Abstract
The Ric-8 gene encodes a guanine exchange factor (GEF) that modulates G protein-mediated signaling, exhibiting a relevant role during regulation of cell division. In mammals, two Ric-8 homologues have been reported (Ric-8A and Ric-8B), and recent studies indicate equivalent roles for each protein. Here, we show that the Ric-8B gene is negatively regulated during osteoblast differentiation by the transcription factor C/EBPβ. Only the larger C/EBPβ isoform (C/EBPβ-LAP*) downregulates Ric-8B gene promoter activity in osteoblastic cells. Accordingly, knockdown of C/EBPβ expression by small intefering RNA in osteoblastic cells results in a significant increase of Ric-8B gene expression. Transient overexpression of Brg1 or Brm, the catalytic subunits of the SWI/SNF chromatin-remodeling complex, inhibits Ric-8B promoter activity. Also, the presence of inactive SWI/SNF complexes in osteoblastic cells results in increased endogenous Ric-8B transcription, indicating that SWI/SNF activity negatively regulates Ric-8B expression. During osteoblast differentiation, Ric-8B gene repression is accompanied by changes in nucleosome placement at the proximal Ric-8B gene promoter and reduced accessibility to regulatory sequences.
INTRODUCTION
Guanine nucleotide exchange factors (GEFs) are important regulators of the function of heterotrimeric G protein complexes at the plasma membrane in eukaryotic cells (35). GEFs stimulate the replacement of the GDP bound to the Gα subunit by GTP, thus leading to an active form of this protein and therefore enhancing the G-mediated signaling cascades (35). Ric-8 was originally identified in Caenorhabditis elegans as a gene that confers resistance to inhibitors of cholinesterase in a mutant strain (RIC-8 [resistance to inhibitors of cholinesterase 8]) (22, 23). Using Gα proteins as bait during yeast two-hybrid screening assays of rat and human brain cDNA libraries (15, 40), two highly homologous Ric-8 genes, Ric-8A and Ric-8B, were later identified. The Ric-8A and Ric-8B proteins have been proposed to function as GEFs, as both proteins can interact with several members of the Gα family, including Gqα, Gi/oα, and G13α, and can modulate G protein-dependent signaling in response to different ligands (15, 20, 28, 33, 40, 47).
It has been recently reported that in mammalian cells, Ric-8 has an important role during asymmetric and symmetric cell division (39). Reduced levels of Ric-8A expression altered the mitotic spindle alignment as well as the correct localization of cortical proteins, including NuMA, LGN, and dynein (52). This was accompanied by a significantly prolonged mitosis, caused occasional mitotic arrest, and decreased mitotic spindle movements. In agreement with this phenotype, recent evidence supports a relevant role of Ric-8 proteins during the initial stages of C. elegans, Drosophila melanogaster, and mouse embryonic development (5, 23, 45). Despite these data, the molecular mechanisms involved in Ric-8 protein function still remain largely undetermined.
Bone formation during development and skeletal remodeling requires the temporal and interdependent expression of cell growth and phenotypic genes. There are three major stages of osteoblastogenesis: proliferation, matrix maturation, and mineralization (29), which are characterized by sequentially expressed genes that support the progression of osteoblast differentiation through developmental transition points. The first transition requires exit from the cell cycle and commitment to the osteoblast phenotype, hence involving downregulation of genes associated with proliferation and upregulation of genes associated with the osteoblast lineage (38). Changes in the expression pattern of bone-related genes are controlled at the transcriptional level by transcription factors, such as Runx2, ATF4, osterix, and C/EBPβ, among others (21). However, whether these specific transcription factors are also associated with the transcriptional repression of genes involved in the preosteoblast active proliferation period remains undetermined.
CCAAT/enhancer binding proteins (C/EBPs) belong to the CREB/ATF family of transcription factors (27). C/EBPβ can be found as three protein isoforms, which are generated by alternative translation initiation: C/EBPβ-1 (also known as liver activating protein* [LAP*]), C/EBPβ-2 (LAP), and C/EBPβ-3 (liver inhibitory protein [LIP]). All three isoforms contain a b-zip (basic leucine zipper) DNA-binding domain and may retain either a complete (C/EBPβ-LAP*) or partial (C/EBPβ-LAP) N-terminal sequence, which contains a transcription regulatory domain. The C/EBPβ-LIP protein isoform completely lacks this N-terminal regulatory domain and therefore can function as a negative regulator of both C/EBPβ-LAP*- and C/EBPβ-LAP-mediated gene transcription (2, 53). Several reports have confirmed a key role for C/EBPβ factors regulating gene expression during osteogenesis (21). C/EBPβ null mice show delayed bone formation and decreased expression of bone phenotypic markers (36, 43). In addition, osteoblast-targeted overexpression of C/EBPβ-LIP in mice generates osteopenia as a result of reduced bone formation (10). C/EBPβ can interact with SWI/SNF chromatin remodeling complexes (16) and can recruit them to bone-related target genes (46). C/EBPβ can also interact with key components of the Mediator complex (19, 25), as well as with transcriptional coregulators, like CBP/p300 (24), G9a (32), and PRMT4/CARM1 (17), which contain histone-modifying activities. Interestingly, a recent report indicated that the ability of C/EBPβ-LAP* to interact with some of these coregulators is dictated at least in part by posttranslational modifications occurring at the N-terminal domain, which contains a key transcription activation region (17). Hence, asymmetric dimethylation of R3 catalyzed by PRMT4/CARM1 inhibits the interaction between C/EBPβ-LAP* and SWI/SNF, affecting the expression of target genes during cell differentiation.
It has been shown recently that C/EBPβ can also downregulate gene transcription (50). Therefore, it is important to define its role (and that of its associated coregulators) as an activator and/or repressor during early stages of osteoblast differentiation. Here, we report that Ric-8B gene expression is downregulated during osteoblast differentiation in a C/EBPβ-LAP*- and SWI/SNF-dependent manner. This transcriptional inhibition involves nucleosome enrichment and reduced accessibility at the Ric-8B promoter regulatory regions.
MATERIALS AND METHODS
Expression constructs.
Constructs containing the mouse Ric-8B promoter fused to the firefly luciferase gene (p2.1Ric8B-Luc, p1.1Ric8B-Luc, p0.56Ric8B-Luc, and p0.33Ric8B-Luc) were obtained by cloning different versions of the Ric-8B promoter into the pGL3-basic plasmid (Promega, Madison, WI). The p2.1Ric8B-Luc and p1.1Ric8B-Luc sequences were obtained by PCR amplification of genomic human DNA, using primers that incorporated SacI and XhoI restriction sites at the 5′ and 3′ ends, respectively (forward −2124SacI, 5′-AGAGCTCCTGGACACAGAGCAGGCACCC-3′; forward −1128SacI, 5′-AGAGCTCGCTTCGGGGGGAAAGGCCTGG-3′; reverse −58XhoI, 5′-ACTCGAGCCCAAGCCGCTGCCTCCAGG-3′). The p0.56Ric8B-Luc construct was obtained after digesting p1.1Ric8B-Luc with the KpnI enzyme to eliminate a 5′-end segment (SacI/KpnI) of the p1.1Ric8B-Luc promoter. p0.33Ric8B-Luc was obtained by direct PCR amplification of human genomic DNA, using primers that incorporate the KpnI and XhoI restriction sites at the 5′ and 3′ ends, respectively (forward −327KpnI, 5′-GGTACCGTCTGTGAAAGCCCCCTTAAAAG-3′; reverse −58XhoI, 5′-ACTCGAGCCCAAGCCGCTGCCTCCAGG-3′). The mutated version of the p2.1Ric8B-Luc construct (C/EBP site III mutated) was obtained using the QuikChange site-directed mutagenesis kit (Stratagene-Agilent, Santa Clara, CA) according with the manufacturer's instructions and confirmed by automatic sequencing. Expression of the Renilla luciferase plasmid was under the control of the simian virus 40 (SV40) constitutive promoter (pRL-SV40). Constructs encoding rat C/EBPβ isoforms pcDNA3.0-C/EBPβ-LAP*, pcDNA3.0-C/EBPβ-LAP, and pcDNA3.1-C/EBPβ-LIP were donated by Jose L. Gutierrez (University of Concepcion, Concepcion, Chile). pCGhBRM and pBJ5-BRG1 plasmids, which encode the human BRM and BRG1 catalytic subunits, respectively, of the ATP-dependent chromatin remodeling complex SWI/SNF, were donated by Anthony N. Imbalzano (University of Massachusetts Medical School, Worcester, MA).
Cell cultures.
Rat osteosarcoma-derived ROS 17/2.8 cells were maintained in F-12 medium supplemented with 5% fetal bovine serum (FBS), 1.176 g/liter NaHCO3, 0.118 g/liter CaCl2 · 2H2O, and 6.670 g/liter HEPES. C2C12 skeletal muscle progenitor cells were maintained in Dulbecco's modified Eagle's medium with F-12 (DMEM/F-12) supplemented with 10% FBS and 1.2 g/liter NaHCO3. To induce osteoblastic differentiation, proliferating C2C12 cells were treated with 300 ng/ml BMP-2 (R&D Biosystems, Minneapolis, MN) for up to 72 h, as described before (3). To induce myoblastic differentiation, confluent C2C12 cells were cultured in medium supplemented with 10% horse serum (14). Mouse preosteoblastic MC3T3 cells (kindly donated by Rafael Burgos, Universidad Austral de Chile, Valdivia, Chile) were maintained in α-MEM without ascorbic acid (AA) and supplemented with 10% FBS and 2.29 g/liter NaHCO3. When required, MC3T3 cells were grown to confluence and induced to differentiate into osteoblasts by supplementing the medium with AA (50 μg/ml) from day 3 of culture. To generate differentiated MC3T3 osteoblastic cells with a mineralized extracellular matrix, the cells were grown in medium supplemented with β-glycerophosphate from day 13. At day 24, cells were washed with ice-cold phosphate-buffered saline (PBS), fixed with 70% ethanol, and stained with 1% Alizarin Red, pH 4.1, for 5 min (room temperature). The ROSBRG1TA cell line was generated, and it has been characterized previously (46). The cells were maintained in 50 μg/ml hygromycin, 100 μg/ml Geneticin, and 10 μg/ml tetracycline. ROSBRG1TA cells were evaluated for their ability to express Flag-tagged mutant BRG1 proteins (Flag–BRG1K-R) by Western blotting using an anti-Flag antibody (Covance, Princeton, NJ).
Nuclear extracts and protein expression analyses.
