Abstract
In multiple tumor types, activation of the transcription factor NF-κB increases the resistance of tumor cells to anticancer therapies and contributes to tumor progression. Genotoxic stress induced by chemotherapy or radiation therapy triggers the ATM-dependent translocation of NF-κB essential modifier (NEMO), also designated IκB kinase γ (IKKγ), from the nucleus to the cytosol, resulting in IκB kinase activation by mechanisms not yet fully understood. RIP1 has been implicated in this response and found to be modified in cells with damaged DNA; however, the nature of the RIP1 modification and its precise role in the pathway remain unclear. Here, we show that DNA damage stimulates the formation of a cytosolic complex containing ATM, NEMO (IKKγ), RIP1, and TAK1. We find that RIP1 is modified by SUMO-1 and ubiquitin in response to DNA damage and demonstrate that modified RIP1 is required for NF-κB activation and tumor cell survival. We show that ATM activates TAK1 in a manner dependent on RIP1 and NEMO. We also reveal TAK1 as a central mediator of the alternative DNA damage response pathway mediated by the p38 mitogen-activated protein kinase (MAPK)/MAPK-activated protein 2 (MAPKAP-2) kinases. These findings have translational implications and reveal RIP1 and TAK1 as potential therapeutic targets in chemoresistance.
INTRODUCTION
The DNA damage response activates cell cycle checkpoint and survival pathways that function to prevent DNA replication until damaged DNA is repaired. These pathways include the well-characterized ATM (ataxia telangiectasia mutated)/CHK2 and ATR (ataxia telangiectasia and Rad-3 related)/CHK1 pathways and the more recently identified ATM/ATR/p38 mitogen-activated protein kinase (MAPK)/MAPK-activated protein 2 (MAPKAP-2; hereinafter called MK2) checkpoint that is active in p53-deficient tumor cells (15, 19). The transcription factor NF-κB regulates apoptosis induced by genotoxic stress and is an attractive therapeutic target in tumor cells whose response to DNA-damaging agents is impaired due to compromised p53 function. Moreover, constitutive NF-κB activity is a hallmark of several cancers, and mutations in NF-κB pathway components have been associated with the activated B cell (ABC) subtype of diffuse large B cell lymphoma (DLBCL), breast cancer, and multiple myeloma (3, 6). Thus, the inclusion of NF-κB inhibitors in cancer therapy could have antioncogenic activities as well as augment the tumor chemotherapeutic response.
NF-κB is normally held in the cytoplasm in an inactive form bound to inhibitor proteins, such as IκBα. Diverse stimuli, such as infection, proinflammatory cytokine production, or treatment with agents that induce DNA damage elicit NF-κB-mediated transcriptional activity by activating the cytosolic IκB kinase (IKK) complex, consisting of IKKα and IKKβ and a regulatory subunit designated IKKγ or NF-κB essential modifier (NEMO) (hereinafter referred to as NEMO). IKK activation results in IκBα phosphorylation, ubiquitination, and subsequent degradation. The NF-κB (p65/p50) heterodimer is then free to enter the nucleus and stimulate gene expression (5).
DNA double-strand breaks are also sensed by the poly(ADP)-ribosylating enzyme poly(ADP-ribose) polymerase 1 (PARP-1). Upon recognition of DNA double-strand breaks, PARP-1 catalyzes the attachment of PAR chains to glutamic acid residues on PARP-1 itself, as well as other substrates. PAR-modified PARP-1 recruits the ATM kinase and the inhibitor of activated STATy (PIASy), which sumoylates the IKK regulatory subunit NEMO at lysines K277 and K309 (16, 22, 27). DNA damage also stimulates interactions in the nucleus between NEMO, receptor-interacting protein 1 (RIP1), and p53-induced protein with death domain (PIDD) (11). PIDD has been shown to translocate to the nucleus in response to genotoxic stress, and either PIDD or RIP1 depletion abolishes DNA damage-induced NF-κB activation and NEMO sumoylation (11). Once sumoylated, NEMO is phosphorylated by ATM and monoubiquitinated potentially by the inhibitor of apoptosis protein cIAP1 in the nucleus (9, 12, 27). These events trigger the translocation of ATM and NEMO into the cytosol (27); however, the mechanism by which NEMO monoubiquitination leads to IKK activation is unknown.
In the present study, we demonstrate that the NF-κB response to DNA damage involves the ubiquitin modification of RIP1 and the recruitment of the ubiquitin-activated kinase TAK1. We reveal that RIP1, like NEMO, is SUMO-1 modified by the SUMO E3 ligase PIASy and that modified RIP1 forms a complex with NEMO, TAK1, and IKKβ. RIP1 is also ubiquitin modified in response to DNA damage, and modified RIP1 signals TAK1 recruitment. We provide evidence that a RIP1 or TAK1 deficiency abrogates NF-κB activity and sensitizes mouse fibroblasts and multiple human tumor cell lines to DNA-damaging agents. This study also reveals novel roles for TAK1 as a critical mediator of the p38 MAPK/MK2 checkpoint in p53-deficient human tumor cells. Thus, TAK1 silencing or inhibition in human tumor cells interferes with both the NF-κB- and p38 MAPK/MK2-mediated survival pathways, raising TAK1 as a potential therapeutic target in chemotherapeutic resistance.
MATERIALS AND METHODS
Cell culture and biological reagents.
The targeted disruption of the Rip1 loci and the generation of Rip1−/− mouse embryonic fibroblasts (MEFs) have been described previously (13). Nemo- and Tak1-deficient fibroblasts and fibroblasts from wild-type littermate controls were generously provided by Manolis Pasparakis (Institute for Genetics, University of Cologne, Cologne, Germany) and Sankar Ghosh (Columbia University, New York, NY), respectively. Human U2OS, HeLa, MCF7, and H1299 cell lines were obtained from ATCC and grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum. For some experiments, U2OS and H1299 cells were infected with a lentivirus expressing a control green fluorescent protein (GFP)- or TAK1-specific short hairpin RNA (shRNA) (Open Biosystems) and selected with 2.0 μg/ml puromycin, and TAK1 protein levels were determined by immunoblotting. Doxorubicin was obtained from Sigma, and etoposide purchased from Alexis. The TAK1 kinase inhibitor 5Z-7-oxozeaenol (5Z7) was obtained from Bioaustralis. Recombinant tumor necrosis factor alpha (TNF-α) and interleukin-1β (IL-1β) were obtained from Sigma and eBioscience, respectively. The C-terminally Flag-tagged RIP1 lysine mutant in which lysines at positions 105, 140, 305, and 565 were mutated to arginines (designated 4KR) was generated using PCR-mediated mutagenesis, and the mutations were confirmed by sequencing. A SUMO-1 moiety devoid of the C-terminal diglycine residues to prevent processing was then fused in frame to the N terminus of the RIP1 4KR-Flag mutant using PCR, and the integrity of the fusion confirmed by sequencing.
Preparation and transfection of siRNA.
Small interfering RNA (siRNA) oligonucleotides against human RIP1 (CCGACATTTCCTGGCATTGAA and CAGCTTGATTTACGTCAGCCA), PIASy (AACAAGACAGGTGGAGTTGAT and AAAAGCTGCCGTTCTTTAATA), UBC13 (GATCCGCACAGTTCTGCTATC), or TAK1 (UGGCUUAUCUUACACUGGA) were purchased from Qiagen and Dharmacon, respectively. A scrambled siRNA was purchased from Dharmacon. U2OS or HeLa cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions and as described previously (30). HEK293T cells were transfected by calcium phosphate precipitation as described in reference 16.
Coimmunoprecipitation and Western blotting.
RIP1 and NEMO antibodies were obtained from BD Bioscience. ATM antibody was obtained from Calbiochem. UBC13 and SUMO-1 antibodies were purchased from Zymed, and β-actin antibody from Sigma. Tak1, PIASy, IKKβ, PARP-1, ubiquitin, IκBα, p38 MAPK, and Jun N-terminal protein kinase (JNK) antibodies were all obtained from Santa Cruz Biotechnology. The phospho-specific antibodies for TAK1, MKK6, MK2, CDC2, IκBα, p38 MAPK, and JNK were obtained from Cell Signaling Technology. Anti-total MK2, CDC2, caspase-3, and MKK6 antibodies were obtained from Cell Signaling Technology. K63-ubiquitin antibody was obtained from Millipore.
For coimmunoprecipitation, cells were lysed in an endogenous lysis buffer (20 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 30 mM NaF, and 2 mM sodium pyrophosphate) supplemented with Complete protease inhibitor cocktail (Roche).
