Abstract
Formation of facultative heterochromatin at specific genomic loci is fundamentally important in defining cellular properties such as differentiation potential and responsiveness to developmental, physiological, and environmental stimuli. By the nature of their formation, heterochromatin and repressive histone marks propagate until the chain reaction is broken. While certain active promoters can block propagation of heterochromatin, there are also specialized DNA elements, referred to as chromatin barriers, that serve to demarcate the boundary of facultative heterochromatin formation. In this study, we identified a chromatin barrier that specifically limits the formation of repressive chromatin to a distal enhancer region so that repressive histone modifications cannot reach the promoter and promoter-proximal enhancer regions of reaper. Unlike all of the known boundary elements identified for Drosophila melanogaster, this IRER (irradiation-responsive enhancer region) left barrier (ILB) does not exhibit enhancer-blocking activity. Not only has the ILB been conserved in different Drosophila species, it can also function as an effective chromatin barrier in vertebrate cells. This suggests that the mechanism by which it functions to spatially restrict the formation of repressive chromatin marked by trimethylated H3K27 has also been conserved widely during evolution.
INTRODUCTION
Posttranslational modifications of histones play a fundamental role in determining the accessibility and expression status of the underlying genetic information. Distinct histone modifications are associated with chromatin that is open (euchromatin) or closed (heterochromatin) (16). Recent high-resolution “epigenomic” analyses have further revealed that even within a gene, distinct patterns of histone modifications are associated with different anatomic parts of the gene, such as the promoter, enhancer, transcribed region, etc. (51). However, the mechanisms that regulate the temporal and spatial patterns of a particular histone modification remain largely enigmatic.
Chromatin boundary elements are regulatory DNA sequences that help to organize the genome into distinct domains (14, 33, 50). Most of the previously characterized boundary elements in higher eukaryotes harbor two activities. One is enhancer-blocking activity, which prevents enhancer-promoter interaction when the boundary is positioned between them. The other is chromatin barrier activity, which blocks the spread of heterochromatin formation into euchromatic regions (13).
The molecular mechanisms underlying these activities of insulator/boundary elements are not well understood. Much of our current knowledge about the mechanism of enhancer blocking came from studying Drosophila melanogaster insulators, which all have enhancer-blocking activity (14). Several models have been proposed for the mechanism underlying their enhancer-blocking function, including the promoter decoy model, the physical barrier model, and the loop domain model (9, 14, 35).
A few model systems have been exploited to understand the mechanism of chromatin barrier activity. In the Saccharomyces cerevisiae mating type locus, the binding of specific transcription factors adjacent to a silent mating cassette creates a nucleosome-free region that prevents the propagation of heterochromatin into the surrounding genomic regions, thereby confining it to the silent mating cassette (5). For higher eukaryotes, much of what we know about chromatin barrier activity came from studying cHS4, the insulator in the chicken β-globin locus. The complete cHS4 has both enhancer-blocking and barrier activities. However, these activities were found to be separable and are carried out by distinct regions of cHS4 (3, 13, 36, 52). Deletion of the sole CTCF binding site, which is responsible for the enhancer-blocking activity, did not affect the barrier activity of cHS4 (36). Its barrier activity requires a binding site for USF1 (upstream stimulatory factor 1), which recruits chromatin-modifying enzymes that catalyze euchromatin-specific histone modifications incompatible with heterochromatin formation (15, 52).
Although enhancer-blocking and barrier activities are clearly separable and are mediated by distinct cis elements and trans factors in the case of cHS4, it is not clear whether this is common for other metazoan insulators. Many insulators have been characterized for Drosophila and fall into at least 5 types distinguished by their associated binding proteins (14, 25). All of these insulators were originally identified by their enhancer-blocking activity. Several of them, such as the Su(Hw)/gypsy insulator, also have strong barrier activity (17, 38). However, there is no evidence that the two functions are mediated by distinct cis elements. In the case of Su(Hw)/gypsy, it appears that different domains of the DNA binding protein Su(Hw) can interact with different proteins that are required for enhancer-blocking or barrier activity (19).
In searching to understand the mechanism that controls the transcriptional responsiveness of proapoptotic genes to cytotoxic stress, we found that the irradiation-responsive enhancer region (IRER) is subject to epigenetic regulation (54). Remarkably, this IRER, located approximately 5 kb 5′ of the reaper transcription start site (TSS), is also required for irradiation-induced expression of the adjacent proapoptotic genes sickle, located ∼40 kb 5′ of reaper, and hid, located ∼210 kb 3′ of reaper, with five additional annotated genes lying between them. All three genes are transcribed in the same direction. The IRER is in an “open” state in undifferentiated proliferating cells during early embryogenesis, conferring high sensitivity to ionizing radiation-induced apoptosis. However, by late embryogenesis, when most cells have completed proliferation, the IRER has formed a heterochromatin-like structure that is inaccessible to DNase I. Consequently, the epigenetic repression of the IRER renders all three proapoptotic genes unresponsive to irradiation in these cells. This epigenetic blocking, signified by enrichment of H3K27me3 and H3K9me3 and by binding of Polycomb group (PcG) proteins, is strictly limited to the IRER. The promoter and transcribed regions of reaper do not acquire repressive histone marks in older (post-stage 12) embryos and remain open. This restriction of heterochromatin formation to the IRER is functionally significant. While reaper becomes unresponsive to irradiation in post-stage-12 embryos, it is activated in response to developmental signals in neuroblasts (27) and differentiated motor neurons (37) and is required for programmed neuroblast cell death in late embryogenesis.
In this study, we identified a DNA segment at the left (promoter-proximal) edge of the IRER that has strong barrier activity that can block the propagation of repressive histone marks. However, unlike previously characterized boundary elements, such as the Su(Hw)/gypsy insulator and the Fab7 element, the IRER left barrier (ILB) lacks enhancer-blocking activity. Thus, it appears that the ILB is a barrier-only element that restricts the formation of facultative heterochromatin to the IRER, preventing it from spreading toward the promoter and more promoter-proximal enhancers. We show that the ILB binds the DNA-binding protein Cut and that Cut binding is required for its barrier activity. Cut is highly conserved from insects to humans, and correspondingly, the Drosophila ILB also functions as an effective barrier in vertebrate cells.
MATERIALS AND METHODS
Construction of transgenes.
The polylinker sequence was synthesized and cloned into pBluescript KS, between EcoRV and BamHI sites. The multiclonal site of the reconstructed vector contained the following restriction sites: KpnI, HindIII, EcoRV, NdeI, PstI, NruI, NcoI, BamHI, XbaI, and NotI sites. The 3′P-end and 5′P-end sequences were PCR amplified from pP{EndsOut2} (a gift from Jeff Sekelsky), verified by sequencing, and then inserted into the vector restricted with KpnI/HindIII and BamHI/XbaI, respectively. The bacterial attachment (attB) site was amplified from P(24) vector (49) and subcloned at the NdeI site. The 3×P3-DsRed fragment was amplified from M{3xP3-RFPattP} and subcloned at the BamHI site. A 416-bp fragment containing two FRT sequences and an SpeI restriction site between them was inserted at the NcoI site.
(i) pBT1.
A 661-bp NdeI-PstI fragment containing the Ubx Polycomb response element (PRE) sequence was kindly provided by V. Pirrotta. This fragment was subcloned between NdeI and PstI sites in the reconstructed vector mentioned above.
(ii) pBT3.
A fragment containing an MluI restriction site flanked by two loxP sites was synthesized and inserted into the pBT1 vector at the PstI site, between the UBX PRE and FRT sequences. The PCR-amplified Su(Hw) binding region was subcloned into the MluI site between the two loxP sequences.
(iii) pIT1.
The UBX PRE sequence in pBT1 was replaced by five tandem optimized GAL4 binding sites amplified from pUAST (7).
The experimental ILB fragments were amplified by PCR from w1118 flies, using primers containing an SpeI site, and were inserted between two FRT sequences in the vectors described above.
Fly strains, germ line transformation, and genetic crosses.
Flies were grown on standard food medium and maintained at 25°C. The transgenic flies were generated by either P element-mediated transformation or use of a ΦC31 integration system (Rainbow Transgenic Flies, Inc., CA). The w1118 Drosophila melanogaster strain was used for P element insertion. ΦC31 lines 9752 and 9724 (PBac{y[+]-attP-3B}VK00037 and PBac{y[+]-attP-3B}VK00003a) were used for site-specific integration into the 2nd chromosome (49). The transformants were verified by PCR analysis, and the transformation efficiency was recorded (data not shown).
