Abstract
Wnt/β-catenin signaling is fundamental in embryogenesis and tissue homeostasis in metazoans. Upon Wnt stimulation, cognate coreceptors LRP5 and LRP6 ([LRP5/6] low-density lipoprotein receptor-related proteins 5 and 6) are activated via phosphorylation at key residues. Although several kinases have been implicated, the LRP5/6 activation mechanism remains unclear. Here, we report that transmembrane protein 198 (TMEM198), a previously uncharacterized seven-transmembrane protein, is able to specifically activate LRP6 in transducing Wnt signaling. TMEM198 associates with LRP6 and recruits casein kinase family proteins, via the cytoplasmic domain, to phosphorylate key residues important for LRP6 activation. In mammalian cells, TMEM198 is required for Wnt signaling and casein kinase 1-induced LRP6 phosphorylation. During Xenopus embryogenesis, maternal and zygotic tmem198 mRNAs are widely distributed in the ectoderm and mesoderm. TMEM198 is required for Wnt-mediated neural crest formation, antero-posterior patterning, and particularly engrailed-2 expression in Xenopus embryos. Thus, our results identified TMEM198 as a membrane scaffold protein that promotes LRP6 phosphorylation and Wnt signaling activation.
INTRODUCTION
Canonical Wnt signaling plays an essential role in embryonic development and adult homeostasis (10, 26, 50). Wnt signaling dysregulation is implicated in numerous human diseases including cancer (8, 13, 27). Two types of cell surface receptors, low-density lipoprotein receptor-related proteins 5 and 6 (LRP5/6) (41, 47, 52) and Frizzled (Fz) (4, 54), are required for canonical Wnt signal transduction. Upon Wnt ligand binding to both receptors, LRP6, the single transmembrane protein, is clustered and phosphorylated with the assistance of Dishevelled (Dsh; Dvl in mammals) and the Axin complex (5, 32, 57). With mechanisms not fully understood, phosphorylated LRP6 prevents β-catenin degradation and activates β-catenin-dependent Wnt signaling (1, 27, 37). In this process, LRP6 phosphorylation is considered a key event for receptor activation.
LRP5/6 phosphorylation upon Wnt stimulation was first reported in 2004 (48), and several phosphorylation sites have since been identified. Among them, Thr-1479, Ser-1490, and Thr-1493 are the most extensively studied residues (37). The motif containing phospho-Ser-1490/Thr-1493 configures a docking site for Axin, and the phosphorylation status is influenced by an upstream Ser/Thr cluster including Thr-1479 (56). Thr-1479/1493 are typical casein kinase targets and are confirmed to be regulated by casein kinase 1 (CK1) family members, particularly CK1γ (17, 58). Glycogen synthase kinase-3 (GSK3) represents another intracellular component of the Wnt pathway, which directly interacts with and phosphorylates Ser/Thr residues in the LRP6 receptor cytoplasmic tail, including Ser-1490 (33, 58). Recently, another two kinases, G protein-coupled receptor kinase 5 (GRK5) and PFTAIRE protein kinase 1 (Pftk1), have been implicated (9, 16, 58). Further upstream, the Frizzled proteins are required via an unknown mechanism while Dvl proteins provide a platform for LRP6 aggregation and phosphorylation (5, 14, 32). Furthermore, LRP6 phosphorylation occurs in acidic vesicles where vacuolar H+-ATPase is an indispensable component (7, 14, 36). Other regulators are also involved such as Caprin-2 (20), a cytoplasmic protein, and phosphatidylinositol (PtdIns) lipid phosphatidylinositol 4,5-bisphosphate (PIP2) (38).
The precise mechanism that triggers LRP6 phosphorylation by its kinases remains elusive. To identify LRP6 regulators, we screened a Xenopus tropicalis cDNA library and identified transmembrane protein 198 (XtTMEM198) as a novel regulator. We found that TMEM198 can specifically activate LRP6 in canonical Wnt signaling by promoting aggregation and phosphorylation. Epistatic analysis indicated that TMEM198 and casein kinases are interdependent in LRP6 phosphorylation. Therefore, TMEM198 likely provides a membrane scaffold that recruits and facilitates kinases phosphorylating LRP6. We further demonstrated that TMEM198 is required for neural patterning during Xenopus embryogenesis, supporting a role in Wnt/β-catenin signaling in vivo. Thus, our results identify a new modulator for canonical Wnt signaling and provide the first biological activity of this functionally unknown transmembrane protein family.
MATERIALS AND METHODS
Constructs and small interfering RNA (siRNA).
Full-length X. tropicalis TMEM198 (NM_001005013) was identified from a cDNA library as described previously (17). A Wnt-responsive reporter screen was carried out as described previously (25) except that LRP6 and Fz5 plasmids were cotransfected as baits. tmem198 and CK1 constructs were generated using PCR and subcloned into pCS2+ vectors with a FLAG, Myc, or V5 tag. N-terminally tagged TMEM198 constructs were generated by adding the signal peptide sequence from Kremen protein (28) to the beginning of the coding region. To generate loss-of-function mutations, we mutated clusters of conserved amino acids, especially serine and threonine residues, using the QuikChange strategy (QuikChange Site-Directed Mutagenesis Kit; Stratagene, La Jolla, CA). TMEM198-M2, in which four amino acids in intercellular loop 3 were mutated (T168P, S171A, T172A, and T174R), was selected for further investigation because almost no activity remained while the expression level and cellular distribution were similar to those of the wild-type protein. Deletion constructs of tmem198 were generated using PCR (X. tropicalis TMEM198-ΔC encoding amino acids 1 to 232, X. tropicalis TMEM198-M2ΔC encoding amino acids 1 to 232 with mutations as in M2, and human TMEM198-C encoding amino acids 239 to 360). All constructs were confirmed using DNA sequencing.
Human TMEM198 (NM_001005209) siRNA target sequences were the following: siRNA-1, GCGTGCAACTGATGCGGAT; siRNA-2, GCCCATCAAACGCTTCAAT. The human TMEM198 short hairpin RNA (shRNA) target sequence was GCTGTTTGTTTGGAGTCGTCT. Fz and Dvl siRNAs were synthesized as described previously (38).
Cell culture, transfection, and reporter assay.
HEK293T and HeLa cells were maintained at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum (FCS). Plasmid DNA was transfected using Fugene-6 transfection reagent (Roche, Basel, Switzerland), and siRNAs were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). The working concentration of siRNAs was 100 nM.
For the luciferase reporter assay, HEK293T cells were seeded in 96-well plates and transfected in triplicates with plasmids or siRNAs, together with Super TOP-FLASH or CAGA reporters and pRL-TK as the internal control. Luciferase activity was determined at 36 h posttransfection. All experiments were repeated at least three times. Plasmid DNAs used per well were as follows: 15 ng of Super TOP-FLASH or CAGA, 0.5 ng of pRL-TK, 5 ng of LRP6, 3 ng of Fz, 5 ng of Wnt, 3 ng of CK1, 5 ng of Dvl, 10 ng of β-catenin, 5 ng of TMEM198, and 15 ng of constitutively active transforming growth factor beta type I receptor (CA-TGFβRI).
Antibodies and immunoblotting.
For immunoblotting, cells were seeded in 24-well plates and at 36 h posttransfection were lysed on ice with lysis buffer (20 mM Tris, pH 7.4, 140 mM NaCl, 10% glycerol, 1% NP-40, 10 mM EDTA, 2 mM sodium vanadate, 25 mM sodium fluoride, and a protease inhibitor cocktail). After centrifugation, the supernatant was subjected to SDS-PAGE and Western blotting using the following antibodies: FLAG (F3165; Sigma, St. Louis, MO), Myc (sc-40; Santa Cruz Biotechnology, Santa Cruz, CA), V5 (A190-119A; Bethyl Laboratories, Montgomery, TX), HA (sc-7392; Santa Cruz Biotechnology), β-catenin (610154; BD Biosciences, San Jose, CA), Dvl2 (3216; Cell Signaling Technology, Danvers, MA), Dvl3 (sc-8027; Santa Cruz Biotechnology), total LRP6 (2560; Cell Signaling Technology), LRP6 Sp-1490 (2568; Cell Signaling Technology), LRP6 Tp-1479 (17), and LRP6 Tp-1493 (produced according to reference 58). Sp-1490, Tp-1479, and Tp-1493 represent antibodies that specifically recognize phosphorylated Ser-1490, Thr-1479, and Thr-1493, respectively.
Immunoprecipitation, immunofluorescence, and biotinylation.
For immunoprecipitation, at 40 h posttransfection, cells in six-well plates were lysed on ice with TNE buffer (10 mM Tris, pH 7.4, 150 mM NaCl, 0.5% NP-40, 1 mM EDTA) supplemented with 2 mM sodium vanadate, 25 mM sodium fluoride, and a protease inhibitor cocktail. FLAG- or Myc-tagged proteins were recovered using anti-FLAG agarose beads (M2; Sigma) or protein A/G plus Myc antibody, respectively. After samples were washed with TNE buffer, the bound proteins were eluted with loading buffer and analyzed using immunoblotting.
For immunofluorescence, cells on coverslips in six-well plates were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100 in phosphate-buffered saline (PBS) and then blocked with 3% bovine serum albumin (BSA)-PBS before primary antibodies were applied. Secondary antibodies (donkey anti-mouse-Alexa Fluor 568 and goat anti-rabbit-Alexa Fluor 488 antibodies) were purchased from Molecular Probes/Invitrogen. If necessary, cell nuclei were visualized with 4′,6′-diamidino-2-phenylindole (DAPI) staining. Plasmid DNAs used for immunofluorescence were as follows: 2 μg of LRP6, 0.35 μg of Mesd, 0.5 μg of TMEM198 or Frizzled, and 1 μg of TGFβRI.
Cell surface biotinylation was carried out in six-well plates, using 0.5 mg/ml sulfo-NHS-LC-biotin (sulfosuccinimidyl-6-biotinamido-hexanoate; Thermo Scientific, Waltham, MA) according to the manufacturer's instructions. After biotin labeling, immunoprecipitation was performed with FLAG-M2 beads, and then the samples were subjected to SDS-PAGE and immunoblotted using the FLAG antibody and streptavidin-horseradish peroxidase (HRP) (3999; Cell Signaling Technology).
In vitro kinase assay and GST pulldown.
