Abstract
Eukaryotic RNA polymerase III (Pol III) relies on a transcription factor TFIIF-like Rpc37/53 subcomplex for promoter opening, elongation, termination, and reinitiation. By incorporating the photoreactive amino acid p-benzoyl-l-phenylalanine (BPA) into Rpc37, Rpc53, and the Rpc2 subunit of Pol III, we mapped protein-protein interactions, revealing the position of Rpc37/53 within the Pol III preinitiation complex (PIC). BPA photo-cross-linking was combined with site-directed hydroxyl radical probing to localize the Rpc37/53 dimerization module on the lobe/external 2 domains of Rpc2, in similarity to the binding of TFIIF on Pol II. N terminal to the dimerization domain, Rpc53 binds the Pol III-specific subunits Rpc82 and Rpc34, the Pol III stalk, and the assembly factor TFIIIC, essential for PIC formation. The C-terminal domain of Rpc37 interacts extensively with Rpc2 and Rpc34 and contains binding sites for initiation factor Bdp1. We also located the C-terminal domain of Rpc37 within the Pol III active center in the ternary elongation complex, where it likely functions in accurate termination. Our work explains how the Rpc37/53 dimer is anchored on the Pol III core and acts as a hub to integrate a protein network for initiation and termination.
INTRODUCTION
RNA polymerase III (Pol III) catalyzes the RNA synthesis of diverse untranslated transcripts, including precursor tRNAs, 5S rRNA, and certain small nuclear RNAs, such as U6 and 7SL RNAs. The other two nuclear RNA polymerases, Pol I and Pol II, transcribe pre-rRNA and mRNA, respectively. In humans, Pol III also participates in the transcription of a number of microRNAs (19) and plays a role in tumorigenesis (41, 58). Among the nuclear RNA polymerases, Pol III is the largest enzyme, comprising 17 subunits with a total molecular mass of approximately 0.7 MDa; its core structure of 12 subunits is conserved among all nuclear Pols (18). The remaining five subunits are specific to Pol III and form two subcomplexes with distinct functions in transcription, Rpc82/34/31 and Rpc37/53. The former subcomplex interacts with the initiation transcription factor TFIIIB for the recruitment of polymerase and also participates in promoter opening and transcription initiation (6, 50, 55, 56), whereas the latter participates in promoter opening and transcription termination and reinitiation (34, 38). Previous yeast two-hybrid screenings provided an overview of protein-protein interactions among Pol III subunits (26), and electron microscopy analysis of Pol III suggested the location of the subcomplexes on the Pol III core structure (24, 25, 32, 52). However, much more information on protein-protein interactions between these subcomplexes and the initiation factors is necessary to understand the mechanisms of initiation, elongation, and termination and to determine how the mechanisms used by the Pol III machinery differ from those used by Pol I and II.
A conserved dimerization module has recently been detected in the Pol I, II, and III machineries as part of Rpa49/34.5 of Pol I, the general transcription factor TFIIF of Pol II, and Rpc37/53 of Pol III (8, 13, 18, 36). The presence of the TFIIF-like complexes is unique to the eukaryotic transcription systems. However, other than the conserved dimerization module and a winged-helix (WH) fold that is present as a single copy in each of the Tfg1 and Tfg2 subunits of TFIIF and as tandem repeats in Rpa49, the overall structural characteristics of the three TFIIF-like complexes differ; thus, their functions in transcription may also differ. For example, although all three of these TFIIF-like complexes are involved in transcription and elongation, they exert opposite effects on elongation, with Rpa49/34.5 and TFIIF increasing and Rpc37/53 reducing the elongation rate (36, 38, 59).
Structural and biochemical analyses have suggested how individual TFIIF-like complexes establish protein interactions for their respective transcriptional activities. The TFIIF dimerization module, which resides in subunits Tfg1 and Tfg2, has been mapped to the Pol II lobe domain above the active site cleft (15, 21, 52) to determine the transcription start site(s). In contrast, the Rpa49/34.5 dimerization module has been assigned near the Pol I funnel on the opposite side of the cleft (36) to anchor the dimer on the core for 3′-RNA nucleotide cleavage, transcription, and elongation (27). Investigations of the cryo-electron microscopic (cryo-EM) structures of Pol III, in combination with three-dimensional reconstruction based on the X-ray structure of the Rpc25/17 dimer comprising the Pol III stalk and homology modeling of the remaining core subunits, have suggested that Rpc37/53 lies on the Rpc2 (homologous to Rpb2 of Pol II and the β subunit of bacterial Pol) lobe domain of the Pol III core, in similarity to the binding of TFIIF on Pol II (24, 25, 52). Furthermore, a recent biochemical study indicated that Rpc53 is likely positioned near the Pol III active center to function in transcription termination and DNA melting based on protein and 3′-RNA cross-linking and protein depletion analyses (34). However, direct evidence that would reveal the positions of the conserved dimerization module and the remaining structural regions on the core as well as on other transcription factors during transcription is lacking.
In this study, we incorporated the site-specific photo-cross-linker p-benzoyl-l-phenylalanine (BPA) as a nonnatural amino acid into the second largest subunit of Pol III (Rpc2) and Pol III-specific subunits Rpc37 and Rpc53 to topologically map protein-protein interactions within the Pol III preinitiation complex (PIC). In combination with molecular modeling, positional mapping derived from site-directed hydroxyl radical probing, and further mutational and binding analyses, we located the Rpc37/53 dimerization module on the lobe/external surface above the active site cleft of the core within the PIC. In addition, we found that the Rpc53 N-terminal segment contacts the Rpc82 and Rpc34 subunits of the other Pol III-specific subcomplex and the Rpc17 and Rpc25 subunits of the Pol III stalk as well as the transcription initiation factor TFIIIC. Furthermore, we determined that an essential segment in the C-terminal region of Rpc37 is located near several highly conserved motifs within the Pol III active center and also interacts with Bdp1 of the transcription factor TFIIIB. These newly uncovered protein-protein interactions for regions in Rpc53 and Rpc37 participate in stabilizing the PIC and determining accurate transcription termination sites. We propose a structural model for the binding of the Rpc37/53 dimerization module and a network of interactions and roles for the Rpc37/53 dimer throughout the transcription cycle.
MATERIALS AND METHODS
Construction of yeast plasmids and strains.
For BPA incorporation into three subunits of Pol III, Saccharomyces cerevisiae RPC2, RPC37, and RPC53 were cloned into the yeast 2μm plasmid pRS425 with a LEU2 selection marker (17). A 414-bp ADH1 promoter sequence was inserted upstream of each gene, and either a 13Myc epitope tag (for RPC2 and RPC53) or a single V5 epitope tag (for RPC37) was inserted at the C terminus by the use of a QuikChange site-directed mutagenesis kit (Stratagene), yielding pYL1 (RPC2-Myc 2μm LEU2), pCW1 (RPC53-Myc 2μm LEU2), and pCW2 (RPC37-V5 2μm LEU2). Individual Rpc2, Rpc53, and Rpc37 amber plasmids containing the TAG codon at a single specific site were constructed by oligonucleotide-directed phagemid mutagenesis.
To analyze growth defects of Rpc37/53 truncation mutants, RPC37 and RPC53 were individually cloned into CEN/ARS plasmid pRS315 (LEU2+) with a single V5 epitope inserted in their C termini and were driven by their endogenous promoters, yielding pCW4 (RPC53-V5) and pCW5 (RPC37-V5). Similarly, each mutant was derived from pCW4 or pCW5 by oligonucleotide-directed phagemid mutagenesis.
Yeast shuffle strains for BPA incorporation into Rpc37, Rpc53, and Rpc2 were all derived from BY4705 (3) with the chromosomal knockout of individual genes by the KanMX4 cassette, yielding YLy1 {MATα ade2::his3G his3Δ200 leu2Δ met15Δ lys2Δ trp1Δ63 ura3Δ [rpc2::KanMX4] RPC2-pRS316(URA3+)}, CWy1 {MATα ade2::his3G his3Δ200 leu2Δ met15Δ lys2Δ trp1Δ63 ura3Δ [rpc37::KanMX4] RPC37-pRS316(URA3+)}, and CWy2 {MATα ade2::his3G his3Δ200 leu2Δ met15Δ lys2Δ trp1Δ63 ura3Δ0 [rpc53::KanMX4] RPC53-pRS316(URA3+)}. The amber strains harboring BPA at specific residues in Rpc37, Rpc53, or Rpc2 were generated from each shuffle strain by cotransforming the pLH157 plasmid containing the tRNA/aminoacyl tRNA synthetase pair for suppressing the TAG (amber) codon (13, 16, 43) and the amber plasmid of each gene into respective shuffle strains, followed by selection on medium containing 5-fluoroorotic acid (5-FOA) and BPA. For identifying the cross-linking targets, candidate genes were tagged with either Flag or hemagglutinin (HA) epitope at their C termini in the shuffle strains through double-crossover homologous recombination with PCR-generated DNA containing the selectable marker cassette hphMX4 resistant to hygromycin B.
PIC isolation by the immobilized template assay and BPA photo-cross-linking.
