Abstract
The ErbB3 receptor tyrosine kinase contributes to a variety of developmental processes, and its overexpression and aberrant activation promote tumor progression and therapeutic resistance. Accumulating evidence suggests that tumor overexpression may be mediated by the loss of posttranscriptional negative regulatory mechanisms, such as protein degradation, that normally keep receptor levels in check. Our previous studies indicate that the RING finger E3 ubiquitin ligase Nrdp1, a protein lost in breast and other tumor types, suppresses ErbB3 levels by mediating ligand-independent receptor ubiquitination and degradation. Here we demonstrate that Nrdp1 preferentially associates with the nascent form of ErbB3 to accelerate its degradation, and we show that the two proteins colocalize at the endoplasmic reticulum (ER). Blocking the exit of ErbB3 from the ER does not affect the ability of Nrdp1 to mediate receptor ubiquitination or degradation, while functional disruption of the conserved ER-associated degradation (ERAD) pathway ATPase VCP/p97 leads to the Nrdp1-dependent accumulation of ubiquitinated ErbB3 but blocks receptor degradation. Further evidence indicates that the ErbB3 targeted by Nrdp1 for degradation is properly folded and fully functional. Collectively, these observations point to a novel mechanism of receptor tyrosine kinase quantity control wherein steady-state levels of signaling-competent receptor are dictated by an ER-localized degradation pathway.
INTRODUCTION
The maintenance of steady-state levels of ErbB family receptor tyrosine kinases in cells is critical for the fidelity of signal transduction. Cells must maintain sufficient receptor numbers to efficiently receive and propagate survival, proliferation, and differentiation signals while limiting receptor numbers to prevent hypersignaling that can lead to dysplasia or malignancy. While gene expression certainly contributes to receptor homeostasis, accumulating evidence suggests that posttranscriptional mechanisms play key roles in determining cellular receptor tyrosine kinase levels (12). Importantly, protein degradation is emerging as a primary mechanism by which cells govern receptor quantity (6).
The canonical protein degradation mechanism governing ErbB receptor content involves the endosomal sorting of mature receptors following their internalization from the cell surface (26, 28, 30), a process thought to be dysregulated in tumors (26). Upon internalization, receptors are trafficked to endosomal compartments, where they are sorted for either lysosomal degradation or recycling to the cell surface. The efficiency with which receptors are sorted for degradation in the absence of growth factor stimulation plays a major role in determining receptor protein half-life and ultimately steady-state receptor levels (33). This efficiency is often dramatically increased upon growth factor stimulation, leading to negative-feedback regulation of receptors and the suppression of further signaling (30). Endosomal sorting of receptors is thought to be carried out by E3 ubiquitin ligases, which physically associate with receptors to mediate their ubiquitination and ultimate trafficking to lysosomes (6). An example is Cbl, a particularly well characterized ubiquitin ligase whose recruitment to and ubiquitination of phosphorylated epidermal growth factor receptor (EGFR) is thought to be responsible for receptor downregulation following ligand stimulation (10, 32).
Signaling by the ErbB3 receptor tyrosine kinase is crucial for the proper development of several tissue types. The overexpression and aberrant activation in tumor cells of ErbB3, a component of the ErbB2-ErbB3 oncogenic axis, are thought to play a significant role in tumor malignancy (7, 18, 31) and also to contribute to the resistance of tumors to targeted therapeutics (2, 16, 19). Analyses of tumor samples from breast cancer patients, as well as tumors derived from a mouse model of ErbB receptor-driven breast cancer, suggest that a significant proportion of the overexpressed ErbB3 observed in tumors results from dysregulation of the posttranscriptional mechanisms that normally keep ErbB3 protein levels in check (20, 25, 29). Nrdp1 is a RING finger E3 ubiquitin ligase that associates with and ubiquitinates ErbB3 in a ligand-independent manner to regulate steady-state levels of the receptor (9, 27). Nrdp1 protein is suppressed in more than half of breast cancer patient tumors, and its levels are strongly inversely correlated with ErbB3 protein levels (36). Abrogation of Nrdp1 function in cultured breast cancer cells elevates ErbB3 levels 5- to 10-fold and potentiates ErbB3 tyrosine phosphorylation, signaling pathway usage, and cellular proliferation and motility in response to the ErbB3 ligand neuregulin-1 (NRG1) (36). These observations indicate that Nrdp1 is a key component of a posttranscriptional pathway that mediates the degradation of the vast majority of signaling-competent ErbB3 receptors.
In the present study, we set out to analyze the cellular and molecular mechanisms by which Nrdp1 regulates ligand-independent ErbB3 levels in cells. Surprisingly, we find that this process does not involve lysosome-mediated degradation of the internalized mature receptor but instead acts on newly synthesized receptors at the endoplasmic reticulum (ER).
MATERIALS AND METHODS
Reagents and cell culture.
The antibodies used include mouse monoclonal anti-Flag antibody M2 (Stratagene), antitubulin (Sigma), anti-ErbB3 antibodies Ab-4 and Ab-6 (Thermo Scientific), anti-myc antibody 9E10 (Calbiochem), anti-calnexin (BD Transduction Laboratories), anti-protein disulfide isomerase (anti-PDI; Enzo Biosciences), rabbit polyclonal antibodies against ErbB3 (C-17) and EGFR (1005) (Santa Cruz Biotechnology), and antibodies against green fluorescent protein (GFP) and hemagglutinin (HA) (Invitrogen). The goat polyclonal antibody against glutathione S-transferase (GST) was from GE Healthcare. Horseradish peroxidase (HRP)-conjugated secondary antibodies and fluorophore-conjugated secondary antibodies were purchased from Molecular Probes (Invitrogen). Brefeldin A (BfA) was purchased from Sigma, and HRP-conjugated streptavidin was purchased from Zymed (Invitrogen). The HEK-293T, MCF7, T47D, and MDA-MB-453 cell lines were purchased from ATCC and were maintained under an atmosphere of 10% CO2 in Dulbecco's modified Eagle medium (DMEM) with 10% fetal calf serum (FCS) and 1% penicillin-streptomycin (all medium components were from Invitrogen). Tran35S-label (containing 35S-labeled l-cysteine and l-methionine) was from MP Biomedicals.
Constructs, transient transfection, and viral transduction.