For ubiquitination and sumoylation assays, cells were boiled for 10 min in 1% SDS before immunoprecipitation. Boiled lysates were diluted to 0.1% SDS with a modified radioimmunoprecipitation (RIPA) buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% NP-40, 0.25% deoxycholic acid, 1 mM EDTA, supplemented with protease inhibitors and 5 μM N-ethylmaleimide [Sigma]). Cleared lysates were incubated overnight with RIP1, NEMO, Flag, or anti-K63-ubiquitin antibodies. Immunoprecipitates were washed 3 times with lysis buffer, separated by SDS-PAGE, transferred to nitrocellulose, and analyzed by immunoblotting with an antiubiquitin, anti-SUMO-1, or anti-RIP1 antibody.
For Western blot analysis to detect either total protein or phosphorylated protein, cells were lysed in kinase lysis buffer (10 mM/1 mM KPO4-EDTA, pH 7.05, 5 mM EGTA, pH 7.2, 10 mM MgCl2, 50 mM β-glycerophosphate, pH 7.2, 0.5% NP-40, 0.1% Brij-35) supplemented with a protease inhibitor mixture (Roche), 1 mM dithiothreitol (DTT), 1 mM Na3VaO4, 1 mM phenylmethylsulfonyl fluoride (PMSF).
In vitro kinase and sumoylation assays.
For the TAK1 in vitro kinase assays, cell lysates were immunoprecipitated with an anti-TAK1 antibody and then incubated with protein A-conjugated beads. Immunoprecipitates were washed 3 times with lysis buffer, and kinase reactions were allowed to proceed for 60 min at 30°C in 20 μl kinase buffer (20 mM HEPES, pH 7.2, 10 mM MgCl2, 0.1 mg/ml bovine serum albumin, 3 mM 2-mercaptoethanol) containing 1 μg of His-tagged, kinase-inactive MKK6(K82A). The reaction samples were separated by SDS-PAGE, and MKK6 phosphorylation determined by immunoblotting with a phospho-specific MKK6 antibody (Cell Signaling). In vitro sumoylation of RIP1 and determination of the effects of PIASy on RIP1 sumoylation were performed by in vitro sumoylation assays described previously (16).
Biochemical fractionation.
Cytoplasmic and nuclear fractions were prepared as described in reference 4. Briefly, treated or untreated U2OS cells were lysed in a cytosolic lysis buffer containing 10 mM HEPES (pH 7.9), 1.5 mM MgCl2, 300 mM sucrose, 0.5% NP-40, 10 mM KCl, supplemented with DTT and protease inhibitors. Nuclei were pelleted by a short centrifugation and lysed in a nuclear buffer containing 20 mM HEPES (pH 7.9), 100 mM NaCl, 0.2 mM EDTA, 20% glycerol, 100 mM KCl, supplemented with DTT and protease inhibitors. Nuclear pellets were subjected to a freeze-thaw cycle, sonicated, and centrifuged to obtain a solubilized nuclear fraction. In coimmunoprecipitation and gel filtration chromatography experiments, both cytosolic and nuclear fractions were added or diluted to 150 mM NaCl.
For gel filtration chromatography, U2OS cytosolic extracts were fractionated using a fast protein liquid chromatography (FPLC) protein purification system (Pharmacia) on a Superose 6 column (Amersham Biosciences), at 4°C, equilibrated with buffer A (20 mM HEPES-KOH, 10 mM KCl, 1 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, pH 7.9, 150 mM NaCl, and 0.5% NP-40). Cytosolic (5 mg) extract was loaded and eluted from the column in buffer A at a flow rate of 0.4 ml/min, and 500-μl fractions were collected. The column was calibrated with gel filtration standards (Bio-Rad Laboratories).
Flow cytometry.
For cell cycle analysis, human U2OS or H1299 cells infected with lentiviruses expressing a GFP- or TAK1-specific shRNA were left untreated or treated with etoposide for 24 and 48 h. The cells were then fixed in 70% ethanol, stained with propidium iodide (PI), and analyzed for DNA content.
Real-time quantitative PCR analysis.
Total RNA was isolated from untreated and etoposide-treated mouse wild-type, RIP1-deficient, or NEMO-deficient cells or U2OS cells using Trizol reagent (Invitrogen), and cDNA was generated with Moloney murine leukemia virus reverse transcriptase (M-MLV RT; Invitrogen). The real-time PCR primers used in this study are available upon request. Product accumulation was monitored by SYBR green fluorescence, with the relative expression levels determined from a standard curve of serial dilutions of cDNA. All samples were normalized to β-actin values, and results are displayed as the fold induction compared to that in the untreated control.
RESULTS
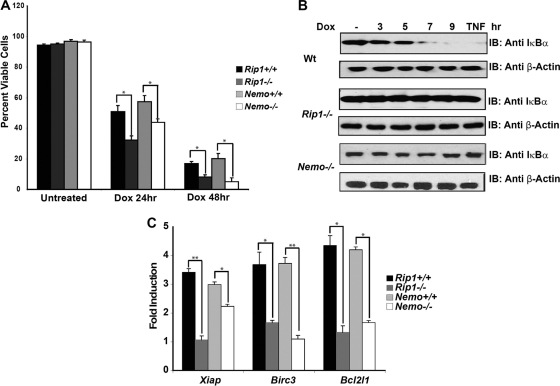
A Rip1 or Nemo deficiency impairs the NF-κB response and sensitizes MEFs to genotoxic stress.
The RIP1 protein has been implicated in the NF-κB response to DNA damage, and its role in the initial DNA damage response has been shown to be independent of its kinase activity and effects on the TNF pathway (10, 11). Although implicated in DNA damage-induced NF-κB activity, the exact function and relative position(s) of RIP1 in this NF-κB pathway remain unclear. Previous studies suggested a nuclear role for RIP1, demonstrating that RIP1 is required for the formation of the PIDDosome and the SUMO modification of NEMO in cells with DNA damage (11). Although not tested, these findings predict that cells deficient for RIP1 or NEMO would be equivalently sensitive to DNA damage. To test this prediction, we compared the levels of sensitivity of Rip1-deficient and Nemo-deficient murine embryonic fibroblasts to doxorubicin treatment. We found that Rip1-deficient and Nemo-deficient cells were equivalently hypersensitive to doxorubicin treatment (Fig. 1A), supporting the idea that both RIP1 and NEMO contribute to the DNA damage response. We also found that IκBα degradation was impaired in both Rip1- and Nemo-deficient MEFs treated with doxorubicin (Fig. 1B), consistent with published studies (8, 10, 22). We also examined NF-κB activity by electrophoretic mobility shift assay (EMSA) in HEK293 cells transfected with control or RIP1-specific siRNAs and observed decreased NF-κB binding in RIP1-silenced cells treated with TNF-α or etoposide (data not shown).
Fig. 1.
A Rip1 or Nemo deficiency impairs NF-κB response and sensitizes cells to genotoxic stress. (A) Wild-type, Rip1-deficient, or Nemo-deficient MEFs were left untreated or treated with doxorubicin (Dox; 2 μg/ml), and cell viability was determined at 24 and 48 h. Samples were done in triplicate; error bars represent the standard deviations. Statistical evaluation was performed using the unpaired Student's t test. *, P < 0.05. (B) DNA damage-induced NF-κB activation is impaired in the absence of Rip1 and Nemo. Wild-type (Wt), Rip1-deficient, or Nemo-deficient MEFs were left untreated, treated with TNF-α for 10 min, or treated with doxorubicin (10 μg/ml) for the time periods indicated. NF-κB activity was monitored by immunoblotting (IB) with an IκBα antibody. (C) NF-κB-dependent survival gene expression is impaired in Rip1- and Nemo-deficient MEFs. Wild-type, Rip1-deficient, and Nemo-deficient MEFs were left untreated or treated with etoposide (40 μM) for 6 h, and Birc3 (cIap2), Xiap, and Bcl2l1(Bcl-xL) mRNA levels were quantified using real-time PCR. Gene expression was normalized to the expression of β-actin and then normalized to the expression in the untreated control to estimate fold induction. Samples were assayed in triplicate; error bars represent the standard deviations. Statistical evaluation was performed using the unpaired Student t test. P values, from left to right, are as follows: P < 0.0008, P < 0.029, P < 0.024, P < 0.0005, P < 0.024, and P < 0.013.
To determine why a Rip1 deficiency results in sensitivity to DNA-damaging agents, we used gene expression profiling to compare the responses of wild-type and Rip1-deficient MEFs to etoposide treatment. Ingenuity pathway analysis revealed a failure to induce survival gene expression when cells lacking Rip1 were exposed to DNA damage. Specifically, the induction of Xiap, Bcl2l1, and Birc3 mRNAs was impaired in Rip1-deficient and in Nemo-deficient cells (Fig. 1C). Thus, the absence of Rip1 or Nemo sensitizes cells to DNA damage due to a failure to activate NF-κB and correlates with an inability to induce survival gene expression.