Yeast cultures were grown in yeast extract-peptone-dextrose (YPD) medium containing BPA (Bachem), and the preparation of yeast cell extract was conducted as described previously (29). For preparation of the DNA fragments used in the immobilized template assay, SUP4 tRNA, U6 snRNA, and 5S rRNA gene sequences were amplified by PCR and cloned into pBluescript SK+/−. Each DNA fragment was then individually amplified with 5′-biotinylated oligonucleotides, yielding 603-bp, 531-bp, and 601-bp DNA templates of SUP4, U6, and 5S promoters, respectively. Details of all constructs and primer sequences are available upon request. The procedures of the immobilized template assay were principally as described by Hahn and Roberts, with minor modification (29). Unless otherwise specified, Pol III PICs were formed on type II promoter genes (LEU3 or SUP4 tRNA). For a typical BPA photo-cross-linking experiment, 800 μg of whole-cell protein extract was incubated with 4 μg of DNA template immobilized on Streptavidin magnetic beads (Dynal). After being washed three times with transcription buffer (20 mM HEPES [pH 7.9], 80 mM KCl, 5 mM MgCl2, 1 mM EDTA, 2% glycerol) containing 0.01% Tween 20, reaction mixtures were divided into two fractions, one for UV irradiation (+UV) and the other for use as the control (−UV). UV irradiation was conducted with a Spectrolinker XL-1500 UV oven (Spectronics) with a total energy of 7,500 μJ cm−2. The isolated PICs were then resuspended in NuPAGE loading buffer (Invitrogen) for Western blot analysis. Bands were visualized by the use of a LI-COR Bioscience Odyssey infrared imaging system. In parallel, PIC activity was confirmed by the addition of nucleoside triphosphates (NTPs) and usage of the transcription start site was verified by primer extension (data not shown). Given that only crude protein extracts were used in the immobilized template assay, the preparation of PIC isolation should not be considered homogeneous and contaminants might have remained bound. The percentage of active PICs capable of producing correct RNA transcripts from immobilized templates was not assayed.
In vitro transcription.
Pol III PICs isolated by the immobilized template assay as described above were resuspended in 17 μl of transcription buffer containing 200 ng of α-amanitin, 4 units of RNase inhibitor (Roche), and 1 mM dithiothreitol (DTT). Transcription was then initiated by adding ATP (500 μM), UTP (500 μM), CTP (500 μM), GTP (50 μM), and [α-32P]GTP (0.16 μM [3,000 Ci/mmol]) to achieve a total volume of 20 μl, and reaction mixtures were incubated for 30 min at 30°C. Reactions were stopped by adding 180 μl of 0.1 M sodium acetate, 10 mM EDTA, 0.5% sodium dodecyl sulfate (SDS), and 200 μg/ml glycogen. After phenol-chloroform extraction and ethanol precipitation, RNA transcripts were analyzed on 6% denaturing polyacrylamide gels and quantitated by PhosphorImager analysis. Sizes of transcripts were estimated by comparison to an RNA ladder (Ambion). For mapping of the SUP4 tRNA start site, transcription was carried out as described above, except that all four NTPs (at 500 μM) were used. After phenol-chloroform extraction and ethanol precipitation, the transcripts were subjected to primer extension using a 5′-end 32P-labeled oligonucleotide, GATAAATTAAAGTCTTGC. To analyze the Pol III ternary elongation complex (TEC), ATP (125 μM), CTP (125 μM), and [α-32P]UTP (0.42 μM [3,000 Ci/mmol]) were added to the isolated PICs to initiate transcription for 5 min at 30°C. After being washed with transcription buffer, the bound transcripts were analyzed on 16% denaturing polyacrylamide gels.
Pol III immunoprecipitation.
Yeast cell extracts containing the Rpc2-Flag and Rpc37 wild type (WT) or mutants with a C-terminal V5 epitope were used for immunoprecipitation (IP). Yeast cell extract (1 mg) from each Rpc37 mutant was mixed with 50 μl of anti-Flag agarose (Sigma) in extract dialysis buffer containing 20 mM HEPES (pH 7.9), 100 mM KCl, 5 mM MgCl2, 1 mM EDTA, and 20% glycerol and incubated at 4°C for 1 h. The immunoprecipitated Pol III was washed 3 times with 400 μl of extract dialysis buffer. The bound Pol III was eluted in 100 μl of extract dialysis buffer containing 0.1 mg/ml triple-Flag peptide (Sigma) and subjected to Western analysis by probing with anti-V5 antibody.
Structure modeling of the Rpc37/53 dimerization module and Pol III 11-subunit models.
On the basis of HHpred analysis and the method described by Geiger et al. (27, 48), sequence alignments of dimerization modules conserved in Rpc37/53 and RAP74/30 were built. The dimerization module model was constructed using Modeler software (23). The Pol III 11-subunit core model was also built using Modeler according to the sequence alignment (32), Pol II (PBD 1Y1V) as the template, and the X-ray structure of Rpc17/25 (PDB 2CKZ). The model for the Rpc37/53 dimerization module on the Pol III-11-subunit core was constructed using ZDOCK and distance constraints derived from the hydroxyl radical cleavage of Rpc2. The strongest cleavage data from FeBABE conjugation at Gly150 and Gly157 of Rpc37 and Gly383 of Rpc53 were utilized to restrict the distances between individual Rpc37/53 residues and their corresponding Rpc2 cut sites. The calculation was conducted by using the default settings of the program. The Rpc37/53-Rpc2 binding interface of the generated model satisfies the cutting pattern determined from all FeBABE derivatives. The coordinates of the Pol III core with Rpc37/53 dimerization module model are available at http://www.imb.sinica.edu.tw/∼htchen/p.html.
FeBABE conjugation of Rpc37/53.
Gene sequences encoding Rpc37 and Rpc53 were cloned into pET21a (Novagen) and expressed in Escherichia coli. The recombinant Rpc37/53 heterodimer was purified with multiple ion exchange columns to a condition approaching homogeneity. Details of the purification procedures are available upon request. The FeBABE conjugation procedures were similar to that described for TFIIF (13). To remove DTT in the protein sample buffer, purified rRpc37/53 single-cysteine variants were loaded onto a preequilibrated NAP-5 desalting column (GE Health Care) and subsequently eluted with storage buffer (20 mM HEPES [pH 7.5], 400 mM KCl, 5 mM MgCl2, 1 mM EDTA, 10% glycerol). DTT-free rRpc37/53 was then incubated with FeBABE (Dojindo) at room temperature for 30 min and subjected to loading on a second NAP-5 desalting column to remove the unincorporated FeBABE. Conjugated proteins were stored in storage buffer at −80°C for further use.
FeBABE cleavage assay and determination of cleavage sites.
In a typical FeBABE cleavage experiment, 400 μg of yeast cell extract containing Rpc2-Flag and Rpc53 Δ(2–280) was incubated with 4 μg of rRpc37/53-FeBABE derivative in a 200-μl reaction mixture before PIC isolation. Pol III PICs were isolated with immobilized SUP4 template as described above. After being washed to remove nonspecifically bound proteins, FeBABE cleavage assay was conducted as previously described (13). For FeBABE cleavage in ternary elongation complex, Pol III TEC was isolated as described above except for the use of nonradioactive UTP. After the TEC isolation, FeBABE cleavage was conducted. The Rpc2 cleavage fragments were detected by probing with anti-Flag antibody and visualized as described above for BPA photo-cross-linking experiments. The cleavage sites were determined by calculating the mobility shift compared to an in vitro-translated ladder as previously described (12). Each cut site was highlighted as an 11-residue patch (centered at the calculated cleavage site) on the Pol III 11-subunit model.
RESULTS
Rpc2 lobe and external 2 domain cross-linked to Rpc37/53.