Cells were transiently transfected using FuGene-6 (Roche) according to the manufacturer's instructions. In those experiments where two or more plasmids were cotransfected, equal amounts of each plasmid were used, and total amounts of DNA transfected were 500 ng/well for 12-well plates and 5 μg/plate for 10-cm-diameter dishes. Plasmids encoding wild-type Nrdp1, dominant negative Nrdp1 (DN-Nrdp1), and full-length ErbB3 have been described previously (9). Plasmids encoding HA-labeled ubiquitin and myc-labeled Lrig1 have been described elsewhere (24). Wild-type valosin-containing protein (VCP) and dominant negative VCP (VCP-QQ) cDNAs were obtained from Addgene and were directionally subcloned into pcDNA3.1 using the KpnI and EcoRV insertion sites. The KDEL-GFP plasmid has been described previously (8), and the ΔF508-CFTR-GFP plasmid was a gift from Ron Kopito. The pSuper-Nrdp1-shRNA1 (KD1) plasmid for the silencing of Nrdp1 has been described previously (36). For pSuper-Nrdp1-shRNA4 (KD4), oligonucleotides GATCCCC GA CTA GAC AGA TGA ACC GAC G TTCAAGAGA CG TCG GTT CAT CTG TCT AGT C TTTTTA and AGCTT AAAAA GA CTA GAC AGA TGA ACC GAC G TCTCTTGAA CG TCG GTT CAT CTG TCT AGT C GGG (underlining indicates the targeting sequence of the shRNA) were first annealed and then ligated into BglII/HindIII-cut pSuper.retro.neo.gfp. pSuper-scramble was designed not to target any mRNA in the human transcriptome and was constructed in the same manner as that described above by using oligonucleotides GATCCCC GG AAA CGA GGT TGA ATA CA T TCAAGAGA T GTA TTC AAC CTC GTT TCC TTTTTA and AGCTTAAAAA GG AAA CGA GGT TGA ATA CA TCTCTTGAA T GTA TTC AAC CTC GTT TCC GGG. Vectors pMX-pie and pMX-pie-Nrdp1 have been described previously, and mass populations of stably transduced cells using these vectors were created as described previously (36).
Immunoprecipitation and immunoblotting.
Following treatments, cell monolayers were harvested using a cell scraper, pelleted, and snap-frozen in liquid nitrogen. Pellets were resuspended in 1 ml of ice-cold lysis buffer containing 4 μg/ml each of the protease inhibitors aprotinin, pepstatin, leupeptin, and aminoethyl benzenesulfonyl fluoride. For the biotinylation and pulse-chase experiments, radioimmunoprecipitation assay (RIPA) lysis buffer (50 mM Tris [pH 7.5], 0.1% sodium dodecyl sulfate [SDS], 1% NP-40, 0.5% sodium deoxycholate, 150 mM NaCl, 0.5 mM EDTA) containing protease inhibitors was used. For ubiquitination experiments, RIPA buffer was used along with protease inhibitors, 5 mM N-ethylmaleimide, and 2 μM MG132. For coimmunoprecipitation experiments, pellets were lysed in 1 ml co-IP buffer (20 mM Tris [pH 7.5], 150 mM NaCl, 1 mM MgCl2, 1% NP-40, 10% glycerol) containing protease inhibitors and 1 mM each sodium orthovanadate and NaF. Fifty microliters of lysate was used for immunoblotting, while the remainder was precleared with 50 μl of protein G-agarose beads (Millipore). The precleared lysate was incubated for 2 h at 4°C with either anti-ErbB3 antibody Ab-6 or anti-FLAG antibody M2, followed by the addition of 50 μl of protein G-Sepharose beads and an additional 2 h of incubation. Beads were washed three times with lysis buffer, and bound proteins were released from beads by boiling in 100 μl of 2× Laemmli sample buffer. Immunoprecipitates and input samples were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and were electrophoretically transferred to nitrocellulose membranes. Membranes were blocked using 5% nonfat dry milk in Tris-buffered saline plus 0.1% Tween 20 (TBST) for 1 h, followed by overnight incubation in the primary antibody. Membranes were washed three times with TBST and were incubated for 1 h in HRP-conjugated secondary antibodies diluted in TBST. Washed membranes were developed using Pierce SuperSignal West chemicals, and immunoreactive bands were visualized and quantified using an Alpha Innotech imaging station with AlphaView SA software.
Immunofluorescence microscopy.
For subcellular localization of Nrdp1 and ErbB3, COS7 or HEK-293T cells were plated onto coverslips in 6-well dishes and were transfected with the indicated constructs. Twenty-four hours posttransfection, cells were fixed with 4% paraformaldehyde and were stained using anti-Flag antibody M2, anti-ErbB3 antibody C-17, anti-calnexin, or anti-PDI as the primary antibody. Images were acquired on an Olympus FV1000 Spectral Scan confocal microscope and were analyzed using Olympus FluoView software (Olympus, Center Valley, PA). For surface-labeling experiments, HEK-293T cells on glass coverslips were transfected with ErbB3 for 48 h and were then treated with either a vehicle control or 1 μg/ml brefeldin A for 6 h. Coverslips were rinsed three times in ice-cold phosphate-buffered saline (PBS) and were transferred to ice-cold serum-free DMEM containing 1 μg/ml mouse antibody Ab-4, directed against the extracellular domain of ErbB3, for 45 min. Coverslips were rinsed three times in ice-cold PBS and were fixed for 20 min with 4% paraformaldehyde in PBS at room temperature. Cells were then permeabilized, blocked overnight in blocking solution (1% bovine serum albumin [BSA], 0.02% sodium azide, 0.2% NP-40, 5% goat serum in PBS), and incubated for 1 h at room temperature with rabbit anti-ErbB3 antibody C-17 to label total ErbB3. Following incubation with fluorescent secondary antibodies and 4′,6-diamidino-2-phenylindole (DAPI), mounted slides were analyzed using an Olympus BX61 fluorescent microscope and SlideBook, version 4.1, imaging software (Intelligent Imaging Innovations).
Biotinylation.
HEK-293T cells in 10-cm-diameter plates were transfected with 5 μg ErbB3, and after 48 h, cells were treated with either a vehicle control or 1 μg/ml brefeldin A for 6 h. Rinsed cells were then incubated for 45 min with 0.5 mg/ml biotin-X-N-hydroxysuccinimide (NHS) (Calbiochem) in borate buffer (10 mM boric acid, 150 mM NaCl [pH 8.0]) at 4°C. Biotinylated cells were washed three times in ice-cold PBS containing 15 mM glycine to quench uncoupled biotin and were harvested for immunoprecipitation of ErbB3.
Pulse-chase metabolic labeling.