Nuclear extracts were prepared as reported previously (30). The protein levels were quantified by Bradford's assay using bovine serum albumin as a standard. For Western blot analyses, 5 to 10 μg of total protein was subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and then transferred to nitrocellulose. Immunoblotting was performed with secondary antibodies conjugated to horseradish peroxide and enhanced chemiluminescence solutions (Perkin-Elmer, Waltham, MA). Primary antibodies used were as follows: Flag epitope antibody PRB-132C (Covance), C/EBPβ C-19 (sc-150; Santa Cruz Biotechnology, Santa Cruz, CA), Brg1 G-7 (sc-17796; Santa Cruz Biotechnology), and TFIIB C-18 (sc-225; Santa Cruz Biotechnology).
ChIP.
Chromatin immunoprecipitation (ChIP) studies were performed as described previously (37). Precleared chromatin fragments (200 to 300 bp) obtained from ROS 17/2.8 or MC3T3 cell cultures (100-mm-diameter plates) were immunoprecipitated overnight with agitation using the following antibodies: anti-C/EBPβ C-19 polyclonal antibody (Santa Cruz Biotechnology), anti-histone H3 polyclonal antibody (Upstate Biotechnology, Millipore, Billerica, MA), and anti-Brm or anti-Brg1 rabbit antisera kindly donated by Anthony Imbalzano (University of Massachusetts Medical School, Worcester, MA). The PCR primers used to evaluate the distal upstream region of the mouse Ric-8B gene (−4,426/−4,421) were 5′-CATGGACAGGGTTTTGGGAGAC-3′ (forward) and 5′-ACCTGTAGGTTCTGTGCATCTC-3′ (reverse). To evaluate the region −636/−426 of the mouse and rat Ric-8B promoters, the primers were 5′-TGGTTTCCGGCCTTTAGGGAAC-3′ (forward) and 5′-GACGACAACTGGCGGGCTGTTC-3′ (reverse). To evaluate the Ric-8B promoter region −396/−284, the primers used were 5′-GGAGAGACAGTTCTGCTCGTGG-3′ (forward) and 5′-GGAGCCACCAGAGACTGAGTCA-3′ (reverse).
Quantitative PCR.
Quantitative PCR (QPCR) was performed using the Brilliant SYBR green QPCR core reagent kit in an MX3000P spectrofluorometric thermal cycler (Stratagene-Agilent, Santa Clara, CA) according to the manufacturer's recommendations. Efficiencies for each primer pair were adjusted to nearly 100%, modifying the Mg2+ concentration in the amplification mix.
RT-PCR.
Total cellular RNA was isolated from 35-mm plates by using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Reverse transcriptase PCR (RT-PCR) was performed using an RT-PCR kit (Promega). Each sample was denatured at 70°C for 5 min in the presence of 0.25 μg oligo(dT)12–18. The mix was chilled and then incubated at 37°C for 1 h with 100 U Moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega), 1× buffer M-MLV RT (Promega), 20 U of RNase inhibitor (New England BioLabs, Beverly, MA), and 0.5 mM deoxynucleside triphosphates (Invitrogen). The reaction was stopped by bringing the samples to 4°C and diluting five times with nuclease-free water. cDNA samples were amplified by conventional PCR or quantified by QPCR. Expression of endogenous genes for osteocalcin, Ric-8B, myogenin, osterix, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was determined by reverse transcription and conventional PCR or by QPCR, using the following specific primers: rat osteocalcin (forward, 5′-CAGACCTAGCAGACACCATAGG-3′; reverse, 5′-CGTCCATACTTTCGAGGCAG-3′), mouse osteocalcin (forward, 5′-CGTAGTCTGACAAAGCCTTC-3′; reverse, 5′-CTGGTCTGATAGCTCGTCAC-3′), Ric-8B (forward, 5′-GGGGCTATCGAGCGGGTCCTG-3′; reverse 5′-GTCTCTGGAGAGAATGCGGAG-3′), osterix (forward, 5′-CTCTGGCTATGCCAATGACTAC-3′; reverse, 5′-ACATATCCAAGGACGTGTAGA-3′); myogenin (forward, 5′-TGGAGCTGTATGAGACATCCC-3′; reverse, 5′-TGGACAATGCTCAGGGGTCCC-3′), C/EBPβ (forward, 5′-TCTACTACGAGCCCGACTGC-3′; reverse, 5′-AGGTAGGGGCTGAAGTCGAT-3′), and GAPDH (forward, 5′-CCTGCTTCACCACCTTCTTGAT-3′; reverse, 5′-CATGGCCTTCCGTGTTCCTA-3′).
Transient-transfection assays.
Subconfluent MC3T3 cells were transfected for 24 h using FuGENE reagent (Roche, Indianapolis, IN), according to the manufacturer's recommendations. Cells cultured in 6- or 24-well plates were transfected with up to 5.0 or 1.0 μg of DNA, respectively. When required, the total DNA concentration was adjusted using purified plasmid DNA. The analysis of luciferase activity was performed in a GloMax-20/20 luminometer according to the Promega dual luciferase reporter assay protocol.
Silencing of C/EBPβ.
ROS 17/2.8 cells were cultured in 6 well-plates and transiently transfected at 50% confluence with either C/EBPβ small interfering RNA (siRNA; Cebpb Silencer Select predesigned siRNAs, siRNA-1 ID s127565 and siRNA-2 ID s127566; Ambion) or a random siRNA mix (Silencer Select negative control 1 siRNA; catalog number 4390844; Ambion) using 4 μl Oligofectamine per well (Invitrogen), according to the manufacturer's recommendations. Transfections were carried out in F-12 medium without serum and antibiotics, during 4 h. Subsequently, F-12 medium supplemented with 15% FBS (3×) was added to the cells and incubated for 48 h.
Restriction endonuclease accessibility analyses.
Restriction endonuclease accessibility analyses were performed in both MC3T3 and ROSBRG1TA cells. Differentiating MC3T3 cells were cultured for up to 13 days in medium supplemented with 50 μg/ml ascorbic acid from day 3. At the indicated times (see the figure legends), cells were washed with ice-cold PBS, scraped, and collected. Cell suspensions were centrifuged at 1,000 × g for 5 min, and the pellets were resuspended in 10 ml of buffer RSB plus NP-40 (10 mM Tris-Cl [pH 7.4], 10 mM NaCl, 5 mM MgCl2, and 0.5% Nonidet NP-40). Cells were disrupted using a Dounce homogenizer, and lysis was confirmed by trypan blue staining. Then, 10 ml of buffer RSB without NP-40 was added, the suspension was centrifuged at 1,000 × g for 5 min, and the supernatant was discarded. The nuclear pellet was resuspended in 3 ml of buffer RSB without NP-40 and then quantified by measuring absorbance at 260 nm. Two A260 units of DNA were incubated with 500 U of EcoRI, BanII, or XmaI (MC3T3 samples) and with BanII or AvaI (ROSBRG1TA samples) restriction enzymes at 37°C for 30 min in the corresponding buffer (New England BioLabs), in a final volume of 1 ml. Digestion was stopped by addition of EDTA to a 25 mM final concentration and 0.5% SDS. The samples were then digested with 100 μg/ml proteinase K for 16 h at 37°C, and DNA was recovered by phenol-chloroform extraction and subsequent ethanol precipitation. The DNA was then resuspended in Tris-EDTA and analyzed by QPCR using specific sets of primers, according to the type of DNA sample (mouse or rat). For the mouse (MCT3T3) samples the following primers were used: forward −636 (5′-TGGTTTCCGGCCTTTAGGGAAC-3′) and reverse −426 (5′-GACGACAACTGGCGGGCTGTTC-3′) (Ric-8B promoter); forward −4,624 (5′-CATGGACAGGGTTTTGGGAGAC-3′) and reverse −4,421 (5′-ACCTGTAGGTTCTGTGCATCTC-3′) (mouse sequence upstream of the Ric-8B gene promoter). For the rat DNA samples the primers were the following: forward −636 and reverse −426 (Ric-8B promoter); forward −118 (5′-GTGGTAGGCAGTCCCACTTT-3′) and reverse +29 (5′-TGTTTGTGAGGCGAATGAAG-3′) (rat Runx2 gene promoter sequence). The results are expressed as the ratio of the DNA sequence amplified with primers −636/−426 (mouse and rat samples) versus that of the DNA amplified with primers −4,626/−4,421 (mouse samples) or with the primers −118/+29 (rat samples).
Ric-8B expression in SWI/SNF-deficient ROS 17/2.8 cells.
To determine whether Ric-8B expression requires the activity of the SWI/SNF chromatin remodeling complex, we used the ROSBRG1TA cell line, which expresses an ATPase-deficient Flag-tagged Brg1 mutant (K-R) protein under the control of a tetracycline-inducible promoter system (46). ROSBRG1TA cells were maintained in F-12 medium supplemented with 50 μg/ml hygromycin, 100 μg/ml Geneticin, and 10 μg/ml tetracycline (46). The mutant Brg1 protein is expressed in these cells when cultured in medium lacking tetracycline for 3 days. Cells were then collected and processed for Western blotting, QPCR, endonuclease accessibility, and ChIP analyses.
RESULTS
Ric-8B expression is downregulated during osteoblast differentiation.
Osteoblast differentiation involves downregulation of genes associated with active proliferation and upregulation of genes involved in establishing the osteoblastic lineage (38). The Ric-8B gene encodes a GEF that has been shown to modulate G protein-dependent signaling in mitotic cells and control cell division (26, 44, 49, 52). We found that the Ric-8B gene is strongly expressed in several osteoblastic and preosteoblastic cells that are actively proliferating. As shown in Fig. 1A, rat (ROS17/2.8 and ROB), mouse (MC3T3), and human (SaOS-2) cell cultures actively transcribed the Ric-8B gene at comparable levels. Similarly, nonosteoblastic cell lines, like the mouse pluripotential C2C12 cells (Fig. 1A), also expressed high levels of Ric-8B mRNA while proliferating. Importantly, we found that Ric-8B expression was significantly reduced when the cells stopped proliferating and became engaged in differentiation processes. Thus, when C2C12 cells were induced to differentiate toward the osteoblastic (Fig. 1B) or myoblastic (Fig. 1C) lineage, Ric-8B mRNA levels were downregulated concomitantly with the increase in expression of osteoblast- and myoblast-specific markers (osterix and myogenin, respectively).
Fig. 1.
