Abstract
Class IA phosphoinositide 3-kinases (PI3Ks) are heterodimeric enzymes composed of a p85 regulatory and a p110 catalytic subunit that induce the formation of 3-polyphosphoinositides, which mediate cell survival, division, and migration. There are two ubiquitous PI3K isoforms p110α and p110β that have nonredundant functions in embryonic development and cell division. However, whereas p110α concentrates in the cytoplasm, p110β localizes to the nucleus and modulates nuclear processes such as DNA replication and repair. At present, the structural features that determine p110β nuclear localization remain unknown. We describe here that association with the p85β regulatory subunit controls p110β nuclear localization. We identified a nuclear localization signal (NLS) in p110β C2 domain that mediates its nuclear entry, as well as a nuclear export sequence (NES) in p85β. Deletion of p110β induced apoptosis, and complementation with the cytoplasmic C2-NLS p110β mutant was unable to restore cell survival. These studies show that p110β NLS and p85β NES regulate p85β/p110β nuclear localization, supporting the idea that nuclear, but not cytoplasmic, p110β controls cell survival.
INTRODUCTION
The phosphoinositide 3-kinase (PI3K) family is divided into four groups (IA, IB, II, and III) according to structural features and substrate specificity. Of these, only class I enzymes catalyze the production of PI(3,4,5)P3 and PI(3,4)P2 in vivo. Class IA PI3Ks are heterodimeric proteins consisting of a p110 catalytic subunit (p110α, p110β, and p110δ) and an associated p85 regulatory subunit (p85α, p85β, and p55γ) (14, 18, 21, 22, 53). p110γ (class IB PI3K) is structurally similar but associates with a distinct class of regulatory subunits. The catalytic subunits p110α and p110β are expressed ubiquitously, whereas p110δ and p110γ are more abundant in hematopoietic cells (14, 44, 53).
Despite the similarity in sequence, expression patterns, and regulatory subunits, p110α and p110β have distinct functions in cell proliferation, cell cycle progression, and development (5, 6, 12, 26, 32–35, 47). p110α has a key role in insulin action and cell cycle entry (12, 13), whereas p110β is reported to play a pivotal role in DNA replication, S phase progression, and DNA repair (32, 34, 35). Activating mutations of p110α, but not of p110β, have been found in human cancer; nonetheless, p110β drives tumorigenesis in PTEN-defective cells and induces focus formation in fibroblasts (8, 9, 26, 29). Moreover, overexpression of p110β is found in specific tumor types (7, 54, 58). Previous studies showed that part of the specific functions of p110α and p110β result from their distinct subcellular localization and activation requirements (34, 35), highlighting the emergence of subcellular localization as a major mechanism to govern cell responses (30). Previous reports showed that p85/p110 complex can translocate to the nucleus regulating cell survival, particularly in neuronal cell lines (37). In addition, p110β, but not p110α, localizes to the nucleus in several cell types. The mechanisms controlling p110β intracellular localization nonetheless remain elusive. We studied here the mechanism by which p110β localizes to the nucleus. p110β is unable to enter the nucleus as a monomer and requires association with the p85β regulatory subunit. We identified a nuclear localization signal (NLS) in the p110β C2 domain that controls the translocation of p85β/p110β complexes to the nucleus. Conversely, the export of the p85β/p110β heterodimer from the nucleus is regulated by a nuclear export sequence (NES) in p85β. We show that nuclear, but not cytoplasmic, p110β regulates cell viability.
MATERIALS AND METHODS
Cell lines and cell culture.
Murine embryonic fibroblasts (MEFs) were prepared as reported elsewhere (15). The cells were maintained in Dulbecco modified Eagle medium (Gibco-BRL, Auckland, New Zealand) supplemented with 10% fetal bovine serum, 2 mM glutamine, 10 mM HEPES, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. PC12, U2OS, NIH 3T3, SAOS-2, and HeLa cell lines were maintained as described previously (35).
Plasmids.
Untagged wild-type (WT) p110β was donated by B. Vanhaesebroeck (Institute of Cancer, London, United Kingdom). pSG5-myc-p110α, pSG5-myc-p110β, and mutant myc-K805R-hp110β have been described in another study (34). NLS-myc-p110β-mutant1, -mutant2, and -mutant3, as well as NESmut rp85β, were generated by using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) with appropriate oligonucleotides. pSG5-p85α and -HA-p85β are described elsewhere (2). The p85β-α chimera was prepared by replacing p85β residues 77 to 351 with the corresponding p85α sequence. Short hairpin RNA (shRNA) against murine PI3K subunits and control-scrambled shRNA were custom-made (Origene Technologies, Rockville, MD). shRNA-resistant WT and mutant p110β were human cDNA.
Antibodies and reagents.
Blots were probed with the following antibodies (Abs): anti-Myc tag (9B11), anti-p-PKB Ser473, and anti-p-PKB Thr308 (Cell Signaling, Beverly, MA); anti-pan-p85, anti-p85α, and anti-histones (Upstate Biotechnology; Millipore, Billerica, MA); and anti-tubulin (GTU-88; Sigma, St. Louis, MO). anti-p110α was donated by A. Klippel (Merck, Boston, MA). Anti-cytochrome c was purchased from Santa Cruz (Santa Cruz, CA), anti-HA was from Covance (Emeryville, CA), and anti-p85β is described elsewhere (I. Cortés and A. C. Carrera, unpublished data). Alexa 488- and Cy3-labeled Abs were from Molecular Probes (Eugene, OR), horseradish peroxidase-conjugated secondary Abs were from Dako (Glostrup, Denmark), and ECL was from GE Healthcare (Buckinghamshire, United Kingdom). Leptomycin B and cycloheximide were from Sigma. Platelet-derived growth factor (PDGF) and nerve growth factor (NGF) were purchased from PeproTech (Rocky Hill, NJ).
Immunofluorescence, WB, and immunoprecipitation.
Western blotting (WB) and immunoprecipitation were performed as described previously (39). For immunofluorescence (IF), cells were plated on coverslips and fixed with 4% formaldehyde (10 min, room temperature [RT]), permeabilized with 0.3% Triton X-100 in phosphate-buffered saline (PBS) staining buffer (10 min), and incubated with blocking buffer (0.1% Triton X-100-3% bovine serum albumin in PBS; 30 min), followed by incubation with primary antibody (1 h, RT, with end-to-end rocking). Cells were washed three times with blocking buffer to remove unbound antibody and incubated with the appropriate secondary antibody (1:500, 1 h, RT). Samples were washed three times with blocking buffer, followed by incubation with the mounting medium Vectashield (Vector Laboratories, Inc., Burlingame, CA). DAPI (4′,6′-diamidino-2-phenylindole) was used to stain the DNA. Images were captured in a Leica Leitz DMRB microscope (Wetzlar, Germany) using an Olympus DP70 charge-coupled device camera or by using a confocal fluorescence microscope with an Olympus FluoView (Olympus, Tokyo, Japan).
In vitro transcription translation and PI3K assay.
Human myc-p110β WT or mutant 1 (C2 domain) and mouse HA-p85β cDNA were transcribed and translated in vitro in the presence of [35S]methionine using the TNT T7-coupled reticulocyte lysate system (Promega, Southampton, United Kingdom). In vitro binding of proteins was analyzed by immunoprecipitation of hemagglutinin (HA) or myc tags. The kinase assays were performed as described previously (27).
Transfection, subcellular fractionation, and apoptosis analysis.