Forty-eight hours after transfection, HEK-293T cells were incubated for 30 min in cysteine- and methionine-free DMEM containing 10% dialyzed FCS. Tran35S-label (250 μCi) was then added to the cells for the pulse period indicated in the figure legends, and cells were washed once in warm PBS and were chased for various times with intact DMEM containing 10% full FCS and 2 mM supplemental “cold” l-methionine and l-cysteine. Cells were then harvested for ErbB3 immunoprecipitation, SDS-PAGE, and transfer to nitrocellulose filters, and labeled precipitated proteins were imaged using a Molecular Dynamics Storm PhosphorImager and ImageQuant analysis software. In all radiolabeling and immunoblotting experiments, estimates of the molecular weight of ErbB3 were based on its mobility relative to biotinylated molecular weight markers purchased from Cell Signaling Technology. To assess the impact of wild-type and dominant negative Nrdp1 on the half-lives of mature and nascent ErbB3, data from four independent pulse-chase experiments were quantified and averaged. Quantitative analysis was carried out using nonlinear regression analysis. Curves were fitted by the least-squares method to a single-phase exponential decay function, and curve slopes were compared by an F-test to obtain P values.
Neuregulin-binding assay.
Transfected HEK-293T cells were incubated with 25 nM GST-NRG1 (13) for 2 min. Cells were then washed twice in ice-cold PBS (supplemented with NaCl to a final concentration of 500 mM), and cleared lysates were analyzed by immunoblotting.
Subcellular fractionation.
HEK-293T cells cotransfected with wild-type Nrdp1 and ErbB3 were fractionated as described previously (11). Briefly, cell monolayers were washed, harvested into ice-cold PBS, pelleted at 200 × g for 10 min, and then resuspended in ice-cold lysis buffer (100 mM KCl, 5 mM MgCl2, 1 mM ATP, 25 mM Tris-HCl [pH 9.6], and 2 μM MG-132, plus protease inhibitors). Cells were mechanically disrupted with a tight-fitting 7-ml Dounce homogenizer (50 strokes), and this homogenate was centrifuged at 1,000 × g for 5 min to obtain a postnuclear supernatant (PNS). The PNS was then centrifuged at 100,000 × g for 65 min in an SW41 Ti rotor to obtain the cytosolic fraction (supernatant) and cell membranes (pellet). The membrane pellet was resuspended in a Percoll-containing gradient solution as described previously (11), and subcellular membranes were separated by ultracentrifugation at 100,000 × g for 21 min in an SW41 Ti rotor. Membrane fractions were collected from the top of the gradient, lysed in 1% NP-40 on ice for 10 min, and then centrifuged at 100,000 × g for 1 h 45 min in a Beckman type 50.2 Ti fixed-angle rotor to pellet the Percoll gradient medium. Membrane proteins were then precipitated out of the solution using trichloroacetic acid and were washed twice in ice-cold acetone before being resuspended in Laemmli sample buffer.
Real-time PCR analysis.
Total RNA was collected using the RNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. The High-Capacity cDNA reverse transcription kit (Applied Biosystems, Carlsbad, CA) was used to convert 5 μg of total RNA into cDNA. Quantitative PCR amplifications were conducted in a Bio-Rad iCycler real-time PCR system using Applied Biosystems TaqMan gene-specific primers and probes and TaqMan Gene Expression Master Mix. Message levels for Nrdp1 and ErbB3 were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels for each sample.
RESULTS
Nrdp1 suppresses the levels of active ErbB3 in cells but does not accelerate the degradation of the major receptor species.
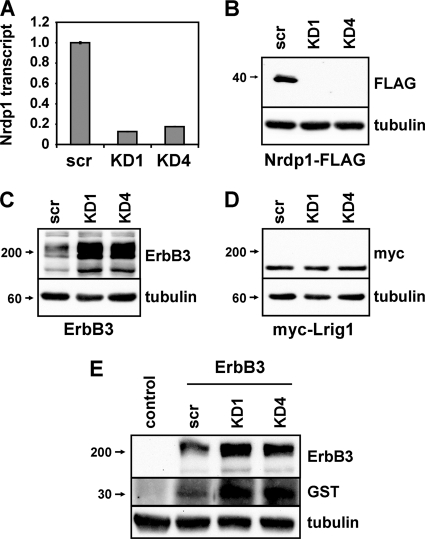
To begin to examine the molecular and cellular mechanisms underlying Nrdp1-mediated ErbB3 degradation, we first developed reagents that efficiently and reliably knock down Nrdp1 in cells. While the low abundance of endogenous Nrdp1 protein in many cell lines makes detection difficult (34), we observed that the short hairpin oligonucleotides KD1 and KD4, derived independently from retroviral vectors, very efficiently suppressed endogenous Nrdp1 transcripts (Fig. 1A) and cotransfected FLAG-tagged protein (Fig. 1B) in HEK-293T cells. Both short hairpin RNAs (shRNAs) elevated levels of cotransfected ErbB3 at least 5-fold (Fig. 1C) but had no effect on cotransfected Lrig1 (Fig. 1D), an unrelated transmembrane protein (15, 24). Importantly, Nrdp1 knockdown concomitantly elevated ErbB3 levels and enhanced the ability of cells to bind GST-tagged NRG1 growth factor (Fig. 1E), indicating that the receptor population eliminated by Nrdp1 action is capable of ligand binding at the cell surface. Taken with our previous observations that Nrdp1 knockdown or dominant negative expression markedly potentiates NRG1-induced signaling, proliferation, and motility in MCF7 breast cancer cells (36), these observations indicate that Nrdp1 very efficiently mediates the degradation of properly folded, signaling-competent ErbB3 receptor.
Fig. 1.
Nrdp1 suppresses levels of the receptor tyrosine kinase ErbB3. (A to D) HEK-293T cells were transiently transfected with Nrdp1-directed shRNAs (KD1 or KD4) or scrambled (scr) constructs either alone (A) or along with Nrdp1-FLAG (B), ErbB3 (C), or myc-Lrig1 (D). (A) Real-time PCR analysis was carried out using total RNA, and the relative quantities of the Nrdp1 transcript were plotted. Error bars represent the standard errors for triplicate samples. (B to D) Lysates were immunoblotted with antibodies to the indicated proteins. (E) Cells transfected without or with ErbB3 and Nrdp1 shRNA constructs, as indicated, were treated for 2 min with the GST-NRG1 growth factor and were washed extensively prior to lysis. Lysates were immunoblotted with the indicated antibodies.