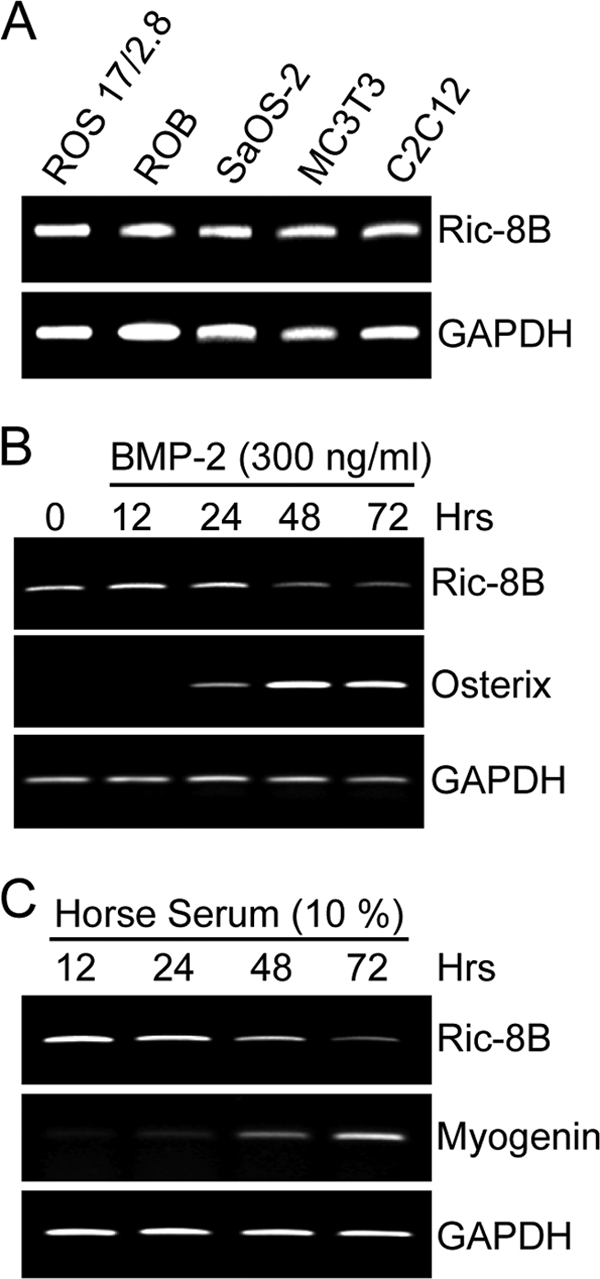
The Ric-8B gene is expressed in proliferating cells and downregulated during differentiation. (A) mRNA levels of Ric-8B were determined by RT-PCR from total RNA samples obtained from different osteoblastic and myoblastic cell lines. ROB, primary rat diploid osteoblasts; ROS17/2.8, rat osteosarcoma cells; SaOS-2, human osteosarcoma cells; MC3T3, mouse preosteoblastic cells; C2C12, mouse promyoblastic cells. (B and C) C2C12 cells were cultured for up to 72 h in the presence of BMP-2 (300 ng/ml) to induce osteoblast differentiation (B) or in medium supplemented with 10% horse serum to induce myoblast differentiation (C). The cells were collected at the indicated times, and the Ric-8B mRNA levels were determined by RT-PCR using specific primers. Osterix, an osteoblastic differentiation marker; myogenin, a myoblastic differentiation marker.
MC3T3 cells are a preosteoblastic cell line that has been shown to progressively differentiate to mineralized osteoblasts when grown in the presence of AA. Therefore, this process represents a well-established model system to study gene regulation during osteogenesis (48). As shown in Fig. 2A, MC3T3 cells exhibited a heavily mineralized extracellular matrix after 24 days of culture in differentiating medium compared to cells grown under control conditions (absence of AA). This differentiation process was reflected in the progressive transcriptional activation of the osteocalcin (OC) gene (Fig. 2C), a well-established bone phenotypic marker (9, 46), which was paralleled by a gradual decrease in Ric-8B mRNA expression (Fig. 2B). Interestingly, Ric-8B transcription was slightly increased between days 2 and 3 of differentiation, reflecting the elevated proliferative activity of these cells at that initial stage of the differentiation process.
Fig. 2.
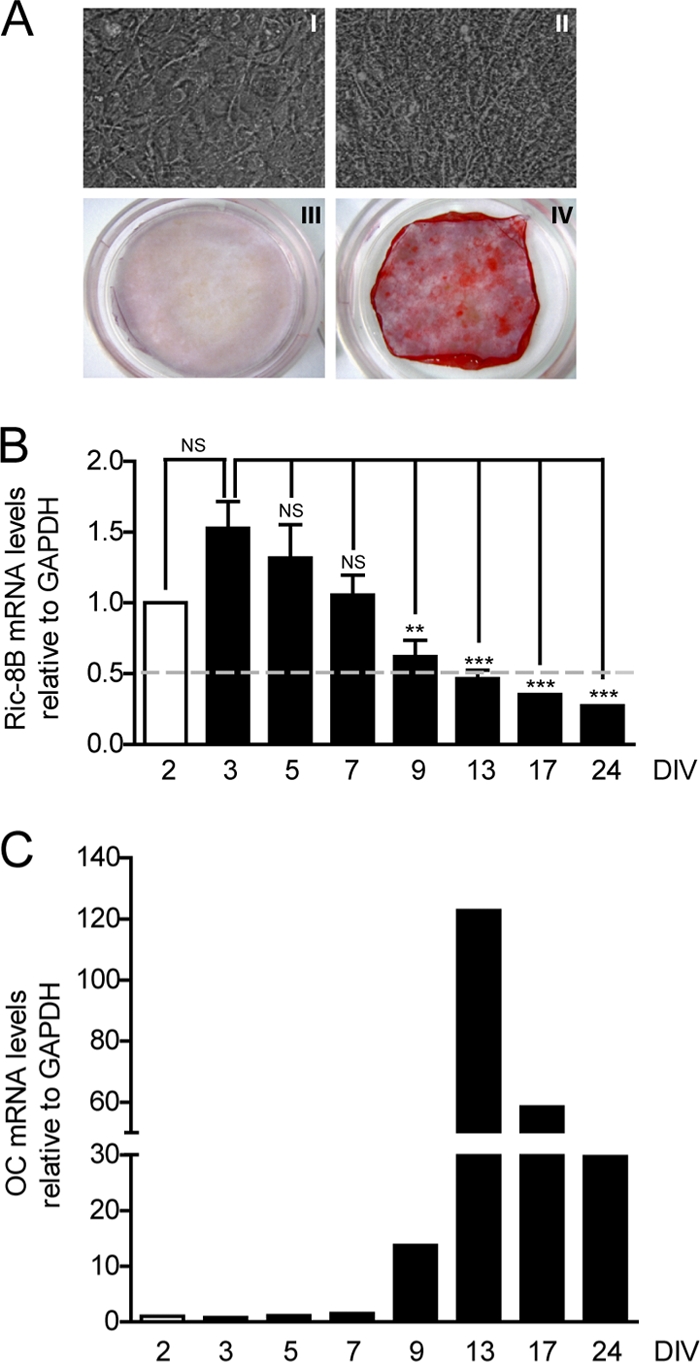
The Ric-8B gene is repressed during osteoblast differentiation. (A) MC3T3 cells were cultured for 24 days in the presence (II and IV) or absence (I and III) of 50 μg/ml AA and 3 mM β-glycerophosphate. The cells were then photographed using an optical microscope (I and II) or stained with Alizarin Red to visualize mineralized nodules (III and IV). (B) MC3T3 cells were cultured for up to 24 days in medium supplemented with 50 μg/ml AA from day 3. The cells were collected at the indicated times, and Ric-8B mRNA levels were determined by QPCR, normalizing with respect to GAPDH mRNA expression. DIV, days of differentiation in vitro. Bars represent means ± the standard errors of the means of three independent experiments. Statistically significant differences were determined by an analysis of variance test. NS, not significant; **, P < 0.01; ***, P < 0.001. (C) To control for osteoblast differentiation, OC gene mRNA expression was determined by QPCR.
The C/EBPβ transcription factor downregulates Ric-8B gene promoter activity.
Previous studies have established that the C/EBPβ transcription factor is a key regulator of bone-related genes (21). Importantly, a recent report indicated that C/EBPβ factors may be important regulators of the proliferative state of mammalian cells (12) by modulating the expression of proliferation-promoting genes. Sequence analysis of the human and mouse Ric-8B gene promoters (first 2.1-kb region) revealed the presence of multiple putative C/EBP-binding sites (Fig. 3A and data not shown). Moreover, the sequence within the proximal 650 bp of this promoter is highly conserved between the human and mouse Ric-8B genes (79% conservation, based on analysis with Vista Tools [http://www.genome.ibl.gov/vista/index.shtml] and ClustalW [http://www.ebi.ac.uk/tools/clustalw2/index.html]), suggesting the presence of evolutionarily conserved transcriptional regulatory mechanisms. As this region contains 3 C/EBP sites (designated site I, site II, and site III, according to their distance from the region including the transcriptional start sites of this gene [Fig. 3A]), we explored the hypothesis that C/EBPβ promotes the osteoblast phenotype not only by upregulating bone-related genes but also by controlling genes (like Ric-8B) associated with the proliferative activity of preosteoblasts.
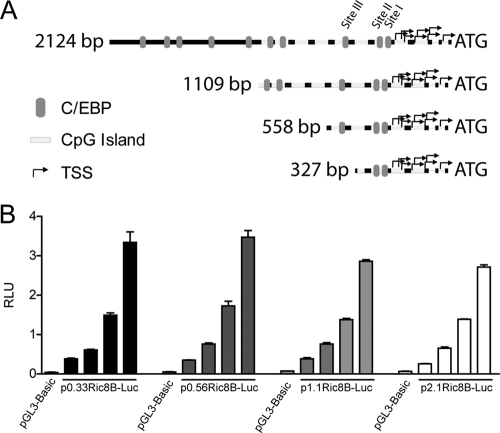
Fig. 3.
Characterization of Ric-8B gene promoter activity in osteoblastic cells. (A) Schematic diagram representing different segments of the Ric-8B gene promoter sequence evaluated through transient-transfection and luciferase activity assays. Gray ovals indicate putative C/EBP-binding sites. White boxes indicate CpG-rich regions, and black arrows represent different transcription start sites (TSS) described for this gene. Each of these promoter segments was cloned into the pGL3-Basic vector, upstream of the firefly luciferase gene. (B) Subconfluent MC3T3 cells were transiently transfected with increasing amounts (100 to 800 ng) of each of the constructs shown in panel A. Twenty-four hours later, luciferase activity was determined. p2.1Ric8B-Luc, p1.1Ric8B-Luc, p0.56Ric8B-Luc, and p0.33Ric8B-Luc correspond to constructs containing segments of the Ric-8B gene promoter of 2,124, 1,109, 558, and 327 bp, respectively. The bars represent the means ± standard errors of the means of two independent experiments, each performed in triplicate.