Germ line excision of the ILB 9-kb barrier sequence was performed by crossing the BT1-ILB9kb line 47-2 with flies carrying a heat shock-inducible FLP transposase (y1 w1118 hsFLP). The progeny were heat shocked for 1.5 h at 37°C on 3 to 5 successive days during larval growth. The female progeny were collected and crossed with TM3/TM6 males. In the following generation, flies were selected for a change in the eye-specific DsRed signal, and PRE excision was confirmed by PCR analysis with the following primers: a1, 5′-CGCCAGCAACAAAGAACTAA-3′; a2, 5′-GGCCGCTCTAGTGGATCTTG-3′; b1, 5′-GATAGGACTACGCGCACCAT-3′; and b2, 5′-TGTTCAGCTGCGCTTGTTTA-3′. The BT3 lines with gypsy excisions were obtained by crossing the flies with the Cre line (y1 w67c23; Sco/CyO Crew1). The female progeny were crossed with Sco/CyO males, and the desired flies were selected from the next generation based on the eye-specific DsRed signals. The excisions were verified by PCR analysis.
Chromatin immunoprecipitation (ChIP).
Preparation of fixed chromatin from adult flies was based on the protocol of Cavalli et al. (11), with some modifications. Adult flies (150 to 200 mg) were collected at 5 days posteclosure and cross-linked in 5 ml of buffer A1 (60 mM KCl, 15 mM NaCl, 4 mM MgCl2, 15 mM HEPES [pH 7.6], 0.5% Triton X-100, 0.5 mM dithiothreitol [DTT], 10 mM sodium butyrate, and 1% protease inhibitor cocktail [PIC]) in the presence of 1.8% formaldehyde by homogenization in a Potter homogenizer and then in a Dounce homogenizer with a “loose” pestle (three strokes each), followed by incubation for 20 min at room temperature. Cross-linking was stopped by adding glycine to 225 mM for 5 min on ice. The homogenate was centrifuged for 5 min at 4,000 × g at 4°C, and then the supernatant was discarded and the crude nuclear pellet was washed three times in 3 ml buffer A1 and once in 3 ml buffer A2 (140 mM NaCl, 15 mM HEPES, pH 7.6, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.5 mM DTT, 0.1% sodium deoxycholate, 10 mM sodium butyrate, 1% PIC) at 4°C. After the washes, nuclei were resuspended in 3 ml buffer A2 in the presence of 0.1% SDS and 0.5% N-lauroylsarcosine and then incubated for 10 min in a rotating wheel at 4°C. At the end of the incubation, 0.3 g of acid-washed glass beads (Sigma) was added, and the samples were sonicated on ice, using a Branson Sonifier 450 instrument, to obtain fragmented DNAs with an average size of approximately 500 bp. Immunoprecipitation and quantitative real-time PCR (Q-RT-PCR) were performed as previously described (54).
Gene expression analysis.
mRNA was extracted with RNeasy minikits (Qiagen). cDNA was prepared by reverse transcription of mRNA with a high-capacity cDNA archive kit (Applied Biosystems). Q-RT-PCR followed protocols provided by the manufacturer. The real-time PCR step used 10 ng cDNA/PCR well, with triplicate wells per gene per sample. The following primers were used for Q-RT-PCR: DsRed_f, 5′-GAAGCTGAAAGACGGTGGTC-3′; DsRed_b, 5′-CGTCCCTCGGTTCTTTCATA-3′; corto_f, 5′-CAACAGCACCAGCAGATGTC-3′; and corto_b, 5′-CTCTGGCTCTGGCTCTGACT-3′.
Barrier assay in vertebrate cells.
Chicken 6C2 erythroleukemia cells carrying interleukin-2 receptor (IL-2R) reporter transgenes (clones 9 and 809) were cultured as described previously (34). For the ILB-flanked IL-2R construct, the 294-bp ILB fragment was cloned into both sides of the {βA-promoter-IL2R-βA/ε enhancer} cassette. 6C2 cells were transfected with 1 μg of hygromycin resistance gene from pREP7 (Invitrogen) and with 1 μg of the IL-2R construct (linearized) by electroporation using a Nucleofector machine (program G16) and a Nucleofector kit V (Amaxa). Individual clones were selected in medium containing 1 μg/μl hygromycin.
For fluorescence-activated cell sorter (FACS) analysis, harvested cells were washed in phosphate-buffered saline (PBS) plus 2% fetal bovine serum (FBS), stained with phycoerythrin (PE)-conjugated anti-human IL-2R antibody (eBioscience, San Diego, CA), and analyzed on an Accuri C6 flow cytometer (Accuri Cytometers, Ann Arbor, MI). The unstained cells were used to set the autofluorescence negative control.
RESULTS
Epigenetic blocking of the IRER is restricted to the upstream regulatory region of reaper.
In our previous work, we showed that in late-stage embryos, the IRER forms a facultative heterochromatin structure that is resistant to DNase I, enriched for both H3K27me3 and H3K9me3, and bound by PcG proteins and HP1 (54). While PcG genes are required for epigenetic blocking of the IRER, the spatial characteristics of epigenetic regulation of the IRER are rather different from those of canonical PcG-mediated silencing of the homeotic genes. The transition to the radiation-resistant state during embryogenesis is associated with the acquisition of repressive histone marks and binding of the PcG proteins that are strictly limited to the IRER, which is located more than 2 kb 5′ of the reaper promoter. The reaper promoter (and transcribed region) remains free of repressive histone marks and PcG proteins (Fig. 1 A).
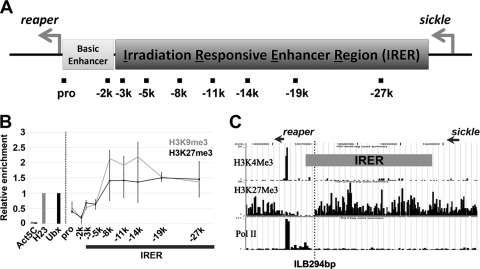
Fig. 1.
Formation of facultative heterochromatin is restricted to the IRER, without reaching the reaper promoter and proximal enhancer regions. (A) Schematic diagram of the intergenic region between reaper and sickle. The IRER is required to mediate irradiation-induced expression of the proapoptotic genes reaper, hid, and sickle (54). Accessibility of the IRER is controlled by a PcG protein-dependent mechanism, which forms a nonpermissive structure in irradiation-resistant cells in post-stage-12 embryos. The decrease of DNA accessibility, accompanied by enrichment of repressive histone marks and binding of PcG proteins, was specifically limited to the IRER without affecting the reaper promoter and proximal enhancer region (54). (B) ChIP assays performed with wild-type adult fly tissues. Pericentromeric heterochromatin locus H23 (H23) and the Ubx promoter region (Ubx) were used as positive controls for repressive histone marks H3K9me3 and H3K27me3, respectively. The coding region of the housekeeping gene Act5C was used as a background control. Enrichment of the repressive histone marks in the IRER was normalized against the respective positive controls and is presented as means ± standard deviations (SD). The high-level enrichment of both H3K27me3 and H3K9me3 in the central part of the IRER dropped significantly at the left boundary of the IRER, about −2 kb to −5 kb relative to the reaper TSS. (C) Distribution of histone marks H3K4me3 and H3K27me3 and binding of Pol II in Drosophila S2 cells revealed by ChIP-Seq. The relative locations of reaper and sickle are indicated by arrows. The dotted vertical line denotes the region that might possess putative chromatin barrier activity.
ChIP analysis with adult flies confirmed that these repressive histone modifications remain restricted to the IRER. The levels of H3K27me3 and H3K9me3 in the central part of the IRER are comparable to or higher than those in the respective positive-control regions of the Ubx promoter and H23 (Fig. 1B). The homeotic gene Ubx is silenced by PRE-mediated repression in most cells. H23 is located within the pericentromeric heterochromatin of chromosome 2. The levels of both repressive marks decrease substantially at the left boundary of the IRER, approximately 5 kb upstream of the reaper TSS.
The restriction of the repressive histone marks to the IRER and their absence from the reaper transcribed region and the immediate enhancer region were confirmed by independent ChIP-sequencing (ChIP-Seq) analysis of cultured Drosophila S2 cells (Fig. 1C). Neither reaper nor hid was responsive to irradiation in the S2 cells (N. Lin et al., unpublished data). The distribution of H3K27me3 upstream of the reaper transcription unit resembles what we observed in adult flies (Fig. 1C).
High-resolution ChIP-Seq analysis also confirmed that the reaper transcription start site is located as previously annotated and is enriched for H3K4me3 and engaged by RNA polymerase II (Pol II) (Fig. 1C). Since reaper mRNA is barely detectable in S2 cells, the significant enrichment of H3K4me3 and RNA Pol II binding suggest that similar to heat shock genes, reaper has an engaged but “paused” RNA polymerase II, ready for quick induction (39, 53).