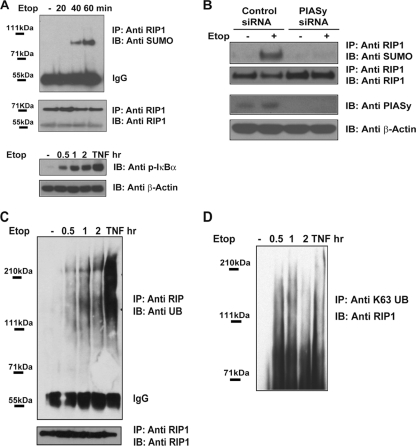
DNA damage stimulates the SUMO-1 and K63-ubiquitin modification of RIP1.
Previous studies detected a modified form of RIP1 in etoposide-treated cells (11). However, the precise nature and functional significance of the RIP1 modification(s) remain unclear. We detect a modified form of RIP1 in etoposide-treated cells that migrates at approximately 95 to 100 kDa. The 20- to 25-kDa increase in the apparent molecular mass suggested that RIP1, like NEMO, might be SUMO-1 (small ubiquitin-like modifier) modified in cells with DNA damage. To determine whether DNA damage leads to RIP1 sumoylation, we treated U2OS cells with etoposide, immunopre-cipitated RIP1, and examined the immunoprecipitates for the presence of sumoylated forms. We detected sumoylated forms of RIP1 40 min following etoposide treatment (Fig. 2A, upper panel). SUMO-1-modified RIP1 could also be detected in etoposide-treated cells immunoprecipitated with an anti-SUMO-1 antibody, followed by immunoblotting with a RIP1 antibody (data not shown). The detection of sumoylated RIP1 appears coincident with phospho-IκBα reactivity detected at 30 min following etoposide exposure (Fig. 2A, lower panel).
Fig. 2.
DNA damage stimulates the SUMO and ubiquitin modification of RIP1. (A) DNA damage stimulates the SUMO-1 modification of RIP1. Human U2OS cells were left untreated or treated with etoposide (Etop; 40 μM) for the time periods indicated. Protein extracts were boiled and then immunoprecipitated (IP) with a RIP1 antibody, and sumoylated proteins were detected by immunoblotting (IB) with a SUMO-1 antibody. (B) A PIASy deficiency reduces SUMO-RIP1 levels in etoposide-treated cells. Human U2OS cells were transfected with control or PIASy-specific siRNAs and then left untreated or treated with etoposide (40 μM) for 1 h. Protein extracts were boiled and then immunoprecipitated with a RIP1 antibody, followed by immunoblotting with a SUMO-1 antibody. (C) DNA damage stimulates RIP1 polyubiquitination. Human U2OS cells were left untreated or treated with TNF-α (10 ng/ml) for 10 min or with etoposide for the time periods indicated. Protein extracts were boiled and then immunoprecipitated with a RIP1 antibody, followed by immunoblotting with antiubiquitin (Anti UB). (D) The experiment as described for panel C was done using a K63 polyubiquitin antibody for immunoprecipitation and a RIP1 antibody for immunoblotting.
The SUMO ligase PIASy mediates NEMO sumoylation in response to genotoxic stress, and RIP1 and NEMO interact in the nucleus of cells with DNA damage (11, 16). To determine whether PIASy mediates RIP1 sumoylation, we introduced two individual PIASy siRNA oligonucleotides into U2OS cells and tested whether the etoposide-induced SUMO modification of RIP1 was affected. We were unable to detect sumoylated RIP1 (or NEMO) in the etoposide-treated, PIASy-depleted cells, indicating that in response to genotoxic stimuli, PIASy may modify both NEMO and RIP1 (Fig. 2B). To examine whether PIASy can function as a direct SUMO E3 ligase for RIP1, we transfected HEK293 cells with Flag-tagged versions of the wild type or a catalytically inactive form of PIASy C342/347A and assayed each for their ability to SUMO-1 modify in vitro-translated RIP1 protein. SUMO-modified RIP1 was detected when wild-type but not mutant PIASy was expressed (data not shown).
In addition to the SUMO-1 modification, RIP1 was also found to be stably ubiquitin modified 30 min following etoposide treatment and in steadily increasing amounts for 2 h (Fig. 2C), consistent with the induction and maintenance of NF-κB activity in cells with DNA damage (Fig. 2A, lower panel). However, the amount of polyubiquitinated RIP1 was significantly less in cells with DNA damage than in those treated with TNF-α for 10 min (Fig. 2C), suggesting that only a fraction of the total RIP1 is modified in cells with DNA damage. As in TNF-treated cells, RIP1 appeared to be stably K63-ubiquitin modified in etoposide-treated cells (Fig. 2D). Consistent with this finding, silencing of the E2-conjugating enzyme UBC13 that is responsible for K63-linked polyubiquitination diminishes the NF-κB response to doxorubicin and sensitizes human tumor cells to DNA-damaging agents (data not shown). These findings indicate that in addition to its nuclear role(s), RIP1 and, potentially, K63-ubiquitin-modified RIP1 may function in the cytosol to assemble an IKK-activating complex in response to genotoxic stress.
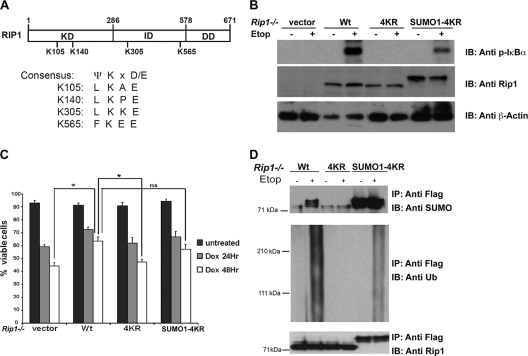
RIP1 is a novel SUMO-1 substrate, and the SUMO-1 modification of RIP1 is required for the NF-κB response to DNA damage.
To address a requirement for the posttranslational modification of RIP1 in the NF-κB response to DNA damage, we identified 4 putative sumoylation sites in the RIP1 protein based on the SUMO-1 modification consensus motif (Ψ-K-X-D/E) and mutated each of these 4 lysines (K105, 140, 305, and 565) to arginines (Fig. 3A); we designated this mutant 4KR. To determine whether the SUMO-1 modification of RIP1 was essential for the NF-κB response to DNA damage, we reconstituted Rip1-deficient fibroblasts with wild-type RIP1, the RIP1 lysine mutant (4KR), or a SUMO-1-conjugated version of the RIP1 4KR mutant (SUMO-1-4KR). The Rip1-reconstituted cells were left untreated or exposed to etoposide and NF-κB activity, and cell viability was measured. The expression of wild-type RIP1 restored the NF-κB response and protected cells from the effects of DNA damage. In contrast, DNA damage-induced NF-κB activation remained impaired in cells expressing the RIP1 4KR lysine mutant, and the cells remained sensitive to etoposide treatment (Fig. 3B and C). The expression of a SUMO-1-conjugated form of the RIP1 lysine mutant partially rescued the NF-κB response, increased survival, and stimulated the ubiquitin modification of RIP1 (Fig. 3B to D). These studies provide genetic evidence that RIP1 sumoylation is required for the activation of NF-κB and protection of cells from genotoxic stimuli and suggest that sumoylation triggers RIP1 ubiquitination.
Fig. 3.
The SUMO-1 modification of RIP1 mediates the NF-κB response, cell survival, and ubiquitin modification of RIP1 in the DNA damage pathway. (A) Predicted sumoylation consensus sites on RIP1. RIP1 contains four putative SUMO sites based on a SUMO modification consensus motif (Ψ-K-X-D/E). Kinase domain is indicated by KD, intermediate domain by ID, and death domain by DD. (B) RIP1 sumoylation is required for DNA damage-induced NF-κB activation. Rip1-deficient mouse embryonic fibroblasts reconstituted with wild-type RIP1 (Wt), a RIP1 lysine mutant called 4KR, or a SUMO-conjugated RIP1 4KR mutant were left untreated or exposed to etoposide (Etop; 40 μM) for 2 h. The NF-κB pathway response was measured by immunoblotting (IB) the cell lysates with a phospho-IκBα antibody. (C) An inability to SUMO-1 modify RIP1 sensitizes cells to DNA damage. Rip1-deficient cells reconstituted with wild-type RIP1 or mutant forms of RIP1 were left untreated or treated with doxorubicin (Dox; 2 μg/ml) for 24 or 48 h. Viable cells were quantified by trypan blue exclusion. Samples were assayed in triplicate; error bars represent the standard deviations. Statistical evaluation was performed using the unpaired Student t test. *, P < 0.05; ns, no statistical difference. (D) Sumoylation is required for the ubiquitin modification of RIP1 in the genotoxic stress response. Rip1−/− cells reconstituted with Flag-tagged RIP1, RIP1 4KR, and SUMO-1-RIP1 4KR plasmids. Cells were then left untreated or treated with etoposide (40 μM) for 2 h, and lysates were immunoprecipitated (IP) with anti-Flag antibodies, followed by immunoblotting with SUMO-1, ubiquitin (Anti UB), or RIP1 antibodies.