To begin examining the contribution of C/EBPβ to the transcriptional regulation of the Ric-8B gene in osteoblastic cells, we cloned a 2.1-kb segment of the Ric-8B gene promoter region in a vector controlling the expression of the luciferase reporter gene (see Materials and Methods). We found that this promoter construct is expressed at high rates and in a dose-dependent manner when transiently transfected in actively proliferating MC3T3 cells (Fig. 3B). Interestingly, constructs carrying deleted versions of the Ric-8B promoter (1.1 kb, 0.56 kb, and 0.33 kb) showed equivalent promoter activities (Fig. 3B), indicating that likely most of the regulatory elements required for Ric-8B transcription in these preosteoblastic cells reside within the highly conserved proximal 600 bp of the promoter. Because of these results, most of our experiments were concentrated in this region of the Ric-8B gene promoter.
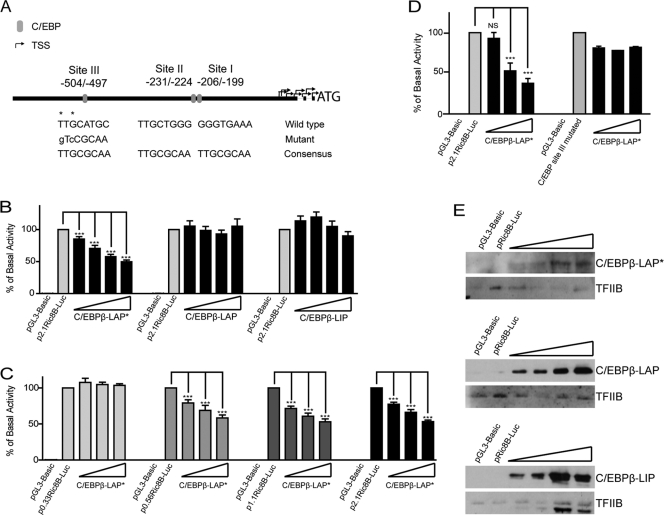
We next evaluated whether the C/EBPβ transcription factor contributes to the regulation of Ric-8B gene promoter activity in osteoblastic cells. As shown in Fig. 4, forced expression of C/EBPβ-LAP* resulted in a significant inhibition of the Ric-8B promoter activity (Fig. 4B). Interestingly, overexpression of C/EBPβ-LAP or C/EBPβ-LIP neither inhibited nor stimulated the activity of the Ric-8B promoter (Fig. 4B), indicating that the effect of C/EBPβ-LAP* is specific. In addition, the C/EBPβ-LAP*-mediated inhibition occurred when reporter constructs carrying 2.1 kb (p2.1Ric8B-Luc), 1.1 kb (p1.1Ric8B-Luc), and 0.56 kb (p0.56Ric8B-Luc) of the Ric-8 gene promoter sequence were evaluated (Fig. 4C). No inhibitory effect, however, was detected when a shorter promoter segment (p0.33Ric8B-Luc) was cotransfected with C/EBPβ-LAP* (Fig. 4C), indicating that this C/EBPβ isoform requires the Ric-8B promoter sequence included between 0.56 and 0.30 kb to downregulate Ric-8B promoter activity. As this promoter region contains the putative C/EBP site III, our results indicated that this site is relevant for C/EBPβ-LAP*-mediated repression of the Ric-8B gene in osteoblasts. To assess this possibility, we mutated by site-directed mutagenesis the C/EBP site III (Fig. 4A), such that it was no longer recognized by the C/EBPβ factor (11). This mutation was introduced in the p2.1Ric8B-Luc construct, which allowed the evaluation of its effect within the context of the full Ric-8B promoter sequence. As shown in Fig. 4D, it was found that the absence of a functional C/EBP site III prevents C/EBPβ-LAP*-mediated repression of Ric-8B promoter activity.
Fig. 4.
C/EBPβ-LAP* negatively regulates the Ric-8B promoter activity in osteoblastic cells. (A) Schematic diagram of putative C/EBP elements within the first 600 bp of the human, mouse, and rat Ric-8B promoters. Negative values indicate the position of these sites relative to ATG. To evaluate the contribution of C/EBP site III, nucleotides marked with asterisks were replaced by the nucleotides showed in lowercase letters by using site-directed mutagenesis. (B) Subconfluent MC3T3 cells were cotransfected with the construct p2.1Ric-8B-Luc (100 ng) and increasing amounts (100 to 800 ng) of the plasmids encoding each C/EBPβ isoform: C/EBPβ-LAP*, C/EBPβ-LAP, and C/EBPβ-LIP. Twenty-four hours later, luciferase activity was determined. (C) MC3T3 cells were cotransfected with the different constructs containing the Ric-8B promoter sequences (2.1, 1.1, 0.56, and 0.33 kb) and increasing amounts of the plasmid encoding C/EBPβ-LAP* (200 to 800 ng). Twenty-four hours after transfection, luciferase activity was determined. (D) MC3T3 cells were cotransfected with the p2.1Ric-8B-Luc plasmid, including a mutated putative C/EBP site III (p2.1Ric-8B-MutC/EBPIII-Luc; 200 ng) and increasing amounts (100 to 800 ng) of the plasmid encoding for C/EBPβ-LAP*. At 24 h after transfection, luciferase activity was determined and compared with that of the wild-type p2.1Ric-8B-Luc promoter construct. (E) Ten micrograms of nuclear extracts obtained from MC3T3 cells transfected under the conditions described for panel B was analyzed by Western blotting to control for overexpression of C/EBPβ. Reblotting against TFIIB was used to control for equal loading. The graphs show the percentage of basal activity, calculated as the ratio between firefly and Renilla luciferase activities. The bars represent the means ± standard errors of the means of three independent experiments, each performed in triplicate. Statistically significant differences were determined by the analysis of variance test. NS, not significant; ***, P < 0.001.
Taken together, these results indicate that C/EBPβ-LAP* inhibits Ric-8B promoter activity by directly recognizing a sequence element (site III) located within the proximal 560 bp of the promoter.
C/EBPβ interacts with the Ric-8B gene promoter during osteoblast differentiation.
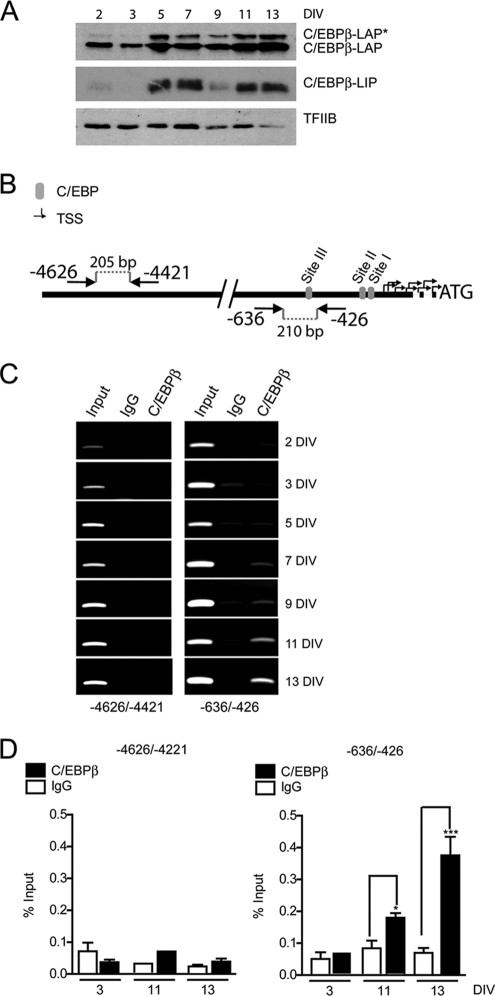
We next assessed whether C/EBPβ directly binds to the endogenous Ric-8B gene promoter in osteoblastic cells. As shown in Fig. 5A, during MC3T3 osteoblast differentiation, the C/EBPβ-LAP* protein content in nuclear extracts increased at day 5 and remained elevated through day 13 of differentiation. This expression pattern accompanied the gradual decrease of Ric-8B mRNA expression as well as the gradual increase in OC mRNA levels (Fig. 2B and C). C/EBPβ-LAP protein levels were also found slightly elevated at day 5 of differentiation (Fig. 5A), which is in agreement with previous reports indicating that C/EBPβ expression is increased during osteoblast differentiation (9). The C/EBPβ-LIP protein isoform was also found elevated at day 5 of differentiation (Fig. 5A). As this protein can function as an inhibitor of C/EBPβ-LAP*- and C/EBPβ-LAP-mediated transcriptional control (2, 53), our results indicate that, under ex vivo culture conditions that reproduce osteoblast differentiation, C/EBPβ-LIP does not prevent the transcriptional effects of the two larger C/EBPβ isoforms.
Fig. 5.
C/EBPβ binds to the Ric-8B promoter in differentiating osteoblastic cells. (A) MC3T3 cells were cultured between days 3 and 13 (DIV, days of differentiation in vitro) in the presence of 50 μg/ml ascorbic acid. Nuclear extracts were prepared on the indicated days, and the levels of C/EBPβ protein isoforms were determined by Western blotting using an anti-C/EBPβ polyclonal antibody. Detection of TFIIB was used to control for equal loading. (B) Diagram illustrating the location of the primers used in the ChIP experiments. Arrows indicate the direction of each primer, and the negative values indicate their position relative to ATG. (C) MC3T3 cells were cultured as described for panel A. At the indicated times, cells were cross-linked with 1% formaldehyde, and the sonicated chromatin fragments were immunoprecipitated using specific polyclonal antibodies against C/EBPβ. The enrichment of Ric-8B promoter sequences in the precipitated chromatin fragments was visualized by conventional PCR using the primers described for panel B. (D) Quantification of this enrichment by QPCR. Bars represent the means ± the standard errors of the means of three independent experiments. Statistically significant differences were determined by the analysis of variance test. ***, P < 0.001; *, P < 0.05.
ChIP analyses revealed that the C/EBPβ factor is gradually recruited to the Ric-8B promoter during MC3T3 osteoblast differentiation (Fig. 5C, right panels). This interaction is restricted to the highly conserved proximal promoter region of the Ric-8B gene, as distal upstream sequences (between 4,626 and 4,421 bp from the translational start codon) were not enriched in the precipitated DNA samples (Fig. 5C, left panels). The C/EBPβ interaction was found to be high at days 11 and 13 of differentiation (Fig. 5C, with quantification by QPCR shown in D), which paralleled the repression of the Ric-8B gene (Fig. 2). Together, these results demonstrate that during osteoblast differentiation, increased expression of C/EBPβ (and especially of C/EBPβ-LAP*) accompanies binding of the factor to the Ric-8B gene promoter and the inhibition of Ric-8B gene transcription.