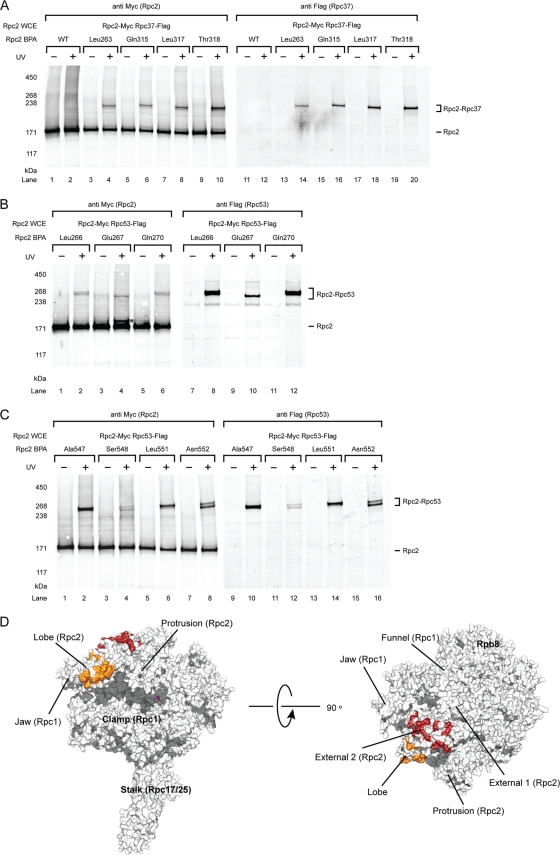
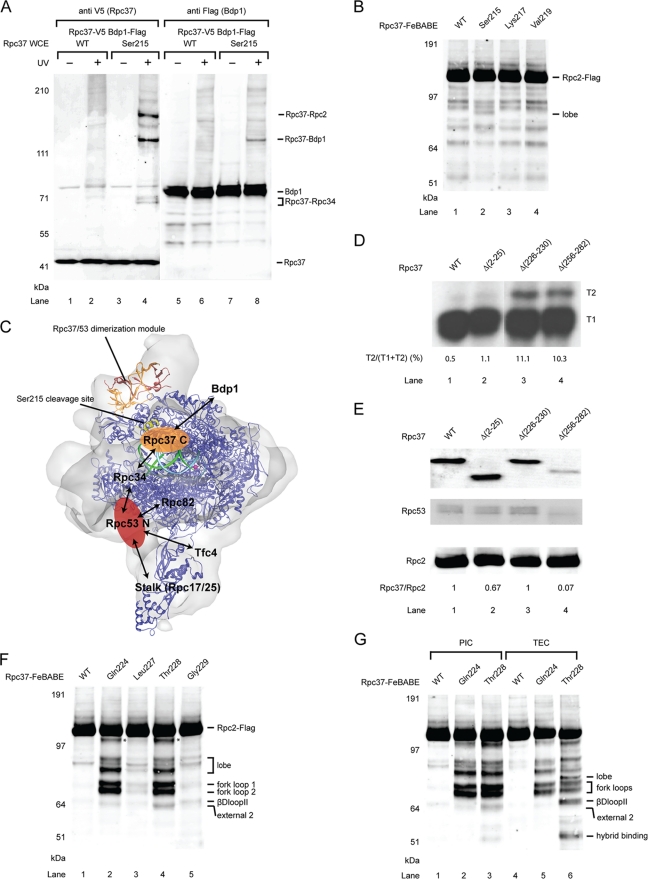
To probe protein-protein interactions for Rpc2 within the PIC, we incorporated BPA at surface-exposed residues of the Rpc2 lobe based on the previously reported Pol III core model and the cryo-EM structure of Pol III (32, 52). We utilized the whole-cell extracts from the Rpc2-BPA strains in the immobilized template assay for PIC isolation and photo-cross-linking. We observed BPA cross-links to other polypeptides with estimated molecular weights ranging from 40 to 60 kDa, close to the size of Rpc37 or Rpc53 (Fig. 1A and B). To identify these cross-linked products, we generated Rpc2-BPA cell extracts that also contained a C-terminally Flag epitope-tagged Rpc37. By conducting duplicate Western blot probing with anti-Myc (Rpc2) and anti-Flag (Rpc37) antibodies, Rpc37 was confirmed to be a cross-linked polypeptide (Fig. 1A). The cross-links corresponding to Rpc2-Rpc53 interaction in the Rpc2 lobe domain were identified in a similar manner by utilizing Flag-tagged Rpc53 cell extracts (Fig. 1B). In addition, Rpc2-Rpc53 cross-links were observed in the adjacent external 2 domain of Rpc2 (Fig. 1C). The identified Rpc2 cross-links with Rpc37 and Rpc53 are summarized in Table S1 in the supplemental material, and the Rpc2-Rpc37 and Rpc2-Rpc53 cross-linking sites on the 11-subunit model of the Pol III core (missing the less-conserved Rpc11 subunit) are displayed in Fig. 1D.
Fig. 1.
Photo-cross-linking maps Rpc37/53 on Rpc2 lobe and external 2 domains within the Pol III PIC. (A) Western analysis of Rpc2 cross-links to Rpc37 on the lobe domain. The BPA-substituted Rpc2 residues are indicated above the lanes. Cross-links were visualized by probing with anti-Myc antibody (left panel). The identity of fusion bands was verified by probing with Flag-tagged Rpc37 (right panel). Rpc2 WCE, Rpc2 whole-cell extract. UV + or −, with or without UV irradiation. WT, wild-type Rpc2 without BPA incorporation. (B) Representative cross-links on Rpc2 lobe to Rpc53. Rpc2 and cross-links were visualized by probing with anti-Myc antibody on the left panel. The identity of the cross-linked polypeptide was verified by probing with Flag-tagged Rpc53 (right panel). Asterisk, unidentified cross-linked polypeptide. (C) Representative cross-links on Rpc2 external 2 domain to Rpc53. (D) Molecular model of the Pol III 11-subunit core. The model was built using Modeler on the basis of the homology model of Pol II core (PBD 1Y1V) and the X-ray structure of Rpc25/17 (PDB 2CKZ) (32, 35). Rpc2-Rpc37 cross-links are shown in orange and Rpc2-Rpc53 cross-links are shown in red. The magenta sphere represents magnesium at the active center.
Our cross-linking data revealed extensive contacts between the Rpc37/53 dimer and Rpc2 on the lobe and the external 2 domains, suggesting that the location of the Rpc37/53 dimer is above the central cleft of the Pol III core (Fig. 1D). This Rpc2-Rpc37/53 binding is in good agreement with the assignment of the Rpc37/53 subcomplex to the additional electron density measured above the Rpc2 lobe in the EM study (24, 25, 52). Furthermore, as recent studies have shown that TFIIF makes contact with the similar Pol II structural domains in Rpb2 (15, 21), our results demonstrate a conserved binding mechanism between the TFIIF-like complex and the Rpb2 homolog in the two transcription systems.
Rpc37/53 photo-cross-linking within the PIC.
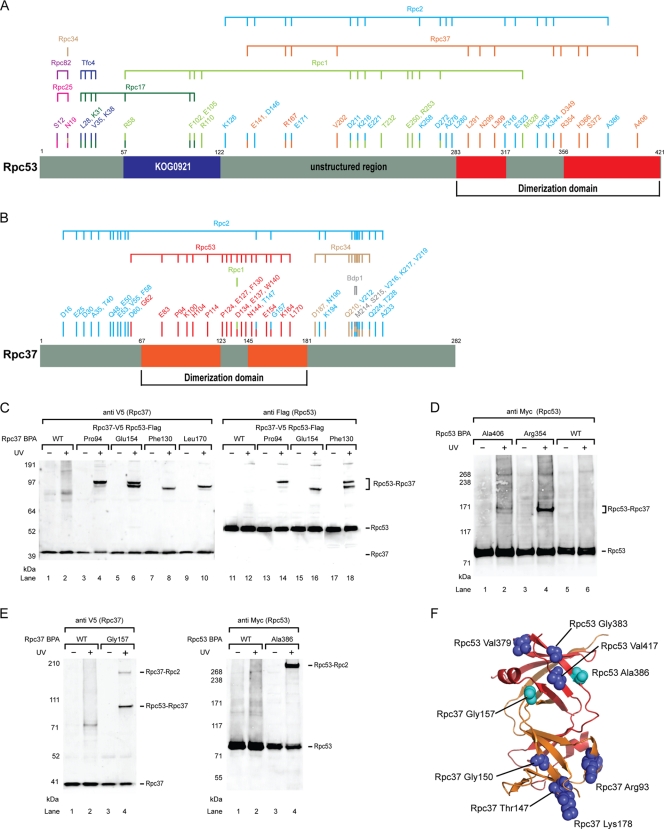
As the Rpc37/53 subcomplex was revealed to be in direct contact with Rpc2 lobe and external 2 domains, we next incorporated the photo-cross-linker BPA into these two subunits to further map interactions of the subcomplex. As summarized in Fig. 2A and B and documented in Table S2 and Table S3 in the supplemental material, Rpc37/53 showed numerous UV-dependent cross-links in the immobilized template assay. As with the identification of Rpc2 cross-links, Rpc37 and Rpc53 cross-linked polypeptides were verified by Western analysis of photo-cross-linking experiments using cell extracts containing epitope-tagged cross-linking candidates. To better define Rpc37/53 functional regions derived from our cross-linking analysis, we applied homology modeling to identify potential structural domains based on multiple sequence alignments generated by HHpred and the proposed alignment by Geiger et al. (27, 48). We then built a structural model of the yeast Rpc37/53 dimerization module by the use of Modeler software (23) and used the model in subsequent studies.
Fig. 2.
Photo-cross-links of Rpc37/53. (A) Schematic representation of BPA incorporation in Rpc53 and cross-linked polypeptides. The region of Rpc53 homologous to the dimerization domain of human TFIIF small subunit RAP30 is shown in red. Numbers labeled above the schematic bars represent BPA-substituted residues generating cross-links. Color coding of each residue corresponds to the identity of cross-linked polypep-tide as indicated above the horizontal connecting lines. (B) Schematic representation of BPA incorporation in Rpc37 and cross-linked polypeptides. The region of Rpc37 homologous to the dimerization of human TFIIF large subunit Rap74 is shown in orange. Like in panel A, BPA-substituted residues and the corresponding cross-linked polypeptides are indicated. (C) Representative Rpc37-Rpc53 cross-links in the Rpc37 dimerization domain. Positions of BPA incorporation in Rpc37 are indicated. Anti-V5 antibody was used to reveal C-terminally V5-tagged Rpc37 and its cross-linking fusion bands (left panel). Rpc37-Rpc53 cross-links were verified by Flag epitope tagging at the C terminus of Rpc53 and probing with anti-Flag antibody (right panel). WT, wild-type without BPA incorporation. (D) Representative Rpc53-Rpc37 cross-links in the Rpc53 dimerization domain. C-terminally 13Myc-tagged Rpc53 and its cross-links are shown. Rpc53-Rpc37 cross-links were verified by Flag-epitope tagging in Rpc37 and probing with anti-Flag antibody (data not shown). (E) Rpc37- and Rpc53-Rpc2 cross-links in the Rpc37/53 dimerization domain (Rpc37 Gly157 and Rpc53 Ala386). (F) Structural model of the Rpc37/53 dimerization module, with Rpc37 shown in orange and Rpc53 shown in red. Light blue spheres indicate residues Ala386 in Rpc53 and Gly157 in Rpc37 that were shown to contact Rpc2 by photo-cross-linking. These two residues and additional residues (blue spheres) of the dimerization domain were subjected to single-cysteine substitution for FeBABE-conjugation to probe their interaction with Rpc2.