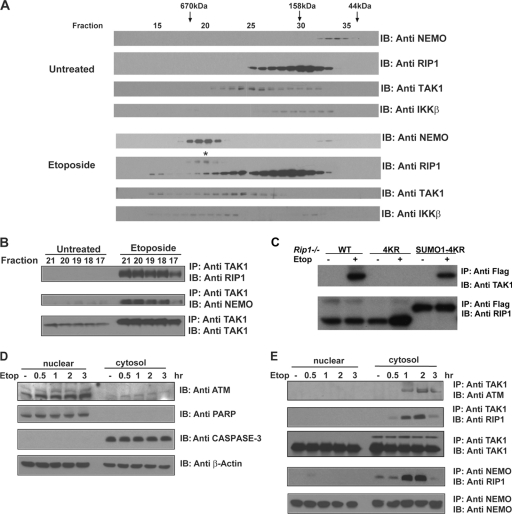
DNA damage induces the formation of a large cytosolic complex containing ATM, NEMO, RIP1, TAK1, and IKKβ.
DNA damage stimulates the formation of complexes in the nucleus consisting of ATM, PARP-1, PIASy, and NEMO, as well as a PIDD, RIP1, and NEMO complex (11, 22). However, cytosolic complexes induced by DNA damage have been less well studied, and as a result, how a nuclear damage signal is conveyed to the cytosolic IKK and MAK kinases remains unresolved. We utilized size exclusion chromatography and coimmunoprecipitation to uncover the cytosolic complexes induced in cells with DNA damage. Size exclusion chromatography of cytosolic lysates from untreated or etoposide-treated cells revealed that DNA damage causes a shift in the migration of both NEMO and RIP1 into overlapping fractions corresponding to a molecular mass of ∼500 to 700 kDa (Fig. 4A, fractions 17 to 21). In contrast to NEMO, only a fraction of the total RIP1 protein appears to shift to the higher-molecular-mass complex. We then examined the complexes for the presence of TAK1. TAK1 migrates in three apparent complexes in cells with DNA damage. One complex (fractions 17 to 21) migrates with an apparent molecular mass that overlaps with the NEMO- and RIP1-containing complex. To determine whether this complex contains NEMO, RIP1, and TAK1 in one complex, we immunoprecipitated TAK1 from fractions 17 to 21 and then examined the immunoprecipitates for the presence of NEMO and RIP1. In the untreated cells, interactions between RIP1, NEMO, and TAK1 were not observed (Fig. 4B). However, when these column fractions were immunoprecipitated from etoposide-treated cells, RIP1 and TAK1 and TAK1 and NEMO associations were readily detected in the cytosol (Fig. 4B). Interestingly, the DNA damage-induced cytosolic complex that contains NEMO and TAK1 appears to also contain a modified form of RIP1 (Fig. 4A, marked with an asterisk). Further analysis revealed that the RIP1 associated with this complex ran as a smear on a 3 to 8% SDS-PAGE gel, suggesting that the complex contains modified RIP1 (data not shown). The results of additional coimmunoprecipitation experiments confirmed that NEMO interacts with modified RIP1 in etoposide-treated cells (data not shown). We also examined the column fractions for the presence of the catalytic IKK subunit, IKKβ. DNA damage induced a shift in IKKβ into a similar-size complex, suggesting that DNA double-strand breaks stimulate the formation of a cytosolic complex consisting of RIP1, NEMO, TAK1, and IKKβ (Fig. 4A). These data are consistent with the hypothesis that DNA damage stimulates the SUMO and ubiquitin modification of RIP1, which in turn recruits and activates TAK1 and IKKβ to achieve IKK activation.
Fig. 4.
DNA damage induces the formation of a large cytosolic complex containing ATM, NEMO, RIP1, TAK1, and IKKβ. (A) A RIP1/NEMO/TAK1/IKKβ complex is detected in the cytosol of cells with DNA damage. U2OS cytosolic extracts were fractionated using an FPLC protein purification system (Pharmacia) on a Superose 6 column (Amersham Biosciences). Fractions 10 to 37 were separated by SDS-PAGE and immunoblotted (IB) with NEMO, Rip1, Tak1, and IKKβ antibodies. The column was calibrated with gel filtration standards (Bio-Rad Laboratories) (B). Fractions 17 to 21 from untreated or etoposide (40 μM)-treated cells were immunoprecipitated (IP) with a Tak1 antibody, followed by immunoblotting with a RIP1, NEMO, or Tak1 antibody. (C) RIP1 sumoylation is required for TAK1 recruitment. Rip1−/− cells reconstituted with Flag-tagged versions of wild-type RIP1 (WT), the RIP1 lysine mutant (4KR), or RIP1 4KR conjugated with a SUMO-1 moiety (SUMO1-4KR) were left untreated or treated with etoposide (40 μM) for 1 h. Cell lysates were immunoprecipitated with a Flag antibody, followed by immunoblotting with a TAK1 antibody. (D) ATM levels in the nuclear and cytosolic extracts were examined by immunoblotting with an ATM antibody. Lysates were also probed with anti-PARP1, anti-caspase-3, and β-actin antibodies to monitor the purity of the nuclear/cytosolic lysates and the total protein levels. (E) DNA damage stimulates interactions between ATM and TAK1, RIP1 and TAK1, and RIP1 and NEMO. Human U2OS cells were left untreated or treated with etoposide (40 μM) for the time periods indicated. Nuclear and cytosolic extracts were immunoprecipitated with a TAK1 or NEMO antibody, followed by immunoblotting with either an ATM or RIP1 antibody.
To investigate whether RIP1 sumoylation stimulates TAK1 recruitment, we treated Rip1−/− reconstituted cells with etoposide and assayed for TAK1 recruitment. We found that the RIP1 4KR mutant was unable to recruit TAK1 in response to DNA damage, whereas the addition of a SUMO-1 moiety to the RIP1 4KR mutant restored the ability of RIP1 to respond to DNA damage and recruit TAK1 (Fig. 4C). Taken together, RIP1 sumoylation triggers its ubiquitination and the subsequent recruitment of TAK1.
We also prepared nuclear and cytoplasmic fractions from cells treated with etoposide for various times and examined each for the presence of ATM, PARP1, and caspase-3. Although ATM is predominantly a nuclear kinase, a small quantity of ATM can be detected in the cytosol (Fig. 4D) (29). When TAK1 was immunoprecipitated from the cytosolic fractions, we found that etoposide treatment induced interactions between TAK1 and cytosolic ATM (Fig. 4E). We then examined the nuclear and cytosolic extracts for the presence of RIP1. A transient and weak RIP1/NEMO interaction was observed in the nucleus of cells treated with etoposide for 30 min, similar to previously published work (11). In contrast, a more robust and sustained RIP1/NEMO interaction was readily detected in the cytosol of etoposide- or doxorubicin-treated cells (Fig. 4E and data not shown). Moreover, RIP1 also associated with TAK1, by coimmunoprecipitation (Fig. 4E). Thus, TAK1 associates with ATM and RIP1, while RIP1 also interacts with NEMO about 1 h following DNA damage (Fig. 4E). The results of the cytosolic gel filtration experiment suggest that these proteins associate in one complex. These interactions are coincident with the induction of NF-κB activity as measured by phospho-IκBα reactivity or IκBα degradation (Fig. 2A, lower panel), suggesting that a cytosolic ATM, RIP1, NEMO, and TAK1 complex recruits and activates IKKβ.
DNA double-strand breaks stimulate TAK1 kinase activity, and TAK1 inhibition impairs the NF-κB response and sensitizes cells to genotoxic stress.
