Abstract
More than 200 proteins associate with human spliceosomes, but little is known about their relative abundances in a given spliceosomal complex. Here we describe a novel two-dimensional (2D) electrophoresis method that allows separation of high-molecular-mass proteins without in-gel precipitation and thus without loss of protein. Using this system coupled with mass spectrometry, we identified 171 proteins altogether on 2D maps of stage-specific spliceosomal complexes. By staining with a fluorescent dye with a wide linear intensity range, we could quantitate and categorize proteins as present in high, moderate, or low abundance. Affinity-purified human B, Bact, and C complexes contained 69, 63, and 72 highly/moderately abundant proteins, respectively. The recruitment and release of spliceosomal proteins were followed based on their abundances in A, B, Bact, and C spliceosomal complexes. Staining with a phospho-specific dye revealed that approximately one-third of the proteins detected in human spliceosomal complexes by 2D gel analyses are phosphorylated. The 2D gel electrophoresis system described here allows for the first time an objective view of the relative abundances of proteins present in a particular spliceosomal complex and also sheds additional light on the spliceosome's compositional dynamics and the phosphorylation status of spliceosomal proteins at specific stages of splicing.
INTRODUCTION
The spliceosome is a highly complex and dynamic megadalton RNP machine. It is comprised of the five snRNPs U1, U2, U4, U5, and U6 and a large number of non-snRNP protein factors (reviewed in reference 42). Spliceosomes assemble de novo in a stepwise manner on each new intron to be spliced and thus pass through a series of distinct complexes (42). Initially, the U1 snRNP binds the pre-mRNA, forming the E complex, and after stable U2 snRNP interaction, the A complex is generated. Subsequently, the U4/U6 and U5 snRNPs associate, as part of the U4/U6.U5 tri-snRNP, and the precatalytic B complex is formed. Through a series of compositional and structural rearrangements, the B complex is activated, first yielding the Bact complex. After the action of the DEXH box protein Prp2, the B* complex is formed, which catalyzes step 1 of splicing. This involves cleavage at the 5′ splice site (ss) of the pre-mRNA and the ligation of the 5′ end of the intron to the so-called branch site to form a lariat-like structure. After the first step, the spliceosomal C complex is formed, and it catalyzes the step 2 of splicing, during which the intron is excised and the exons are ligated together to form mRNA.
Mass spectrometry (MS) analyses have shown that more than 200 proteins copurify with mixtures of human spliceosomal complexes (31, 47). Individual spliceosomal complexes contain many fewer proteins (e.g., ∼125 for B, Bact, and C complexes) and differ from each other considerably in composition (3, 6, 7, 10). However, the relative abundances of all of the proteins present within a given spliceosomal complex are presently not clear. Spliceosomes contain snRNP-associated and non-snRNP proteins, the latter consisting of, among others, members of the SR, hnRNP, PRP, and DEXH/D box protein families. Many non-snRNP spliceosomal proteins associate predominantly with a particular complex. For example, more than 30 non-snRNP proteins are recruited during the A-to-B-complex transition (3, 10), and ∼20 proteins during the transition from the B to Bact complex (6). Similarly, ∼30 non-snRNP proteins are recruited during the formation of the C complex. Interestingly, many of these C-complex-specific proteins do not contain homologues in yeast. The role, if any, of a large number of these non-snRNP spliceosome-associated proteins in splicing is currently unclear. Information about the relative abundances of these proteins would provide an initial indication as to which of them are bona fide spliceosomal proteins that likely play functional/structural roles during splicing. This information is essential in order to decide which of the numerous proteins of unknown function that associate with spliceosomes should be characterized in more detail. In addition, information about the relative abundances of proteins in a given spliceosomal complex is also relevant for deciding which of these complexes is the most homogeneous on the compositional level and thus most amenable to high-resolution structure studies.
Splicing not only involves dramatic changes in the composition of the spliceosome but also changes in the phosphorylation status of individual spliceosomal proteins. Indeed, reversible protein phosphorylation plays a key role during spliceosome assembly and the catalytic steps of splicing (reviewed in references 28 and 37). Several spliceosomal proteins are known to be phosphorylated, including members of the SR protein family, the U1-70K protein, and the U2-associated SF3b155 protein (8, 39, 40). In addition, several human tri-snRNP proteins, including hPrp28/100K, hPrp6/102K, and hPrp31/61K, are also phosphorylated, and their phosphorylation was linked to the stable integration of the U4/U6.U5 tri-snRNP during B complex formation (26, 35). In addition to these studies, MS studies of nuclear phosphoproteins in HeLa cells (2) indicated that additional spliceosomal proteins (including, among others, SF3b145 and U4/U6.U5-110K/SART1) are phosphorylated. However, whether they are also phosphorylated in the spliceosome and, if so, whether their phosphorylation status changes during splicing are currently not known in most cases. Indeed, insight into the dynamics of protein posttranslational modifications throughout the splicing process is currently very limited.
The lack of more quantitative data regarding proteins associating with different spliceosomal complexes is mainly a consequence of the limitations of those methods currently available. Although the number of peptides sequenced by MS for a given protein can provide information about its abundance when different complexes are compared (e.g., relative quantification through spectral count reference), this method has several shortcomings, in particular when analyzing small proteins (<25 kDa), and is not entirely reliable. Likewise, the emPAI (exponential modified protein abundance index), which is calculated from the experimentally observed and theoretically expected numbers of peptides of a protein detected by MS, is proportional (on average) to within 63% of the protein content and thus provides only a rough estimate of absolute protein amounts (19). Protein quantification via MS using stable isotope-labeled standard peptides is not amenable for analyzing complexes with more than 100 proteins (18, 34). Since these MS approaches have limitations, there is clearly a need for other methods to determine the stoichiometry of proteins in very large protein or RNP complexes.
Two-dimensional (2D) gel electrophoresis was used previously to characterize spliceosomal complexes (4, 15, 27, 29) and has the added advantage that visual inspection of the 2D protein maps affords a more rapid means to compare side by side the complexity and purity of various complexes. However, isoelectric focusing (IEF), which was used in these studies to separate proteins in the first dimension, leads to in-gel precipitation of proteins, since they are focused at high concentration close to electroneutrality (30). The tendency of proteins to precipitate increases dramatically with their molecular mass and prevents the separation of large proteins. Since the human spliceosome contains many proteins above 100 kDa, 2D electrophoresis has so far been of only limited value, mainly due to protein precipitation problems. Over 30 years ago, a 2D electrophoresis method was developed to analyze the ribosome (20). In contrast to IEF, where proteins reach zero charge, in this so-called Kaltschmidt-Wittmann system proteins remain charged throughout the procedure and are therefore less prone to in-gel precipitation. Since this original 2D system used to analyze ribosomal proteins was technically complicated, novel 2D systems which nonetheless follow this principle (i.e., preventing proteins from reaching zero charge) were developed (1, 22) and most recently allowed the reliable analysis of proteins associated with ribosomes without protein loss (1).
Here we have established a novel 2D electrophoresis system and demonstrated its applicability for characterizing spliceosomal proteins. Using this system, proteins with a wide range of isoelectric points (IEPs) and with masses greater than 300 kDa can be separated without in-gel precipitation. We used this system to analyze both spliceosomal snRNPs and various human spliceosomal complexes affinity purified under native conditions. By staining proteins with a fluorescent dye with a wide range of signal linearity, we have demonstrated that only approximately one-half of the proteins previously shown to be associated with a given human spliceosomal complex, as determined by MS, are present in stoichiometric or near-stoichiometric amounts. Our data also provide new insights into the compositional dynamics of the spliceosome and changes in the phosphorylation status of proteins during the splicing process. This system thus represents an important advance toward an objective appraisal of core/abundant components of the human spliceosome.
MATERIALS AND METHODS
Purification of human snRNPs, SR proteins, and spliceosomal complexes.
HeLa nuclear extract was prepared essentially as described previously (11). A mixture of spliceosomal snRNPs was isolated from HeLa nuclear extract via anti-m3G immunoaffinity purification (21). 12S U1 snRNPs and 25S U4/U6.U5 tri-snRNPs were fractionated on a 10 to 30% glycerol gradient (21). U1 snRNPs sedimenting in the 12S region of the gradient (together with 12S U2 snRNPs) were further purified using a heparin column (41) prior to 2D analysis. Tri-snRNPs peaking in the 25S region of the glycerol gradient were used without further purification. Native 17S U2 snRNPs were affinity purified from HeLa nuclear extract via anti-SF3a66 immunoaffinity chromatography essentially as described previously (43). SR proteins were isolated as previously described (46). Human spliceosomal B, Bact, and C complexes were affinity purified under physiological conditions (150 mM salt in the absence of heparin) via MS2 affinity selection as previously described (6, 7, 10). To purify A complexes via MS2 affinity selection, a splicing reaction incubated under conditions optimal for B complex formation was separated on a 10 to 30% glycerol gradient. The 40 to 45S peak of the gradient was used for the isolation of B complexes, whereas complexes peaking in the 30S region were used to isolate A complexes. The MINX pre-mRNA substrate was used for the isolation of A and B complexes (3, 10), and the PM5 pre-mRNA (or a truncated version thereof, termed PM5-20) was used for the isolation of C and Bact complexes, respectively. The MINX substrate contains a 5′ exon, intron, and 3′ exon plus MS2 aptamers. In order to stall splicing at the C complex stage, the PM5 pre-mRNA lacks a 3′ exon (and 3′ ss) and contains a long polypyrimidine tract plus MS2 aptamers (7). By truncating the latter to within 20 nucleotides (nt) of the branch site (PM5-20), human spliceosomes can also be stalled at the Bact stage (6). The purity of the B, Bact, and C complexes obtained was confirmed based on their snRNA composition and the presence of either unspliced pre-mRNA alone or the intermediates of the splicing reaction (6, 7, 10; data not shown). In the case of the B complex, the U1 snRNP was less abundant than all other spliceosomal snRNPs, consistent with previous results (6, 10).