The restriction of repressive histone marks and DNase-resistant heterochromatin formation to the IRER and their absence from the reaper promoter and transcription unit are functionally important. Although reaper is no longer responsive to irradiation in embryos after stage 12, it is expressed in late embryogenesis and required for the developmentally programmed cell death of selected neuroblasts (4, 32). Thus, the restricted formation of repressive chromatin over the IRER significantly attenuates the transcriptional responsiveness of reaper to environmental stresses but not to other developmental signals. Since the transition to repressive histone modifications occurs about 5 kb 5′ of the reaper TSS, it is unlikely that the transition is due to TSS-associated histone modifications. Rather, we hypothesized that a chromatin boundary element is responsible for preventing the repressive histone marks from spreading to the reaper promoter and the promoter-proximal enhancer regions.
Identification and characterization of ILB.
Due to the large size of the candidate region to be tested, we developed the construct “barrier tester 1” (pBT1), which allows us to test barrier activity in DNA fragments of up to 9 kb via either P insertion or phiC31-mediated docking (Fig. 2 A). With this construct, the compact 3×P3-DsRed reporter/marker gene gives strong DsRed (red fluorescent protein [RFP]) expression in the eye (6, 43). The PRE from the Ubx locus was placed upstream of 3×P3-DsRed. This PRE has been shown to function as a general silencer that can initiate PcG-mediated silencing in different loci (10, 42). Restriction sites between the PRE and the reporter allow insertion of DNA fragments to be tested for barrier activity. The inserted DNA fragment is flanked by two FRT sites to allow FLP-mediated excision. Similar designs with different (larger) reporter genes have been used successfully to demonstrate that the Su(Hw)/gypsy and other insulators can block PRE-mediated silencing when placed between the PRE and the promoter (26, 44). We reasoned that if the tested DNA sequence contains a barrier activity that is able to counteract the repressive effect of the PRE, then the DsRed reporter will be expressed and allow the recovery of insertion events.
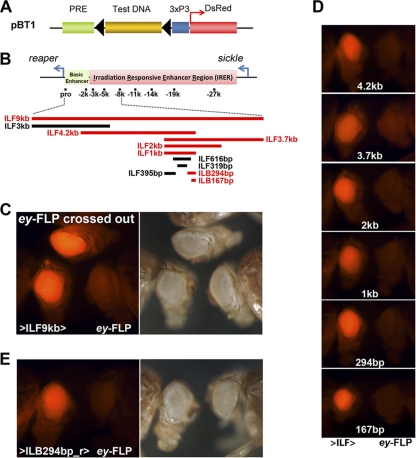
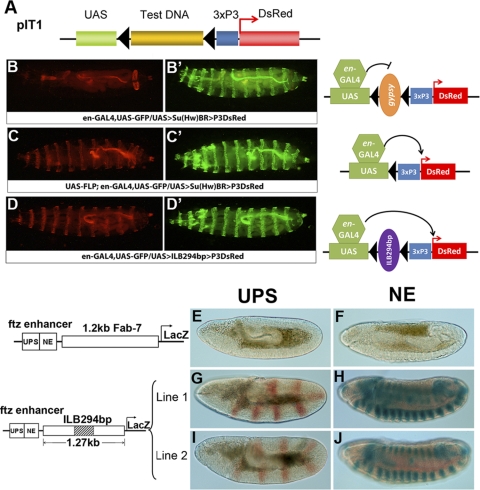
Fig. 2.
Verification of barrier activity and narrowing down the IRER left barrier. (A) Barrier tester construct pBT1. Eye-specific 3×P3-DsRed served as the reporter/marker. The PRE from the Ubx promoter was placed upstream of 3×P3-DsRed to initiate the formation of facultative heterochromatin. The tested fragments (ILFs) were cloned between the reporter gene and the PRE and were flanked by two FRT sequences. The transgenic flies were generated by either P-mediated insertion or ΦC31-mediated integration. (B) A series of fragments within the IRER left boundary region were tested with the pBT1 vector for barrier activity. The fragments with and without barrier activity are shown as red and black bars, respectively. The essential barrier region was narrowed down to the ILB167bp region. (C) Example of verification of barrier activity. The left and right panels show the same group of flies under the RFP fluorescence channel and bright-field microscopy, respectively. Transgenic line 47-2, carrying one copy of pBT1-ILF9kb (>ILF9kb>; fly head on the left), had a strong eye-specific DsRed signal, which was diminished when the ILF9kb fragment was removed by crossing with an ey-FLP strain (ey-FLP; fly head on the right). The DsRed signal was fully restored when ey-FLP was crossed out (top fly head). (D) pBT1 constructs carrying subfragments of ILF9kb were integrated into the same attP docking site on the 2nd chromosome. Barrier activity was verified as mentioned above. The fly heads of the original transgenic strains are on the left, while those that also have ey-FLP are on the right side of each panel. This series of tests indicated that the ILB167 fragment possessed full barrier activity compared to longer fragments. (E) The barrier activity was not affected when the ILB294bp fragment was inserted into pBT1 in the reverse direction, indicating that ILB is orientation independent.
To test the feasibility of this approach, a 9-kb fragment (ILF9kb; Chr3L positions 18,391,265 to 18,399,880 [D. melanogaster genome release 5]) that covers the region from the left side of the IRER to the reaper promoter was cloned into pBT1 (Fig. 2B). P-mediated insertions were recovered by monitoring the 3×P3 DsRed expression in the adult eye (Fig. 2C, left panel). The expression of DsRed suggests that the reporter gene was protected from PRE-mediated silencing by the inserted fragment. To verify that the inserted fragment was indeed responsible for blocking PRE-mediated silencing, the ILF9kb fragment was excised from the transgene in the eye by crossing the transgenic lines to a fly strain carrying ey-FLP. This resulted in a great reduction of the DsRed signal in the eye, indicating that the reporter gene was silenced/suppressed in the absence of ILF9kb (Fig. 2C). The excision of ILF9kb in flies carrying ey-FLP was verified by PCR analysis. The level of DsRed expression was fully restored when ey-FLP was crossed out (Fig. 2C, top fly head). Similar findings were confirmed with all 7 independent insertion lines, suggesting that these observations are independent of the genomic insertion site. These results indicate that ILF9kb is capable of blocking PRE-mediated silencing of the DsRed reporter gene.
To locate the essential barrier sequences within the 9-kb sequence and to rule out the possibility that the observed suppression of PRE-mediated silencing was due simply to the length of the fragment, we tested a series of DNA fragments within the 9-kb region (Fig. 2B). In order to make the expression levels comparable, the test constructs for the subfragments were inserted into the same attP docking site on the 2nd chromosome (line 9752; PBac{y[+]-attP-3B}VK00037). The barrier activities of these fragments were initially screened based on the expression of the marker gene. Positive fragments were then verified by comparing the relative expression levels of the reporter in the absence and presence of ey-FLP (Fig. 2D).
To safeguard against false-negative results due to problems associated with generating the transgenic insertion, a random sample of individual F1 progeny of each injected embryo was assayed for the presence of the transgene by PCR using a pair of primers flanking the two FRT sequences (Fig. 3 A). This PCR analysis confirmed that the transgenic efficiencies for obtaining transgenic animals from the negative and positive constructs were about the same (Fig. 3B). Some of the negative fragments from the original screen were further verified as negative by using a modified version of barrier tester 1. This construct, pBT3, contains a gypsy insulator flanked by two loxP sequences between the PRE and the tested fragment (Fig. 3C). In the presence of the gypsy insulator, both BT3-ILF395bp and BT3-ILF1kb transgenic adults had similar DsRed expression levels in the eye. However, after the removal of the gypsy insulator by Cre-mediated recombination, BT3-ILF395bp flies lost the eye expression of DsRed. In contrast, BT3-ILF1kb flies were not affected by the removal of the gypsy insulator (Fig. 3D). This indicates that without the gypsy element, only the ILF1kb sequence, not the ILF395bp sequence, could counteract the silencing effect of the PRE.
Fig. 3.