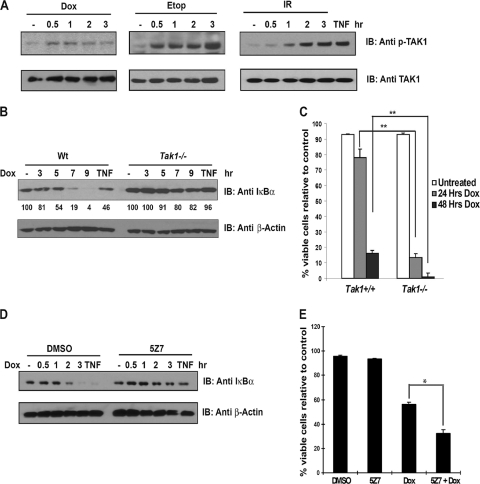
Treatment with proinflammatory cytokine TNF-α or IL-1β has been shown to stimulate TAK1 kinase activity, which can be detected using an in vitro kinase assay or by probing cell lysates with an anti-phospho-TAK1 antibody to measure TAK1 autophosphorylation (21, 29). To determine whether DNA damage stimulates TAK1 kinase activity, we treated the human tumor cell line U2OS with TNF-α, doxorubicin, etoposide, or gamma irradiation for the time periods indicated in Fig. 5 and measured anti-phospho-TAK1 levels. We detected a modest increase in phospho-TAK1 reactivity in doxorubicin-treated cells (Fig. 5A). Treatment of U2OS cells with etoposide or gamma irradiation, treatments known to induce numerous DNA double-strand breaks, significantly increased and sustained phospho-TAK1 reactivity, indicating that, like TNF-α treatment, DNA double-strand breaks are potent inducers of TAK1 kinase activity (Fig. 5A).
Fig. 5.
DNA double-strand breaks stimulate TAK1 kinase activity, and TAK1 inhibition impairs the NF-κB response and sensitizes cells to genotoxic stimuli. (A) Multiple DNA-damaging agents stimulate TAK1 kinase activity. Human U2OS tumor cells were left untreated or treated with TNF-α (10 ng/ml) for 10 min, doxorubicin (Dox; 10 μg/ml), or etoposide (Etop; 40 μM) or exposed to 10 Gy gamma irradiation (IR) for the time periods indicated. TAK1 activity was measured by immunoblotting with a (IB) phospho-specific TAK1 antibody. Total TAK1 levels were measured by immunoblotting with a TAK1 antibody. (B) The NF-κB response to genotoxic stress is impaired in Tak1-deficient MEFs. SV40T-immortalized wild-type (Wt) or Tak1-deficient murine embryonic fibroblasts were left untreated, treated with TNF-α, or treated with doxorubicin for the time periods indicated, and NF-κB activity was measured by monitoring IκBα degradation. Cell lysates were probed with anti-β-actin antibody to ensure that equal amounts of total protein were loaded. IκBα degradation was quantified using densitometry, and values are shown for each lane. (C) Tak1-deficient MEFs are hypersensitive to doxorubicin treatment. Wild-type or Tak1-deficient MEFs were treated with doxorubicin for 24 and 48 h, and viable cells were quantified by trypan blue exclusion. Samples were assayed in triplicate; error bars represent the standard deviations. The experiment whose results are shown is representative of at least three. Statistical evaluation was performed using the unpaired Student t test. **, P < 0.01. (D) Treatment with a TAK1 kinase inhibitor impairs the NF-κB response to DNA damage. Human U2OS cells were pretreated with the TAK1 kinase inhibitor 5Z7 (Bioaustralis) (1 μM) or dimethyl sulfoxide (DMSO) for 2 h. The cells were left untreated or treated with TNF or doxorubicin for the time periods indicated. NF-κB activation was measured by immunoblotting the cell lysates with an IκBα antibody. Total protein levels were determined by immunoblotting with an anti-β-actin antibody. (E) TAK1 kinase inhibition sensitizes human tumor cells to doxorubicin treatment. Human MCF7 tumor cells were pretreated with the TAK1 kinase inhibitor 5Z7 (1 μM) or DMSO control for 2 h. Cells were then left untreated or treated with doxorubicin for 48 h. Viable cells were quantified by trypan blue exclusion. Samples were assayed in triplicate; error bars represent the standard deviations. The experiment whose results are shown is representative of at least three. Statistical evaluation was performed using the unpaired Student t test. *, P < 0.05.
To further test the contribution of TAK1 to the genotoxic stress response, we examined NF-κB activity and sensitivity to doxorubicin in Tak1-deficient, simian virus 40 T antigen (SV40T)-immortalized MEFs. A Tak1 deficiency results in an inability to activate NF-κB in response to TNF-α, IL-1β, or Toll-like receptor 3/4 (TLR3/4) ligands (20). We found that TNF- and doxorubicin-induced IκBα degradation was impaired in the absence of Tak1 (Fig. 5B) and that Tak1-deficient cells were hypersensitive to doxorubicin treatment compared to the results for wild-type controls (Fig. 5C) (P < 0.01).
We then tested whether TAK1 kinase activity was required for the NF-κB response to DNA damage. To examine a requirement for the kinase activity of TAK1, we pretreated cells with vehicle or with 5Z7, a potent and highly selective inhibitor of TAK1 (Bioaustralis) (18, 23, 26). The cells were left untreated or exposed to the proinflammatory cytokine TNF-α for 10 min or to doxorubicin for the time periods indicated in Fig. 5. We examined the cell lysates for evidence of IκBα degradation and measured cell viability. As expected, TNF-α or doxorubicin treatment induced IκBα degradation in the vehicle-treated cells. Pretreatment with the TAK1 kinase-selective inhibitor 5Z7 reduced the IκBα degradation observed in cells treated with TNF-α or doxorubicin (Fig. 5D). Consistent with the impaired NF-κB response, TAK1 kinase inhibition additionally sensitized the cells to doxorubicin-induced cell death (Fig. 5E) (P < 0.05), indicating that the kinase activity of TAK1 is required for NF-κB activation and survival in response to DNA damage.
DNA damage fails to activate TAK1 or NF-κB in the absence of ATM, NEMO or RIP1.
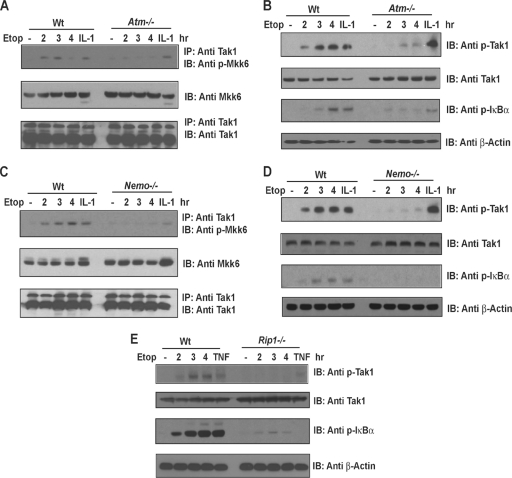
The results of our biochemical studies suggest that ATM, NEMO, and RIP1 are sensors and transducers of the DNA damage signal and lie upstream of TAK1 and IKK-β. To genetically test this model, we asked whether Atm, Nemo, and Rip1 are each required for DNA damage-induced Tak1 activation. Wild-type, Atm−/−, Nemo−/−, and Rip1−/− mouse embryonic fibroblasts were treated with proinflammatory cytokine IL-1β or TNF-α for 10 to 15 min or exposed to etoposide for the time periods indicated in Fig. 6. We measured Tak1 kinase activity using an in vitro kinase assay with a His-tagged, kinase-inactive MKK6 as a substrate. Additionally, the cell lysates were examined for phospho-Tak1 and phospho-IκBα levels.
Fig. 6.
DNA damage fails to activate TAK1 or NF-κB in the absence of ATM, NEMO, or RIP1. Murine embryonic fibroblasts from Atm-, Nemo-, or Rip1-deficient mice and control littermates (wild type [Wt]) were left untreated, treated with TNF-α (10 ng/ml) or IL-1β (10 ng/ml), or treated with etoposide (Etop; 40 μM) for the time periods indicated. TAK1 was immunoprecipitated (IP) from the cell lysates, and TAK1 kinase activity measured using an in vitro kinase assay with a His-tagged, kinase-inactive MKK6 as a substrate. MKK6 phosphorylation was then measured using a phospho-specific MKK6 antibody as described in reference 29. Total MKK6 input levels were measured by immunoblotting (IB) with an anti-MKK6 antibody (A and C). Alternatively, TAK1 activity was measured by immunoblotting cell lysates with a phospho-specific TAK1 antibody (B, D, and E). Total TAK1 levels were measured by immunoblotting with a TAK1 antibody.
