Abstract
Recent data demonstrate that small synthetic compounds specifically targeting bromodomain proteins can modulate the expression of cancer-related or inflammatory genes. Although these studies have focused on the ability of bromodomains to recognize acetylated histones, it is increasingly becoming clear that histone-like modifications exist on other important proteins, such as transcription factors. However, our understanding of the molecular mechanisms through which these modifications modulate protein function is far from complete. The transcription factor GATA1 can be acetylated at lysine residues adjacent to the zinc finger domains, and this acetylation is essential for the normal chromatin occupancy of GATA1. We have recently identified the bromodomain-containing protein Brd3 as a cofactor that interacts with acetylated GATA1 and shown that this interaction is essential for the targeting of GATA1 to chromatin. Here we describe the structural basis for this interaction. Our data reveal for the first time the molecular details of an interaction between a transcription factor bearing multiple acetylation modifications and its cognate recognition module. We also show that this interaction can be inhibited by an acetyllysine mimic, highlighting the importance of further increasing the specificity of compounds that target bromodomain and extraterminal (BET) bromodomains in order to fully realize their therapeutic potential.
INTRODUCTION
Covalent posttranslational modifications (PTMs) on the histone proteins that package eukaryotic DNA have been shown to be essential for normal gene expression. The histone code hypothesis (28) posits that specific combinations of PTMs specify distinct transcriptional outcomes, and although it has been suggested that such a one-to-one correspondence is not supported by existing data (56), the biological significance of these PTMs in gene regulation is clear. The molecular mechanisms underlying the addition, removal, and recognition of these modifications are also now beginning to be understood. For example, the acetylation of lysine side chains in histone N-terminal tails has been strongly linked to the activation of nearby genes (32), and it has been shown that acetyllysine-containing sequences are specifically recognized by bromodomains, helical bundles that carry an acetyllysine-specific binding pocket (45).
A substantial body of work has also supported the hypothesis that inhibition of the addition, removal, or recognition of these PTMs could constitute a powerful method to treat a number of human disorders. Recently, for example, much effort has been focused on the specific inhibition of bromodomains from the bromodomain and extraterminal (ET) domain (BET) family, and several compounds based on a thienotriazolodiazepine platform (JQ1 and I-BET) have been demonstrated to mimic bromodomain targets and show considerable promise as therapeutic agents (17, 49).
Emerging data (reviewed in references 2, 15, 20, 35, and 57) indicate that certain transcription factors (TFs), in addition to histones, are subject to PTMs, such as acetylation and methylation. The tumor suppressor p53 was the first transcription factor that was shown to be acetylated (36); the enzymes responsible, p300 and p300/CREB-binding protein (CBP)-associated factor (P/CAF), had already been identified as p53 coactivators (22), and the structural basis of this interaction has been described recently (44). Numerous other transcription factors have since been shown to be functionally acetylated. Hormone-dependent activation by androgen receptor depends on the acetylation of several lysines (18, 19), and acetylation of the architectural DNA protein HMG 1(Y) is essential for the induction of beta interferon that takes place following viral infection (46). The human immunodeficiency virus type 1 (HIV-1) trans-activator protein Tat can also be acetylated and bind to the host bromodomain P/CAF, competing effectively against the binding of HIV-1 TAR RNA to Tat (43). K288 of erythroid Krüppel-like factor (EKLF) is acetylated in vitro and in erythroid cells by CBP (62), and a K288R mutant, when introduced into EKLF-null cells by retroviral infection, does not localize to the promoter of the target gene β-globin (55) and cannot activate β-globin expression. In addition, recent mass spectrometric data indicate that at least 1,000 cytosolic/mitochondrial proteins, many of which are gene-regulatory proteins, are acetylated in human cells (63).
These and other data point clearly to the existence of an additional level of regulatory control in gene transcription that revolves around PTMs to nonhistone proteins. Although ∼40 to 50 high-resolution structures containing modified histone peptides have been reported (Protein Data Bank, October 2010), little is known about transcription factor modifications at a biochemical and structural level.
The transcription factor GATA1 regulates the expression of all erythroid and megakaryocyte-specific genes. Mutations in GATA1 are associated with congenital anemias and megakaryoblastic leukemias (16, 40). We showed previously that CBP acts as a GATA1 coactivator that can bind to the zinc finger region of GATA1 (3), and we and others subsequently demonstrated that GATA1 can be acetylated by CBP in vitro at specific lysine residues adjacent to each of the two zinc finger domains (4, 26). Further, mutation of the lysines identified by in vitro acetylation with CBP prevented retrovirally introduced GATA1 from driving erythroid differentiation in G1E cells, a GATA1-null cell line which is poised in a preerythroid state (26), whereas binding of this GATA1 mutant to DNA in vitro was unperturbed. Importantly, we have recently shown that acetylation of the lysine residues located adjacent to the C-terminal zinc finger of GATA1 regulates the chromatin occupancy of the protein (33). It has further been demonstrated that GATA2 (24), -3 (61), and -4 (30) are all functionally acetylated at specific lysines.
We have recently identified the BET family protein Brd3 (58) as a cofactor that is able to bind to GATA1 (Fig. 1) in an acetylation-dependent manner and have demonstrated that the localization of GATA1 at erythroid gene promoters is contingent on the ability of a specific region of GATA1 to be acetylated (33). BET family proteins contain two bromodomains and a C-terminal ET domain (the function of which is not known). The best-characterized members of the family are Brd2 and Brd4, and one striking attribute is that they have been shown to associate with chromatin during mitosis (12), perhaps acting as molecular “bookmarks” to aid in the restoration of the parental gene expression pattern following cell division.
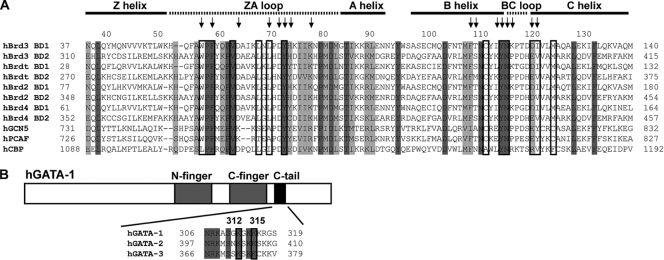
Fig. 1.
Sequence alignments. (A) Sequence alignment of human bromodomains that are part of the BET family (Brd2, Brd3, Brd4, and Brdt), as well as GCN5, P/CAF, and CBP. Dark-gray and light-gray bars indicate regions that are completely conserved and highly similar regions, respectively. Arrows indicate residues that were significantly perturbed in 15N-HSQC titrations with tetra-acetylated (4Ac) GATA1 peptides (Fig. 3); boxed residues are binding residues, as identified from the structure (Fig. 6). The names of the different regions of the BD1 structure (Fig. 6) are shown above. (B) Sequence alignment of the C tail regions of GATA1, GATA2, and GATA3. Completely conserved residues are indicated with gray shading, and the acetylation sites in GATA1 that are important for Brd3 binding are boxed. h, human.