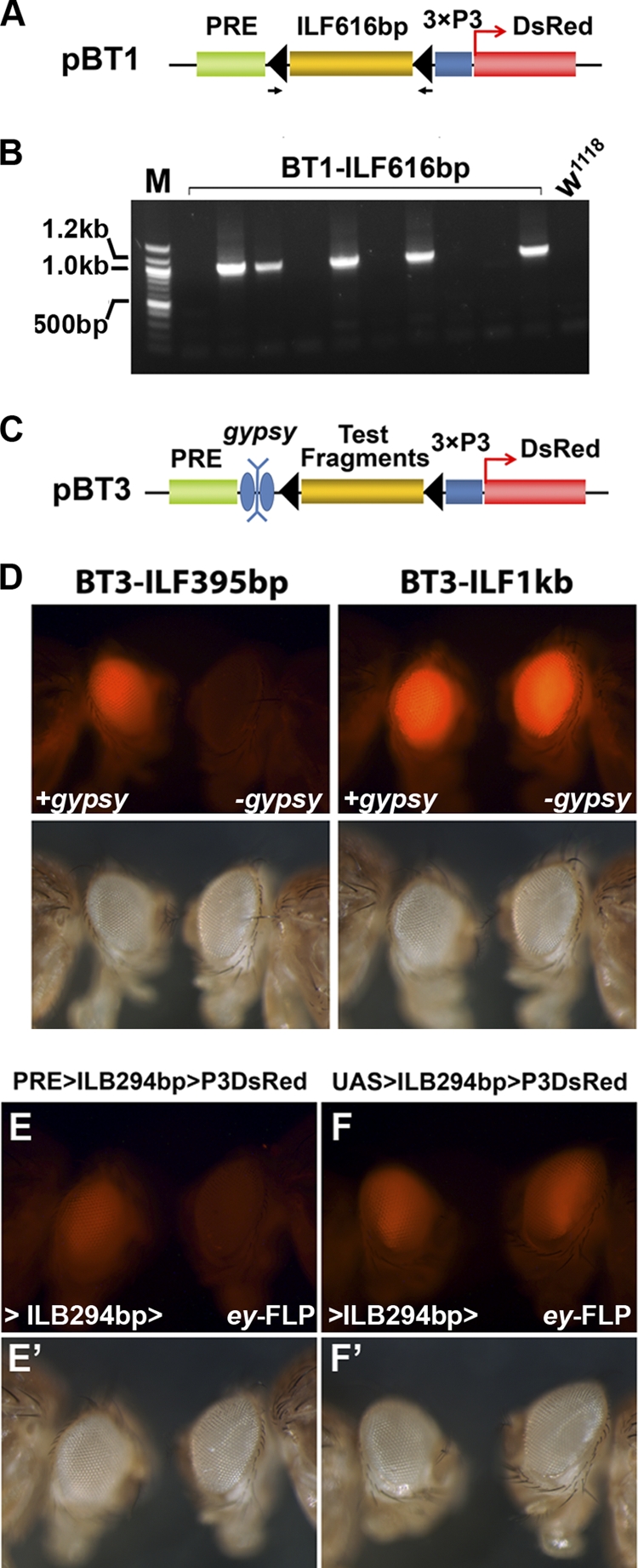
Verification of tested fragments. (A and B) PCR verification of the transformation events demonstrated with BT1-ILF616bp, from which no DsRed-positive flies were recovered. Randomly selected progenies from each individual vial were collected for genomic DNA extraction, and PCR analysis was performed with a pair of primers flanking the two FRT sequences. The genomic DNAs from 5 of 10 tested vials (each established with 2 or 3 injected adults) showed PCR products of around 1 kb, indicating an ∼50% transformation rate. This evidence indicates that the failure to recover BT1-ILF616bp (and other negative fragments) was not due to potential problems associated with transformation but rather was due to the silencing of the reporter gene by the PRE, i.e., the lack of barrier activity of the tested fragment. (C) Some of the negative fragments were further verified with the reporter construct pBT3, which contains a gypsy element flanked by two loxP sequences between the PRE and the test DNA fragment. Transgenic flies were generated with the ΦC31 line 9752. Germ line excision of the gypsy element was performed by crossing the transgenic flies to a strain providing the source of Cre recombinase (y w; Sco/CyO Crew1). ILF395bp was negative, while ILF1kb tested positive in the original BT1-mediated assay. (D) Both BT3-ILF395bp and BT3-ILF1kb transgenic files had similar levels of DsRed in the presence of the gypsy insulator (+gypsy). However, the level of DsRed in the BT3-ILF395bp line diminished after the excision of gypsy (left panel, −gypsy), indicating that ILF395bp does not have barrier activity. In contrast, the excision of the gypsy insulator from BT3-ILF1kb did not lead to any detectable decrease of the DsRed signal (right panel, −gypsy), indicating that ILF1kb is sufficient to block heterochromatin formation initiated by the PRE. To verify that ILB294bp does not have eye-specific enhancer activity, transgenic lines carrying BT1-ILB294 (PRE>ILB294bp>P3DsRed) (E and E′) or IT1-ILB294 (UAS>ILB294bp>P3DsRed) (F and F′) were crossed to flies carrying ey-FLP. The BT1-ILB294 transformant line showed a significant reduction of DsRed signal after the somatic excision mediated by ey-FLP. In contrast, flies carrying IT1-ILB294bp had little change after being crossed with ey-FLP. This indicates that there is no eye-specific enhancer activity associated with the 294-bp fragment.
Testing of progressively shorter fragments led to the identification of a 294-bp fragment that had full barrier activity compared to the original longer fragments. The barrier activity associated with this 294-bp fragment is orientation independent, as the barrier function was not affected when this sequence was inserted in the reverse orientation between the reporter and the PRE (Fig. 2E).
The expression of the reporter in the presence of ILF294bp and the almost complete suppression of DsRed signal in the absence of it strongly suggest that ILF294bp functions as a chromatin barrier against PRE-mediated silencing. However, one alternative explanation is that the tested ILF fragments contained a strong eye-specific enhancer and that the excision of this enhancer resulted in the downregulation of 3×P3-DsRed expression in the eye. To rule out this possibility, we replaced the PRE with a yeast GAL4 upstream activating sequence (UAS) and performed the same assay. As shown in Fig. 3E and F, excision of ILF294bp from UAS>ILF294bp>P3DsRed flies did not cause any reduction of the DsRed signal, indicating that the ILF294bp fragment does not have enhancer activity. With this validation, we are reasonably confident in concluding that there is a chromatin barrier activity within this 294-bp sequence. We refer to this activity as the IRER left barrier (ILB). Further verification indicated that most of the barrier activity resides in a smaller, 167-bp fragment (Fig. 2B). We refer to the two fragments as ILB294bp and ILB167bp, respectively.
ILB prevents PRE-mediated transcriptional silencing of nearby genes.
Q-RT-PCR verified that the strong attenuation of the DsRed signal following removal of the ILB-containing fragments was due to transcriptional silencing of the 3×P3-DsRed reporter gene (Fig. 4A and B). Crossing the positive barrier tester lines with ey-FLP reduced the level of DsRed mRNA to less than 10% of that of the original lines that carried one copy of the barrier tester. This reduction was largely independent of the insertion site, since lines with different original expression levels showed similar relative reductions following removal of the ILB-containing sequence. Considering that ey-FLP may not lead to the excision of the tested ILB-containing fragment in every cell of the eye disc, the observed reductions in mRNA levels assayed by Q-RT-PCR suggest that the Ubx PRE is very efficient in silencing transcription of the 3×P3-DsRed reporter gene. In addition, the data indicate that the ILB element blocks PRE-mediated epigenetic silencing.
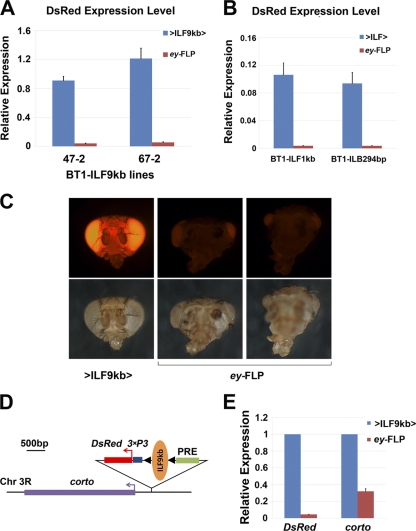
Fig. 4.
ILB prevents transcriptional silencing mediated by PRE. (A and B) The mRNA level of the 3×P3-DsRed reporter gene, detected by Q-PCR, was significantly reduced after the excision of the ILB-containing fragments by ey-FLP. BT1-ILF9kb transgenic lines 47-2 and 67-2 were generated by P insertions (A), while the BT1-ILF1kb and BT1-ILB294bp lines were generated by ΦC31-mediated integration (B). (C) When crossed to ey-FLP, in addition to the decreased DsRed signal, the BT1-ILF9kb transgenic line 67-2 showed an eye ablation phenotype similar to that of a corto mutant. (D) Inverse PCR identified that the BT1-ILF9kb transgene in line 67-2 was inserted about 400 bp upstream of the corto gene by P insertion. (E) For line 67-2, the level of corto expression was significantly reduced, to less than 50% of the original level, after the excision of ILF9kb by ey-FLP.