Intracellular domains of LRP6 and TMEM198 were fused with maltose binding protein (MBP) and glutathione S-transferase (GST) tags, respectively, and purified from Escherichia coli. The kinase (casein kinase 1ε) was purified from transfected HEK293T cells using FLAG-M2 beads (Sigma). For the in vitro kinase assay, substrates and kinases were mixed and incubated in kinase buffer (50 mM Tris, pH 7.4, 10 mM MgCl2, 1 mM ATP, 2 mM dithiothreitol [DTT], 150 mM NaCl) for 1 h at 25°C, and then SDS loading buffer was added to stop the reaction. The samples were then subjected to SDS-PAGE.
For the GST pulldown assay, GST-TMEM198-C was first incubated with GS4B beads for 30 min at 4°C. After beads were washed with PBS, they were further incubated in TNE buffer for 2 h at 4°C with in vitro translated or overexpressed CK1ε from transfected HEK293T cells. After samples were washed with PBS, the bound proteins were eluted with SDS loading buffer and subjected to SDS-PAGE.
Cell fractionation and sucrose gradient centrifugation.
Fractionation of the cell membrane and cytosol was performed as previously described with minor modifications (17). Cells in six-well plates were collected in a low-salt buffer (5 mM HEPES, pH 7.0, 1 mM MgCl2, 10 mM sodium pyrophosphate, 10 mM sodium fluoride, 5 mM sodium vanadate, and a protease inhibitor cocktail) and homogenized with 40 Dounce strokes. After centrifugation (500 × g for 10 min), the supernatant was subjected to ultracentrifugation (100,000 × g for 1 h) using a Beckman TLA100.3 rotor. The resultant supernatant was taken as the cytosol fraction. The pellet was solubilized with cholate buffer (20 mM HEPES, pH 7.4, 150 mM NaCl, 1% sodium cholate, 10 mM EDTA, 10 mM sodium fluoride, 5 mM sodium vanadate, and a protease inhibitor cocktail) and analyzed as the membrane fraction.
Sucrose gradient centrifugation was carried out as previously described with minor modifications (5). In brief, HEK293T cells in 10-cm dishes were transfected with 4 μg of LRP6 and 0.7 μg of Mesd with or without 2 μg of TMEM198. After 36 h, cells were washed and pelleted in PBS and then lysed for 30 min in extraction buffer (30 mM Tris, pH 7.4, 140 mM NaCl, 1% Triton X-100, 25 mM sodium fluoride, 5 mM sodium vanadate, and a protease inhibitor cocktail). Lysates were clarified using centrifugation at 14,000 rpm for 10 min at 4°C, and then the supernatant was layered on top of a 15 to 40% sucrose gradient prepared in 30 mM Tris, pH 7.4, 140 mM NaCl, 0.02% Triton X-100, 25 mM sodium fluoride, 5 mM sodium vanadate, and protease inhibitors. Ultracentrifugation was performed with a Beckman MLS50 rotor at 240,000 × g for 4 h at 4°C. After centrifugation, fractions were collected from the bottom of the tube and analyzed using immunoblotting. For coimmunoprecipitation, fractions were pooled, and the sucrose was diluted by adding 800 μl of extraction buffer to a 400-μl sample and then incubated for 4 h with FLAG-M2 beads (Sigma) at 4°C. After samples were washed with extraction buffer, bound proteins were eluted with SDS loading buffer and subjected to SDS-PAGE.
Xenopus embryo assays.
Xenopus embryos were cultured under standard conditions. mRNA was synthesized using a mMESSAGE mMACHINE SP6 kit (Ambion, Austin, TX) according to the manufacturer's instructions. Antisense and standard control morpholino (MO) oligonucleotides were purchased from Gene Tools, LLC (Philomath, OR). The MO sequences of Xenopus laevis tmem198 (NM_001094258) were as follows: MO-1, CGGTGGGACAACAGACGATAGATCA; MO-2, GAATCTGGTGTTAAGTAAGCATGTC. Whole-mount in situ hybridization and LacZ staining were performed as described previously (51). RNA probes were hybridized at 60°C, and BM purple substrate (Roche) was applied for the chromogenic reaction.
Total RNA isolation and reverse transcription-PCR (RT-PCR).
Total RNA was isolated using TRIzol reagent (Invitrogen), and reverse transcription was performed using the Reverse Transcription System (Promega, Fitchburg, WI). Primers used for human tmem198 were 5′-GCAAGGAGAAAAGGCGGAAAA-3′ and 5′-CTGTGGGTGAGGCCATGAAG-3′. For X. laevis tmem198, the primers were 5′-TGAACAGTTTTATCACCCGCC-3′ and 5′-TATTATGACATCAGTATGAGAGAA-3′. Other primers were used as previously described (51).
RESULTS
TMEM198 promotes LRP6 signaling and is required for Wnt signaling upstream of the β-catenin destruction complex.
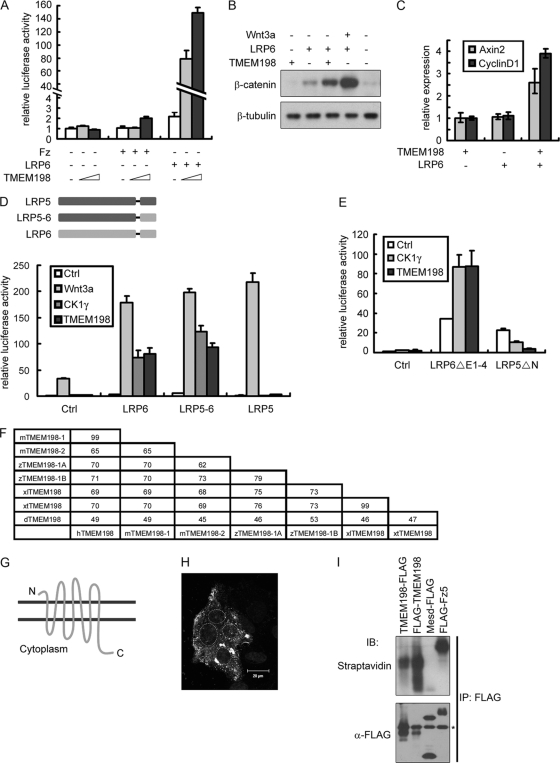
We screened a Xenopus tropicalis embryonic cDNA library and identified a novel 7-transmembrane protein, TMEM198, that specifically and strongly cooperated with the Wnt coreceptor LRP6 in activating TOP-FLASH reporter expression (Fig. 1A). Among many components in the canonical Wnt pathway, TMEM198 cooperated only with LRP6, and the cooperation was dose dependent (Fig. 1A and data not shown). In addition, TMEM198 and LRP6 cooperated to induce β-catenin accumulation in the cytosol of HEK293T cells (Fig. 1B), a characteristic feature of Wnt signaling activation, and to enhance the expression of direct Wnt target genes axin2 and cyclin D1 (Fig. 1C). These results suggest that TMEM198 is able to activate LRP6 in Wnt signaling.
Fig. 1.
TMEM198 specifically promotes LRP6 signaling in the canonical Wnt/β-catenin pathway. (A) Wnt-responsive TOP-FLASH reporter assay in HEK293T cells with indicated transfections. TMEM198 plasmid DNA was used at 1 ng and 10 ng. (B) Cytosol fractions from HEK293T cells transfected with indicated DNA samples were immunoblotted with β-catenin or β-tubulin (loading control) antibodies. Cells in six-well plates were transfected with 100 ng of TMEM198, 100 ng of Wnt3a, or 300 ng of LRP6 plasmid DNA as indicated. (C) The expression levels of Axin2 and Cyclin D1 from transfected HEK293T cells were quantified using real-time PCR. Cells in six-well plates were transfected with 100 ng of TMEM198 and/or 300 ng of LRP6 plasmid DNA. (D) Wnt reporter assay in HEK293T cells transfected with the indicated plasmids. LRP5-6 is a chimeric protein in which the C terminus of LRP5 was replaced with that of LRP6 (schematically showing in the upper panel). Plasmid amounts were as follows: 5 ng LRP6, LRP5, and LRP5-6; 5 ng of Wnt3a; 5 ng of TMEM198; and 3 ng of CK1γ. Note that TMEM198, like CK1γ, activated LRP6 and LRP5-6 but not LRP5. Wnt3a was used as a positive control to demonstrate that the LRP5 was functional. (E) Wnt-responsive reporter experiment in HEK293T cells transfected with the indicated plasmids. Plasmid amounts were as follows: 1 ng of LRP6ΔE1-4, 1 ng of LRP5ΔN, 5 ng of TMEM198, and 3 ng of CK1γ. Note that TMEM198, like CK1γ, further enhanced LRP6ΔE1-4 but not LRP5ΔN, while each truncated receptor itself was similarly active. (F) Percent similarities of amino acid sequences among TMEM198 proteins in different species. Two TMEM198 homologues in mouse (mTMEM198-1 and mTMEM198-2) and zebrafish (zTMEM198-1A and zTMEM198-1B) and one homologue in X. laevis (XlTMEM198), X. tropicalis (XtTMEM198), Drosophila (dTMEM198), and human (hTMEM198) were analyzed. (G) Predicted topology of TMEM198 protein. Dark lines represent the plasma membrane, and the N or C stands for N or C terminus of TMEM198 protein, respectively. (H) Confocal microscopy images of HeLa cells transfected with TMEM198-FLAG. The dashed lines indicate the nuclei. Note that overexpressed TMEM198 was localized at the plasma membrane as well as in intracellular vesicle-like structures. (I) Cell surface biotinylation experiment from HEK293T cells transfected as indicated. Note that both TMEM198-FLAG and FLAG-TMEM198 were biotinylated and therefore cell surface localized. Mesd-FLAG (an endoplasmic reticulum retention chaperon protein) and FLAG-Fz5 were used as a negative and positive control, respectively. The asterisk indicates the heavy chain of FLAG antibody. IP, immunoprecipitation; IB, immunoblotting.
LRP5 is another Wnt coreceptor closely related to LRP6 in terms of structure and function (23). However, in contrast to LRP6, LRP5 was not activated by coexpressed TMEM198, whereas Wnt3a was able to activate both coreceptors (Fig. 1D). More interestingly, when the intracellular domain of LRP5 was replaced with that of LRP6, the chimeric protein (LRP5-6) could be activated by TMEM198 as strongly as LRP6 (Fig. 1D). These results suggest that TMEM198 selectively cooperates with LRP6 but not LRP5, and the specificity relies on the intracellular domain of the LRP5/6 receptors. Corroborating this conclusion, the constitutively active form of LRP6, LRP6ΔE1-4, was further activated by TMEM198, whereas that of LRP5, LRP5 with a deletion of the N terminus (LRP5ΔN), was not activated (Fig. 1E). Of note, casein kinase 1γ (CK1γ), a known kinase responsible for LRP6 phosphorylation, displayed similar selectivity on full-length as well as constitutively active LRP5/6 receptors (Fig. 1D and E). The reason for this selectivity is unknown; however, we cannot rule out a quantitative difference between LRP5 and LRP6 in transducing Wnt signaling.