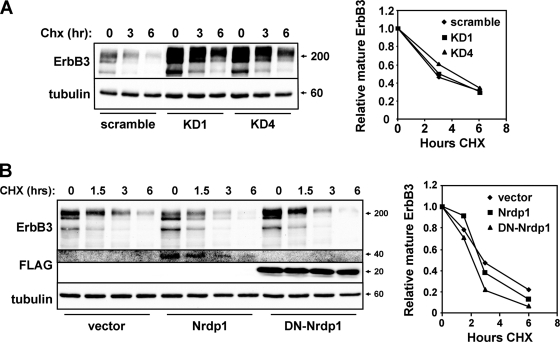
If Nrdp1 behaves canonically and mediates the sorting of internalized ErbB3 receptors to lysosomes for degradation, we predict that the ligase would accelerate the degradation of the fully glycosylated mature form of the protein. Surprisingly, Nrdp1 knockdown had no effect on the stability of the major mature receptor species after inhibition of translation by cycloheximide (Fig. 2A). Likewise, while overexpression of wild-type or dominant negative Nrdp1 suppressed or elevated steady-state ErbB3 levels, respectively, neither affected the rate of mature receptor degradation (Fig. 2B). These observations strongly suggest that Nrdp1 influences receptor quantity through a mechanism independent of the endosomal sorting of mature receptors.
Fig. 2.
Nrdp1 does not accelerate the degradation of the major ErbB3 receptor species. (A) HEK-293T cells transiently cotransfected with ErbB3 and the indicated knockdown constructs were treated for various times with 100 μg/ml cycloheximide (CHX). Lysates were immunoblotted for the indicated proteins (left); the mature (upper) ErbB3 band was quantified, and the amount relative to that at the zero time point was plotted (right). (B) 293T cells were transiently transfected with ErbB3 and Nrdp1 constructs and were analyzed as described for panel A.
Nrdp1 interacts with nascent ErbB3 at the endoplasmic reticulum.
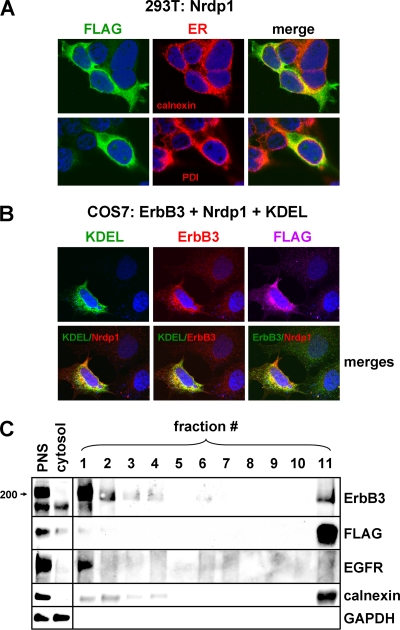
To determine the cellular site of Nrdp1 action, we used confocal immunofluorescence microscopy to localize FLAG-tagged Nrdp1 with organelle markers in transiently transfected cells. Unexpectedly, we observed extensive colocalization of Nrdp1 with the ER markers calnexin and protein disulfide isomerase (PDI) in transiently transfected 293T cells (Fig. 3A). Moreover, in transiently transfected COS7 cells, ErbB3 and Nrdp1 colocalized with each other and with GFP-KDEL (Fig. 3B), a commonly employed ER marker consisting of green fluorescent protein fused to the KDEL endoplasmic reticulum retention signal downstream of the prolactin leader sequence (8). For the quantitative assessment of ErbB3 and Nrdp1 localization at the ER, we employed Percoll density centrifugation to fractionate membranous organelles after cellular disruption (see reference 11). As shown in Fig. 3C, the plasma membrane marker EGFR was present at the top of the gradient (fraction 1) while the ER marker calnexin was present largely at the bottom of the gradient (fraction 11). Interestingly, the majority of the Nrdp1 and 16% of the total ErbB3 cofractionated with calnexin. Moreover, the ErbB3 present in the ER-containing fraction was an undersized form of the receptor (see below). Together, the observations illustrated in Fig. 3 point to the possibility that Nrdp1 acts on ErbB3 at the endoplasmic reticulum.
Fig. 3.
Nrdp1 and ErbB3 colocalize at the endoplasmic reticulum. (A) 293T cells were transiently transfected with Nrdp1-FLAG, and its colocalization with endogenous calnexin (top) and PDI (bottom) was examined by confocal indirect immunofluorescence microscopy. (B) COS7 cells were cotransfected with GFP-KDEL, ErbB3, and Nrdp1-FLAG. (Top) The localization of each was determined by confocal microscopy. (Bottom) The colocalization of proteins was demonstrated by merging red and green pseudocolored channels. (C) 293T cells were transiently transfected with Nrdp1-FLAG and ErbB3, and cellular fractionation was carried out by differential centrifugation and Percoll gradient centrifugation. The postnuclear supernatant (PNS), cytosolic, and Percoll gradient fractions were immunoblotted with the indicated antibodies.
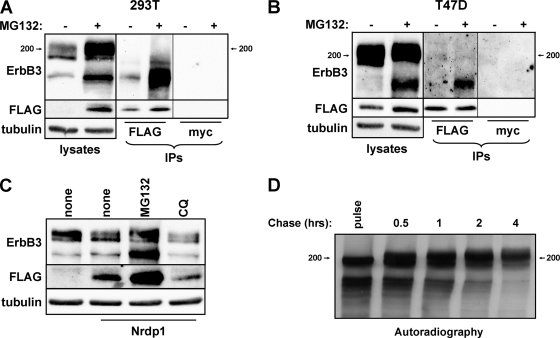
ErbB3 exists as multiple species in many cell types: the major high-molecular-weight species (210 kDa) analyzed in the experiments for which results are shown in Fig. 2, as well as minor lower-molecular-weight species (170 and 190 kDa). The stabilization of both the lower- and higher-molecular-weight forms of ErbB3 by the proteasome inhibitor MG132 (Fig. 4A to C), together with the lack of effect of the lysosome inhibitor chloroquine (Fig. 4C), suggests that proteasomal degradation plays the predominant role in suppressing cellular ErbB3 levels. Interestingly, despite a 10-fold greater prevalence of the high-molecular-weight form of ErbB3, Nrdp1-FLAG specifically associated with the lower-molecular-weight species by coimmunoprecipitation in HEK-293T cells (Fig. 4A). Likewise, Nrdp1-FLAG specifically associated with the endogenous low-molecular-weight ErbB3 species in T47D (Fig. 4B) and MDA-MB-453 (not shown) breast cancer cells.
Fig. 4.