We noticed that for two of the P-mediated insertion lines carrying BT1-ILF9kb, crossing with ey-FLP not only led to suppression of the DsRed signal in the eye but also led to an eye ablation phenotype (Fig. 4C). Using inverse PCR, we mapped the insertion site in the 67-2 line to about 400 bp 5′ of the corto gene. The orientation of the insertion was such that 3×P3-DsRed was located more proximal to the corto promoter than the PRE (Fig. 4D). When the ILB-containing sequence was excised by ey-FLP, not only was the DsRed mRNA level reduced to less than 10% that of the original line, but the level of corto mRNA was also reduced to about 30% of the original level (Fig. 4E). Homozygous corto mutants exhibit a similar eye ablation phenotype (18), suggesting that the eye ablation phenotype associated with the removal of ILB was due to silencing of the endogenous corto gene by the PRE present within the transgene. The reduction of the corto mRNA level by more than 50% was surprising, given that the 67-2 insertion was heterozygous. However, this was likely due to the known ability of PRE-mediated silencing to also function in trans in such situations, provided that the two copies of the gene targeted by the single PRE undergo homologous pairing (44).
These results indicate that ILB can effectively block robust silencing of the DsRed reporter and both cis and trans silencing of both copies of the adjacent endogenous corto gene by the Ubx PRE.
ILB prevents the propagation of H3K27 trimethylation.
To understand the mechanism of ILB activity, we examined the changes in chromatin structure of the reporter gene before and after the removal of ILB. Removal of the ILB-containing sequence in the 47-2 transgenic line was done by hs-FLP-induced germ line recombination and verified by PCR analysis. Flies without ILB showed a complete loss of DsRed signal in the eye (Fig. 5A). Adult flies of the same age were subjected to ChIP analysis. In the absence of ILB, all four loci within the tester construct had high levels of H3K27me3 enrichment that were comparable to that of the Ubx PRE locus (Fig. 5B, −ILB bars). In contrast, in the presence of the barrier sequence (+ILB), only the locus to the left of ILB had a high level of H3K27me3, while the P3 promoter and DsRed coding regions had significantly lower levels of the repressive histone mark (Fig. 5B, +ILB bars). This indicates that ILB is capable of blocking PRE-initiated propagation of H3K27me3.
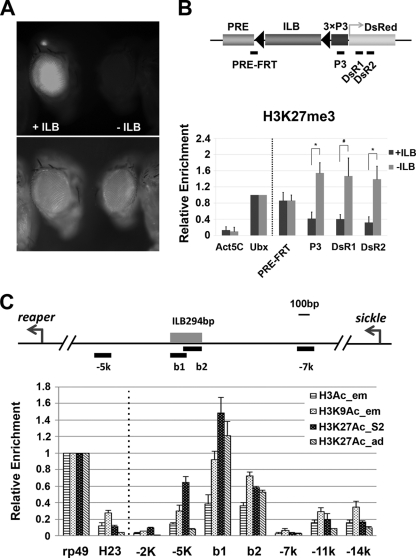
Fig. 5.
ILB blocks the propagation of repressive histone marks initiated by the PRE. (A) hs-FLP was used to remove the ILB-containing fragment through germ line recombination. No DsRed signal was detectable in the resulting PRE>3×P3-DsRed flies (−ILB). (B) Enrichment of H3K27me3 in and around the reporter gene before (+ILB) and after (−ILB) the removal of ILB via germ line recombination. Targeted loci for primer pairs for the ChIP assays are indicated by black bars below the schematic map of the transgene. Removal of ILB led to significant enrichment of H3K27me3 in the reporter gene loci P3, DsR1, and DsR2 (*, P < 0.05; #, P = 0.06847). Note that the level of H3K27me3 remained about the same for the PRE-FRT locus, which is not shielded by ILB. (C) Higher levels of histone H3 acetylation at the barrier site. The ILB294bp region (b1 and b2, approximately 6 kb from the reaper TSS) has a significantly higher level of H3 acetylation than the surrounding region. Specifically, both H3K9 and H3K27 are hyperacetylated in the ILB294 region. ChIP assays were performed with late-stage embryos (H3Ac and H3K9Ac), S2 cells (H3K27Ac), and adult flies (H3K27Ac). Data were normalized against the recovery rate for the rp49 locus before statistical analysis.
The cHS4 barrier activity has been shown to be mediated by the deposition of histone modifications at the barrier site that are characteristic of euchromatin and incompatible with repressive histone marks. We first monitored the distribution of characteristic euchromatic marks in and around the endogenous ILB locus in late-stage embryos, in which the IRER is heavily trimethylated on H3K9 and H3K27. We found that there was a significantly higher level of H3 acetylation within the ∼300-bp region encompassing ILB294bp (b1 and b2 loci in Fig. 5C) than in the immediately adjacent regions (−5k and −7k regions). Specifically, the levels of acetylated H3K9 and H3K27 (H3K9ac and H3K27ac) within ILB were as high as and higher than that of the positive-control rp49 locus, respectively. Not surprisingly, the region immediately to the right of ILB (−7k) had very low levels of H3K9ac and H3K27ac. However, the H3K9ac and H3K27ac levels within ILB were also significantly higher than those at loci immediately to the left of ILB (such as −5k). These observations strongly suggest that the barrier activity of ILB may be mediated by specific histone acetylation activities associated with it.
ILB lacks enhancer-blocking activity.
As mentioned above, all of the known Drosophila boundaries/insulators have enhancer-blocking activity. To test whether ILB has enhancer-blocking activity, we modified the barrier tester construct by replacing the PRE with the GAL4 UAS (Fig. 6A). When a gypsy insulator was inserted between the UAS and the DsRed reporter, the interaction between UAS/GAL4 and the DsRed promoter was totally blocked, as indicated by the absence of epidermal DsRed expression in the presence of the en-GAL4 driver transgene (Fig. 6B and B′). When loss of the gypsy insulator was induced by the presence of a UAS-FLP transgene, DsRed became expressed in the engrailed pattern (Fig. 6C and C′). This confirmed that this construct is sensitive and suitable for testing the enhancer-blocking activity of insulators. However, insertion of ILB294bp between the UAS and the DsRed reporter had no effect on the engrailed expression pattern of DsRed (Fig. 6D and D′), indicating that it does not possess enhancer-blocking activity.
Fig. 6.
The IRER left barrier does not contain enhancer-blocking activity. (A) The reporter construct pIT1 was designed to test enhancer-blocking activity. DNA fragments flanked by FRT were inserted between a UAS sequence and the 3×P3-DsRed reporter. Transgenic flies carrying pIT1 can be crossed to an engrailed (en)-GAL4 UAS-GFP strain. (B) If the DNA fragment has enhancer-blocking activity, such as the gypsy insulator, DsRed cannot be expressed in the engrailed pattern. (B′) GFP channel showing the same larva representing expression of en-GAL4. (C and C′) When the gypsy insulator was removed by FLP, DsRed was expressed in the same engrailed pattern as GFP. (D) With this testing scheme, the ILB294bp barrier element, which had complete barrier activity, did not display any detectable enhancer-blocking activity. (E to J) Potential enhancer-blocking activity of ILB was also assayed with the pCfhL reporter system. The expression of lacZ was placed under the control of ftz UPS and NE enhancers for expression in early and later embryogenesis, respectively. While the 1.2-kb Fab-7 insulator completely blocked the enhancer function of UPS and NE (E and F), the 1.27-kb fragment encompassing the essential ILB294bp barrier sequences did not block the enhancer-promoter interaction for either UPS or NE (G to J). lacZ expression was detected by X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining in germ band-extended (stage 9/10) embryos (E, G, and I) or germ band-retracted (stage 13) embryos (F, H, and J). Five independent pCfhl-ILB transgenic lines were tested, and none blocked UPS or NE enhancer function.
To verify that ILB indeed lacks enhancer-blocking activity, we also sought to test it with previously validated reporter systems. The pCfhl construct designed by Schweinsberg et al. was used successfully to demonstrate the enhancer-blocking activity of Fab-7 (Frontabdominal-7), a boundary element in the bithorax complex (40). Since it contains both an early embryonic ftz enhancer (UPS) and a later embryonic enhancer (NE), it was also used to show that distinct parts of Fab-7 are responsible for enhancer-blocking activity at different development stages (41). In contrast to the Fab-7 insulator, which blocks the ftz enhancer activity (Fig. 6E and F), a 1.27-kb sequence encompassing ILB294bp with ∼500 bp of flanking sequence on each side, inserted between the ftz enhancers and lacZ, failed to block the function of either enhancer (Fig. 6G to J). The levels of expression of 5 independent strains were about the same as or higher than those of two lines carrying a random sequence control (41). This strongly demonstrates that ILB lacks the kind of enhancer-blocking activity seen with fly insulator elements such as gypsy or Fab-7. Notably, not only did the 294-bp ILB lack enhancer-blocking function, but there was no detectable enhancer-blocking function in the 1.27-kb region flanking ILB294bp. This indicates that ILB is also not in close proximity to any enhancer-blocking element.