Knockdown of C/EBPβ activates Ric-8B expression in osteoblastic cells.
It was necessary to demonstrate that C/EBPβ is an important component of the regulatory machinery that modulates transcription of the Ric-8B gene in osteoblastic cells. Our experimental approach was to reduce the expression of C/EBPβ in osteoblastic cells by using specific siRNA (see Materials and Methods) and then to determine the effect on Ric-8B gene transcription. Because C/EBPβ is required for the expression of key bone phenotypic genes (9, 21, 46), C/EBPβ knockdown in MC3T3 cells would prevent the analyses at later stages of osteoblast differentiation (e.g., day 13). Therefore, we carried out the knockdown experiments in ROS 17/2.8 cells, which represent an actively proliferating osteoblastic cell line that constitutively expresses several bone phenotypic genes, including Runx2 and OC, among others (34). Treatment of these cells during 48 h with a specific siRNA against C/EBPβ significantly reduced C/EBPβ mRNA (>70%) and protein (>50%) levels without affecting the expression of C/EBPβ-independent genes (e.g., GAPDH and TFIIB) (Fig. 6A and B). Importantly, knockdown of C/EBPβ resulted in a 2-fold increase of Ric-8B mRNA levels. As a necessary control we showed that treatment of these osteoblastic cells with a nonspecific siRNA mix (see Materials and Methods) affected the expression of neither C/EBPβ nor of Ric-8B (Fig. 6A and B). Together, these results demonstrate that in osteoblastic cells, C/EBPβ negatively regulates Ric-8B gene transcription.
Fig. 6.
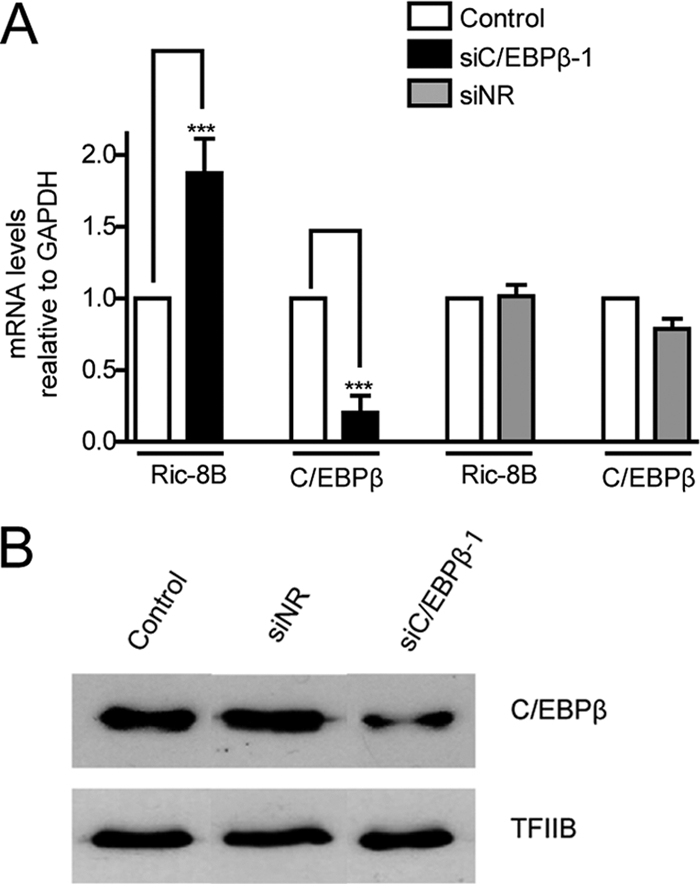
C/EBPβ downregulates the endogenous expression of Ric-8B in intact osteoblastic cells. (A) ROS17/2.8 osteoblastic cells were transfected with a specific siRNA against C/EBPβ (siC/EBPβ-1). At 48 h later, mRNA levels of Ric-8B and C/EBPβ were determined by QPCR. The bars represent the means ± the standard errors of the means of two independent experiments. Statistically significant differences were determined by the analysis of variance test. ***, P < 0.001. (B) Nuclear extracts from ROS17/2.8 cells, treated as described for panel A, were analyzed by Western blotting to determine C/EBPβ protein levels. Control, cells transfected with vehicle; siNR, cells transfected with an unrelated siRNA mix (for siRNA information, see Materials and Methods).
SWI/SNF binds to the Ric-8B gene and contributes to Ric-8B repression.
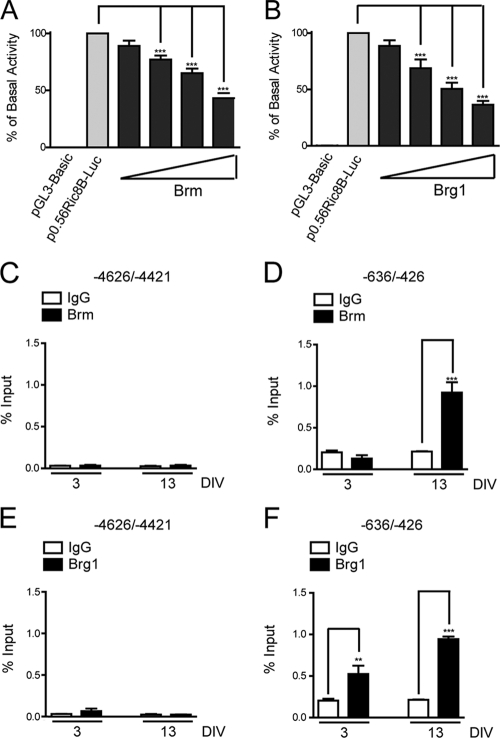
It has been reported that C/EBPβ can interact with SWI/SNF, recruiting the complex to target genes to modulate transcription (16, 17, 46). Both Brm- and Brg1-containing SWI/SNF complexes can be important components of the repressive machinery that downregulates transcription of target genes (4, 7). Therefore, we examined whether SWI/SNF complexes were involved in the transcriptional repression of the Ric-8B gene during osteoblast differentiation. As shown in Fig. 7, independent transient overexpression of both SWI/SNF catalytic subunits, Brm and Brg1, resulted in a dose-dependent reduction of Ric-8B gene promoter activity in transfected MC3T3 cells (Fig. 7A and B, respectively). In agreement with these results, we found by ChIP analyses that during osteoblast differentiation, binding of both Brm and Brg1 to the Ric-8B promoter is significantly enhanced (Fig. 7D and F, respectively), indicating that the SWI/SNF complex is enriched at the Ric-8B promoter concomitant with C/EBPβ binding (Fig. 7D) and the transcriptional repression of the Ric-8B gene (Fig. 2B). This SWI/SNF enrichment at the proximal Ric-8B promoter is specific, as no significant association of the complex (or C/EBPβ factor) was detected at upstream regions of the Ric-8B gene (Fig. 7, compare left and right panels). To examine the contribution of C/EBPβ to SWI/SNF binding to the Ric-8B gene promoter, we performed ChIP experiments in osteoblastic cells in which C/EBPβ had been knocked down. As shown in Fig. 8, reduced expression of C/EBPβ factor by using specific siRNAs (Fig. 8B) resulted in the absence of C/EBPβ binding at the Ric-8B promoter as well as in a significant reduction of Brm and Brg1 association with promoter sequence (Fig. 8A). Together these results indicate that the SWI/SNF interaction with the Ric-8B gene promoter in osteoblastic cells requires C/EBPβ.
Fig. 7.
Brm and Brg1 bind to the Ric-8B promoter during osteoblast differentiation and downregulate its activity. (A and B) Proliferating MC3T3 cells were cotransfected with the construct p0.56Ric8B-Luc (100 ng) and increasing amounts of the plasmids coding for the SWI/SNF catalytic subunits Brm (A) or Brg1 (B) (100 to 800 ng). At 24 h after transfection, luciferase activity was determined. The graphs show the percentage of basal activity, calculated as described in for Fig. 3. Overexpression of Brm and Brg1 proteins was confirmed by Western blotting using specific antibodies (data not shown). The bars represent the averages ± standard errors of the means of three independent experiments, each performed in triplicate. (C to F) MC3T3 cells were cultured for up to 13 days in the presence of 50 μg/ml AA, starting at day 3 (DIV, days of differentiation in vitro). At the indicated times, cells were cross-linked with 1% formaldehyde, and the fragmented chromatin was immunoprecipitated using specific antibodies against Brm (C and D) and Brg1 (E and F). The enrichment levels of Ric-8B promoter sequences in the precipitated chromatin were determined by QPCR using specific primers. The DNA sequences analyzed are indicated at the top of each graph. The bars represent the means ± standard errors of the means of two independent experiments performed in duplicate. Statistically significant differences were determined by the analysis of variance test. ***, P < 0.001; **, P < 0.01.
Fig. 8.
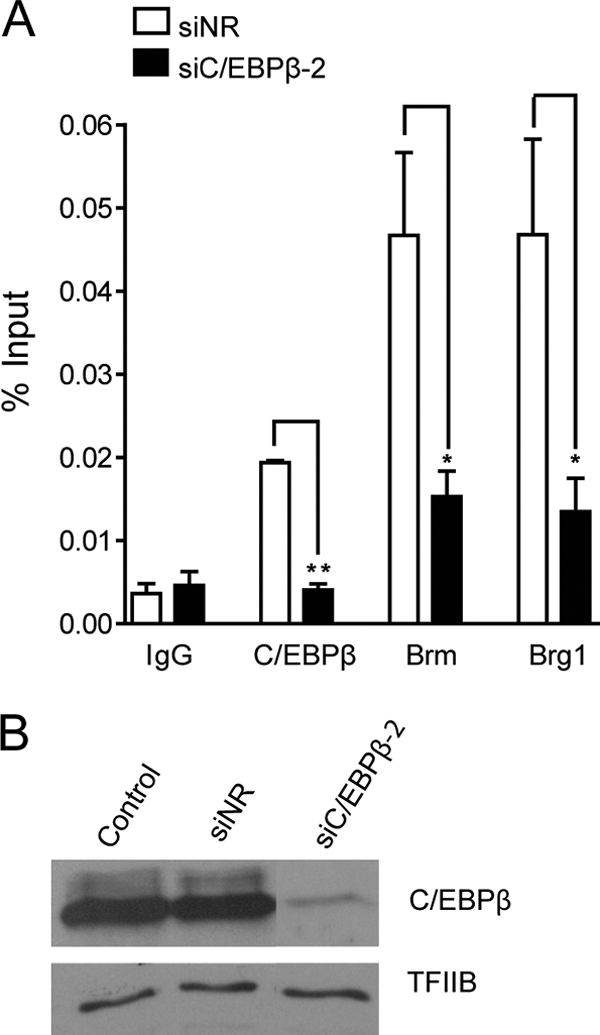
Binding of Brm and Brg1 to the Ric-8B gene promoter decreases in the absence of C/EBPβ. (A) ROS 17/2.8 osteoblastic cells were transfected with a specific siRNA against C/EBPβ (siC/EBPβ-2). At 48 h later, cells were cross-linked with 1% formaldehyde, and the fragmented chromatin was immunoprecipitated using specific antibodies against C/EBPβ, Brm, and Brg1. The enrichment of Ric-8B promoter sequences in the precipitated chromatin fragments was quantified by QPCR using specific primers to amplify the sequence −636 to −426. Bars represent the means ± standard errors of the means of three independent experiments. Statistically significant differences were determined by the analysis of variance test. *, P < 0.05; **, P < 0.01. (B) Nuclear extracts from ROS17/2.8 cells treated as described for panel A were analyzed by Western blotting to determine C/EBPβ protein levels. Control, cells transfected with vehicle; siNR, cells transfected with an unrelated siRNA mix (for siRNA information, see Materials and Methods).