As expected, the IL-1β and TNF-α-induced Tak1 kinase activities appeared unaffected by an Atm deficiency (Fig. 6A and B) and, consistent with the role of Nemo downstream of Tak1 in the IL-1β pathway, similar phospho-Tak1 levels were detected in the IL-1β-treated Nemo-deficient cells (Fig. 6D). However, Tak1 kinase and NF-κB activities were significantly reduced in the etoposide-treated fibroblasts lacking Atm, Nemo, or Rip1 (Fig. 6A to E). A Rip1 deficiency impaired the NF-κB response to both TNF-α and etoposide, as reduced IκBα and Tak1 phosphorylation levels were observed when Rip1-deficient MEFs or RIP1-silenced human tumor cells were treated with TNF-α or etoposide (Fig. 6E and data not shown). Collectively, these data support a model whereby Atm/Nemo and Rip1 are each required for Tak1 kinase activation and for an optimal NF-κB response to DNA damage.
RIP1 or TAK1 silencing sensitizes tumor cells to DNA damage due to an inability to induce NF-κB-dependent survival gene expression.
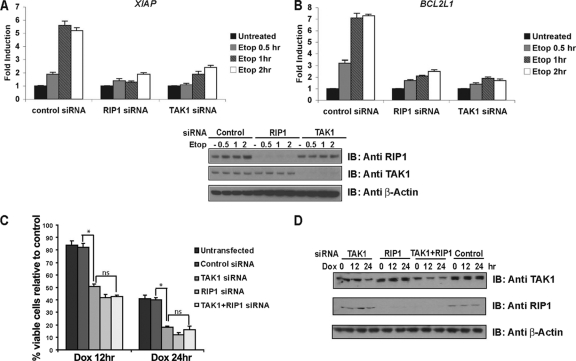
The results of our studies in mouse fibroblasts implicate Tak1 in the NF-κB response to genotoxic stimuli and raise the possibility that the TAK1 kinase may protect human tumor cells from the effects of DNA-damaging agents and contribute to chemotherapeutic resistance. To determine whether TAK1 or RIP1 silencing in human tumor cells affects NF-κB-dependent survival gene expression, we quantified XIAP and BCL2L1 mRNA levels in human tumor cells transfected with control or TAK1- or RIP1-specific siRNA oligonucleotides. Etoposide treatment of cells expressing the control siRNA stimulated a 2.5-fold increase in XIAP and BCL2L1 mRNA levels at 30 min, followed by a 5-fold increase in mRNA levels at 1 and 2 h. However, we observed little to no induction of XIAP or BCL2L1 expression in the etoposide-treated RIP1- or TAK1-silenced cultures (Fig. 7A and B), revealing that both RIP1 and TAK1 contribute to NF-κB-mediated survival gene expression in tumor cells exposed to DNA damage. TAK1 silencing also impaired the NF-κB response to DNA damage (not shown), and TAK1-depleted human tumor cells exhibited significantly more cell death than doxorubicin-treated control cells (Fig. 7C). Similarly, RIP1 depletion sensitized human tumor cells to doxorubicin or etoposide treatment (Fig. 7C and data not shown).
Fig. 7.
RIP1 or TAK1 silencing sensitizes tumor cells to DNA damage due to an inability to induce NF-κB-dependent survival gene expression. (A and B) DNA damage fails to induce the expression of NF-κB-dependent survival genes in TAK1- or RIP1-silenced human tumor cell lines. Human U2OS cells were transfected with a control siRNA, a TAK1 siRNA, or a RIP1 siRNA for 48 h. Cells were then left untreated or treated with etoposide (Etop; 40 μM) for 0.5, 1, or 2 h. XIAP and BCL2L1(BCL-xL) mRNA levels were quantified using real-time PCR. Gene expression is reported as copy number per 1,000 copies of β-actin. Samples were assayed in triplicate; error bars represent the standard deviations. Lower panels show RIP1 and/or TAK1 knockdown efficiency as monitored by immunoblotting the cell lysates with RIP1 or TAK1 antibodies. (C) A RIP1 or TAK1 deficiency does not result in increased doxorubicin (Dox) sensitivity. Human HeLa cells were left untransfected or transfected with control siRNA, TAK1 siRNA, RIP1 siRNA, or TAK1 plus RIP1 siRNA oligonucleotides for 48 h. Cells were then left untreated or treated with doxorubicin (2 μg/ml) for 12 and 24 h. Viable cells were quantified by trypan blue exclusion. Samples were assayed in triplicate; error bars represent the standard deviations. Statistical evaluation was performed using the unpaired Student t test. *, P < 0.05; ns, not significant. (D) RIP1 and/or TAK1 knockdown efficiency was monitored by immunoblotting the cell lysates with RIP1 or TAK1 antibodies.
Although our biochemical studies support a model whereby TAK1 lies downstream of the ATM/NEMO/RIP1 complex in this NF-κB pathway, it remains plausible that RIP1 and TAK1 mediate independent survival pathways. To perform an epistatic analysis, we silenced RIP1 and/or TAK1 expression in HeLa cells and then measured sensitivity to DNA-damaging agents. Silencing both TAK1 and RIP1 had no added effect on tumor cell viability following doxorubicin treatment (Fig. 7C), supporting a model where RIP1 and TAK1 function in the same pathway to mediate tumor cell survival.
TAK1 mediates the p38 MAPK/MK2 checkpoint in p53-deficient human tumor cells.
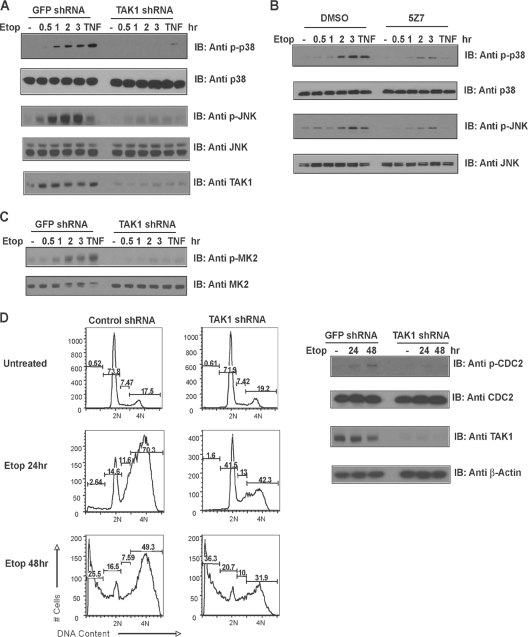
The DNA damage response is primarily mediated by the kinases ATR and ATM, which signal to the checkpoint kinases CHK1 and CHK2. Yet in the absence of a functional ATM/CHK2 or an ATR/CHK1/TSP53 pathway, DNA damage activates a third pathway that involves ATM/ATR signaling to p38 MAPK, which activates a newly identified checkpoint mediated by MK2 (19). Tumor cells that inactivate p53 depend on this alternative pathway to mediate cell cycle arrest and survival following treatment with chemotherapeutic drugs that cause DNA damage. Pretreatment with the p38 MAPK-selective inhibitor SB203580 prior to the administration of DNA-damaging agents abolishes MK2 activation and fails to result in cell cycle arrest (19). Although this alternative DNA damage pathway is active in most p53-deficient human tumor cells, how DNA damage results in p38 MAPK activation was unknown. To investigate whether this alternative survival and repair pathway may be mediated by TAK1, we infected the p53-deficient human lung adenocarcinoma cell line H1299 with a control GFP or a TAK1-specific shRNA lentivirus. Alternatively, we pretreated H1299 tumor cells with a TAK1-selective kinase inhibitor prior to etoposide exposure. Both TAK1 depletion and TAK1 kinase inhibition impaired the p38 MAPK and JNK responses to DNA damage (Fig. 8A and B and data not shown). We then directly examined the cell lysates for evidence of MK2 activation. DNA damage-induced MK2 phosphorylation was significantly impaired in the TAK1-silenced cells but not in the control cultures (Fig. 8C), suggesting that TAK1 contributes to the p38 MAPK/MK2-mediated DNA damage response pathway.
Fig. 8.