Transfection assays were performed by using JetPei-NaCl according to the manufacturer's protocols (Qbiogene, Irvine, CA). Transfected cells were cultured 48 h prior to analysis. For subcellular fractionation (see Fig. 1 and 4), cells were cultured in exponential growth and then collected. Cytoplasmic, nuclear, and chromatin fractions were isolated as described previously (40). Buffer A, used for cytoplasmic extraction, consisted of 10 mM HEPES (pH 7.9), 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, and 1 mM dithiothreitol (DTT). The nonsalt buffer for nuclear extraction was composed of 3 mM EDTA, 0.2 mM EGTA, and 1 mM DTT; for chromatin, proteins were extracted after boiling and sonicating samples in Laemmli buffer. In all chases, samples were quantified with a BCA protein assay kit (Pierce, Rockford, IL), and the same amount of protein was analyzed by WB. For apoptosis and cytochrome c release, we transfected cells with different shRNAs in combination with rp85β and either WT p110β or NLS-p110β-mutant1 (24 h). Cells were gamma-irradiated (MARK 1; Shephard, Louisville, KY) using a 137Cs probe, collected after 24 h, and analyzed by flow cytometry in a Cytomics FC500 (Beckman-Coulter, Fullerton, CA) using annexin V and propidium iodide. Cytochrome c release was examined by using WB.
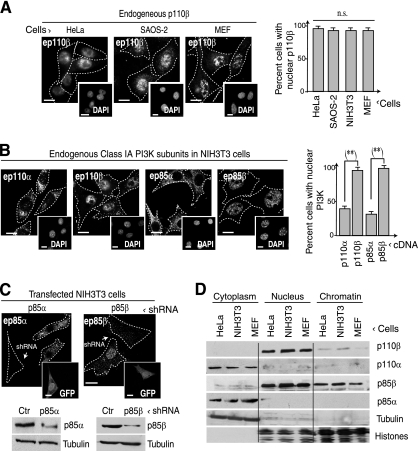
Fig. 1.
Class IA PI3K subunits p85β and p110β concentrate in the nucleus. (A) HeLa and SAOS-2 cells and freshly isolated MEFs were cultured in exponential growth and analyzed by IF using anti-p110β Ab. DNA was stained with DAPI (insets). The graph shows the percentage of cells with predominant p110β nuclear staining (n = 30). (B) Endogenous p110β, p110α, p85α, and p85β localization in NIH 3T3 cells was analyzed by IF using specific Ab; DNA was stained with DAPI (insets). The graph is as described in panel A. (C) NIH 3T3 cells were cotransfected with GFP plus p85α- or p85β-specific shRNA (48 h), and the cells were fixed and analyzed by IF using specific Abs. WB shows the downregulation of p85α and p85β after shRNA transfection. Insets show transfected (GFP+) cells. (D) HeLa cells, MEFs, and NIH 3T3 cells were fractionated into cytoplasmic nuclear and chromatin extracts, which were analyzed by WB using the indicated Abs. Tubulin and histone were used as cytoplasmic and nuclear/chromatin controls. Bar, 10 μm. Dashed lines depict cell membrane. n.s., not statistically significant; **, P < 0.001 (Student t test).
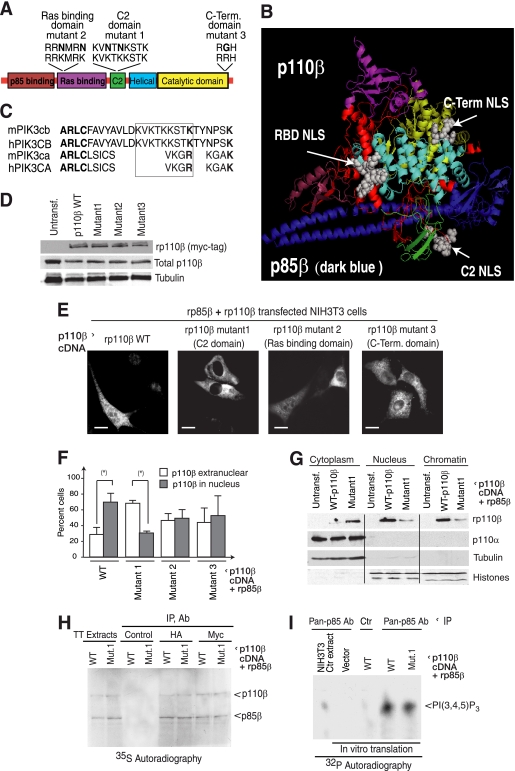
Fig. 4.
p110β contains an NLS motif in the C2 domain. (A) Domain structure of p110β, potential NLS sequences, and replacement of basic with nonbasic residues in mutants 1 to 3. (B) Computational model of the p85β/p110β complex. The p85β fragment (containing the nSH2 and iSH2 domains) is indicated in blue. The following p110β domains are indicated: the p85-binding domain (brown), the Ras-binding domain (purple), the catalytic domain (yellow), the C2 domain (green), and the helical domain (cyan). NLS sequences (located in the RBD, C2 domain, and C terminus) are shown as gray spheres; intermediate sequences are indicated in red. (C) Alignment of human and mouse p110β and p110α sequences at the region surrounding the C2 domain NLS of p110β (boxed). (D) NIH 3T3 cells were transfected with rp85β and WT or rp110β mutants 1 to 3. Expression levels were examined by WB. (E) The cellular localization of rp110β mutants was analyzed by IF using anti-Myc tag Ab. (F) Percent cells with the indicated phenotypes (n = 30). (G) NIH 3T3 cells were cotransfected with rp85β with WT-rp110β or NLS-p110β-mutant1. Cytoplasmic, nuclear, and chromatin fractions were examined by WB. Tubulin and histones were used as controls. Bar, 10 μm. (H) cDNA encoding mouse rp85β and WT- or C2 mutant1-rp110β were transcribed or translated in vitro in the presence of [35S]methionine. The association of rp85β with WT- or mutant1-rp110β was analyzed by HA- or Myc-tag IP. The extract composition (TT extracts) and p85/p110 complex formation were examined by SDS-PAGE and autoradiography. (I) cDNA encoding mouse rp85β and WT- or mutant1-rp110β were transcribed or translated as in panel A. rp85β/rp110β complexes were purified with anti-pan-p85 Ab and tested in an in vitro kinase assay using PtdIns(4,5)P2 as a substrate. *, P < 0.01 (Student t test).
Modeling of the p85β/p110β complex.
Models of the full-length p110β associated with the p85β fragment containing nSH2 and iSH2 domains were independently created by using I-TASSER (60), and their qualities were evaluated with the Swiss-MODEL server (3). The two models were structurally aligned to the corresponding chains in the crystal structure of the p85α(nSH2-iSH2)/p110α complex (PDB 3hhm [33]) in order to generate a draft model of the complex. This structural alignment was generated with the Dali system (19). Finally, the model of the complex was refined by molecular dynamics to remove clashes between chains, etc. Molecular dynamics analysis was performed using GROMACS (52).
Statistical analyses and databases.
The fluorescence intensity was quantitated using ImageJ software; to determine the nuclear signal, we selected the area and calculate the pixels referred to those found in the entire cell. Error bars represent the standard deviations of the mean values compared. Statistical significance was evaluated with a Student t test and the chi-square test calculated using Prism5V.5.0 software. For NES and NLS sequence identification, we used online databases (one at http://www.cbs.dtu.dk/databases/NESbase, CBS [Technical University of Denmark], and one at http://cubic.bioc.columbia.edu/db/NLSdb, Columbia University, respectively).