A Cut binding site is required for ILB activity.
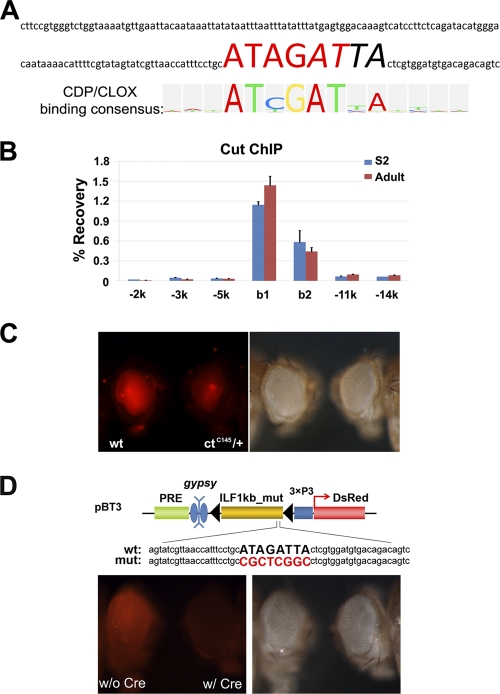
It was reported recently that the Drosophila ortholog of CREB-binding protein (dCBP or Nej) specifically acetylates H3K27 and antagonizes PcG-mediated silencing (47). The analysis of potential binding sites in the 167-bp ILB region identified a site that conforms to the V$CDP_02 matrix (TRANSFAC accession no. M00102) (Fig. 7A). The matrix was generated based on systematic evolution of ligands by exponential enrichment (SELEX) using the human CCAAT displacement protein [CDP; HGNC:Cutl1(Cut-like 1)] (1). CDP is the human ortholog of Drosophila Cut. CDP and Cut share exceptional sequence similarity and display similar DNA binding specificities (31). The Drosophila gene cut, originally named for the notched wing phenotype associated with a viable hypomorph allele, was later found to be an essential gene required for the development of a variety of tissues and organs, such as the central and peripheral nervous systems, muscles, ovarian follicle cells, etc. (reviewed in reference 30). Although it has not been demonstrated for the Cut protein, human CDP/Cutl1 interacts directly with CBP (20).
Fig. 7.
Cut is required for ILB activity. (A) The ILB167bp fragment contains a putative CDP/Cut binding site (large capital letters). The logo representation of the V$CDP_02 matrix is aligned on the bottom and is composed of a palindromic ATCGAT motif (highlighted in red in the corresponding putative binding site) overlapping with the homeodomain binding motif ATTA (italic sequence). (B) Cut protein was highly enriched in the 300-bp region encompassing ILB294bp (b1 and b2), as shown by ChIP analysis of both S2 cells and adult flies. (C) BT1-ILB294bp homozygous females were crossed to either w1118 males or ctC145 males, and the DsRed levels of their female progeny (aged for 2 days) are shown. Significantly decreased levels of DsRed were found in animals heterozygous for ctC145 compared to wild-type (wt) animals. (D) The Cut/CDP binding site in ILB is required for the barrier activity of ILB. When the binding site was mutated (in red), ILB_1k no longer had barrier activity following the removal of the gypsy insulator (for negative and positive examples, refer to Fig. 3). For both panels C and D, the left and right panels are pictures of the same flies taken with the DsRed filter set and no filter (regular light), respectively.
In order to determine whether Cut binds to ILB, we performed ChIP with a monoclonal antibody against Cut. In both S2 cells and adult flies, Cut bound specifically to ILB (Fig. 7B). We then tested whether cut activity is required for ILB function. CutC145 is a null allele of cut, and homozygous animals die during embryogenesis (28). In animals heterozygous for CutC145, reporter expression from BT1-ILB294bp (Fig. 2A and B) was significantly lower than that in the wild-type background (Fig. 7C), indicating that cut function is essential for the barrier activity of ILB.
We further tested whether the Cut binding site is required for ILB activity by using the BT3 construct, which contains the ILB element and a copy of the gypsy insulator flanked by loxP sites (Fig. 7D). Following the Cre-mediated excision of the gypsy element, we found that when the putative Cut binding site in ILB was mutated, the ILB_1K construct could no longer prevent the silencing initiated by the PRE present in the construct (Fig. 7D). In contrast, ILB_1k with the wild-type binding site fully prevented PRE-mediated silencing in the absence of the gypsy insulator (Fig. 3). This result unequivocally indicates that the Cut binding site is required for ILB activity.
ILB is evolutionarily conserved.
The 294-bp ILB sequence (Chr3L positions 18,369,207 to 18,369,500 [genome release 5]) is conserved even in distantly related Drosophila species such as D. virilis and D. mojavensis, both of which diverged from D. melanogaster approximately 40 to 60 million years ago (an alignment is available at the Vista genome browser). To test whether the function of ILB is conserved, we extracted a 2-kb D. pseudoobscura genomic sequence encompassing the orthologous ILB294bp region and tested its activity in pBT1 (Fig. 2A and B). We found that this orthologous sequence had complete barrier activity in D. melanogaster (Fig. 8A).
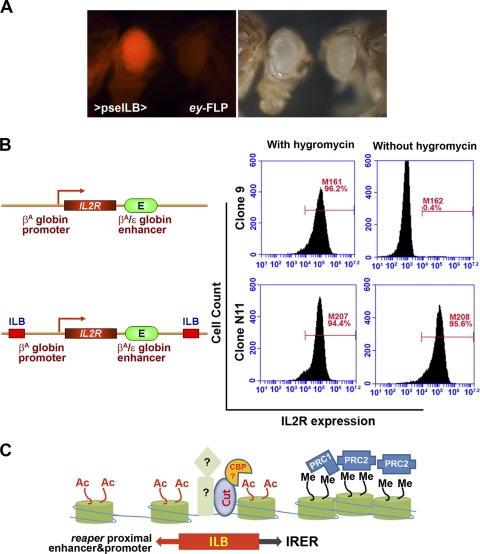
Fig. 8.
ILB is evolutionally conserved. (A) The 2-kb D. pseudoobscura genomic sequence encompassing the ILB294bp orthologous region (pseILB) displayed a similar level of barrier activity in D. melanogaster to that of native ILB. The transgenic flies were generated by ΦC31-mediated integration. (B) The 294-bp ILB protected the reporter from silencing in chicken erythroid 6C2 cells. The designs of the reporters used for generating stable transfection lines are shown on the left. Following hygromycin withdrawal, the reporter without barrier protection was silenced in most cells, as indicated by FACS analyses of IL-2R expression (top). However, when the reporter was flanked on both sides by one copy of ILB294bp (bottom), the expression of the reporter was maintained. Data for representative clones are shown. (C) Diagram summarizing the function of ILB. The binding of Cut likely recruits a histone acetyltransferase such as dCBP, which catalyzes a euchromatic histone modification that is incompatible with heterochromatin formation. The potential interaction between Cut and dCBP remains to be verified. The presence of other DNA binding proteins in addition to Cut is suggested by the conservation of other DNA motifs/binding sites within ILB.
To determine whether ILB function is conserved in vertebrates, we introduced ILB into a barrier tester construct that has been used to investigate the barrier activity of the 5′ HS4 boundary element of the chicken beta-globin gene (Fig. 8B, left panel) (15, 52). A total of 21 stable clones of 6C2 cells were established. Interestingly, even during the selection stage, 3 clones had no detectable level of IL-2R expression, suggesting that the expression of IL-2R may have been suppressed by nearby transcription suppressors. However, in all 18 clones that had detectable IL-2R expression with hygromycin, the expression was maintained 19 days after hygromycin withdrawal (Fig. 8B). In contrast, the IL-2R reporter was silenced in all of the control clones generated with the IL-2R reporter without ILB 19 days following antibiotic withdrawal. For most of the clones with ILB, the expression of the reporter was still maintained at 45 days post-hygromycin withdrawal, indicating that the Drosophila ILB element exhibits similar, if not stronger, barrier activity to that of cHS4 in vertebrate cells.
