Abstract
CPEB is a sequence-specific translational regulatory RNA binding protein that mediates cellular senescence in primary mouse and human cells. CPEB knockout mouse embryo fibroblasts (MEFs) bypass senescence and synthesize large amounts of interleukin-6 (IL-6) and many other cytokines, which is not the case with either wild-type MEFs immortalized by prolonged culture or p53-deficient MEFs. CPEB regulates the production of IL-6 at both the translational and transcriptional levels; in CPEB-depleted cells, aberrant IL-6 transcription is mediated by improper NF-κB p65 phosphorylation and nuclear localization. Although IL-6 strengthens the senescence of wild-type cells, it has no effect on CPEB-deficient cells, even though they produce prodigious amounts of the cytokine. IL-6-promoted entry into senescence requires p53; CPEB knockout MEFs, however, synthesize only ∼50% of the p53 of wild-type MEFs, which is insufficient to respond to IL-6. Thus, CPEB deficiency not only increases IL-6 production but also renders the cell incapable of a senescence-promoting response.
INTRODUCTION
Senescence is a stress-induced cell cycle arrest that limits the proliferation of mitotic cells grown in culture (21) and suppresses tumor progression in vivo (5, 12). The p53 and Rb pathways mediate the senescence response; p53 induces the expression of p21, an inhibitor of cyclin-dependent kinase (CDK), while Rb induces the expression of p16, another CDK inhibitor that prevents Rb phosphorylation and consequent inactivation. Rb inhibits cell proliferation by suppressing the expression of genes required for cell cycle progression by the transcription factor E2F (4, 7). E2F can also activate ARF, which elevates p53 levels by repressing the activity of HDM2, an E3 ubiquitin-protein ligase (7). The degree of activation of either or both pathways varies, depending on the cellular stress that induces senescence, the cell type, and the species.
Recently, a connection between senescence and cytokines has been elucidated. During oncogene-induced senescence in human cells, increased levels of interleukin-6 (IL-6) and IL-8 help to reinforce the senescence response (1, 25). In vivo, these cytokines help promote the senescence of premalignant cells, leading to their growth arrest. In addition, they recruit inflammatory immune cells that may help to clear premalignant cells and prevent tumor formation (1, 7, 25). Therefore, the balance of senescence and immune clearance may be critical for slowing malignant transformation of preneoplastic lesions (34).
CPEB is a sequence-specific RNA binding protein that regulates the translation of mRNAs important for development, neuronal synaptic plasticity, and senescence (6, 9, 18, 19, 33, 35). It binds the cytoplasmic polyadenylation element (CPE; UUUUUAU) in the 3′ untranslated regions (UTRs) of mRNAs; in concert with several other factors, it controls poly(A) tail length and translation (3, 8, 23, 24, 30, 31). MEFs (mouse embryo fibroblasts) derived from CPEB knockout (KO) animals bypass senescence and are immortal, at least partially due to an overexpression of myc mRNA (18). Human primary cells that are deficient in CPEB also bypass senescence, but in this case, it is caused by reduced polyadenylation and translation of p53 mRNA (6). The myc and p53 mRNAs contain CPEs in their 3′ UTRs and are direct targets of CPEB regulation. We noticed that the 3′ UTRs of IL-6 and several other cytokine mRNAs that are involved in senescence also contain conserved CPEs, and thus, we considered that CPEB might modulate entry into senescence by controlling cytokine mRNA translation. We find that IL-6 reinforces senescence in murine cells and that its mRNA is regulated at the translational level by CPEB. However, IL-6 production in CPEB KO cells far exceeds that which can be accounted for by translational control. The vast majority of IL-6 expression in CPEB-deficient cells occurs at the level of transcription and is mediated by prolonged NF-κB nuclear localization by improper S276 phosphorylation. Although CPEB KO MEFs uniquely secrete large amounts of IL-6, senescence is not reinforced because robust p53 levels are compromised. Thus, a regulatory axis consisting of CPEB, IL-6, NF-κB, and p53 is essential for cellular senescence.
MATERIALS AND METHODS
Reagents and cell culture.
The mouse IL-6 3′ UTR was amplified by reverse transcription (RT)-PCR and cloned into the XbaI and BamHI sites of the pRL-TK vector (Promega), replacing the simian virus 40 late poly(A) signal. NF-κB promoter luciferase assays used the vectors pGL4.32[luc2P/NFKB-RE/Hygrgo] and pRL-TK (for normalization) (Promega). WT and CPEB KO MEFs were derived as described previously (18). Cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and penicillin-streptomycin according to a 3T3 protocol. p53 heterozygous and null MEFs were cultured in the same manner. Some cells were infected with retrovirus expressing FLAG alone or FLAG-CPEB; this was followed by puromycin selection for 3 to 5 days. Where indicated, the cells were transfected with equal amounts of DNA plasmids using Lipofectamine (Invitrogen) with Opti-MEM medium. Cells were also treated with 1 μg/ml neutralizing IL-6 antibody (clone MP5-20F3; eBioscience) or 1 ng/ml recombinant IL-6 (eBioscience) at each passage. Neutralization of IL-6 activity was confirmed by T1165 growth assays (29).
Subcellular fractionation and binding assays.
