Abstract
Bromodomain protein 4 (Brd4) plays critical roles in development, cancer progression, and virus-host pathogenesis. To gain mechanistic insight into the various biological functions of Brd4, we performed a proteomic analysis to identify and characterize Brd4-associated cellular proteins. We found that the extraterminal (ET) domain, whose function has to date not been determined, interacts with NSD3, JMJD6, CHD4, GLTSCR1, and ATAD5. These ET-domain interactions were also conserved for Brd2 and Brd3, the other human BET proteins tested. We demonstrated that GLTSCR1, NSD3, and JMJD6 impart a pTEFb-independent transcriptional activation function on Brd4. NSD3 as well as JMJD6 is recruited to regulated genes in a Brd4-dependent manner. Moreover, we found that depletion of Brd4 or NSD3 reduces H3K36 methylation, demonstrating that the Brd4/NSD3 complex regulates this specific histone modification. Our results indicate that the Brd4 ET domain through the recruitment of the specific effectors regulates transcriptional activity. In particular, we show that one of these effectors, NSD3, regulates transcription by modifying the chromatin microenvironment at Brd4 target genes. Our study thus identifies the ET domain as a second important transcriptional regulatory domain for Brd4 in addition to the carboxyl-terminal domain (CTD) that interacts with pTEFb.
INTRODUCTION
One mechanism underlying the regulation of gene expression is the targeting of multiprotein complexes to modified histones, which then alters the chromatin microenvironment to stimulate or inhibit gene expression. The bromodomains and extraterminal (BET) domain family of proteins are characterized by the presence of two conserved domains, the tandem, amino-terminal bromodomains (BDI and BDII), which bind acetylated chromatin, and an extraterminal (ET) domain, whose function is unknown. The BET family is conserved from yeast to mammals and includes Saccharomyces cerevisiae bromodomain factor 1 (bdf1) and bromodomain factor 2 (bdf2), Drosophila melanogaster female sterile homeotic [fs(1)h], and mammalian Brd2, Brd3, Brd4, and testes/oocyte-specific BrdT/Brd6. In yeast, deletion of bdf1 leads to a reduced growth rate and deletion of both bdf1 and bdf2 is lethal (27). Mutations of fs(1)h cause segmental abnormalities, including missing organs and homeotic transformations in the progeny of mutant females in Drosophila (13). Knockout of Brd4 or Brd2 in mice results in early embryonic lethality (18, 21).
The BET proteins have been shown to be important players in human disease, including viral infections and cancer. Several different viruses target the individual BET proteins for a variety of purposes but often to regulate viral and cellular transcription (4, 7, 31, 37, 41, 45, 57, 60). The papillomavirus E2 proteins bind to Brd4, and some utilize this interaction in tethering the viral genomes to mitotic chromosomes (1, 3, 57, 59). The papillomavirus E2 transcriptional activation functions are also mediated through Brd4 (35, 41, 42). With regard to human cancer, the Brd4-NUT and Brd3-NUT fusion oncoproteins are causally linked to a lethal form of undifferentiated carcinoma (15–17). The fusion protein is targeted to chromatin by the BET protein moiety, and the NUT component recruits p300/CBP, creating hyperacetylated chromatin domains (38). The gene expression signature resulting from ectopic expression of Brd4 predicts the clinical prognosis of patients with estrogen receptor-positive breast carcinoma (8). Additionally, Brd2 also has a role in lymphomagenesis (reviewed in reference 51).
A number of studies highlight the conserved roles of the BET proteins in gene expression regulation. The yeast bdf1 regulates the expression of a large subset of genes and has an antisilencing effect on euchromatin-heterochromatin boundaries (27). Bdf1 interacts with the general transcriptional machinery component TATA-binding protein (TBP)-associated factor 67 (34). Brd2 and Brd3 allow transcription by RNA polymerase II (RNA pol II) through hyperacetylated chromosomes using in vitro systems (29). Brd2 associates with various transcription regulatory proteins, including E2F transcription factors, histone deacetylases, and the RNA polymerase II Mediator complex (9, 10, 25, 44). Brd4 regulates gene expression and is unique among the human BET family proteins in having a carboxyl-terminal domain (CTD) not present in the other BET family proteins. The Brd4 CTD interacts with the cyclin T1 and Cdk9 subunits of positive transcription elongation factor b (pTEFb) complex (24, 56). Brd4 displaces negative regulators, the HEXIM1 and 7SKsnRNA complex, from pTEFb, thereby transforming it into an active form that can then phosphorylate RNA pol II. This recruitment of pTEFb by Brd4 stimulates the transcription of primary responsive genes (19). Moreover, papillomavirus E2-mediated viral transcriptional activation and repression functions are dependent on Brd4 (22, 35, 41, 42, 45, 52).
While all the members of the BET family have been linked to transcriptional regulation, it is only a subset of these proteins, specifically Brd4 and fs(1)h, that possess the CTD, which recruits pTEFb. This suggests that there must be a CTD-independent mechanism by which BET family proteins mediate transcriptional regulation. To gain further insights into the mechanisms of transcription regulation by Brd4, we took an unbiased proteomic approach to identify and characterize the cellular binding partners of Brd4. Affinity purification and mass spectrometry analysis led to the identification of a number of new binding partners of Brd4. By utilizing various truncation mutants, the interactions were mapped to the different domains of Brd4, including the ET domain, a region of Brd4 whose binding partners have yet to be determined. Specific binding partners of the ET domain included GLTSCR1, ATAD5, NSD3, JMJD6, and CHD4. Investigation of the significance of these interactions in the transcriptional regulation functions of Brd4 revealed that the ET domain interactors GLTSCR1, JMJD6, and NSD3 were important for transcription activation. The ET domain is highly conserved among the BET family of proteins, and the interaction of the ET domains with these different cellular proteins appears to also be conserved. Functional studies with Brd4 indicate that the ET domain mediates pTEFb-independent transcriptional activation through a subset of these associated factors, including NSD3.
MATERIALS AND METHODS
Plasmids.
The hemagglutinin (HA)-Brd4 FL expression construct, pcDNA4-TO-HA-Brd4FL (p#6344), was engineered by subcloning from pcDNA4C-Brd4FL (57) into pcDNATO (Invitrogen) that was modified to express an N-terminal HA tag. HA tag was inserted using the QiaQuik mutagenesis protocol (Stratagene). The constructs for expressing Brd4 fragments included MSCV-CMV-Flag-HA-Brd4-1–722 (p#6345), MSCV-CMV-Flag-HA-NLS-Brd4-1–444 (p#6346), MSCV-CMV-Flag-HA-NLS-Brd4-444–722 (p#6347), MSCV-CMV-Flag-HA-NLS-Brd4-1047–1362 (p#6348), and MSCV-CMV-Flag-HA-NLS-Brd4-1224–1362 (p#6349). ATAD5, NSD3, and JMJD6 were expressed using MSCV-CMV-Flag-HA-ATAD5 (p#6350), MSCV-CMV-Flag-HA-NSD3 (p#6351), and MSCV-CMV-Flag-HA-JMJD6 (p#6352), respectively. GLTSCR1 was expressed using PHAGE-C-HA-GLTSCR1 (p#6355) or MSCV-CMV-C-FLAG-HA-GLTSCR1 (p#6356). CHD4 was expressed from MSCV-CMV-Flag-HA-CHD4 (p#6353), which was subcloned from pLP-X7-CHD4 (43). EGFPC1-Brd4, -Brd3, and -Brd2 were previously described (37). pGL4.20-BPV1LCR-luc (p#5191) was described previously (40). Bovine papillomavirus type 1 (BPV1) E2TA was previously described (28, 57).