Here, we have elucidated the molecular basis for the GATA1-Brd3 interaction by identifying the lysine residues in GATA1 that are critical for the Brd3 interaction and then using solution nuclear magnetic resonance (NMR) spectroscopy to determine the three-dimensional (3D) structure of a GATA1-Brd3 complex. Our data show that nonhistone proteins can take on multiple posttranslational modifications that can be recognized by a single recognition module and point to what is likely to be an intimate relationship between the regulation of gene expression via reversible histone modifications and via modifications made to other gene-regulatory proteins and indicate that bromodomain inhibitors, such as JQ1 and I-BET, are likely to need optimization to prevent unwanted side effects in essential processes, such as erythropoiesis.
MATERIALS AND METHODS
Preparation of proteins.
A construct encoding the first bromodomain (BD1) of Brd3 (residues 25 to 147) (Fig. 1) was codon optimized by GENEART (Regensburg, Germany) for expression in Escherichia coli and cloned into pGEX-6P (GE Healthcare). Brd3 BD2 (residues 307 to 419) (Fig. 1) and BD1 mutants were all cloned into the same expression vector. All constructs were overexpressed as fusions with glutathione (GSH) S-transferase (GST) at 37°C upon induction with IPTG (isopropyl-β-d-thiogalactopyranoside) under standard conditions; isotopically labeled BD1 and BD2 constructs were overexpressed using the protocol described in reference 7. Proteins were purified using GSH affinity chromatography and subjected to PreScission protease cleavage and gel filtration (Superdex-75 in NMR buffer [20 mM Tris, 100 mM NaCl, 1 mM dithiothreitol, pH 7.0]). Protein concentrations were verified by determining absorbances at 215, 225, and 280 nm (60). The correct folding of BD1 point mutants was confirmed by one-dimensional (1D) 1H NMR spectroscopy.
Preparation of peptides.
All acetylated and nonacetylated GATA1 peptides were synthesized by Peptide 2.0 Inc. (Chantilly, VA). Peptides were subjected to high-pressure liquid chromatography (HPLC) purification, resulting in a purity of at least 90%. They were dissolved in NMR buffer prior to usage, and their concentrations were verified by determining absorbances at 215 and 225 nm (60). The sequence used for the monoacetylated peptides [K(Ac)312, K(Ac)314, K(Ac)315, and K(Ac)316], diacetylated peptides [K(Ac)312/314, K(Ac)314/315, and K(Ac)312/315], and a triacetylated peptide [K(Ac)312/314/315] was CRKASGKGKKKRGSNL, that for the tetra-acetylated (4Ac) peptide was KASGKGKKKRGSN, that for the peptide used in the NMR structure determination (see Fig. 6) [K(Ac)312/315] was KASGKGKKKRGSN, that for competition experiments (see Fig. 7A) [K(Ac)308/312/315] was RNRKASGKGKKKRGS, and that for sequence specificity experiments [K(Ac)312/315 with a flanking K] was KASKKKKKKRGSN. All sequences are based on the mouse GATA1 sequence and were acetylated at the positions indicated in their corresponding names (see above; acetylated lysines are in italics).
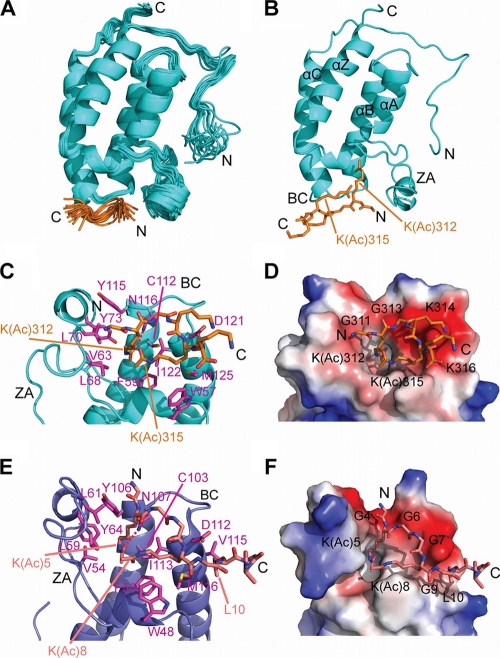
Fig. 6.
NMR solution structure of a BD1:GATA1 K(Ac)312/315 complex. GATA1 residues 308 to 310 and 317 to 320 were disordered in solution and therefore not included in the structure calculations. (A) Overlay of the family of the 20 best structures (RMSD, backbone = 0.61 Å). The GATA1 peptide is shown in orange. (B) Lowest-energy structure from panel A showing the acetylated lysines of GATA1 that bind BD1. The four helices and the two loops of the bromodomain are named according to the work of Dhalluin et al. (13). (C) Detailed view of the GATA1 binding site of Brd3-BD1, with GATA1-binding residues colored in magenta (PDB code 2L5E). (D) Space-filling representation of the Brd3-BD1 surface, with the GATA1 peptide shown in orange. Blue, white, and red areas indicate positive, neutral, and negative charges, respectively. (E) Structure of diacetylated histone H4 peptide (orange) bound to the first bromodomain (BD1) of Brdt (blue), with H4-binding residues colored in magenta (PDB code 2WP2). (F) Space-filling representation analogous to the representation in panel D. The H4 peptide is colored salmon in panels E and F.
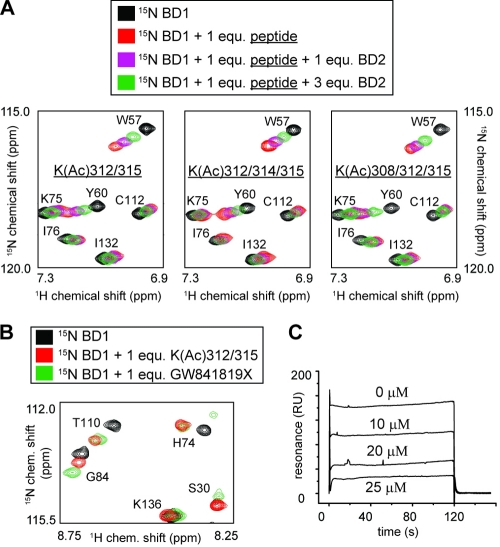
Fig. 7.
(A) Portions of 15N-labeled HSQC spectra of [15N]BD1 (∼300 μM) before (black) and after (red) addition of GATA1 peptide are shown. Spectra obtained in the presence of both GATA1 peptide (1 molar equivalent) and different amounts of unlabeled BD2 (purple and green) are also shown. The GATA1 peptide used in the three different experiments is indicated in each spectrum. (B) Portion of a 15N-HSQC spectrum of BD1 alone (black) and in the presence of GATA1 peptide (red) or compound GW841819X (green). (C) SPR curves showing BD1 binding (25 μM) to the 4Ac-GATA1 peptide immobilized on a Biacore SA chip at increasing concentrations of GW841819X.
Peptide affinity assays.