Cells were washed with phosphate-buffered saline (PBS) and resuspended in buffer (20 mM Tris, pH 7.4, 100 mM KCl, 20 mM β-glycerol phosphate, 1 mM dithiothreitol [DTT], 0.25 mM Na3VO4, 10 nM NaF, 1 mM EDTA, 1 mM EGTA, 10 nM okadaic acid, 1 mM phenylmethylsulfonyl fluoride, 1% NP-40). Some of the cells were treated with Benzonase (5 U) and SDS (0.1% final concentration) and stored as the whole-cell lysate. Nuclei were pelleted from other cells by centrifugation at 10,000 × g for 10 min at 4°C. For the NF-κB binding assay, nuclear lysates were prepared by cell fractionation as described in the manufacturer's instructions (TransFactor extraction kit; Clontech). Briefly, cells were washed with PBS, resuspended in buffer 1 (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 1 mM DTT, and EDTA-free protease inhibitor cocktail [Roche]), and left on ice for 15 min. Cells were pelleted, resuspended in fresh buffer 1, and lysed using a 28-gauge syringe. The preparation was centrifuged at 10,000 × g, and the pellet was resuspended in buffer 2 (20 mM HEPES [pH 7.9], 1.5 mM MgCl2, 0.42 M NaCl, 0.2 mM EDTA, 25% glycerol, 1 mM DTT, and EDTA-free protease inhibitor cocktail [Roche]) and lysed with a 28-gauge needle. The suspension was agitated for 30 min at 4°C and pelleted at 20,000 × g. The supernatant was taken as the nuclear extract, and the total protein was measured by Bradford analysis. SDS-PAGE and Western blotting were done according to standard protocols. The antibodies used were as follows: p53 Ab1 and Ab3 at a 1:1 ratio from Calbiochem; Flag and tubulin from Sigma; p105/50 from eBioscience; p65 from Santa Cruz Biotechnology; P65-Pi, S6, and IKBα from Cell Signaling.
NF-κB analysis.
Cells were incubated with the NF-κB inhibitor JSH-23 (5 μM; EMD) solubilized in dimethyl sulfoxide for the times indicated. The cells were then washed and incubated with fresh medium without JSH-23. For the NF-κB binding assay, NeutrAvidin-coated 96-well plates (Thermo Scientific) were primed with 100 μl 33 nM biotinylated, double-stranded oligonucleotides containing the NF-κB binding consensus or control sequence. After washing and blocking (1× assay diluent; eBioscience), equal total protein amounts of nuclear lysate diluted in 4 mM HEPES (pH 7.5)-100 mM KCl-8% glycerol-5 mM DTT-0.2% bovine serum albumin were added for 1 h of incubation at room temperature. The plates were washed, and a 1:500 dilution of NF-κB p65 antibody in blocking buffer was added to each well, followed by washing and addition of a horseradish peroxidase-conjugated secondary antibody. After washing, 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) was added for color development and quantification at 405 nm.
Statistical analysis.
Statistically significant differences between two groups of data were determined with a two-tailed unpaired Student t test.
Cytokine array.
Conditioned media from equivalent numbers of WT and CPEB KO MEFs at early (passage 2 [P2]), senescent (P10), and late (P20) passages were added to cytokine arrays (each antibody was spotted in duplicate) from Raybiotech and processed according to the manufacturer's recommendations.
Microscopy.
Cells grown on coverslips in 30-mm dishes were fixed with 3% formaldehyde-PBS, permeabilized with 0.3% Triton X-100-PBS, and blocked with 10% fetal bovine serum-PBS. Primary antibody and secondary antibody conjugated with Alexa Fluor 594 (Molecular Probe) in blocking buffer were added sequentially. Coverslips were mounted in ProLong Gold with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen) and imaged using a Nikon Eclipse fluorescence microscope.
Senescence-associated β-galactosidase activity was determined at pH 6.0 as described previously (18).
RT-PCR.
RNA from 100-mm plates of each cell type was extracted with Trizol (Invitrogen). Equal total RNA amounts were used in RT reactions using an oligo(dT) primer and Superscript II (Invitrogen); equivalent amounts of the RT reaction products were used for PCRs.
Luciferase assays.
Luciferase detection was performed using a Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions on a Tecan system luminometer.
ELISA.
Equivalent numbers of cells were plated, and 3 days later, equivalent portions of media were assayed for IL-6 levels by enzyme-linked immunosorbent assay (ELISA; eBioscience). Where appropriate, cells corresponding to the medium sample were also counted on the day of harvest.
Primers.
The primers used for IL-6 were as follows: forward, TTGCCTTCTTGGGACTGATG; reverse, CTGAAGGACTCTGGCTTTGT. Those used for the IL-6 intron were as follows: forward, TCTTGTTCCAGCAGGGTCTT; reverse, GTAAGGTCCAGAGGTCAGC. Those used for KC were as follows: forward, CTGGGATTCACCTCAAGAACATC; reverse, CAGGGTCAAGGCAAGCCTC. Those used for the KC intron were as follows: forward, TCAGGAGGTCGGAAAGTTGT; reverse, GGGAAATCTCACTGGCAAAA. Those used for firefly luciferase were as follows: forward, GTCGCTCTGCCTCATAGAACTGCCTG; reverse, TCAGAGTGCTTTTGGCGAAGAAGG. Those used for Renilla luciferase were as follows: forward, GGAGAATAACTTCTTCGTGGAAACCA; reverse, GAGAACTCGCTCAACGAACGATTTG. Those used for glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were as follows: forward, AACTTTGGCATTGTGGAAGG; reverse, GGAGACAACCTGGTCCTCAG. Those used for NF-κB binding assays were as follows: BT-WT:/5biosg/, AGTTGAGGGGACTTTCCCAGG; WT reverse, GCCTGGGAAAGTCCCCTCAACT; BT MUT:/5BIOSG/, AGTTGAGGCAACGGTCCCAGG; MUT reverse, GCCTGGGACCGTTGCCTCAACT; COMP, AGTTGAGGGGACTTTCCCAGGC.