RESULTS
p110β concentrates in the nucleus.
Most of the research on inositide-dependent signal transduction pathways has focused on events that take place at the plasma membrane. Nonetheless, PI3K is also found in the nucleus (36, 38, 43); we previously reported that p110β, but not p110α, localizes at the cell nucleus concentrating at this site in S phase in NIH 3T3 cells (35). HeLa cells, primary MEFs, and SAOS-2 cells also contained nuclear p110β, as determined by IF analysis (Fig. 1A).
We also analyzed the localization of the other endogenous ubiquitous PI3K subunits in NIH 3T3 cells. Whereas p110β was predominantly nuclear, p110α localized mainly in the cytoplasm (Fig. 1B), as reported previously (35). The Abs used were shown to be specific, since the p110α or p110β IF signal decreased following depletion of the corresponding isoform (35). As for the p85 ubiquitous regulatory subunits, the majority of the p85α localized in the cytoplasm but p85β was more abundant in the nuclear compartment (Fig. 1B). To control antibody specificity, we cotransfected NIH 3T3 cells with shRNA for p85α or p85β and a green fluorescent protein (GFP) transfection reporter; cells transfected with p85α shRNA showed reduction of the p85α signal, whereas p85β-specific shRNA reduced the p85β signal (Fig. 1C).
In a complementary experiment, we confirmed intracellular localization for p85 and p110 subunits by cell fractionation and WB. We tested whether the distinct ubiquitous class IA PI3K subunits appeared in cytoplasmic, nuclear, or chromatin fractions (MEFs, HeLa cells, and NIH 3T3 cells) (Fig. 1D). Although a proportion of the different subunits appeared in the nucleus and the cytoplasm (visible in long exposures [data not shown]), p110β and p85β concentrated in the nucleus, in contrast to the cytoplasmic localization of p110α and p85α (Fig. 1D).
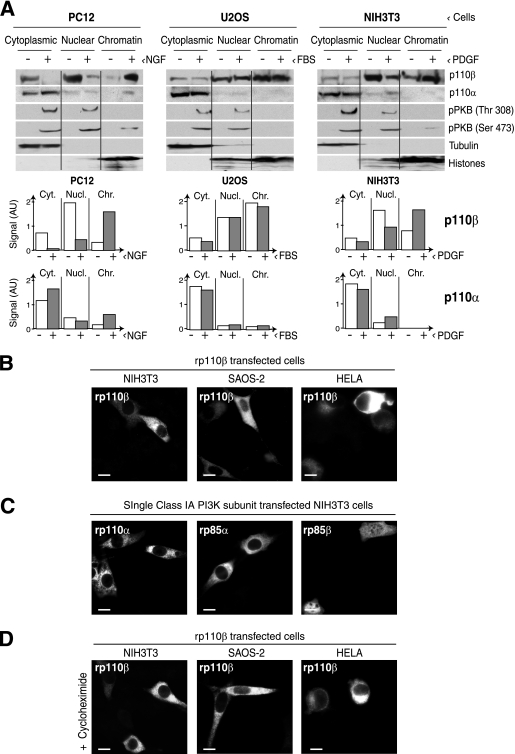
To determine the contribution of cell activation for p110β translocation to the nucleus, we examined various cell types (PC12, U2OS, and NIH 3T3) upon serum deprivation or after stimulation with growth factors (NGF, serum, and PDGF, respectively). Whereas p110α localization showed minor changes after cell stimulation in the three cell types, p110β was mainly nuclear even in quiescence and stimulation of PC12 and NIH 3T3 cells increased p110β fraction bound to chromatin (Fig. 2A).
Fig. 2.
Overexpressed p110β localizes in cytoplasm. (A) PC12, U2OS, and NIH 3T3 cells were cultured alone or treated with NGF (100 ng/ml), fetal bovine serum (20%), or PDGF (50 ng/ml), respectively (30 min). Cells were fractionated, and cytoplasmic nuclear and chromatin fractions were analyzed by WB using the indicated Abs. Graphs show the p110β and p110α signal intensities in arbitrary units (AU). (B) NIH 3T3, HeLa, and SAOS-2 cells were transfected with rp110β (48 h). rp110β localization was examined by IF using anti-p110β Ab. (C) Myc-tagged rp110α, HA-rp85α, and HA-rp85β were transfected individually into NIH 3T3 cells and processed for indirect IF with appropriate tag-specific antibodies. (D) rp110β-transfected NIH 3T3, HeLa, and SAOS-2 cells were treated with cycloheximide (10 μg/ml, 5 h) before IF staining as described in panel A. Bar, 10 μm.
p110β overexpression results in cytoplasmic retention.
To elucidate the structural features that determine p110β nuclear localization, we transfected full-length p110β into NIH 3T3 cells. Recombinant (r)p110β overexpression resulted in cytoplasmic accumulation of this protein (Fig. 2B). Transfection of Myc-tagged-rp110β yielded a similar result using anti-tag Ab for IF (data not shown). We confirmed that the entire sequence of the rp110β cDNA clones was correct. We also examined the localization of recombinant p110α, p85α, and p85β (rp110α, rp85α, and rp85β) in NIH 3T3 cells; rp110α and rp85α concentrated in the cytoplasm (Fig. 2C), similar to their endogenous counterparts (Fig. 1B). Overexpressed p85β showed diffuse cytoplasmic and nuclear staining (Fig. 2C), with a larger proportion of cytoplasmic protein compared to the endogenous protein (Fig. 1).
The ectopic cytoplasmic localization of recombinant p110β in cells could result from the accumulation of newly translated protein in the endoplasmic reticulum prior to translocation to the nuclei. To exclude this possibility, we transfected the rp110β and tested whether inhibition of de novo protein synthesis by cycloheximide treatment (5 h prior to IF analysis) facilitated the accumulation of rp110β to the nucleus. This was not the case; rp110β remained cytoplasmic after cycloheximide treatment, excluding that this fraction represents newly translated protein (Fig. 2C).
p85β promotes p110β nuclear localization.
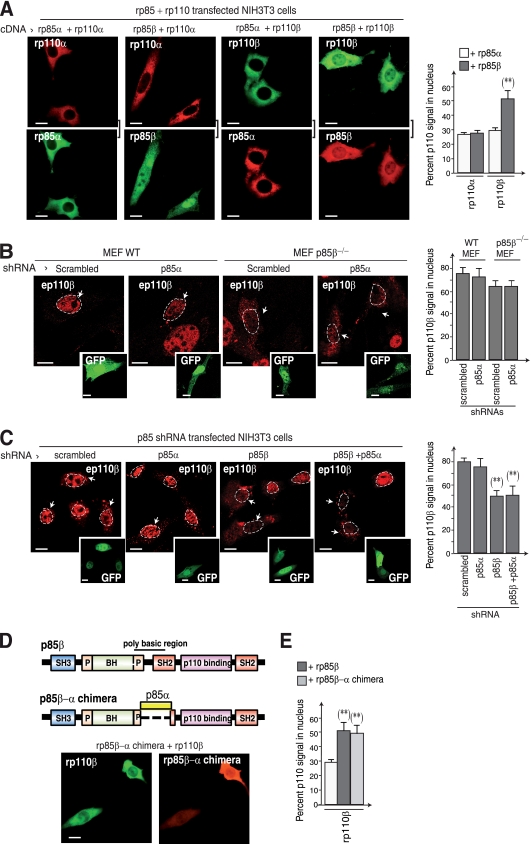
Class IA catalytic and regulatory subunits normally form heterodimers (16). We confirmed biochemically that p85α and p85β form complexes with either p110α or p110β (data not shown), as reported earlier (16). We examined the possibility that cytoplasmic accumulation of rp110β might result from the lack of sufficient associated regulatory subunit. To determine whether the ubiquitous regulatory subunits (p85α or p85β) were necessary for p110β nuclear localization, we cotransfected combinations of the ubiquitous catalytic and regulatory subunits.