Sample preparation for 2D gel electrophoresis.
Affinity-purified spliceosomal complexes or snRNPs (10 to 15 pmol) were concentrated by sedimentation at 700,000 × g and 4°C for 6 h. Pellets were resuspended in 20 μl of buffer containing 1 M NaCl, 2 mM MgCl2, and 0.1 mM EDTA in 50 mM HEPES-KOH (pH25 7.9) and incubated with 500 U of RNase T1 (Ambion), 0.5 μg RNase A (Ambion), and 50 U of RNase I (Ambion) for 1 h at 37°C. In order to achieve complete RNA digestion, the samples were incubated stepwise with 1 M, 2 M, 4 M and 7 M urea. Each step was followed by a 40-min incubation at 37°C. A fresh aliquot of RNases was added before incubation in the presence of 7 M urea. In order to prevent oxidation and subsequent protein loss during 2D electrophoresis, the sample was treated with iodoacetamide as described previously (1). In brief, samples were incubated in 10 mM β-mercaptoethanol for 30 min at 37°C, followed by the addition of 120 mM Tris-HCl (pH25 9.0) and 50 mM iodoacetamide. After incubation for 3 min at 37°C, unreacted iodoacetamide was quenched by adding 500 mM β-mercaptoethanol, and the pH was restored to 4 to 4.5 by addition of 4% (vol/vol) acetic acid (AcOH). The sample was dialyzed against 7 M urea, 2 M thiourea, 40 mM cysteamine, and 50 mM methyl ester of l-cysteine in 10 mM bis-Tris-acetate (OAc) (pH25 4.0). The sample was concentrated by dialysis against Slide-A-Lyser Concentration solution (Pierce) and loaded onto the first-dimension gel in 17% (vol/vol) AcOH. Typically, 3 pmol of a given spliceosomal complex was loaded. The volume of the loaded sample did not exceed 10 μl. Basic fuchsin was added to the sample as a leading dye.
First-dimension gel electrophoresis.
Electrophoresis in the first dimension was performed in 260-mm-long glass tubes with a 4-mm inner diameter. The separating gel contained 2% (wt/vol) acrylamide, 1.4% (wt/vol) methylene bisacrylamide, 7 M urea, 4 M thiourea, 40% (vol/vol) formamide in 40 mM Bis-Tris-OAc (pH25 5.7). Gel polymerization was induced by methylene blue as described in reference 23. The polymerized separating gel was overlaid by an ∼5-mm stacking gel. The latter, which contained the same components as the separating gel plus 5% (vol/vol) AcOH, was also polymerized using methylene blue and was overlaid with 50% (vol/vol) AcOH prior to polymerization. The lower electrode buffer was 10 mM Bis-Tris-OAc (pH25 6.0), and the upper electrode buffer was prepared just prior to use and consisted of 20 mM cysteamine and 1% (vol/vol) β-mercaptoethanol in 10 mM Bis-Tris-OAc (pH25 3.8). Electrophoresis was initially carried out at 0.125 mA per tube for 1 h, followed by 0.375 mA per tube for approximately 46 h at 15°C.
Second-dimension gel electrophoresis.
Electrophoresis in the second dimension was performed as described previously (33) with the following modifications. The separating gel contained a linear 6 to 12.5% acrylamide gradient that was prepared using ammonium persulfate (APS)-N,N,N′,N′-tetramethylethylenediamine (TEMED) for polymerization. The resulting gel was overlaid by approximately 15 mm of a 4% acrylamide stacking gel that was also polymerized with APS/TEMED. The first-dimension gel was placed on the stacking gel without equilibration of the buffer and sealed with the stacking gel with 7% (wt/vol) acrylamide and 0.23% (wt/vol) methylene bisacrylamide in 40 mM Bis-Tris-OAc (pH 5.7) polymerized using methylene blue. The cathode buffer contained 0.3% (wt/vol) SDS and 0.08% (vol/vol) mercaptoacetic acid. After electrophoresis, gels were fixed with 10% AcOH in 60% methanol.
2D electrophoresis of proteins with molecular masses less than 25 kDa.
To improve the resolution of smaller proteins, two modifications were introduced. First, the separating gel contained 4% (wt/vol) acrylamide and 0.15 (wt/vol) methylene bisacrylamide. Furthermore, the second-dimension gel consisted of a 10 to 15% acrylamide gradient.
Gel staining and mass spectrometry.
Gels were stained first for phosphoproteins with Pro-Q Diamond (Invitrogen) according to the manufacturer's instructions. Spot fluorescence was monitored by a FujiFilm FLA-7000 scanner, and phosphorylated proteins were separated into two groups according to spot intensity as determined by eye. Weakly stained proteins (barely over background fluorescence) are not included in Table 1. Gels were stained subsequently for total protein with Sypro Ruby (Invitrogen), also according to the manufacturer's instructions, with subsequent fluoroimaging. Finally, for mass spectrometry, gels were stained with silver essentially as described previously (36), except that the silver concentration was lowered to 6 mM and the developing step was performed with 190 mM sodium carbonate. Visible spots were cut out of the gel and digested with trypsin, and peptides were analyzed on a liquid chromatography (LC)-coupled ESI-LTQ-Orbitrap (ThermoFisherScientific) mass spectrometer under standard conditions, except that the separation time of the applied liquid chromatography gradient (7% to 38% solvent B) was shortened to 17 min, where solvent A was 0.1% (vol/vol) formic acid and solvent B consisted of 80% (vol/vol) acetonitrile and 0.1% formic acid. Proteins were identified by searching fragment spectra against the NCBI nonredundant (nr) database using Mascot as a search engine.
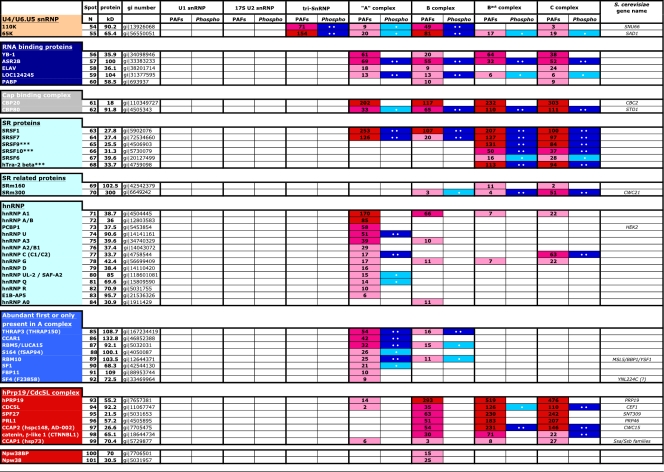
Table 1.
Abundances and phosphorylation statuses of proteins detected in the major spliceosomal snRNPs and spliceosomal A, B, Bact, and C complexesa
SnRNPs and spliceosomal complexes were purified, and their proteins were analyzed by 2D gel electrophoresis as described in Materials and Methods. A and B complexes were formed on MINX pre-mRNA, whereas Bact and C complexes were formed on PM5-20 and PM5 pre-mRNA, respectively. Proteins (common name given at the left) are grouped according to snRNP/spliceosomal complex association or function and are followed by the number of their corresponding spot on the 2D gels. The detection of a given protein is indicated by a number (or “+”) which represents its protein abundance factor (PAF) value. The PAF values shown are the averages of multiple determinations. They were determined by staining with Sypro Ruby and represent staining intensity (in linear arbitrary units) divided by the molecular mass. Proteins with PAF values >75 (indicating that a protein is highly abundant) are highlighted in red, those with PAF values from 30 to 75 (indicating moderate abundance) are highlighted in magenta, and those with values <30 (indicating low abundance) are shown in pink. Phosphorylated proteins are indicated in the lanes right of those showing PAF values by one (highlighted in light blue) or two (highlighted in purple) dots based on their relative Pro-Q Diamond staining intensity, where use of one and two dots indicates moderate or strong phosphorylation, respectively. NF, not found. *, SmB and its isoform SmB′ migrate as distinct spots but are designated by one spot number, and only one PAF value for both was determined. **, due to its highly basic nature, SmD3 runs out of the first-dimension gel. ***, these proteins are also abundant in A and/or B complexes formed on the PM5 pre-mRNA. ****, acinus is visible on the 2D gels only if the fluorescence intensity is greatly increased.