RESULTS
CPEB controls IL-6 production.
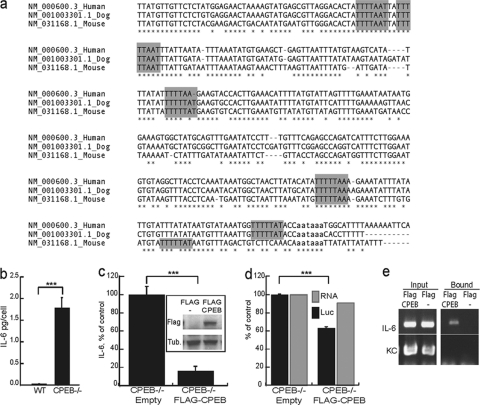
The 3′ UTRs of IL-6 (Fig. 1a) and several other cytokine mRNAs that are involved in senescence contain conserved CPEs, suggesting that CPEB might modulate entry into senescence by controlling cytokine mRNA translation. Indeed, examination of IL-6 production demonstrates that in CPEB KO MEFs, it was elevated by 1.7-fold compared to that in WT MEFs (Fig. 1b). Moreover, ectopic expression of CPEB in the KO MEFs reduced IL-6 levels by ∼80% (Fig. 1c). In addition, ectopic CPEB reduced the translation of a CPE-containing reporter RNA without affecting transcript levels (Fig. 1d). These results, coupled with the observation that FLAG-CPEB coimmunoprecipitated IL-6 mRNA but not KC (IL-8) mRNA (Fig. 1e), indicate not only the specificity of CPEB interaction with mRNA but also that CPEB inhibits IL-6 mRNA translation.
Fig. 1.
CPEB regulation of IL-6 mRNA translation. (a) Sequence comparison of a portion of the IL-6 3′ UTR in humans, dogs, and mice. The putative CPEs are highlighted in gray, and the polyadenylation hexanucleotide is in lowercase. (b) ELISA determination of levels of IL-6 secreted into the medium of cultured WT and CPEB KO immortalized MEFs. (c) IL-6 levels secreted by CPEB KO MEFs transduced with virus expressing FLAG-CPEB or FLAG only. The Western blot shows FLAG-CPEB expression. (d) CPEB KO MEFs were cotransfected with FLAG-CPEB or the empty vector together with a Renilla luciferase reporter plasmid containing the 3′ UTR of mouse IL-6 and a firefly luciferase (Luc) vector as a normalization control. The black bar refers to the ratio of Renilla to firefly luciferase expression as a percentage of the control, and the gray bar indicates the ratio of Renilla to firefly luciferase RNA as measured by quasiquantitative RT-PCR. (e) CPEB KO MEFs transduced with FLAG-CPEB- or FLAG-expressing virus were used for RNP coimmunoprecipitation with FLAG-tagged antibody. RNA was extracted from the precipitate and examined for IL-6 and KC RNAs by RT-PCR. Asterisks indicate P values of <0.001.
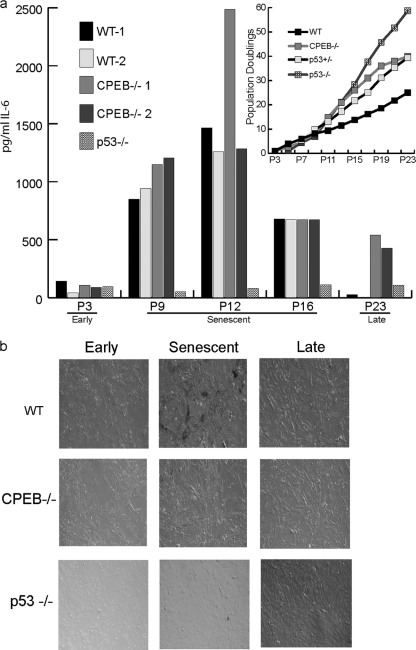
To further investigate the relationship between CPEB control of IL-6 production and senescence, IL-6 was measured in the culture medium of 2 different WT and CPEB KO MEF lines (Fig. 2a). Early-passage MEFs of both genotypes produced very low levels of IL-6, which increased as the cells approached the time when senescence would normally occur (P9 to P12). At P16, IL-6 production was reduced in all cells and by very late passage (>P16), the IL-6 production in WT cells had fallen to nearly undetectable levels. However, IL-6 continued to be synthesized by the KO MEFs up to at least P29 (Fig. 2a, cf. inset denoting population doublings). Cellular senescence was confirmed by staining the cells for β-galactosidase activity at acid pH (Fig. 2b) (13).
Fig. 2.