TMEM198 is predicted to be a transmembrane protein and is conserved from fruit fly to human (Fig. 1F); however, its orthologs have not been characterized in any organism including Drosophila (Drosophila tmem198; accession number CG14234). Thus, we cloned TMEM198 homologues from Xenopus laevis (GeneID 447551), human (GeneID 130612), and mouse (GeneIDs 319998 and 73827) and confirmed that all retained similar activity levels in promoting LRP6 signaling (data not shown). Topology prediction using the SMART program (EMBL) suggested that TMEM198 consists of a very short extracellular domain (31 amino acids for X. tropicalis TMEM198), seven transmembrane domains, and a cytoplasmic tail with ∼110 amino acids (Fig. 1G). To experimentally confirm this prediction, we transplanted the signal peptide from the Kremen protein (28) to the N terminus of TMEM198 and found that this fusion protein was as active as the wild-type in promoting LRP6 signaling (data not shown). Moreover, TMEM198 became inactive when green fluorescent protein (GFP) was fused at the N terminus while retaining full activity when GFP was fused at the C terminus (data not shown). Immunofluorescent analysis of transfected HeLa cells indicated that a large amount of TMEM198 localized intracellularly in vesicle-like structures (Fig. 1H), suggesting that TMEM198 may not be a typical plasma membrane protein. However, the plasma membrane-localized TMEM198 was readily detected by the cell surface biotinylation assay, confirming that at least part of the overexpressed TMEM198 proteins were at the plasma membrane (Fig. 1I).
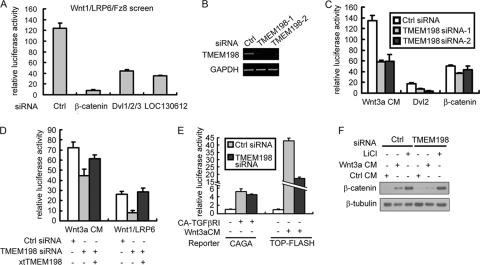
In parallel, TMEM198 was identified in a genome-wide siRNA screen for genes required for Wnt/β-catenin signaling in HEK293T cells (Fig. 2A) (14). To further address the involvement of TMEM198 in canonical Wnt signaling, we designed two specific siRNAs to knock down TMEM198 in HEK293T cells (Fig. 2B). As shown in Fig. 2C, Wnt3a signaling was significantly reduced when TMEM198 siRNAs were applied, while β-catenin signaling was largely unaffected. Signaling induced by Dvl2 was also markedly downregulated by TMEM198 siRNA (Fig. 2C), consistent with the model of Dvl involvement in promoting LRP6 activation and signaling via LRP6 (5, 32, 44, 57). The siRNA effect was specific because the activity was rescued by cotransfected X. tropicalis TMEM198 (XtTMEM198) (Fig. 2D). Further confirming specificity, TMEM198 siRNA affected Wnt3a-induced TOP-FLASH expression but had no effect on the expression of the CAGA-luciferase reporter induced by constitutively active TGFβ type I receptor (CA-TGFβRI) (Fig. 2E). Epistatically, TMEM198 was required for β-catenin accumulation induced by Wnt3a but not LiCl, a potent GSK3 inhibitor (Fig. 2F), suggesting that TMEM198 functions upstream of β-catenin accumulation. Moreover, the TMEM198/LRP6 signal was completely blocked with cotransfection of Axin, the scaffold protein of the β-catenin degradation complex (data not shown). Taken together, these results indicate that TMEM198 is able to activate LRP6 and is required for canonical Wnt signaling upstream of the β-catenin destruction complex.
Fig. 2.
TMEM198 is required for canonical Wnt signaling upstream of the β-catenin destruction complex. (A) Wnt-responsive reporter assay in HEK293T cells. LOC130612 (human TMEM198) was identified in a genome-wide siRNA screen for genes required for Wnt/β-catenin signaling (14). Wnt signaling was activated by cotransfection of Wnt1 (5 ng), LRP6 (3.3 ng), and Fz8 (1 ng). (B) The siRNA knockdown efficiency of TMEM198 in HEK293T cells determined by RT-PCR. Control (Ctrl) or two different TMEM198 siRNAs (TMEM198-1 and TMEM198-2) were transfected in HEK293T cells. At 60 h posttransfection, total RNA was isolated, and RT-PCR was performed. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. (C to E) Luciferase reporter assay in HEK293T cells transfected with the indicated plasmid DNAs and siRNA samples. Wnt3a CM, Wnt3a-conditioned medium. In panel D, 1 ng and 3 ng of X. tropicalis TMEM198 (XtTMEM198) plasmid DNA were used to rescue the signaling of Wnt3a CM and Wnt1/LRP6, respectively. (F) Western blots of cytosol fractions from HEK293T cells transfected with indicated siRNAs and then treated with control, Wnt3a CM, or LiCl (20 mM) for 4 h before cell fractionation.
TMEM198 associates with LRP6 and promotes phosphorylation.
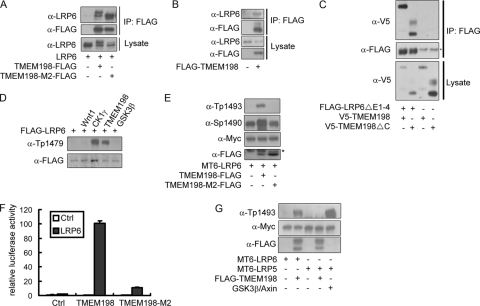
Next, we addressed whether TMEM198 was able to interact with LRP6. In coimmunoprecipitation (co-IP) assays with transfected HEK293T cells, TMEM198 associated with LRP6 (Fig. 3A). Since an antibody recognizing endogenous TMEM198 protein was lacking, we transfected FLAG-TMEM198 into HEK293T cells and detected coimmunoprecipitated endogenous LRP6 (Fig. 3B). The interaction is likely mediated by the transmembrane domains because the TMEM198 protein lacking the cytoplasmic portion (TMEM198-ΔC) was also coimmunoprecipitated with LRP6 (data not shown) and LRP6ΔE1-4, an extracellular truncated form of LRP6 (Fig. 3C). The interaction between overexpressed TMEM198 and LRP5 was also detected (data not shown) although the latter was not activated.
Fig. 3.
TMEM198 associates with LRP6 and promotes phosphorylation. (A) Western blots of immunoprecipitates (IP) or initial lysates from HEK293T cells transfected as indicated. TMEM198-M2-FLAG is a mutant form of TMEM198. Note that the precipitated LRP6 from a wild-type TMEM198-cotransfected sample migrated more slowly than that from TMEM198-M2 because of heavy phosphorylation induced by the wild-type but not mutant TMEM198. (B and C) Western blots of immunoprecipitates or initial lysates from HEK293T cells transfected as indicated. Endogenous LRP6 was detected in panel B. The asterisk in panel C indicates the antibody heavy chain. (D and E) Western blots of total cell lysates from HEK293T cells transfected as indicated. Tp-1479, Sp-1490, and Tp-1493 represent antibodies that specifically recognize phosphorylated Thr-1479, Ser-1490, and Thr-1493 forms of human LRP6 protein, respectively. MT6-LRP6, Myc6-tagged LRP6. The asterisk in panel E indicates a nonspecific band. Cells in 24-well plates were transfected with 200 ng of LRP6 and 50 ng of CK1γ, GSK3β, or TMEM198 plasmid. (F) Wnt-responsive reporter experiment in HEK293T cells transfected with the indicated plasmids. Note that TMEM198-M2 was much weaker in activating LRP6. (G) Western blots of total cell lysates from HEK293T cells transfected as indicated. MT6-LRP6/MT6-LRP5, Myc6-tagged LRP6 or Myc6-tagged LRP5. Cells in 24-well plates were transfected with 200 ng of tagged LRP6/LRP5 and 50 ng of GSK3β, Axin, or TMEM198 plasmid. α, anti.
Activation of LRP6 in canonical Wnt signaling requires phosphorylation; therefore, the status of LRP6 phosphorylation at three key residues was monitored using phospho-specific antibodies. As expected, LRP6 phosphorylation at Thr-1479, Ser-1490, and Thr-1493 was dramatically increased when TMEM198 was coexpressed (Fig. 3D and E). As a positive control, CK1γ induced massive phosphorylation at Thr-1479, whereas GSK3β did not induce phosphorylation (Fig. 3D). A mutant form of TMEM198 (TMEM198-M2) was also tested in this assay and showed no effect (Fig. 3E). Consistently, this mutant had greatly reduced activity in promoting LRP6 signaling (Fig. 3F). TMEM198-M2 was expressed at an equal level (Fig. 3A and E) and distributed in a similar pattern in HeLa cells as the wild type (data not shown) and retained binding to LRP6 (Fig. 3A). As another control of the seven-transmembrane protein, Frizzled 7 was cotransfected with LRP6, and no enhancement of LRP6 phosphorylation was observed (data not shown), suggesting that TMEM198 specifically promoted LRP6 phosphorylation. Further demonstrating the specificity and consistent with the results of the activity assay (Fig. 1D) were our findings that TMEM198 did not enhance Thr-1493 phosphorylation of LRP5 while overexpression of GSK3/Axin did enhance phosphorylation (Fig. 3G). Together, these results indicate that TMEM198 specifically activates LRP6 by promoting phosphorylation.
TMEM198 facilitates casein kinase 1 family members in phosphorylating LRP6.