Peptides were synthesized by Rockefeller University and Peptide 2.0 Inc. (Chantilly, VA). An N-terminal cysteine was added to allow coupling to Sulfo-link resin (Pierce). Peptides were coupled according to the manufacturer's instructions. Peptide affinity assays were performed by incubating 25 to 50 ng immobilized peptide with nuclear extracts prepared from 8 million to 10 million G1E-ER4 cells stably expressing hemagglutinin (HA)-Brd3. Nuclear extracts were prepared as described previously (3) and diluted to 150 mM NaCl. Following stringent NaCl washes, resin was boiled in sodium dodecyl sulfate (SDS) sample buffer, separated on a 10% SDS-PAGE gel, transferred to a nitrocellulose membrane, and assayed by anti-HA Western blotting. Affinity assays were performed in the presence of 10 mM sodium butyrate, 1 mM phenylmethylsulfonyl fluoride (PMSF), and protease inhibitor cocktail (Sigma), which was added according to the manufacturer's recommendation. For the GST-pulldown assays, 1 μg of purified GST Brd3 BD1 protein was incubated with 25 to 50 ng immobilized peptide overnight in the presence of 10 mM sodium butyrate, 1 mM PMSF, and protease inhibitor cocktail (Sigma). Resin was washed five times with buffer containing 450 mM NaCl, 50 mM Tris, pH 7.5, and 0.5% Igepal and eluted by boiling it in SDS sample buffer. Western blotting was performed using anti-GST antibodies (sc-138; Santa Cruz).
SPR measurements.
Kinetic analysis was carried out on a Biacore 3000 surface plasmon resonance (SPR) instrument (Biacore AB, Uppsala, Sweden). Biotinylation of tetra-acetylated GATA1 peptides was performed by chemical synthesis as follows. The biotin labeling reagent EZ-Link maleimide-polyethylene glycol 2 (PEG2)-biotin (Thermo Scientific, Rockford, IL), dissolved in NMR buffer to a final concentration of 20 μM, was added to the peptides in a 20-fold excess. The reaction mixture was then left at 4°C for 5 h. Labeled peptide was then separated from excess biotin using a Superdex peptide HR 10/30 column operating on a BioLogic fast protein liquid chromatography (FPLC) system by monitoring the absorbance at 215 nm. Biotinylated 4Ac GATA1 peptides were immobilized on a streptavidin-coated SA sensor chip (Biacore AB, Uppsala, Sweden). The buffer used for all experiments was NMR buffer with 0.005% P20 detergent. The chip was pretreated according to the manufacturer's instructions with conditioning solution (three 1-min injections at 20 μl/min with 50 mM NaOH, 1 M NaCl). The biotinylated GATA1 peptide was diluted to 100 nM and injected onto one of the sensor chip channels (Fc-2 or Fc-4) at a flow rate of 20 μl/min for 2 min, resulting in an immobilization level of approximately 50 to 100 response units (RU). The sensor chip was then washed with running buffer. Upstream, unmodified channel surfaces were used for reference subtraction. Kinetic measurements with Brd3 BD1 protein concentrations across the range of 1 μM to 200 μM (40 μl) were performed at 25°C with a KINJECT protocol and a flow rate of 20 μl/min. Wild-type and mutant protein samples were sampled alternately, zero-concentration samples were included for double-referencing, and 3 to 5 cycles were performed. Data analysis was initially performed with the BIAevaluation software (Biacore). However, no reliable off rates could be obtained due to the fast off-kinetics (measurement was limited by the data collection rate); therefore, a steady-state analysis (48) using Origin 7.0 (OriginLab Corp., Northampton, MA) was performed. Interactions for which an injection of 50 μM BD1 did not result in an SPR signal of >10 RU were considered not detectable (ND) (see Fig. 4). For the Biacore competition experiments (see Fig. 7C), the published methods (50, 52) for the estimation of the binding constant were used.
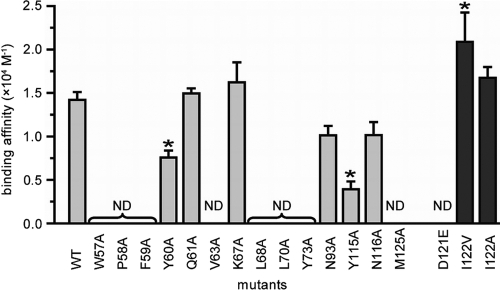
Fig. 4.
A mutational analysis reveals critical GATA1-binding residues in Brd3. Summary of affinity constants (±standard errors) for BD1 and various alanine mutants (gray bars), as well as microchimeric D121/I122 mutants (dark-gray bars), for binding to GATA1, as measured by SPR. Asterisks indicate a significant (P < 0.05) difference in affinity from that of the wild type (WT). ND, the binding was not detectable.
NMR spectroscopy.
NMR samples contained 0.5 to 1.5 mM purified 15N and 13C-labeled, 15N-labeled, and unlabeled Brd3 BD1 in NMR buffer (1 μl 10 μM 2,2-dimethyl-2-silapentane-5-sulfonic acid [DSS]) as a chemical shift reference and either 5 to 10% (vol/vol) D2O or 100% D2O. Samples of [15N]Brd3 BD2 and BD1 mutants were prepared similarly. BD1-GATA1 complex samples were prepared by adding 1.0 to 1.1 molar equivalents of GATA1 K(Ac)312/315 peptide to Brd3 BD1 at 0.5 to 1.5 mM for structure determination (see Fig. 6) and at 300 μM for all HSQC titrations (see Fig. 2, 3, 5, and 7). Spectra were recorded at 298 K on Bruker 600-MHz and 800-MHz spectrometers equipped with cryoprobes. All homonuclear 2D data were collected and analyzed as described previously (37). Mixing times were 60 and 150 ms for all total correlation spectroscopy (TOCSY) and nuclear Overhauser enhancement spectroscopy (NOESY) spectra, respectively. 15N and 13C chemical shift assignments were made from the standard suite of triple-resonance experiments as described previously (11). NOE-derived distance restraints were obtained from 3D 13C-separated NOESY and 3D 15N-separated NOESY spectra. Intermolecular NOEs were also obtained from 3D 13C-separated 15N13C-filtered (in F1 or F3) NOESY experiments (5, 27, 65). GATA1 peptide assignments were made based on a series of 2D 15N13C-filtered (in F1 and F2) NOESY and TOCSY experiments recorded in the absence and presence of increasing amounts of BD1. All NMR data were processed using TOPSPIN (Bruker, Karlsruhe, Germany) and analyzed with SPARKY 3 (21). The weighted chemical shift changes in Fig. 3 and 5 were calculated according to the protocol of Ayed et al. (1).
Fig. 2.