Nrdp1 preferentially interacts with newly synthesized ErbB3. (A) HEK-293T cells cotransfected with Nrdp1-FLAG and ErbB3 were treated with the vehicle dimethyl sulfoxide (−) or 5 μM MG132 (+) for 8 h. Cleared lysates and anti-FLAG or control anti-myc immunoprecipitates (IPs) were immunoblotted with antibodies to the indicated proteins. (B) T47D cells stably transduced with the Nrdp1-FLAG retrovirus were similarly treated without or with MG132 and were analyzed as described for panel A. (C) 293T cells transiently transfected either with ErbB3 alone or with ErbB3 together with Nrdp1-FLAG were treated with the vehicle, MG132, or chloroquine (CQ), and lysates were blotted with the indicated antibodies. (D) HEK-293T cells transiently transfected with ErbB3 were pulse-labeled for 20 min with [35S]methionine-cysteine and were then chased with excess unlabeled methionine-cysteine. Cleared lysates from cells harvested at the indicated times were immunoprecipitated with anti-ErbB3 antibodies and were analyzed by autoradiography.
The simplest explanation for these observations is that Nrdp1 interacts with newly synthesized ErbB3 at the ER prior to its transport to the Golgi apparatus, where additional glycosylation increases its molecular weight. To determine whether the low-molecular-weight species are indeed precursors of the higher-molecular-weight form, we used pulse-chase experiments with [35S]methionine-cysteine metabolic labeling to follow the fate of newly synthesized ErbB3. As shown in Fig. 4D, a 20-min pulse with radiolabeled amino acids labeled the low-molecular-weight (170- and 190-kDa) forms of ErbB3. A chase as long as 2 h with unlabeled amino acids resulted in a shift in the label to a higher form, consistent with the notion that the lower-molecular-weight ErbB3 species are precursors of the heavier mature species. With longer chase times, such as 4 h, the abundance of label in the higher-molecular-weight form subsided, presumably the result of its normal degradation. Overall, these observations are consistent with the notion that Nrdp1 preferentially interacts with the immature, newly synthesized ErbB3 population within the endoplasmic reticulum.
Nrdp1 shortens the half-life of newly synthesized ErbB3.
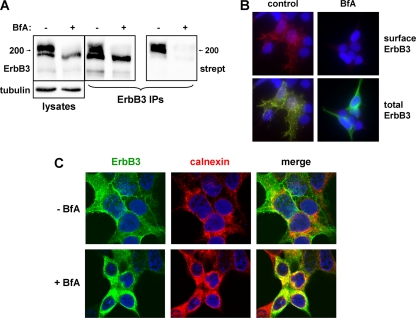
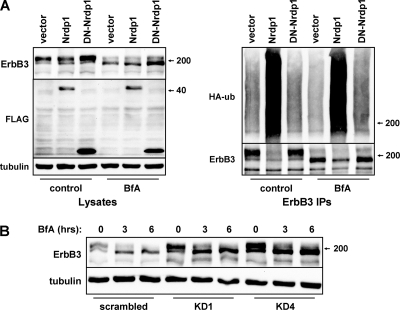
If Nrdp1 regulates ErbB3 levels by acting at the ER, we would predict that blocking receptor exit from the ER would have little impact on Nrdp1-mediated ErbB3 ubiquitination or degradation. To examine the impact of Nrdp1 on ER-localized ErbB3, we employed the macrocyclic lactone antibiotic brefeldin A (BfA), which inhibits the transport of newly synthesized membrane proteins from the ER to the Golgi apparatus (23). As expected, treatment of cells for 6 h with BfA very efficiently inhibited the biotinylation of cell surface ErbB3 (Fig. 5A) and caused the accumulation of receptors in a perinuclear compartment (Fig. 5B) that contains the ER marker calnexin (Fig. 5C). Mature receptors were degraded over the course of BfA treatment, leaving only the newly synthesized ErbB3 form (Fig. 5A). However, BfA-mediated blockade of ErbB3 exit from the ER had no impact on the ability of Nrdp1 to mediate receptor ubiquitination or suppression (Fig. 6A). Moreover, elevation of ErbB3 levels upon Nrdp1 knockdown occurred in the presence of BfA (Fig. 6B), indicating that the Nrdp1-mediated degradation of ErbB3 occurs at the endoplasmic reticulum.
Fig. 5.
Brefeldin A (BfA) blocks the exit of ErbB3 from the ER. (A) HEK-293T cells transfected with ErbB3 were treated without or with BfA; surface proteins were biotinylated; and cleared lysates (left) were immunoprecipitated (IP) with anti-ErbB3 antibodies (right). Precipitates were blotted with streptavidin (strept) and anti-ErbB3. (B) 293T cells transiently transfected with ErbB3 were treated either with the vehicle control (left) or with 1 μg/ml brefeldin A (right) for 6 h and were immunostained for surface (red) (top) or both surface and total (green) ErbB3 (bottom). DAPI staining of nuclei is shown in blue. (C) 293T cells transfected with ErbB3 were treated without (top) or with (bottom) BfA, and confocal immunofluorescence microscopy was used to examine the colocalization of ErbB3 with endogenous calnexin.
Fig. 6.
Disruption of ErbB3 exit from the ER does not disrupt Nrdp1-mediated receptor ubiquitination or degradation. (A) HEK-293T cells were transiently cotransfected with ErbB3 and HA-ubiquitin (HA-ub), along with vector control or Nrdp1 constructs. Cells were then treated with the vehicle control or 1 μg/ml brefeldin A for 6 h; ErbB3 was immunoprecipitated (IP) from cleared lysates; and lysates (left) and precipitates (right) were blotted with the indicated antibodies. (B) HEK-293T cells transiently transfected with ErbB3 and the indicated knockdown constructs were either left untreated (0 h) or treated for 3 or 6 h with 1 μg/ml brefeldin A. Lysates were blotted for the indicated proteins.
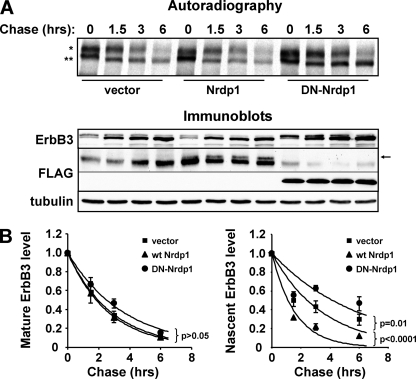
To rigorously assess the impact of Nrdp1 on the degradation of the nascent and mature ErbB3 forms, we coupled metabolic pulse-chase labeling with brefeldin A treatment. In the experiment for which results are shown in Fig. 7A, HEK-293T cells transiently cotransfected with ErbB3 and either an empty vector, wild-type Nrdp1, or DN-Nrdp1 were pulse-labeled for 2 h and were then chased for the indicated times in a medium containing BfA. The presence of BfA in the chase medium prevented the conversion of the immature forms (Fig. 7A, lower band) to the mature form (upper band) during the chase period, allowing simultaneous and independent assessment of the degradation of both ErbB3 species. Quantification of the bands (Fig. 7B) revealed that while neither construct affected the rate of degradation of mature ErbB3, wild-type Nrdp1 and DN-Nrdp1 accelerated and suppressed the degradation rate of immature ErbB3, respectively.