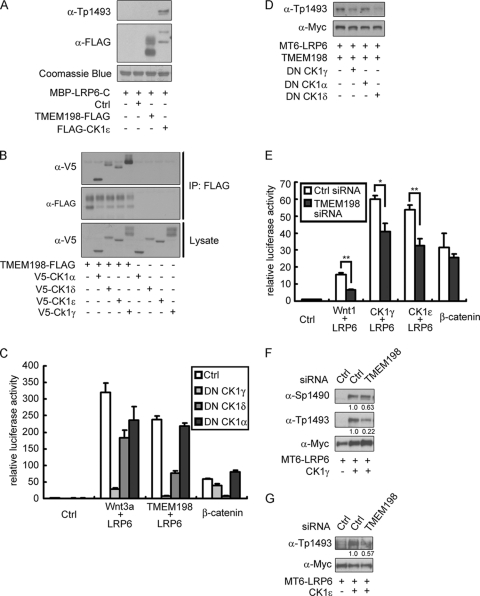
Because there is no predicted kinase domain within TMEM198, we speculate that TMEM198 cannot phosphorylate LRP6 directly. As shown in the in vitro kinase assay, unlike CK1ε, TMEM198 was not able to phosphorylate purified LRP6-C, the cytoplasmic domain of LRP6 (Fig. 4A). This failure to phosphorylate the cytoplasmic domain confirmed that TMEM198 per se is not a kinase; however, it may assist kinases. As casein kinase 1 (CK1) family members and GSK3 have been implicated in LRP6 phosphorylation (17, 58), we verified the potential interaction between TMEM198 and casein kinase 1 or GSK3β. As shown in Fig. 4B, all four CK1s (CK1α/ε/δ/γ) were coimmunoprecipitated with TMEM198, whereas GSK3β was not coimmunoprecipitated (data not shown). In the Wnt-responsive reporter assay, LRP6/TMEM198 signaling was inhibited by dominant negative CK1γ ([DN-CK1γ] which specifically blocks CK1γ activity [17]) and dominant negative CK1δ ([DN-CK1δ] which blocks the activity of both CK1ε and CK1δ [58]) (Fig. 4C). The inhibition was specific because dominant negative CK1α ([DN-CK1α] which blocks CK1α activity [58]) had no effect (Fig. 4C). Moreover, DN-CK1γ and DN-CK1δ, but not DN-CK1α, significantly blocked TMEM198-induced LRP6 phosphorylation (Fig. 4D). These results indicate that casein kinase 1 family members are required for TMEM198-induced LRP6 phosphorylation and activation.
Fig. 4.
Casein kinase 1 is involved in TMEM198 stimulated LRP6 phosphorylation. (A) Western blots and Coomassie blue staining of SDS-PAGE samples after the in vitro kinase assay. Coomassie blue staining indicated MBP-LRP6-C protein. Tp-1493 staining indicated phosphorylated LRP6 and FLAG antibody-detected purified TMEM198-FLAG or FLAG-CK1ε from transfected HEK293T cells. Ctrl, FLAG immunoprecipitates from pCS2+ vector-transfected cells. (B) Western blots of immunoprecipitates (IP) or initial lysates from HEK293T cells transfected as indicated. (C) Wnt reporter assay in HEK293T cells with indicated transfections. DN, dominant negative. Five nanograms of each DN-CK1 was used. (D) Western blots of total cell lysates from HEK293T cells transfected as indicated. MT6-LRP6, Myc6-tagged LRP6. Cells in 24-well plates were transfected with 200 ng of LRP6, 50 ng of TMEM198, and 200 ng of each DN-CK1. (E) Wnt reporter assay in HEK293T cells transfected as indicated (*, P < 0.05; **, P < 0.01; Student's t test, n = 3). (F and G) Western blots of total cell lysates from HEK293T cells transfected as indicated. A total of 200 ng of LRP6 and 50 ng of CK1γ/CK1ε plasmid DNAs were used. MT6-LRP6, Myc6-tagged LRP6. Band intensities were quantified, and phospho-signals were normalized against the signal of the corresponding total LRP6. The signal of control siRNA-transfected cells was designated 1.0, and the relative rates of TMEM198 siRNA-transfected cells are shown.
CK1 overexpression is able to induce LRP6 phosphorylation. Therefore, we addressed whether endogenous TMEM198 was involved in this process. As shown in Fig. 4E, knockdown of TMEM198 using siRNA significantly reduced Wnt signaling activated by LRP6/CK1γ or LRP6/CK1ε. Consistently, LRP6 phosphorylation induced by either CK1γ or CK1ε was also reduced with TMEM198 depletion (Fig. 4F and G). Taken together, these results suggest that TMEM198 and casein kinases are mutually dependent for LRP6 phosphorylation and activation.
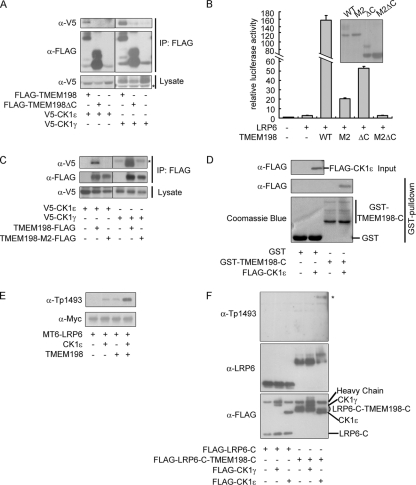
TMEM198 is a multitransmembrane protein, and the C terminus likely extends toward the cytosol while the casein kinases are either membrane associated (CK1γ, via lipid modification) or cytosolic. The topology suggested that TMEM198 might interact with CK1s via its C-terminal domain (C domain). Indeed, TMEM198 with a deletion of the C terminus (TMEM198ΔC) exhibited greatly reduced interaction with all CK1s (Fig. 5A and data not shown) as well as significantly decreased Wnt promoting activity (Fig. 5B). This result suggested that the TMEM198 C-terminal domain was crucial for CK1 binding. Similarly, TMEM198-M2, a mutant with greatly reduced activities in Wnt signaling and in promoting LRP6 phosphorylation (Fig. 3E and F), bound undetectable CK1ε or CK1γ in the co-IP assay (Fig. 5C). Furthermore, although either TMEM198-M2 or TMEM198ΔC retained some activity, TMEM198-M2ΔC was completely inactive (Fig. 5B), suggesting that the CK1 binding capacity is indispensable for promoting LRP6 signaling. Direct interactions between the C domains of TMEM198 (GST-TMEM198-C purified from E. coli) and CK1ε that was either in vitro translated (Fig. 5D) or overexpressed in HEK293T cells (data not shown) were detected in GST pulldown experiments. These results suggest that the TMEM198 cytoplasmic domain interacts with CK1 directly.
Fig. 5.
TMEM198 recruits casein kinase via the cytoplasmic domain. (A) Western blots of immunoprecipitates (IP) or initial lysates from HEK293T cells transfected as indicated. The asterisk indicates a nonspecific band. (B) Wnt-responsive reporter assay in HEK293T cells transfected with the indicated plasmids. Plasmid amounts used were as follows: 5 ng of LRP6, 5 ng of TMEM198, and mutants. The inset shows the expression levels of wild-type and various TMEM198 mutations. (C) Western blots of immunoprecipitates or initial lysates from HEK293T cells transfected as indicated. Note that the mutant TMEM198 (TMEM198-M2) lost interaction with CK1γ/ε. The asterisk indicates the antibody heavy chain. (D) Western blots and Coomassie blue staining of samples before (input) or after the GST pulldown assay as indicated. Coomassie blue staining indicated GST fusion proteins. (E) Western blots of total cell lysates from HEK293T cells transfected as indicated. A total of 200 ng of LRP6 and 30 ng of TMEM198 or CK1ε plasmid DNAs were used. MT6-LRP6, Myc6-tagged LRP6. (F) Western blots of immunoprecipitates from HEK293T cells transfected as indicated. FLAG-tagged LRP6-C or FLAG-LRP6-C-TMEM198-C (LRP6-C fused with TMEM198-C) was immunoprecipitated using FLAG-M2 beads and then probed with the indicated antibodies. Note that the fusion protein became a smear, another indication of phosphorylation. The asterisk indicates the phosphorylation signal of FLAG-LRP6-C-TMEM198-C.
The above results suggested that TMEM198 likely activated LRP6 via recruitment of casein kinase 1 family members. Therefore, we investigated whether TMEM198 was able to assist CK1 with phosphorylating LRP6. When cotransfected, minimal doses of TMEM198 and CK1ε indeed synergistically promoted LRP6 phosphorylation (Fig. 5E). To demonstrate a cofactor feature of TMEM198 for CK-mediated LRP6 phosphorylation, we fused the LRP6 cytoplasmic domain (LRP6-C) with the TMEM198 C domain (TMEM198-C) and tested whether this fusion protein was an improved substrate for casein kinases in transfected cells. As shown in Fig. 5F, the fusion protein (LRP6-C-TMEM198-C) was, indeed, more strongly phosphorylated than LRP6-C. This result strongly suggests that TMEM198 is able to facilitate CK1-mediated LRP6 phosphorylation. Taken together, these results suggest that TMEM198 promotes LRP6 phosphorylation and activation partially via recruiting and facilitating CK1.
TMEM198 promotes LRP6 aggregation.
TMEM198 promoted LRP6 phosphorylation at all three amino acid sites tested, Thr-1479, Ser-1490, and Thr-1493. Among them, Thr-1479 and Thr-1493 are typical casein kinase targets while Ser-1490 could be phosphorylated by GSK3, GRK5, or Pftk1 (9, 16, 17, 58). Our results indicated that TMEM198 does not interact with GSK3β or Pftk1 (data not shown), suggesting that the effect on Ser-1490 by TMEM198 is secondary. One explanation could be that the phosphorylation on Thr-1479 enhanced that on Ser-1490, and this possibility is supported by a previous report (56). Another possibility is that TMEM198 affects LRP6 phosphorylation globally and provides a microenvironment that facilitates phosphorylation.