Calculation of the PAF.
The fluorescent images of equally loaded gels, obtained with scanner settings such that all spot intensities were in the linear range, were analyzed by the Fujifilm Science Lab 2003 Image Gauge software program, version 4.22. The linear concentration range was initially determined by analyzing several different amounts of a given spliceosomal complex after staining with Sypro Ruby. Individual protein spots were encircled manually, and the corresponding value of total intensity in linear arbitrary units (LAU) was determined. The background value of intensity in LAU was determined using the very same shape used for quantification of a spot containing a protein but quantitating an area of the gel lacking any protein spots, and this value was deducted from the individual protein spot intensity. The resulting value was divided by the protein molecular mass in kDa to generate the protein abundance factor (PAF). The results were reproduced in at least three independent 2D electrophoretic separations with at least two independently purified spliceosomal complexes/snRNPs. PAF values for a given protein varied maximally by ±20%.
RESULTS
Establishment of a novel 2D electrophoresis system.
To characterize the composition of human spliceosomes on a more quantitative basis, we set out to establish a 2D electrophoresis system suited for the separation of spliceosomal proteins. We initially based our method on a recent 2D system used to analyze ribosomal proteins which employs charge-driven electrophoresis (as opposed to IEF) (1). However, under these conditions, separation of proteins from the U4/U6.U5 tri-snRNP resulted in extensive precipitation of the high-molecular-mass U5 proteins (i.e., U5-220K, U5-200K, and U5-116K) at the edge of the first-dimension gel (see Fig. S1A in the supplemental material). Similar precipitation problems were also observed when proteins from human spliceosomal B complexes (see Fig. S1B) or HeLa nuclear extract were analyzed (data not shown).
We thus optimized/changed several parameters to improve protein solubility during the electrophoresis run in the first dimension (see Fig. S2 in the supplemental material). These included the following: (i) increasing the gel pore size by decreasing the amount of acrylamide and increasing the cross-linker methylene-bisacrylamide (MBA) (32), (ii) using different hydro-organic solvents (e.g., dimethyl sulfoxide [DMSO] and formamide) in which the gel was prepared, and (iii) using a wide variety of powerful protein denaturants, including thiourea. For polypeptides above 25 kDa, the best separation in the first dimension was observed when electrophoresis was carried out under strongly denaturing conditions at acidic pH in a gel matrix with large pores (i.e., when 2% acrylamide-1.4% methylene bisacrylamide was used). Under these conditions, polymerization could not be initiated with the commonly used TEMED-persulfate but instead required photopolymerization with methylene blue (23). The best results were obtained with gels containing 7 M urea, 4 M thiourea, and 40% (wt/vol) formamide. Similar conditions were used to analyze smaller proteins (i.e., below 25 kDa), except that the size of the gel pores was reduced. In the second dimension, proteins were separated on a linear 6 to 12.5% or 10 to 15% (for proteins under 25 kDa) polyacrylamide gradient gel. Using this system, proteins with a wide range of isoelectric points (IEPs) and with masses up to 300 kDa could be separated without in-gel protein precipitation.
2D gel analyses of spliceosomal snRNPs.
To test the reliability of our 2D gel system, we first analyzed the major spliceosomal snRNPs (12S U1 and 17S U2 snRNPs and the 25S U4/U6.U5 tri-snRNP), whose protein compositions are generally well documented. After extensive RNase treatment, RNA-free protein samples were separated by 2D gel electrophoresis. The gel was then stained consecutively for phosphoproteins (Pro-Q Diamond), for all proteins (Sypro Ruby), and last, to identify proteins via LC-tandem MS (LC-MSMS), the gel was stained with silver to enable spots to be visualized and excised from the gel. The fluorescent dye Sypro Ruby has a wide (more than 3 orders of magnitude) linear signal intensity range (5), and thus it is particularly useful for quantitating the relative amounts of proteins within an snRNP or spliceosomal complex. For semiquantitative comparison of protein composition, we calculated the protein abundance factor (PAF), which is defined as the single-spot intensity in linear arbitrary units (LAU) divided by the protein's molecular mass (in order to compensate for the difference in staining for large versus small proteins) (Table 1). On the basis of their PAF values, proteins identified were divided into three groups: (i) abundant proteins (PAF > 75), meaning that the protein is likely present in at least one copy per complex, (ii) proteins present in moderate amounts (PAF of 30 to 75), and (iii) proteins present in small amounts (PAF < 30). This last group may include loosely associated proteins that are partially lost during purification or proteins that simply associate with only a small subset of snRNPs or spliceosomal complexes. In the case of the U1, U2, and U4/U6.U5 snRNPs, only predominant spots (after silver staining) were analyzed by MS; thus, protein components present in small amounts were not analyzed.
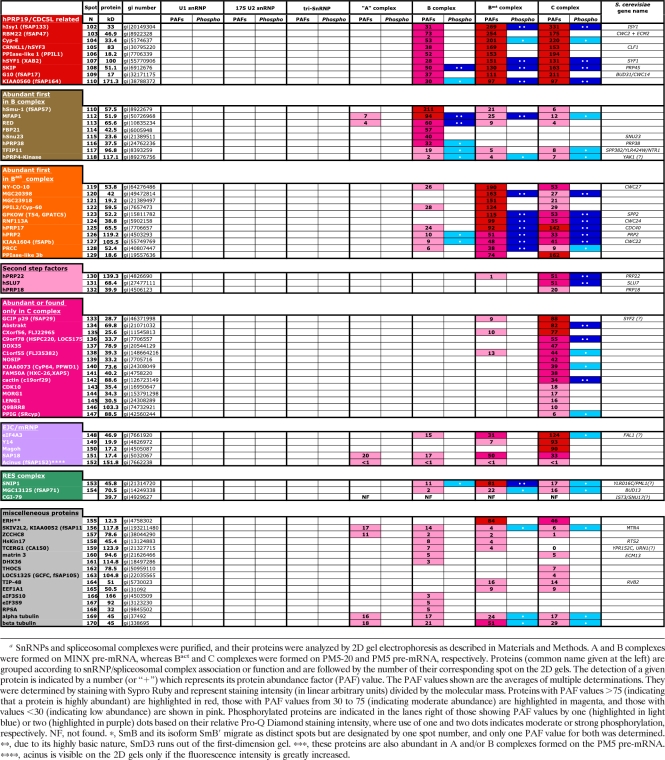
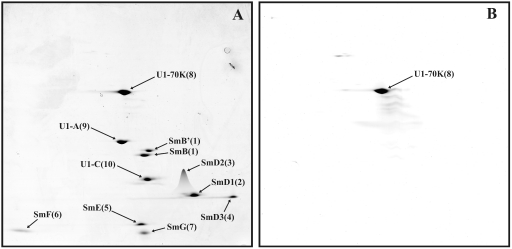
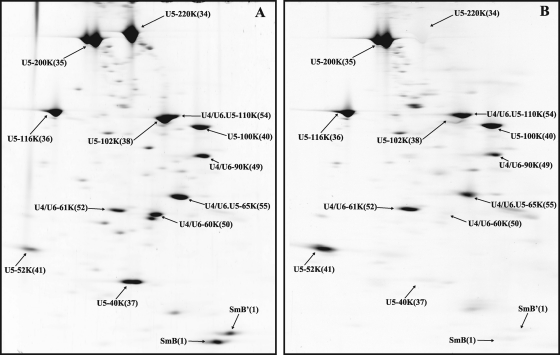
When human U1 snRNPs were analyzed, all known U1-associated proteins (i.e., 70K, A, C, and the seven Sm proteins) were clearly visible in predominant, well-defined spots (with little or no smearing) (Fig. 1A). As expected, the aforementioned proteins were found to be highly abundant components of the U1 snRNP based on their PAFs (Table 1). Significantly, the U1-70K protein, which is known to be phosphorylated at multiple sites and, upon IEF of U1 snRNP proteins, migrates as 11 to 13 different spots (44), is present in a single spot in our 2D gels. Staining with Pro-Q Diamond confirmed that U1-70K is indeed phosphorylated (Fig. 1B). Thus, the migration of this phosphoprotein is independent of its degree of phosphorylation. This indicates that our 2D system is also suited for analyzing phosphoproteins and thus for studying the dynamics of protein phosphorylation during splicing. 2D analyses of human U4/U6.U5 tri-snRNPs showed that all known components of the tri-snRNP, with the exception of the 27K protein (which, due to its highly basic pI [reference 13], runs out of the first-dimension gel), are abundant components (Fig. 2 and Table 1; also data not shown). Slightly lower PAFs were found for the LSm5, LSm8, 61K/hPrp31, and 110K/hSnu66 proteins. In contrast, U5-40K exhibited a very high PAF value, suggesting it is present in more than one copy.
Fig. 1.
2D gel electrophoresis of human U1 snRNP proteins. U1 snRNPs were purified as described in Materials and Methods, and U1-associated proteins were separated by 2D gel electrophoresis using the conditions for the analysis of low-molecular-mass proteins and then stained with Sypro Ruby (A) or with Pro-Q Diamond to detect phosphoproteins (B). Spots were visualized by fluoroimaging. The major spots visible after silver staining were analyzed by mass spectrometry, and the identified proteins are indicated by their common name and a number subsequently used to identify them in Fig. 4 to 8 (see also Table 1).