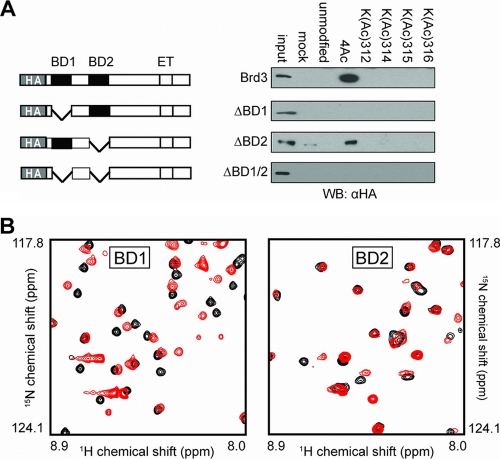
The first bromodomain of Brd3 is required for GATA1 binding. (A) Peptide affinity assays using nuclear extracts from cells expressing bromodomain mutant forms of Brd3. (B) Portion of 15N-HSQC spectra of BD1 (left) and BD2 (right) of Brd3, in the absence (black) and presence (red) of one molar equivalent of 4Ac GATA1 peptide. WB, Western blot.
Fig. 3.
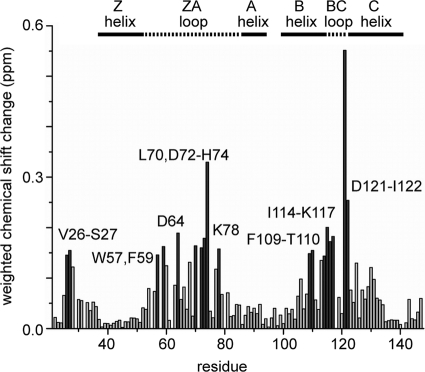
Summary of chemical shift changes of 15N-labeled BD1 upon addition of 1 molar equivalent of 4Ac GATA1. Changes larger than the average plus 1 standard deviation are depicted as dark-gray bars. The names of the different regions of the BD1 structure (Fig. 6) are shown above the graph.
Fig. 5.
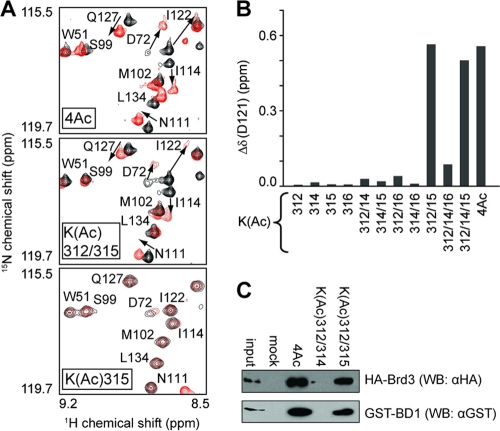
Two acetylated lysines with specific spacing are required for optimal binding. (A) Portions of 15N-HSQC spectra of 15N-labeled BD1 (black) and in the presence of 1 molar equivalent of tetra-acetylated (upper part), diacetylated (K312 and K315) (middle), and mono acetylated (K315) (lower part) GATA1 peptides (all in red). (B) Weighted chemical shift changes for BD1 residue D121 as a function of GATA1 acetylation (following the addition of 1 molar equivalent of peptide) obtained from 15N-HSQC spectra with initial BD1 concentrations of ∼300 μM. (C) Peptide binding assays carried out by incubating the indicated GATA1 peptides with nuclear extracts from G1E-ER4 cells expressing HA Brd3. Bound material was analyzed by Western blotting using anti-HA antibodies.
Structure calculations.
Initial structures of Brd3 BD1 without the GATA1 peptide were calculated in CYANA (23) from manually assigned unambiguous NOEs from 15N and 13C NOESY spectra. Φ and Ψ restraints for BD1 were included on the basis of an analysis of backbone chemical shifts in the program TALOS (10). GATA1 residues 308 to 310 as well as 317 to 320 were disordered in solution and therefore not included in the structure calculations. Final calculations of the BD1-GATA1 complex structure were then carried out using ARIA 1.2 (38) implementing CNS 1.1 (6), using the standard protocols provided, with the experimentally determined tautomeric state of the histidine side chains fixed (51) and the preassigned intermolecular NOEs present. Final assignments made by ARIA 1.2 were checked manually and corrected where necessary. In the final set of calculations, the 20 lowest-energy structures were refined in a 9-Å shell of water using standard ARIA 1.2 water refinement modules (minimization and dynamics steps; for details, see references 29 and 39). The 20 conformers with the lowest value of total energy were analyzed and visualized using MOLMOL (31), PYMOL (Schrödinger, NY), and PROCHECK-NMR (34) (for structure calculation statistics, see Table 1).
Table 1.
NMR structure statistics
| Parameter | Value |
|---|---|
| NMR distance constraints | |
| Total no. of intramolecular constraints | 1,617 |
| No. of intraresidue constraints | 539 |
| No. of interresidue constraints | 1,078 |
| No. of sequential constraints (|i − j| = 1) | 414 |
| No. of medium-range constraints | |
| (|i − j| ≤ 4) | 251 |
| No. of long-range constraints (|i − j| > 4) | 413 |
| No. of intermolecular constraints | 39 |
| TALOS dihedral constraints | |
| Phi | 55 |
| Psi | 55 |
| Ramachandran analysis (20 structures) | |
| Most favorable region (%) | 85.2 |
| Additional allowed region (%) | 12.9 |
| Generously allowed region (%) | 1.1 |
| Disallowed region (%) | 0.9 |
| Structure statistics (20 structures) | |
| Violations (means ± SD) | |
| No. of distance constraints (Å) | |
| (no violations were >0.5 Å) | 0.32 ± 0.02 |
| Dihedral constraints (°) | |
| (no violations were >5°) | 0.36 ± 0.07 |
| Maximal distance constraint (Å) | 0.36 |
| Maximal dihedral angle constraints (°) | 0.50 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.00338 ± 0.00007 |
| Bond angles (°) | 0.508 ± 0.009 |
| Impropers (°) | 1.19 ± 0.05 |
| Average pairwise RMSD (Å) | |
| (residues 25–147 [BD1]; residues | |
| 311–316 [GATA1]) | |
| Heavy | 0.965 |
| Backbone | 0.612 |
Protein Data Bank accession number.
The protein structure of Brd3 BD1 bound to GATA1 has been deposited into the Protein Data Bank (PDB identifier [ID], 2L5E, BMRB 17270).
RESULTS
BD1 of Brd3 recognizes acetylated GATA1.
In recent work, we demonstrated that a peptide corresponding to residues 302 to 324 of GATA1 in which lysines K312, K314, K315, and K316 bear acetylated side chain amino groups (Fig. 1B) can pull the bromodomain protein Brd3 from G1E cell nuclear lysates (33a). In order to probe the molecular basis for this interaction, we first determined which of the Brd3 bromodomains is responsible for the interaction with acetylated GATA1. We carried out pulldown experiments using biotinylated tetra-acetylated (4Ac) and monoacetylated GATA1 peptides bound to streptavidin-coated agarose beads and nuclear extracts from G1E-ER4 cells that expressed mutant forms of HA-tagged Brd3 (Fig. 2A). A mutant lacking the first bromodomain (BD1) was not able to bind, whereas in the absence of the second bromodomain (BD2), an interaction could still be detected, indicating that BD1 but not BD2 is essential for GATA1 binding.