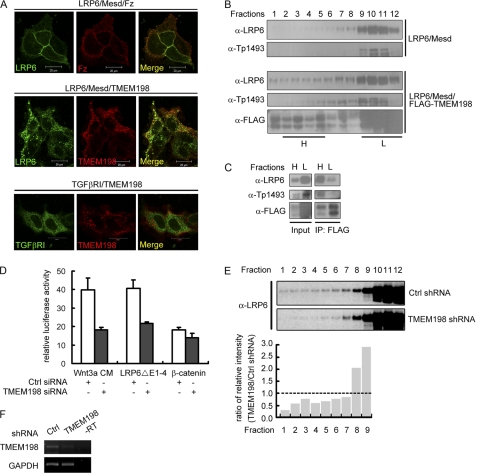
Dishevelled (Dvl)-mediated LRP6 clustering or aggregation has been proposed as a key step in LRP6 phosphorylation and activation, and upon Wnt ligand stimulation the LRP6 signalosome is induced (5, 32, 38). We therefore addressed whether TMEM198 was able to affect the subcellular distribution of LRP6. As shown in Fig. 6A, LRP6 was detected at the cell membrane and colocalized with cotransfected Frizzled 5 (90% of cotransfected cells). In contrast, when TMEM198 was cotransfected, LRP6 formed cytosolic punctate structures together with TMEM198 (95% of cotransfected cells), suggesting the formation of signalosome-like structures (Fig. 6A). To rule out unspecific aggregation with overexpressed transmembrane proteins, we coexpressed TMEM198 together with the TGFβ type I receptor (TGFβRI) and observed no colocalization although TMEM198 itself was punctate (100% of cotransfected cells) (Fig. 6A). The LRP6 signalosome has been shown to consist of caveolin-containing acidic vesicles with an unclear identity (5) but which are thought to correspond to multivesicular bodies (MVBs) (46). We therefore performed immunofluorescence analysis and observed that TMEM198/LRP6 structures were partially colocalized with caveolin but not with clathrin or EEA (an early endosome marker) (data not shown). To further confirm that TMEM198 was able to promote LRP6 aggregation, we performed sucrose sedimentation experiments using LRP6- or LRP6/TMEM198-transfected HEK293T cells. As shown in Fig. 6B, significantly more total LRP6 as well as phosphorylated LRP6 was detected in high-molecular-weight (HMW) fractions when TMEM198 was coexpressed. Importantly, when we separately harvested the low-molecular-weight (LMW) and HMW fractions and performed immunoprecipitation against FLAG-TMEM198, much more LRP6 was coprecipitated from the HMW fractions although less LRP6 was contained in the HMW fractions before precipitation (Fig. 6C). Similarly, in the HMW precipitates, phospho-LRP6 was enriched (Fig. 6C). These results suggest that TMEM198 is likely capable of promoting LRP6 aggregation, thus facilitating LRP6 phosphorylation.
Fig. 6.
TMEM198 promotes LRP6 aggregation and is required for the spontaneous aggregation and activation of LRP6ΔE1-4. (A) Confocal microscopy images of HeLa cells transfected as indicated. LRP6-GFP (green) was cotransfected with Mesd and Fz5 (red) or TMEM198 (red). TGFβRI (green) was used as a control. Yellow signals indicate colocalization. (B) Western blots of fractions from sucrose gradient sedimentation. HEK293T cells were transfected as indicated, and total cell lysates were separated in a sucrose gradient using ultracentrifugation. The fractions were collected from the bottom up. Note that TMEM198 promoted high-molecular-weight aggregation of LRP6. H and L (high- and low-molecular-weight fractions, respectively) indicate the fractions that were pooled together for immunoprecipitation in panel C. (C) Western blots of input or immunoprecipitates of pooled fractions from panel B. Note that significantly more LRP6 as well as phospho-LRP6 was detected in the input from low-molecular-weight fractions (input, L), while LRP6 and phospho-LRP6 were specifically coimmunoprecipitated from the high-molecular-weight fractions (IP, H). (D) Wnt reporter assay in HEK293T cells transfected with indicated plasmids and siRNAs. Wnt3a CM, Wnt3a-conditioned medium. Plasmid amounts were 1 ng of LRP6ΔE1-4 and 10 ng of β-catenin. (E) Western blots of fractions from sucrose gradient sedimentation experiments. Four micrograms of LRP6ΔE1-4 was transfected into control (Ctrl) or TMEM198 shRNA-expressing HEK293T cells. Note that in TMEM198 knockdown cells, high-molecular-weight aggregation of LRP6ΔE1-4 was slightly reduced (upper panel). The lower panel indicates the quantification of the Western blots. Intensity ratio of each LRP6ΔE1-4 band from the TMEM198 shRNA cells against the corresponding one from the control shRNA cells (TMEM198/Ctrl shRNA) in fractions 1 to 9 was calculated using Quantity One software (Bio-Rad) and plotted. The dashed line represents a 1:1 ratio. Note that in fractions 1 to 7, there was more LRP6ΔE1-4 in the control shRNA sample while in fractions 8 and 9 there was more in the TMEM198 shRNA sample, suggesting that knockdown of TMEM198 reduced the formation of high-molecular-weight aggregation of LRP6ΔE1-4. (F) RT-PCR result showing that in specific-shRNA-expressing HEK293T cells, which were used in the experiment shown in panel E, the TMEM198 mRNA level was significantly downregulated. GAPDH was used as loading control. −RT, minus reverse transcription control.
It has been proposed that LRP6ΔE1-4, the constitutively active form, signals independently of Dvl because of spontaneous self-aggregation (5). However, how this ligand- and Dvl-independent self-aggregation occurs was unknown. We found that TMEM198 was able to associate with LRP6ΔE1-4 (Fig. 3C) to further enhance its activity (Fig. 1E) and was required for its activation in the Wnt-responsive reporter assay (Fig. 6D). Moreover, in sucrose sedimentation experiments, a significant portion of LRP6ΔE1-4 protein was detected in the HMW fractions (Fig. 6E), as previously reported (5). However, in TMEM198 knockdown cells (Fig. 6F) this HMW distribution was slightly but consistently reduced (Fig. 6E). These results suggest that TMEM198 is probably involved in self-aggregation and spontaneous activation of LRP6ΔE1-4.
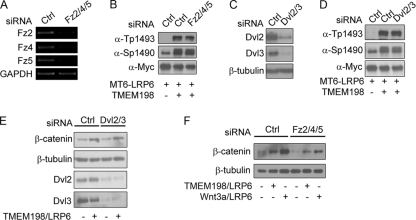
Previous studies revealed that Frizzled and Dishevelled proteins are required for Wnt-dependent LRP6 phosphorylation (5, 57). We next investigated whether they were involved in TMEM198-induced LRP6 phosphorylation and activation. In HEK293T cells, Fz2/Fz4/Fz5 (Fz2/4/5) and Dvl2/Dvl3 (Dvl2/3) are expressed at detectable levels, and previous reports have confirmed that siRNA-mediated knockdown of these genes efficiently blocks Wnt signaling or LRP6 receptor activation (38). We therefore used these siRNA combinations (Fig. 7A and C) and found that knockdown of either Fz or Dvl proteins did not significantly affect TMEM198-induced LRP6 phosphorylation (Fig. 7B and D). Moreover, TMEM198/LRP6-induced β-catenin accumulation was not affected by Dvl siRNAs (Fig. 7E). These results suggest that TMEM198 promotes LRP6 phosphorylation probably independent or downstream of Dvl-mediated receptor aggregation. However, we noted that when Fz genes were knocked down with siRNAs, TMEM198/LRP6-induced β-catenin accumulation was slightly reduced (Fig. 7F). An explanation for this observation is that besides leading to LRP6 phosphorylation, Fz may have another activity required for β-catenin stabilization.
Fig. 7.
TMEM198 promotes LRP6 phosphorylation independent of Fz and Dvl. (A to D) RT-PCR result shows that in specific-siRNA-expressing HEK293T cells, which were used in the experiments shown in panels B and F, Fz2, Fz4 and Fz5 mRNA levels were significantly downregulated. GAPDH was used as a loading control. Western blots of total cell lysate from HEK293T cells transfected with the indicated plasmids or siRNAs. Control (Ctrl) and Fz2/4/5 (A) or Dvl2/3 siRNAs were transfected 24 h before plasmid DNA transfection. At 36 h after the plasmid transfection, cells were lysed and probed with the indicated antibodies. The knockdown efficiencies of Dvl2 and Dvl3 were monitored by specific antibodies, as shown in panel C. MT6-LRP6, Myc6-tagged LRP6. (E and F) Western blots of cytosol fractions from HEK293T cells transfected with indicated plasmids and siRNAs. Control (Ctrl) and Fz2/4/5 (A) or Dvl2/3 siRNAs were transfected 24 h before plasmid DNA transfection. At 36 h after the plasmid transfection, cells were lysed in low-salt buffer, and the cytosol fractions were isolated and probed with β-catenin or β-tubulin (as loading control) antibodies. The knockdown efficiencies of Dvl2 and Dvl3 were monitored by specific antibodies as shown in panel E. Note that whereas Fz2/4/5 siRNAs (F) slightly inhibited TMEM198/LRP6-induced β-catenin accumulation in the cytosol, Dvl2/3 siRNAs did not (E).
TMEM198 activates LRP6 in Xenopus embryos and is involved in neural patterning.
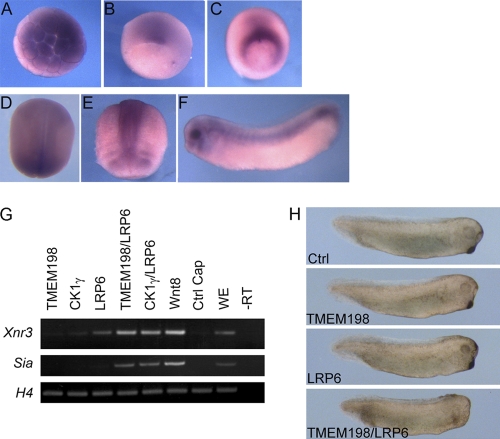
Canonical Wnt signaling plays a vital role during early embryonic development (26, 27, 50). To address TMEM198 function in Xenopus embryos, we analyzed its expression pattern by RT-PCR and in situ hybridization. By RT-PCR, tmem198 mRNA was detected constantly from the two-cell stage to tadpole embryos (data not shown). By in situ hybridization, tmem198 mRNA was detected in cleavage embryos with enrichment in the animal blastomeres, indicative of maternal distribution (Fig. 8A and data not shown). At stage 10+, tmem198 mRNA was detected mainly in the organizer region, just above the dorsal blastopore lip (Fig. 8B). Its expression extended toward the ventral side along the blastopore formation (Fig. 8C). During gastrulation and neuralization, the expression domain of tmem198 extended anteriorly and covered the entire neural plate as well as the underlying mesodermal cells (Fig. 8D and E and data not shown). At stage 14, a gradual enhanced expression was detected toward the posterior pore (Fig. 8D). At the late neural stage, tmem198 was expressed in the neural tube, brain subdomains, branch arches, and quite strongly in the eye regions (Fig. 8F).
Fig. 8.
TMEM198 activates LRP6 in Xenopus embryos. (A to F) Whole-mount in situ hybridization of tmem198 mRNA in developing X. laevis embryos. (A) Stage 6.5, animal view. (B) Stage 10+, dorsal-vegetal view. (C) Stage 11.5, vegetal view with dorsal up. (D) Stage 14, dorsal-posterior view. (E) Stage 17, dorsal-anterior view. (F) Stage 28, lateral view. (G) Four-cell-stage embryos were injected animally into each blastomere with 100 pg of tmem198, 100 pg of CK1γ, 500 pg of LRP6, 8 pg of Wnt8 mRNA, or in combination as indicated. At stage 8 to 9, animal caps were dissected and cultured until stage 11 for expression analyses of the indicated marker genes using RT-PCR. −RT, minus reverse transcription control; WE, whole embryo; H4, histone 4. (H) Eight-cell-stage embryos were injected in the animal blastomeres or in the animal pole regions of blastomeres with 400 pg of tmem198 or 2 ng of LRP6 mRNA as indicated.