We previously reported the generation of ROS17/2.8 cell lines (ROSBRG1TA) that inducibly express (tetracycline-inducible Tet-off system) Flag-tagged Brg1 mutated in the ATP-binding site (46). This mutant Brg1 protein is competent for assembling nonfunctional SWI/SNF complexes that bind to the bone-specific OC gene promoter and inhibit both chromatin remodeling and OC gene transcription (46). Cells grown in the presence of tetracycline express Ric-8B and OC mRNAs at levels comparable to those detected in wild-type ROS17/2.8 (data not shown). As previously reported (3, 46), removal of tetracycline from the cell medium for 3 to 4 days induced expression of the mutant Flag-tagged Brg1 (Fig. 9A) and a significant reduction of OC mRNA levels (Fig. 9B). In contrast, we found that Ric-8B mRNA levels were significantly increased in cells grown in the absence of tetracycline (Fig. 9B), indicating that the presence of inactive SWI/SNF complexes in the osteoblastic cells reverts repressive mechanisms operating at the Ric-8B gene. As expected, the mRNA levels of control genes, such as GAPDH, were unaffected by the presence of inactive SWI/SNF complexes (data not shown). We next examined whether these inactive SWI/SNF complexes were interacting with the Ric-8B gene promoter. As shown in Fig. 9C, ChIP analyses using an anti-Flag antibody (3, 46) demonstrated that only in the osteoblastic cells grown in the absence of tetracycline was the mutant Flag-tagged Brg1 found to interact with the promoter region of Ric-8B.
Fig. 9.
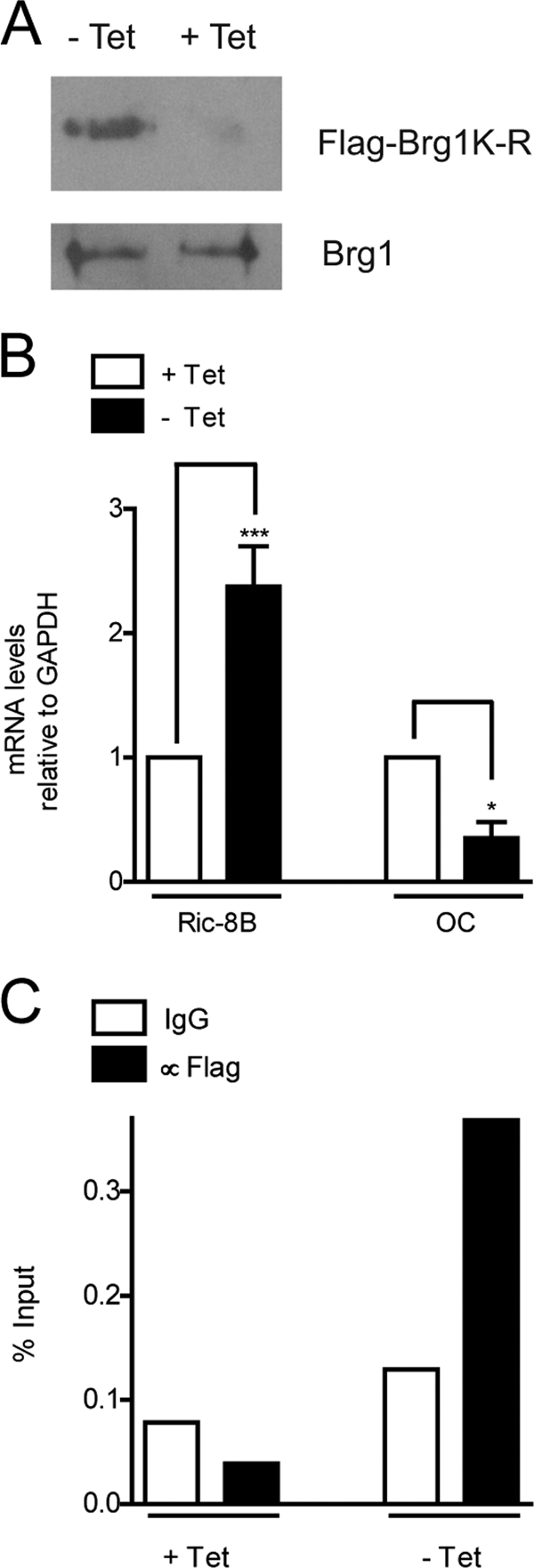
SWI/SNF represses Ric-8B gene expression in osteoblastic cells in an ATP-dependent manner. ROSBRG1TA cells expressing Flag-tagged Brg1 protein mutated in the ATP-binding site (Flag-Brg1K-R) and under the control of the tetracycline-inducible Tet-off system were cultured for 3 days in the presence (+Tet) or absence (-Tet) of tetracycline (10 μg/ml). The cells were then collected and processed to confirm Flag-Brg1K-R protein expression, using an anti-Flag antibody, as reported previously (46). Endogenous Brg1 was detected using a specific antibody (A). Ric-8B and OC mRNA levels were determined by QPCR (B). Binding of the Flag-Brg1K-R mutant protein to the proximal Ric-8B promoter was analyzed by ChIP using the anti-Flag antibody (C). Statistically significant differences were determined by the analysis of variance test. ***, P < 0.001; *, P < 0.05.
Transcriptional downregulation of the Ric-8B gene during osteoblast differentiation is reflected by a selective increase in nuclease accessibility and nucleosome enrichment at the promoter region.
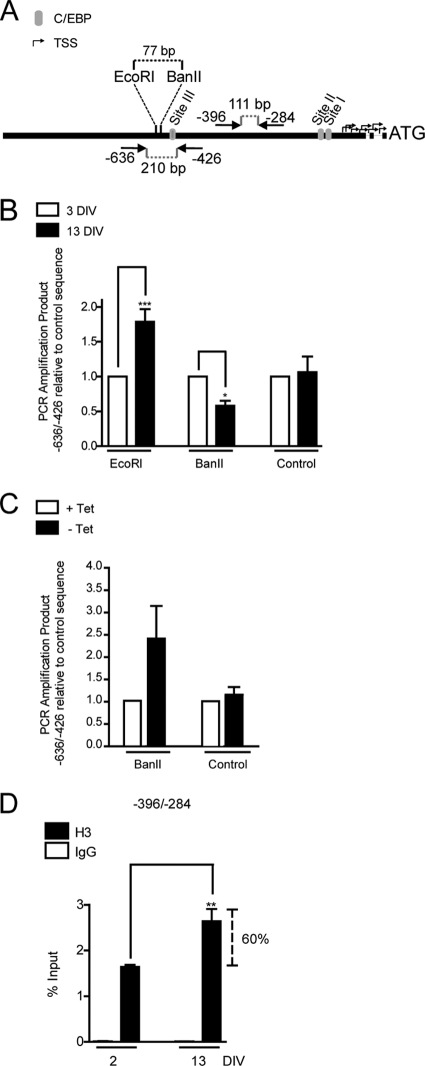
SWI/SNF activity alters nucleosome structure and nucleosome positioning at target sites in chromatin. This repositioning of histone octamers may occur along the same DNA segment by sliding (18, 51) or to a nucleosome-free DNA segment by octamer transfer (8, 31). Therefore, we examined whether binding of C/EBPβ and SWI/SNF at the Ric-8B gene promoter during osteoblast differentiation resulted in nucleosome reorganization, which may reflect the transcription inhibition of this gene. We first assessed alterations in chromatin structure associated with Ric-8B gene repression by analyzing changes in restriction endonuclease accessibility at the C/EBPβ-targeted promoter region. As shown in Fig. 10A, we determined accessibility to the restriction enzymes EcoRI and BanII, which cleave the Ric-8B promoter within the region spanning the putative C/EBP site III. Therefore, the degree of accessibility to these two enzymes, especially to BanII, can directly indicate changes in chromatin organization associated with the C/EBPβ- and SWI/SNF-dependent Ric-8B gene repression process. Careful analysis of the mouse Ric-8B promoter sequence indicated that the EcoRI site includes a noncanonical nucleotide (GGGAAgTCT instead of GGGAATTCT). However, this change does not prevent the specific recognition and subsequent cleavage by the enzyme (reference 42 and our results). We isolated nuclei from differentiating MC3T3 cell cultures at days 3 and 13, incubated them with each enzyme, and determined the degree of digestion relative to a noncleavable distal upstream sequence (see Materials and Methods). It was found that while accessibility to EcoRI digestion was significantly decreased during osteoblast differentiation, cleavage by BanII was significantly enhanced (Fig. 10B). As a control, we showed that incubation of nuclei from either proliferating or differentiated cells with XmaI, which can cleave the Ric-8B promoter in two sites located proximal to the sequence spanning the transcription start sites (control cleavage) that are not included in the sequence amplified by this set of primers (Fig. 10A), did not affect the specific amplification of the −636/−426 sequence (Fig. 10B). Together, these results indicated that transcriptional repression of the Ric-8B gene during osteoblast differentiation involves a chromatin reorganization process that includes increased accessibility at the C/EBP site III (increased cleavage by BanII) and reduced accessibility upstream (77 bp) of this element (reduced cleavage by EcoRI). We next assessed whether this increased BanII accessibility at the Ric-8B promoter is dependent on SWI/SNF activity. We isolated nuclei from ROSBRG1TA cells grown in the presence or absence of tetracycline and incubated them with BanII. As shown in Fig. 10C, chromatin from cells that express mutant Brg1 (cells cultured in the absence of tetracycline [Fig. 9A]) exhibited reduced accessibility to the restriction enzyme at the Ric-8B promoter. Therefore, this result indicates that increased accessibility to BanII in the sequence that includes C/EBP binding site III during Ric-8B gene repression requires SWI/SNF activity.