To corroborate this finding, we expressed and purified uniformly 15N-labeled BD1 and BD2 and recorded the 15N-heteronuclear single-quantum-coherence (HSQC) spectrum of each protein before and after the addition of one molar equivalent of a GATA1 peptide (residues 308 to 320) acetylated at K312, K314, K315, and K316 (4Ac GATA1). As shown in Fig. 2B and C, the magnitude of the observed chemical shift changes is substantially larger in the case of BD1, indicating that stronger binding to this domain occurs. These data corroborate our in vivo observations that recruitment of Brd3 to GATA1-occupied sites requires BD1 but not BD2 (33a).
By making resonance assignments for BD1 in the absence and presence of one molar equivalent of the 4Ac peptide, we could further see that the largest chemical shift changes are restricted to the regions predicted to be the interhelical ZA and BC loops (Fig. 1A and 3). These regions are known to be important for acetyllysine recognition in histone-binding bromodomains, such as that of the p300/CBP-associated factor (P/CAF) (13).
Hydrophobic residues are critical for the interaction.
In order to further characterize the GATA1-BD1 binding interface, we made a series of point mutations in BD1 and tested their ability to bind to GATA1. Biotinylated 4Ac peptide was immobilized on streptavidin-coated chips and treated with solutions of BD1 mutants with single-alanine mutations in surface plasmon resonance (SPR) experiments. Dissociation constants were calculated from equilibrium binding data (Fig. 4). Replacement of any of the hydrophobic residues W57, V63, L68, Y73, and M125 with alanine completely eliminated binding. Y60A and Y115A mutations also significantly reduced binding, whereas changes to the charged residues Q61 and K67, as well as N93 (which is distant from the GATA1-binding surface), did not affect the interaction (Fig. 4, light-gray bars). Interestingly, mutation of the conserved asparagine N116 did not yield a significant affinity change. This residue is conserved within all bromodomain family proteins, and a hydrogen bond to the oxygen of an acetyl group is generally observed in the structures of bromodomain-acetyllysine complexes. Overall, these results indicate that hydrophobic interactions are the strongest determinant of binding affinity to 4Ac GATA1.
Comparison of the BD1 sequence with BD2 (Fig. 1) reveals that most of the residues that contact GATA1 are conserved. Conservative mutations exist in D121 and I122, which are changed to E396 and V397 in BD2. We created microchimeric point mutations at these positions in BD1, creating D121E, I122V, and I122A mutants (Fig. 4, dark-gray bars), and tested their ability to bind to GATA1 by SPR. Intriguingly, I122V binds slightly more strongly than the wild type, whereas the conservative change D121E eliminates binding. This result indicates that the size of residue 121 is critical for optimal binding and explains why BD1 is the bromodomain responsible for mediating the interaction between Brd3 and acetylated GATA1.
The BD1-GATA1 interaction requires two acetylated lysines.
Although K312 has been identified as the major acetylation target by CBP in vitro and in vivo, lysines 308, 314, and 315 in the C tail of GATA1 can all be modified by CBP (26, 33). To establish which lysines in GATA1 must be acetylated for maximal Brd3 binding activity, we titrated a range of mono-, di-, and triacetylated GATA1 peptides into 15N-labeled BD1 (Fig. 5B) and quantified the resulting chemical shift changes. Surprisingly, acetylation of any single lysine resulted in very small chemical shift changes. In contrast, the diacetyl combination K(Ac)312/315 mimics the binding mode of the 4Ac peptide (Fig. 5A), whereas any other combination of acetylated residues that does not include K312 and K315 results in measurably weaker binding. Figure 5B shows the weighted chemical shift changes of the backbone HN and N atoms of D121 upon addition of one molar equivalent of GATA1 peptide for each acetylation combination. To confirm these findings, we incubated GATA1 peptides bearing K(Ac)312/314 or K(Ac)312/315 modifications with nuclear extracts from G1E cells (26) expressing full-length HA-Brd3 or recombinant GST-BD1 protein (Fig. 5C). As expected, only K(Ac)312/315 was able to interact with Brd3 and BD1. Overall, these data indicate that diacetylated GATA1 is the target of Brd3 BD1 and that a specific diacetylated combination is recognized preferentially.
Structure of a BD1-GATA1 K(Ac)312/315 complex.
We next used NMR spectroscopy to determine the solution structure of the complex formed by Brd3 BD1 and the same 13-residue GATA1 peptide that has been used for the SPR studies acetylated at lysines 312 and 315 [K(Ac)312/315]. The family of structures was calculated from using a total of 1,657 intramolecular and 39 intermolecular NOEs (Table 1 and Fig. 6) and displayed a backbone root mean square deviation (RMSD) of 0.61 Å (PDB ID, 2L5E). Brd3-BD1 exhibits the classical bromodomain fold consisting of a left-handed bundle of four helices (αZ, αA, αB, and αC) with a peptide-binding pocket defined by the short BC and the longer ZA loops (Fig. 6B). The majority of intermolecular NOEs were observed between the acetyl group of K312 or K315 of GATA1 and residues on the binding surface of BD1, underscoring the importance of the diacetyl modification for binding. The side chain of K312 is located in a deep hydrophobic pocket made up of BD1 residues F59, V63, L68, L70, Y73, Y115, and C112 (Fig. 6C). A hydrogen bond is formed between the carbonyl oxygen of the acetyl group and the side chain amide of N116, as expected. The acetyl group of GATA1 K(Ac)315 is situated approximately 7.3 ± 1.1 Å away from K(Ac)312 (average distance between the carbonyl atoms of the acetyl group) and is recognized by BD1 residues W57, P58, and I122 as well as D121 and M125; these residues lie somewhat outside the main hydrophobic pocket that accommodates K(Ac)312. The substantial number of hydrophobic residues that contact GATA1 corroborates the results of our mutational analysis (Fig. 4).
The side chain of I122 contacts both residue 312 and residue 315 of GATA1, consistent with the large chemical shift changes seen for this residue in 15N-HSQC titrations (see, for example, Fig. 5A). Intermolecular NOEs were also observed between the backbone of positively charged K314 and the side chain of negatively charged D121 in Brd3, suggesting that this aspartic acid plays a role in GATA1 recognition. In keeping with this observation, acetylation of K314 results in reduced chemical shift changes for D121 (Fig. 5B). Mutation of D121 to a larger glutamic acid would most likely lead to steric clashes between this residue and the backbone of the GATA1 peptide, consistent with our SPR results (Fig. 4). Overall, except for D121, the interaction is predominantly mediated by hydrophobic contacts (Fig. 4 and 6C), with significant contributions made by the aromatic residues W57 and Y73.
BD1 and BD2 do not bind cooperatively to the acetylated C tail of GATA1.