Immortalized CPEB KO MEFs produce high levels of IL-6. (a) MEFs derived from WT, CPEB KO (all from different founders), or p53 KO mice were cultured according to a 3T3 protocol; the medium was harvested on the third day prior to splitting, and the IL-6 levels were determined by ELISA. The inset shows the growth curves of the 4 genotypes. (b) Acidic β-galactosidase staining of the indicated cell types at the early (presenescent), senescent, and late stages.
Because p53 is a critical regulator of cellular senescence (4), we also measured IL-6 production in MEFs lacking this tumor suppressor. Interestingly, p53 KO MEFs never expressed high levels of IL-6, even at very late passages (Fig. 2a), indicating that the elevated levels of IL-6 in immortal CPEB KO MEFs was specifically due to a lack of CPEB and was not a general characteristic of immortalization.
Cytokine expression in CPEB knockout MEFs.
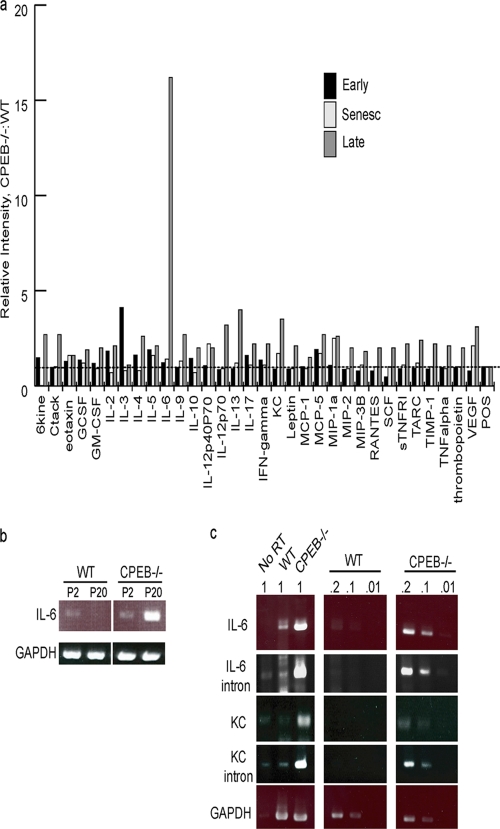
Many cytokine mRNAs contain CPEs in their 3′ UTRs, and we thus surmised that several might be regulated by CPEB. Consequently, the levels of cytokines produced by presenescent, senescent, and immortalized WT and CPEB KO MEFs were monitored by spotting culture medium from an equivalent number of cells onto membranes containing a variety of cytokine antibodies (Fig. 3a). The intensities of the spots were quantified, and the amounts were plotted as a ratio of CPEB KO to WT values. With the exception of IL-3, which was ∼4-fold higher in CPEB KO cells, there was little difference in any cytokine between the two cell types during the early and senescent stages. However, following immortalization, the levels of several cytokines were elevated in the KO MEFs, particularly IL-6, which was increased by ∼15-fold. IL-13, IL-12, KC, and vascular endothelial growth factor (VEGF) were all increased by at least 3-fold in the CPEB KO MEFs; several other cytokines were increased more modestly. Because the expression of several CPE-lacking (e.g., Ctack, Il-13), as well as CPE-containing (e.g., KC, VEGF), RNAs was elevated in the CPEB KO MEFs, we surmised that the broad changes in cytokine production probably did not occur by CPEB-regulated translation but possibly occurred through a more general alteration of transcription.
Fig. 3.
Cytokine production in WT and CPEB KO MEFs. (a) Cytokine levels produced by immortalized CPEB KO MEFs. A mouse cytokine antibody array was used to measure the levels of the indicated cytokines produced by WT and CPEB KO MEFs at the early (presenescent), senescent, and late stages. The values indicate the ratios of CPEB KO to WT levels. (b) Determination of relative IL-6 RNA levels from early (P2)- or late (P20)-passage immortalized MEFs by RT-PCR. GAPDH served as a control. (c) RNA from WT or CPEB KO immortalized MEFs was used for quasiquantitative RT-PCR analysis using intron- or exon-specific primers for IL-6 and KC RNAs. GAPDH served as a control. The values above the panels indicate the degrees of dilution of the starting RNA.
Quasiquantitative RT-PCR analysis of RNA derived from WT and KO MEFs shows that although there were similar but very low levels of IL-6 RNA in early-passage cells of both genotypes, by P20, only the KO MEFs produced large amounts of IL-6 RNA (Fig. 3b). Further analysis of IL-6 and KC (IL-8) late-passage CPEB KO MEF RNAs using intron-specific (reflecting nuclear pre-mRNA) and exon-specific (reflecting mostly cytoplasmic RNA) (24) primers for quasiquantitative RT-PCR demonstrated that the nuclear pre-mRNA and cytoplasmic mRNA levels were elevated proportionately in KO and WT MEFs, indicating that the relatively large amount of IL-6 and IL-8 RNAs in KO MEFs was due to transcriptional enhancement and not increased RNA stability (Fig. 3c). These results show that cytokine RNA synthesis is enhanced in immortalized KO MEFs and suggest that CPEB modulates the activity or level of a factor(s) that promotes the transcription of these genes.
Aberrant NF-κB nuclear localization and phosphorylation in CPEB knockout MEFs.