Fig. 10.
Ric-8B gene repression during osteoblast differentiation is associated with chromatin remodeling at the Ric-8B promoter. (A) Diagram of the location of the primers used in panels B to D, the putative C/EBP sites I, II, and III (gray ovals), and the location of target sites for the restriction endonucleases EcoRI and BanII. (B) MC3T3 cells were cultured for up to 13 days in medium containing AA (50 μg/ml) from day 3. At the indicated times, nuclei were isolated and a nuclease accessibility assay was performed, with incubation for 30 min at 37°C with EcoRI, BanII, or a restriction enzyme cutting away from the amplified sequence (control cleavage, XmaI or AvaI). Cleaved genomic DNA was then purified, and the Ric-8B promoter region −636/−426, as well as control sequences (that were not cleaved by any of the enzymes [see Materials and Methods]) were amplified and quantified by QPCR. (C) ROSBRG1TA cells were cultured as described for Fig. 9. After harvesting, nuclei were isolated from cells, and nuclease accessibility assays were performed as described for panel B. (D) MC3T3 cells were grown as described for panel B. At the indicated times, cells were cross-linked with 1% formaldehyde, and the fragmented chromatin was immunoprecipitated using specific antibodies against histone H3. The enrichment of Ric-8B promoter sequences in the precipitated chromatin was determined by QPCR using specific primers to amplify the region −396/−284. The bars represent the means ± standard errors of the means of at least two independent experiments, each performed in duplicate. Statistically significant differences were determined by the analysis of variance test. ***, P < 0.001; **, P < 0.01; *, P < 0.05.
To determine whether Ric-8B gene repression is also reflected by nucleosome enrichment at the Ric-8B gene promoter, we carried out ChIP analyses using an antibody against total histone H3. As shown in Fig. 10D, transcriptional repression of Ric-8B during osteoblast differentiation was accompanied by a significant increase (more than 60% enrichment) in the nucleosomal histone H3 bound to the proximal promoter region of this gene. Taken together, our results indicate that during osteoblast differentiation, C/EBPβ-mediated repression of the Ric-8B gene involves nucleosome enrichment in areas flanking the C/EBP site III element.
DISCUSSION
Here, we report that the Ric-8B gene is downregulated during osteoblast differentiation by the transcription factor C/EBPβ. We found that only the larger C/EBPβ isoform (C/EBPβ-LAP*) inhibited Ric-8B gene promoter activity in osteoblastic cells, while the C/EBPβ-LAP isoform did not exhibit a significant transcriptional effect. In agreement with the inhibitory effect of C/EBPβ-LAP*, siRNA-mediated knockdown of C/EBPβ expression in osteoblastic cells resulted in a significant increase in Ric-8B gene expression. Transient overexpression of Brg1 or Brm, the catalytic subunits of the SWI/SNF chromatin remodeling complex, also inhibited Ric-8B promoter activity. Moreover, the presence of inactive SWI/SNF complexes resulted in increased endogenous Ric-8B gene transcription, indicating that SWI/SNF activity downregulates Ric-8B expression in proliferating osteoblastic cells. Importantly, the SWI/SNF interaction with the Ric-8B promoter requires C/EBPβ expression. Finally, we demonstrated that during osteoblast differentiation, repression of the Ric-8B gene involves nucleosome enrichment at the proximal Ric-8B gene promoter together with changes in nuclease accessibility at regulatory regions of this promoter. These chromatin remodeling events require SWI/SNF activity.
Recent reports support differential functions for the two long C/EBPβ protein isoforms. Thus, C/EBPβ-LAP* is found exclusively in normal mammary epithelial cells, while C/EBPβ-LAP is found in mitotic cells, both normal and tumor derived (6). Also, and in agreement with our results, actively proliferating mouse preosteoblastic cells express C/EBPβ-LAP, while cells engaged in osteoblast differentiation increasingly express C/EBPβ-LAP* (36). Other groups have also demonstrated that C/EBPβ-LAP overexpression activates the promoter activity of the cyclin D1 gene and induces an invasive phenotype in MCF10A cells (1, 6). This condition also favors mitotic clonal expansion of MEFs from C/EBPβ null mice (41), as well as increasing proliferation and inhibiting differentiation of preosteoblastic MC3T3-E1 cells (13). On the other hand, overexpression of C/EBPβ-LAP* in chondrocytes from C/EBPβ null mice results in a block of proliferation, increased expression of the cell cycle-dependent kinase inhibitor p57, and induced cell differentiation (12). Despite these findings, it was only recently established that C/EBPβ-LAP* and C/EBPβ-LAP can also have different effects when regulating transcription at C/EBPβ target genes. Thus, C/EBPβ-LAP*, but not C/EBPβ-LAP, is capable of repressing transcription of the COX-2 gene in human cells (50). In agreement with these results, we observed that transient overexpression of C/EBPβ-LAP* in proliferating MC3T3 preosteoblasts resulted in decreased endogenous Ric-8B gene transcription (data not shown). Although a definitive molecular mechanism for this differential effect has not been determined, recent studies point to the role of the N-terminal domain of C/EBPβ-LAP*, which is missing in C/EBPβ-LAP. This domain has been shown to be important for the interaction of C/EBPβ-LAP* with HDAC4, a histone deacetylase that is a key component in the nuclear complex that represses the COX-2 gene (50). This N-terminal domain also contributes to the interaction of C/EBPβ-LAP* with the SWI/SNF chromatin remodeling complex during C/EBPβ-mediated recruitment of SWI/SNF to target genes (16, 17). Moreover, the ability of C/EBPβ-LAP* to interact with SWI/SNF may be dictated, at least in part, by covalent modifications occurring at this N-terminal domain; dimethylation of the R3 residue by the methylase PRMT4/CARM1 inhibits the C/EBPβ-LAP*–SWI/SNF association, thereby decreasing the expression of C/EBPβ-stimulated genes during cell differentiation (17).
Interestingly, we also found that SWI/SNF downregulates the expression of the Ric-8B gene in osteoblastic cells. This transcriptional repression occurs concomitantly with our previously described C/EBPβ (C/EBPβ-LAP)- and SWI/SNF-dependent upregulation of the bone phenotypic gene osteocalcin (9, 46). Together, these results not only suggest that during osteoblast differentiation the C/EBPβ transcription factor can inhibit and activate the expression of target genes, but also that it can perform these opposite regulatory functions by forming specific C/EBPβ-LAP*–SWI/SNF and C/EBPβ-LAP–SWI/SNF regulatory complexes, respectively. The transcriptional repression by the C/EBPβ-LAP*–SWI/SNF complex that we propose here is in agreement with results from other groups that have also identified SWI/SNF as an important component of the regulatory complexes that downregulate transcription during cell differentiation (7).
We found that downregulation of Ric-8B gene transcription involves nucleosome enrichment and changes in nuclease accessibility at specific regions of the Ric-8B promoter. Our results also indicate that the increased association of SWI/SNF with this promoter sequence supports a chromatin remodeling process that contributes to transcriptional repression of the Ric-8B gene. Interestingly, this change in chromatin structure is not reflected by a generalized reduction in nuclease accessibility at the Ric-8B gene promoter. Thus, BanII nuclease cleavage at a site that overlaps the C/EBP site III is significantly increased upon Ric-8B gene repression. As our transient-transfection and site-directed mutagenesis data point to this C/EBP site III as the principal contributor to C/EBPβ-LAP*-mediated repression of the Ric-8B promoter activity, we propose that the increased accessibility at this regulatory sequence reflects the enhanced association of C/EBPβ with this promoter region during Ric-8B gene repression.
The role of the Ric-8B protein during proliferation of preosteoblastic cells remains to be established, as does determining the effect of its downregulation during osteoblast differentiation. Preliminary results from our group indicate that the expression of this GEF is necessary at early stages of craniofacial development (unpublished data), in particular during proliferation and clonal expansion of preosteogenic precursor cells at the neural crest. Current studies in vivo using animal models are addressing this important issue.
ACKNOWLEDGMENTS
We thank Brigitte van Zundert (Universidad Andres Bello, Santiago, Chile) for critical reading and interesting comments of the manuscript. We also thank Anthony N. Imbalzano (University of Massachusetts Medical School, Worcester, MA) for providing the Brg1 and Brm expression vectors as well as the anti-Brg1 and anti-Brm antibodies and José Gutiérrez (Universidad de Concepcion, Concepcion, Chile) for providing the different C/EBPβ expression constructs. In addition, we thank Rafael Burgos (Universidad Austral de Chile, Valdivia, Chile) for providing the MC3T3 cells.
This work was supported by grants FONDECYT 1095075 (to M.M.) and 1090150 (to J.O.) and FONDAP 15090007 (to M.M.). R.G., H.S., and R.A. have been supported by doctoral fellowships from CONICYT. R.G. was also supported by a doctoral thesis grant from CONICYT. B.H. is supported by a postdoctoral fellowship grant from FONDECYT 3110138.
Footnotes
Published ahead of print on 23 May 2011.