BPA-substituted residues in the respective Rpc37 and Rpc53 dimerization domains showed extensive intersubunit cross-links (Fig. 2C and D), in good agreement with the domain prediction and structural model. In addition, as displayed in Fig. 2E, we found that residues such as Gly157 in Rpc37 and Ala386 in Rpc53, which are located on the same side in the dimerization model (Fig. 2F), cross-linked with Rpc2, suggesting the existence of a protein surface on the dimerization module for Rpc2 binding.
The Rpc37/53 dimerization module contacts Rpc2 lobe and external 2 domains.
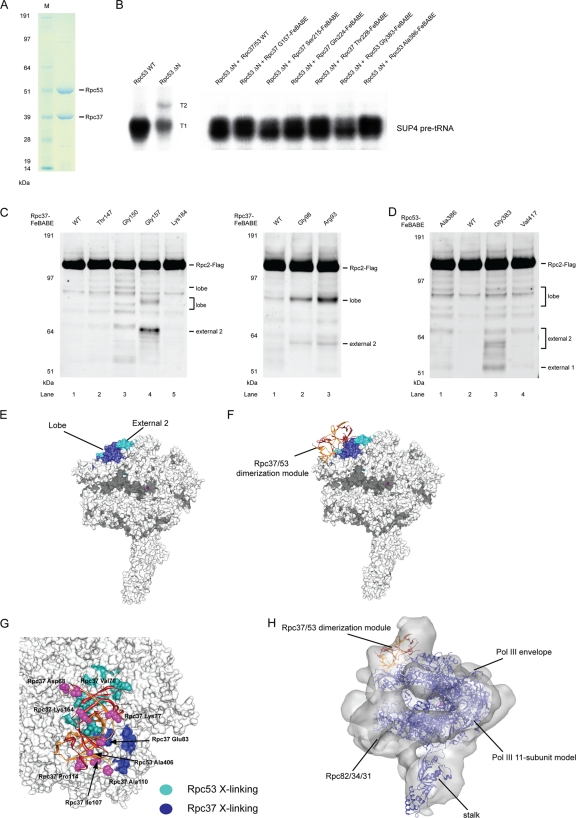
The exact binding interface between Rpc37/53 dimerization module and Rpc2 remained unknown, even though the dimerization module was previously modeled on the surface of Rpc2 lobe based on cryo-EM results. The Rpc37/53-Rpc2 cross-linking data obtained here revealed the first direct evidence for the suggested protein-protein interactions. To provide a more detailed insight into the Rpc37/53 binding mode, the hydroxyl radical-generating probe FeBABE was tethered to specific cysteine residues on the surface of the Rpc37/53 dimerization module, particularly on the surface predicted to contact Rpc2 (Fig. 2F). To permit hydroxyl radical probing in the Pol III PIC, we utilized the FeBABE-tethered recombinant Rpc37/53 in the immobilized template assay containing the cell extract from a mutant strain lacking the Rpc53 N-terminal fragment (Δ2–280). All Rpc37/53-FeBABE derivatives were observed to compensate the transcriptional defect caused by the Rpc53 truncation, indicating that incorporation of the Rpc37/53-FeBABE derivatives into the Pol III PIC was successful (Fig. 3A and B). We utilized these FeBABE derivatives to analyze hydroxyl radical protein cleavage and map binding sites for the respective Rpc37/53 residues on other Pol III subunits, especially Rpc2.
Fig. 3.
Directed hydroxyl radical probing localizes Rpc37/53 dimerization module on Rpc2 lobe/external 2. (A) SDS-PAGE analysis and Coomassie staining of the purified recombinant Rpc37/53 dimer. (B) Representative transcription assays of Rpc37/53-FeBABE derivatives. Whole-cell extracts (Rpc53 WT or ΔN) and the added Rpc37/53-FeBABE derivatives are indicated above. Each derivative in transcription assay with Rpc53 ΔN, deletion of Ser2-Leu280, whole-cell extract was shown to restore full transcription level and termination property on the SUP4 tRNA gene as the wild type extract. (C) Western analysis of hydroxyl radical cleavage of Rpc2 by FeBABE conjugated to Rpc37 surface residues in its dimerization domain. Rpc2 and peptide fragments generated from hydroxyl radical cleavage were visualized by probing with anti-Flag antibody against the C-terminally Flag epitope-tagged Rpc2. The single cysteine substitutions used for FeBABE conjugation in Rpc37 are indicated on the top, and the positions of the substituted residues are displayed in the dimerization domain model in Fig. 2F. Rpc2-Flag and the corresponding sites of cleavage fragments are indicated on the right. (D) Rpc2 cleavage from Rpc53-FeBABE derivatives in the dimerization domain. (E) Hydroxyl radical cleavage sites on the Rpc2 surface from tethered FeBABE in the Rpc37/53 dimerization domain. Calculated cleavage sites are mapped on the Pol III 11-subunit model. Based on the accuracy of cleavage site calculation, individual cleavage sites are represented by 11-residue patches centered at the cut sites and are colored in blue and cyan for cleavages from Rpc37- and Rpc53-FeBABE derivatives, respectively. (F) Model of the Rpc37/53 dimerization module on the Pol III 11-subunit core. The model was built by applying FeBABE cleavage patterns as structural restraints for ZDOCK modeling. Ribbon model represents the dimerization module, with Rpc37 in orange and Rpc53 in red. Same as in E, the hydroxyl radical cleavage sites in Rpc2 from FeBABE derivatives are colored. (G) Cross-validation of the Rpc37/53 dimerization domain-Pol III core model by the photo-cross-linking pattern from Rpc2-BPA. Blue and cyan residues on the Pol III 11-subunit (core) surface model indicate BPA-substitutions in the Rpc2 lobe/external 2 domains that produce cross-links to Rpc37 and Rpc53, respectively. The Rpc37/Rpc53 dimerization domain is shown as ribbons with Rpc37 in orange and Rpc53 in red. Magenta spheres represent BPA-substituted residues that are predicted from our model of Rpc37/53 dimerization domain on the Pol III core (shown as the white molecular surface below) to face away from Rpc2. None of these outward-facing residues in the dimerization domain cross-links to Rpc2. (H) Fit of the Rpc37/53 dimerization module-Pol III core model into the cryo-EM envelope of Pol III (25). The Pol III core is shown in blue ribbons. Rpc37 and Rpc53 are shown in orange and red, respectively.
Rpc2 cleavage fragments generated by Rpc37-FeBABE derivatives were detected by Western blotting; representative data are shown in Fig. 3C (data are summarized in Table S4 in the supplemental material). The approximate positions of Rpc2 cleavages were deduced by calculating the mobility shift of each fragment based on the molecular-weight ladder generated using in vitro-translated, C-terminally Flag-tagged Rpc2 (data not shown) (12). The determined cleavage sites were distributed in the lobe and external 2 domains of Rpc2. The Rpc2 cleavage from Rpc53-FeBABE derivatives indicated the presence of similar cut sites with additional positions mapped to the external 1 domain of Rpc2 (Fig. 3D). The cleavage sites were then mapped onto the Pol III 11-subunit model to localize the interactions between the dimerization module and Rpc2 (Fig. 3E). Both the Rpc2 cross-linking and FeBABE cleavage data are highly consistent (compare Fig. 1D and 3E), revealing that the Rpc37/53 dimerization module contacts the Rpc2 lobe and external 2 domains.
Because FeBABE is known to produce hydroxyl radical cleavage within a range of 30 Å from the Fe-EDTA center (31), we utilized the Rpc2 cleavage positions from each specific FeBABE derivative as distance restraints to model the binding of the Rpc37/53 dimerization module on the Pol III 11-subunit core model using ZDOCK (Fig. 3F) (14). The model was cross-validated by inspecting individual Rpc2 BPA cross-links in the lobe and external 2 domains as shown in Fig. 3G; all cross-linking and cleavage results were internally consistent. Additionally, BPA substitutions of residues on the Rpc37/53 dimerization module facing away from the RNA polymerase did not yield Rpc2 cross-links (Fig. 3G), strongly supporting our model of the Rpc37/53 dimerization module on the Pol III core. Furthermore, when the model was fitted into a Pol III cryo-EM envelope (Fig. 3H), the dimerization module and the rest of the model were almost entirely enclosed by the envelope, except for Rpc17/25 of the Pol III stalk (25, 32, 52).
Rpc53 extends to the Pol III stalk and contacts Tfc4.
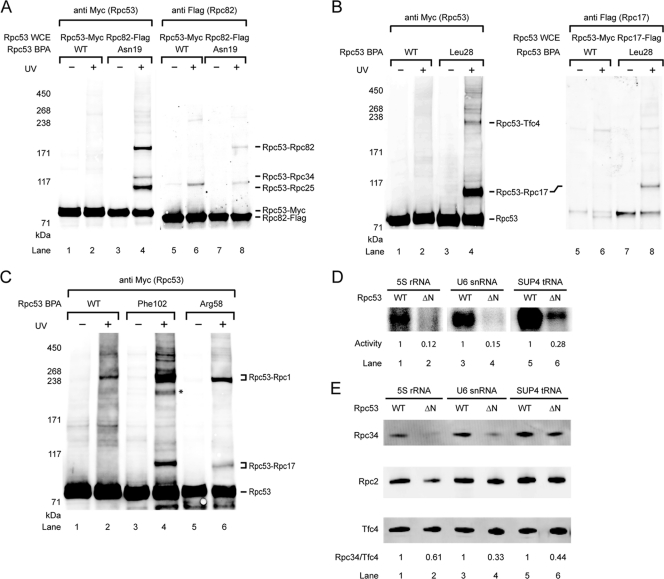
Further expansion of photo-cross-linker incorporation into Rpc53 revealed previously uncharacterized interactions of the N-terminal region of Rpc53 with the Pol III-specific Rpc82/34/31 subcomplex and with the Rpc17/25 dimer that forms the stalk (Fig. 2A). As shown in Fig. 4A, BPA substitution of Asn19 in Rpc53 produced a cross-link confirmed to be Rpc82 as well as two other cross-links with Rpc34 and Rpc25 (data not shown). Located a few residues away, Ser12-BPA also cross-linked to Rpc82 and Rpc25 (data not shown). Cross-linking to another stalk subunit, Rpc17, was observed for a BPA substitution of Leu28 (Fig. 4B). The Rpc53-Rpc17 cross-link was additionally observed for BPA substitutions of Arg58, Phe102, and Glu105, and these three residues also showed another high-molecular-weight cross-link verified to be an Rpc53-Rpc1 product (Fig. 4C).