Fig. 2.
2D gel electrophoresis of human U4/U6.U5 tri-snRNP proteins. Tri-snRNPs were purified as described in Materials and Methods, and their associated proteins were separated by 2D gel electrophoresis under conditions used for high-molecular-mass proteins and analyzed as described for Fig. 1. (A) Sypro Ruby staining. (B) Pro-Q Diamond staining. Note that the tri-snRNPs analyzed here are not highly pure, and thus multiple minor spots, which represent contaminants, are also visible.
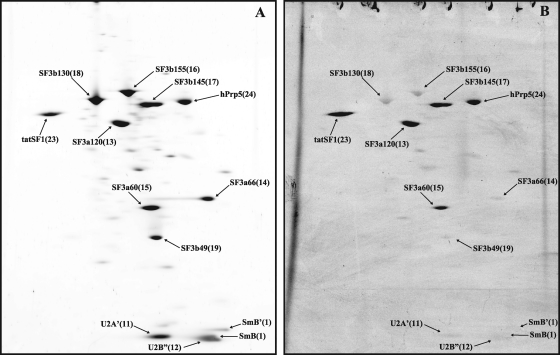
Similar analyses were performed with immunoaffinity-purified 17S U2 snRNPs (Fig. 3; for proteins of less than 25 kDa in mass, data not shown). As expected, in addition to the Sm proteins, U2-A′ and U2-B″ and all subunits of SF3a and SF3b were abundant. Previous MS analyses of purified human 17S U2 snRNPs revealed the presence of several additional proteins (denoted “17S U2 related” in Table 1), but their abundance in the 17S U2 snRNP was not clear (43). Significantly, most of these proteins, with the exception of hPrp5 and tat-SF1, are apparently present in very small amounts, based on the fact that they were not among those proteins identified by MS in the moderately to intensely stained spots on the 2D gel (Fig. 3A; Table 1). Interestingly, tat-SF1, which previously escaped detection by MS in human 17S U2 snRNPs (43), is the human homologue of the CUS2 protein in the yeast Saccharomyces cerevisiae, where it is thought to facilitate proper folding of the yeast U2 snRNA (45). Taken together, these results demonstrate the power of this method for distinguishing between core components of a complex and those that are present in substoichiometric amounts.
Fig. 3.
2D gel electrophoresis of human 17S U2 snRNP proteins. 17S U2 snRNPs were purified as described in the Materials and Methods and their associated proteins were separated by 2D gel electrophoresis under conditions used for high molecular mass proteins and analyzed as described for Fig. 1. (A) Sypro Ruby staining. (B) Pro-Q Diamond staining.
Affinity purification of stage-specific human spliceosomal complexes.
To characterize the relative abundances of spliceosomal proteins associated at different stages of the splicing process, we affinity purified human A, B, Bact, and C complexes formed under splicing conditions in HeLa nuclear extract (3, 6, 7, 10). All complexes were first subjected to glycerol gradient centrifugation, and gradient fractions containing the respective peak of each complex were then subjected to MS2 affinity selection under physiological conditions. The purity of the B, Bact, and C complexes obtained was confirmed based on their RNA composition. Pure A complexes, in contrast, are difficult to obtain in preparative amounts (3). Analysis of the RNA composition of complexes purified from the 30S gradient peak that we designate A complex indicated that they consist predominantly (∼70%) of A complexes (with equimolar amounts of unspliced pre-mRNA, U1 and U2 snRNA) but also contain H complexes (pre-mRNA complexed with general RNA binding proteins) and traces of B complexes (data not shown).
2D gel analyses of purified spliceosomal complexes.
Sypro Ruby staining of the 2D maps of proteins associated with partially purified A complexes (i.e., the 30S gradient peak preceding the B complex peak) and affinity-purified B complexes revealed ∼105 and ∼145 distinct spots, respectively, that varied considerably in intensity (Fig. 4A and 5A and Table 2; see also Fig. 8). Of these, 90 proteins (in A complexes) and 116 proteins (in B complexes) were detectable by silver staining and subsequently could be identified by MS. Approximately 127 and 136 distinct spots were observed upon Sypro Ruby staining of the 2D maps of Bact and C complexes, respectively (Fig. 6A, 7A, and 8; Table 2), with 100 (Bact) and 116 (C complex) proteins subsequently identified by MS. The additional spots detected by Sypro Ruby staining were generally much less intense and either were proteolytic fragments of major proteins or were proteins present in very small amounts and thus not detected by silver staining (and therefore not analyzed by MS). In previous studies, proteins copurifying with a given spliceosomal complex were separated by one-dimensional (1D) SDS-PAGE, and entire lanes of the gel were analyzed; thus, even proteins not visible by silver staining were analyzed by MS, leading to the identification of proteins present in extremely small amounts. In a very limited number of cases, two proteins overlapped on the 2D maps, e.g., Prp2 (spot 126) and KIAA1604 (spot 127) or 61K (spot 52) and SF3a60 (spot 15), leading to uncertainties in the PAF values calculated for each individual protein.
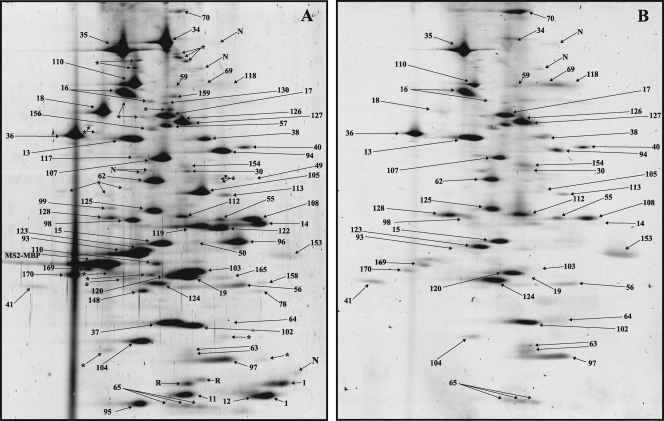
Fig. 4.
2D gel electrophoresis of partially purified human spliceosomal A complexes. Spliceosomal complexes were allowed to form on MINX pre-mRNA and then purified via MS2 affinity selection. Proteins were separated by 2D gel electrophoresis using the conditions for the analysis of high-molecular-mass proteins (see Fig. 8A for analysis of spliceosomal proteins with molecular masses less than 25 kDa) and then stained with Sypro Ruby (A) or with Pro-Q Diamond (B). Spots were visualized by fluoroimaging. All spots visible after silver staining were analyzed by mass spectrometry, and the identified proteins are indicated by a number (see Table 1 for the identities of the numbered spots). MS2-MBP (present in ∼6 copies per complex) is labeled. Spots containing proteolytic fragments are indicated by an asterisk, those not identified are indicated by “N,” and those corresponding to RNases are indicated by “R.”
Fig. 5.
2D gel electrophoresis of affinity-purified human spliceosomal B complexes. Spliceosomal complexes were allowed to form on MINX pre-mRNA and then purified and analyzed by 2D gel electrophoresis as described for Fig. 4. (A) Sypro Ruby staining. (B) Pro-Q Diamond staining.
Table 2.
Protein spots detected by 2D gel electrophoresis in spliceosomal complexes
| Complex | Total no. of spots (Sypro Ruby) | No. of protein fragmentation spots | No. of unique proteins identified by MS (silver) | No. of highly/ moderately abundant proteins |
|---|---|---|---|---|
| “A” complex | 105 | 11 | 90 | 60 |
| B complex | 145 | 25 | 116 | 69 |
| Bact complex | 127 | 20 | 100 | 63 |
| C complex | 136 | 18 | 116 | 72 |
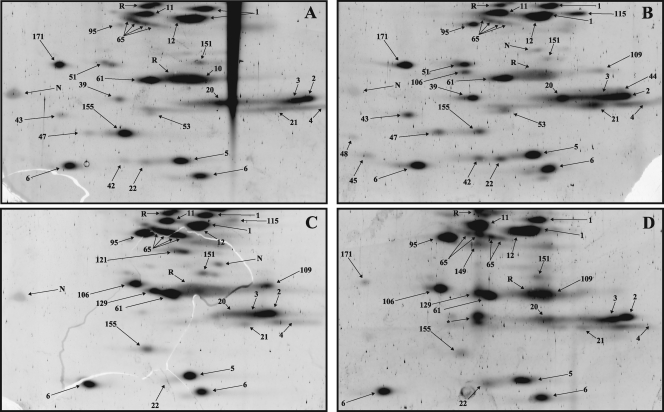
Fig. 8.
2D gel electrophoresis of low-molecular-mass proteins from affinity-purified spliceosomal A (A), B (B), Bact (C), and C (D) complexes. Spliceosomal complexes formed on PM5-based pre-mRNA were analyzed by 2D gel electrophoresis using conditions optimal for the separation of proteins under 25 kDa in mass, and spots were visualized by staining with Sypro Ruby. None of the low-molecular-mass proteins were detected at levels significantly above background upon staining with Pro-Q Diamond (data not shown). Spot no. 171 corresponds to ALG-2/PDCD6 (gi∣121948367) and was abundant only in B complexes formed on the PM5 pre-mRNA.