We also identified K308 and K314 in the C tail of murine GATA1 as additional candidate acetylation sites in vitro (26). In light of this and the recent finding that two molecules of the second bromodomain of the highly related BET family protein Brd2 (Brd2 BD2) can bind diacetylated (K5/K12) histone H4 (59) simultaneously, we used NMR spectroscopy to investigate the possibility that BD2 might act together with BD1 in binding the C tail of GATA1 by binding to either acetylated K308 or K314. Unlabeled BD2 was added to a complex of 15N-labeled BD1 bound to one of several acetylated GATA1 peptides: K(Ac)312/315 (Fig. 7A, left), K(Ac)312/314/315 (middle), or K(Ac)308/312/315 (right). However, in all three cases, only weak competitive binding of BD2 was observed, as evidenced by shifts in the positions of GATA1-binding residues (e.g., W57 and Y60) back toward their unbound positions (black peaks in Fig. 7A). Thus, it appears that BD2 most likely does not contribute to the binding of the GATA1 C tail.
Binding specificity of BD1 is in part due to steric constraints.
In our structure of BD1 bound to GATA1, specific interactions were not observed for any residues outside the sequence that is flanked by the two acetylated lysines. We therefore asked whether the sequence context in which each acetylated lysine is found has a bearing on which lysine occupies the canonical acetyllysine-binding pocket or whether either lysine could occupy the pocket. We tested by NMR the ability of a GATA1 C tail peptide containing a G311K G313K double mutation [so that K(Ac)312 is placed in the same sequence context as K(Ac)315] (Fig. 1) to bind to BD1. 15N-HSQC titration data indicate that this peptide binds in the same region of BD1 but with significantly lower affinity ([2.5 ± 1.0] × 103 M−1 compared to [1.1 ± 0.6] × 104 M−1 for the wild-type peptide). Thus, the sequence context of the lysine that binds in the canonical recognition pocket is clearly important for optimal binding and larger residues are not as well tolerated in the positions that flank K312.
Specific inhibition of GATA1 binding by an acetyllysine mimic.
The addition of the acetyllysine mimic GW841819X (GlaxoSmithKline), which is chemically almost identical to JQ1 and I-BET (17, 49), to 15N-labeled BD1 gives rise to chemical shift changes in essentially the same set of resonances that are involved in GATA1 binding (Fig. 7B), indicating structurally similar binding modes. The addition of increasing concentrations of this compound in SPR experiments (Fig. 7C) in which immobilized 4Ac peptide is treated with BD1 shows clearly that GW841819X competes directly with GATA1 for BD1 binding. The affinity constant for the GW841819X-Brd3 interaction is estimated to be around 70 nM based on the results of these experiments, which is in agreement with the published data for JQ1 and I-BET (17, 49).
DISCUSSION
Structural differences and similarities between BET family bromodomain acetyllysine recognition.
We have shown here that Brd3 interacts directly with GATA1 and that the interaction takes place in a region of GATA1 that is not involved in binding to DNA or any other protein that is known to be important for the proper association of GATA1 with its genomic targets (40). The recognition is sequence specific and involves two distinct acetylated lysines that are also conserved in GATA2 and GATA3, suggesting that Brd3 might also be able to bind these transcription factors. Indeed, preliminary pulldown experiments performed with acetylated GATA2 show an interaction with Brd3 (data not shown).
We have determined the structural basis for this interaction, and our data represent the first structural insight into the recognition of nonhistone proteins carrying multiple acetylation marks. Our data show that two acetylated lysines in a highly basic sequence adjacent to the DNA-binding domain of GATA1 are recognized by the BD1 bromodomain of Brd3. The recognition is specific, in that both K312 and K315 must be acetylated for maximum binding, and other combinations of acetylated lysines (e.g., K312 and K314) do not bind with the same affinity.
A number of structural studies over the last 10 to 15 years have established a paradigm for acetyllysine recognition by bromodomains (45), in which a pocket formed at one end of the bromodomain helical bundle by the ZA and BC loops is shaped to fit a single acetyllysine side chain. An asparagine residue at the base of the pocket provides specificity by forming a hydrogen bond with the acetyl group.
The recognition mode exhibited by GATA1 in complex with Brd3 displays substantial similarities with this binding mode, but the BD1 bromodomain additionally accommodates a second acetyllysine in a nearby surface site. This arrangement was recently observed in the structure of the closely related Brdt protein bound to a diacetylated peptide derived from histone H4 (acetylated at K5 and K8) (42), and overall, this structure closely resembles the Brd3-GATA1 structure (Fig. 6E and F). Thus, the means of recognition of the two acetylated lysines are similar and the same bromodomain residues are engaged in binding the acetylated lysines. However, closer inspection reveals that aspects of the specificity of peptide binding in the two structures differ. In the H4-Brdt complex, specificity is created in part by recognition of the histone H4 residue L10, which makes contact with three hydrophobic residues in Brdt (F46, W48, and V115) (Fig. 6E). There is no corresponding leucine in the GATA1 sequence, and instead we find that the size of side chains flanking the two acetyllysines creates a degree of specificity. That is, K(Ac)315 is flanked by two lysines, whereas K312 is flanked by two glycines, and replacement of these glycines with lysines reduces binding. Additional specificity is achieved from an electrostatic interaction between the backbone of K314 in GATA1 and D121 in BD1. It is notable in this regard that K314 is completely conserved between GATA1, -2, and -3 (Fig. 1) but is not observed in GATA4, -5, and -6. Further, with the exception of K16, all lysines in H4 that are acetylated (K5, K8, and K12) are those that are flanked by two glycines (64), perhaps indicating that the size of these neighboring residues plays a role in driving specificity, by determining which bromodomains can recognize which acetylated target proteins.
Our data also indicate that the BD2 bromodomain of Brd3 does not play a significant role in the recognition of the GATA1 C tail. It remains possible that this domain instead contacts a different target; this target could perhaps be either acetylated lysines in other parts of GATA1 (such as those found immediately C terminal to the other zinc finger in GATA1 [26]) or acetylated N-terminal histone tails.
It is further notable that Brd3 BD1 exists as a monomer in solution, in contrast to the related bromodomain protein Brd2 (47). A comparison between the two structures reveals that the hydrophobic cavity that exists in Brd2 and which is important for self-association is not present in Brd3, providing a possible explanation for this observation.
The spacing between the two acetylated lysines is conserved in other transcription factors.
Our data demonstrate that the correct spacing between the two acetylated lysines in the Brd3-GATA1 complex is essential for optimal binding. Intriguingly, the myogenic transcription factor MyoD is also functionally acetylated at two lysines that are spaced three residues apart: K99 and K102 (53). This acetylation event is essential for normal myoblast differentiation in adult mice (14). Acetylation of MyoD also increases its affinity for CBP (54), the enzyme that also acetylates GATA1 (26). The transcription factors E2F1 (41) and RelA (8, 9) have also been demonstrated to be functionally acetylated, and in both cases, acetylation takes place on two lysines that are three residues apart (K117 and K120 for E2F1 and K318 and K321 for RelA). Moreover, the more N terminal of the two lysines is flanked by two small residues (cysteine and alanine for MyoD and two glycines for E2F1), strongly suggesting that these acetylated sequences will be recognized by bromodomain-containing proteins. Interestingly, RelA is also monoacetylated at lysine 310, and this acetylation has been shown to be essential for binding to Brd4 (25), perhaps indicating that certain transcription factors can recruit different bromodomains through different lysines, leading to distinct functional outcomes.