DISCUSSION
Unlike constitutive centromeric heterochromatin, which is consistent and rarely changes, facultative heterochromatic regions are subject to cell-specific regulation and may adopt a euchromatic formation in certain cells or under specific conditions. What controls the formation of facultative heterochromatin is not fully understood. However, both centromeric and facultative heterochromatin has an intrinsic property to spread until its propagation is blocked. For instance, PRE could recruit PRC2, which has the enzymatic activity to catalyze the formation of the repressive histone mark H3K27me3 in nearby nucleosomes. The H3K27me3 thus formed could in turn recruit PRC1 and PRC2 complexes, which lead to the spread of heterochromatin formation until this cyclic reaction is stopped by a barrier. Several kinds of epigenomic landmarks, such as an active transcribing promoter or a strong enhancer, may serve as a natural barrier to the spread of heterochromatin (35). Yet, under many circumstances, specific boundary elements are needed to demarcate the range of heterochromatin formation.
Using a strategy designed for testing barrier activity against PRE-induced heterochromatin formation, we verified the existence of a chromatin barrier at the left boundary of the IRER and narrowed it down to a 167-bp DNA region. This boundary element is very efficient in blocking the spread of PRE-initiated heterochromatin formation. Similar to what was described for USF-mediated chromatin barrier activity in the cHS4 insulator (15, 52), the endogenous ILB element is associated with high levels of histone acetylation that are incompatible with the H3K27 trimethylation catalyzed by PcG repressive complexes. Unlike any previously identified boundary elements in Drosophila, ILB does not display enhancer-blocking activity.
ILB is a barrier-only boundary element.
Although the barrier activity and enhancer-blocking activity of the cHS4 insulator are mediated by distinct cis elements, these elements interpose with each other in close proximity. To our knowledge, ILB is the only boundary element that neither contains nor is in close proximity to any enhancer-blocking activity. It is noteworthy that there is a putative CTCF binding site (and CTCF binding detected by ChIP-chip/ChIP-Seq) about 1.5 kb upstream of ILB (distal to the reaper TSS) (29). Currently, we do not have evidence to exclude the possibility that the binding of CTCF there could have an enhancer-blocking function. However, it is clear that at least at the vicinity (±500 bp) of ILB294, there is no enhancer-blocking activity detectable by the systems we used. This distinction of the ILB element, although a little surprising, may be required for its role in demarcating the enhancer-specific epigenetic regulation of the IRER.
The IRER is an enhancer region that controls the stress responsiveness of not one but three proapoptotic genes located in the same synteny (21, 54). This synteny contains four IAP antagonist genes, hid, grim, reaper, and sickle, which together are required for most developmental cell death as well as for cell death in response to a variety of environmental stimuli (46). Coordinated expression of reaper and hid is observed during development and is required to eliminate obsolete cells (55). These two genes and sickle are induced within 15 min following ionizing irradiation (8). When the IRER is deleted, none of the three genes can be induced by irradiation (54). While the IRER is required for stress responsiveness, it is not required for other aspects of transcriptional regulation, such as the expression of reaper in differentiated motor neurons or neuroblasts (4, 32, 37). Thus, the formation of DNase I-resistant heterochromatin at the IRER serves specifically to block or downregulate the responsiveness of the three proapoptotic genes to environmental stress. However, at the same time, the basic promoter and other enhancer regions remain open, and the genes can still be expressed under other developmental control.
A barrier-only boundary may be necessary for this type of enhancer-specific epigenetic regulation. Using a fluorescent reporter knocked into the IRER via homologous recombination, we found that epigenetic blocking of the IRER is dynamic and reversible. In specific cells of the developing larval imaginal disc, the IRER could change from a closed conformation (lack of reporter expression) to an open conformation (high-level reporter expression) following environmental stress such as heat shock or irradiation. Consequently, cells with an open IRER are sensitive to stress-induced reaper/hid expression. When the IRER is open, a boundary with enhancer-blocking activity will block the stress-responsive enhancers in the IRER to interact with the reaper promoter. Indeed, our earlier work has shown that several gypsy-containing p and piggyBac insertions between the IRER and the reaper promoter totally block irradiation-induced reaper expression (54).
Our survey of H3K27me3 modifications in Drosophila S2 cells indicated that intergenic islands enriched for this repressive mark account for about 15% of all H3K27me3-positive regions. However, whole-genome surveys of mammalian cells indicated that the majority (about two-thirds) of H3K27me3-enriched islands were in intergenic regions (12). Many of the intergenic H3K27me3 modifications may be associated with enhancers. In the case of the retinoic acid-responsive enhancer (RARE) upstream of the RET (rearranged during transfection) gene, the level of H3K27me3 could be reduced significantly within hours of retinoic acid (RA) treatment (2). It remains to be identified what kind of boundary element is responsible for limiting the repressive histone mark within the RARE upstream of RET. However, it is likely that the responsible boundary element may lack enhancer-blocking activity, since the gene is regulated by other enhancers and the enhancer function of RARE is required for mediating RA-induced RET expression. Given the prevalence of intergenic H3K27me3 islands in mammalian genomes, it is possible that the ILB-like chromatin barrier without enhancer-blocking activity is actually more common in vertebrates and mammals than it is in insects.
Is the barrier element conserved from flies to humans?
In searching for the potential trans-factors responsible for the barrier activity of ILB, we checked the modENCODE database for the occupancy of all of the known Drosophila insulator/boundary-associated proteins, including Su(Hw), CTCF, BEAF32, GAF, CP190, and Mod(mdg4). Despite the fact that many data sets are available for a variety of tissues and cultured cells, there is no indication that any of these known insulator proteins is enriched in the vicinity (±500 bp) of ILB294bp.
Our analysis indicated that ILB is bound by Cut, a highly conserved DNA binding protein (Fig. 7B). Not only can human Cutl1 and Drosophila Cut bind to the same DNA sequences (31), but human Cutl1 could rescue the developmental defect of a Drosophila cut mutant (22). Cut and Cutl1 have 4 DNA binding motifs, including a homeodomain and three cut repeats. The expression of many genes can be affected by Cut/Cutl1. The function of Cutl1 (and Cut) has been described as transcription repression or activation, depending on the affected gene (reviewed in reference 30). The actual functional mechanism of Cutl1/Cut has not been elucidated. However, analysis of different classes of cut mutants in Drosophila indicated that it is an essential gene required for cellular differentiation of a variety of tissues. A similar role was attributed to mammalian cutl1 (30), as mice homozygous for hypomorphic cutl1 alleles display defects in a variety of tissues (23, 45, 48). Our finding that Cut binds to the barrier element ILB and prevents the spread of facultative heterochromatin could potentially explain the pleiotropic phenotypes associated with cut/cutl1 mutants. However, it is completely possible that a complex protein such as Cut/Cutl1 may have multiple functions in different contexts and that the barrier function is but one of many tasks undertaken by this protein.
Our analysis indicated that cut function is required for the chromatin barrier activity of ILB. While it is clear that the barrier function of ILB is conserved not only in Drosophila but also in vertebrates, it remains to be seen how general this mechanism is for delimiting the formation of facultative heterochromatin. It also remains to be verified whether other trans-factors, with or without direct interaction with Cut/CBP, are also involved in carrying out the barrier function of ILB (Fig. 8C).
ACKNOWLEDGMENTS
We are very grateful to Paul Schedl at Princeton University for providing us the pCfhl vector and transgenic lines carrying pCfhl and pCfhl-Fab7_1.2k. The H3K27me3 and H3K9me3 antibodies used by the Zhou group were kindly provided by Thomas Jenuwein at the Max Planck Institute of Immunobiology. The 2B10 (Cut) antibody developed by Gerry Rubin was obtained from the Developmental Studies Hybridoma Bank, developed under the auspices of the NICHD and maintained by the University of Iowa.
This work was supported by grants CA095542 (L.Z.) and HL091929 and HL090589 (S.H.).
Footnotes
Published ahead of print on 25 April 2011.