DNA transfections.
The day before performance of transfections, cells were seeded such that they would reach 70% confluence at the time of transfection. C33A cells were transfected with the indicated plasmids using a 3:1 FuGENE (μl)/DNA (μg) ratio according to the instructions of the manufacturer (Roche). 293T cells were transfected using TransIT-293 (Mirus) with the indicated plasmids using a 3:1 TransIT-293 reagent (μl)/DNA (μg) ratio according to the protocol recommended by the manufacturer. Transfection efficiency was monitored using EGFPC1 (Clontech).
Cell culture and cell lines.
C33A and 293T cells were grown in Dulbecco modified Eagle medium (DMEM) (Invitrogen) supplemented with 10% (vol/vol) fetal bovine serum (FBS; SH3008803; HyClone) and 1% penicillin-streptomycin (Gibco/Invitrogen). To generate the 293T cell lines used for the HA immunoprecipitation (IP)/mass spectrometry analysis, 293T/Phoenix cells were cotransfected with the retroviral vector MSCV-CMV-Flag-HA, expressing the Brd4 fragments or full-length NSD3 or JMJD6, along with Gag-Pol and vesicular stomatitis virus G protein (VSV-G) expression vectors. 293T cells were then infected with the resulting virus using a standard protocol and selected with 0.75 μg/ml puromycin for 10 days. To generate single-cell clones that stably expressed BPV1LCR-luciferase (p5191) and BPV1 E2TA (p#5066), the reporter plasmid was linearized and transfected using FuGENE6 (Roche) into the C33A/BPV1E2TA cell line as described in reference 57. Cells were placed under selection with 0.75 μg/ml puromycin from day 1 posttransfection. After 10 days of selection, multiple single-cell clones of each cell line were established and maintained as subconfluent monolayers under puromycin selection. Cell lines coexpressing HPV18LCR-luciferase (p5194) and BPV1 E2TA (p#5066) have been previously described (45).
Antibodies.
Primary antibodies used for immunoprecipitations and immunoblotting were against actin (Millipore), Brd4 (41), HA (Roche catalog no. 12 013 819 001), NSD3 (Abcam catalog no. ab4514), JMJD6 (Abcam catalog no. ab10526), CHD4 (Abcam catalog no. ab72418), ATAD5 (Abcam catalog no. ab72111), green fluorescent protein (GFP) (Clontech), CDK-9 (Santa Cruz catalog no. sc-8338), and cyclin T1 (Santa Cruz catalog no. sc-10750). Secondary antibodies used were donkey anti-rabbit (Amersham/GE Healthcare) and Alexa Fluor 680-conjugated goat anti-mouse (Molecular Probes/Invitrogen) IgG secondary antibody. Chromatin immunoprecipitations (ChIPs) were performed with anti-IgG (Abcam catalog no. ab46540), anti-HA (Abcam catalog no. ab9110), anti-H3 (Abcam catalog no. ab1791), anti-Brd4 (Bethyl catalog no. A301-985A), and anti-H3 (trimethyl K36) (Abcam catalog no. ab9050).
Mass spectrometry and CompPASS analysis.
HA immunoprecipitations (IPs) were performed from 293T cells stably expressing Flag-HA-tagged bait proteins and were prepared for mass spectrometry (liquid chromatography-tandem mass spectrometry [LC-MS/MS]) as previously described (46). Data for BRD4 fragments 444–772, 1047–1362, and 1224–1362 were obtained on an LTQ instrument, while all remaining data were obtained using an LTQ Velos (Thermo Scientific). All peptide identifications were made using Sequest, and the resulting data were analyzed using CompPASS with LTQ data analyzed against previous data (46) and LTQ Velos data analyzed using a database containing IP-MS/MS data for 73 unrelated bait proteins analyzed on this machine (unpublished data). Interaction network analysis was performed using the STRING (http://string-db.org/) and BioGRID (http://thebiogrid.org/) databases with software included in CompPASS and visualized using Cytoscape software (www.cytoscape.org). The entire data sets for proteins identified by mass spectrometry are provided in Table S1 in the supplemental material.
Immunoprecipitations.
Large-scale HA IPs were performed for mass spectrometry and CompPASS analysis using 293T cells stably expressing the respective proteins with a Flag-HA tag on the amino-terminal end. Briefly, four 15-cm plates of 85% confluent cells were harvested in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% Nonidet P-40, 1 mM EDTA) supplemented with protease inhibitor cocktail (Roche). Cell lysates were sonicated at 35% intensity for 10 s with a Branson digital Sonifier and clarified by spinning at 17,000 × g for 15 min and passing through a 0.45-μm spin filter (Millipore UFC4 0HV 00). The lysate was then incubated with anti-HA–agarose resin (Sigma; A2095) overnight at 4°C. After 5 washes with the lysis buffer, the resin was exchanged into phosphate-buffered saline (PBS) and eluted with 250 μg/ml HA peptide (Sigma) at room temperature. The eluted proteins were concentrated by trichloroacetic acid (Sigma) precipitation, followed by acetone (Sigma) washes as previously described (46).
Small-scale anti-HA and anti-Flag (Flag M2 resin; Sigma A2220) IPs were performed similarly, but with cell lysates from a single 10-cm plate. Anti-GFP (Clontech) and endogenous Brd4 immunoprecipitations (anti-Brd4 antibody [41]) were also performed similarly from 10-cm plates, except that the lysates were precleared with protein A–Sepharose CL-4B (GE Healthcare) for 1 h at 4°C. The supernatant was subsequently incubated overnight at 4°C with antibody. Protein A beads were added to the samples and incubated for another 1 h at 4°C. In each case, the bound protein was eluted in SDS Laemmli buffer and size fractionated on Bis-Tris gels (Invitrogen) prior to blotting with the indicated antibodies.
Chromatin immunoprecipitations.