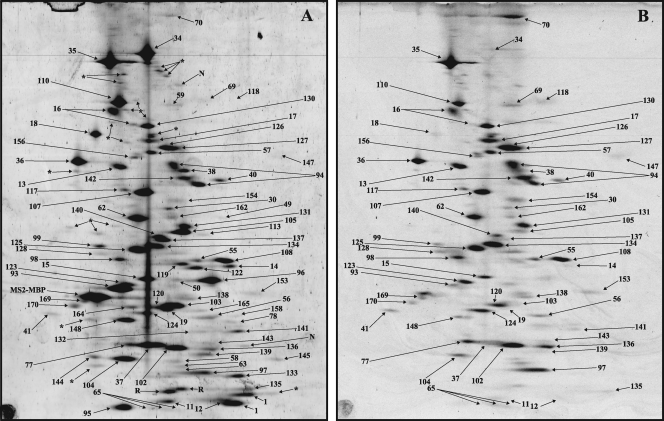
Fig. 6.
2D gel electrophoresis of affinity-purified human spliceosomal Bact complexes. Spliceosomal complexes were allowed to form on PM5-20 pre-mRNA and then purified and analyzed by 2D gel electrophoresis as described for Fig. 4. (A) Sypro Ruby staining. (B) Pro-Q Diamond staining.
Fig. 7.
2D gel electrophoresis of affinity-purified human spliceosomal C complexes. Spliceosomal complexes were allowed to form on PM5 pre-mRNA and then purified and analyzed by 2D gel electrophoresis as described for Fig. 4. (A) Sypro Ruby staining. (B) Pro-Q Diamond staining.
Protein composition of the human A complex.
Proteins associated with U1 (70K, A, and C) and U2 (all SF3a/b subunits and A′ and B″) plus the Sm proteins were found to be highly abundant in our partially purified A complexes, as evidenced by the intensity of their individual spots on the 2D gels (Fig. 4A and 9 ; see Table 1 for the identity of each spot) and based on their calculated PAF values (Table 1). Tri-snRNP proteins were also identified but can be attributed to the small amounts of contaminating B complexes present in the A complex preparation. Most U2-related proteins are abundant components of A complexes, exhibiting moderate to high PAF values (Table 1). Interesting exceptions are hPrp5 and tat-SF1, which are present in small amounts or entirely absent in our affinity-purified A complexes despite being major components of the 17S U2 snRNP. Thus, they appear to dissociate for the most part upon association of U2 with the pre-mRNA. The majority of non-snRNP proteins reproducibly identified previously in purified A complexes by MS (3) were also found after 2D analyses (Table 1, “Abundant first in A complex,“ “RNA binding,” and “hnRNP” or “SR” proteins). A large number of hnRNP proteins with various abundances were detected in our partially purified A complexes. However, since most of them are no longer present in later spliceosomal complexes, it is likely that the majority of hnRNP proteins identified are present solely in the H complexes that contaminate the A complex preparation.
Fig. 9.
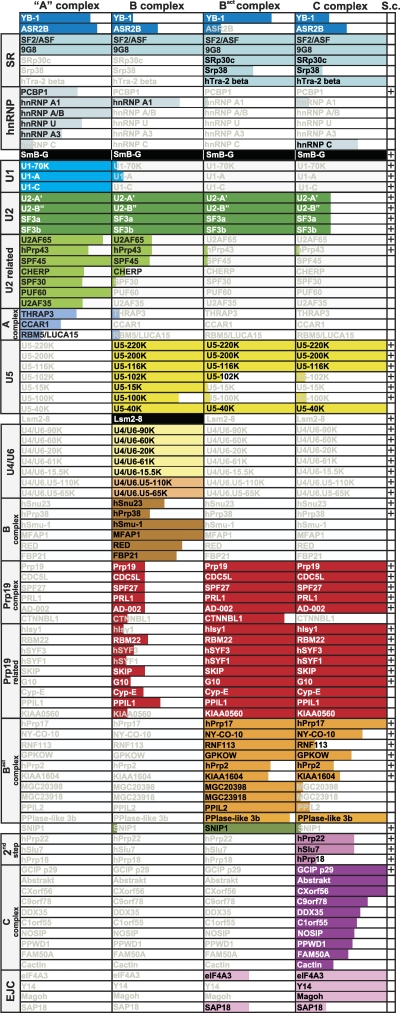
Major components of the human A, B, Bact, and C spliceosomal complexes. Only proteins with PAF values above 30 are shown. Note that CBP20/CBP80, as well as individual Sm, LSm, SF3a, and SF3b proteins, are not shown (see also Table 1). The extent of color shading in each column reflects the abundance of each protein, where the entire box is colored for proteins with PAFs above 75. Note that SRSF9, SRSF10, and hTra-2 beta are abundant in A and/or B complexes formed on PM5 but not on MINX pre-mRNA. In the “S.c.” column, a “+” indicates that a homolog is present in the yeast S. cerevisiae.
Only a subset of the SR proteins previously detected by MS in purified A complexes formed on the MINX pre-mRNA were identified by our 2D analyses, namely, SRSF1 and SRSF7. In addition to these last two SR proteins, A complexes (as well as later complexes) formed on the PM5 pre-mRNA substrate additionally contained SRSF9, hTra-2 beta, and SRSF6 (data not shown) (Table 1); SRSF10 was also found solely in complexes formed on PM5 pre-mRNA but was detected first in the B complex. Thus, some SR proteins bind in a pre-mRNA substrate-specific manner. All of the SR proteins identified in A complexes (with the exception of SRSF6) were highly abundant and remained associated with the spliceosome throughout the first catalytic step of splicing (Table 1). To test whether the missing SR proteins (e.g., SRSF3, SRSF2, and SRSF4) are truly absent in the spliceosomal complexes analyzed here or simply cannot be detected via our 2D gel system, we purified a mixture of SR proteins from HeLa nuclear extract and analyzed them using the 2D gel conditions used to identify proteins with small molecular masses. SRSF3, SRSF1, SRSF2, SRSF10, SRSF6, and SRSF4 could be identified (see Fig. S3 in the supplemental material), and thus the lack of detection of SRSF3, SRSF2, and SRSF4 in the various spliceosomal complexes indicates that they are indeed absent or are present in very small amounts.
Protein composition of the human precatalytic B complex.
Essentially all U1 and U2 snRNP and U4/U6-U5 tri-snRNP proteins were identified on the 2D map of the B complex, consistent with its snRNA composition. The most abundant proteins (i.e., PAF > 75) were 17S U2 snRNP proteins, the tri-snRNP proteins (including LSm proteins 2 to 8), and several splicing factors (e.g., MFAP, Smu-1, and SF2/ASF). These proteins probably represent the structural core of the B complex. Compared to findings for the A complex, the U1 snRNP proteins are dramatically reduced in the B complex (Table 1), consistent with the reduced levels of U1 snRNA present (data not shown). All proteins of the hPrp19/CDC5L complex and those proteins operationally defined as Prp19 related are found in moderate amounts in purified B complexes; since they were for the most part absent in A complexes, it is clear that they are first recruited at the time of B complex formation. Several additional non-snRNP factors (Table 1, “Abundant first in B”), such as FBP21, RED, hSmu23, and hPrp38, were found in moderate amounts. U2-related proteins were generally less abundant in B complexes than in A (Fig. 5A and Table 1). Similarly, proteins denoted “abundant or only present in A complexes” (e.g., CCAR1, RBM5, SF1, FBP11, and SF4) were less abundant or not detected in the B complex, indicating that they dissociate completely or are further destabilized during the A-to-B-complex transition. Taken together, our data indicate that numerous non-snRNP proteins previously detected in purified B complexes by MS are present in very small amounts and thus unlikely to play functional/structural roles in the spliceosome at this stage.
Protein composition of the human Bact spliceosomal complex.
The most abundant Bact proteins (i.e., PAF > 75) included all 17S U2 snRNP proteins, a subset of U5 proteins (220K, 200K, 116K, and 40K), and most proteins of the Prp19/CDC5L complex and Prp19-related proteins (Table 1). Since the Prp19 complex and related proteins were less abundant in the B complex, it can be concluded that they are more stably associated with the spliceosome first at the time of its activation, as suggested by previous studies (6, 7, 24). Prp19 itself has the highest PAF (i.e., 519) of all spliceosomal proteins, suggesting more than one copy is present per complex, which correlates well with recent data suggesting it is present in the human Prp19 complex as a tetramer (17). Consistent with the loss of U1 and U4 snRNA during spliceosome activation, U1, U4/U6, and tri-snRNP-specific proteins are absent or strongly underrepresented in the Bact complex (Fig. 6A and 8C; Table 1). U2-related proteins and the group of proteins designated “Abundant first in A complex” are also for the most part no longer detected. Also, those proteins designated “Abundant first in B complex” are also much less abundant in the Bact complex, indicating that they dissociate or are destabilized during spliceosome activation. The 2D analysis of the Bact complex also revealed that at this stage a specific set of proteins is recruited or is more abundant (Table 1, “Abundant first in Bact”). Most of these proteins are present in stoichiometric amounts, and with the exception of KIAA1604 and Prp17, they are less abundant in the C complex as judged by their PAF values (Table 1), suggesting that they are stably associated specifically during activation. Several proteins were previously found by MS in purified Bact complexes, albeit with very low peptide numbers (6). The vast majority of these were not detected by our 2D system, confirming that they are present in negligible amounts in the human Bact complex.
