Abstract
Cellular transformation induced by oncogenic tyrosine kinases is a multistep process involving activation of growth-promoting signaling pathways and inactivation of suppressor molecules. Dok-1 is an adaptor protein that acts as a negative regulator of tyrosine kinase-initiated signaling and opposes oncogenic tyrosine kinase-mediated cell transformation. Findings that its loss facilitates transformation induced by oncogenic tyrosine kinases suggest that Dok-1 inactivation could constitute an intermediate step in oncogenesis driven by these oncoproteins. However, whether Dok-1 is subject to regulation by oncogenic tyrosine kinases remained unknown. In this study, we show that oncogenic tyrosine kinases, including p210bcr-abl and oncogenic forms of Src, downregulate Dok-1 by targeting it for degradation through the ubiquitin-proteasome pathway. This process is dependent on the tyrosine kinase activity of the oncoproteins and is mediated primarily by lysine-dependent polyubiquitination of Dok-1. Importantly, restoration of Dok-1 levels strongly suppresses transformation of cells expressing oncogenic tyrosine kinases, and this suppression is more pronounced in the context of a Dok-1 mutant that is largely refractory to oncogenic tyrosine kinase-induced degradation. Our findings suggest that proteasome-mediated downregulation of Dok-1 is a key mechanism by which oncogenic tyrosine kinases overcome the inhibitory effect of Dok-1 on cellular transformation and tumor progression.
INTRODUCTION
Protein tyrosine kinases (PTKs), of which more than 90 are encoded by the human genome, are key regulators of intracellular signal transduction pathways that control a variety of cellular processes, such as proliferation, differentiation, survival, cell movement, and cytoskeletal organization (46, 52). Under physiological conditions, the activity of these kinases, including receptor and cytoplasmic PTKs, is tightly controlled to maintain cell and tissue homeostasis. However, when deregulated, due to, for example, mutations, gene amplifications, or impaired deactivation of the PTK, this control is lost, leading to aberrant downstream signaling, which can result in malignant transformation. To date, deregulation of more than 50 human PTKs has been implicated in the pathogenesis of various solid tumors and hematologic malignancies (2, 7, 36).
Significant progress has been made toward unraveling the mechanisms of oncogenic activation of PTKs. Also, numerous studies have identified key signaling molecules and pathways that are activated by oncogenic tyrosine kinases (OTKs) and participate in mediating their effects on malignant transformation (45, 55, 57, 71, 72, 75). However, besides triggering positive signaling events, OTKs also need to overcome negative regulatory constraints to drive tumor initiation and/or progression (7, 24, 42, 57). These include, for instance, constraints brought about by tumor suppressors or inhibitory molecules that counteract the signaling activity triggered by the OTKs (17, 69). The nature of these “negative regulators” and, importantly, the mechanisms by which OTKs overcome their inhibitory effects remain largely elusive.
Members of the Dok family of adaptor proteins have emerged as key modulators of PTK signaling. Dok-1, also known as p62dok, is the prototypical family member. It was first identified as a substrate of OTKs p210bcr-abl and v-Abl (13, 93) and found to be a substrate of many endogenous PTKs (9, 18, 19, 23, 34, 51, 53, 59, 60, 64, 78, 91, 92, 97); hence, it was termed Dok, for downstream of tyrosine kinases. Since the identification of Dok-1, six additional family members, Dok-2 to Dok-7, have been identified in human and mice, all of which are substrates of various receptor or cytoplasmic PTKs, with some of them having partially overlapping functions while others having distinct functions (15, 16, 20, 30, 40, 49, 61, 66). All Dok family members share structural similarities, characterized by an N-terminal pleckstrin homology (PH) domain, a central phosphotyrosine binding (PTB) domain, and a C-terminal region containing multiple tyrosine residues and proline-rich motifs (PXXP) (53).
Within the Dok family, Dok-1, Dok-2, and Dok-3 comprise a closely related subfamily. Ample evidence indicates that they function as negative regulators of antigen receptor- and growth factor receptor-mediated cell proliferation and/or survival and that they act by interfering with the activation of distinct signaling pathways downstream of these receptors (31, 33, 39, 53, 61, 82, 86, 90, 94, 95, 98). In particular, upon tyrosine phosphorylation elicited by receptor stimulation, the Dok proteins dock to the plasma membrane in a PH domain-dependent manner, where they recruit inhibitory effector molecules via interactions involving their phosphorylated tyrosine residues and PxxP motifs and the SH2 and SH3 domains of the interacting partners, respectively (31, 53, 81, 94, 99). Through their ability to coordinate the formation of these signaling complexes, Dok-1, -2, and -3 establish negative feedback loops that antagonize receptor-initiated signaling pathways (33, 39, 53, 86, 90, 98, 99). For example, Dok-1 interferes with platelet-derived growth factor (PDGF) receptor-mediated activation of the Ras/MAPK and Src/c-Myc pathways by enhancing the recruitment of RasGAP to Ras and tethering Csk to active Src kinases, respectively (98). Dok-1's actions on both of these signaling pathways contribute to its inhibitory effect on PDGF receptor-mediated cell proliferation (98).
Importantly, Dok-1, -2, and -3 also emerged as negative regulators of OTK-induced transformation. For instance, Dok-1 and Dok-3 were shown to antagonize v-Src- and/or v-Abl-evoked transformation (15, 67, 80), and Dok-1 and Dok-2 were shown to oppose p210bcr-abl-mediated transformation in vivo (21, 63, 95). The p210bcr-abl chimeric protein, which displays deregulated tyrosine kinase activity, originates from a chromosomal translocation between the BCR gene on chromosome 22 and the ABL gene on chromosome 9. The latter is the causative mutation found in 95% of cases of chronic myelogenous leukemia (CML), a myeloproliferative disorder of the hematopoietic stem cell (57, 74). CML typically evolves in distinct clinical phases, starting with a chronic phase, which spans approximately 3 to 5 years, followed by an accelerated phase, which eventually leads to an acute malignant phase known as blast crisis (57, 74). Strikingly, inactivation of Dok-1 in mice markedly accelerates the onset of both the chronic and fatal blastic phases of the CML-like myeloproliferative disease induced by p210bcr-abl (63, 95). Similar observations have been made for Dok-2, whose inactivation also accelerates leukemia and blast crisis onset in Tec-p210bcr-abl transgenic mice (63, 95). Thus, these data indicate that Dok-1 and Dok-2 oppose p210bcr-abl-induced leukemogenesis and suggest that they possess tumor suppressive activity in the context of myeloid leukemia. In support of this are the findings that mice lacking both Dok-1 and Dok-2 spontaneously develop CML-like myeloproliferative disease (63, 95). Furthermore, more recent studies have shown that mice with combined Dok-1, Dok-2, and Dok-3 knockouts also succumb to aggressive histiocytic sarcoma (54) or develop lung adenocarcinoma with penetrance and latency dependent on the number of lost Dok family members (4). Finally, DOK-2 has been reported to be a target of frequent copy number loss in human lung cancer (4).
Together, these studies indicate that Dok-1, Dok-2, and Dok-3 possess tumor suppressive activities and that their inactivation can contribute to disease/tumor progression associated with deregulated PTK signaling, for example, as in the case of p210bcr-abl-driven CML-like disease in mice. In extension, these findings also suggest that inactivation of Dok function(s) is likely a critical component in the progression of p210bcr-abl-driven leukemogenesis and, probably, of other OTK-driven tumors as well. Consistent with this notion, low levels of Dok-1 and/or Dok-2 have been reported in a number of leukemic cell lines established from patients with myeloid leukemia (65, 95). To date, however, very little is known about the regulation of Dok proteins by OTKs. Here, we have focused on Dok-1 and mechanisms of its regulation by p210bcr-abl and other OTKs. We demonstrate that OTKs, including p210bcr-abl and oncogenic forms of Src, significantly downregulate Dok-1 expression levels in several cell types by targeting it for ubiquitin-proteasome-mediated degradation and that this process is dependent on the tyrosine kinase activity of the oncoproteins. Importantly, restoration of Dok-1 levels results in suppression of cellular proliferation and transformation induced by p210bcr-abl and oncogenic Src kinases, and this suppression is even more effective in the context of a Dok-1 mutant that is largely refractory to OTK-induced polyubiquitination and degradation. Together, our data support a model in which proteasome-mediated degradation of Dok-1 is an important contributive step toward tumor development and/or progression driven by OTKs.
MATERIALS AND METHODS
DNA constructs.
pMSCVpuro retroviral vectors expressing p210bcr-abl and the STI571-resistant mutant p210bcr-ablT351I were gifts from H. G. Wendel (MSKCC). p210bcr-ablK1172R (p210bcr-ablKD [where KD means “kinase deficient”]) and v-Abl-expressing retroviral vectors were constructed by subcloning p210bcr-ablKD and v-Abl coding sequences from pSRαMSVTKneo-p210bcr-ablKD and pSRαMSVTKneo-v-Abl (gifts from A. M. Pendergast, Duke University), respectively, into the EcoRI site of pMSCVpuro (Clontech). Mammalian expression vectors pUSE-p210bcr-abl and pUSE-p210bcr-ablKD were generated by subcloning p210bcr-abl and p210bcr-ablKD coding sequences into the EcoRI site of pUSE (Upstate Biotechnology). pBABEpuro-v-Src and pBABEpuro-H-RasV12 were described previously (8, 73). v-Src and H-RasV12 cDNAs were cloned as NheI-EcoRI fragments into pUSE. pUSE vectors encoding constitutively active SrcY529F (SrcKA [where KA means “kinase active”]) and kinase-deficient SrcK297R (SrcKD) were gifts from R. Karni (IMRIC Institute). SrcKA and SrcKD cDNAs were cloned as XhoI-BamHI fragments into XhoI-BglII sites of pBABEpuro(linker), which contains an extended multicloning site (a gift from J. Rodriguez, Rockefeller University). The His6-ubiquitin mammalian expression vector was a gift from W. Tansey (Vanderbilt University) and was described previously (85). pBABEpuro/FLAG-His retroviral vector carrying full-length human Dok-1 cDNA was described previously (98). A sequence encoding Dok-1 tagged at its C terminus with a peptide containing a FLAG epitope was inserted as a BglII-SalI fragment into pWZLhygro retroviral vector or into BglII-XhoI sites of pMSCVhygro (Clontech). cDNA encoding a lysineless mutant of human Dok-1 (Dok-1K0), which carries 23 lysine-to-arginine substitutions, was generated by GenScript Corporation (Piscataway, NJ). A hemagglutinin (HA) tag-encoding sequence was added to the C terminus of Dok-1K0 and wild-type Dok-1 (Dok-1WT) by PCR, and the resulting fragments were cloned into BglII-EcoRI sites of pMSCV-IRES-GFP (a gift from S. Lowe, CSHL) or into BamHI-EcoRI sites of pBABEhygro (product no. 1765; Addgene). All constructs used in this study were sequence verified.
Cell culture and gene transfer.