In Xenopus embryos, TMEM198 cooperated with LRP6 in activating Wnt-responsive reporter gene expression (data not shown) and inducing the expression of siamois (sia) and Xnr3, two direct Wnt target genes, in animal caps (Fig. 8G). In addition, tmem198 mRNA injection into animal blastomeres inhibited head formation (data not shown), a hallmark of overactivated Wnt signaling (11, 35), and a suboptimal dose of TMEM198 cooperated with LRP6 in the same assay (n = 37 embryos; 92%) (Fig. 8H). These results suggest that TMEM198 is able to activate LRP6-mediated Wnt signaling in Xenopus embryos.
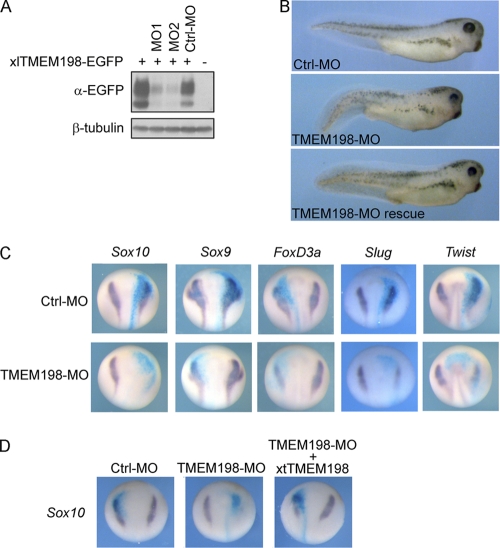
Two independent morpholino antisense oligonucleotides (MO) were both effective in blocking mRNA translation of Xenopus laevis TMEM198 (Fig. 9A) and were used in combination to assess whether TMEM198 is necessary for the normal development of Xenopus embryos. Embryos injected with TMEM198-MOs developed largely unaffected at the gastrula and early neurula stages. However, at the tadpole stage, morphants were often less pigmented (n = 94; 67%) with ventrally bent tails (n = 94; 47%) (Fig. 9B). Anteriorly, these embryos developed smaller eyes (often only the dorsal half of the retina was retained) and forebrain structures (n = 94; 62%) than the control-MO-injected embryos (Fig. 9B). The overall phenotype resembled that of the LRP6 morphants (22) as well as embryos that overexpressed Dkk1, a Wnt antagonist (21). TMEM198 morphants were specific because in X. tropicalis tmem198 mRNA-coinjected embryos, the defects in pigmentation, tail, and anterior structures were reduced to 11%, 9%, and 13%, respectively (Fig. 9B lower panel) (n = 45). Loss of pigmentation is an indication of defective neural crest formation, a process regulated by Wnt/β-catenin signaling (42, 43). We therefore investigated the role of tmem198 in neural crest development. Control- or TMEM198-MO, together with LacZ mRNA as a lineage tracer, was injected into one of the two blastomeres in two-cell-stage embryos, and at the early neurula stage, the expression of neural crest markers was examined using in situ hybridization. In the TMEM198-MO-injected side, expression levels of Sox10 (n = 46; 96%), Sox9 (n = 28; 50%), FoxD3a (n = 25; 60%), Slug (n = 20; 40%), and Twist (n = 26; 63%) were markedly reduced (Fig. 9C), confirming that TMEM198 is required for neural crest formation. In contrast, the pan-neural marker Sox3 was not affected (data not shown), suggesting that the deficient formation of neural crests was not due to the defective neural induction. The downregulation of neural crest marker expression by TMEM198-MO was specific because the expression of Sox10 was rescued in 63% of embryos coinjected with X. tropicalis tmem198 mRNA, which is refractory to MO targeting X. laevis tmem198 (n = 28) (Fig. 9D).
Fig. 9.
TMEM198 functions in neural crest development. (A) Western blot of total cell lysate from Xenopus embryos injected with indicated RNA or morpholino oligonucleotides (MO). Four nanograms of X. laevis TMEM198-EGFP mRNA (XlTMEM198-EGFP) and 40 ng of control or two different TMEM198 MOs (MO1 and MO2) were injected into two- to four-cell-stage embryos, and at stage 11, the embryos were lysed and analyzed with enhanced GFP (EGFP) and β-tubulin (as the loading control) antibodies. (B) Four-cell-stage embryos were injected animally with 40 ng of control morpholino oligonucleotide (Ctrl-MO), TMEM198-MO, or together with 100 pg of XtTMEM198 mRNA. (C and D) Two-cell-stage embryos were injected animally into one blastomere with 40 ng of control morpholino oligonucleotides (Ctrl-MO) or TMEM198-MO as indicated, together with 400 pg of LacZ RNA as a lineage tracer, and analyzed at stage 15 using in situ hybridization for the indicated genes. A total of 100 pg of XtTMEM198 mRNA was coinjected to rescue Sox10 expression in the experiment shown in panel D.
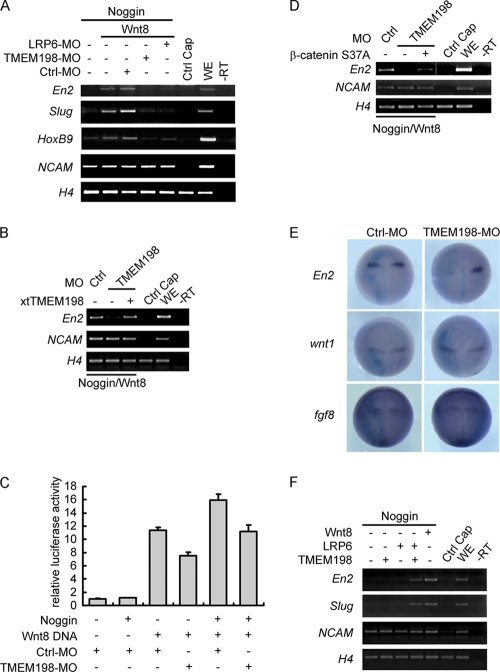
Wnt/β-catenin signaling is involved in antero-posterior (A-P) body axis determination of both vertebrates and invertebrates (18, 34, 35, 39). To uncover a role of TMEM198 in Wnt-mediated A-P patterning, we used Xenopus animal caps that were neuralized with injected Noggin and further posteriorized with coinjected Xenopus Wnt8. In this experiment, neuralized animal cap cells were transformed with Wnt/β-catenin signaling into posterior type neural tissues as indicated by the expression of engrailed 2 (En2), a midbrain-hindbrain-boundary (MHB) marker, and HoxB9, a spinal cord marker (Fig. 10A). The neural crest marker gene Slug was also induced, which confirmed Wnt signaling involvement during neural crest induction (Fig. 10A). As expected, depletion of Wnt coreceptor LRP6 using a specific MO abolished the induction of all three genes by Wnt. TMEM198-MO also blocked the activity of Wnt in inducing the expression of En2, HoxB9, and Slug, indicating that TMEM198 is required downstream of Wnt ligand in this experimental setting (Fig. 10A). Coinjection of X. tropicalis tmem198 mRNA rescued En2 expression, suggesting that the effect of TMEM198-MO was specific (Fig. 10B). To further confirm that TMEM198 acted through the β-catenin-dependent Wnt signaling, we monitored the expression of a Wnt-responsive reporter gene in these animal caps and found that the reporter was consistently downregulated by TMEM198-MO (Fig. 10C). Moreover, the blocking effect of TMEM198-MO on Wnt-induced En2 expression was reversed by coinjection of β-catenin S37A, a constitutively active form (Fig. 10D). These results indicate that TMEM198 is required for Wnt/β-catenin-mediated posteriorization of neural tissues.
Fig. 10.
TMEM198 is required for Wnt-mediated antero-posterior patterning in Xenopus embryos. (A and B) Four-cell-stage embryos were injected animally in each blastomere with 100 pg of noggin and 50 pg of Wnt8 mRNA or 10 ng of control-MO (Ctrl-MO), 5 ng of LRP6-MO, and 10 ng of TMEM198-MO as indicated. At stage 8 to 9, animal caps were dissected and cultured until equivalent to stage 15 for expression analysis of the indicated marker genes using RT-PCR. −RT, minus reverse transcription control; WE, whole embryo; H4, histone 4. In panel B, 200 pg of XtTMEM198 plasmid DNA was coinjected to rescue En2 expression. (C) Wnt reporter assay in Xenopus animal caps. Each blastomere of four-cell-stage embryos was injected animally with 100 pg of noggin mRNA, 75 pg of Wnt8 DNA, 10 ng of control-MO (Ctrl-MO), or 10 ng of TMEM198-MO as indicated. Twenty-five picograms of TOP-FLASH and 5 pg of pRL-TK plasmid DNAs were coinjected in all samples. At stage 8 to 9, animal caps were dissected, cultured until equivalent to stage 15, and then lysed for luciferase activity determination. (D) The animal cap assay was performed as described for panel B, except that 100 pg of β-catenin S37A (the constitutively active form of β-catenin) plasmid DNA was coinjected to rescue En2 expression. (E) One blastomere of two-cell-stage embryos was injected animally with 40 ng of control morpholino oligonucleotides (Ctrl-MO) or TMEM198-MO, as indicated, together with 400 pg of LacZ RNA as a lineage tracer and analyzed at stage 14 using in situ hybridization for the indicated genes. Note that En2 expression was downregulated (n = 42; 87%) by TMEM198-MO injection while that of wnt1 (n = 36; 100%) and fgf8 (n = 27; 100%) was not affected. (F) Each blastomere of four-cell-stage embryos was injected animally with 100 pg of noggin and 100 pg of tmem198 mRNA or 500 pg of LRP6 mRNA as indicated. At stage 8 to 9, animal caps were dissected, cultured until equivalent to stage 15, and analyzed for expression of the indicated marker genes using RT-PCR. −RT, minus reverse transcription control; WE, whole embryo; H4, histone 4. Fifty picograms of Wnt8 mRNA was coinjected with noggin as a positive control.