Protein composition of the human spliceosomal C complex.
The most abundant C complex proteins included a subset of U5 proteins (220K, 200K, 116K, and 40K) and most proteins of the Prp19/CDC5L complex and Prp19-related proteins (Table 1), which were also abundant in Bact complexes. Thus, these proteins remain stable core components of the spliceosome through the first catalytic step of splicing. In contrast, all 17S U2 snRNP proteins were reduced by ∼70% compared with the Bact complex (Table 1). In addition, several second-step factors are moderately/highly abundant, including hPrp22 (spot 130), hPrp17 (spot 125), and hSlu7 (spot 131). In contrast, hPrp18 was present in very small amounts, and hPrp16 was not detected (Table 1), which is in good agreement with the low numbers of peptides sequenced previously for these proteins by MS in purified C complexes (6, 7). Several non-snRNP proteins previously designated C complex specific (i.e., GCIP p29, Abstrakt, CXorf56, and PPIase-like 3B [Fig. 7A and 8D]) are present in stoichiometric amounts, whereas several other proteins, including C9orf78, DDX35, C1orf 55, NOSIP, PPWD1, FAM50A, and cactin, are moderately abundant (Table 1). Since these C complex proteins were not detected or had only very low PAFs in the Bact complex, our results provide further evidence that they are indeed mainly associated with the C complex.
Protein phosphorylation detected by 2D electrophoresis.
To investigate the phosphorylation status of proteins present in human snRNPs and spliceosomal complexes, 2D gels were stained with the phosphate-specific, fluorescent dye Pro-Q Diamond (Fig. 1B to 7B), and those proteins with signals clearly above the background level were classified as moderately or strongly phosphorylated. The efficiency of phospho staining was not proportional to protein molecular mass (compare U5-220K [spot 34] and U5-200K [spot 35]) or to protein abundance (Fig. 5, compare SF3b130 [spot 18] and THRAP3 [spot 85]); rather, it reflects the total number of phosphate groups attached to the protein in the spot (38). In nearly all cases, the position of the protein's phosphate signal coincided exactly with the polypeptide signal, confirming that the phospho isoforms were not present in separate spots. However, in some cases (MFAP1, spot 112; LOC124245, spot 59), protein spots were elongated (as observed with SR proteins), which could represent unresolved phosphorylated isoforms of the same protein.
A number of proteins are phosphorylated in purified human snRNPs. These include U1-70K (Fig. 1B) and the U2-associated proteins SF3a120, SF3a60, SF3b145, Prp5, and tat-SF1 (Fig. 3B). A large number of proteins in the tri-snRNP are also heavily phosphorylated, including U5-200K/Brr2, U5-116K/hSnu114, U5-110K, hPrp28/100K, hPrp3/U4/U6-90K, 65K, hPrp31, and the 52K protein, making it a major potential regulatory target during splicing. The phosphorylation status of these proteins does not change significantly during splicing, although one cannot rule out that the loss of some of these proteins during splicing may be triggered by their dephosphorylation/hyperphosphorylation or that the phosphorylation sites of a given protein change during splicing (see Discussion). Note that quantitative changes in the phosphorylation of each of these snRNP proteins from one complex to the next correlate with changes in their abundances and thus do not appear to reflect different levels of phosphorylation. A notable exception is the U2-associated SF3b155 protein, whose phosphorylation increases significantly upon catalytic activation of the spliceosome. Phosphorylation leads to a change in the spot position of SF3b155, with it initially being present in a lower migrating spot (spot 16), both in 17S U2 (Fig. 3A) and the A complex (Fig. 4A), which exhibits very little phosphopeptide signal (Fig. 3B), and later upon activation, all of the SF3b155 protein is present in an “upper” heavily phosphorylated spot (Fig. 6B).
Numerous non-snRNP proteins are also significantly phosphorylated in the spliceosome. In total, approximately one-third of spliceosomal proteins were found to be phosphorylated as judged by Pro-Q Diamond staining. Similar to the vast majority of phosphorylated snRNP proteins, the phosphorylation status of the non-snRNP spliceosomal proteins does not appear to change significantly during splicing (note that the observed changes for the most part reflect changes in the abundances of these proteins). A notable exception is the CDC5L protein, a component of the Prp19/CDC5L complex. Although it is equally abundant in the Bact and C complex, it is first heavily phosphorylated in the C complex (Fig. 6 and 7 and Table 1). Taken together, these results indicate that a large number of spliceosomal proteins are phosphorylated, but in most cases their phosphorylation status does not appear to change significantly during splicing.
DISCUSSION
Recent MS analyses of affinity-purified human spliceosomal complexes identified more than 200 copurifying proteins, suggesting an unparalleled complexity and molecular masses exceeding 10 MDa for the splicing machinery. However, based on the sedimentation coefficient of spliceosomes (∼40 to 50S) and their molecular mass predicted from three-dimensional (3D) electron microscopy studies, e.g., ∼5 MDa for the C complex (14), it is clear that a large number of the proteins identified previously must be present in very small amounts. Here we developed a novel 2D electrophoresis system that has allowed for the first time an objective view of the relative abundances of the various proteins present in a particular spliceosomal complex. This method allowed us to compare (i) on a near-quantitative level the amounts of a given protein in different spliceosomal complexes and (ii) on a semiquantitative basis the amounts of different proteins within the same splicing complex.
Reliability of our 2D gel electrophoresis method.
The general reliability of the 2D gel electrophoresis method described here for identifying and providing quantitative information about components of the spliceosome is evidenced by several observations. First, when purified human snRNPs were analyzed on our 2D gels and their proteins subsequently identified by MS and quantitated by Sypro Ruby staining, nearly all known snRNP proteins were found, as expected, to be abundant components (Table 1). A comparison of previous MS and Western blot analyses of purified spliceosomal complexes with the results of our 2D analyses also speaks for the general reliability of this method for identifying and quantitating proteins copurifying with spliceosomal complexes. Though not strictly quantitative, a comparison of the number of peptides identified by MS for a certain protein in different complexes provides a good indication, in most cases, of the relative amounts of that particular protein present in these complexes when samples are directly, sequentially analyzed. The relative intensities (i.e., PAF values) of individual spliceosomal proteins observed via our 2D gel analyses correlates in the vast majority of cases with protein abundance estimations made based on peptide numbers and with the results of Western blotting, which are available for only a limited number of spliceosomal proteins (3, 6, 7, 10). Again here only a few clear discrepancies appear to exist. For example, based on the number of peptides sequenced, previous MS analyses of affinity-purified B, Bact, and C complexes strongly suggested that all three proteins of the human RES complex are specifically enriched in the Bact complex, with SNIP1 and MGC13125 being abundant components (6). Surprisingly, CGI-79 was not detected at all on 2D gels, and MGC13125 had a low PAF value in B, Bact, and C complexes. One possible explanation for this discrepancy could be that these proteins were selectively lost during sample preparation for 2D gel electrophoresis. KIAA1604 was also found with only a very low PAF value in the Bact complex, although based on peptides sequenced by MS, it appeared in previous studies to be very abundant in both Bact and C complexes (6). Since the vast majority of the KIAA1604 protein comigrates with hPrp2 on our 2D gels (and it is thus difficult to distinguish between the signals from each protein), its PAF value may be artificially low. Finally, previous MS studies also suggested that the SR-related protein SRm300 is abundant in both Bact and C complexes (6). However, based on its PAF value, SRm300 appears to be present in small amounts in the Bact complex, although the amounts detected in individual Bact complex preparations varied considerably.
A few spliceosomal proteins that are expected to be present in large amounts, but for which very low peptide numbers (relative to their molecular masses) were previously sequenced by MS, exhibited high PAF values. One such example is the SF3b49 protein, which as a component of the highly stable SF3b complex is expected to be highly abundant in A, B, and Bact complexes, as is observed for other SF3b components. Other examples include several of the LSm proteins. Thus, in these cases, our 2D gel electrophoresis system clearly is a much more reliable method for providing quantitative information about spliceosome-associated proteins.
Human spliceosomal complexes contain only ∼60 to 70 highly/moderately abundant proteins.