Baf3 cells (68) and Baf3-derived TonB210.1 cells expressing p210bcr-abl under the control of a tetracycline-inducible promoter (a gift from G. Q. Daley, Harvard University) (43) were cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS; HyClone) and 10% WEHI-3 conditioned medium as a source of interleukin-3 (IL-3). Mo7e cells and R10− cells (gifts from B. Clarkson, MSKCC) (5) were maintained in RPMI 1640 containing 10% FBS and 20% FBS, respectively. For Mo7e cells, the medium was supplemented with 10 ng/ml of recombinant human granulocyte-macrophage colony stimulating factor (R&D Systems). K562 cells were cultured in RPMI 1640 containing 10% FBS. IMR90 human fibroblasts and HEK293 and HEK293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% FBS. NIH 3T3 cells were cultured in DMEM containing 10% calf serum (Invitrogen). For growth factor stimulation, cells were serum starved for 24 h in DMEM containing 0.2% calf serum and then exposed to PDGF (12 ng/ml) or epidermal growth factor (EGF; 50 ng/ml) (R&D Systems) for the indicated times. In a number of experiments, the cells were treated with Abl-specific tyrosine kinase inhibitor STI571 (LC Laboratories), proteasome inhibitors MG132 or ALLN (Calbiochem), or cycloheximide (Sigma). For transient expression, HEK293 and HEK293T cells were transfected using the calcium phosphate method as described previously (38). To establish stable cell lines, viral particles were produced by cotransfecting retroviral vectors and the packaging-defective helper plasmid pCl-Eco into HEK293T cells. Some of the viruses were pseudotyped with the vesicular stomatitis virus G (VSV-G)-expressing plasmid (38). Infected cell populations were selected for 2 days in medium containing 2 μg/ml of puromycin (Sigma) (0.5 μg/ml in the case of R10− and Mo7e cells) or for 10 to 14 days in medium containing 300 μg/ml of hygromycin (Calbiochem) (1 mg/ml in the case of Baf3 cells). To limit long-term mutagenic effects of OTKs, the cells were used within a week after selection.
Cell lysis and Western blotting.
Cells were washed with phosphate-buffered saline (PBS), and total cell lysates were prepared in a buffer containing 75 mM Tris-HCl (pH 6.8), 3.8% SDS, 4 M urea, and 20% glycerol (29). The lysates were resolved on SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to an Immobilon polyvinylidene difluoride membrane (Millipore). Primary antibodies (Abs) used were monoclonal antiphosphotyrosine (PY20) and monoclonal anti-RasGAP (BD Transduction Laboratories), monoclonal anti-γ-tubulin and polyclonal anti-FLAG (Sigma), horseradish peroxidase (HRP)-conjugated antiphosphotyrosine, polyclonal anti-Abl (K-12), and anti-c-Myc (N-262) (Santa Cruz Biotechnology), polyclonal Erk2 (Cell Signaling Technology), monoclonal anti-v-Src (Ab-1; Calbiochem), monoclonal anti-HA (HA.11; Covance), anti-OPHN1 Ab (29), monoclonal anti-Dok-1 (Dok-1m), and polyclonal anti-Dok-1 (Dok-1PH) Abs (99). Immune complexes were detected using secondary HRP-conjugated anti-mouse or anti-rabbit Abs (Bio-Rad) and enhanced chemiluminescence (GE Healthcare).
Immunoprecipitation.
For the analysis of the tyrosine phosphorylation status of Dok-1 proteins, cells were lysed in modified RIPA buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 5% glycerol, 50 mM NaF, 40 mM β-glycerophosphate, 2 mM Na3VO4, complete protease inhibitors [Roche]), and equal amounts of cell extracts were incubated with anti-Dok-1m or anti-HA Ab precoupled to protein G-agarose (Roche) for 2 to 5 h at 4°C. Beads were then washed with RIPA buffer, and immunoprecipitates were eluted by boiling in 2× Laemmli buffer at 95°C for 10 min and subjected to Western blotting with the indicated Abs.
For coimmunoprecipitation experiments, NIH 3T3 cells transduced with pBABEhygro containing HA-tagged Dok-1WT (Dok-1WTHA), Dok-1K0HA, or a control empty vector or HEK293T cells transiently transfected with pMSCV-IRES-GFP containing Dok-1WTHA, Dok-1K0HA, Dok-1WTFLAG, Dok-1K0FLAG, or a control empty vector were serum starved and either left unstimulated or stimulated with PDGF (12.5 ng/ml) or EGF (50 ng/ml) for 10 min. Cells were lysed at 4°C in lysis buffer (LB) (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Triton X-100, 10% glycerol, 50 mM NaF, 40 mM β-glycerophosphate, 2 mM Na3VO4, and complete protease inhibitors), and cell extracts were subjected to immunoprecipitation with anti-FLAG M2 affinity resin (Sigma) or anti-HA Ab precoupled to protein G-agarose (Roche) for 8 h at 4°C. Anti-FLAG or anti-HA immunoprecipitates were eluted with LB containing FLAG peptide (0.2 mg/ml) (Sigma) or Laemmli buffer, respectively, and resolved by SDS-PAGE, followed by immunoblotting with the indicated Abs.
Alkaline phosphatase treatment.
Cells were washed with ice-cold PBS and lysed in buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1% Triton X-100, 5% glycerol, 10 mM MgCl2, 1 mM dithiothreitol, and EDTA-free complete protease inhibitors (Roche). Equal amounts of lysates were then diluted in lysis buffer containing either phosphatase inhibitors (to the final concentration of 50 mM NaF, 40 mM β-glycerophosphate, and 2 mM Na3VO4,) or 10 units of calf intestinal phosphatase (CIP; New England BioLabs) and incubated for 1 h at 37°C. Laemmli buffer was added to terminate the reaction, and lysates were subjected to Western blot analysis.
Reverse transcription-PCR (RT-PCR) and Q-PCR.
Total RNA was isolated using TRIzol (Invitrogen), and 2 μg of each RNA sample was reverse transcribed using a TaqMan kit (Applied Biosystems) and oligo(dT) primers (Applied Biosystems), according to the manufacturers' recommendations. For quantitative real-time PCR (Q-PCR) analyses, Dok-1 and endogenous reference hypoxanthine-guanine phosphoribosyltransferase (HPRT) cDNAs were quantified using SYBR green PCR master mix (Applied Biosystems) and the 7900HT sequence detection system (Applied Biosystems) as follows. Aliquots (0.5 to 1 μl) of the reverse-transcribed mixtures were amplified in 12-μl reaction volumes containing 0.15 μM primers specific for mouse Dok-1 (mDok-1) and HPRT (mHPRT) (see below). Cycling conditions used were 50°C for 2 min and 95°C for 10 min, followed by 40 amplification cycles at 95°C for 15 s and 58°C for 1 min. Both sets of primers shared comparable amplification efficiencies, and melting curve analysis at the end of each run confirmed amplification of single PCR products. Each experiment was performed with three separate determinations for each amplicon. Data were analyzed using SDS RQ manager v1.2 software (Applied Biosystems), which utilizes the comparative 2−ΔΔCT method (50).
For RT-PCR analyses, PCR was performed using 1-μl aliquots of the reverse-transcribed mixtures in 25-μl reaction mixtures containing 0.2 mM deoxynucleoside triphosphate (dNTP) mix (Roche), 1× ThermoPol buffer (NEB), 0.05 U/μl of Taq DNA polymerase (NEB), and 0.4 mM each primer. Primers specific for mouse Dok-1 and endogenous reference HPRT were the following: mDok-1 forward primer, 5′ TCTGGCCCTACACTCTGTTG 3′; mDok-1 reverse primer, 5′ CGACCAGCTTCAAAGGAGAA 3′; mHPRT forward primer, 5′ TTCCTCATGGACTGATTATGGA 3′; and mHPRT reverse primer, 5′ TCCCATCTCCTTCATGACATC 3′. Primers specific for human Dok-1 and endogenous reference β-actin were the following: hDok-1 forward primer, 5′ AGAGTCAGCGCTTTGGGAC 3′; hDok-1 reverse primer, 5′ AGGTGGGTTATCGGTAGGC 3′; hβ-actin forward primer, 5′ GCATGGGTCAGAAGGATTC 3′; and hβ-actin reverse primer, 5′ CATCTCTTGCTCGAAGTCC 3′. PCR conditions used for reaction mixtures containing cDNAs prepared from Baf3 cells were 95°C for 5 min, then 32 cycles (for mDok-1) or 26 cycles (for mHPRT) of 95°C for 30 s, 57°C for 30 s, and 72°C for 15 s, followed by 10 min at 72°C. For cDNAs prepared from NIH 3T3 cells, the number of cycles used for the mDok-1 and mHPRT primers was 35 cycles and 32 cycles, respectively. PCR parameters used for cDNAs prepared from IMR90 cells were 95°C for 5 min, followed by 30 cycles (for hβ-actin) or 35 cycles (for hDok-1) at 95°C for 30 s, 55°C for 30 s, and 72°C for 45 s, and a final step at 72°C for 10 min. PCR products were separated on 2% agarose gels or on 12% nondenaturing polyacrylamide gels and visualized by ethidium bromide staining using the U:Genius imaging system (Syngene).
35S labeling and pulse-chase analysis.
Logarithmically growing Baf3 cells transduced with pMSCV-Dok-1FLAG retroviral vector were washed twice with warm PBS and starved in methionine/cysteine-free RPMI medium (Sigma) supplemented with 10% dialyzed FBS (HyClone) and 10 ng/ml of murine IL-3 (R&D Systems) for 1 h. Methionine/cysteine-starved cells were incubated with 200 μCi/ml of EXPRE35S35S protein labeling mix [35S] (PerkinElmer) for 1 h. The cells were then washed twice with warm PBS and resuspended in chase medium (Baf3 culture medium supplemented with 2 mM methionine and 2 mM cysteine). At indicated time points, cell aliquots were removed, washed with ice-cold PBS, and lysed in buffer containing 50 mM Tris-HCl (pH 7.5), 200 mM NaCl, 1% Triton X-100, 10% glycerol, 50 mM NaF, 40 mM β-glycerophosphate, 2 mM Na3VO4, and EDTA-free complete protease inhibitors (Roche). Cell extracts were subjected to immunoprecipitation with anti-FLAG M2 affinity resin (Sigma) for 1 h at 4°C, and bound proteins were eluted by boiling in 2× Laemmli buffer at 95°C for 10 min. The immunoprecipitates were resolved by SDS-PAGE followed by blotting. Membranes were exposed to a PhosphorImager screen, followed by immunoblotting with anti-FLAG Ab and Alexa Fluor 562-conjugated goat anti-rabbit fluorescent secondary Ab (Invitrogen). Radioactive and fluorescent signals were detected using a Fujifilm FLA-5100 instrument (Fuji Medical Systems USA, Inc.), and band intensities were quantified using MultiGauge v2.3 software (FujiFilm).
Affinity precipitation of ubiquitin conjugates.