Using in situ hybridization in whole embryos, we next verified the expression of Otx2, Gbx2, En2, Krox20, and HoxB9, which are often used to demarcate different compartments of the central nervous system along the A-P axis. Surprisingly, none except En2 was consistently downregulated by TMEM198-MO injection, suggesting a rather specific role of TMEM198 in the A-P patterning of the central nervous system (Fig. 10E and data not shown). En2 has been demonstrated as a direct target of Wnt/β-catenin signaling (30), and its expression in the MHB was induced/maintained directly by Wnt1 (31) and further upstream by fibroblast growth factor 8 (FGF8) (24, 55). We observed that the expression of neither wnt1 nor fgf8 was altered in TMEM198-MO-injected embryos (Fig. 10E), suggesting that TMEM198 likely functions downstream of Wnt1. We also verified the expression of Otx2 and Gbx2, two transcription factors that antagonistically regulate formation of the MHB and wnt1 expression. Our results revealed that the expression levels of these two genes were also largely unaffected (data not shown), indicating that the specification of the MHB was not defective and suggesting again that TMEM198 was required more downstream for Wnt1 signaling. In line with this conclusion, we were able to demonstrate that in Noggin-neutralized animal caps, TMEM198 and LRP6 together could mimic the activity of Wnt8 to activate the expression of En2 and Slug (Fig. 10F). These results suggest that TMEM198 is required for Wnt-mediated induction of En2 expression in Xenopus embryos.
DISCUSSION
The present study identified the first cellular activity of the TMEM198 family proteins. Our results suggest that TMEM198 is a potent regulator of LRP6 phosphorylation and activation. These data also demonstrate that TMEM198 functions in Wnt/β-catenin signaling-mediated neural patterning of Xenopus embryos.
Function of TMEM198 during Xenopus embryogenesis.
Sequence analysis indicates that there are two tmem198 homologues in the mouse genome while we identified only one in the Xenopus genome. Both maternal and zygotic tmem198 transcripts were widely distributed in developing embryos. During gastrulation, stronger tmem198 expression was detected in the posterior end around the blastopore. Consistent with the results from mammalian cells, TMEM198 was able to activate canonical Wnt signaling together with LRP6 in Xenopus embryos (Fig. 8G and H). On the other hand, TMEM198-MO blocked Wnt-induced posteriorization in animal cap cells (Fig. 10A and B) and En2 expression (Fig. 10A, B, D, and E), an MHB marker gene and known direct target of Wnt/β-catenin signaling (30) in neurula embryos. These gain- and loss-of-function results together suggest that TMEM198 participates in the neural patterning of Xenopus embryos by affecting Wnt/β-catenin signaling.
TMEM198 morphants also had defects in neural crest formation, as indicated by the reduction of neural crest marker expression in gastrula embryos and less pigmented tail bud embryos (Fig. 9B to D). In addition, craniofacial defects were consistently observed when TMEM198-MO was injected unilaterally at the two-cell stage (data not shown). Neural crest cells are multipotent progenitors that are induced at the border of the neural plate and nonneural ectoderm (12, 29). Canonical Wnt signaling has been extensively documented to play important roles during neural crest induction (3, 42, 43), and LRP6, as a coreceptor, is also indispensable (22, 45). The expression of Sox9, Slug, and Sox10 is under the control of Wnt/β-catenin signaling (2, 6, 49), and the promoters of Slug and Sox9 contain T-cell factor (TCF)-responsive-elements (6, 49), which is indicative of direct regulation. Our results indicated that Xenopus tmem198 was essential for Slug expression in neurula embryos (Fig. 9C), was required for Slug induction by Wnt in animal caps (Fig. 10A), and was capable of promoting Slug expression together with LRP6 (Fig. 10F). These results suggest that TMEM198 is involved in Wnt-mediated neural crest induction during Xenopus embryogenesis and further support a role for TMEM198 in LRP6 activation.
In the cleavage embryos, maternal Wnt/β-catenin signaling determines the dorsal-ventral axis and formation of the organizer (19). Consistent with the role of TMEM198 in early Wnt signaling, tmem198 mRNA is maternally distributed. However, the organizer formation, as indicated by goosecoid (gsc) and chordin expression, was hardly affected in TMEM198 morphants (data not shown). This may be because of the functional redundancy of another TMEM198-like protein. Alternatively, maternal depletion is required to demonstrate a role of TMEM198 in pre-midblastula transition (pre-MBT) Wnt signaling, especially considering that LRP6 activation occurs very early following fertilization.
Regulation of LRP6 phosphorylation and activation during canonical Wnt signaling.
As a coreceptor, LRP6 is indispensable for Wnt/β-catenin signaling (23). LRP6 phosphorylation is regulated by the extracellular ligand and is required for transducing stimuli further down to the β-catenin degradation complex (1, 27, 37). The phosphorylated motifs provide docking sites for the Axin complex (17, 48, 58) and may, in addition, function as direct inhibitors of GSK3β, thus preventing β-catenin phosphorylation and degradation (15, 33, 40, 53). Although several kinases, including casein kinase 1γ, GSK3, Pftk1, and GRK5, have been implicated in this process (9, 16, 17, 37, 58), the phosphorylation-triggering mechanism is still unknown. Our work also implicates TMEM198, a previously functional unknown protein, in LRP6 phosphorylation and activation. We provide evidence showing the following: (i) that TMEM198 is able to promote LRP6 phosphorylation and activation, (ii) that TMEM198 is able to recruit and facilitate casein kinases for LRP6 phosphorylation, (iii) that TMEM198 is able to promote LRP6 aggregation, and (iv) that TMEM198 is required for Wnt signal transduction and CK1-mediated LRP6 phosphorylation. These results support the hypothesis that TMEM198 functions as a membrane scaffold protein assembling kinases and substrates into a higher-molecular-weight complex, thereby facilitating the phosphorylation event.
We believe that TMEM198-promoted LRP6 activation is specific. First, LRP6 phosphorylation is a tightly regulated event not triggered by coexpression with other seven-transmembrane proteins like Fz (data not shown) or TMEM198-M2 (Fig. 3E). Second, TMEM198 selectively promotes the phosphorylation and activation of LRP6 but not LRP5 (Fig. 1D and 3G). Third, TMEM198-promoted LRP6 aggregation is not an overexpression artifact because this did not occur with TGFβRI (Fig. 6A), another single-transmembrane protein. Our results suggest that at least a part of TMEM198 activity is to recruit casein kinases through its cytoplasmic domain. Supporting this conclusion, the C domain is indeed required for CK1 binding and full activity (Fig. 5A and B). The lack of CK1 binding and LRP6 activation of TMEM198-M2 suggests that the third intracellular loop (where the mutations reside) may have regulative roles affecting conformation or modification of the TMEM198 protein.
Previous reports proposed that Dvl functions in assembling LRP6 signalosomes (5, 32, 44). Upon Wnt stimulation, Frizzled recruits Dvl, and primarily phosphorylated LRP6 in turn recruits the Axin complex. When Frizzled and LRP6 are brought together by Wnt, Dvl assembles oligomeric receptor-ligand complexes via an interaction with Axin. Our results indicate that TMEM198 also promotes the formation of active LRP6 aggregations but through a different mechanism. TMEM198 associates with LRP6 but exhibits no direct interaction with Dvl2 or Axin (data not shown). Therefore, TMEM198 is probably another assembling factor functioning independently or downstream of Dvl. Corroborating this conclusion, TMEM198 is required for the spontaneous aggregation and signaling of LRP6ΔE1-4 (Fig. 6D and E), which occurs without Dvl involvement (5).
We prefer to speculate that TMEM198 functions as a tissue-specific modulator rather than as a core component of the canonical Wnt signaling pathway. TMEM198 cooperated specifically with LRP6 but not LRP5 (Fig. 1D and E), which is also essential for Wnt signaling. Moreover, knockdown of TMEM198 in HEK293T cells did not significantly reduce Wnt3a-induced LRP6 phosphorylation (at least at Ser-1490 and Thr-1493 sites) (data not shown). However, TMEM198 was required for CK1-induced LRP6 phosphorylation when both the kinase and receptor were overexpressed (Fig. 4F and G). Our results did not rule out the possibility that TMEM198 may also be implicated in other signaling pathways. Indeed, microinjecting higher doses of tmem198 mRNA into Xenopus embryos inhibited Xbra expression and proper gastrulation (data not shown). This could explain why TMEM198/LRP6 did not induce the secondary body axis (data not shown). Inhibition of Xbra expression might be because of its activity on TGFβ or FGF signals. However, our loss-of-function studies did not support a role of TMEM198 in either of these pathways. Identification of TMEM198 as a potent modulator of Wnt/β-catenin signaling raises intriguing questions about its role in other developmental processes as well as in human diseases, especially considering the broad expression of human TMEM198 detected using expressed sequence tag (EST) analysis in the UniGene database.
ACKNOWLEDGMENTS
We thank Randy Moon, Dianqing Wu, and Ye-Guang Chen for reagents and Roel Nusse, Xi He, and Weijun Pan for helpful discussions and suggestions.
This work was supported by grants to W.W. from the National Natural Science Foundation of China (30730048 and 30921004), Major Science Programs of China (2006CB943402, 2011CB943803), and Tsinghua University Initiative Scientific Research Program (2010THZ0). Work in the laboratories of M.B. and C.N. was supported by the FOR1036 research group of the Deutsche Forschungsgemeinschaft.
Footnotes
Published ahead of print on 2 May 2011.