Based on the results presented here, human B, Bact, and C complexes affinity purified under physiological conditions are comprised of approximately 63 to 72 abundant/moderately abundant proteins. Based on these results, the molecular masses of human B, Bact, and C complexes are approximately 4.3, 4.0, and 5.1 MDa, respectively. These values correlate well with the molecular masses of Bact and C complexes estimated from analytical ultracentrifugation studies (D. Agafonov, A. Schomburg, and R. Lührmann, unpublished data), with measurements made by quantitative scanning transmission electron microscopy (STEM) of the C complex (H. Stark, S. Müller, and R. Lühr-mann, unpublished data) and with 3D electron microscopy of human C complexes whose molecular mass was calculated to be approximately 5 MDa (14). Figure 9 summarizes the major components of human A, B, Bact, and C complexes purified under physiological conditions. These proteins should account for the mass of these complexes that is observed under the electron microscope, and many of them likely play key structural and/or functional roles.
The human A complex is comprised of U1-, U2-, and U2-related proteins plus only a few non-snRNP proteins (Fig. 9). The B complex additionally contains the tri-snRNP proteins and a unique set of 6 non-snRNP proteins (Fig. 9, brown boxes) but no longer most U2-related proteins (light green boxes) or A-complex-specific non-snRNP proteins. Prp19 complex proteins and related proteins (red boxes) appear to be present in smaller amounts. However, it is more likely that our B complex preparation consists of two subpopulations, one lacking Prp19 and related proteins and another containing abundant amounts of them. Indeed, the calculated PAF values for these proteins suggest they interact in a concerted manner, and STEM analyses of our B complex preparations have revealed two distinct populations of the B complex with different molecular weights (H. Stark, S. Müller, and R. Lührmann, unpublished data). Furthermore, it is clear from previous studies that B complex formation does not require the Prp19 complex (9, 25).
The human Bact complex is comprised of 17S U2 proteins, a subset of U5 proteins, stoichiometric amounts of Prp19 complex proteins and related proteins, non-snRNP proteins common to all complexes analyzed here, plus 10 non-snRNP proteins (Fig. 9, orange boxes) first abundant at this stage. The human C complex additionally contains step II factors and a new subset of 10 non-snRNP proteins (Fig. 9, dark purple boxes) but reduced amounts of all U2 proteins and also of most of those non-snRNP proteins first abundant in the Bact complex (Fig. 9, orange boxes). Previously, it was not clear in many cases which complex-specific spliceosomal proteins are indeed abundant components. During the transition from the human Bact complex to the C complex, a large number of non-snRNP proteins are recruited (Fig. 9, dark purple boxes). Most of these proteins do not have homologs in S. cerevisiae, and thus they appear to be recruited to the spliceosome solely in higher eukaryotes. It is also unclear what their role in splicing may be. Data obtained from our 2D gel electrophoresis studies demonstrate that most of these proteins are indeed highly abundant and thus attractive candidates for future functional studies to elucidate their potential roles at the late stages of splicing or in subsequent cellular processes linked to splicing.
Recent MS analyses of purified S. cerevisiae B, Bact, and C complexes demonstrated that the yeast spliceosome is less complex than the splicing machinery of higher eukaryotes (12). A comparison of abundant proteins present in the human B, Bact, and C complex, as summarized in Fig. 9, with the corresponding yeast spliceosomal complexes indicates that homologues of most of these human proteins are also present in yeast spliceosomes and that the dynamics of protein recruitment and release are also largely conserved. However, some notable differences exist, including the absence of SR and hnRNP proteins and cyclophilins (which for the most part are completely absent in yeast). S. cerevisiae homologs of several human B- and Bact-complex-specific non-snRNP proteins and, excluding step II factors, nearly all C-complex-specific non-snRNP proteins (Fig. 9, brown, orange, and purple boxes; see also Table 1) also appear to be absent. The function of the later groups of human spliceosomal proteins is unclear, but based on their absence in yeast it is tempting to speculate that some of them may be involved in regulated splicing events which are seldom observed in S. cerevisiae.
Our data indicate that a large number of proteins previously detected by MS after 1D SDS-PAGE of affinity-purified spliceosomal complexes are present in negligible amounts. While the vast majority of proteins not detected by our 2D gel system are most likely truly low-abundance components and thus in many cases contaminants, a handful may escape detection (or appear to be present in small amounts) due to their potential (i) smearing or separation into multiple spots (potentially due to posttranslational modifications), (ii) comigration with a highly abundant protein, and (iii) selective loss during sample preparation. Since nearly all snRNP/spliceosomal proteins that are expected to be abundant were in fact shown to have high PAF values, such cases appear to be very rare. Comigration of a few spliceosomal proteins was indeed observed, and thus this method may not be ideal for the analysis of even more highly complex mixtures of proteins.
The majority of proteins with low PAF values likely do not play important functional/structural roles in the splicing process. It should be noted, however, that some of them may nonetheless be required for splicing, in particular those with enzymatic activity. For example, Prp4 kinase is present in small amounts in B, Bact, and C complexes (Table 1) but has been shown to be required for tri-snRNP addition during B complex formation (35). The low level of some of these factors may also reflect the fact that they are very loosely associated with the spliceosome and thus readily lost during the purification process. This low affinity could reflect their potential involvement in the splicing of only a subset of pre-mRNAs. Likewise, they could be involved in alternative splicing events whose regulation would be aided by the ease with which they can be recruited or released in response to qualitative or quantitative changes in the splicing environment. Finally, some of the missing or low-abundance proteins may interact transiently and be present in high abundance at a spliceosome assembly or functional stage not analyzed here (i.e., in an intermediate splicing complex). Future studies aimed at isolating novel assembly intermediates of the human spliceosome may lead to an even finer dissection of the spliceosome's compositional dynamics.
Taken together, the data presented here regarding the relative abundances of proteins within various spliceosomal complexes are highly valuable for deciding which proteins are good candidates for more detailed functional studies. They also provide a rational basis for future structural studies, including studies of the following: (i) which of the non-snRNP spliceosome-associated proteins should be targeted for immuno-electron microscopy (EM) studies and (ii) which of the human spliceosomal complexes (due to lower numbers of nonstoichiometric components) is best suited for crystallization studies. The 2D gel electrophoresis system described here also allows a visual comparison of the purity and complexity of different spliceosomal complexes and thus is highly useful for rapid screening of the effects of splicing inhibitors or different buffer conditions on spliceosome composition.
A large number of spliceosomal proteins are phosphorylated.
Reversible protein phosphorylation plays a key role during spliceosome assembly and the catalytic steps of splicing (reviewed in refs 28, 37). Approximately one-third of the spliceosomal proteins identified in the present study were found to be moderately to heavily phosphorylated, including all whose phosphorylation or dephosphorylation is known to be functionally relevant during splicing, such as SR proteins (reviewed in references 28 and 37), U1-70K (39), and CDC5L (16). Although the intensity of phosphostaining of some proteins varied, in the vast majority of cases these changes simply reflected changes in the abundance of that protein from one spliceosomal complex to the next. Thus, differential phosphorylation was not observed for most of the phosphoproteins detected in spliceosomes. However, small changes in the number of phosphorylated sites could escape detection. Likewise, since Pro-Q Diamond has a limited linear signal range (i.e., the staining of phosphoproteins is no longer linear in those cases where more than 12 sites are phosphorylated [38]), the hyperphosphorylation of already highly phosphorylated proteins may also elude detection. Furthermore, our studies provide no information about which sites are phosphorylated. Thus, qualitative changes in phosphorylation that do not dramatically effect the overall number of phospho sites would not be detected by this method.
Consistent with previous observations (6, 40), spliceosome activation was accompanied by a significant increase in the phosphorylation of SF3b155, whereas CDC5L, a component of the hPrp19 complex, was found to be intensively phosphorylated specifically upon C complex formation (Table 1). The precise function of the stage-specific phosphorylation of each of these proteins is unclear. Given that the SF3a and SF3b proteins are stably bound to the Bact complex and SF3b155 is already fully phosphorylated at this stage, it is unlikely that phosphorylation of SF3b155 leads to the dissociation of SF3a and SF3b observed during the Bact-to-C-complex transition. Since the Prp19 complex is stably bound to the spliceosome at the Bact complex stage and at the same time the CDC5L protein is not phosphorylated, it is also unlikely that this phosphorylation event leads to Prp19 complex stabilization within the spliceosome. However, in purified human Prp19 complexes, the CDC5L protein lacks any phospho signal (data not shown), supporting the idea that its phosphorylation plays a regulatory role during splicing.
The role of phosphorylation of most of the other phosphoproteins detected in human spliceosomal complexes (with the exception of SR proteins) is not known. Interestingly, several transiently associated non-snRNP proteins are heavily phosphorylated. This suggests that phosphorylation may act as a binding trigger for some of the proteins recruited at a specific stage, whereas dephosphorylation potentially might lead to their release from the spliceosome. Taken together, we conclude that the 2D electrophoresis system described here is a powerful new tool for investigating complex and dynamic cellular multiprotein complexes.
Supplementary Material
ACKNOWLEDGMENTS
We thank I. Öchsner for excellent technical assistance. We are grateful to T. Conrad and H. Kohansal for preparing HeLa cell nuclear extract, to M. Raabe, J. Lehne, and U. Plessmann for excellent help in MS analysis, and to G. Weber and S. Trowitzsch for providing purified human U1 snRNPs and U4/U6.U5 tri-snRNPs and G. Heyne for preparing 17S U2 snRNPs. We also thank B. Kastner for help in constructing 2D gel electrophoresis chambers and for constructive comments on the manuscript.