HEK293T cells, and HEK293 cells transduced with pWZLhygro-Dok-1FLAG, were transfected with indicated expression vectors. A total of 48 h after transfection, the cells were washed with ice-cold PBS and lysed under denaturing conditions in buffer containing 6 M guanidine-HCl, 0.1 M NaH2PO4/Na2HPO4 (pH 7.0), 1% Triton X-100, 900 mM NaCl, 40 mM imidazole, and 20 mM β-mercaptoethanol. Samples were sonicated, cleared by centrifugation, and incubated with Ni-nitrilotriacetic acid (NTA) resin (Qiagen) for 3 h at room temperature. The beads were washed five times with lysis buffer and three times with wash buffer containing 25 mM Tris-HCl (pH 6.8) and 20 mM imidazole. Bound proteins were eluted by being boiled in 2× Laemmli buffer supplemented with 300 mM imidazole for 10 min at 95°C and analyzed by SDS-PAGE and Western blotting.
Cell proliferation and transformation assays.
For cell growth curves, R10− cells transduced with pBABEpuro-Dok-1FLAG-His, or with empty control vector, were seeded at a density of 5 × 104 cells per well in 6-well plates. The number of viable cells was monitored daily with the aid of a Neubauer hemocytometer over a period of 14 days. Cells were subcultured every 3 days into fresh medium to maintain exponential growth over the time course of the experiment. Cellular transformation was scored by a focus formation assay. NIH 3T3 cells stably transduced with pBABEhygro-Dok-1WTHA, pBABEhygro-Dok-1K0HA, or empty control vector were infected with pBABEpuro-SrcKA retrovirus or empty control vector. Two days postinfection, 1 × 104 infected cells were mixed with 3 × 105 uninfected cells and plated on 10-cm dishes. The cell medium was replaced every 4 days, and the appearance of transformed foci was quantified after 8 to 12 days. Three replicate dishes were plated for each sample, and each experiment was repeated three times.
RESULTS
p210bcr-abl downregulates Dok-1 protein expression.
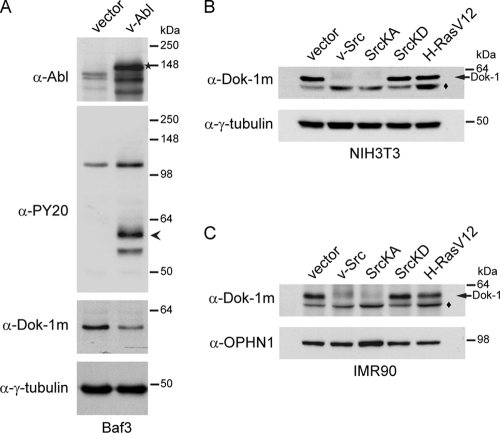
Previous studies suggested that the abrogation of Dok-1 function is an important component in the progression of p210bcr-abl-driven leukemogenesis (63, 95). This prompted us to assess whether p210bcr-abl modulates Dok-1 function during oncogenesis. We began by investigating whether p210bcr-abl alters the expression levels of Dok-1 protein in hematopoietic cells. The p210bcr-abl gene was introduced into the growth factor-dependent human leukemic cell line Mo7e, which has been widely used as a model system for hematopoietic progenitors (32, 48), and into the bone marrow-derived murine Baf3 pro-B cell line (68) by retroviral transduction. As expected, the p210bcr-abl-transformed cells displayed elevated tyrosine phosphorylation of cellular proteins, as visualized by immunoblotting with an antiphosphotyrosine-specific antibody (Fig. 1 A and B). Interestingly, we observed a significant reduction in the levels of Dok-1 protein in the p210bcr-abl-transformed Mo7e and Baf3 cells compared to those in the parental, nontransformed cells (Fig. 1A and B). These results were obtained with antibodies that recognize two different Dok-1 epitopes (Fig. 1C). Furthermore, we noticed that the extent of Dok-1 downregulation inversely correlates with the levels of p210bcr-abl expression. This was clearly seen when Baf3/TonB210.1 cells expressing p210bcr-abl under the control of a tetracycline-inducible promoter (43) were treated with increasing concentrations of doxycycline for 48 h. As shown in Fig. 1D, increasing p210bcr-abl expression levels correlated with gradual decreases in Dok-1 expression levels. Thus, the degree of Dok-1 downregulation is dose dependent on p210bcr-abl expression levels.
FIG. 1.
p210bcr-abl induces downregulation of Dok-1 protein levels in a tyrosine kinase activity-dependent manner. (A, B) Mo7e cells (A) and Baf3 cells (B) were stably transduced with empty control vector or a p210bcr-abl-, p210bcr-ablKD-, or p210bcr-ablT315I-expressing retroviral vector. Where indicated, the cells were cultured in the presence of 1 μM STI571. Tyrosine phosphorylation of cellular proteins was determined in total cell lysates by Western blot analysis with antiphosphotyrosine (α-PY20) Ab. The blots were reprobed with anti-Abl Ab, a polyclonal Ab raised against the PH domain of Dok-1 (α-Dok-1PH), and anti-γ-tubulin Ab as a loading control. The arrowhead indicates the position of tyrosine-phosphorylated Dok-1, as confirmed by anti-Dok-1m immunoprecipitation (IP), followed by Western blot analysis with antiphosphotyrosine (α-PY20) and anti-Dok-1PH Abs (B, right). HC, Ig heavy chain. (C) Total cell lysates prepared from Mo7e cells stably transduced with empty control vector or a p210bcr-abl- or p210bcr-abl KD-expressing retroviral vector were analyzed by Western blotting with anti-Abl Ab and a monoclonal anti-Dok-1 (α-Dok-1m) Ab. The blots were reprobed with anti-Dok-1PH Ab and anti-γ-tubulin Ab as a loading control. (D) Baf3/TonB210.1 cells expressing p210bcr-abl under the control of the tetracycline-inducible promoter were treated with increasing concentrations of doxycycline to induce p210bcr-abl expression. Total cell lysates were analyzed by Western blotting with antiphosphotyrosine (α-PY20) Ab, anti-Abl Ab, anti-Dok-1m Ab, and anti-γ-tubulin Ab as a loading control. (E) Total cell lysates prepared from Mo7e cells and K562 cells treated with the indicated concentrations of STI571, or a control vehicle, were analyzed by Western blotting with antiphosphotyrosine (α-PY20) Ab, anti-Abl Ab, anti-Dok-1PH Ab, and anti-γ-tubulin Ab as a loading control. Asterisks indicate the positions of endogenous c-Abl.
Next we asked whether the kinase activity of p210bcr-abl is required for its ability to downregulate Dok-1. To this end, the expression of Dok-1 protein in Mo7e and Baf3 cells transduced with a kinase-deficient mutant of p210bcr-abl, p210bcr-ablK1172R (p210bcr-ablKD), was examined. We found that p210bcr-ablKD failed to downregulate Dok-1 expression (Fig. 1A to C). To further determine the requirement of p210bcr-abl's tyrosine kinase activity, we investigated the effect of the Abl-specific tyrosine kinase inhibitor STI571 (also known as imatinib mesylate [Gleevec]) on the p210bcr-abl-induced reduction in Dok-1 levels. The STI571 inhibitor competitively binds to the ATP-binding site of p210bcr-abl tyrosine kinase, thereby inhibiting its activity (76). As expected, treatment of Baf3/p210bcr-ablcells with STI571 (1 μM) strongly reduced p210bcr-abl-elicited tyrosine phosphorylation of cellular proteins (Fig. 1B). Importantly, we observed that in the presence of STI571, p210bcr-abl did not downregulate Dok-1 protein levels (Fig. 1B). However, when examining Dok-1 expression in Baf3 cells transduced with an STI571-resistant mutant of p210bcr-abl, p210bcr-ablT351I (28), we found that this p210bcr-abl mutant was still able to efficiently downregulate Dok-1 even in the presence of STI571 (Fig. 1B). Furthermore, we observed that K562 cells, which express endogenous p210bcr-abl, treated with STI571 displayed higher levels of Dok-1 than untreated K562 cells (Fig. 1E). Thus, our data indicate that the tyrosine kinase activity of p210bcr-abl is critical for its ability to downregulate Dok-1.
Oncogenic Abl and Src tyrosine kinases, but not oncogenic H-Ras, downregulate Dok-1.
We next investigated whether Dok-1 downregulation is an event triggered specifically by p210bcr-abl or is a more general event that can also be evoked by other oncogenic tyrosine kinases (OTKs). To this end, we analyzed the expression of Dok-1 protein in cells transduced with retroviral vectors encoding v-Abl or oncogenic forms of Src tyrosine kinase. Similar to p210bcr-abl, expression of v-Abl led to a significant reduction in total Dok-1 protein levels in Baf3 cells (Fig. 2 A). To determine the effect of oncogenic Src on Dok-1 downregulation, we chose to use fibroblasts, as Dok-1 had previously been shown to inhibit Src-induced transformation in these cells (80). We found that expression of v-Src or a constitutively active mutant of Src kinase, SrcY519F (SrcKA), but not a kinase-deficient mutant of Src, SrcK297R (SrcKD), resulted in a strong reduction in Dok-1 protein levels in both NIH 3T3 mouse embryonic fibroblasts and human IMR90 fibroblasts (Fig. 2B and C). Thus, oncogenic Src can also downregulate Dok-1 in a tyrosine kinase activity-dependent manner.
FIG. 2.
v-Abl and oncogenic Src kinases, but not H-RasV12, induce Dok-1 downregulation. (A) Baf3 cells were stably transduced with empty control vector or v-Abl-expressing retroviral vector. Total cell lysates were analyzed by immunoblotting with antiphosphotyrosine (α-PY20) Ab, anti-Abl Ab, anti-Dok-1m Ab, and anti-γ-tubulin Ab as a loading control. The arrowhead indicates the position of tyrosine-phosphorylated Dok-1. The asterisk indicates the position of v-Abl. (B, C) NIH 3T3 cells (B) and IMR90 cells (C) were stably transduced with empty control vector or a v-Src-, SrcKA-, SrcKD-, or H-RasV12-expressing retroviral vector. Total cell lysates were analyzed by Western blotting with anti-Dok-1m Ab and anti-γ-tubulin Ab (B) or anti-OPHN1 Ab (C) as loading controls. The diamonds denote nonspecific bands.
To determine whether oncogenes that lack tyrosine kinase activity affect the expression levels of Dok-1, we examined the levels of Dok-1 protein in cells expressing an oncogenic mutant form of H-Ras. NIH 3T3 and IMR90 fibroblasts were transduced with H-RasV12, and the levels of Dok-1 expression were analyzed by Western blotting. Expression of H-RasV12 did not cause a detectable reduction in Dok-1 protein levels, as the amount of Dok-1 protein in these cells was essentially identical to that found in cells transduced with empty control vector (Fig. 2B and C). Together, these results suggest that oncogenes with constitutive tyrosine kinase activity (including p210bcr-abl and oncogenic forms of Src), but not nontyrosine kinase oncogenes (such as H-RasV12), downregulate Dok-1.
p210bcr-abl reduces Dok-1 protein stability.