Cotransfection of rp110α with rp85α or rp85β did not alter the cytoplasmic localization of rp110α, although rp85α was cytoplasmic and rp85β was cytoplasmic and nuclear (Fig. 3A). In contrast, cotransfection of rp110β with rp85β, but not with rp85α, yielded a significant proportion of nuclear rp110β (Fig. 3A). To analyze the contribution of endogenous p85 regulatory subunits in the nuclear localization of p110β, we analyzed its localization in WT or p85β-deficient MEFs. A moderate reduction in nuclear p110β was seen in p85β−/− MEFs (Fig. 3B), suggesting that other p110β-associated nuclear proteins (such as PCNA or Nbs1 [32, 35]) might facilitate p110β translocation to the nucleus in p85β-deficient cells. In contrast, acute reduction of p85β levels with shRNA (as in Fig. 1C) induced a significant decrease in p110β nuclear levels (Fig. 3C). Endogenous p85β thus regulates the nuclear entry of p110β.
Fig. 3.
The p85β regulatory subunit controls p110β nuclear translocation. (A) NIH 3T3 cells cotransfected with Myc-rp110β or -rp110α in combination with HA-rp85β or -rp85α (48 h) were fixed and analyzed by IF. Catalytic subunits were stained with anti-Myc-tag Ab and p85 with anti-HA Ab; square brackets indicate channels from the same image. Graphs show the percentage of p110 nuclear signal relative to the total (100%) (n = 30). (B) WT or p85β-deficient MEFs were cotransfected with GFP plus scrambled or p85α shRNA (48 h), and the cells were fixed and analyzed by IF using specific Abs. Insets show transfected (GFP+) cells. The graph shows the percentage of cells with predominant p110β nuclear staining (n = 30). (C) NIH 3T3 cells were cotransfected with GFP plus p85α- or p85β-specific shRNA or both (48 h), the cells were fixed, and nuclear p110β was analyzed by IF using specific Abs. Insets show transfected (GFP+) cells. Dashed lines depict the cell nuclei. Graphs are as described in panel A. (D) Scheme of the rp85β-α chimera. The p85β region between amino acids 78 to 351 was replaced with amino acids 77 to 363 from p85α. NIH 3T3 cells cotransfected with the HA-rp85β-α chimera plus rp110β were stained with anti-p110β and -HA Ab. (E) Graphs are as described in panel A. Bar, 10 μm. **, P < 0.001.
A polybasic region of p85β does not act as an NLS.
The finding that p85β expression, but not that of p85α, induced p110β nuclear localization led us to examine the primary sequence of p85α and p85β to search for potential nuclear localization sequences (NLSs). We found a polybasic region between the BCR (Bcr homologous region) and the N-SH2 region of p85β (residues 77 to 351); this sequence was not present in p85α. To establish the contribution of this p85β region in the nuclear localization of p85β/p110β complexes, we constructed a p85β-α chimera, replacing p85β amino acids 77 to 351 with the corresponding residues in the p85α sequence (amino acids 77 to 363; see Fig. 3D). We cotransfected the rp85β-α chimera with rp110β and examined rp110β subcellular distribution. No difference was observed in rp110β nuclear localization when cotransfected with the p85β-α chimera or WT p85β; the p85β-α chimera continued to localize with p110β in the nucleus (Fig. 3D), similar to WT-rp85β (Fig. 3A). Quantification of the proportion of p110α and p110β nuclear signal (Fig. 3E) confirmed that rp110β can transit to the nucleus when cotransfected with rp85β; nonetheless, the polybasic sequence located between BCR and N-SH2 domains in p85β is not a NLS.
The p110β C2 domain contains an NLS.
We sought potential NLSs in p110β that could explain the nuclear localization of p85β/p110β complexes and identified three putative NLS polybasic motifs in p110β, one in the C2 domain (residues 310 to 318; KVKTKKSTK), one in the Ras-binding domain (RBD; residues 149 to 154; RRKMRK), and one at the C terminus (residues 994 to 996; RRH) (Fig. 4A). To establish which of these motifs might be functional, we generated a structural model of the p85β(nSH2iSH2)/p110β complex (Fig. 4B) based on the p85α (nSH2iSH2)/p110α structure (33). This model showed that the basic motif in the C2 domain is located in a loop in close proximity to p85β; only the residues at the beginning and the end of the NLS are resolved in this structure (Fig. 4B). Alignment of this region in p110β and p110α primary structure (Fig. 4C), as well as examination of p85α/p110α structure (24, 33, 42), showed that most of this motif is lost in p110α. The other candidate motifs are not found near p85β and seem less likely to be affected by interaction with this protein (Fig. 4B).
We replaced several basic residues in each of the three motifs with nonbasic residues to generate the C2 domain NLS-p110β-mutant1 (KVNTTKSTK), RBD NLS-p110β-mutant2, and C-terminal NLS-p110β-mutant3 (RGH) (Fig. 4A). The expression levels of these constructs were similar (Fig. 4D). We tested whether any of these mutants, in combination with rp85β, was excluded from the nucleus. NIH 3T3 cells transfected with rp85β and the rp110βNLS mutants in the RBD and C-terminal domain showed minor differences compared to WT-rp110β; in contrast, the C2 domain NLS-p110β-mutant1 was cytoplasmic (Fig. 4E and F). This suggested that the KVKTKRSTK motif in the C2 domain acts as an NLS for p110β. Separation of cells expressing the NLS-p110β-mutant1 plus rp85β into cytoplasmic, nuclear, and chromatin fractions showed that WT-rp110β localized in nuclear and chromatin fractions and confirmed that mutation of the NLS-p110β-mutant1 is mainly cytoplasmic, similar to p110α (Fig. 4G).
We confirmed that mutation in the C2 domain does not affect association of in vitro-transcribed translated purified p110β to purified p85β (Fig. 4H). A similar association of rp110β or NLS-p110β-mutant1 with rp85β was confirmed in transfected NIH 3T3 cells (not shown). Moreover, there was no difference in kinase activity between WT or mutant1-rp110β (Fig. 4I). Thus, the C2 mutant associates with p85β similarly to WT-p110β and shows kinase activity but does not translocate to the nucleus.
p85β regulates p110β nuclear exit.
We previously observed changes in the relative amount of nuclear p110β during cell cycle progression, suggesting that this molecule shuttles in and out of the nucleus (35). We studied the mechanism that controls p110β nuclear export. Various means of nuclear export have been documented (28); the most common mechanism is a conserved leucine-rich NES that binds the nuclear export protein Crm1 (11, 31). We used leptomycin B to inhibit Crm1 binding to the cargo proteins; this treatment results in retention of NES-containing proteins in the nucleus (11). NIH 3T3 cells were transfected with rp110α, rp110β, rp85α, or rp85β constructs and, after 24 h, the cells were treated with leptomycin B (5 ng/ml, 2 h). After leptomycin B treatment, only rp85β showed a notable increase in the amount of nuclear protein (Fig. 5A). This suggests that nuclear exit of p85β/p110β complex is mediated by an NES located in p85β via Crm1. The moderate enhancement of p110β nuclear localization after leptomycin B treatment might result from association to endogenous p85β.