Fig. 7.
Nrdp1 specifically accelerates the degradation of immature ErbB3. Cells transiently transfected with ErbB3 along with either an empty vector, wild-type (wt) Nrdp1, or dominant negative Nrdp1 (DN-Nrdp1) were metabolically pulse-labeled for 2 h with [35S]methionine-cysteine (at time point zero) and were chased in the presence of 1 μg/ml brefeldin A for the indicated times. (A) Cleared lysates were immunoprecipitated with anti-ErbB3 antibodies (bottom) and were analyzed by autoradiography (top). *, mature ErbB3; **, immature ErbB3. Wild-type Nrdp1 in lysates is observed immediately above a nonspecific band (arrow). (B) Quantification of the radioactivity associated with mature and immature ErbB3 bands from four independent pulse-chase experiments. Curves show the fits of average values to an exponential decay function. P values for the differences between the vector and wt Nrdp1, and between the vector and DN-Nrdp1, are given.
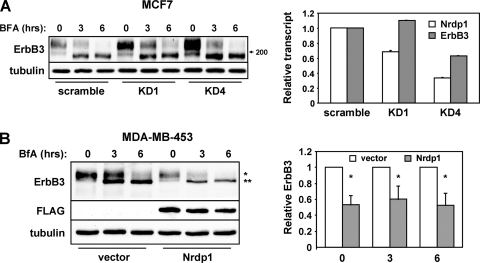
Our observations with transiently transfected HEK-293T cells indicate that the Nrdp1 ubiquitin ligase specifically accelerates the degradation of the newly synthesized ErbB3 population. To confirm that endogenous ErbB3 is targeted by Nrdp1 at the ER, we addressed the impact of BfA treatment on Nrdp1-mediated ErbB3 loss in MCF7 and MDA-MB-453 breast cancer cells. MCF7 cells express modest levels of ErbB3 protein, and knockdown of endogenous Nrdp1 markedly elevated steady-state receptor levels even in the presence of BfA (Fig. 8A). Likewise, Nrdp1 expression suppressed endogenous ErbB3 in MDA-MB-453 cells independently of BfA treatment (Fig. 8B), indicating that ER-localized Nrdp1 effects on the receptor are not an outcome of ErbB3 receptor transfection or overexpression.
Fig. 8.
Nrdp1 acts on the nascent form of endogenous ErbB3. (A) MCF7 breast cancer cells stably transduced with scrambled or Nrdp1-directed shRNAs were treated for various times with brefeldin A. (Left) Lysates were immunoblotted with antibodies to ErbB3 and tubulin. (Right) Real-time PCR was used to measure the levels of Nrdp1 and ErbB3 transcripts. (B) MDA-MB-453 cells stably transduced with an empty vector or Nrdp1 were treated for various times with brefeldin A. (Left) Cleared lysates were immunoblotted with the indicated antibodies. *, mature ErbB3; **, immature ErbB3. (Right) The relative intensities of mature ErbB3 (at time zero) or immature ErbB3 (at 3 and 6 h) bands are plotted for four independent experiments. *, P < 0.05.
VCP activity is necessary for Nrdp1-mediated ErbB3 degradation.
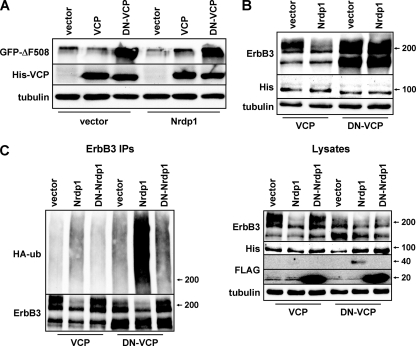
The localization of Nrdp1 function to the ER, coupled with the dependence of ErbB3 levels on the proteasome, suggests that the ER-associated degradation (ERAD) pathway could be involved in regulating ErbB3 stability. To begin to explore the relationship between ErbB3 degradation at the ER and the ERAD pathway, we asked whether a dominant negative form of the necessary ERAD component VCP (DN-VCP) that lacks ATPase activity interferes with Nrdp1-induced ErbB3 degradation. In accordance with the findings of previous studies (35), we observed that expression of DN-VCP, but not that of wild-type VCP, stabilized a canonical ERAD substrate (GFP-tagged CFTR-ΔF508) (Fig. 9A), underscoring the requirement for VCP in the ERAD pathway. Expression of Nrdp1 had no effect on CFTR-ΔF508 stability, indicating that the ligase likely does not play a universal role in ERAD pathway-mediated degradation.
Fig. 9.
Nrdp1 acts upstream of VCP to regulate the quantities of ErbB3 in cells. (A) HEK-293T cells were transiently transfected with GFP-tagged CFTR-ΔF508 without or with wild-type Nrdp1, and with a vector control or wild-type or dominant negative VCP. Lysates were immunoblotted with the indicated antibodies. (B) Cells were cotransfected with ErbB3, a vector control or Nrdp1, and His-tagged versions of either wild-type or dominant negative VCP, as indicated. Lysates were blotted with the indicated antibodies. (C) Cells were cotransfected with ErbB3, HA-ubiquitin (HA-ub), either wild-type or dominant negative Nrdp1, and His-tagged versions of either VCP or DN-VCP, as indicated. Cleared lysates (right) and ErbB3 immunoprecipitates (IPs) (left) were immunoblotted with the indicated antibodies.
We further observed that DN-VCP markedly stabilized ErbB3 (Fig. 9B), particularly the nascent form, suggesting that newly synthesized ErbB3 could be an ERAD substrate. Importantly, Nrdp1-mediated ErbB3 loss was abrogated by DN-VCP, indicating that VCP acts downstream of Nrdp1 in the ErbB3 degradation pathway. Wild-type VCP had no effect on ErbB3 levels relative to the vector control (not shown), suggesting that endogenous VCP levels are sufficient for full ErbB3 degradation activity. Interestingly, Nrdp1-mediated ErbB3 ubiquitination was markedly potentiated by DN-VCP (Fig. 9C), suggesting that the ErbB3 population ubiquitinated by Nrdp1 is normally degraded in a VCP-dependent manner and that receptor ubiquitination by Nrdp1 precedes receptor translocation by VCP. These observations indicate that VCP is an essential component of the Nrdp1-mediated ErbB3 degradation pathway.