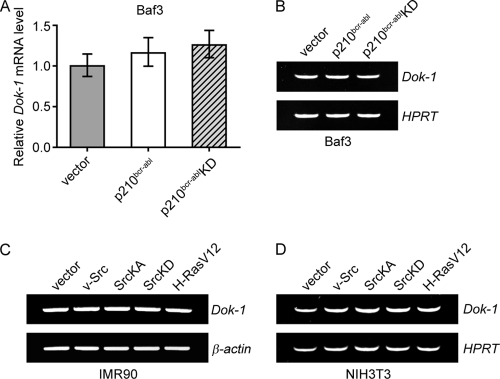
There are several possible mechanisms by which OTKs could downregulate Dok-1. For instance, they could interfere with Dok-1 gene transcription or, alternatively, affect the stability of Dok-1 protein. To gain insight into the underlying mechanism, we began by testing whether p210bcr-abl, or oncogenic forms of Src kinase, affects the levels of Dok-1 mRNA. Q-PCR and RT-PCR analyses revealed that the levels of Dok-1 transcripts in p210bcr-abl-transformed Baf3 cells and cells transduced with p210bcr-ablKD, or empty control vector, were not significantly different (Fig. 3 A and B). Similarly, no significant changes in the levels of Dok-1 mRNA were detected upon v-Src or SrcKA expression in IMR90 and NIH 3T3 cells (Fig. 3C and D). These results imply that OTKs downregulate Dok-1 posttranscriptionally.
FIG. 3.
Oncogenic tyrosine kinases do not affect the levels of Dok-1 mRNA. (A, B) Total RNA was isolated from Baf3 cells stably transduced with empty control vector or a p210bcr-abl- or p210bcr-ablKD-expressing retroviral vector, and the levels of Dok-1 transcripts were measured by Q-PCR (A) and RT-PCR (B) using gene-specific primers as described in Materials and Methods. The hypoxanthine-guanine phosphoribosyltransferase (HPRT) gene was amplified as an internal control. (C, D) RT-PCR analysis of Dok-1 mRNA levels in IMR90 cells (C) and NIH 3T3 cells (D) stably transduced with the indicated retroviral vectors. β-actin (C) and HPRT (D) genes were used as internal controls.
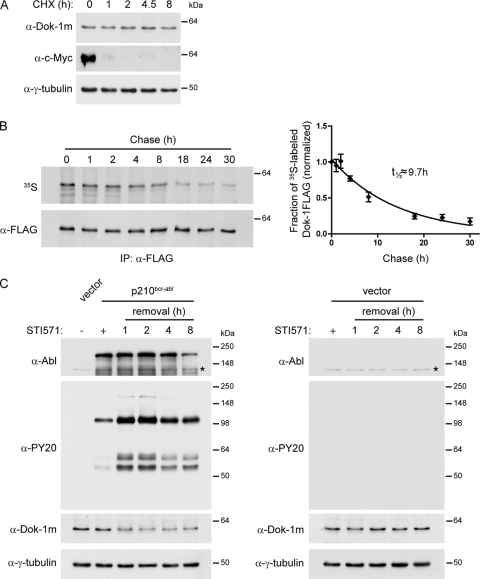
We next explored whether OTKs affect Dok-1 protein stability. We first examined the stability of Dok-1 protein in the absence of OTKs. Baf3 cells were treated with the translation inhibitor cycloheximide (CHX), and Dok-1 protein levels were analyzed by Western blotting at the indicated times of CHX treatment (Fig. 4 A). c-Myc, which is a relatively short-lived protein, was included as a positive control. In contrast to c-Myc, no major changes in Dok-1 protein levels were detected during the course of the experiment (Fig. 4A), indicating that Dok-1 is a relatively stable protein. To corroborate these findings, we performed [35S]methionine/cysteine pulse-chase experiments using Baf3 cells stably expressing FLAG-tagged Dok-1. These experiments revealed that the estimated half-life of Dok-1 in nontransformed cells is approximately 10 h (Fig. 4B).
FIG. 4.
p210bcr-abl downregulates Dok-1 by reducing Dok-1 protein stability. (A) Baf3 cells were treated with protein synthesis inhibitor cycloheximide (CHX; 200 μg/ml). At indicated time points, cells were lysed, and total cell lysates were analyzed by Western blotting for Dok-1, c-Myc, and γ-tubulin as a loading control. (B) Half-life of Dok-1 in Baf3 cells. Baf3 cells stably expressing FLAG-tagged Dok-1 were pulse-labeled with [35S]methionine/cysteine for 1 h, chased in fresh medium, and harvested at the indicated times postchase. Cell lysates were subjected to anti-FLAG immunoprecipitation (IP), and the immunoprecipitates were resolved by SDS-PAGE followed by blotting (left). 35S-labeled Dok-1FLAG was quantified by phosphorimaging, and total levels of Dok-1FLAG protein were quantified by Western blotting with anti-FLAG Ab. The fraction of radiolabeled Dok-1FLAG remaining is shown for each chase time point (right). Error bars represent the standard deviations (SD) from three independent experiments. (C) Baf3 cells transduced with an empty control vector or p210bcr-abl-expressing retroviral vector were cultured in the absence (−) or presence (+) of 1 μM STI571. The STI571-treated p210bcr-abl-expressing cells (left) or control vector-expressing cells (right) were lysed at 0 to 8 h after STI571 removal, and total cell lysates were analyzed by Western blotting with antiphosphotyrosine (α-PY20) Ab, anti-Abl Ab, anti-Dok-1m Ab, and anti-γ-tubulin Ab as a loading control. Asterisks indicate the positions of endogenous c-Abl.
We then examined the kinetics of Dok-1 downregulation by p210bcr-abl. Baf3 cells expressing p210bcr-abl were maintained in the presence of STI571 to prevent downregulation of Dok-1 by p210bcr-abl. To determine the kinetics of Dok-1 downregulation, STI571 was removed, and Dok-1 protein levels were analyzed at different time points after drug removal by Western blotting. Notably, within 1 h of STI571 removal, we observed a considerable increase in tyrosine phosphorylation of cellular proteins, indicating a prompt reactivation of p210bcr-abl kinase activity (Fig. 4C, left). Interestingly, a significant reduction in Dok-1 protein levels was also detected within 1 h after STI571 withdrawal (Fig. 4C, left). Treatment of nontransformed Baf3 cells with STI571 inhibitor did not alter cellular tyrosine phosphorylation and did not affect the levels of Dok-1 (Fig. 4C, right). Thus, combined with our finding that Dok-1 is a relatively long-lived protein, the rapid (within 1 h) decrease in Dok-1 protein levels upon p210bcr-abl activation strongly indicates that p210bcr-abl decreases Dok-1 protein stability.
Oncogenic tyrosine kinases downregulate Dok-1 through the ubiquitin-proteasome pathway.
The ubiquitin-proteasome proteolytic pathway is a major pathway that controls protein stability in the cell (41). Therefore, we tested whether p210bcr-abl and other OTKs trigger degradation of Dok-1 through this pathway. We first analyzed the effect of the proteasome inhibitor MG132 on Dok-1 protein levels in p210bcr-abl-transformed Baf3 cells. We observed that MG132 treatment stabilized Dok-1 protein in these cells (Fig. 5 A). However, we also found that proteasome inhibition caused a strong reduction in p210bcr-abl detection, which was accompanied by a significant decrease in tyrosine phosphorylation of cellular proteins (Fig. 5A), indicating a loss of p210bcr-abl tyrosine kinase activity. While the reason for the reduction in p210bcr-abl detection upon MG132 treatment is unclear, this finding prevented us from distinguishing whether MG132-induced stabilization of Dok-1 resulted from inhibition of proteasome activity or was a direct consequence of loss of p210bcr-abl tyrosine kinase activity.
FIG. 5.
Oncogenic tyrosine kinase-induced downregulation of Dok-1 requires proteasome activity. (A) Baf3 cells transduced with p210bcr-abl or empty control vector were treated with proteasome inhibitor MG132 (10 μM) for the indicated times. Total lysates were analyzed by Western blotting using antiphosphotyrosine (α-PY20) Ab, anti-Abl Ab, anti-Dok1PH Ab, and anti-γ-tubulin Ab as a loading control. The asterisk indicates the position of endogenous c-Abl. (B, C) IMR90 cells stably transduced with empty control vector or a v-Src-, SrcKA-, SrcKD-, or H-RasV12-expressing retroviral vector were treated with proteasome inhibitors MG132 (10 μM), ALLN (50 μM), or control vehicle for 8 h. Total protein and RNA were isolated. (B) The lysates were analyzed by Western blotting with antiphosphotyrosine (α-PY20) Ab, anti-Dok-1m Ab, and anti-γ-tubulin Ab as a loading control. The diamonds denote nonspecific bands. (C) The levels of Dok-1 transcript were measured by RT-PCR, as described for Fig. 3C. (D) IMR90 cells stably transduced with empty control vector or a v-Src- or SrcKA-expressing retroviral vector were treated with proteasome inhibitors, as described for panel B. Total cell lysates were analyzed by Western blotting using anti-Src Ab and anti-γ-tubulin Ab as a loading control.
To overcome this problem, we decided to use other OTKs to assess the link between OTK-induced downregulation of Dok-1 and the proteasome-dependent proteolytic pathway. In particular, we tested the effect of the MG132 and ALLN proteasome inhibitors on Dok-1 levels in IMR90 cells expressing oncogenic Src. Neither of the two inhibitors appreciably affected the kinase activity and expression levels of v-Src or SrcKA, as assessed by antiphosphotyrosine and anti-Src immunoblotting (Fig. 5B and D). Importantly, we found an increase in Dok-1 protein levels in v-Src- and SrcKA-expressing IMR90 fibroblasts treated with MG132 or ALLN (Fig. 5B). Notably, MG132 and ALLN treatment did not alter the levels of Dok-1 transcript in these cells (Fig. 5C and data not shown). These observations imply that the observed increase in Dok-1 levels reflects Dok-1 protein stabilization elicited by inhibition of proteasomal proteases.
To examine whether Dok-1 is a direct target of the ubiquitination machinery, we analyzed the status of Dok-1 ubiquitination in HEK293 cells stably expressing FLAG-tagged Dok-1. These cells were transfected with a His-tagged ubiquitin-encoding plasmid. The ubiquitin conjugates were then purified by Ni-affinity chromatography and subjected to Western blot analysis using an antibody specific for FLAG-tagged Dok-1. We observed the presence of a heterogeneous population of high-molecular-weight species of Dok-1. Notably, these slower-migrating species were detected only in cells expressing His-tagged ubiquitin and became more prominent upon inhibition of proteasome activity by MG132 (Fig. 6 A, left). Thus, these data strongly suggest that Dok-1 is polyubiquitinated. Importantly, we found that coexpression of v-Src, SrcKA, or p210bcr-abl significantly increased the amount of ubiquitinated Dok-1 (Fig. 6A to C), which could be further elevated upon treatment with proteasome inhibitor MG132 (Fig. 6A). As seen with Dok-1 protein levels (Fig. 1D), the extent of Dok-1 ubiquitination was dose dependent on the expression levels of SrcKA and p210bcr-abl (Fig. 6B and C). In addition, the ability of OTKs to induce Dok-1 ubiquitination required their tyrosine kinase activity, since the kinase-deficient p210bcr-ablKD and SrcKD mutants failed to induce Dok-1 ubiquitination (Fig. 6B and C). Together, our data indicate that OTKs destabilize Dok-1 by triggering its polyubiquitination and subsequent degradation by the proteasome.