Brd3 and antibromodomain drugs.
We show here that the acetyllysine mimic GW841819X can specifically block the interaction between Brd3 and acetylated GATA1. In addition, we have shown by chromatin immunoprecipitation (ChIP) experiments (33a) that the same drug substantially reduces the chromatin occupancy of both Brd3 and GATA1 from positively acting GATA1 target sites. GW841819X is chemically almost identical to the recently reported compounds JQ1 and I-BET (17, 49), which have been shown to specifically inhibit bromodomains from the BET family and which subsequently lead, for example, to tumor regression and improved overall survival in NUT midline carcinoma patient-derived xenograft model cell lines (17). Since the modes of recognition of such compounds are structurally similar, their administration could potentially interfere with Brd3 recruitment by GATA1 to important target genes, thereby leading to unwanted side effects. Our data therefore indicate a need for higher selectivity in such compounds if they are to be used for therapeutic purposes.
ACKNOWLEDGMENTS
We thank Ann Kwan for expert maintenance of the NMR spectrometers.
This work was supported in part by a program grant from the National Health and Medical Research Council of Australia to J.P.M., by an NIH grant (RO1 DK054937) to G.A.B., and by an NIH predoctoral training grant (T32 HL007971-07) to J.M.L.
Footnotes
Published ahead of print on 9 May 2011.
REFERENCES
- 1. Ayed A., et al. 2001. Latent and active p53 are identical in conformation. Nat. Struct. Biol. 8:756–760 [DOI] [PubMed] [Google Scholar]
- 2. Bannister A. J., Miska E. A. 2000. Regulation of gene expression by transcription factor acetylation. Cell. Mol. Life Sci. 57:1184–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blobel G. A., Nakajima T., Eckner R., Montminy M., Orkin S. H. 1998. CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc. Natl. Acad. Sci. U. S. A. 95:2061–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boyes J., Byfield P., Nakatani Y., Ogryzko V. 1998. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature 396:594–598 [DOI] [PubMed] [Google Scholar]
- 5. Breeze A. L. 1999. Isotope-filtered NMR methods for the study of biomolecular structure and interactions. Prog. Nucl. Magn. Reson. Spectrosc. 36:323–372 [Google Scholar]
- 6. Brunger A. T., et al. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905–921 [DOI] [PubMed] [Google Scholar]
- 7. Cai M., et al. 1998. An efficient and cost-effective isotope labeling protocol for proteins expressed in Escherichia coli. J. Biomol. NMR 11:97–102 [DOI] [PubMed] [Google Scholar]
- 8. Chen L., Fischle W., Verdin E., Greene W. C. 2001. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science 293:1653–1657 [DOI] [PubMed] [Google Scholar]
- 9. Chen L. F., Mu Y., Greene W. C. 2002. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J. 21:6539–6548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cornilescu G., Delaglio F., Bax A. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13:289–302 [DOI] [PubMed] [Google Scholar]
- 11. Deane J. E., et al. 2003. Structural basis for the recognition of ldb1 by the N-terminal LIM domains of LMO2 and LMO4. EMBO J. 22:2224–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dey A., et al. 2000. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G2-to-M transition. Mol. Cell. Biol. 20:6537–6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dhalluin C., et al. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399:491–496 [DOI] [PubMed] [Google Scholar]
- 14. Duquet A., et al. 2006. Acetylation is important for MyoD function in adult mice. EMBO Rep. 7:1140–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Egorova K. S., Olenkina O. M., Olenina L. V. 2010. Lysine methylation of nonhistone proteins is a way to regulate their stability and function. Biochemistry (Mosc). 75:535–548 [DOI] [PubMed] [Google Scholar]
- 16. Ferreira R., Ohneda K., Yamamoto M., Philipsen S. 2005. GATA1 function, a paradigm for transcription factors in hematopoiesis. Mol. Cell. Biol. 25:1215–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Filippakopoulos P., et al. 2010. Selective inhibition of BET bromodomains. Nature 468:1067–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fu M., et al. 2000. p300 and p300/cAMP-response element-binding protein-associated factor acetylate the androgen receptor at sites governing hormone-dependent transactivation. J. Biol. Chem. 275:20853–20860 [DOI] [PubMed] [Google Scholar]
- 19. Gaughan L., Logan I. R., Cook S., Neal D. E., Robson C. N. 2002. Tip60 and histone deacetylase 1 regulate androgen receptor activity through changes to the acetylation status of the receptor. J. Biol. Chem. 277:25904–25913 [DOI] [PubMed] [Google Scholar]
- 20. Glozak M. A., Sengupta N., Zhang X., Seto E. 2005. Acetylation and deacetylation of non-histone proteins. Gene 363:15–23 [DOI] [PubMed] [Google Scholar]
- 21. Goddard T., Kneller D. 2006. SPARKY 3. University of California, San Francisco, CA [Google Scholar]
- 22. Gu W., Roeder R. G. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90:595–606 [DOI] [PubMed] [Google Scholar]
- 23. Guntert P., Mumenthaler C., Wuthrich K. 1997. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273:283–298 [DOI] [PubMed] [Google Scholar]
- 24. Hayakawa F., et al. 2004. Functional regulation of GATA-2 by acetylation. J. Leukoc. Biol. 75:529–540 [DOI] [PubMed] [Google Scholar]
- 25. Huang B., Yang X. D., Zhou M. M., Ozato K., Chen L. F. 2009. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol. Cell. Biol. 29:1375–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hung H. L., Lau J., Kim A. Y., Weiss M. J., Blobel G. A. 1999. CREB-binding protein acetylates hematopoietic transcription factor GATA-1 at functionally important sites. Mol. Cell. Biol. 19:3496–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Iwahara J., Wojciak J. M., Clubb R. T. 2001. Improved NMR spectra of a protein-DNA complex through rational mutagenesis and the application of a sensitivity optimized isotope-filtered NOESY experiment. J. Biomol. NMR. 19:231–241 [DOI] [PubMed] [Google Scholar]
- 28. Jenuwein T., Allis C. D. 2001. Translating the histone code. Science 293:1074–1080 [DOI] [PubMed] [Google Scholar]
- 29. Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L. 1983. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79:926–935 [Google Scholar]
- 30. Kawamura T., et al. 2005. Acetylation of GATA-4 is involved in the differentiation of embryonic stem cells into cardiac myocytes. J. Biol. Chem. 280:19682–19688 [DOI] [PubMed] [Google Scholar]