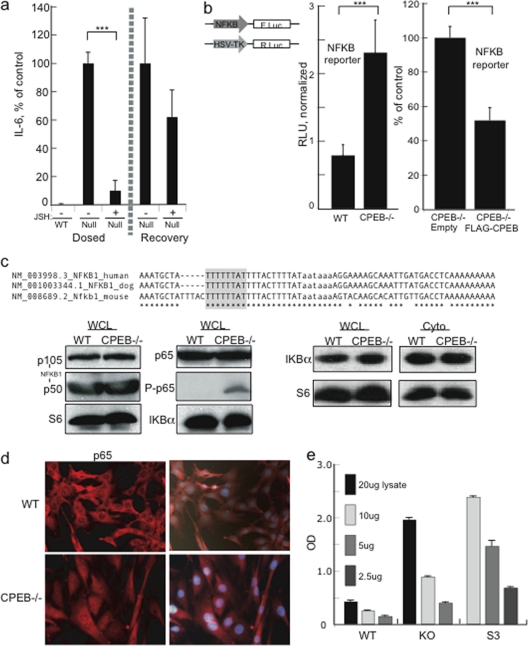
Two factors that both activate IL-6 transcription and mediate senescence are C/EBP and NF-κB (1, 25). Of the RNAs encoding these proteins, only NF-κB1 mRNA contains a conserved 3′ UTR CPE (Fig. 4c). To investigate whether NF-κB was at least partly responsible for the increased IL-6 transcription seen in immortalized CPEB KO MEFs, these and WT cells were incubated with the NF-κB inhibitor JSH-23 (22); ELISA was then used to measure IL-6 in the culture medium (Fig. 4a). JSH-23 reduced IL-6 production to 10% of that of untreated cells at 48 h; washout of the inhibitor resulted in the recovery of normal IL-6 levels.
Fig. 4.
NF-κB p65 is constitutively active in CPEB KO MEFs. (a) WT and CPEB KO MEFs were incubated with the NF-κB inhibitor JSH-23 or the vehicle alone for 48 h. After 48 h, the cells were washed and cultured in fresh medium without JSH-23. Samples of the medium were taken at 48 h, and IL-6 was determined by ELISA. (b) MEFs were transfected with a firefly NF-κB-promoter reporter and a Renilla luciferase reporter for normalization into WT, CPEB KO, or CPEB KO cells transduced with FLAG-CPEB or the empty vector. Five days later, the luciferase expression was determined. (c) Alignment of a portion of the 3′ UTR of human, dog, and mouse NF-κB1. The putative CPE is highlighted, and the polyadenylation hexanucleotide is in lowercase. WCL, whole-cell lysate; Cyto, cytoplasm. The Western blots show the relative levels of various NF-κB family members, as well as the regulatory protein IKBα. P-p65 refers to p65 phosphorylated on residue 276. S6 served as a loading control. (d) Localization of p65 in WT and CPEB KO MEFs determined by immunocytochemistry analysis. The cells were also stained with DAPI and merged. (e) NF-κB p65 transcription factor binding assay. The indicated amounts of nuclear lysate from WT, CPEB KO, and HeLa S3 cells were mixed with a biotinylated oligonucleotide containing the NF-κB consensus binding site. Antibody against p65 was then used to determine the amount of this protein captured by streptavidin. Asterisks indicate P values of <0.001. OD, optical density.
To further assess NF-κB activity, a firefly luciferase reporter gene under the control of an NF-κB-dependent promoter was transfected into WT or CPEB KO MEFs together with a Renilla luciferase gene that was used for normalization. Activation of the NF-κB-sensitive reporter was elevated by ∼3-fold in CPEB KO versus WT MEFs. Moreover, ectopic expression of FLAG-CPEB in the KO MEFs reduced luciferase reporter activity by 50% (Fig. 4b). These results show that CPEB control of NF-κB activity mediates IL-6 production.
NF-κB consists of a family of genes: NF-κB1 (p50), NF-κB2 (p52), RELA (p65), RELB, and c-REL (11). Generally, p65, c-Rel, and/or RelB form hetero- or homodimers with other NF-κB family members, bind to promoters, and activate specific gene transcription (20). The activity of NF-κB is regulated at multiple levels, but in the prototypical pathway, the subunits are retained in the cytoplasm by the IKBα inhibitory complex; when IKBα is destroyed, NF-κB translocates to the nucleus and stimulates transcription (11). To gain insight into which NF-κB family member(s) might be regulated by CPEB, we examined their 3′ UTR sequences and found that only NF-κB1 contains a putative CPE (Fig. 4c). However, Western blot analysis of WT and CPEB KO MEFs did not show any change in the level of the NF-κB1 precursor (p105) or mature form (p50) (Fig. 4d). Additionally, neither p65 and nor IKBα protein levels were changed in WT versus CPEB KO MEFs. An examination of whole-cell or cytoplasmic lysates also did not show a change in IKBα levels in WT versus KO cells (Fig. 4d), indicating that CPEB did not regulate the canonical NF-κB pathway.