REFERENCES
- 1. Andres V., Chiara M. D., Mahdavi V. 1994. A new bipartite DNA-binding domain: cooperative interaction between the cut repeat and homeo domain of the cut homeo proteins. Genes Dev. 8:245–257 [DOI] [PubMed] [Google Scholar]
- 2. Angrisano T., et al. 2011. Chromatin and DNA methylation dynamics during retinoic acid-induced RET gene transcriptional activation in neuroblastoma cells. Nucleic Acids Res. 39:1993–2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bell A. C., West A. G., Felsenfeld G. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98:387–396 [DOI] [PubMed] [Google Scholar]
- 4. Bello B. C., Hirth F., Gould A. P. 2003. A pulse of the Drosophila Hox protein Abdominal-A schedules the end of neural proliferation via neuroblast apoptosis. Neuron 37:209–219 [DOI] [PubMed] [Google Scholar]
- 5. Bi X., Broach J. R. 2001. Chromosomal boundaries in S. cerevisiae. Curr. Opin. Genet. Dev. 11:199–204 [DOI] [PubMed] [Google Scholar]
- 6. Bischof J., Maeda R. K., Hediger M., Karch F., Basler K. 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. U. S. A. 104:3312–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brand A. H., Perrimon N. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401–415 [DOI] [PubMed] [Google Scholar]
- 8. Brodsky M. H., et al. 2004. Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol. Cell. Biol. 24:1219–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bushey A. M., Dorman E. R., Corces V. G. 2008. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol. Cell 32:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chan C. S., Rastelli L., Pirrotta V. 1994. A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J. 13:2553–2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chanas G., Lavrov S., Iral F., Cavalli G., Maschat F. 2004. Engrailed and polyhomeotic maintain posterior cell identity through cubitus-interruptus regulation. Dev. Biol. 272:522–535 [DOI] [PubMed] [Google Scholar]
- 12. Cui K., et al. 2009. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell 4:80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gaszner M., Felsenfeld G. 2006. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 7:703–713 [DOI] [PubMed] [Google Scholar]
- 14. Gurudatta B. V., Corces V. G. 2009. Chromatin insulators: lessons from the fly. Brief. Funct. Genomic. Proteomic. 8:276–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang S., Li X., Yusufzai T. M., Qiu Y., Felsenfeld G. 2007. USF1 recruits histone modification complexes and is critical for maintenance of a chromatin barrier. Mol. Cell. Biol. 27:7991–8002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jenuwein T., Allis C. D. 2001. Translating the histone code. Science 293:1074–1080 [DOI] [PubMed] [Google Scholar]
- 17. Kahn T. G., Schwartz Y. B., Dellino G. I., Pirrotta V. 2006. Polycomb complexes and the propagation of the methylation mark at the Drosophila ubx gene. J. Biol. Chem. 281:29064–29075 [DOI] [PubMed] [Google Scholar]
- 18. Kodjabachian L., et al. 1998. Mutations in ccf, a novel Drosophila gene encoding a chromosomal factor, affect progression through mitosis and interact with Pc-G mutations. EMBO J. 17:1063–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kurshakova M., et al. 2007. Evolutionarily conserved E(y)2/Sus1 protein is essential for the barrier activity of Su(Hw)-dependent insulators in Drosophila. Mol. Cell 27:332–338 [DOI] [PubMed] [Google Scholar]
- 20. Li S., Aufiero B., Schiltz R. L., Walsh M. J. 2000. Regulation of the homeodomain CCAAT displacement/cut protein function by histone acetyltransferases p300/CREB-binding protein (CBP)-associated factor and CBP. Proc. Natl. Acad. Sci. U. S. A. 97:7166–7171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lin N., Zhang C., Pang J., Zhou L. 2009. By design or by chance: cell death during Drosophila embryogenesis. Apoptosis 14:935–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ludlow C., Choy R., Blochlinger K. 1996. Functional analysis of Drosophila and mammalian cut proteins in flies. Dev. Biol. 178:149–159 [DOI] [PubMed] [Google Scholar]
- 23. Luong M. X., et al. 2002. Genetic ablation of the CDP/Cux protein C terminus results in hair cycle defects and reduced male fertility. Mol. Cell. Biol. 22:1424–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. MacManus J. P., Buchan A. M. 2000. Apoptosis after experimental stroke: fact or fashion? J. Neurotrauma 17:899–914 [DOI] [PubMed] [Google Scholar]
- 25. Maeda R. K., Karch F. 2007. Making connections: boundaries and insulators in Drosophila. Curr. Opin. Genet. Dev. 17:394–399 [DOI] [PubMed] [Google Scholar]
- 26. Mallin D. R., Myung J. S., Patton J. S., Geyer P. K. 1998. Polycomb group repression is blocked by the Drosophila suppressor of Hairy-wing [su(Hw)] insulator. Genetics 148:331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maurange C., Cheng L., Gould A. P. 2008. Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cell 133:891–902 [DOI] [PubMed] [Google Scholar]
- 28. Micchelli C. A., Rulifson E. J., Blair S. S. 1997. The function and regulation of cut expression on the wing margin of Drosophila: Notch, Wingless and a dominant negative role for Delta and Serrate. Development 124:1485–1495 [DOI] [PubMed] [Google Scholar]
- 29. Negre N., et al. 2010. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 6:e1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nepveu A. 2001. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene 270:1–15 [DOI] [PubMed] [Google Scholar]
- 31. Neufeld E. J., Skalnik D. G., Lievens P. M., Orkin S. H. 1992. Human CCAAT displacement protein is homologous to the Drosophila homeoprotein, cut. Nat. Genet. 1:50–55 [DOI] [PubMed] [Google Scholar]
- 32. Peterson C., Carney G. E., Taylor B. J., White K. 2002. Reaper is required for neuroblast apoptosis during Drosophila development. Development 129:1467–1476 [DOI] [PubMed] [Google Scholar]
- 33. Phillips J. E., Corces V. G. 2009. CTCF: master weaver of the genome. Cell 137:1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pikaart M. J., Recillas-Targa F., Felsenfeld G. 1998. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 12:2852–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Raab J. R., Kamakaka R. T. 2010. Insulators and promoters: closer than we think. Nat. Rev. Genet. 11:439–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Recillas-Targa F., et al. 2002. Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. Proc. Natl. Acad. Sci. U. S. A. 99:6883–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rogulja-Ortmann A., Renner S., Technau G. M. 2008. Antagonistic roles for Ultrabithorax and Antennapedia in regulating segment-specific apoptosis of differentiated motoneurons in the Drosophila embryonic central nervous system. Development 135:3435–3445 [DOI] [PubMed] [Google Scholar]
- 38. Roseman R. R., Pirrotta V., Geyer P. K. 1993. The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO J. 12:435–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rougvie A. E., Lis J. T. 1988. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54:795–804 [DOI] [PubMed] [Google Scholar]
- 40. Schweinsberg S., et al. 2004. The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics 168:1371–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schweinsberg S. E., Schedl P. 2004. Developmental modulation of Fab-7 boundary function. Development 131:4743–4749 [DOI] [PubMed] [Google Scholar]
- 42. Sengupta A. K., Kuhrs A., Muller J. 2004. General transcriptional silencing by a Polycomb response element in Drosophila. Development 131:1959–1965 [DOI] [PubMed] [Google Scholar]
- 43. Sheng G., Thouvenot E., Schmucker D., Wilson D. S., Desplan C. 1997. Direct regulation of rhodopsin 1 by Pax-6/eyeless in Drosophila: evidence for a conserved function in photoreceptors. Genes Dev. 11:1122–1131 [DOI] [PubMed] [Google Scholar]
- 44. Sigrist C. J., Pirrotta V. 1997. Chromatin insulator elements block the silencing of a target gene by the Drosophila polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics 147:209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sinclair A. M., et al. 2001. Lymphoid apoptosis and myeloid hyperplasia in CCAAT displacement protein mutant mice. Blood 98:3658–3667 [DOI] [PubMed] [Google Scholar]
- 46. Steller H. 2008. Regulation of apoptosis in Drosophila. Cell Death Differ. 15:1132–1138 [DOI] [PubMed] [Google Scholar]
- 47. Tie F., et al. 2009. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 136:3131–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tufarelli C., Fujiwara Y., Zappulla D. C., Neufeld E. J. 1998. Hair defects and pup loss in mice with targeted deletion of the first cut repeat domain of the Cux/CDP homeoprotein gene. Dev. Biol. 200:69–81 [DOI] [PubMed] [Google Scholar]
- 49. Venken K. J., He Y., Hoskins R. A., Bellen H. J. 2006. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314:1747–1751 [DOI] [PubMed] [Google Scholar]
- 50. Wallace J. A., Felsenfeld G. 2007. We gather together: insulators and genome organization. Curr. Opin. Genet. Dev. 17:400–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang Z., Schones D. E., Zhao K. 2009. Characterization of human epigenomes. Curr. Opin. Genet. Dev. 19:127–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. West A. G., Huang S., Gaszner M., Litt M. D., Felsenfeld G. 2004. Recruitment of histone modifications by USF proteins at a vertebrate barrier element. Mol. Cell 16:453–463 [DOI] [PubMed] [Google Scholar]
- 53. Zeitlinger J., et al. 2007. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 39:1512–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang Y., et al. 2008. Epigenetic blocking of an enhancer region controls irradiation-induced proapoptotic gene expression in Drosophila embryos. Dev. Cell 14:481–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhou L., et al. 1997. Cooperative functions of the reaper and head involution defective genes in the programmed cell death of Drosophila central nervous system midline cells. Proc. Natl. Acad. Sci. U. S. A. 94:5131–5136 [DOI] [PMC free article] [PubMed] [Google Scholar]