Fig. 5.
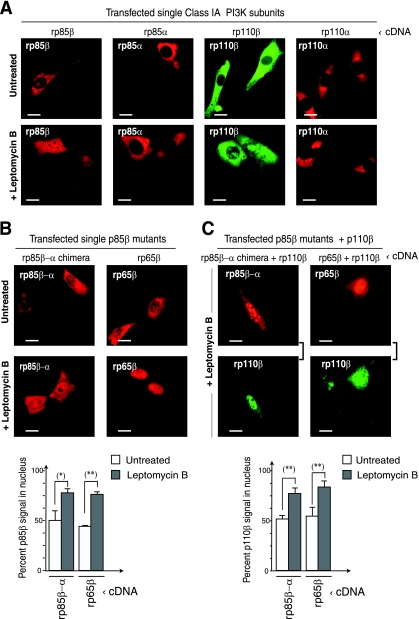
p85β regulates p110β nuclear export. (A) NIH 3T3 cells were transfected with HA-rp85β, HA-rp85α, Myc-rp110β, or Myc-rp110α (48 h). Transfected cells were untreated or leptomycin B treated (5 ng/ml; 2 h) before fixing. Samples were stained for IF using anti-HA or -Myc tag Ab. (B) NIH 3T3 cells were transfected with the HA-rp85β-α chimera or HA-rp65β (48 h). Cells were untreated or pretreated with leptomycin B 2 h prior to fixing, and then stained as described above. The graph shows the percentage of nuclear signal relative to total cell signal (100%) (n = 30). (C) rp85β/rp110β or p65β/p110β were expressed in NIH 3T3 cells (48 h). The cells were treated with leptomycin B as described above and examined by IF; square brackets indicate channels from the same image. The graph is as described in panel B. Bar, 10 μm. *, P < 0.01; **, P < 0.001.
To define the putative region containing the NES in p85β, we transfected the rp85β-α chimera described above and tested whether leptomycin B treatment affected its intracellular localization. Overexpressed p85β-α chimera localized to the cytoplasm and nucleus and responded to leptomycin B treatment by increasing its nuclear localization (Fig. 5B), similar to rp85β (Fig. 5A). A C-terminal deletion mutant in p85α (p65α) behaves as an oncogene (27); a similar deletion in p85β was reported in a tumor cell line (25). We prepared a similar C-terminal deletion mutant in p85β (p65β) lacking residues 562 to 723 of the C terminus and tested the effect of leptomycin B treatment on its subcellular localization; p65β behaved as did WT p85β (Fig. 5A and B). Indeed, transfection of rp110β plus rp85β, the p85β-α chimera, or rp65β, followed by leptomycin B treatment of cells, led to a comparable increase in p110β nuclear localization (Fig. 5A and C).
Thus, p85β regulates p110β nuclear import and export; however, neither p85β residues 77 to 351 nor the p85β C-terminal region (amino acids 562 to 723) control p85β/p110β nuclear exit.
The p85β N-terminal region has an NES.
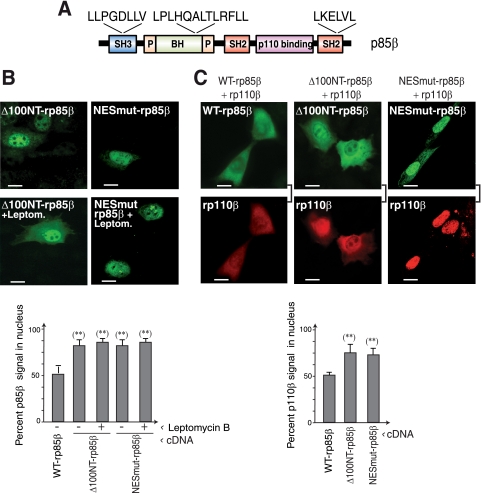
To determine the p85β region involved in nuclear export, we used specific NES databases to search for conserved leucine-rich regions; this search rendered three potential NES motifs (Fig. 6A). One of these was found at residues 683 to 688, although these residues are absent in rp65β, a mutant that behaves like WT p85β after leptomycin B treatment. An alternative high score region was found at residues 214 to 229, which are absent in the rp85β-α chimera; since this chimera remains sensitive to leptomycin B treatment (Fig. 5), this motif is not a functional NES for p85β. Finally, a potential motif was indicated at residues 25 to 32. We generated a 100-amino-acid N-terminal deletion mutant of p85β (Δ100Np85β), as well as a double point mutation in this Leu-rich motif (L25 and L30; NESmut-p85β). Deletion or mutation of this region rendered p85β predominantly at the nucleus and unaffected by leptomycin B treatment (Fig. 6B), confirming that this region contains a functional Crm1-regulated NES sequence.
Fig. 6.
The p85β subunit has a Crm1-dependent nuclear export signal. (A) Scheme of Leu-rich regions in the p85β sequence. (B) NIH 3T3 cells were transfected with cDNA encoding Δ100Np85β or NESmut-rp85β (48 h). Transfected cells were untreated or leptomycin B treated (5 ng/ml, 2 h) and processed for IF using anti-HA Ab. The graph shows the percentage of nuclear signal relative to total cell signal (100%) (n = 30). (C) NIH 3T3 cells were cotransfected with Myc-rp110β and either Δ100Np85β or NESmut-rp85β (48 h); protein localization was analyzed by IF using anti-HA or -p110β Ab. The graph is as described in panel B. Bar, 10 μm. **, P < 0.001.
We examined the role of this region in p110β nuclear export. NIH 3T3 cells transfected with rp110β in combination with Δ100NT-p85β or with NESmut-p85β showed an increase in rp110β nuclear localization (Fig. 6C), confirming a contribution of p85β residues 25 to 32 in the regulation of p85β/p110β nuclear export.
Reconstitution of p110β-deficient cells with nuclear but not cytoplasmic p110β restores cell survival.
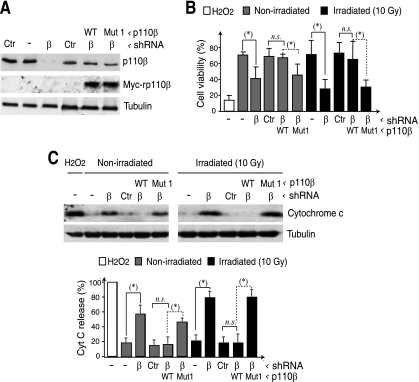
Mice deficient in p110β die at embryonic days 2 to 3 (5). We previously showed that p110β is mainly nuclear and controls DNA replication and repair (32, 34, 35); in the course of these studies, we observed that efficient p110β knockdown reduced cell survival (34). To test whether p110β nuclear localization influences cell survival, we depleted NIH 3T3 cells of p110β using shRNA and reconstituted p110β expression with WT-rp110β or cytoplasmic NLS-p110β-mutant1. WB was used to confirm p110β silencing with specific shRNA, as well as the expression of WT or mutant rp110β (Fig. 7A). We cotransfected cells with p110β-specific shRNA and shRNA-resistant humanWT-p110β or shRNA-resistant human NLS-p110β-mutant1; the second combination was more sensitive to spontaneous and gamma-irradiation-induced apoptosis than untransfected cells or rp110β WT-expressing cells (Fig. 7B).