NF-κB can also be activated in an alternative manner leading to phosphorylation of p65 and enhancement of promoter binding and transcription (20, 27). In KO but not WT MEFs, p65 was phosphorylated, indicating that atypical activation of NF-κB might be controlled by CPEB. Indeed, Fig. 4d shows substantial p65 nuclear staining in KO but not WT MEFs, suggesting an increased activity of this transcription factor in the absence of CPEB. To assess this directly, various amounts of lysate derived from WT and CPEB KO MEF nuclei were mixed with biotinylated oligonucleotides containing the NF-κB consensus binding site. Antibody against p65 was then used to determine the amount captured by streptavidin. A HeLa cell (S3) control demonstrates the efficacy of the assay (Fig. 4e). KO but not WT MEF lysates showed substantial concentration-dependent p65 binding (Fig. 4f). Taken together, these results indicate that NF-κB p65 is constitutively active in immortalized CPEB KO MEFs and that expression of ectopic CPEB can partially rescue this defect.
IL-6 reinforcement of senescence requires CPEB and p53.
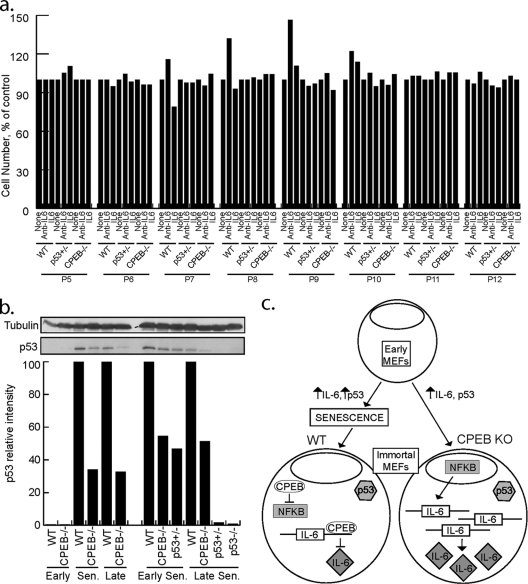
Although IL-6 reinforces oncogene-induced senescence in primary human cells (1, 25), its role in mouse cell senescence is unclear. To determine whether IL-6 is involved in senescence in MEFs and, if so, whether it is connected to CPEB, neutralizing antibody to IL-6 or recombinant IL-6 was added to early-passage WT, CPEB KO, or p53 heterozygous MEFs. p53 heterozygous MEFs were included in the analysis because CPEB-mediated senescence in human cells occurs via regulation of p53 mRNA polyadenylation and translation (i.e., CPEB depletion in foreskin fibroblasts reduces p53 by ∼50%, which elicits senescence bypass [6]). Consequently, the effects of CPEB deficiency on senescence were mimicked by p53 heterozygous MEFs even though CPEB levels were normal (Fig. 5a).
Fig. 5.
A senescence regulatory pathway composed of IL-6, CPEB, and p53. (a) WT, p53 heterozygous, or CPEB KO MEFs were incubated in the presence of IL-6 neutralizing antibody or recombinant IL-6. The agents were added to the culture medium and replenished at each passage, and this was followed by determination of cell numbers. (b) p53 protein levels are reduced in CPEB KO MEFs. Lysates prepared from WT, CPEB KO, or p53 heterozygous MEFs at various passage points were analyzed by Western blotting for p53 and quantified. β-Tubulin served as a loading control. Sen, senescence. (c) Model illustrating interplay among CPEB, IL-6, NF-κB, and p53 in controlling senescence. Early WT and CPEB-null MEFs exhibit low levels of p53 and IL-6. As WT and CPEB KO MEFs increase in passage number, IL-6 levels increase. In WT MEFs, higher levels of IL-6 reinforced the senescence response, p53 protein levels increase, and cells enter senescence and have reduced division rates. In contrast, CPEB KO MEFs do not enter senescence and do not exhibit decreased division and p53 levels are 50% of those of passage-matched WT MEFs, likely through lack of CPEB-mediated polyadenylation of p53 mRNA. After WT cells become immortalized, IL-6 levels are decreased and NF-κB is not active. In contrast, passage-matched immortalized CPEB KO MEFs continue to secrete high levels of IL-6 and NF-κB is activated. In immortalized MEFs, p53 is likely mutated. Immortalized CPEB KO MEFs continue to secrete IL-6 for many passages.
IL-6 neutralizing antibody or recombinant IL-6 protein was added to the culture medium after each passage of WT, CPEB KO, and p53 heterozygous MEFs. Prior to senescence (P5, P6), all cells were insensitive to either IL-6 or IL-6 antibody (Fig. 5a). However, at P7 and P8, fewer WT cells entered senescence in the presence of neutralizing IL-6 antibody and more cells senesced in the presence of recombinant IL-6. As WT cells continued through senescence, the effect of IL-6 became less pronounced and once senescence was established, addition of IL-6 or neutralizing antibody had no effect on the cell number (P11 and P12). These results indicate that, similar to human cells, IL-6 reinforces the senescence response in mouse cells. Because neither the CPEB KO MEFs nor the p53 heterozygous MEFs responded to IL-6 or neutralizing IL-6 antibody, we propose that p53 is essential for IL-6 reinforcement of senescence.