Fig. 4.
The N-terminal region of Rpc53 contacts multiple Pol III subunits and TFIIIC for Pol III recruitment. (A) Western blot analysis showing that Rpc53 cross-links with the Rpc82/34/31 subcomplex. Rpc53 Asn19-BPA cross-linking was visualized by probing with anti-Myc antibody in the left panel. The cross-linked polypeptide Rpc82 was validated by Flag tagging Rpc82 as indicated in the right panel (anti Flag). Rpc34 and Rpc25 cross-links were also separately verified by Flag tagging (data not shown). (B) Rpc53 cross-links with the stalk protein Rpc17 and the Tfc4 subunit of the transcription factor TFIIIC. Rpc53 Leu28-BPA cross-linking was visualized by probing with anti-Myc antibody in the left panel. Right panel: the shift of Rpc53-Rpc17 fusion band was detected after Flag-tagging Rpc17 (compare lane 4 and 8), and Rpc17 in the cross-linking band was identified by probing with anti-Flag antibody (lane 8). Similarly, the Rpc53-Tfc4 cross-link in the upper fusion band was verified by Flag tagging Tfc4 (data not shown). (C) Representative Western analysis showing that BPA substitutions at Rpc53 residues Phe102 and Arg58 cross-link to Rpc1 and Rpc17 simultaneously. Asterisk, unidentified cross-linked polypeptide. (D) Rpc53 Δ(11–40) mutant compromises transcription activity. The autoradiogram shows transcription activities from the indicated Pol III promoters after PIC formation on the immobilized DNA templates and subsequent addition of NTPs spiked with 32P-labeled GTP. The relative activity of WT and Rpc53 N-terminal (aa 11 to 40) deletion (ΔN) mutant is listed below each lane. (E) The Rpc53 Δ(11–40) mutant affects PIC formation. Immobilized assay was conducted using Rpc53 mutant or Rpc53 WT whole-cell extracts with three types of Pol III promoters. Anti-Tfc4 and anti-Rpc34 antibodies were used to quantify Tfc4 and Rpc34 by Western analysis. The relative intensity of Rpc34, normalized to Tfc4, is listed below each lane. Additionally, Rpc2 was visualized by probing with anti-Flag antibody against C-terminally Flag tagged Rpc2 in the whole-cell extract.
As determined on the basis of a homology search using HHpred (48), the residues cross-linking simultaneously to Rpc17 and Rpc1 lie within an N-terminal segment homologous to KOG0921 (Fig. 2A), a conserved domain present in protein homologs of the MLE RNA helicase (8). The segment C terminal to KOG0921 showed extensive interactions with Rpc2; in particular, one segment (amino acids [aa] 211 to 272) concurrently cross-linked to Rpc1 and Rpc2 (data are summarized in Fig. 2A). This result suggests that this segment is located between Rpc1 and Rpc2. Our cross-linking data thus indicate a multisubunit binding mode for the N-terminal region of Rpc53 that includes interactions with the stalk, the Rpc82/34/31 subcomplex, and likely the active site cleft of Rpc1/Rpc2.
In addition to the interactions between Pol III subunits, one of the two distinct cross-links for Rpc53 Leu28-BPA was verified to be the Tfc4 subunit of transcription factor TFIIIC (Fig. 4B, lane 4; upper band). Tfc4-Rpc53 cross-links were also observed at residues Lys31 and Val35 (Fig. 2A). Among the three types of Pol III promoters in yeast, the type I 5S rRNA promoter contains an internal control region (ICR) to which TFIIIA binds to recruit TFIIIC. The type II promoter, exemplified by tRNA genes, contains intragenic A and B boxes for TFIIIC recognition, whereas the type III U6 snRNA promoter contains a canonical TATA box, an A box, and an unusual extragenic B box located about 120 bp downstream of the termination site (5, 9, 22, 33, 45). To examine whether the interaction between Rpc53 and Tfc4 is specific with respect to the promoter type, we performed additional cross-linking experiments using BPA-substituted Leu28 and immobilized DNA containing either the U6 or 5S promoter. Interestingly, the Rpc53-Tfc4 cross-link also formed at both the U6 and 5S promoters (data not shown), indicating that the Rpc53-Tfc4 contact is conserved in the three classes of Pol III promoters. This result is consistent with the notion that recruitment of Pol III to the U6 promoter is dependent on the presence of TFIIIC (5), although it has been shown that TFIIIB alone can suffice for U6 snRNA transcription in vitro (7, 20).
The N-terminal region of Rpc53 contributes to PIC formation.
On the basis of our BPA photo-cross-linking results, we wondered which regions in Rpc53 and in Rpc37 are essential in vivo. A series of yeast strains containing internal deletions or terminal truncations of either Rpc53 or Rpc37 were generated by plasmid shuffling, and cell viability was scored on YPD plates (see Table S5 in the supplemental material). Unexpectedly, our results showed that the N-terminal region of Rpc53 was not required for cell viability (see Table S5B in the supplemental material [del S2-L280]); the corresponding yeast strain with this deletion displayed reduced growth at 18°C but normal growth at permissive temperature. In contrast, the predicted C-terminal dimerization domain of Rpc53 and, analogously, the dimerization domain of Rpc37 are essential for growth. A similar phenomenon was observed for the TFIIF dimerization domain but not for the Rpa49/34.5 dimerization domain (36, 40). In line with the photo-cross-linking results and the structural prediction, the conserved Rpc53 segment (aa 281 to 315) within the dimerization domain is required for cell viability; by contrast, the nonconserved insertion (aa 316 to 350) that exhibited cross-linking to Rpc1 or Rpc2 was found to be inessential (see Table S5B in the supplemental material and Fig. 2A).
To gain more insight into the functional role of the Rpc53 N-terminal region, we assayed in vitro transcription activity of the Rpc53 Δ(11–40) mutant that displayed a cold-sensitive phenotype (see Table S5B in the supplemental material). As revealed by BPA photo-cross-linking, the deleted segment contains multiple binding sites for Rpc82, Rpc34, the Pol III stalk, and Tfc4 (Fig. 2A). The transcriptional activity of a whole-cell extract bearing Rpc53 Δ(11–40) was reduced to ∼30% of the activity of the wild-type extract (Fig. 4D). We further investigated whether the mutant affected PIC formation. In the immobilized template assay, Rpc53 Δ(11–40) reduced Rpc34 levels on the three types of promoter DNA to 33% to 61% of the levels obtained with the wild type (Fig. 4E). These results indicated that the decrease in transcription activity may have been caused, at least in part, by the reduction of Rpc34 of the Pol III-specific Rpc82/34/31 subcomplex on promoters, suggesting that the protein interaction network of the Rpc53 N-terminal segment (aa 11 to 40) is important for a complete assembly of the PIC.
The C-terminal region of Rpc37 contains binding sites for Rpc2, Bdp1, and Rpc34.
Despite extensive cross-linking of the region N terminal to the dimerization domain in Rpc37 with Rpc2, our mutation studies showed that deletion of this region did not hamper cell growth (see Table S5A in the supplemental material). In contrast, several internal deletion mutations in the region C terminal to the dimerization domain resulted in lethality. Further photo-cross-linking experiments in this region showed widespread cross-links with Rpc2 and Rpc34 (Fig. 2B), and two consecutive residues, Met214 and Ser215, revealed an unexpected cross-link to Bdp1 (Fig. 5A and data not shown). Interestingly, Ser215-BPA yielded multiple bands that were identified as cross-links with three polypeptides, Rpc2, Rpc34, and Bdp1.
Fig. 5.