Fig. 7.
Nuclear localization of p110β is necessary for cell survival. (A) NIH 3T3 cells were cotransfected with p110β-specific shRNA and rp85β plus shRNA-resistant WT-p110β or -NLS-p110β mutant1 (48 h); protein expression was examined by WB. (B) NIH 3T3 cells were transfected as described in panel A. After 24 h, irradiated (10 Gy) or unirradiated cells were cultured for a further 24 h. As a positive control, cells were treated with H2O2. Cell viability was determined by flow cytometry using annexin V and propidium iodide (mean ± the standard deviation [SD]; n = 3). (C) Transfected NIH 3T3 cells from panel A were lysed, and the cytoplasmic fraction was analyzed by WB. The graph shows the percentage of cytochrome c relative to the maximal signal from H2O2-treated cells (100%) (mean ± the SD; n = 3). n.s., not statistically significant; *, P < 0.01.
As an alternative method to examine apoptosis, we monitored cytochrome c release in WB. Cytochrome c was present in the cytoplasmic fractions of apoptotic positive control cells (H2O2 treated), as well as in cells lacking p110β expression, but not in controls (Fig. 7C). Expression of shRNA-resistant WT-p110β nonetheless rescued cell death, since it decreased cytochrome c release; in contrast, expression of the shRNA-resistant cytoplasmic C2-domain NLS-p110β-mutant1 did not reduce cytochrome c release (Fig. 7C). The results indicate that nuclear localization of p110β is necessary for cell viability and that its expression in the cytoplasm does not prevent apoptotic events.
DISCUSSION
We examined the structural features that determine p110β nuclear localization. Whereas overexpressed recombinant p110β remains mainly cytoplasmic, transfected rp110β in combination with rp85β, but not rp85α, localizes to the nucleus. Although the ubiquitous catalytic and regulatory subunits form all possible heterodimeric combinations (16), only p85β/p110β complexes localize efficiently in the nucleus. The search for nuclear localization motifs in p85β and p110β yielded several candidate sequences, but only mutation of the NLS located within the p110β C2 domain significantly reduced p110β nuclear localization. The fact that p110β alone does not enter the nucleus suggests that p110β must associate with p85β for its NLS motif to be functional; the predicted quaternary structure of p85β/p110β reported here supports this possibility (see below). p110β nuclear PI3K activity is maximal in S phase (35), suggesting that p85β/p110β complexes shuttle in and out of the nucleus. We identify here a functional NES in p85β which, when deleted, increases p85β/p110β nuclear localization.
Proteins enter the nucleus through nuclear pores, large macromolecular complexes composed of nucleoporins. Understanding of macromolecular transport processes across the nuclear envelope has increased in recent years, and many transport receptors have been identified. Most of these receptors are similar to the import receptor importin β (karyopherin β). Members of this family have been classified as importins or exportins, and both types are regulated by the GTPase Ran. Importins recognize their substrates in cytoplasm and transport them to the nucleus; once in the nucleus, RanGTP binds to importins, inducing the release of import cargoes. In contrast, exportins interact with their substrates only in the nucleus in the presence of RanGTP and release them after GTP hydrolysis in the cytoplasm (reviewed in reference 49). Nuclear import and export are multistep processes initiated by the recognition of NLSs or NESs. The most thoroughly examined import signal (“classical” and bipartite NLS) contains multiple basic residues. Their transport is mediated by importin β, which directly associates these NLS via the adaptor protein importin α (49). The functional NLS in p110β is homologous to that found in class II PI3KC2α, which also transits to the nucleus (10), suggesting potential conservation of structural features for nuclear import between PI3K classes.
The best-studied exportins are Crm1/Xpo1, which recognizes leucine-rich NES. Crm1 forms a stable ternary complex with Ran-GTP and with NES cargoes that can exit the nucleus. Studies of Crm1-mediated export were aided by the discovery of the antifungal agent leptomycin B, a highly specific and potent inhibitor of Crm1 function (11). Of the three potential NES sequences in p85β, only the one located at the N terminus regulated the nuclear localization of p85β/p110β. In the p85β/p110β complex, p110β therefore contributes by providing the NLS, whereas p85β supplies a functional NES, showing that this complex acts as a single entity for nuclear transport. Indeed, the predicted structure of p85β/p110β described here (based on that in reference 33) shows that the NLS sequence in the C2 domain is in close proximity to p85β, supporting the possibility that p110β association to p85β alters p110β structure in this region to yield a functional NLS.
Neither p110β nor p85β is exclusively nuclear; the cytoplasmic forms might represent complexes with p85α and p110α, respectively, or p85β/p110β complexes in transit from both compartments. In the case of p85β, its overexpression renders a fraction of this protein nuclear, suggesting that it associates with other NLS-containing proteins. In support of this possibility, p85α and, to a greater extent, p85β, associates with X-box binding protein 1 (XBP1), modulating the nuclear localization of this transcription factor (which contains an NLS) (46, 59). Similarly, in the case of p110β, association with p85β is critical for its translocation to the nucleus; however, other p110β-associated nuclear proteins (such as PCNA or Nbs1 [32, 35]) might facilitate the translocation of p110β to the nucleus in p85β-deficient cells.
We focused on a comparison of the class IA PI3K isoforms p110α and p110β; there is nonetheless an additional class IA isoform, p110δ, as well as the closely related class IB p110γ isoform, which associates with p101 and p84 regulatory subunits (50). When overexpressed in HepG2 cells, p110γ localizes to the nucleus after serum treatment; in this case, interference of p110γ association with p101 increases p110γ nuclear localization (41). There is no region homologous to that of the p110β C2 domain in p110γ (45), although we found polybasic motifs in the N terminus, in the helical domain, and at the beginning of the C2 domain (data not shown). Alignment of the NLS region in p110β and p110α (Fig. 4) shows that most of this basic motif in p110β is lost in p110α. Comparison of the p85α/p110α structure (24, 33, 42) to the p85β/p110β structural prediction described here (Fig. 4) also shows that the loop in which p110β NLS localizes is much shorter in p110α. These observations might explain why a large proportion of p110β, but not of p110α, localizes to the nucleus. An in silico search for p110β NLS homologues in p110δ, as well as p110δ structure (4), showed a similar polybasic region in p110β and p110δ C2 domains; further study is needed to define whether the p110δ motif is a functional NLS.
The first report of nuclear PIP3 showed rapid translocation of a PIP3-binding protein (PIP3BP), which is abundant in brain, to the nuclei of the rat pheochromocytoma PC12 cell line after NGF treatment, as well as in PDGF-treated NIH 3T3 cells (51). In human promyelocytic HL60 cells, both retinol and vitamin D3 induced differentiation to granulocytes or monocytes, respectively, and triggered an increase in nuclear p85 staining (reviewed in references 36 and 37). In all of these cases, the authors defined the specific isoform localizing to the nucleus. The negative regulator of PI3K, PTEN, is also reported to transit to the nucleus and regulate cell survival (17). In the case of nuclear PI3K in PC12 cells, a nucleus-specific phospholipase C activates a neuron-specific GTPase, PIKE (phosphoinositide 3-kinase enhancer), which is able to increase nuclear PI3K activity (56, 57). Isolated nuclei from PC12 cells treated with NGF or transfected with active PI3K were resistant to DNA fragmentation factor caspase-activated DNase (DFF40/CAD); interference with p110α diminished NGF protection from apoptosis, supporting p110α control of nuclear PIP3 in PC12 cells (1).