TAK1 mediates the p38 MAPK/MK2 checkpoint in p53-deficient human tumor cells. (A and C) A TAK1 deficiency impairs p38 MAPK, MK2, and JNK activation in response to DNA damage. Human U2OS cells were infected with a control GFP or TAK1 shRNA lentivirus. The cells were left untreated, treated with TNF-α, or treated with etoposide (40 μM) for the time periods indicated. Cell lysates were probed with a phospho-specific p38 MAPK, MK2, or JNK1/2 antibody. Total p38 MAPK, MK2, and JNK1/2 levels were measured by immunoblotting (IB) the cell lysates with total p38 MAPK, MK2, or JNK1/2 antibody. (B) Inhibiting TAK1 kinase activity impairs the p38 MAPK and JNK1/2 response to DNA damage. U2OS cells were pretreated with vehicle (dimethyl sulfoxide [DMSO]) or the TAK1-selective kinase inhibitor 5Z7 (Bioaustralis) (1 μM), and cells were left untreated, treated with TNF-α, or treated with etoposide for various time periods. Levels of JNK and p38 MAPK activation were measured by immunoblotting the cell lysates with phospho-JNK1/2 or phospho-p38 MAPK antibodies. Total p38 MAPK and JNK1/2 levels were measured by immunoblotting the cell lysates with p38 MAPK or JNK1/2 antibodies. (D) Impaired cell cycle arrest upon DNA damage in TAK1-deficient, p53-deficient human tumor cells. H1299 cells were infected with a control GFP shRNA or TAK1 shRNA lentivirus, and selected cells were left untreated or treated with etoposide (40 μM) for 24 or 48 h. Cell cycle analysis was performed by staining the cells with PI, followed by flow cytometry. DNA contents are shown as percentages. Phospho-CDC2 levels were compared by immunoblotting the cell lysates with phospho-specific CDC2 antibodies. Total protein levels were determined by immunoblotting with an anti-β-actin antibody.
To determine whether a TAK1 deficiency interferes with p38 MAPK/MK2 checkpoint signaling, we compared the cell cycle profiles of control or TAK1-silenced cells left untreated or treated with etoposide for 24 or 48 h. Treatment of the control infected cells with etoposide for 24 h led to the accumulation of cells with 4N DNA content, indicative of an intact checkpoint. In contrast, when TAK1 expression is limiting, etoposide-induced cell cycle arrest is impaired, with half as many cells detected with 4N DNA content (Fig. 8D) (70.3% ± 3.7% [mean ± standard deviation] for control cells versus 42.3% ± 5.4% for TAK1 knockdown cells in three independent experiments; P < 0.05). Consistently, an impaired p38 MAPK/MK2 response to DNA damage reduced phospho-CDC2 levels detected in the TAK1-depleted tumor cells compared to the levels in cells expressing the control GFP shRNA. These data demonstrate that TAK1 mediates the NF-κB genotoxic stress response and the p38 MAPK/MK2 checkpoint. These findings have translational implications and raise the TAK1 kinase as a potential therapeutic target for tumors refractory to conventional chemo-/radiation therapy.
DISCUSSION
In the present study, we reveal how nuclear DNA damage signals are conveyed to the cytosolic IKK and p38 MAPK/MK2 kinases to mediate cell survival. We demonstrate crucial cytosolic roles for RIP1 and TAK1 in the NF-κB response, demonstrating that SUMO-1- and ubiquitin-modified RIP1 triggers recruitment of the ubiquitin-activated kinase TAK1. These findings indicate that in addition to its nuclear role(s), RIP1 has cytosolic functions and contributes downstream of the nuclear ATM/NEMO/RIP1 complex. Our studies also reveal a requirement for TAK1 in the NF-κB and the alternative p38 MAPK/MK2 survival pathway that is active in p53-deficient malignancies. We demonstrate that DNA damage stimulates TAK1 kinase activity and, importantly, that TAK1 kinase inhibition impairs both NF-κB and p38 MAP/MK2 kinase activation and thereby sensitizes p53-deficient human tumor cells to DNA-damaging agents. These findings have implications for translational cancer research and lead to the prediction that inhibition of TAK1 and/or RIP1 modification may sensitize p53-deficient, chemoresistant tumors to DNA-damaging agents.
Our coimmunoprecipitation studies reveal that DNA damage stimulates interactions between ATM, NEMO, RIP1, and TAK1 in the cytosol of etoposide-treated cells. These findings led us to interrogate the nature of the cytosolic complexes formed in response to DNA damage. Size exclusion chromatography revealed that DNA damage causes shifts in RIP1, NEMO, TAK1, and IKKβ proteins into overlapping fractions corresponding to a molecular-mass range of 500 to 700 kDa. Coimmunoprecipitation of column fractions confirmed that DNA damage induces the formation of a NEMO, RIP1, and TAK1 complex. Interestingly, these studies also reveal that the NEMO/TAK1 complex contains modified RIP1. Surprisingly, only a fraction of the total RIP1 protein appears to be modified and shifts into the 500- to 700-kDa complex in cells with DNA damage (Fig. 4A and data not shown). This is in contrast to the amount of polyubiquitinated RIP1 detected when the same cells are treated with TNF-α for 10 min (Fig. 2C). The reasons for these differences are unclear but may reflect the nature of the NF-κB responses. Although the TNF-induced NF-κB response is rapid and robust, occurring within minutes of receptor activation, DNA-damaging agents induce a weaker, more sustained NF-κB response, detected at 30 min and lasting several hours. The delay observed in the damage response may reflect the time required to relay the nuclear damage signal to the cytosol, whereas the duration of the DNA damage-induced NF-κB response may reflect how the pathway is negatively regulated. Cytokine-induced NF-κB responses are terminated in part by the rapid deubiquitination of cytosolic components, such as RIP1, TRAF6, and NEMO (1, 2, 14, 24, 25). In contrast, the DNA damage-induced NF-κB pathway may be regulated in the nucleus potentially at the level of the SUMO proteases that control NEMO and/or RIP1 sumoylation.
In addition to RIP1 and NEMO, the p53-induced protein with death domain (PIDD) was also shown to interact with RIP1 to facilitate NEMO sumoylation (11). Yet in contrast to a Rip1, Nemo, or Tak1 deficiency, Pidd-deficient fibroblasts appear to respond normally to DNA damage (17). These findings indicate that either PIDD is not absolutely required for this NF-κB response or that redundant pathways exist in certain cell types. Consistent with this idea, the requirement for PARylation in DNA damage-induced NF-κB activation appears to be cell type dependent (22). In contrast to the study of Janssens et al., which identified PIDD, RIP1, and NEMO in the nucleus of HEK293 cells with damaged DNA (11), Stilmann et al. failed to detect PIDD or RIP1 as components of the nuclear PARP-1 complex that is induced in mouse embryonic fibroblasts following gamma irradiation (22). These seemingly disparate findings could indicate that the composition of the DNA damage-induced nuclear complexes vary among cell types or that the PIDD/RIP1/NEMO nuclear complex functions downstream of the PARP-1 signalsome.
Consistent with this model, the PARP-1 signalsome is detected 10 min after DNA damage, prior to the formation of the cytosolic RIP1, NEMO, and TAK1 complex which becomes detectable between 30 min and 1 h following etoposide treatment (Fig. 4) (22). Hence, a delay exists between PARP-1 signalsome (10 min) formation and RIP1 and NEMO sumoylation (>30 min), suggesting that sumoylation is downstream of the PARP-1 signalsome. This study detected modified RIP1 in the cytosolic complex coincident with the detection of phospho-IκBα reactivity, thereby temporally linking the posttranslational modification(s) of RIP1 to IKK activation (Fig. 2A and 4).
In the present study, we reveal the nature of the RIP1 modifications observed in cells with DNA damage. We find that RIP1 is SUMO and ubiquitin modified and demonstrate that PIASy silencing impairs our ability to detect both sumoylated RIP1 and NEMO in etoposide-treated cells (Fig. 2A and B). Moreover, we detect sumoylated RIP1 and NEMO at similar time points following DNA damage (Fig. 2A and data not shown), suggesting that PIASy modifies both NEMO and RIP1. We provide genetic evidence that modified RIP1 is required for NF-κB activation and the survival of cells exposed to genotoxic stimuli (Fig. 3).
DNA damage also stimulates the ubiquitin modification of RIP1, and sumoylated RIP1 appears to be required for RIP1 ubiquitination and TAK1 recruitment in cells with DNA damage (Fig. 2C, 3D, and 4C). Our preliminary analysis suggests that more than one RIP1 lysine may be modified in cells with DNA damage and that sumoylation may precede the ubiquitin modification of RIP1 (Fig. 3D). Moreover, silencing of the E2-conjugating enzyme UBC13 that is responsible for K63-linked polyubiquitination diminishes the NF-κB response and sensitizes multiple human cancer cell lines to DNA damage (data not shown). Two recent papers also implicate TAK1, UBC13, and K63-linked ubiquitination in the NF-κB response to DNA damage and identify critical roles for an ELKS-XIAP or a TRAF6-cIAP1 module in the genotoxic stress response (7, 28). These complexes could reflect signaling intermediates in a common pathway, since we detect ELKS in our etoposide-induced NEMO/RIP1/TAK1 complex (Y. Yang and M. A. Kelliher, unpublished data), or these findings could indicate that different cell types engage distinct modules. Collectively, these data reinforce crucial roles for K63-ubiquitin in the stabilization and assembly of the cytosolic DNA damage response complex and reveal that this NF-κB response utilizes ubiquitin-dependent signaling mechanisms to mediate cell cycle arrest and survival.