DISCUSSION
Collectively, the observations outlined here define a novel receptor tyrosine kinase protein degradation cascade that we call the Nrdp1–valosin-containing protein (VCP) pathway, where newly synthesized ErbB3 is recognized and ubiquitinated by Nrdp1 to elicit its VCP-mediated proteasomal degradation. Blockage of this pathway by proteasome inhibition, VCP inhibition, or Nrdp1 knockdown leads to the dramatic accumulation of both nascent and mature ErbB3, pointing to the remarkable potency of Nrdp1-VCP in keeping ErbB3 levels in check. To our knowledge, ErbB3 is the first example of a receptor tyrosine kinase whose steady-state levels are critically regulated at the endoplasmic reticulum. These observations challenge the widely held notion that the posttranscriptional regulation of cellular receptor tyrosine kinase levels is mediated predominantly via lysosomal degradation of internalized cell surface receptors.
Roughly one-third of eukaryotic proteins are synthesized in the ER prior to their trafficking via the secretory pathway to their final cellular destinations (4). As a primary site of protein synthesis, the ER harbors a number of pathways collectively called endoplasmic reticulum quality control (ERQC) that ensure the production of intact, active products by facilitating the folding of newly synthesized proteins and promoting the destruction of their misfolded counterparts. While much of the ERQC system is composed of chaperones and components dedicated to facilitating proper folding, the ERAD arm of ERQC is responsible for the degradation of misfolded proteins whose accumulation could cause damage to the cell. ERAD is initiated upon the ubiquitination of a target protein, marking it for retrotranslocation out of the ER and into the cytosol, where it is ultimately degraded by the proteasome. One of the key functional components of the ERAD pathway is VCP (also known as p97), an AAA-ATPase required for the extraction of ERAD substrates from the ER prior to their delivery to cytosolic proteasomes for destruction (21, 35).
The relationship of the Nrdp1-VCP pathway to ERAD remains an interesting question. While a crucial element of ERAD is employed in mediating ErbB3 degradation, the inability of Nrdp1 to act on a canonical ERAD substrate, as well as the action of Nrdp1-VCP toward a properly folded substrate, calls into question the fit of Nrdp1-VCP to the traditional view of ERAD. However, work over the past several years has uncovered roles for ERAD in protein quantity control, where the ERAD machinery is used to control levels of native or properly folded proteins (4, 17). For example, intact apolipoprotein B, the major structural component of low-density and very low density lipoproteins, is efficiently degraded by hepatocyte ERAD if insufficient cholesterol is available for transport (3). In contrast, 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMG CoA reductase), the rate-limiting enzyme in the biosynthesis of cholesterol and its sterol precursors, is efficiently degraded by ERAD upon accumulation of excess sterols (22). ER-localized inositol trisphosphate (IP3) receptors, responsible for the release of Ca2+ from the ER, undergo ERAD-mediated degradation following their activation (1). Finally, the half-life of type 2 iodothyronine deiodinase (D2), an ER-resident enzyme responsible for the conversion of the thyroid prehormone thyroxine (T4) to the active hormone triiodothyronine (T3), is shortened as a result of T4-induced ubiquitination and subsequent ERAD (14).
In each of these instances, ERAD-mediated degradation of the biologically active substrate is a highly regulated process and is influenced by proteins specifically committed to mediating the stability of that substrate. We envision a model where Nrdp1 fulfills the role of the committed regulatory component, plugging into the ERAD machinery to rid the cell of ErbB3 when signaling by the receptor is counterproductive. The factors that regulate Nrdp1-mediated ErbB3 degradation remain a question. It appears that a major mechanism of Nrdp1 regulation is its own lability by autoubiquitination (34), which is often dramatically enhanced in tumor cells over that in nontransformed cells (20). We suspect that normal cells harbor pathways that promote Nrdp1 stability and that key components of these pathways are disrupted in tumors. Efforts are under way to identify cellular components that contribute to Nrdp1 stability and to characterize environmental conditions to which these components respond.
It should be noted that although Nrdp1 colocalizes and cofractionates with the endoplasmic reticulum under the conditions described here, its localization and function may not be restricted to the ER under all circumstances. Our previous studies indicate that NRG1 stimulation of ErbB3 in MCF7 breast tumor cells accelerates the degradation of the mature receptor in an Nrdp1- and lysosome-dependent manner (5). Part of this effect was ascribed to growth factor-stimulated stabilization of the ligase; ligand-induced elevation of Nrdp1 levels accelerates the ubiquitination and degradation of internalized ErbB3. Our results described here suggest that ligand stimulation may also prompt the relocalization of a population of Nrdp1 from the ER to the late endosome, where it can ubiquitinate mature ErbB3 to signal its trafficking to the lysosome. The mechanisms by which growth factor stimulation can coopt the function of a factor that controls basal receptor levels to elicit the downregulation of activated receptors remain to be fully explored.
The advantage of employing the ERAD system to degrade newly synthesized receptors (rather than using the lysosomal system to degrade internalized mature receptors) for the regulation of overall ErbB3 levels is unclear. However, one possibility could be that the destruction of unnecessary receptors prior to their arrival at the cell surface prevents aberrant signaling by any residual ligand. Such a notion would predict that receptor tyrosine kinases whose ligands are deposited on the extracellular matrix are strong candidates for ER-mediated quantity control.
Overall, our observations uncover a previously unappreciated role for the endoplasmic reticulum in regulating receptor tyrosine kinase abundance, and they underscore a role for ER-based processes in tumor development. Whether other receptor tyrosine kinases are similarly regulated at the ER, and the mechanisms by which such processes are regulated in nontransformed cells, remain questions for the future.
ACKNOWLEDGMENTS
We thank Alan Verkman and Ron Kopito for providing reagents.
This research was supported by NIH grants GM068994 and CA123541 (to K.L.C.) and CA118384 (to C.S.).
Footnotes
Published ahead of print on 16 May 2011.