This work was supported by grants from the DFG and the European Commission (EURASNET-518238) to R.L. and a YIP grant from EURASNET to H.U. P.O. was supported by the Stiftung der DeutschenWirtschaft, and E.W. was supported by the Studienstiftung des Deutschen Volkes.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 2 May 2011.
REFERENCES
- 1. Agafonov D. E., Kolb V. A., Nazimov I. V., Spirin A. S. 1999. A protein residing at the subunit interface of the bacterial ribosome. Proc. Natl. Acad. Sci. U. S. A. 96:12345–12349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beausoleil S. A., et al. 2004. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. U. S. A. 101:12130–12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Behzadnia N., et al. 2007. Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J. 26:1737–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bennett M., Michaud S., Kingston J., Reed R. 1992. Protein components specifically associated with prespliceosome and spliceosome complexes. Genes Dev. 6:1986–2000 [DOI] [PubMed] [Google Scholar]
- 5. Berggren K., et al. 2000. Background-free, high sensitivity staining of proteins in one- and two-dimensional sodium dodecyl sulfate-polyacrylamide gels using a luminescent ruthenium complex. Electrophoresis 21:2509–2521 [DOI] [PubMed] [Google Scholar]
- 6. Bessonov S., et al. 2010. Characterization of purified human Bact spliceosomal complexes reveals compositional and morphological changes during spliceosome activation and first step catalysis. RNA 16:2384–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bessonov S., Anokhina M., Will C. L., Urlaub H., Lührmann R. 2008. Isolation of an active step I spliceosome and composition of its RNP core. Nature 452:846–850 [DOI] [PubMed] [Google Scholar]
- 8. Cao W., Jamison S. F., Garcia-Blanco M. A. 1997. Both phosphorylation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA 3:1456–1467 [PMC free article] [PubMed] [Google Scholar]
- 9. Chan S. P., Kao D. I., Tsai W. Y., Cheng S. C. 2003. The Prp19p-associated complex in spliceosome activation. Science 302:279–282 [DOI] [PubMed] [Google Scholar]
- 10. Deckert J., et al. 2006. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell. Biol. 26:5528–5543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dignam J. D., Lebovitz R. M., Roeder R. G. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fabrizio P., et al. 2009. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol. Cell 36:593–608 [DOI] [PubMed] [Google Scholar]
- 13. Fetzer S., Lauber J., Will C. L., Lührmann R. 1997. The [U4/U6.U5] tri-snRNP-specific 27K protein is a novel SR protein that can be phosphorylated by the snRNP-associated protein kinase. RNA 3:344–355 [PMC free article] [PubMed] [Google Scholar]
- 14. Golas M. M., et al. 2010. 3D Cryo-EM structure of an active step I spliceosome and localization of its catalytic core. Mol. Cell 40:927–938 [DOI] [PubMed] [Google Scholar]
- 15. Gozani O., Patton J. G., Reed R. 1994. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 13:3356–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Graub R., et al. 2008. Cell cycle-dependent phosphorylation of human CDC5 regulates RNA processing. Cell Cycle 7:1795–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grote M., et al. 2010. Molecular architecture of the human Prp19/CDC5L complex. Mol. Cell. Biol. 30:2105–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hochleitner E. O., et al. 2005. Protein stoichiometry of a multiprotein complex, the human spliceosomal U1 small nuclear ribonucleoprotein: absolute quantification using isotope-coded tags and mass spectrometry. J. Biol. Chem. 280:2536–2542 [DOI] [PubMed] [Google Scholar]
- 19. Ishihama Y., et al. 2005. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell Proteomics 4:1265–1272 [DOI] [PubMed] [Google Scholar]
- 20. Kaltschmidt E., Wittmann H. G. 1970. Ribosomal proteins. XII. Number of proteins in small and large ribosomal subunits of Escherichia coli as determined by two-dimensional gel electrophoresis. Proc. Natl. Acad. Sci. U. S. A. 67:1276–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kastner B., Lührmann R. 1999. Purification of U small nuclear ribonucleoprotein particles. Methods Mol. Biol. 118:289–298 [DOI] [PubMed] [Google Scholar]
- 22. Knopf U. C., Sommer A., Kenny J., Traut R. R. 1975. A new two-dimensional gel electrophoresis system for the analysis of complex protein mixtures: application to the ribosome of E. coli. Mol. Biol. Rep. 2:35–40 [DOI] [PubMed] [Google Scholar]
- 23. Lyubimova T., Caglio S., Gelfi C., Righetti P. G., Rabilloud T. 1993. Photopolymerization of polyacrylamide gels with methylene blue. Electrophoresis 14:40–50 [DOI] [PubMed] [Google Scholar]
- 24. Makarov E. M., et al. 2002. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298:2205–2208 [DOI] [PubMed] [Google Scholar]
- 25. Makarova O. V., et al. 2004. A subset of human 35S U5 proteins, including Prp19, function prior to catalytic step 1 of splicing. EMBO J. 23:2381–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mathew R., et al. 2008. Phosphorylation of human PRP28 by SRPK2 is required for integration of the U4/U6-U5 tri-snRNP into the spliceosome. Nat. Struct. Mol. Biol. 15:435–443 [DOI] [PubMed] [Google Scholar]
- 27. Michaud S., Reed R. 1993. A functional association between the 5′ and 3′ splice site is established in the earliest prespliceosome complex (E) in mammals. Genes Dev. 7:1008–1020 [DOI] [PubMed] [Google Scholar]
- 28. Misteli T. 1999. RNA splicing: what has phosphorylation got to do with it? Curr. Biol. 9:198–200 [DOI] [PubMed] [Google Scholar]
- 29. Neubauer G., et al. 1998. Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat. Genet. 20:46–50 [DOI] [PubMed] [Google Scholar]
- 30. O'Farrell P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007–4021 [PMC free article] [PubMed] [Google Scholar]
- 31. Rappsilber J., Ryder U., Lamond A. I., Mann M. 2002. Large-scale proteomic analysis of the human spliceosome. Genome Res. 12:1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Righetti P. G., Brost B. C., Snyder R. S. 1981. On the limiting pore size of hydrophilic gels for electrophoresis and isoelectric focusing. J. Biochem. Biophys. Methods 4:347–363 [DOI] [PubMed] [Google Scholar]
- 33. Schaegger H., von Jagow G. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368–379 [DOI] [PubMed] [Google Scholar]
- 34. Schmidt C., Lenz C., Grote M., Luhrmann R., Urlaub H. 2010. Determination of protein stoichiometry within protein complexes using absolute quantification and multiple reaction monitoring. Anal. Chem. 82:2784–2796 [DOI] [PubMed] [Google Scholar]
- 35. Schneider M., et al. 2010. Human PRP4 kinase is required for stable tri-snRNP association during spliceosomal B complex formation. Nat. Struct. Mol. Biol. 17:216–221 [DOI] [PubMed] [Google Scholar]
- 36. Shevchenko A., Wilm M., Vorm O., Mann M. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850–858 [DOI] [PubMed] [Google Scholar]
- 37. Soret J., Tazi J. 2003. Phosphorylation-dependent control of the pre-mRNA splicing machinery. Prog. Mol. Subcell. Biol. 31:89–126 [DOI] [PubMed] [Google Scholar]
- 38. Steinberg T. H., et al. 2003. Global quantitative phosphoprotein analysis using multiplexed proteomics technology. Proteomics 3:1128–1144 [DOI] [PubMed] [Google Scholar]
- 39. Tazi J., et al. 1993. Thiophosphorylation of U1-70K protein inhibits pre-mRNA splicing. Nature 363:283–286 [DOI] [PubMed] [Google Scholar]
- 40. Wang C., et al. 1998. Phosphorylation of spliceosomal protein SAP 155 coupled with splicing catalysis. Genes Dev. 12:1409–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Weber G., Trowitzsch S., Kastner B., Lührmann R., Wahl M. C. 2010. Functional organization of the Sm core in the crystal structure of human U1 snRNP. EMBO J. 29:4172–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Will C. L., Lührmann R. 2011. Spliceosome structure and function, p. 181–203In Atkins J. F., Gesteland R. F., Cech T. R. (ed.), RNA worlds: from life's origins to diversity in gene regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 43. Will C. L., et al. 2002. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J. 21:4978–4988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Woppmann A., Patschinsky T., Bringmann P., Godt F., Lührmann R. 1990. Characterisation of human and murine snRNP proteins by two-dimensional gel electrophoresis and phosphopeptide analysis of U1-specific 70K protein variants. Nucleic Acids Res. 18:4427–4438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yan D., et al. 1998. CUS2, a yeast homolog of human Tat-SF1, rescues function of misfolded U2 through an unusual RNA recognition motif. Mol. Cell. Biol. 18:5000–5009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zahler A. M. 1999. Purification of SR protein splicing factors. Methods Mol. Biol. 118:419–432 [DOI] [PubMed] [Google Scholar]
- 47. Zhou Z., Licklider L. J., Gygi S. P., Reed R. 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419:182–185 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.