Cells were rinsed with PBS and then cross-linked with 5 mM freshly prepared dimethyl-3,3′-dithiobispropionimidate (DTBP) for 30 min at room temperature followed by quenching with 150 mM NaCl, 100 mM Tris, pH 8.0, for 5 min. Cells were then cross-linked further with 1% formaldehyde for 10 min at room temperature and subsequently quenched with 125 mM glycine. Cross-linked cells were lysed in 5 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 8.0, 85 mM KCl, 0.5% NP-40 supplemented with protease and phosphatase inhibitors (Roche protease inhibitor cocktail plus 10 mM β-glycerophosphate, 10 mM NaF, and 1 mM Na3VO4) for 10 min. The resulting pellet was resuspended in SDS (−) buffer A (50 mM HEPES/KOH, pH 7.6, 500 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, 1 mM EDTA) supplemented with protease phosphatase inhibitors and sonicated to yield an average DNA fragment size of 500 bp. The samples were then precleared with 40 μl protein A/G slurry (1:1 protein A-agarose [Pierce] and protein G-agarose [Pierce] containing 1 mg/ml bovine serum albumin and 1 mg/ml sheared salmon sperm DNA). Precleared lysates were incubated with the antibody of interest overnight at 4°C. Immunocomplexes were collected by incubation with 40 μl of the protein A/G slurry for 1 h at 4°C. Complexes were subsequently washed twice with each of the following buffers, SDS (−) buffer A, SDS (−) buffer B (50 mM HEPES/KOH, pH 7.6, 0.3 M LiCl, 1 mM EDTA, 0.5% NP-40, 0.7% sodium deoxycholate), and Tris-EDTA (TE). DNA was eluted with 50 mM NaHCO3, containing 1% SDS, 10 mM dithiothreitol (DTT), 10 mM EDTA, 200 mM NaCl with sheared salmon sperm DNA. Samples were then de-cross-linked by incubating at 65°C for 4 h and treated with RNase A and proteinase K, and the DNA was purified and concentrated using the Qiagen PCR purification kit. Input and immunoprecipitated DNAs were analyzed by quantitative real-time PCR using primers for the promoters and coding regions of CCND1, DCPS, and PIM2 genes.
RNA interference (RNAi).
Small interfering RNAs (siRNAs) were obtained from Dharmacon. A reverse transfection protocol was utilized to deliver siRNAs using DharmaFECT1 (HeLa and 293T cells) or DharmaFECT2 (C33A cells) (Dharmacon/Thermo Scientific). Briefly, siRNAs diluted in 1× siRNA buffer (Dharmacon) and DharmaFECT1 preincubated with Opti-MEM were added to each well/plate and complexed for 20 min. Cells were seeded on top of the siRNA/DharmaFECT mixture at densities to reach ∼85% confluence at the end of the experiment. Medium was replaced 24 h posttransfection, and transfection efficiency was monitored using siGLO Red (Dharmacon). Cells were harvested 72 h posttransfection for bicinchoninic acid (BCA) assays, luciferase assays, and/or immunoblotting. Each experiment was performed in triplicate.
BCA protein and luciferase reporter assays.
Cells were lysed using 1× reporter lysis buffer (Promega), subjected to a freeze-thaw at −80°C, vortexed, and centrifuged to remove cell debris. Lysate protein concentration was quantified using the BCA protein assay kit (Pierce/Thermo Scientific) and measured by a SpectraMax190 (Molecular Devices) according to manufacturer specifications. Luciferase activity was measured using the Promega luciferase assay system (Promega), followed by measurement with an L-Max luminometer (Molecular Devices). For each condition, the relative luciferase units (RLUs) were normalized by the total protein concentration (mg/ml).
Gene expression analysis using RQ-PCR.
Total RNA was isolated from C33A cells 72 h post-siRNA transfection using the NucleoSpin RNA II kit (Clontech) according to the manufacturer's instructions. The concentration of each sample was determined by UV spectrophotometry, and equal amounts of RNA were reverse transcribed using the High-Capacity cDNA reverse transcription kit (Applied Biosystems). Quantitative real-time PCR (RQ-PCR) was performed in an Applied Biosystems ABI 7500 Fast Sequence Detection System using TaqMan Fast Universal PCR Master Mix (Applied Biosystems) and the TaqMan Gene Expression assays (Applied Biosystems) for PIM2 (assay ID: Hs00179139_m1), CCND1 (assay ID: Hs00277039_m1), DCPS (assay ID: Hs00204009_m1), RBPL13A (assay ID: Hs03043885_g1), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (assay ID: Hs99999905_m1). The relative amounts of cDNA in each sample were calculated based on a standard curve prepared using serial dilutions of the reference cDNA. The cDNA amount was then normalized to the housekeeping gene, GAPDH. The fold change in transcription of the gene following siRNA transfection was calculated by comparison with siControl-treated cells.
RESULTS
Proteomic analysis of the Brd4-associated cellular proteins.
To gain further insight into the various functions of Brd4, we employed an unbiased proteomic approach to identify proteins that interact with human Brd4. We generated tagged fragments of Brd4-containing specific regions and domains as shown in Fig. 1 A. To ensure that these fragments of Brd4 localized to the nucleus like endogenous Brd4, nuclear localization signals (NLS) were fused to the N terminus of Brd4 fragments Brd4 444–722, Brd4 1047–1362, and Brd4 1224–1362, which did not have the native NLS present in the N-terminal fragment Brd4 1–444 or Brd4 1–722. 293T cells that stably expressed these N-terminally Flag-HA-tagged fragments of Brd4 or the GFP control were established. Additionally, full-length tagged Brd4 was transiently transfected into 293T cells because of the toxicity associated with stable expression of exogenous full-length Brd4 (58). Cell lysates were affinity purified via HA immunoprecipitation, associated proteins were identified by LC-MS/MS, and the data were analyzed using the proteomics platform CompPASS (Comparative Proteomics Analysis Software Suite) (46). This facilitated the identification of high-confidence candidate-interacting proteins (HCIPs) in parallel mass spectral studies by using DN scores of >1 and/or Z scores of >3.5 based on previously established methods (Fig. 1B; see also Table S1 in the supplemental material). A number of new candidate interactors of Brd4 were identified using this approach along with the previously reported pTEFb components cyclin T1 and Cdk9, which bind Brd4 through its CTD. Among the top-scoring HCIPs, five interacted with both Brd4 1–722 and Brd4 444–722 fragments, each of which contains the ET domain. These were GLTSCR1, ATAD5, NSD3, JMJD6, and CHD4 (Fig. 1B; see also Table S1). In order to understand the possible functions associated with the ET domain, these candidates were selected for further analysis.
Fig. 1.
Human Brd4 associates with multiple epigenetic and transcription regulators. (A) Schematic of the Brd4 fragments covering various domains used for the proteomic analysis. Full-length protein or the indicated fragments were expressed as N-terminally Flag-HA-tagged proteins in 293T cells, large-scale HA IPs were performed with whole-cell lysates, and immunoprecipitates were analyzed by LC-MS/MS. (B) Interaction maps. “Interaction from MS” was identified via mass spectrometry and CompPASS analysis.
Validation of interactions with human Brd4.
To validate the association of ATAD5, NSD3, JMJD6, and CHD4 with Brd4, we utilized the stable 293T cells expressing Flag-HA-tagged Brd4 1–722, Brd4 444–722, and Brd4 1047–1362 as well as the GFP control to perform HA immunoprecipitations followed by Western blotting with antibodies to endogenous NSD3, JMJD6, CHD4, and ATAD5. We were unable to test GLTSCR1 binding to the Brd4 fragments in this manner due to the lack of antibodies to the protein. As shown in Fig. 2 A, we confirmed the binding of endogenous NSD3, JMJD6, CHD4, and ATAD5 to the ET domain containing fragments of Brd4, but not to the C-terminal fragment Brd4 1047-1362 or to the GFP control. To determine if these proteins bound the full-length, endogenous Brd4 protein, 293T cells were transiently transfected with Flag-NSD3 (short isoform), Flag-JMJD6, Flag-CHD4, Flag-GLTSCR1, and Flag-ATAD5. Endogenous Brd4 coimmunoprecipitated with Flag-tagged NSD3, JMJD6, CHD4, ATAD5, and GLTSCR1 (Fig. 2B). In addition, we were able to confirm binding of endogenous JMJD6 and CHD4 to endogenous Brd4 by coimmunoprecipitations (Fig. 2C). We were unable to directly test the binding of endogenous NSD3, GLTSCR1, or ATAD5 with Brd4 due to the lack of commercially available antibodies that effectively immunoprecipitated these proteins. The data presented here, however, confirmed the mass spectrometry analysis and established that endogenous Brd4 does interact with GLTSCR1, ATAD5, NSD3, JMJD6, and CHD4.