The antiapoptotic function of nuclear PIP3 in PC12 cells is proposed to result from PIP3 binding to B23 nucleophosmin, a protein that inhibits DFF40/CAD (20, 55). Other authors have suggested that nuclear PKB function in NGF-treated PC12 cells is mediated by PKB phosphorylation of acinus, resulting in acinus inhibition of apoptotic chromatin condensation (23). A third mechanism has been reported for the function of nuclear PI3K in NGF-treated PC12 cells, PCKζ-PI3K-dependent nuclear translocation, which mediates phosphorylation of nucleolin, a stabilizing agent for the antiapoptotic protein Bcl-2 (48). These results indicate that in some cell types (PC12 cells), the neurotrophin NGF activates nuclear PI3K, which in turn induces cell survival.
We report here that the p85β/p110β complex localizes to the nucleus in several cell types. This translocation is regulated by an NLS sequence in the p110β C2 domain and by an NES in the p85β N-terminal domain. Our results demonstrate that the p85β/p110β complex regulates cell viability only when it is correctly localized at the cell nucleus.
ACKNOWLEDGMENTS
We thank M. White for the myc-p110α plasmid, B. Vanhaesebroeck for the p110β plasmid, A. Klippel for anti-p110 Ab, F. Pazos for support in p85β/p110β structure prediction, and C. Mark for editorial assistance.
A.K. held a predoctoral fellowship associated with a project financed by the Fundación Ramón Areces. J.R.-M. has a JAE postdoctoral fellowship from the Spanish National Research Council (CSIC). V.P.-G received a predoctoral FPI fellowship associated with a project financed by the Spanish Ministry of Science and Innovation (MICINN). This study was financed by grants from the Spanish Association Against Cancer (AECC), the MICINN (SAF2007-63624 and SAF2010-21019 and Network of Cooperative Research in Cancer RD07/0020/2020), the Madrid regional government (S-BIO-0189/06), the Sandra Ibarra Foundation, and Genoma España.
Footnotes
Published ahead of print on 7 March 2011.
REFERENCES
- 1. Ahn J. Y., Rong R., Liu X., Ye K. 2004. PIKE/nuclear PI3K signaling mediates the antiapoptotic actions of NGF in the nucleus. EMBO J. 23:3995–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alcázar I., et al. 2009. p85β PI3K regulates CD28 coreceptor function. Blood 113:3198–3208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arnold, Bordoli K. L., Kopp J., Schwede T. 2006. The Swiss-MODEL Workspace: a web-based environment for protein structure homology modeling. Bioinformatics 22:195–201 [DOI] [PubMed] [Google Scholar]
- 4. Berndt A., et al. 2010. The p110δ structure: mechanisms for selectivity and potency of new PI(3)K inhibitors. Nat. Chem. Biol. 6:117–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bi L., Okabe I., Bernard D. J., Nussbaum R. L. 2002. Early embryonic lethality in mice deficient in the p110β catalytic subunit of PI3K. Mamm. Genome 13:169–172 [DOI] [PubMed] [Google Scholar]
- 6. Bi L., Okabe I., Bernard D. J., Wynshaw-Boris A., Nussbaum R. L. 1999. Proliferative defect and embryonic lethality in mice homozygous for a deletion in the p110α subunit of PI3K. J. Biol. Chem. 274:10963–10968 [DOI] [PubMed] [Google Scholar]
- 7. Carvalho S., Milanezi F., Costa J. L., Amendoeira I., Schmitt F. 2010. PIKing the right isoform: the emergent role of the p110β subunit in breast cancer. Virchows Arch. 456:235–243 [DOI] [PubMed] [Google Scholar]
- 8. Czauderna F., et al. 2003. Functional studies of the PI(3)-kinase signaling pathway employing synthetic and expressed shRNA. Nucleic Acids Res. 31:670–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Denley A., Kang S., Karst U., Vogt P. K. 2008. Oncogenic signaling of class I PI3K isoforms. Oncogene 27:2561–2574 [DOI] [PubMed] [Google Scholar]
- 10. Didichenko S. A., Thelen M. 2001. PI3K c2α contains a nuclear localization sequence and associates with nuclear speckles. J. Biol. Chem. 276:48135–48142 [DOI] [PubMed] [Google Scholar]
- 11. Fornerod M., Ohno M., Yoshida M., Mattaj I. W. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051–1060 [DOI] [PubMed] [Google Scholar]
- 12. Foukas L. C., Berenjeno I. M., Gray A., Khwaja A., Vanhaesebroeck B. 2010. Activity of any class IA PI3K isoform can sustain cell proliferation and survival. Proc. Natl. Acad. Sci. U. S. A. 107:11381–11386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Foukas L. C., et al. 2006. Critical role for the p110α PI3K in growth and metabolic regulation. Nature 441:366–370 [DOI] [PubMed] [Google Scholar]
- 14. Fruman D. A., Meyers R. E., Cantley L. C. 1998. Phosphoinositide kinases. Annu. Rev. Biochem. 67:481–507 [DOI] [PubMed] [Google Scholar]
- 15. García Z., et al. 2006. A PI3K activity-independent function of p85 subunit in control of mammalian cytokinesis. EMBO J. 25:4740–4751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Geering B., Cutillas P. R., Nock G., Gharbi S. I., Vanhaesebroeck B. 2007. Class IA PI3K are obligate p85-p110 heterodimers. Proc. Natl. Acad. Sci. U. S. A. 104:7809–7814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gil A., et al. 2006. Nuclear localization of PTEN by a Ran-dependent mechanism enhances apoptosis: involvement of an N-terminal nuclear localization domain and multiple nuclear exclusion motifs. Mol. Biol. Cell 17:4002–4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hiles I. D., et al. 1992. PI3K: structure and expression of the 110-kDa catalytic subunit. Cell 70:419–429 [DOI] [PubMed] [Google Scholar]
- 19. Holm L., Rosenström P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38:W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hsu C. Y., Yung B. Y. 2000. Over-expression of nucleophosmin/B23 decreases the susceptibility of human leukemia HL-60 cells to retinoic acid-induced differentiation and apoptosis. Int. J. Cancer 88:392–400 [DOI] [PubMed] [Google Scholar]
- 21. Hu P., Mondino A., Skolnik E. Y., Schlessinger J. 1993. Cloning of a novel, ubiquitously expressed human PI3K and identification of its binding site on p85. Mol. Cell. Biol. 13:7677–7688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hu Q., Klippel A., Muslin A. J., Fantl W. J., Williams L. T. 1995. Ras-dependent induction of cellular responses by constitutively active phosphatidylinositol-3 kinase. Science 268:100–102 [DOI] [PubMed] [Google Scholar]
- 23. Hu Y., et al. 2005. Akt phosphorylates acinus and inhibits its proteolytic cleavage, preventing chromatin condensation. EMBO J. 24:3543–3554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang C. H., et al. 2007. The structure of a human p110α/p85α complex elucidates the effects of oncogenic PI3Kα mutations. Science 318:1744–1748 [DOI] [PubMed] [Google Scholar]