Although the data in Fig. 5a demonstrate that an ∼50% reduction of p53 is sufficient to induce senescence bypass in MEFs irrespective of IL-6, our initial investigation of CPEB KO MEFs failed to reveal any change in p53. In that analysis, p53 levels were compared to tubulin over many passages on different Western blots; thus, an ∼50% reduction might not have been detected (18). A reinvestigation of p53 in WT and CPEB KO MEFs by quantitative Western blotting shows that although early-passage cells contained no obvious p53, it was detected in senescing WT cells. In contrast, p53 was reduced by ∼65% at similar passages in the KO MEFs. Moreover, in early senescing cells, p53 levels were similar in CPEB KO and p53 heterozygous MEFs (Fig. 5b). These data indicate that CPEB KO MEFs escape IL-6 reinforcement of senescence because p53 expression was reduced by ∼50%, most likely by translational inefficiency caused by an aberrantly short poly(A) tail (6).
DISCUSSION
Here, we show that IL-6 reinforces senescence in murine cells and its mRNA is regulated at the translational level by CPEB. However, the level of IL-6 production in CPEB KO cells far exceeds that which can be accounted for by translational control. The vast majority of IL-6 expression in CPEB-deficient cells occurs at the level of transcription, as mediated by prolonged NF-κB nuclear localization by improper S276 phosphorylation. Although CPEB KO MEFs uniquely secrete large amounts of IL-6, senescence is not reinforced because robust p53 levels are compromised. Thus, a regulatory axis consisting of CPEB, IL-6, NF-κB, and p53 is essential for cellular senescence.
Model of CPEB, IL-6, and NF-κB activity during senescence.
Figure 5c presents a model integrating CPEB, IL-6, NF-κB, and p53 in cellular senescence. Early WT and CPEB-null MEFs exhibit low levels of p53 and IL-6. As WT and CPEB KO MEFs increase in passage number, IL-6 levels increase as well. In WT MEFs, higher levels of IL-6 reinforce the senescence response, p53 protein levels increase, and cells enter senescence and have reduced division rates. In contrast, CPEB KO MEFs do not enter senescence and do not exhibit decreased cell division and their p53 levels are 50% of those of passage-matched WT MEFs, likely through lack of CPEB-mediated polyadenylation of p53 mRNA (6). After WT cells become immortalized, IL-6 levels are decreased and NF-κB is not active. In contrast, passage-matched immortalized CPEB KO MEFs continue to secrete high levels of IL-6 and NF-κB is activated.
Constitutive NF-κB activation.
Activation of NF-κB generally requires IKBα degradation, which releases cytoplasmic NF-κB dimers to transit to the nucleus and activate specific gene transcription (20). Immortalized CPEB-null MEFs do not exhibit decreased levels of IKBα, and the level of IL-6 secreted by these cells is lower than that seen during the senescence phase or typically seen after stimulation of immune cells (which exhibit reduced IKBα levels) (17). This suggests that in CPEB-null MEFs, an alternative pathway leads to NF-κB activation. NF-κB p65 has an elevated nuclear localization and phosphorylation in CPEB-null MEFs, two activities that are known to increase NF-κB transcriptional activity. Several kinases, including RSK1 and IKK, can directly phosphorylate p65 (32); however, the 3′ UTRs of these kinases do not contain CPE and thus are unlikely to be direct targets of CPEB.
Many other cytokines are also upregulated in immortalized CPEB-null MEFs, perhaps also due to constitutive NF-κB activation in these cells. Constitutive NF-κB activation is a feature of many cancer cells (10, 14, 15, 16, 28), and increased levels of cytokines and other secreted factors during senescence drive the initial senescence response and growth arrest (1, 25). High levels of these secreted factors may also recruit immune cells to the site of senescent cells, helping to clear them and prevent tumor formation (2, 7, 26). This clearance may require a balance between growth arrest of the premalignant cells, as mediated by senescence, and the rate of clearance by immune cells (34). In the case of CPEB-null MEFs, though these cells secrete high levels of IL-6, they do not undergo senescence and thus the balance of growth arrest to immune cell clearance may be altered. A constant output of IL-6 and other growth factors (e.g., VEGF) by CPEB-null immortalized cells could stimulate the growth of surrounding preneoplastic or metastatic cells, leading to an increased cancer incidence (1, 25). While CPEB-null mice do not exhibit an increased incidence of cancer, CPEB-null mice treated with 7,12-dimethyl-benz[a]anthracene-12-O-tetradecanoylphorbol-13-acetate show an increased rate and amount of tumor formation (6). Thus, induction of stress-induced malignant transformation of CPEB-deficient cells may be mediated by enhanced cytokine production.
ACKNOWLEDGMENTS
We thank Elena Filippova and Nemisha Dawra for technical assistance.
R.G. was supported by an institutional postdoctoral training grant (HD07312). This work was supported by grants from the NIH (AG30323 and HD37267). Additional core support from the Diabetes Endocrinology Research Center (DK32520) is gratefully acknowledged.
Footnotes
Published ahead of print on 2 May 2011.