Fig. 2.
Validation of interactions with human Brd4. (A) The indicated Brd4 fragments stably expressed in 293T cells were subjected to small-scale HA IPs. Equal amounts of input (IN) samples along with 50% of IPs were separated by SDS-PAGE and immunoblotted with antibodies specific to HA, ATAD5, NSD3, JMJD6, CHD4, and actin. (B) 293T cells were transiently transfected with Flag-GLTSCR1, Flag-ATAD5, Flag-NSD3, Flag-JMJD6, or Flag-CHD4, and cell lysates were harvested 48 h posttransfection. Proteins were immunoprecipitated with anti-Flag antibody and visualized as described in panel A with antibodies to Flag epitope, Brd4, or actin. (C) Endogenous IPs were performed using whole-cell lysates from 293T cells with antibodies to CHD4 or JMJD6. Bound proteins were captured using protein A beads, eluted, and analyzed by Western blotting with anti-CHD4, anti-JMJD6, anti-Brd4, or actin.
The ET domain mediates protein-protein interactions, which are conserved for the human BET proteins.
To establish whether the ET domain is sufficient to mediate the interactions with the proteins identified, we tested the interaction of two different Brd4 fragments, Brd4 608–671, comprising the ET domain only, and Brd4 608–722, which contains the ET domain and an additional C-terminal 50 amino acids. Endogenous CHD4, NSD3, and JMJD6 coimmunoprecipitated with both fragments tested, demonstrating that the Brd4 ET domain is sufficient for mediating the interaction with these proteins (Fig. 3 B). Interaction with ATAD5 could not be detected in this experiment, while the interaction with GLTSCR1 could not be assessed due to the lack of commercially available antibody to GLTSCR1. Brd4 608–671 appeared to bind NSD3 better than did Brd4 608–722, suggesting that the amino acid sequences C terminal to the ET domain may have an inhibitory effect on interaction with NSD3.
Fig. 3.
ET domain-mediated protein-protein interaction is conserved across the human BET proteins. (A) Amino acid sequence alignment of human BET proteins, Brd2, Brd3, Brd4, and Brd6, demonstrating high sequence similarity. (B) 293T cells were transfected with GFP-tagged constructs expressing the ET domains of Brd4, Brd3, and Brd2 (35). At 48 h posttransfection, anti-GFP immunoprecipitations were performed with whole-cell extract. Equal amounts of input (IN) samples along with 50% of IPs were separated by SDS-PAGE and immunoblotted with antibodies specific to GFP, NSD3, JMJD6, CHD4, and actin. The symbols *1, *2, and *3 denote the GFP band, the Brd4 band, and the Brd2/Brd3 bands, respectively.
The ET domains of human, mouse, Xenopus, Drosophila, and yeast BET proteins are highly conserved, and the human orthologs, Brd4, Brd3, Brd2, and Brd6, share >80% identity (Fig. 3A). Given this high level of amino acid homology among the ET domains, the association of specific cellular proteins with the ET domain might also be conserved across different BET proteins. We therefore examined the ability of the ET domains of the human BET proteins Brd2 and Brd3 to associate with JMJD6, NSD3, and CHD4. As shown in Fig. 3B, recombinant GFP fusion proteins containing fragments of Brd2 and Brd3 that contain the ET domain interacted well with CHD4. The fragment containing the Brd2 ET domain also interacted well with NSD3 and JMJD6, whereas with the Brd3 fragment, a weaker association was detected with NSD3 and no binding was detected with JMJD6 (Fig. 3B). As with the Brd4 ET domain, these data suggest that amino acid residues C terminal to the ET domain of Brd2 and Brd3 may abrogate interactions with NSD3 and JMJD6. These findings demonstrate that some of the ET domain-cellular protein interactions are conserved across the human BET proteins.
Brd4 ET domain interactors form distinct multiprotein complexes.
To determine whether Brd4 and the ET domain interactors form a single or multiple distinct complexes, 293T cells stably expressing HA-tagged forms of GLTSCR1, ATAD5, NSD3, JMJD6, and CHD4 were established and HA immunoprecipitates were analyzed via mass spectrometry and CompPASS analysis (see Table S1 in the supplemental material). The results from these proteomic analyses together with interactions reported in BIOGRID and String databases are displayed as an interaction map in Fig. 4. ATAD5 interacted with Brd4 but not with any of the other ET domain-interacting proteins, whereas NSD3 interacted with Brd4 and GLTSCR1, indicating that Brd4, NSD3, and GLTSCR1 likely exist in a complex together. Furthermore, glycerol gradient analysis of the Brd4 ET complex showed that JMJD6 and CHD4 cofractionate with NSD3 (data not shown), suggesting that JMJD6 could also be a component of this complex. In contrast, ATAD5 appears to form a separate, distinct complex with Brd4. This proteomic analysis also showed that JMJD6 interacts with pre-mRNA spliceosome components in accordance with its documented interaction with factors involved in RNA processing (50). CHD4 was observed to interact with components of the NuRD corepressor complex also as predicted previously (54). The major interactors of NSD3 included Brd4 and GLTSCR1 along with H2AFy, the repressive nucleosome variant of core histone H2A (Fig. 4). GLTSCR1 interacts with NSD3, along with several components of the BRG1-CREST complex, linking Brd4 to a chromatin remodeling complex (Fig. 4). Interestingly, ATAD5 interacted with several proteins associated with the DNA damage repair pathway, including USP1 and BAZ1B (Fig. 4). This analysis indicates that the Brd4 ET domain-associated proteins form multiple, distinct complexes with Brd4, which potentially connect Brd4 to various pathways and distinct functions.
Fig. 4.
The Brd4 ET domain associates with several multiprotein complexes. HA immunoprecipitations were performed with lysates from 293T cells stably expressing HA-GLTSCR1, HA-ATAD5, HA-NSD3, HA-JMJD6, or HA-CHD4, and the bound proteins were subjected to mass spectrometry and CompPASS analysis (MS or IP).
JMJD6, NSD3, and GLTSCR1 are important for viral transcriptional activation of the BPV1 LCR mediated by Brd4.