REFERENCES
- 1. Angers S., Moon R. T. 2009. Proximal events in Wnt signal transduction. Nat. Rev. Mol. Cell Biol. 10:468–477 [DOI] [PubMed] [Google Scholar]
- 2. Aoki Y., et al. 2003. Sox10 regulates the development of neural crest-derived melanocytes in Xenopus. Dev. Biol. 259:19–33 [DOI] [PubMed] [Google Scholar]
- 3. Bastidas F., De Calisto J., Mayor R. 2004. Identification of neural crest competence territory: role of Wnt signaling. Dev. Dyn. 229:109–117 [DOI] [PubMed] [Google Scholar]
- 4. Bhanot P., et al. 1996. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature 382:225–230 [DOI] [PubMed] [Google Scholar]
- 5. Bilic J., et al. 2007. Wnt induces LRP6 signalosomes and promotes Dishevelled-dependent LRP6 phosphorylation. Science 316:1619–1622 [DOI] [PubMed] [Google Scholar]
- 6. Blache P., et al. 2004. SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. J. Cell Biol. 166:37–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buechling T., et al. 2010. Wnt/Frizzled signaling requires dPRR, the Drosophila homolog of the prorenin receptor. Curr. Biol. 20:1263–1268 [DOI] [PubMed] [Google Scholar]
- 8. Cadigan K. M., Peifer M. 2009. Wnt signaling from development to disease: insights from model systems. Cold Spring Harb. Perspect. Biol. 1:a002881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen M., et al. 2009. G protein-coupled receptor kinases phosphorylate LRP6 in the Wnt pathway. J. Biol. Chem. 284:35040–35048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chien A. J., Conrad W. H., Moon R. T. 2009. A Wnt survival guide: from flies to human disease. J. Invest. Dermatol. 129:1614–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Christian J. L., Moon R. T. 1993. Interactions between Xwnt-8 and Spemann organizer signaling pathways generate dorsoventral pattern in the embryonic mesoderm of Xenopus. Genes Dev. 7:13–28 [DOI] [PubMed] [Google Scholar]
- 12. Christiansen J. H., Coles E. G., Wilkinson D. G. 2000. Molecular control of neural crest formation, migration and differentiation. Curr. Opin. Cell Biol. 12:719–724 [DOI] [PubMed] [Google Scholar]
- 13. Clevers H. 2006. Wnt/beta-catenin signaling in development and disease. Cell 127:469–480 [DOI] [PubMed] [Google Scholar]
- 14. Cruciat C. M., et al. 2010. Requirement of prorenin receptor and vacuolar H+-ATPase-mediated acidification for Wnt signaling. Science 327:459–463 [DOI] [PubMed] [Google Scholar]
- 15. Cselenyi C. S., et al. 2008. LRP6 transduces a canonical Wnt signal independently of Axin degradation by inhibiting GSK3's phosphorylation of beta-catenin. Proc. Natl. Acad. Sci. U. S. A. 105:8032–8037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Davidson G., et al. 2009. Cell cycle control of Wnt receptor activation. Dev. Cell 17:788–799 [DOI] [PubMed] [Google Scholar]
- 17. Davidson G., et al. 2005. Casein kinase 1 gamma couples Wnt receptor activation to cytoplasmic signal transduction. Nature 438:867–872 [DOI] [PubMed] [Google Scholar]
- 18. De Robertis E. M. 2010. Wnt signaling in axial patterning and regeneration: lessons from planaria. Sci. Signal. 3:pe21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. De Robertis E. M., Kuroda H. 2004. Dorsal-ventral patterning and neural induction in Xenopus embryos. Annu. Rev. Cell Dev. Biol. 20:285–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ding Y., et al. 2008. Caprin-2 enhances canonical Wnt signaling through regulating LRP5/6 phosphorylation. J. Cell Biol. 182:865–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Glinka A., et al. 1998. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature 391:357–362 [DOI] [PubMed] [Google Scholar]
- 22. Hassler C., et al. 2007. Kremen is required for neural crest induction in Xenopus and promotes LRP6-mediated Wnt signaling. Development 134:4255–4263 [DOI] [PubMed] [Google Scholar]
- 23. He X., Semenov M., Tamai K., Zeng X. 2004. LDL receptor-related proteins 5 and 6 in Wnt/beta-catenin signaling: arrows point the way. Development 131:1663–1677 [DOI] [PubMed] [Google Scholar]
- 24. Joyner A. L., Liu A., Millet S. 2000. Otx2, Gbx2 and Fgf8 interact to position and maintain a mid-hindbrain organizer. Curr. Opin. Cell Biol. 12:736–741 [DOI] [PubMed] [Google Scholar]
- 25. Kazanskaya O., et al. 2004. R-Spondin2 is a secreted activator of Wnt/beta-catenin signaling and is required for Xenopus myogenesis. Dev. Cell 7:525–534 [DOI] [PubMed] [Google Scholar]
- 26. Klaus A., Birchmeier W. 2008. Wnt signalling and its impact on development and cancer. Nat. Rev. Cancer 8:387–398 [DOI] [PubMed] [Google Scholar]
- 27. MacDonald B. T., Tamai K., He X. 2009. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell 17:9–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mao B., et al. 2002. Kremen proteins are Dickkopf receptors that regulate Wnt/beta-catenin signalling. Nature 417:664–667 [DOI] [PubMed] [Google Scholar]
- 29. Mayor R., Aybar M. J. 2001. Induction and development of neural crest in Xenopus laevis. Cell Tissue Res. 305:203–209 [DOI] [PubMed] [Google Scholar]
- 30. McGrew L. L., Takemaru K., Bates R., Moon R. T. 1999. Direct regulation of the Xenopus engrailed-2 promoter by the Wnt signaling pathway, and a molecular screen for Wnt-responsive genes, confirm a role for Wnt signaling during neural patterning in Xenopus. Mech. Dev. 87:21–32 [DOI] [PubMed] [Google Scholar]
- 31. McMahon A. P., Joyner A. L., Bradley A., McMahon J. A. 1992. The midbrain-hindbrain phenotype of Wnt-1−/Wnt-1− mice results from stepwise deletion of engrailed-expressing cells by 9.5 days postcoitum. Cell 69:581–595 [DOI] [PubMed] [Google Scholar]
- 32. Metcalfe C., Mendoza-Topaz C., Mieszczanek J., Bienz M. 2010. Stability elements in the LRP6 cytoplasmic tail confer efficient signalling upon DIX-dependent polymerization. J. Cell Sci. 123:1588–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mi K., Dolan P. J., Johnson G. V. 2006. The low density lipoprotein receptor-related protein 6 interacts with glycogen synthase kinase 3 and attenuates activity. J. Biol. Chem. 281:4787–4794 [DOI] [PubMed] [Google Scholar]
- 34. Niehrs C. 2010. On growth and form: a Cartesian coordinate system of Wnt and BMP signaling specifies bilaterian body axes. Development 137:845–857 [DOI] [PubMed] [Google Scholar]
- 35. Niehrs C. 2004. Regionally specific induction by the Spemann-Mangold organizer. Nat. Rev. Genet. 5:425–434 [DOI] [PubMed] [Google Scholar]
- 36. Niehrs C., Boutros M. 2010. Trafficking, acidification, and growth factor signaling. Sci. Signal. 3:pe26. [DOI] [PubMed] [Google Scholar]
- 37. Niehrs C., Shen J. 2010. Regulation of Lrp6 phosphorylation. Cell. Mol. Life Sci. 67:2551–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pan W., et al. 2008. Wnt3a-mediated formation of phosphatidylinositol 4,5-bisphosphate regulates LRP6 phosphorylation. Science 321:1350–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Petersen C. P., Reddien P. W. 2009. Wnt signaling and the polarity of the primary body axis. Cell 139:1056–1068 [DOI] [PubMed] [Google Scholar]
- 40. Piao S., et al. 2008. Direct inhibition of GSK3β by the phosphorylated cytoplasmic domain of LRP6 in Wnt/beta-catenin signaling. PloS One 3:e4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pinson K. I., Brennan J., Monkley S., Avery B. J., Skarnes W. C. 2000. An LDL-receptor-related protein mediates Wnt signalling in mice. Nature 407:535–538 [DOI] [PubMed] [Google Scholar]
- 42. Raible D. W. 2006. Development of the neural crest: achieving specificity in regulatory pathways. Curr. Opin. Cell Biol. 18:698–703 [DOI] [PubMed] [Google Scholar]
- 43. Sauka-Spengler T., Bronner-Fraser M. 2008. Evolution of the neural crest viewed from a gene regulatory perspective. Genesis 46:673–682 [DOI] [PubMed] [Google Scholar]
- 44. Schwarz-Romond T., Metcalfe C., Bienz M. 2007. Dynamic recruitment of axin by Dishevelled protein assemblies. J. Cell Sci. 120:2402–2412 [DOI] [PubMed] [Google Scholar]
- 45. Song L., Li Y., Wang K., Zhou C. J. 2010. Cardiac neural crest and outflow tract defects in Lrp6 mutant mice. Dev. Dyn. 239:200–210 [DOI] [PubMed] [Google Scholar]
- 46. Taelman V. F., et al. 2010. Wnt signaling requires sequestration of glycogen synthase kinase 3 inside multivesicular endosomes. Cell 143:1136–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tamai K., et al. 2000. LDL-receptor-related proteins in Wnt signal transduction. Nature 407:530–535 [DOI] [PubMed] [Google Scholar]
- 48. Tamai K., et al. 2004. A mechanism for Wnt coreceptor activation. Mol. Cell 13:149–156 [DOI] [PubMed] [Google Scholar]
- 49. Vallin J., et al. 2001. Cloning and characterization of three Xenopus slug promoters reveal direct regulation by Lef/beta-catenin signaling. J. Biol. Chem. 276:30350–30358 [DOI] [PubMed] [Google Scholar]
- 50. van Amerongen R., Nusse R. 2009. Towards an integrated view of Wnt signaling in development. Development 136:3205–3214 [DOI] [PubMed] [Google Scholar]
- 51. Wang Y., et al. 2010. Xenopus skip modulates Wnt/beta-catenin signaling and functions in neural crest induction. J. Biol. Chem. 285:10890–10901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wehrli M., et al. 2000. Arrow encodes an LDL-receptor-related protein essential for Wingless signalling. Nature 407:527–530 [DOI] [PubMed] [Google Scholar]
- 53. Wu G., Huang H., Garcia Abreu J., He X. 2009. Inhibition of GSK3 phosphorylation of beta-catenin via phosphorylated PPPSPXS motifs of Wnt coreceptor LRP6. PloS One 4:e4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yang-Snyder J., Miller J. R., Brown J. D., Lai C. J., Moon R. T. 1996. A Frizzled homolog functions in a vertebrate Wnt signaling pathway. Curr. Biol. 6:1302–1306 [DOI] [PubMed] [Google Scholar]
- 55. Ye W., et al. 2001. Distinct regulators control the expression of the mid-hindbrain organizer signal FGF8. Nat. Neurosci. 4:1175–1181 [DOI] [PubMed] [Google Scholar]
- 56. Yum S., et al. 2009. The role of the Ser/Thr cluster in the phosphorylation of PPPSP motifs in Wnt coreceptors. Biochem. Biophys. Res. Commun. 381:345–349 [DOI] [PubMed] [Google Scholar]
- 57. Zeng X., et al. 2008. Initiation of Wnt signaling: control of Wnt coreceptor Lrp6 phosphorylation/activation via frizzled, dishevelled and axin functions. Development 135:367–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zeng X., et al. 2005. A dual-kinase mechanism for Wnt coreceptor phosphorylation and activation. Nature 438:873–877 [DOI] [PMC free article] [PubMed] [Google Scholar]