FIG. 6.
p210bcr-abl and oncogenic forms of Src kinase induce polyubiquitination of Dok-1. (A) HEK293 cells stably transduced with FLAG-tagged Dok-1 (HEK293/Dok-1FLAG) were transfected with a His6-tagged ubiquitin (Ub)-encoding vector, or empty control vector, together with either a vector encoding v-Src or empty control vector. A total of 36 h later, the cells were treated with MG132 (10 μM) or control vehicle for 8 h. Ubiquitin conjugates were affinity precipitated under highly denaturing conditions by Ni-NTA chelate chromatography, resolved by SDS-PAGE, and analyzed by immunoblotting with anti-FLAG Ab. The right panel is a lighter exposure of data shown in the left panel. (B, C) HEK293/Dok-1FLAG cells were transiently transfected with empty control vector or His6-tagged ubiquitin-expressing vector and with increasing amounts of expression vectors encoding SrcKA, SrcKD, or empty vector control (B) or with vectors encoding p210bcr-abl, p210bcr-ablKD, or empty control vector (C), as indicated. Cells were lysed 48 h after transfection, and the lysates were subjected to Ni-NTA affinity precipitation and Western blotting with anti-FLAG Ab. The diamond denotes a nonspecific band.
Role of Dok-1 lysine residues in oncogenic tyrosine kinase-induced Dok-1 ubiquitination and degradation.
Protein degradation through the ubiquitin-proteasome pathway usually involves the covalent attachment of polyubiquitin chains to lysine residues of a target protein, which mediates the recruitment of the target protein to the 26S proteasome for its proteolysis (41). Hence, we investigated the requirement for lysines in OTK-induced polyubiquitination and degradation of Dok-1. For this purpose, we constructed a lysineless mutant of Dok-1 (Dok-1K0), in which all 23 lysines were replaced with arginines (see Materials and Methods). We first compared the ubiquitination status of Dok-1K0 with that of wild-type Dok1 (Dok-1WT) in the presence and absence of OTKs. HEK293 cells were cotransfected with an HA-tagged Dok-1WT- or Dok-1K0-expressing vector, a His-tagged ubiquitin construct, and either an empty control vector or an expression vector encoding SrcKA or p210bcr-abl. Ubiquitin conjugates were then purified by Ni-affinity chromatography. As expected, Western blot analyses using an antibody specific for HA-tagged Dok-1 revealed readily detectable amounts of polyubiquitinated Dok-1WTHA, which further accumulated upon proteasome inhibition or coexpression of p210bcr-abl or SrcKA (Fig. 7 A and B). A significant reduction in ubiquitination was observed for the Dok-1K0HA mutant compared to that observed for Dok-1WTHA (Fig. 7A and B), supporting the notion that lysine residues are major Dok-1 ubiquitin acceptor sites. Notably, however, residual ubiquitination of Dok-1K0HA was consistently detected, which could be further increased by coexpression of p210bcr-abl or SrcKA or treatment with the MG132 proteasome inhibitor (Fig. 7A and B). In this regard, several reports have previously demonstrated that nonlysine residues, including cysteines, serines and threonines, can also serve as ubiquitin acceptor sites or that ubiquitination can occur on the N-terminal residue of the substrate (11, 14, 41, 83, 88). Hence, we conclude that while Dok-1 lysine residues are major Dok-1 ubiquitylation sites, other nonlysine Dok-1 residues could also contribute to Dok-1 ubiquitination.
FIG. 7.
Role of lysine residues in oncogenic tyrosine kinase-induced ubiquitination and downregulation of Dok-1. (A, B) HEK293T cells were transiently cotransfected with the indicated expression vectors. A total of 36 h posttransfection, cells were left untreated (A) or were treated with MG132 (10 μM) or control vehicle for 8 h (B). Ubiquitin conjugates were affinity precipitated under denaturing conditions by Ni-NTA chelate chromatography and analyzed by Western blotting with anti-HA Ab. Diamonds denote nonspecific bands. (C) NIH 3T3 cells stably expressing HA-tagged wild-type Dok-1 (Dok-1WTHA), lysineless Dok-1 mutant (Dok-1K0HA), or empty control vector were transduced with a retroviral vector expressing SrcKA or an empty control vector. Total cell lysates were subjected to Western blot analysis with anti-HA Ab and anti-γ-tubulin Ab as a loading control. (Left) A representative example from three independent experiments is shown. (Right) Quantification of Dok-1WTHA and Dok-1K0HA protein levels. Data were normalized to γ-tubulin and then to the value of 1.0 for the non-SrcKA-expressing condition. Error bars represent SD. (D) Lysates prepared from cells described for panel C were subjected to anti-HA immunoprecipitation (IP), followed by Western blotting with antiphosphotyrosine (α-PY20) and anti-HA Abs. (E) Lysates prepared from cells described for panel C were left untreated or were treated with calf intestinal phosphatase (CIP) as described in Materials and Methods, and subjected to Western blotting with antiphosphotyrosine (α-PY20) Ab, anti-HA Ab, and anti-γ-tubulin Ab as a loading control. The arrowhead indicates the position of the tyrosine-phosphorylated Dok-1HA proteins. (F) NIH 3T3 cells stably transduced with Dok-1WTHA, Dok-1K0HA, or control empty vector were serum starved and were either left untreated (−) or treated (+) with PDGF (12.5 ng/ml) for 10 min. Cell extracts were subjected to anti-HA immunoprecipitation (IP). A portion of the final eluates (6%) was subjected to Western blotting with anti-HA Ab (bottom), and the remaining eluted immune complexes were analyzed by Western blotting with anti-RasGAP Ab. HC, Ig heavy chain. (G) HEK293T cells transfected with the indicated expression vectors were serum starved and stimulated with EGF (50 ng/ml) for 10 min. Cell extracts were subjected to anti-FLAG immunoprecipitation (IP), followed by Western blotting with anti-HA and anti-FLAG Abs.
We then examined the impact of mutating Dok-1 lysine residues on Dok-1 downregulation by OTKs. NIH 3T3 cells stably expressing HA-tagged Dok-1WT or Dok-1K0 were transduced with a SrcKA-expressing vector, or empty control vector, and the expression of Dok-1 was analyzed by Western blotting. Consistent with our above-described data, expression of SrcKA triggered a significant downregulation of epitope-tagged Dok-1WTHA, whereas lysineless Dok-1K0HA was downregulated to a considerably lesser extent (Fig. 7C). However, also here a decrease in Dok-1K0 levels was consistently noted (Fig. 7C).
Importantly, we obtained several lines of evidence indicating that the lysine-to-arginine substitutions in Dok-1K0 do not perturb the structure and function of the protein. First, we examined the tyrosine phosphorylation status of the Dok-1K0 mutant in SrcKA-expressing NIH 3T3 cells. We found that, akin to Dok-1WT, Dok-1K0 was efficiently tyrosine phosphorylated by SrcKA (Fig. 7D). Given that an intact PH domain is necessary for the tyrosine phosphorylation of Dok-1 protein (31), these data also imply that the structure and function of Dok-1K0's PH domain is intact. Of note, the band shifts observed for Dok-1WT and Dok-1K0 (in Fig. 7C and D) are due to phosphorylation of the Dok-1 proteins, as treatment of lysates prepared from the SrcKA-expressing cells with calf intestinal phosphatase (CIP) efficiently diminished tyrosine phosphorylation of Dok-1WT and Dok-1K0 and at the same time eliminated the observed band shifts (Fig. 7E). Second, we tested whether Dok-1K0 was still able to bind to RasGAP and found that this is indeed the case (Fig. 7F). Third, we examined the ability of Dok-1K0 to homodimerize, since the PTB domain of Dok-1 has previously been shown to mediate homotypic interactions of Dok-1 (58, 80). We found that Dok-1K0 was able to efficiently homodimerize (Fig. 7G). Thus, based on these data, we infer that the lysine-to-arginine substitutions in Dok-1K0 do not perturb the structure and function of Dok-1.
Taken together, our data indicate that ubiquitination of lysine residues on Dok-1 is critical for its proteasomal degradation but that also a lysine-independent mechanism is to some extent likely involved.
Dok-1 expression levels critically influence cellular proliferation and transformation induced by p210bcr-abl and oncogenic Src kinases.
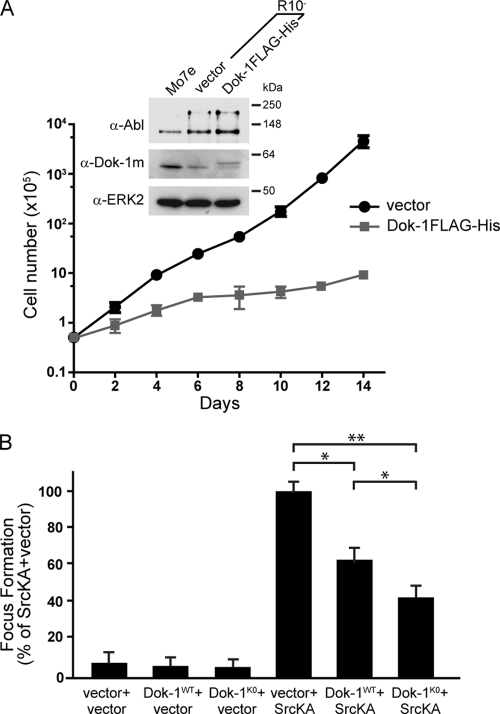
Our data demonstrate that OTKs trigger ubiquitin-proteasome-mediated downregulation of Dok-1 protein. Combined with previous studies demonstrating that Dok-1 opposes OTK-induced transformation and that inactivation of Dok-1 in mice accelerates p210bcr-abl-induced leukemogenesis, we conjectured that Dok-1 degradation via the ubiquitin-proteasome pathway could constitute a mechanism by which OTKs overcome the inhibitory effect of Dok-1 to thereby promote tumorigenesis. We reasoned that if this is the case, restoration of Dok-1 protein levels in OTK-transformed cells, where the levels of Dok-1 are significantly reduced, should diminish the proliferation and transformation potential of these cells. We first tested this idea in the context of p210bcr-abl. For these experiments we used the Mo7e/p210bcr-abl-derived subline R10− (5). As expected, Dok-1 protein levels were significantly lower in R10− cells than in parental Mo7e cells (Fig. 8 A). We then transduced the R10− cells with either an empty control vector or FLAG-tagged Dok-1 so as to elevate Dok-1 protein levels and bring them closer to those found in nontransformed Mo7e cells (Fig. 8A). Importantly, we found that the Dok-1FLAG-expressing R10− cells displayed a significantly lower growth rate than the empty control vector-expressing cells (Fig. 8A), indicating that restoration of Dok-1 levels suppresses proliferation of p210bcr-abl-expressing cells.
FIG. 8.