REFERENCES
- 1. Alzayady K. J., Panning M. M., Kelley G. G., Wojcikiewicz R. J. 2005. Involvement of the p97-Ufd1-Npl4 complex in the regulated endoplasmic reticulum-associated degradation of inositol 1,4,5-trisphosphate receptors. J. Biol. Chem. 280:34530–34537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baselga J., Swain S. M. 2009. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat. Rev. Cancer 9:463–475 [DOI] [PubMed] [Google Scholar]
- 3. Brodsky J. L., Fisher E. A. 2008. The many intersecting pathways underlying apolipoprotein B secretion and degradation. Trends Endocrinol. Metab. 19:254–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brodsky J. L., Wojcikiewicz R. J. 2009. Substrate-specific mediators of ER associated degradation (ERAD). Curr. Opin. Cell Biol. 21:516–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cao Z., Wu X., Yen L., Sweeney C., Carraway K. L., III 2007. Neuregulin-induced ErbB3 downregulation is mediated by a protein stability cascade involving the E3 ubiquitin ligase Nrdp1. Mol. Cell. Biol. 27:2180–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carraway K. L., III 2010. E3 ubiquitin ligases in ErbB receptor quantity control. Semin. Cell Dev. Biol. 21:936–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Citri A., Skaria K. B., Yarden Y. 2003. The deaf and the dumb: the biology of ErbB-2 and ErbB-3. Exp. Cell Res. 284:54–65 [DOI] [PubMed] [Google Scholar]
- 8. Dayel M. J., Hom E. F., Verkman A. S. 1999. Diffusion of green fluorescent protein in the aqueous-phase lumen of endoplasmic reticulum. Biophys. J. 76:2843–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Diamonti A. J., et al. 2002. An RBCC protein implicated in maintenance of steady-state neuregulin receptor levels. Proc. Natl. Acad. Sci. U. S. A. 99:2866–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dikic I., Giordano S. 2003. Negative receptor signalling. Curr. Opin. Cell Biol. 15:128–135 [DOI] [PubMed] [Google Scholar]
- 11. Edwards Y. S., Murray A. W. 1995. Accumulation of phosphatidylalcohol in cultured cells: use of subcellular fractionation to investigate phospholipase D activity during signal transduction. Biochem. J. 308:473–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fry W. H., Kotelawala L., Sweeney C., Carraway K. L., III 2009. Mechanisms of ErbB receptor negative regulation and relevance in cancer. Exp. Cell Res. 315:697–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Funes M., Miller J. K., Lai C., Carraway K. L., III, Sweeney C. 2006. The mucin Muc4 potentiates neuregulin signaling by increasing the cell-surface populations of ErbB2 and ErbB3. J. Biol. Chem. 281:19310–19319 [DOI] [PubMed] [Google Scholar]
- 14. Gereben B., Goncalves C., Harney J. W., Larsen P. R., Bianco A. C. 2000. Selective proteolysis of human type 2 deiodinase: a novel ubiquitin-proteasomal mediated mechanism for regulation of hormone activation. Mol. Endocrinol. 14:1697–1708 [DOI] [PubMed] [Google Scholar]
- 15. Gur G., et al. 2004. LRIG1 restricts growth factor signaling by enhancing receptor ubiquitylation and degradation. EMBO J. 23:3270–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hamburger A. W. 2008. The role of ErbB3 and its binding partners in breast cancer progression and resistance to hormone and tyrosine kinase directed therapies. J. Mammary Gland Biol. Neoplasia 13:225–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hegde R. S., Ploegh H. L. 2010. Quality and quantity control at the endoplasmic reticulum. Curr. Opin. Cell Biol. 22:437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Holbro T., et al. 2003. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc. Natl. Acad. Sci. U. S. A. 100:8933–8938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hsieh A. C., Moasser M. M. 2007. Targeting HER proteins in cancer therapy and the role of the non-target HER3. Br. J. Cancer 97:453–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ingalla E. Q., et al. 2010. Post-transcriptional mechanisms contribute to the suppression of the ErbB3 negative regulator protein Nrdp1 in mammary tumors. J. Biol. Chem. 285:28691–28697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jentsch S., Rumpf S. 2007. Cdc48 (p97): a “molecular gearbox” in the ubiquitin pathway? Trends Biochem. Sci. 32:6–11 [DOI] [PubMed] [Google Scholar]
- 22. Jo Y., Debose-Boyd R. A. 2010. Control of cholesterol synthesis through regulated ER-associated degradation of HMG CoA reductase. Crit. Rev. Biochem. Mol. Biol. 45:185–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Klausner R. D., Donaldson J. G., Lippincott-Schwartz J. 1992. Brefeldin A: insights into the control of membrane traffic and organelle structure. J. Cell Biol. 116:1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Laederich M. B., et al. 2004. The leucine-rich repeat protein LRIG1 is a negative regulator of ErbB family receptor tyrosine kinases. J. Biol. Chem. 279:47050–47056 [DOI] [PubMed] [Google Scholar]
- 25. Miller J. K., et al. 2008. Suppression of the negative regulator LRIG1 contributes to ErbB2 overexpression in breast cancer. Cancer Res. 68:8286–8294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mosesson Y., Mills G. B., Yarden Y. 2008. Derailed endocytosis: an emerging feature of cancer. Nat. Rev. Cancer 8:835–850 [DOI] [PubMed] [Google Scholar]
- 27. Qiu X. B., Goldberg A. L. 2002. Nrdp1/FLRF is a ubiquitin ligase promoting ubiquitination and degradation of the epidermal growth factor receptor family member, ErbB3. Proc. Natl. Acad. Sci. U. S. A. 99:14843–14848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Russell M. R., Nickerson D. P., Odorizzi G. 2006. Molecular mechanisms of late endosome morphology, identity and sorting. Curr. Opin. Cell Biol. 18:422–428 [DOI] [PubMed] [Google Scholar]
- 29. Siegel P. M., Ryan E. D., Cardiff R. D., Muller W. J. 1999. Elevated expression of activated forms of Neu/ErbB-2 and ErbB-3 are involved in the induction of mammary tumors in transgenic mice: implications for human breast cancer. EMBO J. 18:2149–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sorkin A., Goh L. K. 2008. Endocytosis and intracellular trafficking of ErbBs. Exp. Cell Res. 314:3093–3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stern D. F. 2008. ERBB3/HER3 and ERBB2/HER2 duet in mammary development and breast cancer. J. Mammary Gland Biol. Neoplasia 13:215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Waterman H., Yarden Y. 2001. Molecular mechanisms underlying endocytosis and sorting of ErbB receptor tyrosine kinases. FEBS Lett. 490:142–152 [DOI] [PubMed] [Google Scholar]
- 33. Wiley H. S. 2003. Trafficking of the ErbB receptors and its influence on signaling. Exp. Cell Res. 284:78–88 [DOI] [PubMed] [Google Scholar]
- 34. Wu X., Yen L., Irwin L., Sweeney C., Carraway K. L., III 2004. Stabilization of the E3 ubiquitin ligase Nrdp1 by the deubiquitinating enzyme USP8. Mol. Cell. Biol. 24:7748–7757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ye Y., Meyer H. H., Rapoport T. A. 2001. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414:652–656 [DOI] [PubMed] [Google Scholar]
- 36. Yen L., et al. 2006. Loss of Nrdp1 enhances ErbB2/ErbB3-dependent breast tumor cell growth. Cancer Res. 66:11279–11286 [DOI] [PubMed] [Google Scholar]