMed] [Google Scholar]
- 55. Venkitaraman A. R. 2009. Modifying chromatin architecture during the response to DNA breakage. Crit. Rev. Biochem. Mol. Biol. 45:2–13 [DOI] [PubMed] [Google Scholar]
- 56. Wang B., et al. 2007. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 316:1194–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang H., et al. 2004. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431:873–878 [DOI] [PubMed] [Google Scholar]
- 58. Wei J., Zhai L., Xu J., Wang H. 2006. Role of Bmi1 in H2A ubiquitylation and Hox gene silencing. J. Biol. Chem. 281:22537–22544 [DOI] [PubMed] [Google Scholar]
- 59. Wu J., et al. 2009. Histone ubiquitination associates with BRCA1-dependent DNA damage response. Mol. Cell. Biol. 29:849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wu X., et al. 2008. Cooperation between EZH2, NSPc1-mediated histone H2A ubiquitination and Dnmt1 in HOX gene silencing. Nucleic Acids Res. 36:3590–3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang D., Zaugg K., Mak T. W., Elledge S. J. 2006. A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell 126:529–542 [DOI] [PubMed] [Google Scholar]
- 62. Zhu Q., et al. 2009. Chromatin restoration following nucleotide excision repair involves the incorporation of ubiquitinated H2A at damaged genomic sites. DNA Repair (Amsterdam) 8:262–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.