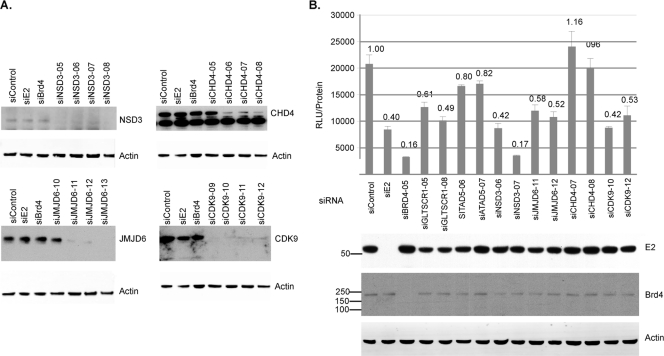
Given the involvement of JMJD6, NSD3, and CHD4 in histone modification and chromatin remodeling, we next asked whether any of the ET domain-associated proteins are important for the transcriptional activation functions of Brd4. We analyzed the effect of siRNA-mediated depletion of GLTSCR1, ATAD5, NSD3, JMJD6, and CHD4 on the transcriptional activation of the bovine papillomavirus (BPV1) long control region (LCR) by the viral E2 protein, whose transcriptional activation function is mediated by Brd4 (35, 41, 42). We generated a transcriptional reporter assay system in which the luciferase gene is under the control of the BPV1 LCR. Linearized BPV1 LCR reporter plasmid was introduced into C33A cells that stably express FLAG-HA-tagged BPV1 E2 (57). Single-cell clones were characterized based on E2 expression level and responsiveness to siRNA-mediated depletion of E2 and/or Brd4. We selected C33A/E2TA/BPV1LCRc2 for our experiments. The two most effective siRNA duplexes for each of the ET domain interactors were utilized in the BPV1 LCR reporter assay (Fig. 5 A; see also Fig. S1 in the supplemental material). Luciferase activity was measured 72 h posttransfection, and the relative luciferase units were normalized to total protein concentration from three replicate experiments. The fold change in reporter activity due to a particular siRNA duplex in comparison to a control siRNA (siGenome NT 1, here called siControl) is depicted in Fig. 5B. Knockdown of GLTSCR1, NSD3, or JMJD6 each decreased reporter gene activity by 2- to 3-fold, which is comparable to the 2-fold effect observed with knockdown of CDK-9, a component of pTEFb. Depletion of GLTSCR1 did lead to a slight decrease in E2 levels, which may have partially contributed to the decrease in E2-mediated activation of the BPV1 LCR. In contrast, depletion of CHD4 or ATAD5 had no significant effect on E2 transactivation of the BPV1 LCR. This experiment demonstrates that a subset of the Brd4 ET domain-associated proteins, specifically GLTSCR1, NSD3, and JMJD6, is necessary for the full activation of the BPV1 LCR by Brd4 and that the ET domain mediates a transcriptional activation function independent of pTEFb.
Fig. 5.
Effect of siRNA-mediated knockdown of Brd4 ET interactors on Brd4-dependent E2-mediated transcriptional activation of the BPV1 LCR. C33A/E2TA/BPV1LCRc2 cells were transfected with the indicated siRNA duplexes. (A) Knockdown of NSD3, JMJD6, CHD4, and CDK9 protein levels shown by resolving equal amounts of lysates by SDS-PAGE and immunoblotting with the indicated antibodies or the control, actin. (B) Effect of knocking down Brd4 ET interactors on BPV1 LCR reporter activity was quantified by measuring luciferase activity 72 h posttransfection and normalizing the relative luciferase units (RLU) to total protein concentrations from three replicate experiments. Equal amounts of lysates were resolved by SDS-PAGE and immunoblotted with the indicated antibodies to determine knockdown levels of the proteins targeted by siRNA or the control, actin.
JMJD6, NSD3, and GLTSCR1 contribute to transcriptional regulation of cellular target genes of Brd4.
We next examined whether the Brd4 ET interactors function in the regulation of cellular genes activated by Brd4. Work in our laboratory has identified a number of cellular genes regulated by Brd4 (M. Ottinger et al., unpublished data). Three of the top-scoring target genes, pim-2 oncogene (PIM2), cyclin D1 (CCND1), and decapping enzyme, scavenger (DCPS), as well as a control gene, ribosomal protein L13A (RPL13A), whose transcription is not affected by Brd4 knockdown, were selected. C33A cells depleted of Brd4, cdk9, or the Brd4 ET domain interactors GLTSCR1, NSD3, and JMJD6 were harvested 72 h post-siRNA transfection. Total RNA was isolated, cDNA was generated by reverse transcription, and mRNA levels were analyzed. Relative mRNA levels were calculated by initially normalizing to GAPDH and subsequently to siControl-treated cells (Fig. 6). The knockdown of Brd4 decreased the expression level of PIM2, CCND1, and DCPS mRNA by 3- to 9-fold and had no effect on the control, RPL13A. Knockdown of JMJD6 did have a slight effect on RPL13A levels, a result that may be due to an mRNA processing defect since JMJD6 has been implicated in this pathway (50). Knockdown of GLTSCR1, NSD3, JMJD6, or the pTEFb component CDK-9 decreased expression of the Brd4 target genes by 2- to 3-fold while not affecting RPL13A expression. These results demonstrate that the Brd4 ET domain-interacting proteins, GLTSCR1, NSD3, and JMJD6, contribute to the transcriptional regulation of cellular target genes of Brd4.
Fig. 6.
GLTSCR1, NSD3, and JMJD6 contribute to transcriptional activation of cellular target genes by Brd4. C33A cells were treated with siRNA to Brd4, GLTSCR1, NSD3, JMJD6, or CDK9, and total RNA was extracted 72 h posttransfection from duplicate experiments. Reverse transcription was then performed to obtain cDNA followed by RQ-PCR. Quantities determined were normalized to the housekeeping gene GAPDH and then compared to siControl sample to calculate relative mRNA levels.
NSD3 is recruited to Brd4 target genes in a Brd4-dependent manner.
To explore whether the ET interactors have a direct effect on the transcription of Brd4 target genes, we employed chromatin immunoprecipitation (ChIP) to analyze the association of NSD3 with the various target genes. To ensure ChIP efficacy, especially given the absence of good commercially available antibody to NSD3, HA ChIPs were performed using 293T cells stably expressing Flag-HA-NSD3. ChIP with anti-IgG antibody was performed as a negative control. NSD3 binding was examined across the Brd4 target genes CCND1, DCPS, and PIM2 using RQ-PCR. The assay's specificity to NSD3 was determined by comparing siNSD3-treated cells to siControl-treated cells, which demonstrated that knockdown of NSD3 led to reduction in the levels of NSD3 at the promoter and coding regions of CCND1, PIM2, and DCPS (Fig. 7 A to C). As expected, knockdown of Brd4 decreased the levels of Brd4 bound to the promoter and coding regions of CCND1, PIM2, and DCPS (Fig. 7D to F). The levels of Brd4 and NSD3 appeared to be higher at the promoter than in coding regions of CCND1 and DCPS, while the converse was true for PIM2 (Fig. 7). The significance of Brd4 in the recruitment of NSD3 to cellular genes was examined by comparing NSD3 ChIP signals in siControl-treated cells to those in siBrd4-treated cells (Fig. 7D to F). This demonstrated that knockdown of Brd4 reduced NSD3 levels at the promoter and coding regions of the genes examined and that this effect was comparable to the reduction observed in siNSD3-treated cells. Thus, NSD3 is recruited to Brd4 target genes and its recruitment is dependent on the presence of Brd4. We also examined recruitment of JMJD6 to these genes in cells stably expressing Flag-HA-JMJD6 (see Fig. S2 in the supplemental material). Like NSD3, JMJD6 is recruited to the promoter and coding regions of CCND1, DCPS, and PIM2 in a Brd4-dependent manner. We were unable to establish cells stably expressing HA-GLTSCR1 at a level sufficient for ChIP analysis.