REFERENCES
- 1. Bundy L. M., Sealy L. 2003. CCAAT/enhancer binding protein beta (C/EBPβ)-2 transforms normal mammary epithelial cells and induces epithelial to mesenchymal transition in culture. Oncogene 22:869–883 [DOI] [PubMed] [Google Scholar]
- 2. Calkhoven C. F., Muller C., Leutz A. 2000. Translational control of C/EBPα and C/EBPβ isoform expression. Genes Dev. 14:1920–1932 [PMC free article] [PubMed] [Google Scholar]
- 3. Cruzat F., et al. 2009. SWI/SNF-independent nuclease hypersensitivity and an increased level of histone acetylation at the P1 promoter accompany active transcription of the bone master gene Runx2. Biochemistry 48:7287–7295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dai Y., Ngo D., Jacob J., Forman L. W., Faller D. V. 2008. Prohibitin and the SWI/SNF ATPase subunit BRG1 are required for effective androgen antagonist-mediated transcriptional repression of androgen receptor-regulated genes. Carcinogenesis 29:1725–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. David N. B., et al. 2005. Drosophila Ric-8 regulates Gαi cortical localization to promote Gαi-dependent planar orientation of the mitotic spindle during asymmetric cell division. Nat. Cell Biol. 7:1083–1090 [DOI] [PubMed] [Google Scholar]
- 6. Eaton E. M., Hanlon M., Bundy L., Sealy L. 2001. Characterization of C/EBPβ isoforms in normal versus neoplastic mammary epithelial cells. J. Cell. Physiol. 189:91–105 [DOI] [PubMed] [Google Scholar]
- 7. Flowers S., Nagl N. G., Jr., Beck G. R., Jr., Moran E. 2009. Antagonistic roles for BRM and BRG1 SWI/SNF complexes in differentiation. J. Biol. Chem. 284:10067–10075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gutierrez J. L., Chandy M., Carrozza M. J., Workman J. L. 2007. Activation domains drive nucleosome eviction by SWI/SNF. EMBO J. 26:730–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gutierrez S., et al. 2002. CCAAT/enhancer-binding proteins (C/EBP) beta and delta activate osteocalcin gene transcription and synergize with Runx2 at the C/EBP element to regulate bone-specific expression. J. Biol. Chem. 277:1316–1323 [DOI] [PubMed] [Google Scholar]
- 10. Harrison J. R., et al. 2005. Col1a1 promoter-targeted expression of p20 CCAAT enhancer-binding protein beta (C/EBPβ), a truncated C/EBPβ isoform, causes osteopenia in transgenic mice. J. Biol. Chem. 280:8117–8124 [DOI] [PubMed] [Google Scholar]
- 11. Henriquez B., et al. 1 February 2011, posting date C/EBPβ binds the P1 promoter of the Runx2 gene and up-regulates Runx2 transcription in osteoblastic cells. J. Cell. Physiol. doi:10.1002/jcp.22652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hirata M., et al. 2009. C/EBPβ promotes transition from proliferation to hypertrophic differentiation of chondrocytes through transactivation of p57. PLoS One 4:e4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Iyer V. V., Kadakia T. B., McCabe L. R., Schwartz R. C. 2004. CCAAT/enhancer-binding protein-beta has a role in osteoblast proliferation and differentiation. Exp. Cell Res. 295:128–137 [DOI] [PubMed] [Google Scholar]
- 14. Katagiri T., et al. 1994. Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J. Cell Biol. 127:1755–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Klattenhoff C., et al. 2003. Human brain synembryn interacts with Gsα and Gqα and is translocated to the plasma membrane in response to isoproterenol and carbachol. J. Cell. Physiol. 195:151–157 [DOI] [PubMed] [Google Scholar]
- 16. Kowenz-Leutz E., Leutz A. 1999. A C/EBP beta isoform recruits the SWI/SNF complex to activate myeloid genes. Mol. Cell 4:735–743 [DOI] [PubMed] [Google Scholar]
- 17. Kowenz-Leutz E., Pless O., Dittmar G., Knoblich M., Leutz A. 2010. Crosstalk between C/EBPβ phosphorylation, arginine methylation, and SWI/SNF/Mediator implies an indexing transcription factor code. EMBO J. 29:1105–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Langst G., Bonte E. J., Corona D. F., Becker P. B. 1999. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell 97:843–852 [DOI] [PubMed] [Google Scholar]
- 19. Li H., et al. 2008. The Med1 subunit of transcriptional mediator plays a central role in regulating CCAAT/enhancer-binding protein-beta-driven transcription in response to interferon-gamma. J. Biol. Chem. 283:13077–13086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Malik S., Ghosh M., Bonacci T. M., Tall G. G., Smrcka A. V. 2005. Ric-8 enhances G protein βγ-dependent signaling in response to βγ-binding peptides in intact cells. Mol. Pharmacol. 68:129–136 [DOI] [PubMed] [Google Scholar]
- 21. Marie P. J. 2008. Transcription factors controlling osteoblastogenesis. Arch. Biochem. Biophys. 473:98–105 [DOI] [PubMed] [Google Scholar]
- 22. Miller K. G., Emerson M. D., McManus J. R., Rand J. B. 2000. RIC-8 (Synembryn): a novel conserved protein that is required for G(q)alpha signaling in the C. elegans nervous system. Neuron 27:289–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miller K. G., Rand J. B. 2000. A role for RIC-8 (Synembryn) and GOA-1 (G(o) alpha) in regulating a subset of centrosome movements during early embryogenesis in Caenorhabditis elegans. Genetics 156:1649–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mink S., Haenig B., Klempnauer K. H. 1997. Interaction and functional collaboration of p300 and C/EBPbeta. Mol. Cell. Biol. 17:6609–6617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mo X., Kowenz-Leutz E., Xu H., Leutz A. 2004. Ras induces mediator complex exchange on C/EBP beta. Mol. Cell 13:241–250 [DOI] [PubMed] [Google Scholar]
- 26. Nagai Y., Nishimura A., Tago K., Mizuno N., Itoh H. 2010. Ric-8B stabilizes the alpha subunit of stimulatory G protein by inhibiting its ubiquitination. J. Biol. Chem. 285:11114–11120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nerlov C. 2008. C/EBPs: recipients of extracellular signals through proteome modulation. Curr. Opin. Cell Biol. 20:180–185 [DOI] [PubMed] [Google Scholar]
- 28. Nishimura A., et al. 2006. Ric-8A potentiates Gq-mediated signal transduction by acting downstream of G protein-coupled receptor in intact cells. Genes Cells 11:487–498 [DOI] [PubMed] [Google Scholar]
- 29. Owen T. A., et al. 1990. Modifications of protein-DNA interactions in the proximal promoter of a cell-growth-regulated histone gene during onset and progression of osteoblast differentiation. Proc. Natl. Acad. Sci. U. S. A. 87:5129–5133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Paredes R., et al. 2004. Bone-specific transcription factor Runx2 interacts with the 1α,25-dihydroxyvitamin D3 receptor to up-regulate rat osteocalcin gene expression in osteoblastic cells. Mol. Cell. Biol. 24:8847–8861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Phelan M. L., Schnitzler G. R., Kingston R. E. 2000. Octamer transfer and creation of stably remodeled nucleosomes by human SWI-SNF and its isolated ATPases. Mol. Cell. Biol. 20:6380–6389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pless O., et al. 2008. G9a-mediated lysine methylation alters the function of CCAAT/enhancer-binding protein-beta. J. Biol. Chem. 283:26357–26363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Romo X., et al. 2008. xRic-8 is a GEF for Gsα and participates in maintaining meiotic arrest in Xenopus laevis oocytes. J. Cell. Physiol. 214:673–680 [DOI] [PubMed] [Google Scholar]
- 34. Shen J., et al. 2002. Histone acetylation in vivo at the osteocalcin locus is functionally linked to vitamin D-dependent, bone tissue-specific transcription. J. Biol. Chem. 277:20284–20292 [DOI] [PubMed] [Google Scholar]
- 35. Siderovski D. P., Willard F. S. 2005. The GAPs, GEFs, and GDIs of heterotrimeric G-protein alpha subunits. Int. J. Biol. Sci. 1:51–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Smink J. J., et al. 2009. Transcription factor C/EBPβ isoform ratio regulates osteoclastogenesis through MafB. EMBO J. 28:1769–1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Soutoglou E., Talianidis I. 2002. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science 295:1901–1904 [DOI] [PubMed] [Google Scholar]
- 38. Stein G. S., et al. 2004. Runx2 control of organization, assembly and activity of the regulatory machinery for skeletal gene expression. Oncogene 23:4315–4329 [DOI] [PubMed] [Google Scholar]
- 39. Tajbakhsh S., Rocheteau P., Le Roux I. 2009. Asymmetric cell divisions and asymmetric cell fates. Annu. Rev. Cell Dev. Biol. 25:671–699 [DOI] [PubMed] [Google Scholar]
- 40. Tall G. G., Krumins A. M., Gilman A. G. 2003. Mammalian Ric-8A (synembryn) is a heterotrimeric Gα protein guanine nucleotide exchange factor. J. Biol. Chem. 278:8356–8362 [DOI] [PubMed] [Google Scholar]
- 41. Tang Q. Q., Otto T. C., Lane M. D. 2003. CCAAT/enhancer-binding protein beta is required for mitotic clonal expansion during adipogenesis. Proc. Natl. Acad. Sci. U. S. A. 100:850–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Thielking V., Alves J., Fliess A., Maass G., Pingoud A. 1990. Accuracy of the EcoRI restriction endonuclease: binding and cleavage studies with oligodeoxynucleotide substrates containing degenerate recognition sequences. Biochemistry 29:4682–4691 [DOI] [PubMed] [Google Scholar]
- 43. Tominaga H., et al. 2008. CCAAT/enhancer-binding protein beta promotes osteoblast differentiation by enhancing Runx2 activity with ATF4. Mol. Biol. Cell 19:5373–5386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tonissoo T., et al. 2010. Nucleotide exchange factor RIC-8 is indispensable in mammalian early development. Dev. Dyn. 239:3404–3415 [DOI] [PubMed] [Google Scholar]
- 45. Tonissoo T., Meier R., Talts K., Plaas M., Karis A. 2003. Expression of ric-8 (synembryn) gene in the nervous system of developing and adult mouse. Gene Expr. Patterns 3:591–594 [DOI] [PubMed] [Google Scholar]
- 46. Villagra A., et al. 2006. Chromatin remodeling and transcriptional activity of the bone-specific osteocalcin gene require CCAAT/enhancer-binding protein beta-dependent recruitment of SWI/SNF activity. J. Biol. Chem. 281:22695–22706 [DOI] [PubMed] [Google Scholar]
- 47. Von Dannecker L. E., Mercadante A. F., Malnic B. 2005. Ric-8B, an olfactory putative GTP exchange factor, amplifies signal transduction through the olfactory-specific G-protein Gαolf. J. Neurosci. 25:3793–3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang D., et al. 1999. Isolation and characterization of MC3T3-E1 preosteoblast subclones with distinct in vitro and in vivo differentiation/mineralization potential. J. Bone Miner. Res. 14:893–903 [DOI] [PubMed] [Google Scholar]
- 49. Wang H., et al. 2005. Ric-8 controls Drosophila neural progenitor asymmetric division by regulating heterotrimeric G proteins. Nat. Cell Biol. 7:1091–1098 [DOI] [PubMed] [Google Scholar]
- 50. Wang W. L., Lee Y. C., Yang W. M., Chang W. C., Wang J. M. 2008. Sumoylation of LAP1 is involved in the HDAC4-mediated repression of COX-2 transcription. Nucleic Acids Res. 36:6066–6079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Whitehouse I., et al. 1999. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature 400:784–787 [DOI] [PubMed] [Google Scholar]
- 52. Woodard G. E., et al. 2010. Ric-8A and Gi alpha recruit LGN, NuMA, and dynein to the cell cortex to help orient the mitotic spindle. Mol. Cell. Biol. 30:3519–3530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xiong W., Hsieh C. C., Kurtz A. J., Rabek J. P., Papaconstantinou J. 2001. Regulation of CCAAT/enhancer-binding protein-beta isoform synthesis by alternative translational initiation at multiple AUG start sites. Nucleic Acids Res. 29:3087–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]