Restoration of Dok-1 expression levels suppresses cellular proliferation and transformation induced by p210bcr-abl and oncogenic Src kinases. (A) Growth curve of R10− cells stably transduced with a Dok-1FLAG-His-expressing retroviral vector or empty control vector. The number of cells was plotted on a logarithmic scale against time on a linear scale. The data represent the means ± SD from triplicate experiments. Data from a representative of three independent experiments are shown. (Inset) Total cell lysates prepared from Mo7e cells and R10− cells expressing Dok-1FLAG-His or empty control vector were analyzed by Western blotting with anti-Abl, anti-Dok-1m Ab, and anti-ERK2 Ab as a loading control. (B) Focus formation assay. NIH 3T3 cells stably transduced with Dok-1WTHA-, Dok-1K0HA-expressing retroviral vector, or empty control vector were infected with a SrcKA-expressing retroviral vector or an empty control vector. Focus-forming activities are presented as percentages of the SrcKA-plus-vector condition. Data represent the means ± SD from three independent experiments performed in triplicate. *, P < 0.05; **, P < 0.01 versus SrcKA plus vector by t test.
Next, we examined the effect of restoring Dok-1 levels in the context of oncogenic Src-induced cell transformation. In particular, we assessed the ability of SrcKA to transform NIH 3T3 cells stably expressing either an empty control vector or Dok-1WTHA by a focus formation assay. As shown in Fig. 8B, expression of Dok-1WT protein reduced SrcKA-induced transformation, as cells coexpressing Dok-1WTHA and SrcKA formed significantly fewer foci than cells coexpressing SrcKA and an empty control vector. Notably, although Dok-1WT considerably suppressed OTK-mediated cell transformation, it was still subject to downregulation by OTKs (Fig. 7C). Therefore, to further determine the importance of ubiquitin-proteasome-mediated degradation of Dok-1 for OTKs to transform cells, we examined the effect of the Dok-1K0 mutant, which is largely refractory to OTK-induced polyubiquitination and degradation (Fig. 7B and C), on SrcKA-induced focus formation. We found that Dok-1K0 exhibited a stronger suppressive effect on the ability of SrcKA to induce foci than that exhibited by Dok-1WT (Fig. 8B), corroborating that ubiquitin-proteasome-mediated degradation of Dok-1 facilitates OTK-mediated cell transformation. Together, these findings suggest that OTKs downregulate Dok-1 levels to overcome its inhibitory effect and to thereby manifest their full transforming activities.
DISCUSSION
The molecular mechanisms by which oncogenic tyrosine kinases (OTKs) bring about tumor formation and/or progression are not fully understood. It is becoming increasingly apparent, however, that besides the activation of growth-promoting signaling pathways, OTKs also need to overcome negative regulatory constraints to drive tumor formation and/or progression. The latter includes, for instance, the inactivation of tumor suppressor or inhibitory molecules that counteract the signaling activity triggered by OTKs (7, 17, 24, 42, 57, 69). Dok-1, Dok-2, and Dok-3 constitute a closely related subfamily of the Dok adaptor proteins, which emerged as negative regulators of PTK-induced signaling and cellular transformation driven by OTKs (15, 21, 63, 67, 80, 95). Findings that their loss promotes transformation or disease progression initiated by deregulated PTKs (4, 21, 54, 63, 67, 95) suggest that their inactivation could constitute an important component of OTK-driven malignant transformation. However, mechanisms by which OTKs could regulate Dok proteins remained unknown. In this study, we demonstrate that OTKs, including p210bcr-abl and oncogenic forms of Src, downregulate the expression of the Dok-1 protein by targeting it for degradation via the ubiquitin-proteasome pathway. This process is dependent on the tyrosine kinase activity of the oncoproteins and is mediated largely by lysine-dependent polyubiquitination of Dok-1. Importantly, we found that restoration of Dok-1 levels suppresses the proliferation and transformation potential of cells expressing p210bcr-abl or oncogenic Src and that this effect is even more pronounced in the context of a Dok-1 mutant that is largely refractory to OTK-induced polyubiquitination and degradation. Thus, our findings support a role for Dok-1 proteasomal degradation in oncogenic transformation driven by OTKs.
Inactivation of growth inhibitory proteins or tumor suppressors is an event often observed in human cancers. It can occur via many mechanisms, including genetic alterations, such as gene mutations or deletions, epigenetic events that lead to gene silencing, and/or proteasomal degradation, as shown, for example, for p53, PTEN, and NF1 (56, 87, 89). Our findings indicate that ubiquitin-proteasome-mediated degradation of Dok-1 is a key mechanism by which OTKs trigger Dok-1 “inactivation.” Expression of OTKs, such as p210bcr-abl or oncogenic forms of Src, elicits a vast increase in the amount of polyubiquitinated Dok-1 and a corresponding decrease in Dok-1 expression, which can be largely restored upon inhibition of proteasome activity. Significantly, this effect seems to be specific for OTKs, as expression of nontyrosine kinase oncogenes, such as H-RasV12, does not affect Dok-1 expression levels.
Our finding that Dok-1 is a target of ubiquitin-mediated proteasomal degradation raises the question as to what signals trigger its polyubiquitination. We found that p210bcr-abl- and oncogenic Src-induced ubiquitination of Dok-1 critically depends on the tyrosine kinase activity of these oncoproteins. For instance, Dok-1 levels remain unaltered upon expression of kinase-deficient mutants of p210bcr-abl or Src, and treatment with the Abl kinase inhibitor STI571 prevents downregulation of Dok-1 in p210bcr-abl-expressing cells. Given that Dok-1 is a substrate of both p210bcr-abl and Src (6, 13, 77, 80, 84), a plausible scenario is that tyrosine phosphorylation of Dok-1 by these kinases creates a signal that is recognized by components of the ubiquitination machinery. PTK-elicited phosphorylation events have previously been shown to trigger protein degradation (35, 47, 96). For example, v-Src-mediated phosphorylation of cofilin at tyrosine 68 leads to its degradation through the ubiquitin-proteasome pathway (96). However, at present, we cannot exclude the involvement of additional and/or alternative posttranslational modifications on Dok-1 that are indirectly induced by OTKs (12, 26, 35). For example, Dok-1 degradation could be elicited by phosphorylation of serine/threonine residues on Dok-1 that are mediated by protein kinases activated downstream of the OTKs. Extensive mutagenesis and proteomic approaches will be needed to determine the precise signals and residues in Dok-1 that are critical for its recognition by the ubiquitin-proteasome machinery.
Regardless of the precise signal(s) involved, our data clearly indicate the requirement of PTK activity for Dok-1 degradation. Moreover, and importantly, this effect seems to be specifically elicited by OTKs displaying constitutive kinase activity, since we did not detect Dok-1 downregulation upon activation of PTKs by physiological stimuli, such as PDGF. Indeed, when we treated NIH 3T3 fibroblasts with PDGF at a concentration of 12.5 ng/ml (98) and assessed Dok-1 protein levels at 15 min, 30 min, 1 h, and 2 h following PDGF addition, no decrease in Dok-1 expression levels during any of the time points was detected (data not shown). Of note, tyrosine phosphorylation of cellular proteins (including Dok-1) reached the maximum after 15 min of PDGF treatment, after which it gradually declined, reaching basal levels at 2 h following PDGF addition. Thus, Dok-1 degradation could be dependent on prolonged activation of tyrosine kinase signaling, as opposed to only transient activation by growth factor stimulation (22). Alternatively, however, growth factors and OTKs may induce distinct patterns of posttranslational modifications on Dok-1, thereby differently influencing its stability. Interestingly, we also found that the degree of downregulation of Dok-1 is dose dependent on the levels of OTK expression. In particular, gradual decreases in Dok-1 levels were observed with increasing levels of p210bcr-abl. As discussed below, this could have significant functional implications, as the expression levels of p210bcr-abl increase with CML progression (3, 25). Thus, Dok-1 levels are expected to decrease as the disease progresses.
In general, proteins are targeted for proteasomal degradation by covalent attachment of polyubiquitin chains to their lysine residues, albeit other residues could also be involved (11, 14, 41, 83, 88). We found that proteasomal degradation of Dok-1 is largely mediated through ubiquitination of its lysine residues. Particularly, we observed that replacement of all Dok-1 lysines with arginines significantly reduces the extent of its polyubiquitination and downregulation by OTKs. It should be noted, however, that a lysine-independent mechanism(s) could also contribute to Dok-1 ubiquitination and degradation. Residual ubiquitination, which becomes more prominent upon proteasome inhibition, and a modest decrease in the expression levels of a lysine-less Dok-1 mutant (Dok-1K0) are consistently observed in the presence of OTKs. In light of this, recent studies have identified cysteines, serines, and threonines, as well as an N-terminal residue, as potential ubiquitination sites (11, 14, 41, 83, 88). However, Dok-1 is subject to cotranslational N-terminal acetylation at its initiator methionine (44) and is predicted to be a poor substrate for methionine aminopeptidases (10, 14, 70); therefore, it is unlikely that Dok-1 is ubiquitinated at its N terminus. Although we cannot per se exclude that replacement of all internal Dok-1 lysines forced the ubiquitination to occur at sites where it would not have normally occurred, ubiquitination of Dok-1 on cysteines, serines, and/or threonines remains a good possibility and thus could contribute to targeting Dok-1 for degradation.
Previous studies have implicated Dok-1 as a negative regulator of OTK-mediated transformation (21, 63, 67, 80, 95). For example, Dok-1 inactivation in mice shortens the chronic phase and accelerates the onset of the fatal blastic phase of the CML-like myeloproliferative disease induced by p210bcr-abl, indicating that Dok-1 opposes p210bcr-abl-driven leukemogenesis (63, 95). Our study provides the first insight into how OTKs can overcome the inhibitory effects of Dok-1. First, we show that OTKs trigger Dok-1 degradation via the ubiquitin-proteasome machinery. Second, we demonstrate that the levels of Dok-1 critically influence the ability of OTKs to promote cell proliferation and induce oncogenic transformation. In particular, we found that restoration of Dok-1 protein levels in p210bcr-abl- and oncogenic Src-expressing cells strongly attenuates the proliferation rate and transformation frequency of these cells. Finally, we show that a lysineless Dok-1 mutant, which is largely refractory to OTK-induced polyubiquitination and degradation, is even more efficient in suppressing OTK-mediated cell transformation. Thus, our data support a model in which ubiquitin-mediated proteasomal degradation of Dok-1 constitutes a mechanism by which OTKs overcome the inhibitory effect of Dok-1, thereby facilitating or promoting oncogenic transformation. Notably, as mentioned above, the level of p210bcr-abl expression has been shown to increase during disease progression, and it is thought to promote secondary molecular and genetic changes essential for transitioning from the chronic phase to the blast crisis phase (57, 74). We observed an inverse correlation between the levels of Dok-1 and those of p210bcr-abl, raising the intriguing possibility that proteasomal degradation of Dok-1 coincides with the progression of CML. Precedence for p210bcr-abl-induced dose-dependent inactivation of negative regulators during CML progression comes, for instance, from studies of the tumor suppressor PP2A. Increasing p210bcr-abl expression was shown to induce an increase in the expression of the phosphoprotein SET, a negative regulator of PP2A, which in turn causes a decrease in the activity of PP2A (62). Moreover, and importantly, the degree of PP2A inactivation reportedly correlates with the progression of CML, as the activity of this phosphatase is only modestly affected in chronic-phase CML progenitors, but is significantly impaired in myeloid cells derived from patients in the blast crisis phase of the disease (62). Thus, it is tempting to speculate that p210bcr-abl dose-dependent degradation of Dok-1 is an important contributive step in the progression from chronic phase to more advanced phases of CML. Similarly, downregulation of Dok-1 could contribute to the development of other tumors whose progression is associated with deregulated tyrosine kinases. Elevated and constitutive Src and Abl tyrosine kinase activities have been implicated in the progression of many cancers, especially breast, colorectal, prostate, and lung (1, 27, 37, 79). Future studies analyzing multiple tumor samples at different stages and grades are required to further assess the general contribution of Dok-1 downregulation to tumor development and progression associated with deregulated tyrosine kinase activity.