REFERENCES
- 1. Acosta J. C., et al. 2008. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell 133:1006–1018 [DOI] [PubMed] [Google Scholar]
- 2. Adams P. D. 2009. Healing and hurting: molecular mechanisms, functions, and pathologies of cellular senescence. Mol. Cell 36:2–14 [DOI] [PubMed] [Google Scholar]
- 3. Barnard D. C., Ryan K., Manley J. L., Richter J. D. 2004. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell 119:641–651 [DOI] [PubMed] [Google Scholar]
- 4. Ben-Porath I., Weinberg R. A. 2005. The signals and pathways activating cellular senescence. Int. J. Biochem. Cell Biol. 37:961–976 [DOI] [PubMed] [Google Scholar]
- 5. Braig M., et al. 2005. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436:660–665 [DOI] [PubMed] [Google Scholar]
- 6. Burns D. M., Richter J. D. 2008. CPEB regulation of human cellular senescence, energy metabolism, and p53 mRNA translation. Genes Dev. 22:3449–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Campisi J., d'Adda di Fagagna F. 2007. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 8:729–740 [DOI] [PubMed] [Google Scholar]
- 8. Cao Q., Kim J. H., Richter J. D. 2006. CDK1 and calcineurin regulate Maskin association with eIF4E and translational control of cell cycle progression. Nat. Struct. Mol. Biol. 13:1128–1134 [DOI] [PubMed] [Google Scholar]
- 9. Cao Q., Richter J. D. 2002. Dissolution of the maskin-eIF4E complex by cytoplasmic polyadenylation and poly(A)-binding protein controls cyclin B1 mRNA translation and oocyte maturation. EMBO J. 21:3852–35862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Catlett-Falcone R., et al. 1999. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity 10:105–115 [DOI] [PubMed] [Google Scholar]
- 11. Chen L. F., Greene W. C. 2004. Shaping the nuclear action of NF-kappaB. Nat. Rev. Mol. Cell Biol. 5:392–401 [DOI] [PubMed] [Google Scholar]
- 12. Collado M., et al. 2005. Tumour biology: senescence in premalignant tumours. Nature 436:642. [DOI] [PubMed] [Google Scholar]
- 13. Dimri G. P., et al. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. U. S. A. 92:9363–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grivennikov S. I., Greten F. R., Karin M. 2010. Immunity, inflammation, and cancer. Cell 140:883–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grivennikov S., et al. 2009. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell 15:103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grivennikov S. I., Karin M. 2010. Dangerous liaisons: STAT3 and NF-kappaB collaboration and crosstalk in cancer. Cytokine Growth Factor Rev. 21:11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Groeneweg M., et al. 2006. Lipopolysaccharide-induced gene expression in murine macrophages is enhanced by prior exposure to oxLDL. J. Lipid Res. 47:2259–2267 [DOI] [PubMed] [Google Scholar]
- 18. Groisman I., et al. 2006. Control of cellular senescence by CPEB. Genes Dev. 20:2701–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hake L. E., Richter J. D. 1994. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell 79:617–627 [DOI] [PubMed] [Google Scholar]
- 20. Hayden M. S., Ghosh S. 2008. Shared principles in NF-kappaB signaling. Cell 132:344–346 [DOI] [PubMed] [Google Scholar]
- 21. Hayflick L. 1965. The limited in vitro lifetime of human diploid cell strains. Exp. Cell Res. 37:614–636 [DOI] [PubMed] [Google Scholar]
- 22. Iliopoulos D., Hirsch H. A., Struhl K. 2009. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 139:693–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jung M. Y., Lorenz L., Richter J. D. 2006. Translational control by neuroguidin, a eukaryotic initiation factor 4E and CPEB binding protein. Mol. Cell. Biol. 26:4277–4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim J. H., Richter J. D. 2006. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol. Cell 24:173–183 [DOI] [PubMed] [Google Scholar]
- 25. Kuilman T., et al. 2008. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell 133:1019–1031 [DOI] [PubMed] [Google Scholar]
- 26. Kuilman T., Peeper D. S. 2009. Senescence-messaging secretome: SMS-ing cellular stress. Nat. Rev. Cancer 9:81–94 [DOI] [PubMed] [Google Scholar]
- 27. Lee H., et al. 2009. Persistently activated Stat3 maintains constitutive NF-kappaB activity in tumors. Cancer Cell 15:283–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lu C., Kerbel R. S. 1993. Interleukin-6 undergoes transition from paracrine growth inhibitor to autocrine stimulator during human melanoma progression. J. Cell Biol. 120:1281–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nordan R. P., Richards C. D., Gauldie J. 1 May 2001, posting date Measurement of interleukin 6. Curr. Protoc. Immunol. Chapter 6. Unit 6.6. doi:10.1002/0471142735.im0606s17 [DOI] [PubMed] [Google Scholar]
- 30. Padmanabhan K., Richter J. D. 2006. Regulated Pumilio-2 binding controls RINGO/Spy mRNA translation and CPEB activation. Genes Dev. 20:199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Richter J. D. 2007. CPEB: a life in translation. Trends Biochem. Sci. 32:279–285 [DOI] [PubMed] [Google Scholar]
- 32. Viatour P., Merville M. P., Bours V., Chariot A. 2005. Phosphorylation of NF-kappaB and IkappaB proteins: implications in cancer and inflammation. Trends Biochem. Sci. 30:43–52 [DOI] [PubMed] [Google Scholar]
- 33. Wu L., et al. 1998. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of alpha-CaMKII mRNA at synapses. Neuron 21:1129–1139 [DOI] [PubMed] [Google Scholar]
- 34. Xue W., et al. 2007. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445:656–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zearfoss N. R., Alarcon J. M., Trifilieff P., Kandel E., Richter J. D. 2008. A molecular circuit composed of CPEB-1 and c-Jun controls growth hormone-mediated synaptic plasticity in the mouse hippocampus. J. Neurosci. 28:8502–8509 [DOI] [PMC free article] [PubMed] [Google Scholar]