- 25. Janssen J. W. G., Schelithoff L., Bartram C. R., Schulz A. S. 1998. An oncogenic fusion product of the PI3K p85β subunit and HUMORF8, a putative deubiquitinating enzyme. Oncogene 16:1767–1772 [DOI] [PubMed] [Google Scholar]
- 26. Jia S., et al. 2008. Essential roles of PI(3)K-p110β in cell growth, metabolism, and tumorigenesis. Nature 454:776–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jiménez C., et al. 1998. Identification and characterization of a new oncogene derived from the regulatory subunit of PI3K. EMBO J. 17:743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kaffman A., O'Shea E. K. 1999. Regulation of nuclear localization: a key to a door. Annu. Rev. Cell Dev. Biol. 15:291–339 [DOI] [PubMed] [Google Scholar]
- 29. Kang S., Bader A. G., Vogt P. K. 2005. PI3K mutations identified in human cancer are oncogenic. Proc. Natl. Acad. Sci. U. S. A. 102:802–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kau T. R., Silver P. A. 2003. Nuclear transport as a target for cell growth. Drug Discov. Today 8:78–85 [DOI] [PubMed] [Google Scholar]
- 31. Kudo N., et al. 1998. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 242:540–547 [DOI] [PubMed] [Google Scholar]
- 32. Kumar A., Fernández-Capetillo O., Carrera A. C. 2010. Nuclear PI3Kβ controls double-strand break DNA repair. Proc. Natl. Acad. Sci. U. S. A. 107:7491–7496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mandelker D., et al. 2009. A frequent kinase domain mutation that changes the interaction between PI3Kα and the membrane. Proc. Natl. Acad. Sci. U. S. A. 106:16996–17001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marqués M., et al. 2008. PI3K p110α and p110β regulate cell cycle entry, exhibiting distinct activation kinetics in G1 phase. Mol. Cell. Biol. 28:2803–2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marqués M., et al. 2009. Specific function of PI3K beta in the control of DNA replication. Proc. Natl. Acad. Sci. U. S. A. 106:7525–7530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martelli A. M., et al. 2007. Nuclear phosphatidylinositol 3,4,5-trisphosphate, phosphatidylinositol 3-kinase, Akt, and PTEN: emerging key regulators of antiapoptotic signaling and carcinogenesis. Eur. J. Histochem. 51(Suppl. 1):125–131 [PubMed] [Google Scholar]
- 37. Martelli A. M., et al. 2006. Intranuclear 3′-phosphoinositide metabolism and Akt signaling: new mechanisms for tumorigenesis and protection against apoptosis? Cell Signal. 18:1101–1107 [DOI] [PubMed] [Google Scholar]
- 38. Martelli A. M., et al. 2006. PI3K /Akt signaling pathway and its therapeutic implications for human acute myeloid leukemia. Leukemia 20:911–928 [DOI] [PubMed] [Google Scholar]
- 39. Martínez-Gac L., Marqués M., García Z., Campanero M. R., Carrera A. C. 2004. Control of cyclin G2 mRNA expression by forkhead transcription factors: novel mechanism for cell cycle control by PI3K and forkhead. Mol. Cell. Biol. 24:2181–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Méndez J., Stillman B. 2000. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20:8602–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Metjian A., Roll R. L., Ma A. D., Abrams C. S. 1999. Agonists cause nuclear translocation of PI3Kγ: a Gβγ-dependent pathway that requires the p110γ amino terminus. J. Biol. Chem. 274:27943–27947 [DOI] [PubMed] [Google Scholar]
- 42. Miled N., et al. 2007. Mechanism of two classes of cancer mutations in the PI3K catalytic subunit. Science 317:239–2342 [DOI] [PubMed] [Google Scholar]
- 43. Neri L. M., Borgatti P., Capitani S., Martelli A. M. 2002. The nuclear PI3K/AKT pathway: a new second messenger system. Biochim. Biophys. Acta 1584:73–80 [DOI] [PubMed] [Google Scholar]
- 44. Okkenhaug K., et al. 2002. Impaired B and T cell antigen receptor signaling in p110δ PI3K mutant mice. Science 297:1031–1104 [DOI] [PubMed] [Google Scholar]
- 45. Pacold M. E., et al. 2000. Crystal structure and functional analysis of Ras binding to its effector PI3Kγ. Cell 103:931–943 [DOI] [PubMed] [Google Scholar]
- 46. Park S. W., et al. The regulatory subunits of PI3K, p85α and p85β, interact with XBP-1 and increase its nuclear translocation. Nat. Med. 16:429–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Roche S., Downward J., Raynal P., Courtneidge S. A. 1998. A function for PI3Kβ (p85α-p110β) in fibroblasts during mitogenesis: requirement for insulin- and lysophosphatidic acid-mediated signal transduction. Mol. Cell. Biol. 18:7119–7129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sengupta T. K., Bandyopadhyay S., Fernandes D. J., Spicer E. K. 2004. Identification of nucleolin as an AU-rich element binding protein involved in bcl-2 mRNA stabilization. J. Biol. Chem. 279:10855–10863 [DOI] [PubMed] [Google Scholar]
- 49. Ström A. C., Weis K. 2001. Importin-beta-like nuclear transport receptors. Genome Biol. 2:3008.1–3008.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Suire S., et al. 2005. p84, a new Gβγ-activated regulatory subunit of the type IB phosphoinositide 3-kinase p110γ. Curr. Biol. 15:566–570 [DOI] [PubMed] [Google Scholar]
- 51. Tanaka K., et al. 1999. Evidence that a phosphatidylinositol 3,4,5-trisphosphate-binding protein can function in nucleus. J. Biol. Chem. 274:3919–3922 [DOI] [PubMed] [Google Scholar]
- 52. van der Spoel D., et al. 2005. GROMACS: fast, flexible, and free. J. Comput. Chem. 26:1701–1718 [DOI] [PubMed] [Google Scholar]
- 53. Vanhaesebroeck B., Leevers S. J., Panayotou G., Waterfield M. D. 1997. PI3K: a conserved family of signal transducers. Trends Biochem. Sci. 22:267–272 [DOI] [PubMed] [Google Scholar]
- 54. Wee S., et al. 2008. PTEN-deficient cancers depend on PIK3CB. Proc. Natl. Acad. Sci. U. S. A. 105:13057–13062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wu M. H., Chang J. H., Yung B. Y. 2002. Resistance to UV-induced cell-killing in nucleophosmin/B23 overexpressed NIH 3T3 fibroblasts: enhancement of DNA repair and up-regulation of PCNA in association with nucleophosmin/B23 overexpression. Carcinogenesis 23:93–100 [DOI] [PubMed] [Google Scholar]
- 56. Ye K., et al. 2002. Phospholipase Cγ1 is a physiological guanine nucleotide exchange factor for the nuclear GTPase PIKE. Nature 415:541–544 [DOI] [PubMed] [Google Scholar]
- 57. Ye K., et al. 2000. Pike. A nuclear GTPase that enhances PI3 kinase activity and is regulated by protein 4.1N. Cell 103:919–930 [DOI] [PubMed] [Google Scholar]
- 58. Yip S. C., et al. 2004. Over-expression of the p110beta but not p110α isoform of PI3K inhibits motility in breast cancer cells. Cell Motil. Cytoskeleton 59:180–188 [DOI] [PubMed] [Google Scholar]
- 59. Yoshida H., Oku M., Suzuki M., Mori K. 2006. pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J. Cell Biol. 172:565–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40. [DOI] [PMC free article] [PubMed] [Google Scholar]