In summary, our findings provide new insight into mechanisms of Dok-1 regulation by OTKs. They reveal that OTKs target Dok-1 for ubiquitin-proteasome-mediated degradation and highlight the importance of elucidating the negative regulatory constraints on OTKs to fully understand the molecular mechanisms that mediate their transforming potential.
ACKNOWLEDGMENTS
We thank J. Skowronski and R. Sordella for their valuable discussions and critical reading of the manuscript. We also thank H. G. Wendel, R. Karni, W. Tansey, G. Q. Daley, and B. Clarkson for providing constructs and/or cell lines.
This work was supported by Public Health Service grant CA-135053 from the National Cancer Institute to L.V.A.
Footnotes
Published ahead of print on 2 May 2011.
REFERENCES
- 1. Abram C. L., Courtneidge S. A. 2000. Src family tyrosine kinases and growth factor signaling. Exp. Cell Res. 254:1–13 [DOI] [PubMed] [Google Scholar]
- 2. Bache K. G., Slagsvold T., Stenmark H. 2004. Defective downregulation of receptor tyrosine kinases in cancer. EMBO J. 23:2707–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barnes D. J., Schultheis B., Adedeji S., Melo J. V. 2005. Dose-dependent effects of Bcr-Abl in cell line models of different stages of chronic myeloid leukemia. Oncogene 24:6432–6440 [DOI] [PubMed] [Google Scholar]
- 4. Berger A. H., et al. 2010. Identification of DOK genes as lung tumor suppressors. Nat. Genet. 42:216–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berman E., et al. 2000. Characterization of two novel sublines established from a human megakaryoblastic leukemia cell line transfected with p210(BCR-ABL). Leuk. Res. 24:289–297 [DOI] [PubMed] [Google Scholar]
- 6. Bhat A., Johnson K. J., Oda T., Corbin A. S., Druker B. J. 1998. Interactions of p62(dok) with p210(bcr-abl) and Bcr-Abl-associated proteins. J. Biol. Chem. 273:32360–32368 [DOI] [PubMed] [Google Scholar]
- 7. Blume-Jensen P., Hunter T. 2001. Oncogenic kinase signalling. Nature 411:355–365 [DOI] [PubMed] [Google Scholar]
- 8. Boettner B., Govek E. E., Cross J., Van Aelst L. 2000. The junctional multidomain protein AF-6 is a binding partner of the Rap1A GTPase and associates with the actin cytoskeletal regulator profilin. Proc. Natl. Acad. Sci. U. S. A. 97:9064–9069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bose R., et al. 2006. Phosphoproteomic analysis of Her2/neu signaling and inhibition. Proc. Natl. Acad. Sci. U. S. A. 103:9773–9778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bradshaw R. A., Brickey W. W., Walker K. W. 1998. N-terminal processing: the methionine aminopeptidase and N alpha-acetyl transferase families. Trends Biochem. Sci. 23:263–267 [DOI] [PubMed] [Google Scholar]
- 11. Cadwell K., Coscoy L. 2005. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science 309:127–130 [DOI] [PubMed] [Google Scholar]
- 12. Caron C., Boyault C., Khochbin S. 2005. Regulatory cross-talk between lysine acetylation and ubiquitination: role in the control of protein stability. Bioessays 27:408–415 [DOI] [PubMed] [Google Scholar]
- 13. Carpino N., et al. 1997. p62(dok): a constitutively tyrosine-phosphorylated, GAP-associated protein in chronic myelogenous leukemia progenitor cells. Cell 88:197–204 [DOI] [PubMed] [Google Scholar]
- 14. Ciechanover A., Ben-Saadon R. 2004. N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 14:103–106 [DOI] [PubMed] [Google Scholar]
- 15. Cong F., Yuan B., Goff S. P. 1999. Characterization of a novel member of the DOK family that binds and modulates Abl signaling. Mol. Cell. Biol. 19:8314–8325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Crowder R. J., Enomoto H., Yang M., Johnson E. M., Jr., Milbrandt J. 2004. Dok-6, a novel p62 Dok family member, promotes Ret-mediated neurite outgrowth. J. Biol. Chem. 279:42072–42081 [DOI] [PubMed] [Google Scholar]
- 17. Dai Z., et al. 1998. Oncogenic Abl and Src tyrosine kinases elicit the ubiquitin-dependent degradation of target proteins through a Ras-independent pathway. Genes Dev. 12:1415–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. DeClue J. E., Vass W. C., Johnson M. R., Stacey D. W., Lowy D. R. 1993. Functional role of GTPase-activating protein in cell transformation by pp60v-src. Mol. Cell. Biol. 13:6799–6809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Demers A., et al. 2009. A concerted kinase interplay identifies PPARgamma as a molecular target of ghrelin signaling in macrophages. PLoS One 4:e7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Di Cristofano A., et al. 1998. Molecular cloning and characterization of p56dok-2 defines a new family of RasGAP-binding proteins. J. Biol. Chem. 273:4827–4830 [DOI] [PubMed] [Google Scholar]
- 21. Di Cristofano A., et al. 2001. p62(dok), a negative regulator of Ras and mitogen-activated protein kinase (MAPK) activity, opposes leukemogenesis by p210(bcr-abl). J. Exp. Med. 194:275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dube N., Cheng A., Tremblay M. L. 2004. The role of protein tyrosine phosphatase 1B in Ras signaling. Proc. Natl. Acad. Sci. U. S. A. 101:1834–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ellis C., Moran M., McCormick F., Pawson T. 1990. Phosphorylation of GAP and GAP-associated proteins by transforming and mitogenic tyrosine kinases. Nature 343:377–381 [DOI] [PubMed] [Google Scholar]
- 24. Fry W. H., Kotelawala L., Sweeney C., Carraway K. L., III 2009. Mechanisms of ErbB receptor negative regulation and relevance in cancer. Exp. Cell Res. 315:697–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gaiger A., et al. 1995. Increase of bcr-abl chimeric mRNA expression in tumor cells of patients with chronic myeloid leukemia precedes disease progression. Blood 86:2371–2378 [PubMed] [Google Scholar]
- 26. Geoffroy M. C., Hay R. T. 2009. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat. Rev. Mol. Cell Biol. 10:564–568 [DOI] [PubMed] [Google Scholar]
- 27. Giaccone G., Zucali P. A. 2008. Src as a potential therapeutic target in non-small-cell lung cancer. Ann. Oncol. 19:1219–1223 [DOI] [PubMed] [Google Scholar]
- 28. Gorre M. E., et al. 2001. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science 293:876–880 [DOI] [PubMed] [Google Scholar]
- 29. Govek E. E., et al. 2004. The X-linked mental retardation protein oligophrenin-1 is required for dendritic spine morphogenesis. Nat. Neurosci. 7:364–372 [DOI] [PubMed] [Google Scholar]
- 30. Grimm J., et al. 2001. Novel p62dok family members, dok-4 and dok-5, are substrates of the c-Ret receptor tyrosine kinase and mediate neuronal differentiation. J. Cell Biol. 154:345–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guittard G., et al. 2009. Cutting edge: Dok-1 and Dok-2 adaptor molecules are regulated by phosphatidylinositol 5-phosphate production in T cells. J. Immunol. 182:3974–3978 [DOI] [PubMed] [Google Scholar]
- 32. Hendrie P. C., Miyazawa K., Yang Y. C., Langefeld C. D., Broxmeyer H. E. 1991. Mast cell growth factor (c-kit ligand) enhances cytokine stimulation of proliferation of the human factor-dependent cell line, M07e. Exp. Hematol. 19:1031–1037 [PubMed] [Google Scholar]
- 33. Honma M., et al. 2006. Dok-3 sequesters Grb2 and inhibits the Ras-Erk pathway downstream of protein-tyrosine kinases. Genes Cells 11:143–151 [DOI] [PubMed] [Google Scholar]
- 34. Hosomi Y., et al. 1994. Characterization of a 60-kilodalton substrate of the insulin receptor kinase. J. Biol. Chem. 269:11498–11502 [PubMed] [Google Scholar]
- 35. Hunter T. 2007. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell 28:730–738 [DOI] [PubMed] [Google Scholar]
- 36. Hunter T. 2009. Tyrosine phosphorylation: thirty years and counting. Curr. Opin. Cell Biol. 21:140–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Irby R. B., Yeatman T. J. 2000. Role of Src expression and activation in human cancer. Oncogene 19:5636–5642 [DOI] [PubMed] [Google Scholar]
- 38. Janas J., Skowronski J., Van Aelst L. 2006. Lentiviral delivery of RNAi in hippocampal neurons. Methods Enzymol. 406:593–605 [DOI] [PubMed] [Google Scholar]
- 39. Jones N., Dumont D. J. 1999. Recruitment of Dok-R to the EGF receptor through its PTB domain is required for attenuation of Erk MAP kinase activation. Curr. Biol. 9:1057–1060 [DOI] [PubMed] [Google Scholar]
- 40. Jones N., Dumont D. J. 1998. The Tek/Tie2 receptor signals through a novel Dok-related docking protein, Dok-R. Oncogene 17:1097–1108 [DOI] [PubMed] [Google Scholar]
- 41. Kerscher O., Felberbaum R., Hochstrasser M. 2006. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22:159–180 [DOI] [PubMed] [Google Scholar]
- 42. Kirisits A., Pils D., Krainer M. 2007. Epidermal growth factor receptor degradation: an alternative view of oncogenic pathways. Int. J. Biochem. Cell Biol. 39:2173–2182 [DOI] [PubMed] [Google Scholar]
- 43. Klucher K. M., Lopez D. V., Daley G. Q. 1998. Secondary mutation maintains the transformed state in BaF3 cells with inducible BCR/ABL expression. Blood 91:3927–3934 [PubMed] [Google Scholar]
- 44. Kobayashi R., Patenia R., Ashizawa S., Vykoukal J. 2009. Targeted mass spectrometric analysis of N-terminally truncated isoforms generated via alternative translation initiation. FEBS Lett. 583:2441–2445 [DOI] [PubMed] [Google Scholar]
- 45. Kolch W., Pitt A. 2010. Functional proteomics to dissect tyrosine kinase signalling pathways in cancer. Nat. Rev. Cancer 10:618–629 [DOI] [PubMed] [Google Scholar]
- 46. Krause D. S., Van Etten R. A. 2005. Tyrosine kinases as targets for cancer therapy. N. Engl. J. Med. 353:172–187 [DOI] [PubMed] [Google Scholar]
- 47. Laszlo G. S., Cooper J. A. 2009. Restriction of Src activity by Cullin-5. Curr. Biol. 19:157–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee Y., Broxmeyer H. E. 2001. Synergistic activation of RSK correlates with c-fos induction in MO7e cells stimulated with GM-CSF plus Steel factor. Biochem. Biophys. Res. Commun. 281:897–901 [DOI] [PubMed] [Google Scholar]