Fig. 7.
The Brd4 ET domain-associated NSD3 is recruited to Brd4 target genes, CCND1, DCPS, and PIM2. (A to C) 293T cells stably expressing Flag-HA-NSD3 were treated with siRNA to NSD3, siNSD3, or with the control siRNA, siControl, for 72 h. ChIP was performed with anti-IgG (IgG), anti-Brd4 (Brd4), or anti-HA (NSD3) as indicated on the x axis. Immunoprecipitated DNA was quantified by real-time PCR in duplicates with primers specific to the promoter or coding regions of CCND1, PIM2, and DCPS genes, and results are presented as enrichment relative to input DNA. Each ChIP was repeated twice with reproducible results. Error bars represent standard deviations from means. (D to F) 293T/Flag-HA-NSD3 cells were treated with siRNA to Brd4, siBrd4, or with the control siRNA, siControl, for 72 h, and ChIP analysis was performed as indicated in panels A to C.
The Brd4/NSD3 complex regulates histone-3 lysine-36 (H3K36) trimethylation levels.
NSD3, also known as WHSC1L1, is a histone methyltransferase that belongs to the mammalian NSD family of SET domain-containing methyltransferases, which have been shown to catalyze H3K36 methylation (30, 36). Studies ranging from yeast to humans suggest that methylated H3K36 is enriched in regions of active transcription and is linked to transcriptional elongation and productive transcription within coding regions (5, 48, 49, 53). To explore the mechanism by which NSD3 contributes to the transcription activation of Brd4 target genes, we next asked whether the Brd4/NSD3 complex regulates H3K36 methylation levels at Brd4 target genes. To this end, we performed ChIPs using anti-H3K36 dimethyl (H3K36me2)- and anti-H3K36 trimethyl (H3K36me3)-specific antibodies using extracts from HeLa cells treated with siControl, siBrd4, or siNSD3. Anti-H3 and anti-IgG antibodies were used as positive and negative controls, respectively. Total H3, H3K36me2, and H3K36me3 levels at the CCND1 promoter and coding regions were quantified by RQ-PCR with primers to the various regions of the CCND1 gene (Fig. 8A). H3K36me2 levels appeared similar at the promoter and coding regions, and no significant change was observed in the levels of H3K36me2 levels upon Brd4 or NSD3 knockdown (data not shown). H3K36me3 levels were higher within the coding region, peaking at the 3′ end, than at the promoter in accordance with published findings (2). More interestingly, we found that knockdown of either NSD3 or Brd4 leads to reduction in H3K36 trimethylation levels, especially within the gene body (Fig. 8B).
Fig. 8.
Knockdown of Brd4 or NSD3 reduces H3K36 trimethylation levels within the gene body of CCND1. (A) Schematic showing locations of primers used for ChIP/RQ-PCR analysis within the CCND1 locus. Nucleotide positions (bp) are depicted with respect to transcription start site (arrow). (B) HeLa cells were treated with siRNA to Brd4 (siBrd4), NSD3 (siNSD3), or the control siRNA (siControl) for 72 h. Chromatin extracts from these cells were subjected to immunoprecipitation with anti-H3 antibody (H3), anti-H3K36me3 antibody (H3K36me3), or the negative-control anti-IgG antibody (IgG). Immunoprecipitated DNA was quantified by RQ-PCR in duplicates with primers specific to the promoter or coding regions of the CCND1 gene. The results from this analysis are presented as percentages of input DNA. The experiment was repeated three times with reproducible profiles. Error bars represent standard deviations from means.
DISCUSSION
Characterization of the gene expression regulatory functions of Brd4 has been previously limited to the chromatin targeting of its two bromodomains and the interactions of its C-terminal domain (CTD) with the basal transcriptional elongation factor, pTEFb. The strong conservation of the ET domain among the various BET family proteins suggests a potentially vital but as-yet-unidentified ET-associated function of Brd4. To further understand the function of the Brd4 ET domain, we performed a proteomic analysis of Brd4 and identified five proteins that interact specifically with the ET domain, NSD3, CHD4, JMJD6, GLTSCR1, and ATAD5. Moreover, the interactions of these proteins are conserved among the human BET proteins Brd4, Brd3, and Brd2 and likely to be a general characteristic of all BET family proteins. We demonstrated that NSD3, JMJD6, and GLTSCR1, a subset of the ET domain interactors, contribute to transcriptional regulatory functions of Brd4, indicating that both the ET and CTD domains of Brd4 recruit distinct regulatory complexes. Quantitative changes in Brd4-dependent transcriptional activation by selective knockdown of the individual components indicated that the ET domain functions as an independent transcriptional regulatory domain for Brd4. This is further supported by our finding that NSD3 as well as JMJD6 is recruited to the promoter and gene bodies of Brd4-regulated genes in a Brd4-dependent manner. This suggests a direct effect of these ET domain-associated proteins on Brd4-mediated transcription activation.
The ET domain is highly conserved among the BET family of proteins from yeast to human. The high sequence homology is strongly predictive of a conserved evolutionary function for the ET domain. Many different BET family members, including the yeast bdf1/bdf2, Drosophila fs(1)h, and the mammalian Brd2, Brd3, and Brd4 proteins, have been implicated in transcription regulation (11, 13, 24, 27, 32). However, only the CTD, present solely in Brd4 and fs(1)h, has been rigorously documented to regulate transcription through its interaction with pTEFb (4, 24, 56). Our finding that the ET domain associates with NSD3, JMJD6, and GLTSCR1 and is important for a pTEFb-independent transcriptional activation function of Brd4 delineates a plausible physiological function for the ET domain across the BET family of proteins.
NSD3, also known as WHSC1L1, is a histone methyltransferase that belongs to the mammalian NSD family of SET domain-containing methyltransferases, which also includes NSD1 and NSD2 (WHSC1/MMSET). The NSD family of proteins is essential in development and is mutated in human acute myeloid leukemia, Sotos syndrome, multiple myeloma, and lung cancer (14, 26, 39, 47, 49). The SET domain of NSD proteins is homologous to the Saccharomyces cerevisiae histone-3 lysine-36 (H3K36)-specific methyltransferase SET2 and is specific for H3K36 dimethylation (H3K36me2) (30). Studies ranging from yeast to humans suggest that methylated H3K36 is enriched in regions of active transcription and is linked to transcriptional elongation and productive transcription within coding regions (5, 48, 49, 53).
We found that depletion of NSD3 or Brd4 leads to a reduction in H3K36 trimethylation within the gene body of CCND1, a Brd4-regulated gene. We did note that levels of NSD3 and Brd4 were higher at the promoter than at the 3′ end of the gene, where the effect of knocking down Brd4/NSD3 on H3K36 trimethylation had the greatest impact. This suggests that the lower levels of Brd4/NSD3 at the 3′ end are sufficient for directly methylating H3K36. Perhaps the high levels of Brd4/NSD3 present at the promoter and 5′ end of the gene influence H3K36 methylation levels downstream of the gene. We predict that reduction of NSD3 and the subsequent decrease in H3K36 methylation within the gene body lead to defects in transcriptional elongation. It is plausible that a reduction of Brd4 and NSD3 at the 5′ end of the gene could lead to abrogation of transcription initiation that could indirectly affect transcription elongation. Future work will focus on whether NSD3 affects transcriptional elongation or other aspects of transcription.