- 31. Koradi R., Billeter M., Wuthrich K. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14:29–32, 51-55 [DOI] [PubMed] [Google Scholar]
- 32. Kuo M. H., Allis C. D. 1998. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 20:615–626 [DOI] [PubMed] [Google Scholar]
- 33. Lamonica J. M., Vakoc C. R., Blobel G. A. 2006. Acetylation of GATA-1 is required for chromatin occupancy. Blood 108:3736–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a. Lamonica J. M., et al. 2 May 2011, posting date. Bromodomain protein Brd3 associates with acetylated GATA1 to promote its chromatin occupancy at erythroid target genes. Proc. Natl. Acad. Sci. U. S. A. doi:10.1073/pnas.1102140108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. 1996. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8:477–486 [DOI] [PubMed] [Google Scholar]
- 35. Lee Y. H., Stallcup M. R. 2009. Protein arginine methylation of nonhistone proteins in transcriptional regulation. Mol. Endocrinol. 23:425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li A. G., et al. 2007. An acetylation switch in p53 mediates holo-TFIID recruitment. Mol. Cell. 28:408–421 [DOI] [PubMed] [Google Scholar]
- 37. Liew C. K., et al. 2000. Solution structures of two CCHC zinc fingers from the FOG-family protein U-shaped that mediate protein-protein interactions. Structure 8:1157–1166 [DOI] [PubMed] [Google Scholar]
- 38. Linge J. P., Habeck M., Rieping W., Nilges M. 2003. ARIA: automated NOE assignment and NMR structure calculation. Bioinformatics 19:315–316 [DOI] [PubMed] [Google Scholar]
- 39. Linge J. P., Williams M. A., Spronk C. A., Bonvin A. M., Nilges M. 2003. Refinement of protein structures in explicit solvent. Proteins 50:496–506 [DOI] [PubMed] [Google Scholar]
- 40. Lowry J. A., Mackay J. P. 2006. GATA-1: one protein, many partners. Int. J. Biochem. Cell Biol. 38:6–11 [DOI] [PubMed] [Google Scholar]
- 41. Martinez-Balbas M. A., Bauer U. M., Nielsen S. J., Brehm A., Kouzarides T. 2000. Regulation of E2F1 activity by acetylation. EMBO J. 19:662–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moriniere J., et al. 2009. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature 461:664–668 [DOI] [PubMed] [Google Scholar]
- 43. Mujtaba S., et al. 2002. Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain. Mol. Cell 9:575–586 [DOI] [PubMed] [Google Scholar]
- 44. Mujtaba S., et al. 2004. Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation. Mol. Cell 13:251–263 [DOI] [PubMed] [Google Scholar]
- 45. Mujtaba S., Zeng L., Zhou M. M. 2007. Structure and acetyl-lysine recognition of the bromodomain. Oncogene 26:5521–5527 [DOI] [PubMed] [Google Scholar]
- 46. Munshi N., et al. 1998. Acetylation of HMG I(Y) by CBP turns off IFNβ expression by disrupting the enhanceosome. Mol. Cell. 2:457–467 [DOI] [PubMed] [Google Scholar]
- 47. Nakamura Y., et al. 2007. Crystal structure of the human BRD2 bromodomain: insights into dimerization and recognition of acetylated histone H4. J. Biol. Chem. 282:4193–4201 [DOI] [PubMed] [Google Scholar]
- 48. Nguyen B., Tanious F. A., Wilson W. D. 2007. Biosensor-surface plasmon resonance: quantitative analysis of small molecule-nucleic acid interactions. Methods 42:150–161 [DOI] [PubMed] [Google Scholar]
- 49. Nicodeme E., et al. 2010. Suppression of inflammation by a synthetic histone mimic. Nature 468:1119–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nieba L., Krebber A., Pluckthun A. 1996. Competition BIAcore for measuring true affinities: large differences from values determined from binding kinetics. Anal. Biochem. 234:155–165 [DOI] [PubMed] [Google Scholar]
- 51. Pelton J. G., Torchia D. A., Meadow N. D., Roseman S. 1993. Tautomeric states of the active-site histidines of phosphorylated and unphosphorylated IIIGlc, a signal-transducing protein from Escherichia coli, using two-dimensional heteronuclear NMR techniques. Protein Sci. 2:543–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Persson B. D., et al. 2009. An arginine switch in the species B adenovirus knob determines high-affinity engagement of cellular receptor CD46. J. Virol. 83:673–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Polesskaya A., et al. 2000. CREB-binding protein/p300 activates MyoD by acetylation. J. Biol. Chem. 275:34359–34364 [DOI] [PubMed] [Google Scholar]
- 54. Polesskaya A., Harel-Bellan A. 2001. Acetylation of MyoD by p300 requires more than its histone acetyltransferase domain. J. Biol. Chem. 276:44502–44503 [DOI] [PubMed] [Google Scholar]
- 55. Sengupta T., Chen K., Milot E., Bieker J. J. 2008. Acetylation of EKLF is essential for epigenetic modification and transcriptional activation of the beta-globin locus. Mol. Cell. Biol. 28:6160–6170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sims R. J., III, D. Reinberg. 2008. Is there a code embedded in proteins that is based on post-translational modifications? Nat. Rev. Mol. Cell Biol. 9:815–820 [DOI] [PubMed] [Google Scholar]
- 57. Spange S., Wagner T., Heinzel T., Kramer O. H. 2009. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int. J. Biochem. Cell Biol. 41:185–198 [DOI] [PubMed] [Google Scholar]
- 58. Thorpe K. L., et al. 1997. Chromosomal localization, gene structure and transcription pattern of the ORFX gene, a homologue of the MHC-linked RING3 gene. Gene 200:177–183 [DOI] [PubMed] [Google Scholar]
- 59. Umehara T., et al. 2010. Structural implications for K5/K12-di-acetylated histone H4 recognition by the second bromodomain of BRD2. FEBS Lett. 584:3901–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wolf P. 1982. A critical reappraisal of Wadell's technique for ultraviolet spectrophotometric protein estimation. Anal. Biochem. 129:145–155 [DOI] [PubMed] [Google Scholar]
- 61. Yamagata T., et al. 2000. Acetylation of GATA-3 affects T-cell survival and homing to secondary lymphoid organs. EMBO J. 19:4676–4687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang W., Kadam S., Emerson B. M., Bieker J. J. 2001. Site-specific acetylation by p300 or CREB binding protein regulates erythroid Kruppel-like factor transcriptional activity via its interaction with the SWI-SNF complex. Mol. Cell. Biol. 21:2413–2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhao S., et al. 2010. Regulation of cellular metabolism by protein lysine acetylation. Science 327:1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zheng C., Hayes J. J. 2003. Structures and interactions of the core histone tail domains. Biopolymers 68:539–546 [DOI] [PubMed] [Google Scholar]
- 65. Zwahlen C., et al. 1997. Methods for measurement of intermolecular NOEs by multinuclear NMR spectroscopy: application to a bacteriophage lambda N-peptide/boxB RNA complex. J. Am. Chem. Soc. 119:6711–6721 [Google Scholar]