The C-terminal region of Rpc37 contains binding sites for Bdp1 and lies near the active center for accurate termination. (A) Cross-links of Rpc37 Ser215-BPA to Bdp1, Rpc2 and Rpc34 identified by Western analysis. Left panel: Rpc37 and its cross-links were detected using anti-V5 antibody (lanes 1 to 4). Right panel: Identification of the cross-linked polypeptide Bdp1 (lanes 5 to 8) was achieved by immunostaining of the Flag tag containing Bdp1 with anti-Flag antibody. Similarly, verification of Rpc37-Rpc2 cross-link was based on probing with anti-Flag antibody for epitope tagged target (data not shown). The Rpc37-Rpc34 cross-link was identified by probing with anti-Rpc34 antibody (data not shown). (B) Position of Rpc37 Ser215 is mapped to the Rpc2 lobe. Western analysis showing Ser215-FeBABE cleaved Rpc2 lobe. The calculated cleavage site is shown in panel C. (C) Localization of the C-terminal domain of Rpc37 (Rpc37 C). Pol III 11-subunit core (blue ribbon) and Rpc37/53 dimerization module (orange and red ribbons) are fitted in Pol III cryo-EM envelope as in Fig. 2F. The yellow ribbon indicates the cleavage site in Rpc2 from the Rpc37 Ser215-FeBABE derivative. The orange oval marks the approximate position of Rpc37 C on the Pol III core. Protein-protein interactions with Bdp1 and Rpc34 are indicated. Template (cyan) and nontemplate DNA (green) are located as observed in the Pol II elongation complex (1Y1V) (35). The magenta sphere shows the active center of Pol III. Possible location of the N-terminal domain of Rpc53 (Rpc53 N; red oval) on the Pol III core is also indicated. Protein-protein interactions with Rpc34, Rpc82, Rpc17, Rpc25, and Tfc4 for Rpc53 N are marked by arrows. (D) Rpc37 C-terminal truncation mutants reduce termination efficiency. Transcription assay was conducted with whole-cell extracts containing indicated Rpc37 mutants following the immobilized template assay on the SUP4 tRNA gene that contains two consecutive T-stretches at the 3′-end of its coding sequence. T1 and T2 transcription products observed in the autoradiogram are generated from termination at the upstream and downstream T stretches, respectively. Mutants with reduced termination efficiency shift termination to site T2, whereas WT is observed to terminate mostly at T1. The relative termination shift, calculated as intensity ratio of T2 over the sum of T1 and T2, is listed below each lane. (E) Rpc37 Δ(226–230) mutant does not affect Rpc37 association with Rpc2. Coimmunoprecipitation was conducted with anti-Flag agarose gel to pull down Flag-tagged Rpc2, and probed for coimmunoprecipiated V5 epitope tagged WT or indicated Rpc37 mutants with anti-V5 antibody. The relative intensity of Rpc37 WT/mutant immunostaining is normalized to Rpc2 and listed below each lane. In addition, coimmunoprecipitated Rpc53 was visualized by probing with anti-Rpc53 antibody. In contrast to the observed coimmunoprecipitation of Rpc37 Δ(226–230) mutant, the association of Rpc37 Δ(256–282) mutant with Rpc2 was severely compromised. Coimmunoprecipitation of Rpc53 correlates with the association of Rpc37 WT/mutant with Rpc2. (F) The C-terminal region of Rpc37 is located near the Pol III active center. FeBABE hydroxyl radical cleavage of Rpc2-Flag by Rpc37-FeBABE derivatives is shown in the Western blot. Amino acid positions of Rpc37-FeBABE derivatives are listed above, and the corresponding Rpc2 cleavage sites for individual Flag epitope containing fragments are indicated on the right. The asterisk indicates an unmapped Rpc2 fragment. (G) The C-terminal region of Rpc37 moves closer to the βDloop2 of Pol III active site in the elongation complex. Rpc2 cleavages from Rpc37-FeBABE derivatives within the Pol III PIC (lanes 1 to 3) and the Pol III TEC (lanes 4 to 6) are shown.
Previous DNA-protein cross-linking studies have shown that Bdp1 cross-links to DNA regions upstream of the SUP4 tRNA gene (−2 or −3 to −37 or −38), whereas Rpc53 cross-links to downstream locations (at +11, +16, and +17); however, no DNA-Rpc37 cross-link was detected in the PIC (1, 2, 39). The Rpc37-Bdp1 interaction might therefore occur at some distance from the promoter DNA. In order to map the interaction surface between Pol III and the transcription factor Bdp1, FeBABE was tethered to cysteine residues replacing those C-terminal residues in Rpc37 that cross-linked to Bdp1. The Rpc37 Ser215-FeBABE derivative produced a cleavage fragment (at aa 317 to 327) within the lobe domain of Rpc2 (corresponding to Rpb2 aa 347 to 356) (Fig. 5B), which is consistent with the Rpc2-Rpc37 cross-links observed in the lobe domain (see Table S1 in the supplemental material). Our biochemical probing data thus reveal important multiprotein interactions near the Rpc2 lobe that include the C-terminal domain of Rpc37, Rpc34, and Bdp1 (Fig. 5C).
The C-terminal region of Rpc37 extends to the Pol III active center.
Direct involvement of Rpc37/53 in transcription termination was observed in previous biochemical analyses in which a Pol IIIΔ mutant lacking the Rpc37/53 dimer exhibited a reduced ability to recognize the regular termination site and generated additional termination read-through transcripts from a downstream termination signal (11, 38). Using the Rpc37 and Rpc53 mutant strains generated in our study that displayed temperature-dependent growth defects, we further investigated the molecular basis of how Rpc37/53 facilitates accurate transcription termination. No viable Rpc53 mutants displayed termination defects except for an N-terminal Δ(2–280) (Rpc53 ΔN) that produced termination read-through transcripts on the SUP4 tRNA gene (Fig. 3B). As reported by Landrieux et al. (38), C-terminal (aa 256 to 282) truncation in Rpc37 results in complete dissociation of the Rpc37/53 dimer after Pol III purification and termination read-through. Utilization of the cell extract from the same Rpc37 C-terminal truncation strain also yielded read-through transcripts (T2) in our assay (Fig. 5D, lane 4); however, shorter transcripts (T1) resulting from normal termination were present as the major transcription products, likely due to inefficient dissociation of the less tightly associated mutant Rpc37/53 after PIC isolation from the immobilized template assay. Interestingly, a smaller internal deletion of the Rpc37 segment (aa 226 to 230) that was identified by cross-linking experiments as binding to Rpc2 (Fig. 2B) influences termination accuracy in a manner similar to that seen with the C-terminal truncation mutant (Fig. 5D, lanes 3 and 4) without affecting the transcription start site (data not shown).
As mentioned above, the Rpc37 segment (aa 226 to 230) interacts with Rpc2 as revealed on the basis of our photo-cross-linking study; therefore, the defective termination resulting from the presence of the Rpc37 Δ(226–230) mutant might be due to a weakened association of Rpc37 or the Rpc37/53 dimer, as seen with the C-terminal (aa 256 to 282) truncation mutant. To test this possibility, whole-cell extracts containing Flag-tagged Rpc2 and wild-type or mutant Rpc37 were immunoprecipitated (IP) with anti-Flag agarose gel and analyzed for IP of Rpc37. In good agreement with previous biochemical analyses (38), Rpc37 Δ(256–282) severely reduced Rpc37 association (Fig. 5E, lane 4); in contrast, Rpc37 Δ(226–230) did not affect the association of Rpc37/53 with Rpc2 (Fig. 5E, lane 3). Thus, the termination defect caused by Rpc37 Δ(226–230) might have been due to an Rpc37-Rpc2 interaction specifically required for termination but not essential for Rpc37-Rpc2 association.
To further localize the Rpc37 (aa 226 to 230) segment on Rpc2, FeBABE was conjugated to cysteine substitutions at Gln224, Leu227, Thr228, and Gly229. Utilization of these Rpc37-FeBABE derivatives in the immobilized template assay produced Rpc2 cleavage sites localized to fork loops 1 and 2 and to a conserved loop lying next to the active site in the E. coli RNA polymerase that was termed βDloopII (49) (Fig. 5F and 6A). To investigate whether these interactions change when Pol III elongates, we isolated the ternary elongation complex (TEC) on the SUP4 template in the presence of ATP, CTP, and UTP (GTP withdrawal) (38). As a fraction of polymerases might fail to initiate transcription (1), the cleavage fragments observed in the PIC could also subsist in the assay for TEC isolation. However, we observed increased cleavage activity in the βDloopII from the Thr228-FeBABE derivative in the TEC (Fig. 5G; compare lanes 3 and 6). Furthermore, we identified two additional cleavage fragments that were mapped to the lobe domain and the hybrid binding domain situated close to βDloopII inside the active site cleft (Fig. 5G and Fig. 6B). Our results thus showed that the C-terminal segment (aa 226 to 230) of Rpc37 lies near the Pol III active center, undergoes a conformational change upon transition to TEC, and further participates in transcription termination.
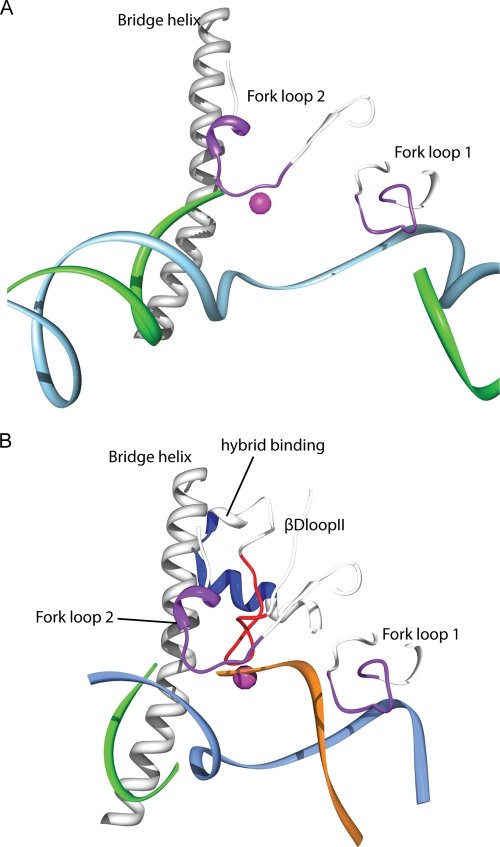
Fig. 6.