JMJD6 and GLTSCR1 also regulate Brd4-dependent transcriptional activation via interaction with the Brd4 ET domain. The specific role of JMJD6 is presently unclear. It was initially reported to be a histone arginine demethylase, responsible for H3R2me and H4R3me modifications (6). However, more recently, it was reported to be important for lysyl hydroxylation of U2AF65, a factor associated with RNA splicing (50). Crystallographic studies support the hydroxylase catalytic activity and suggest that JMJD6 binds single-stranded RNA (20, 33). A role for Brd4 in splicing regulation is plausible. A recent study demonstrates that transcription of primary responsive genes in stimulated macrophages is dependent on Brd4 and is linked to production of mature spliced transcripts (19). Brd4 ET-mediated recruitment of JMJD6 could be important for proper RNA splicing and work synergistically with pTEFb to produce mature, spliced transcripts. The function of GLTSCR1 is unknown. GLTSCR1 polymorphisms have been associated with the development of oligodendrogliomas, broadly suggesting a role as a tumor suppressor (55). Our study reveals that GLTSCR1 contributes to transcriptional activation by Brd4 (Fig. 5 and 6). From mass spectrometry analysis of the Brd4 ET domain-associated proteins, it appears that NSD3 and GLTSCR1 may exist in the same complex (Fig. 3). The other interactors of the ET domain that we identified were CHD4 and ATAD5. CHD4, also known as Mi-2β, is a component of the Mi-2/nucleosome remodeling and deacetylase (NuRD) complex that has been implicated in repression of a number of different genes. Notably, evidence suggests that it is involved in repression of genes that are regulated by estrogen receptor α (12). The role of ATAD5 is not well studied, and it is thought to be involved in ATM/ATR-mediated DNA damage response (23). While we did not observe any effect of CHD4 or ATAD5 knockdown on Brd4-dependent transcriptional activation, these proteins could still impart functions to Brd4 that are relevant to its transcription roles but in other functional settings.
Overall, this study characterizes Brd4-associated proteins and provides new insights into the functional significance of the ET domain. We identified the cellular binding partners of the ET domain as factors that are recruited to acetylated chromatin by Brd4 and show that some of these interactors have a role in a transcriptional activation function of Brd4. Since the interactions are conserved also for the Brd2 and Brd3 ET domains, we conclude that the ET domain is important generally for the transcriptional regulation functions of the BET family of proteins. The exact role of the Brd4 complex(es), however, will require additional study, and the physiologic roles of NSD3 and JMJD6 as well as the other ET domain-interacting proteins should be illuminated in this process. Previously, Brd4 studies have focused on the bromodomains and the CTD to understand the molecular mechanisms of Brd4 functions. This is the first study that links the ET domain to a transcriptional regulatory function and identifies the cellular proteins with which it interacts, highlighting the importance of this domain in the transcriptional activation functions of the BET family of proteins. Specifically, we have demonstrated than NSD3 is recruited to Brd4 target genes in a Brd4-dependent manner where the Brd4/NSD3 complex regulates H3K36 methylation levels, a chromatin modification linked to active transcription. We hypothesize that Brd4 may stimulate transcription through two independent mechanisms, the ET domain recruitment of NSD3 and the CTD interaction with pTEFb.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Doxsey (University of Massachusetts Medical School) for the CHD4 expression construct.
This work has been supported by National Institutes of Health grants R01CA116720 (to P.M.H.), T32CA009361 (to S.R.), and AG011085 and GM054137 (to J.W.H.).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 9 May 2011.
REFERENCES
- 1. Abbate E. A., Voitenleitner C., Botchan M. R. 2006. Structure of the papillomavirus DNA-tethering complex E2:Brd4 and a peptide that ablates HPV chromosomal association. Mol. Cell 24:877–889 [DOI] [PubMed] [Google Scholar]
- 2. Barski A., et al. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823–837 [DOI] [PubMed] [Google Scholar]
- 3. Baxter M. K., McPhillips M. G., Ozato K., McBride A. A. 2005. The mitotic chromosome binding activity of the papillomavirus E2 protein correlates with interaction with the cellular chromosomal protein, Brd4. J. Virol. 79:4806–4818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bisgrove D. A., Mahmoudi T., Henklein P., Verdin E. 2007. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc. Natl. Acad. Sci. U. S. A. 104:13690–13695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carrozza M. J., et al. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123:581–592 [DOI] [PubMed] [Google Scholar]
- 6. Chang B., Chen Y., Zhao Y., Bruick R. K. 2007. JMJD6 is a histone arginine demethylase. Science 318:444–447 [DOI] [PubMed] [Google Scholar]
- 7. Cho W. K., et al. 2007. Modulation of the Brd4/P-TEFb interaction by the human T-lymphotropic virus type 1 tax protein. J. Virol. 81:11179–11186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Crawford N. P., et al. 2008. Bromodomain 4 activation predicts breast cancer survival. Proc. Natl. Acad. Sci. U. S. A. 105:6380–6385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Crowley T. E., Kaine E. M., Yoshida M., Nandi A., Wolgemuth D. J. 2002. Reproductive cycle regulation of nuclear import, euchromatic localization, and association with components of Pol II mediator of a mammalian double-bromodomain protein. Mol. Endocrinol. 16:1727–1737 [DOI] [PubMed] [Google Scholar]
- 10. Denis G. V., et al. 2006. Identification of transcription complexes that contain the double bromodomain protein Brd2 and chromatin remodeling machines. J. Proteome Res. 5:502–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Denis G. V., Vaziri C., Guo N., Faller D. V. 2000. RING3 kinase transactivates promoters of cell cycle regulatory genes through E2F. Cell Growth Differ. 11:417–424 [PMC free article] [PubMed] [Google Scholar]
- 12. Denslow S. A., Wade P. A. 2007. The human Mi-2/NuRD complex and gene regulation. Oncogene 26:5433–5438 [DOI] [PubMed] [Google Scholar]
- 13. Digan M. E., et al. 1986. Genetic and molecular analysis of fs(1)h, a maternal effect homeotic gene in Drosophila. Dev. Biol. 114:161–169 [DOI] [PubMed] [Google Scholar]
- 14. Douglas J., et al. 2005. Evaluation of NSD2 and NSD3 in overgrowth syndromes. Eur. J. Hum. Genet. 13:150–153 [DOI] [PubMed] [Google Scholar]
- 15. French C. A., et al. 2001. BRD4 bromodomain gene rearrangement in aggressive carcinoma with translocation t(15;19). Am. J. Pathol. 159:1987–1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. French C. A., et al. 2003. BRD4-NUT fusion oncogene: a novel mechanism in aggressive carcinoma. Cancer Res. 63:304–307 [PubMed] [Google Scholar]
- 17. French C. A., et al. 2008. BRD-NUT oncoproteins: a family of closely related nuclear proteins that block epithelial differentiation and maintain the growth of carcinoma cells. Oncogene 27:2237–2242 [DOI] [PubMed] [Google Scholar]