The sensitivity to DNA damage observed in mouse embryonic fibroblasts deficient for Nemo, Rip1, or Tak1 prompted us to test whether RIP1 and TAK1 contribute to the DNA damage response in human tumor cells. We demonstrate that RIP1 and/or TAK1 silencing in multiple human tumor cell lines results in an impaired NF-κB-dependent antiapoptotic response and sensitivity to DNA damage (Fig. 7 and data not shown). In addition to the DNA damage-induced NF-κB response, our studies reveal novel functions for TAK1 in the alternative p38 MAPK/MK2 checkpoint found to be active in human tumor cells that lack functional p53 (19). We demonstrate that TAK1 silencing or TAK1 kinase inhibition impairs both the p38 MAPK/MK2 and JNK responses to DNA damage, interferes with cell cycle arrest, and sensitizes human tumor cells to etoposide treatment (Fig. 8).
The present study reveals RIP1 and TAK1 as critical mediators of the genotoxic stress response. Although we provide evidence that the kinase activity of TAK1 is required for NF-κB and p38 MAPK/MK2 activation, the kinase activity of RIP1 has been shown to be dispensable for the initial NF-κB response to DNA damage (10). Our data establish a requirement for modified RIP1 in the NF-κB response to DNA damage, raising the possibility that the SUMO-1 or ubiquitin modifying enzymes that regulate the genotoxic stress response could serve as new therapeutic targets in chemoresistance. Based on the results of our studies, these targets would be predicted to inhibit the NF-κB response and sensitize resistant tumor cells to DNA-damaging agents without the immune-associated side effects associated with complete IKK inhibition.
ACKNOWLEDGMENTS
We thank Sankar Ghosh for the control and Tak1−/− MEFs, Manolis Pasparakis for the control and Nemo−/− MEFs, and Stephen Jones for the control and Atm−/− MEFs used in this study. We thank Zhijian (James) Chen for providing the His-tagged, kinase-inactive MKK6(K82A) substrate. We thank Neal Silverman for critical reading of the manuscript.
We also gratefully acknowledge that Core resources supported by Diabetes Endocrinology Research Center grant DK32520 were also used. This work was supported by grant RO1 AI075118 to M.A.K. and grants RO1 CA77474 and GM083681 to S.M.
Footnotes
Published ahead of print on 23 May 2011.
REFERENCES
- 1. Boone D. L., et al. 2004. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat. Immunol. 5:1052–1060 [DOI] [PubMed] [Google Scholar]
- 2. Brummelkamp T. R., Nijman S. M., Dirac A. M., Bernards R. 2003. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature 424:797–801 [DOI] [PubMed] [Google Scholar]
- 3. Demchenko Y. N., et al. 2010. Classical and/or alternative NF-kappaB pathway activation in multiple myeloma. Blood 115:3541–3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dignam J. D., Martin P. L., Shastry B. S., Roeder R. G. 1983. Eukaryotic gene transcription with purified components. Methods Enzymol. 101:582–598 [DOI] [PubMed] [Google Scholar]
- 5. Hayden M. S., Ghosh S. 2008. Shared principles in NF-kappaB signaling. Cell 132:344–362 [DOI] [PubMed] [Google Scholar]
- 6. Hideshima T., et al. 2009. Biologic sequelae of I{kappa}B kinase (IKK) inhibition in multiple myeloma: therapeutic implications. Blood 113:5228–5236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hinz M., et al. 2010. A cytoplasmic ATM-TRAF6-cIAP1 module links nuclear DNA damage signaling to ubiquitin-mediated NF-kappaB activation. Mol. Cell 40:63–74 [DOI] [PubMed] [Google Scholar]
- 8. Huang T. T., Feinberg S. L., Suryanarayanan S., Miyamoto S. 2002. The zinc finger domain of NEMO is selectively required for NF-kappa B activation by UV radiation and topoisomerase inhibitors. Mol. Cell. Biol. 22:5813–5825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang T. T., Wuerzberger-Davis S. M., Wu Z. H., Miyamoto S. 2003. Sequential modification of NEMO/IKKgamma by SUMO-1 and ubiquitin mediates NF-kappaB activation by genotoxic stress. Cell 115:565–576 [DOI] [PubMed] [Google Scholar]
- 10. Hur G. M., et al. 2003. The death domain kinase RIP has an essential role in DNA damage-induced NF-kappa B activation. Genes Dev. 17:873–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Janssens S., Tinel A., Lippens S., Tschopp J. 2005. PIDD mediates NF-kappaB activation in response to DNA damage. Cell 123:1079–1092 [DOI] [PubMed] [Google Scholar]
- 12. Jin H. S., et al. 2009. cIAP1, cIAP2, and XIAP act cooperatively via nonredundant pathways to regulate genotoxic stress-induced nuclear factor-kappaB activation. Cancer Res. 69:1782–1791 [DOI] [PubMed] [Google Scholar]
- 13. Kelliher M. A., et al. 1998. The death domain kinase RIP mediates the TNF-induced NF-kappaB signal. Immunity 8:297–303 [DOI] [PubMed] [Google Scholar]
- 14. Kovalenko A., et al. 2003. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature 424:801–805 [DOI] [PubMed] [Google Scholar]
- 15. Lavin M. F. 2008. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat. Rev. Mol. Cell Biol. 9:759–769 [DOI] [PubMed] [Google Scholar]
- 16. Mabb A. M., Wuerzberger-Davis S. M., Miyamoto S. 2006. PIASy mediates NEMO sumoylation and NF-kappaB activation in response to genotoxic stress. Nat. Cell Biol. 8:986–993 [DOI] [PubMed] [Google Scholar]
- 17. Manzl C., et al. 2009. Caspase-2 activation in the absence of PIDDosome formation. J. Cell Biol. 185:291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ninomiya-Tsuji J., et al. 2003. A resorcylic acid lactone, 5Z-7-oxozeaenol, prevents inflammation by inhibiting the catalytic activity of TAK1 MAPK kinase kinase. J. Biol. Chem. 278:18485–18490 [DOI] [PubMed] [Google Scholar]
- 19. Reinhardt H. C., Aslanian A. S., Lees J. A., Yaffe M. B. 2007. p53-deficient cells rely on ATM- and ATR-mediated checkpoint signaling through the p38MAPK/MK2 pathway for survival after DNA damage. Cancer Cell 11:175–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shim J. H., et al. 2005. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes Dev. 19:2668–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Singhirunnusorn P., Suzuki S., Kawasaki N., Saiki I., Sakurai H. 2005. Critical roles of threonine 187 phosphorylation in cellular stress-induced rapid and transient activation of transforming growth factor-beta-activated kinase 1 (TAK1) in a signaling complex containing TAK1-binding protein TAB1 and TAB2. J. Biol. Chem. 280:7359–7368 [DOI] [PubMed] [Google Scholar]
- 22. Stilmann M., et al. 2009. A nuclear poly(ADP-ribose)-dependent signalosome confers DNA damage-induced IkappaB kinase activation. Mol. Cell 36:365–378 [DOI] [PubMed] [Google Scholar]
- 23. Takaesu G., et al. 2003. TAK1 is critical for IkappaB kinase-mediated activation of the NF-kappaB pathway. J. Mol. Biol. 326:105–115 [DOI] [PubMed] [Google Scholar]
- 24. Trompouki E., et al. 2003. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature 424:793–796 [DOI] [PubMed] [Google Scholar]
- 25. Wertz I. E., et al. 2004. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 430:694–699 [DOI] [PubMed] [Google Scholar]
- 26. Windheim M., Lang C., Peggie M., Plater L. A., Cohen P. 2007. Molecular mechanisms involved in the regulation of cytokine production by muramyl dipeptide. Biochem. J. 404:179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu Z. H., Shi Y., Tibbetts R. S., Miyamoto S. 2006. Molecular linkage between the kinase ATM and NF-kappaB signaling in response to genotoxic stimuli. Science 311:1141–1146 [DOI] [PubMed] [Google Scholar]
- 28. Wu Z. H., et al. 2010. ATM- and NEMO-dependent ELKS ubiquitination coordinates TAK1-mediated IKK activation in response to genotoxic stress. Mol. Cell 40:75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xia Z. P., et al. 2009. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature 461:114–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang Y., et al. 2007. NOD2 pathway activation by MDP or Mycobacterium tuberculosis infection involves the stable polyubiquitination of Rip2. J. Biol. Chem. 282:36223–36229 [DOI] [PubMed] [Google Scholar]