Rpc37 influences transcription termination by interacting with fork loops and βDloopII. (A) Model of the Pol III active center and residues cleaved by Rpc37 Thr288-FeBABE derivative within Pol III PIC. The Rpc1 bridge helix and Rpc2 fork loops 1 and 2 are shown as white ribbons, and the calculated cleavage sites are shown as 11-residue purple patches. Template (cyan) and nontemplate DNA (green) strands are as observed in 1Y1V (35) by superimposition of Pol III 11-subunit model with Pol II structure. (B) Rpc37 Thr228-FeBABE cleaves βDloopII within Pol III TEC. The bridge helix, fork loops 1 and 2, βDloopII and the helix in hybrid binding domain are shown as white ribbons and the cleavage sites are indicated in red on βDloopII and in blue on hybrid binding domain, respectively. Template, nontemplate DNA and nascent RNA are as observed in 3HOW (49) and are shown as cyan, green and orange ribbons, respectively.
DISCUSSION
Rpc37/53 participates in multiple steps of transcription, including promoter opening, termination, and reinitiation. Although previous biochemical studies revealed some functions of Rpc37/53 (34, 38), the position of Rpc37/53 within Pol III and the mechanism that allows Rpc37/53 to function remained unclear. On the basis of the biochemical characterization of yeast RNA polymerase III lacking Rpc37, Rpc53, and Rpc11 (Pol IIIΔ) and the unassigned additional electron density described in a previous cryo-EM study, Rpc37/53 was proposed to be located at the outer end of the DNA-binding cleft, sensing the incoming DNA (25, 44). In addition, RNA-protein photo-cross-linking revealed that Rpc53 lies near the catalytic site of Pol III (34).
Through incorporation of BPA into Rpc2, the positions of Rpc37 and Rpc53 on the Rpc2 surface in the Pol III 11-subunit core structure have been mapped (Fig. 1). The position of the Rpc37/53 dimerization module within the PIC was further verified in our study by a hydroxyl radical cleavage assay, and a detailed model for the structure of the Rpc37/53 dimerization module on the Pol III 11-subunit core was constructed to illustrate how this conserved module interacts with the core (Fig. 3). We found that in our model the Rpc37/53 dimerization module contacts Rpc2 in a manner similar to that by which the TFIIF dimerization module contacts Rpb2 of Pol II (21). This finding suggests that interactions in these two nuclear transcription machineries are conserved.
Based on our findings that the N-terminal segment (aa 11 to 40) of Rpc53 cross-links with Rpc82, Rpc34, the stalk proteins Rpc25 and Rpc17, and the Tfc4 subunit of TFIIIC (Fig. 2A), this 30-residue segment likely functions to integrate a multiprotein network important for PIC formation. Consistent with the importance of these interactions, deletion of Rpc53 (aa 11–40) compromised transcription activity and reduced the protein level of Rpc34 in the PIC formation assay. Since Rpc34 remained bound in the purification of Pol IIIΔ lacking the Rpc37/53 dimer (38), the Rpc53 (aa 11 to 40) deletion might not influence the association of Rpc34 in the Pol III complex. Therefore, the destabilization of Rpc34 in the PIC assembly might result from a dynamic interaction between Rpc53 (aa 11 to 40) and Rpc34 upon PIC formation. In addition, as the Pol III-specific Rpc82/34/31 subcomplex was previously proposed to connect the stalk and Rpc1 clamp domain, the Rpc53 (aa 11 to 40) segment as well as Tfc4 could localize to the connection of the Rpc82/34/31 subcomplex and the stalk (Fig. 5C). The location for Tfc4 on the Pol III surface is in good agreement with previous DNA-protein cross-linking studies that showed that Tfc4 contacts a region upstream of the box A sequence and the transcriptional start site where Pol III subunits such as Rpc1, Rpc2, Rpc53, Rpc34, Rpc82, and Rpc25 also make some contacts (2, 4).
Transcription termination by eukaryotic RNA Pol III resembles rho-independent termination by E. coli RNA polymerase, because polymerases in both systems directly recognize the termination signal encoded in the template DNA (44). It has been demonstrated that mutations in Rpc2 of yeast Pol III as well as in its counterpart in E. coli RNA polymerase, the β subunit, alter the termination site(s) (37, 46, 47). The defective termination mutations in the Rpc2 lobe segments (aa 300 to 325) overlap with the Rpc2-Rpc37 cross-linking sites (Fig. 1A), while mutations in another segment (aa 489 to 521) are located near the positions yielding Rpc2-Rpc53 cross-links (Lys531, Tyr534, and Val535; data not shown). In addition, our model for binding of the Rpc37/53 dimerization module on the core deduced from FeBABE-directed hydroxyl radical probing also places the domain on the Rpc2 lobe and external 2 domains (Fig. 3F) in close proximity to the aforementioned Rpc2 segments causing termination defects. Therefore, our biochemical probing analyses and proposed binding model adequately link an important functional role for Rpc2 in transcription termination to its structural association with the Rpc37/53 dimerization module.
In addition to the dimerization domain, the C-terminal segment of Rpc37 (aa 226 to 230) contacting Rpc2 is also accountable for accurate termination. Our FeBABE cleavage analyses revealed that the binding site for this segment is near Rpc2 fork loops 1 and 2. Fork loop 1 was previously found to stabilize the DNA-RNA hybrid and set the upstream boundary for DNA-RNA strand separation (57). Interestingly, mutations in Rpc2 fork loop 2 lead to defective termination (46, 47); this structural motif in both E. coli RNA Pol and yeast RNA Pol II has been proposed to stabilize the downstream edge of the transcription bubble critical for polymerase pausing (28, 51, 54). A recent study also reinforced the role of fork loop 2 in stabilizing the downstream edge of the DNA-RNA hybrid by showing that this loop contributes to Pol II pausing when a frayed RNA 3′ end is present (49). As transcription of a stretch of rU contributes to pausing and termination for Pol III (30, 42) and a run of rU:dA base pairs likely causes frayed RNA at the 3′ end that was previously defined as representing the elemental pause (10, 49, 51), Rpc37 might thus influence the pausing and termination intermediation through its contact with the Rpc2 fork loop 2 near the active site.
Our finding that the Rpc37 segment (aa 226 to 230) is repositioned closer to the βDloopII in the stalled elongation complex further supports the notion that Rpc37 is involved in Pol III pausing and termination. This evolutionarily conserved active site motif was previously shown to be another important structural element contacting and stabilizing frayed or backtracked RNA (49, 51, 53). In summary, our findings, together with those of previous crystallographic and biochemical analyses, lead us to propose that Rpc37 participates in transcription bubble maintenance and perhaps in DNA-RNA hybrid base pairing at the 3′ end through its contacts with Pol III fork loops and βDloopII (Fig. 6A and B). Upon encountering a termination signal, Rpc37 contributes to destabilize the contacts among Pol III active site loops and DNA-RNA hybrids for efficient termination. Alternatively, Rpc37 might interact directly with the 3′ frayed RNA or the transcription bubble for subsequent disruption of DNA-RNA base pairs, leading to destabilization of the ternary complex and dissociation of the polymerase.
Supplementary Material
ACKNOWLEDGMENTS
We thank Cheng-Feng Lo and Abhijit Barate for initial cloning and purification of Rpc37 and Rpc53 and members of the Chen laboratory for helpful suggestions during the course of the work. We also thank Steve Hahn, Bruce Knutson, and Sebastian Gruenberg (Fred Hutchinson Cancer Research Center, Seattle, WA) for comments on the manuscript. The figures were prepared using PyMol and Chimera (University of California, San Francisco).
This work was supported by grants from National Science Council and Academia Sinica, ROC.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 2 May 2011.