- 18. Gyuris A., et al. 2009. The chromatin-targeting protein Brd2 is required for neural tube closure and embryogenesis. Biochim. Biophys. Acta 1789:413–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hargreaves D. C., Horng T., Medzhitov R. 2009. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 138:129–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hong X., et al. 2010. Interaction of JMJD6 with single-stranded RNA. Proc. Natl. Acad. Sci. U. S. A. 107:14568–14572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Houzelstein D., et al. 2002. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol. Cell. Biol. 22:3794–3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ilves I., Maemets K., Silla T., Janikson K., Ustav M. 2006. Brd4 is involved in multiple processes of the bovine papillomavirus type 1 life cycle. J. Virol. 80:3660–3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ishii H., et al. 2005. Frag1, a homolog of alternative replication factor C subunits, links replication stress surveillance with apoptosis. Proc. Natl. Acad. Sci. U. S. A. 102:9655–9660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jang M. K., et al. 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19:523–534 [DOI] [PubMed] [Google Scholar]
- 25. Jiang Y. W., et al. 1998. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc. Natl. Acad. Sci. U. S. A. 95:8538–8543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Keats J. J., et al. 2005. Overexpression of transcripts originating from the MMSET locus characterizes all t(4;14)(p16;q32)-positive multiple myeloma patients. Blood 105:4060–4069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ladurner A. G., Inouye C., Jain R., Tjian R. 2003. Bromodomains mediate an acetyl-histone encoded antisilencing function at heterochromatin boundaries. Mol. Cell 11:365–376 [DOI] [PubMed] [Google Scholar]
- 28. Lambert P. F., Spalholz B. A., Howley P. M. 1987. A transcriptional repressor encoded by BPV-1 shares a common carboxy-terminal domain with the E2 transactivator. Cell 50:69–78 [DOI] [PubMed] [Google Scholar]
- 29. LeRoy G., Rickards B., Flint S. J. 2008. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol. Cell 30:51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li Y., et al. 2009. The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. J. Biol. Chem. 284:34283–34295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lin A., Wang S., Nguyen T., Shire K., Frappier L. 2008. The EBNA1 protein of Epstein-Barr virus functionally interacts with Brd4. J. Virol. 82:12009–12019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lygerou Z., et al. 1994. The yeast BDF1 gene encodes a transcription factor involved in the expression of a broad class of genes including snRNAs. Nucleic Acids Res. 22:5332–5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mantri M., et al. 2010. Crystal structure of the 2-oxoglutarate- and Fe(II)-dependent lysyl hydroxylase JMJD6. J. Mol. Biol. 401:211–222 [PubMed] [Google Scholar]
- 34. Matangkasombut O., Buratowski R. M., Swilling N. W., Buratowski S. 2000. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 14:951–962 [PMC free article] [PubMed] [Google Scholar]
- 35. McPhillips M. G., Oliveira J. G., Spindler J. E., Mitra R., McBride A. A. 2006. Brd4 is required for e2-mediated transcriptional activation but not genome partitioning of all papillomaviruses. J. Virol. 80:9530–9543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nimura K., et al. 2009. A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome. Nature 460:287–291 [DOI] [PubMed] [Google Scholar]
- 37. Ottinger M., et al. 2009. The interaction of the gammaherpesvirus 68 orf73 protein with cellular BET proteins affects the activation of cell cycle promoters. J. Virol. 83:4423–4434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Reynoird N., et al. 2010. Oncogenesis by sequestration of CBP/p300 in transcriptionally inactive hyperacetylated chromatin domains. EMBO J. 29:2943–2952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rosati R., et al. 2002. NUP98 is fused to the NSD3 gene in acute myeloid leukemia associated with t(8;11)(p11.2;p15). Blood 99:3857–3860 [DOI] [PubMed] [Google Scholar]
- 40. Schweiger M. R., Ottinger M., You J., Howley P. M. 2007. Brd4-independent transcriptional repression function of the papillomavirus e2 proteins. J. Virol. 81:9612–9622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schweiger M. R., You J., Howley P. M. 2006. Bromodomain protein 4 mediates the papillomavirus E2 transcriptional activation function. J. Virol. 80:4276–4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Senechal H., Poirier G. G., Coulombe B., Laimins L. A., Archambault J. 2007. Amino acid substitutions that specifically impair the transcriptional activity of papillomavirus E2 affect binding to the long isoform of Brd4. Virology 358:10–17 [DOI] [PubMed] [Google Scholar]
- 43. Sillibourne J. E., Delaval B., Redick S., Sinha M., Doxsey S. J. 2007. Chromatin remodeling proteins interact with pericentrin to regulate centrosome integrity. Mol. Biol. Cell 18:3667–3680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sinha A., Faller D. V., Denis G. V. 2005. Bromodomain analysis of Brd2-dependent transcriptional activation of cyclin A. Biochem. J. 387:257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Smith J. A., et al. 2010. Genome-wide siRNA screen identifies SMCX, EP400, and Brd4 as E2-dependent regulators of human papillomavirus oncogene expression. Proc. Natl. Acad. Sci. U. S. A. 107:3752–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W. 2009. Defining the human deubiquitinating enzyme interaction landscape. Cell 138:389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tatton-Brown K., et al. 2005. Genotype-phenotype associations in Sotos syndrome: an analysis of 266 individuals with NSD1 aberrations. Am. J. Hum. Genet. 77:193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tompa R., Madhani H. D. 2007. Histone H3 lysine 36 methylation antagonizes silencing in Saccharomyces cerevisiae independently of the Rpd3S histone deacetylase complex. Genetics 175:585–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tonon G., et al. 2005. High-resolution genomic profiles of human lung cancer. Proc. Natl. Acad. Sci. U. S. A. 102:9625–9630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Webby C. J., et al. 2009. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science 325:90–93 [DOI] [PubMed] [Google Scholar]
- 51. Weidner-Glunde M., Ottinger M., Schulz T. F. 2010. WHAT do viruses BET on? Front. Biosci. 15:537–549 [DOI] [PubMed] [Google Scholar]
- 52. Wu S. Y., et al. 2006. Brd4 links chromatin targeting to HPV transcriptional silencing. Genes Dev. 20:2383–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xiao T., et al. 2003. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 17:654–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xue Y., et al. 1998. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2:851–861 [DOI] [PubMed] [Google Scholar]
- 55. Yang P., et al. 2005. Polymorphisms in GLTSCR1 and ERCC2 are associated with the development of oligodendrogliomas. Cancer 103:2363–2372 [DOI] [PubMed] [Google Scholar]
- 56. Yang Z., et al. 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 19:535–545 [DOI] [PubMed] [Google Scholar]
- 57. You J., Croyle J. L., Nishimura A., Ozato K., Howley P. M. 2004. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell 117:349–360 [DOI] [PubMed] [Google Scholar]
- 58. You J., et al. 2009. Regulation of aurora B expression by the bromodomain protein Brd4. Mol. Cell. Biol. 29:5094–5103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. You J., Schweiger M. R., Howley P. M. 2005. Inhibition of E2 binding to Brd4 enhances viral genome loss and phenotypic reversion of bovine papillomavirus-transformed cells. J. Virol. 79:14956–14961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. You J., et al. 2006. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen interacts with bromodomain protein Brd4 on host mitotic chromosomes. J. Virol. 80:8909–8919 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.