- 49. Lemay S., Davidson D., Latour S., Veillette A. 2000. Dok-3, a novel adapter molecule involved in the negative regulation of immunoreceptor signaling. Mol. Cell. Biol. 20:2743–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 51. Lugo T. G., Pendergast A. M., Muller A. J., Witte O. N. 1990. Tyrosine kinase activity and transformation potency of bcr-abl oncogene products. Science 247:1079–1082 [DOI] [PubMed] [Google Scholar]
- 52. Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. 2002. The protein kinase complement of the human genome. Science 298:1912–1934 [DOI] [PubMed] [Google Scholar]
- 53. Mashima R., Hishida Y., Tezuka T., Yamanashi Y. 2009. The roles of Dok family adapters in immunoreceptor signaling. Immunol. Rev. 232:273–285 [DOI] [PubMed] [Google Scholar]
- 54. Mashima R., et al. 2010. Mice lacking Dok-1, Dok-2, and Dok-3 succumb to aggressive histiocytic sarcoma. Lab. Invest. 90:1357–1364 [DOI] [PubMed] [Google Scholar]
- 55. Masson K., Ronnstrand L. 2009. Oncogenic signaling from the hematopoietic growth factor receptors c-Kit and Flt3. Cell. Signal. 21:1717–1726 [DOI] [PubMed] [Google Scholar]
- 56. McGillicuddy L. T., et al. 2009. Proteasomal and genetic inactivation of the NF1 tumor suppressor in gliomagenesis. Cancer Cell 16:44–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Melo J. V., Barnes D. J. 2007. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat. Rev. Cancer 7:441–453 [DOI] [PubMed] [Google Scholar]
- 58. Mihrshahi R., Barclay A. N., Brown M. H. 2009. Essential roles for Dok2 and RasGAP in CD200 receptor-mediated regulation of human myeloid cells. J. Immunol. 183:4879–4886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Moran M. F., Polakis P., McCormick F., Pawson T., Ellis C. 1991. Protein-tyrosine kinases regulate the phosphorylation, protein interactions, subcellular distribution, and activity of p21ras GTPase-activating protein. Mol. Cell. Biol. 11:1804–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Neet K., Hunter T. 1995. The nonreceptor protein-tyrosine kinase CSK complexes directly with the GTPase-activating protein-associated p62 protein in cells expressing v-Src or activated c-Src. Mol. Cell. Biol. 15:4908–4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nelms K., Snow A. L., Hu-Li J., Paul W. E. 1998. FRIP, a hematopoietic cell-specific rasGAP-interacting protein phosphorylated in response to cytokine stimulation. Immunity 9:13–24 [DOI] [PubMed] [Google Scholar]
- 62. Neviani P., et al. 2005. The tumor suppressor PP2A is functionally inactivated in blast crisis CML through the inhibitory activity of the BCR/ABL-regulated SET protein. Cancer Cell 8:355–368 [DOI] [PubMed] [Google Scholar]
- 63. Niki M., et al. 2004. Role of Dok-1 and Dok-2 in leukemia suppression. J. Exp. Med. 200:1689–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Noguchi T., et al. 1999. Tyrosine phosphorylation of p62(Dok) induced by cell adhesion and insulin: possible role in cell migration. EMBO J. 18:1748–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nowicki M. O., et al. 2003. Chronic myelogenous leukemia molecular signature. Oncogene 22:3952–3963 [DOI] [PubMed] [Google Scholar]
- 66. Okada K., et al. 2006. The muscle protein Dok-7 is essential for neuromuscular synaptogenesis. Science 312:1802–1805 [DOI] [PubMed] [Google Scholar]
- 67. Oki S., et al. 2005. Dok1 and SHIP act as negative regulators of v-Abl-induced pre-B cell transformation, proliferation and Ras/Erk activation. Cell Cycle 4:310–314 [PubMed] [Google Scholar]
- 68. Palacios R., Steinmetz M. 1985. Il-3-dependent mouse clones that express B-220 surface antigen, contain Ig genes in germ-line configuration, and generate B lymphocytes in vivo. Cell 41:727–734 [DOI] [PubMed] [Google Scholar]
- 69. Peng C., et al. 2010. PTEN is a tumor suppressor in CML stem cells and BCR-ABL-induced leukemias in mice. Blood 115:626–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Polevoda B., Sherman F. 2003. N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J. Mol. Biol. 325:595–622 [DOI] [PubMed] [Google Scholar]
- 71. Porter A. C., Vaillancourt R. R. 1998. Tyrosine kinase receptor-activated signal transduction pathways which lead to oncogenesis. Oncogene 17:1343–1352 [DOI] [PubMed] [Google Scholar]
- 72. Rodrigues G. A., Park M. 1994. Oncogenic activation of tyrosine kinases. Curr. Opin. Genet. Dev. 4:15–24 [DOI] [PubMed] [Google Scholar]
- 73. Sabe H., Okada M., Nakagawa H., Hanafusa H. 1992. Activation of c-Src in cells bearing v-Crk and its suppression by Csk. Mol. Cell. Biol. 12:4706–4713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Savona M., Talpaz M. 2008. Getting to the stem of chronic myeloid leukaemia. Nat. Rev. Cancer 8:341–350 [DOI] [PubMed] [Google Scholar]
- 75. Scheijen B., Griffin J. D. 2002. Tyrosine kinase oncogenes in normal hematopoiesis and hematological disease. Oncogene 21:3314–3333 [DOI] [PubMed] [Google Scholar]
- 76. Schindler T., et al. 2000. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science 289:1938–1942 [DOI] [PubMed] [Google Scholar]
- 77. Shah K., Shokat K. M. 2002. A chemical genetic screen for direct v-Src substrates reveals ordered assembly of a retrograde signaling pathway. Chem. Biol. 9:35–47 [DOI] [PubMed] [Google Scholar]
- 78. Shinohara H., Yasuda T., Yamanashi Y. 2004. Dok-1 tyrosine residues at 336 and 340 are essential for the negative regulation of Ras-Erk signalling, but dispensable for rasGAP-binding. Genes Cells 9:601–607 [DOI] [PubMed] [Google Scholar]
- 79. Sirvent A., Benistant C., Roche S. 2008. Cytoplasmic signalling by the c-Abl tyrosine kinase in normal and cancer cells. Biol. Cell 100:617–631 [DOI] [PubMed] [Google Scholar]
- 80. Songyang Z., Yamanashi Y., Liu D., Baltimore D. 2001. Domain-dependent function of the rasGAP-binding protein p62Dok in cell signaling. J. Biol. Chem. 276:2459–2465 [DOI] [PubMed] [Google Scholar]
- 81. Stork B., et al. 2007. Subcellular localization of Grb2 by the adaptor protein Dok-3 restricts the intensity of Ca2+ signaling in B cells. EMBO J. 26:1140–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Suzu S., et al. 2000. p56(dok-2) as a cytokine-inducible inhibitor of cell proliferation and signal transduction. EMBO J. 19:5114–5122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tait S. W., et al. 2007. Apoptosis induction by Bid requires unconventional ubiquitination and degradation of its N-terminal fragment. J. Cell Biol. 179:1453–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Titz B., et al. 2010. The proximal signaling network of the BCR-ABL1 oncogene shows a modular organization. Oncogene 29:5895–5910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Treier M., Staszewski L. M., Bohmann D. 1994. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78:787–798 [DOI] [PubMed] [Google Scholar]
- 86. Van Slyke P., et al. 2005. Dok-R mediates attenuation of epidermal growth factor-dependent mitogen-activated protein kinase and Akt activation through processive recruitment of c-Src and Csk. Mol. Cell. Biol. 25:3831–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wade M., Wang Y. V., Wahl G. M. 2010. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol. 20:299–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wang X., et al. 2007. Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J. Cell Biol. 177:613–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wang X., Jiang X. 2008. Post-translational regulation of PTEN. Oncogene 27:5454–5463 [DOI] [PubMed] [Google Scholar]
- 90. Wick M. J., Dong L. Q., Hu D., Langlais P., Liu F. 2001. Insulin receptor-mediated p62dok tyrosine phosphorylation at residues 362 and 398 plays distinct roles for binding GTPase-activating protein and Nck and is essential for inhibiting insulin-stimulated activation of Ras and Akt. J. Biol. Chem. 276:42843–42850 [DOI] [PubMed] [Google Scholar]
- 91. Wisniewski D., Strife A., Wojciechowicz D., Lambek C., Clarkson B. 1994. A 62-kilodalton tyrosine phosphoprotein constitutively present in primary chronic phase chronic myelogenous leukemia enriched lineage negative blast populations. Leukemia 8:688–693 [PubMed] [Google Scholar]
- 92. Woodring P. J., et al. 2004. c-Abl phosphorylates Dok1 to promote filopodia during cell spreading. J. Cell Biol. 165:493–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Yamanashi Y., Baltimore D. 1997. Identification of the Abl- and rasGAP-associated 62 kDa protein as a docking protein, Dok. Cell 88:205–211 [DOI] [PubMed] [Google Scholar]
- 94. Yasuda T., et al. 2007. Dok-1 and Dok-2 are negative regulators of T cell receptor signaling. Int. Immunol. 19:487–495 [DOI] [PubMed] [Google Scholar]
- 95. Yasuda T., et al. 2004. Role of Dok-1 and Dok-2 in myeloid homeostasis and suppression of leukemia. J. Exp. Med. 200:1681–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yoo Y., Ho H. J., Wang C., Guan J. L. 2010. Tyrosine phosphorylation of cofilin at Y68 by v-Src leads to its degradation through ubiquitin-proteasome pathway. Oncogene 29:263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Zhang Y., et al. 2004. Molecular basis of distinct interactions between Dok1 PTB domain and tyrosine-phosphorylated EGF receptor. J. Mol. Biol. 343:1147–1155 [DOI] [PubMed] [Google Scholar]
- 98. Zhao M., Janas J. A., Niki M., Pandolfi P. P., Van Aelst L. 2006. Dok-1 independently attenuates Ras/mitogen-activated protein kinase and Src/c-myc pathways to inhibit platelet-derived growth factor-induced mitogenesis. Mol. Cell. Biol. 26:2479–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zhao M., et al. 2001. Phosphoinositide 3-kinase-dependent membrane recruitment of p62(dok) is essential for its negative effect on mitogen-activated protein (MAP) kinase activation. J. Exp. Med. 194:265–274 [DOI] [PMC free article] [PubMed] [Google Scholar]