REFERENCES
- 1. Bartholomew B., Durkovich D., Kassavetis G. A., Geiduschek E. P. 1993. Orientation and topography of RNA polymerase III in transcription complexes. Mol. Cell. Biol. 13:942–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bartholomew B., Kassavetis G. A., Geiduschek E. P. 1991. Two components of Saccharomyces cerevisiae transcription factor IIIB (TFIIIB) are stereospecifically located upstream of a tRNA gene and interact with the second-largest subunit of TFIIIC. Mol. Cell. Biol. 11:5181–5189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brachmann C. B., et al. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115–132 [DOI] [PubMed] [Google Scholar]
- 4. Braun B. R., Bartholomew B., Kassavetis G. A., Geiduschek E. P. 1992. Topography of transcription factor complexes on the Saccharomyces cerevisiae 5 S RNA gene. J. Mol. Biol. 228:1063–1077 [DOI] [PubMed] [Google Scholar]
- 5. Brow D. A., Guthrie C. 1990. Transcription of a yeast U6 snRNA gene requires a polymerase III promoter element in a novel position. Genes Dev. 4:1345–1356 [DOI] [PubMed] [Google Scholar]
- 6. Brun I., Sentenac A., Werner M. 1997. Dual role of the C34 subunit of RNA polymerase III in transcription initiation. EMBO J. 16:5730–5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burnol A. F., et al. 1993. Basal promoter and enhancer element of yeast U6 snRNA gene. J. Mol. Biol. 233:644–658 [DOI] [PubMed] [Google Scholar]
- 8. Carter R., Drouin G. 2010. The increase in the number of subunits in eukaryotic RNA polymerase III relative to RNA polymerase II is due to the permanent recruitment of general transcription factors. Mol. Biol. Evol. 27:1035–1043 [DOI] [PubMed] [Google Scholar]
- 9. Challice J. M., Segall J. 1989. Transcription of the 5 S rRNA gene of Saccharomyces cerevisiae requires a promoter element at +1 and a 14-base pair internal control region. J. Biol. Chem. 264:20060–20067 [PubMed] [Google Scholar]
- 10. Chan C. L., Wang D., Landick R. 1997. Multiple interactions stabilize a single paused transcription intermediate in which hairpin to 3′ end spacing distinguishes pause and termination pathways. J. Mol. Biol. 268:54–68 [DOI] [PubMed] [Google Scholar]
- 11. Chédin S., Riva M., Schultz P., Sentenac A., Carles C. 1998. The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes Dev. 12:3857–3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen H. T., Hahn S. 2003. Binding of TFIIB to RNA polymerase II: mapping the binding site for the TFIIB zinc ribbon domain within the preinitiation complex. Mol. Cell 12:437–447 [DOI] [PubMed] [Google Scholar]
- 13. Chen H. T., Warfield L., Hahn S. 2007. The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat. Struct. Mol. Biol. 14:696–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen R., Li L., Weng Z. 2003. ZDOCK: an initial-stage protein-docking algorithm. Proteins 52:80–87 [DOI] [PubMed] [Google Scholar]
- 15. Chen Z. A., et al. 2010. Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 29:717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chin J. W., et al. 2003. An expanded eukaryotic genetic code. Science 301:964–967 [DOI] [PubMed] [Google Scholar]
- 17. Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119–122 [DOI] [PubMed] [Google Scholar]
- 18. Cramer P., et al. 2008. Structure of eukaryotic RNA polymerases. Annu. Rev. Biophys. 37:337–352 [DOI] [PubMed] [Google Scholar]
- 19. Dieci G., Fiorino G., Castelnuovo M., Teichmann M., Pagano A. 2007. The expanding RNA polymerase III transcriptome. Trends Genet. 23:614–622 [DOI] [PubMed] [Google Scholar]
- 20. Dieci G., Percudani R., Giuliodori S., Bottarelli L., Ottonello S. 2000. TFIIIC-independent in vitro transcription of yeast tRNA genes. J. Mol. Biol. 299:601–613 [DOI] [PubMed] [Google Scholar]
- 21. Eichner J., Chen H. T., Warfield L., Hahn S. 2010. Position of the general transcription factor TFIIF within the RNA polymerase II transcription preinitiation complex. EMBO J. 29:706–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eschenlauer J. B., Kaiser M. W., Gerlach V. L., Brow D. A. 1993. Architecture of a yeast U6 RNA gene promoter. Mol. Cell. Biol. 13:3015–3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eswar N., Eramian D., Webb B., Shen M. Y., Sali A. 2008. Protein structure modeling with MODELLER. Methods Mol. Biol. 426:145–159 [DOI] [PubMed] [Google Scholar]
- 24. Fernández-Tornero C., et al. 2010. Conformational flexibility of RNA polymerase III during transcriptional elongation. EMBO J. 29:3762–3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fernández-Tornero C., et al. 2007. Insights into transcription initiation and termination from the electron microscopy structure of yeast RNA polymerase III. Mol. Cell 25:813–823 [DOI] [PubMed] [Google Scholar]
- 26. Flores A., et al. 1999. A protein-protein interaction map of yeast RNA polymerase III. Proc. Natl. Acad. Sci. U. S. A. 96:7815–7820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Geiger S. R., et al. 2010. RNA polymerase I contains a TFIIF-related DNA-binding subcomplex. Mol. Cell 39:583–594 [DOI] [PubMed] [Google Scholar]
- 28. Gnatt A. L., Cramer P., Fu J., Bushnell D. A., Kornberg R. D. 2001. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science 292:1876–1882 [DOI] [PubMed] [Google Scholar]
- 29. Hahn S., Roberts S. 2000. The zinc ribbon domains of the general transcription factors TFIIB and Brf: conserved functional surfaces but different roles in transcription initiation. Genes Dev. 14:719–730 [PMC free article] [PubMed] [Google Scholar]
- 30. Hamada M., Sakulich A. L., Koduru S. B., Maraia R. J. 2000. Transcription termination by RNA polymerase III in fission yeast. A genetic and biochemically tractable model system. J. Biol. Chem. 275:29076–29081 [DOI] [PubMed] [Google Scholar]
- 31. Han H., Dervan P. B. 1994. Visualization of RNA tertiary structure by RNA-EDTA. Fe(II) autocleavage: analysis of tRNA(Phe) with uridine-EDTA. Fe(II) at position 47. Proc. Natl. Acad. Sci. U. S. A. 91:4955–4959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jasiak A. J., Armache K. J., Martens B., Jansen R. P., Cramer P. 2006. Structural biology of RNA polymerase III: subcomplex C17/25 X-ray structure and 11 subunit enzyme model. Mol. Cell 23:71–81 [DOI] [PubMed] [Google Scholar]
- 33. Kassavetis G. A., Braun B. R., Nguyen L. H., Geiduschek E. P. 1990. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell 60:235–245 [DOI] [PubMed] [Google Scholar]
- 34. Kassavetis G. A., Prakash P., Shim E. 2010. The C53/C37 subcomplex of RNA polymerase III lies near the active site and participates in promoter opening. J. Biol. Chem. 285:2695–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kettenberger H., Armache K. J., Cramer P. 2004. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell 16:955–965 [DOI] [PubMed] [Google Scholar]
- 36. Kuhn C. D., et al. 2007. Functional architecture of RNA polymerase I. Cell 131:1260–1272 [DOI] [PubMed] [Google Scholar]
- 37. Landick R., Stewart J., Lee D. N. 1990. Amino acid changes in conserved regions of the beta-subunit of Escherichia coli RNA polymerase alter transcription pausing and termination. Genes Dev. 4:1623–1636 [DOI] [PubMed] [Google Scholar]
- 38. Landrieux E., et al. 2006. A subcomplex of RNA polymerase III subunits involved in transcription termination and reinitiation. EMBO J. 25:118–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lannutti B. J., Persinger J., Bartholomew B. 1996. Probing the protein-DNA contacts of a yeast RNA polymerase III transcription complex in a crude extract: solid phase synthesis of DNA photoaffinity probes containing a novel photoreactive deoxycytidine analog. Biochemistry 35:9821–9831 [DOI] [PubMed] [Google Scholar]
- 40. Liljelund P., Mariotte S., Buhler J. M., Sentenac A. 1992. Characterization and mutagenesis of the gene encoding the A49 subunit of RNA polymerase A in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 89:9302–9305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marshall L., White R. J. 2008. Non-coding RNA production by RNA polymerase III is implicated in cancer. Nat. Rev. Cancer 8:911–914 [DOI] [PubMed] [Google Scholar]
- 42. Matsuzaki H., Kassavetis G. A., Geiduschek E. P. 1994. Analysis of RNA chain elongation and termination by Saccharomyces cerevisiae RNA polymerase III. J. Mol. Biol. 235:1173–1192 [DOI] [PubMed] [Google Scholar]
- 43. Mohibullah N., Hahn S. 2008. Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3. Genes Dev. 22:2994–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Richard P., Manley J. L. 2009. Transcription termination by nuclear RNA polymerases. Genes Dev. 23:1247–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schramm L., Hernandez N. 2002. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 16:2593–2620 [DOI] [PubMed] [Google Scholar]
- 46. Shaaban S. A., Bobkova E. V., Chudzik D. M., Hall B. D. 1996. In vitro analysis of elongation and termination by mutant RNA polymerases with altered termination behavior. Mol. Cell. Biol. 16:6468–6476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shaaban S. A., Krupp B. M., Hall B. D. 1995. Termination-altering mutations in the second-largest subunit of yeast RNA polymerase III. Mol. Cell. Biol. 15:1467–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Soding J., Biegert A., Lupas A. N. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sydow J. F., et al. 2009. Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA. Mol. Cell 34:710–721 [DOI] [PubMed] [Google Scholar]
- 50. Thuillier V., Stettler S., Sentenac A., Thuriaux P., Werner M. 1995. A mutation in the C31 subunit of Saccharomyces cerevisiae RNA polymerase III affects transcription initiation. EMBO J. 14:351–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Toulokhonov I., Zhang J., Palangat M., Landick R. 2007. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol. Cell 27:406–419 [DOI] [PubMed] [Google Scholar]
- 52. Vannini A., et al. 2010. Molecular basis of RNA polymerase III transcription repression by Maf1. Cell 143:59–70 [DOI] [PubMed] [Google Scholar]
- 53. Wang D., et al. 2009. Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution. Science 324:1203–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang D., Bushnell D. A., Westover K. D., Kaplan C. D., Kornberg R. D. 2006. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell 127:941–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang Z., Roeder R. G. 1997. Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev. 11:1315–1326 [DOI] [PubMed] [Google Scholar]
- 56. Werner M., Chaussivert N., Willis I. M., Sentenac A. 1993. Interaction between a complex of RNA polymerase III subunits and the 70-kDa component of transcription factor IIIB. J. Biol. Chem. 268:20721–20724 [PubMed] [Google Scholar]
- 57. Westover K. D., Bushnell D. A., Kornberg R. D. 2004. Structural basis of transcription: separation of RNA from DNA by RNA polymerase II. Science 303:1014–1016 [DOI] [PubMed] [Google Scholar]
- 58. White R. J. 2005. RNA polymerases I and III, growth control and cancer. Nat. Rev. Mol. Cell Biol. 6:69–78 [DOI] [PubMed] [Google Scholar]
- 59. Zhang C., Zobeck K. L., Burton Z. F. 2005. Human RNA polymerase II elongation in slow motion: role of the TFIIF RAP74 alpha1 helix in nucleoside triphosphate-driven translocation. Mol. Cell. Biol. 25:3583–3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.